Abstract
Plasmacytoid dendritic cells (pDCs) are potent producers of type-I and type-III interferons (IFNs) and play a major role in antiviral immunity and autoimmune disorders. The innate sensing of nucleic acids remains the major initiating factor for IFN production by pDCs. Toll-like receptor (TLR)-mediated sensing of nucleic acids via endosomal pathways has been studied and documented in detail, while the sensing of DNA in cytosolic compartment in human pDCs remains relatively unexplored. We now demonstrate the existence and functionality of the components of cytosolic DNA sensing pathway comprising of cGAMP synthetase (cGAS) and stimulator of interferon gene (STING) in human pDCs. cGAS was initially located in the cytosolic compartment of pDCs and time-dependently co-localized with non-CpG double-stranded immuno-stimulatory DNA (ISD). Following the co-localization of ISD with cGAS, the downstream pathway was triggered as STING disassociated from its location at the endoplasmic reticulum. Upon direct stimulation of pDCs by STING agonist 2′3′ cyclic AMP-GMP (cGAMP) or dsDNA, pDCs produced type-I, and type-III interferon. Moreover, we documented that cGAS/STING-mediated IFN production is mediated by nuclear translocation of IRF3 while TLR9-mediated activation occurs through IRF7. Our data also indicate that pDC pre-stimulation of cGAS/STING dampened the TLR9-mediated IFN production. Further, triggering of cGAS-STING induced expression of SOCS1 and SOCS3 in pDCs, indicating a possible autoinhibitory loop that impedes IFN production by pDCs. Thus, our study indicates that the cGAS-STING pathway exists in parallel to the TLR9-mediated DNA recognition in human pDCs with cross-talk between these two pathways.
Introduction:
Plasmacytoid dendritic cells (pDCs) function as key mediators of innate immunity by producing copious amount of type I and III interferons (IFNs) in response to pathogenic stimuli (1–5). To initiate IFN production, pDCs have to successfully sense of the pathogen-associated molecular patterns (PAMP) via innate sensors. One of the principal PAMPs is pathogen-derived DNA that can be recognized by either endosomal or cytosolic DNA sensors present in various immune cells (6). Human pDCs express the endosomal DNA sensor toll-like receptor 9 (TLR9), which is involved both in recognition of pathogen-derived DNA as well as self-DNA from dying cells, mounting a robust immune or autoimmune response by inducing IFN production (7). Although endosomal DNA recognition has been studied extensively in human pDCs, the realm of cytosolic DNA sensors in pDC is less well understood.
The innate DNA sensor cyclic GMP-AMP synthase (cGAS) is a newly found player in the world of innate immunity. Upon sensing of double-stranded DNA, cGAS recruits endogenous ATP and GTP to produce cyclin dinucleotide 2′5′cyclic GMP-AMP (cGAMP) (8, 9). Like bacterial cyclic dinucleotides, the mammalian non-canonical 2′5′cGAMP is also recognized by the endoplasmic resident (ER) protein stimulator of interferon genes (STING) (10, 11). STING (also known as ERIS, MITA, or MPYS) is a transmembrane domain protein (TMEM173) that phosphorylates and activates the TBK1-IKKε mediated downstream pathway to produce Type I IFN (12). Activated STING gets dissociated from the endoplasmic reticulum (ER) and accumulates in the perinuclear space where its N-terminal end works as a scaffold for both TBK1 and IRF3. STING phosphorylates and activates TBK1, which then phosphorylates IRF3 that dimerizes before translocating into the nucleus and inducing type I IFN production (13, 14).
pDCs produce IFN-α in response to both RNA and DNA viruses (2). They also produce other Type I IFNs including IFN-β, IFN-ω, IFN-κ to varying amounts (15). Following the discovery of Type III IFN, it was confirmed that pDCs act as the major immune cell producer of that particular subtype of antiviral protein as well (4, 5). In response to both DNA and RNA viruses, pDCs produce a broad range of IFN-α subtypes as seen at the transcript level (3, 15). Moreover, human pDC also produce the full-range of Type III IFNs: IFN-λ1, −2 and −3, (5). pDCs express both Type I and Type III IFN receptors on their surface and hence, are capable of responding to IFN in both an autocrine and paracrine manner (5). IFN-α can prime pDCs to produce more interferons (along with upregulation of other interferon-stimulated genes (ISGs), thus creating an autocrine loop of antiviral immune reaction (16).
The role of pDCs as the professional producers of type I IFNs renders any innate sensing pathway special in this particular cell type. The TLR9-mediated pathway in human pDCs is well-documented, while few studies have addressed cytosolic DNA sensors in these cells. Putative cytosolic DNA sensor DDX41 was reported to be functional in pDCs via STING, although the pathway was not fully elucidated (17). Other putative DNA sensors such as Z-DNA binding protein (also known as DAI), AIM2, IFI16 have not been studied in pDCs. Recently, the cGAS-STING pathway was documented to be existent and functional in pDCs (18–20), but how it may intersect with the endosomal TLR9 pathway was not clear.
In our current study, we investigated if the cGAS-STING pathway in primary human pDCs runs parallel to the TLR9-mediated pathway and if they intersect with each other. We hypothesized that the molecular machinery used by the cGAS-STING pathway will be expressed in human pDCs and tested to see the sub-cellular localization and movement of the key molecules upon the stimulation of this pathway. We observed the exclusivity of the involvement of IRF3 vs. IRF7 in cGAS-STING and TLR9 pathways, respectively. To our surprise, we did not observe any nuclear translocation of NF-κB or the production of NF-κB-dependent cytokines with cGAS-STING stimulation in pDCs. We have also shown how cGAS-STING pathway stimulation impedes TLR9-mediated interferon production, possibly through the induction of SOCS1 and SOCS3. These data suggest that an interplay between the cGAS-STING pathway and TLR9-mediated pathway exists in human pDCs that can potentially be harnessed and modulated to reap clinical benefits in both host immune response and autoimmunity.
Materials and Methods:
Viruses and cell lines
The THP1 monocytic cell line was grown in RPMI 1640 medium with L-glutamine (Mediatech, Manassas, VA) containing 10% heat-inactivated fetal bovine serum (FBS), 100 U/ml penicillin, 100 mg/ml streptomycin, 100 mg/ml gentamicin (Mediatech, Manassas, VA), and 25 mM HEPES (Sigma-Aldrich) (complete RPMI). HEK293 cells were grown in DMEM containing 10% FBS and same antibiotics.
HSV-1 strain 2931 (HSV) was originally obtained from Dr. C. Lopez, then at the Sloan-Kettering Institute (New York, NY). The HSV stocks were expanded in VERO cells (American Type Culture Collection, Manassas, VA) and titered by a plaque assay on VERO cells. Influenza virus A (IAV) strain PR/8/34 propagated in SPF eggs and Sendai virus (SeV) strain Sendai/Cantell (ATCC VR-907 Parainfluenza 1) grown in amnioallantoic fluid from SPF chicken embryos were purchased from Charles River Laboratory (SPAFAS, N. Franklin, CT). Aldithrion-2 (AT2)–inactivated HIV-1, strain MN, lot P3939, was kindly provided by Dr. Jeff Lifson of the Biological Products Core of the AIDS and Cancer Virus Program of the Frederick National Laboratory for Cancer Research. All viruses were stored at −80 ˚C until use. HSV K26-GFP was originally provided by P. Desai (Johns Hopkins University, Baltimore, MD) and was grown in VERO cells.
Preparation of PBMCs and purified pDCs
PBMCs were isolated by Ficoll–hypaque (Mediatech, Manassas, VA) density centrifugation from fresh, heparinized blood obtained from healthy adult volunteers. Donors included approximately equal representation of males and females, ages 18–65, and were Caucasian, African-American, or Asian. The Institutional Review Board (IRB) of the New Jersey Medical School approved the human studies protocol and informed consent documentation. The PBMCs were resuspended in complete RPMI and counted with a Coulter Particle Counter Z1 (Beckman Coulter).
pDCs were purified by negative selection from prepared PBMCs with the Human Plasmacytoid Dendritic Cell Isolation Kit-II (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. The purity of enriched pDC was determined by flow cytometry (CD123 and BDCA-2 double-positive) and viability was determined by trypan blue exclusion. All experiments utilizing highly enriched pDCs were performed with 90–99% pure pDCs. The major contaminants of negatively selected pDCs are cells within lymphocyte gate with low forward and side scatter.
Preparation of mononuclear cell suspensions from tonsil
Palantine tonsillar tissue from pediatric patients undergoing tonsillectomy for obstructive sleep apnea were obtained with IRB approval and mononuclear cell suspensions were prepared. Briefly, tonsils were cut into smaller sections with cortical and medullary parts and cells are teased out from the tissue into RPMI by separating out the capsule. The cell suspension was filtered through 20 micron mesh, centrifuged, and resuspended into fresh complete RPMI to prepare a single cell suspension.
Reagents
TLR9 agonist CpG-A (ODN 2216) was purchased from Invivogen (San Diego, CA) and used at a final concentration of 5μM. Brefeldin A (Sigma) was used to attain the final concentration of 1 μg/ml. Synthetic 2′3′ cyclic GMP-AMP (cGAMP) was purchased from Invitrogen, reconstituted in nuclease-free water and used at the concentration between 2–50 μg/ml. Lipofectamine 2000 and Lipofectamine siRNAmax (Invitrogen, Thermo Fisher Scientific, Waltham, MA) were used to transfect cGAMP and siRNA, respectively. TBK1/IKKε inhibitor BX795 was purchased from Invivogen (San Diego, CA) and used at a 1μM concentration. TLR9 inhibitor ODN (ttaggg)4 was purchased from Adipogen (San Diego, CA). BD Horizon Brilliant stain buffer from BD Biosciences (San Jose, CA) was used for staining with brilliant violent-conjugated antibodies. The sense and antisense strand of immune-stimulatory DNA (ISD) was purchased with or without fluorescein conjugation (at 3′ end of each strand) and was hybridized before use (Integrated DNA technologies, Coralville, Iowa) by mixing and incubating two strands of equal molar amount at 95°C for 5 minutes followed by gradually cooling down to room temperature. Recombinant human IFN-α−2b, rhIFN-λ1, rhIL-3, and rhIL-10 were purchased from Peprotech Inc. (Rocky Hill, NJ), while IFN-α−2b was purchased from Schering-Plough. siRNA directed to silence STING (Cat#4392420) and a scrambled siRNA (Cat#4390843) were purchased from Thermo Fisher Scientific.
Antibodies
PECY7-antiCD123 (6H6), BV510-antiCD123 (6H6), PE-antiBDCA2 (201A), BV421-antiBDCA2 (201A), PerCPCY5.5-antiBDCA2 (12C2), AF700-antiCD3 (SK7), FITC-antiCD4 (RPA-T4), APC-antiCD8a (RPA-T8), APCCY7-antiCD14 (HCD14), and AF594-antiSTING (O94E12) were purchased from BioLegend (San Diego, CA). Draq5 was purchased from Cell Signaling (Danver, MA). AF488-antiIRF3 was purchased from R&D systems (Minneapolis, MN). AF647-antiSTING (T45–2342) and matching isotype were purchased from BD Biosciences (San Jose, CA). Rabbit polyclonal anti-cGAS (c6orf150) antibody was purchased from Bioorbyt (Cambridge, UK). Anti-IFNAR2 antibody (MMHAR-2), which is used for blocking of the type I IFN receptor, was purchased from EMD Millipore (Temecula, CA) and was used at a concentration of 5 μg/ml. AF647-anti-phospho-IRF3 antibody was purchased from Cell Signaling (Danver, MA). PE-anti-phospho-IRF7 was purchased from BD Biosciences (San Jose, CA).
qRT-PCR measurement of mRNA level of cGAS and STING
Total RNA was isolated from purified pDCs by RNeasy® Plus Micro kit from QIAGEN™ following the manufacturer-provided protocol. The purity and concentration of total RNA were measured with NanoDrop ND-1000 UV-Vis spectrophotometer and ND-1000 V3.3.1 software (ThermoScientific, Wilmington, DE). mRNA, derived from 200,000 pDCs, was reverse transcribed to prepare cDNA using random hexamer primer. Prepared cDNA was amplified via real time PCR to quantify mRNA expression in pDCs using Taqman gene-specific probe-based assays. The following assay mixes were used (ThermoScientific, Wilmington, DE):
cGAS: Hs00403553_m1MB21D1
STING: Hs00736958_m1TMEM173
SOCS1: Hs00864158_g1SOCS1
SOCS3: Hs01000485_g1SOCS3
SOCS5: Hs00367107_m1SOCS5
Expression of the house-keeping gene β-actin was used as the internal control: (Hs99999903_m1ACTB).
Stimulation of PBMCs and pDCs:
2 X 106 PBMC/ml were stimulated with recombinant IFN-α−2b at a dose of 1000 international units/ml and recombinant interferon-λ1 at 25 ng/ml for 8 hours at 37°C with 5% CO2 concentration. For viral stimulation, HSV-1, at an MOI of 1, IAV at 4 HA units/ml, SeV at 16 HA units/ml and HIV-MN at 500 ng of p24 equivalents/ml were used. After 8 hours of stimulation, cells were stained for pDC markers and fixed in 1% PFA in PBS overnight. The next day, the cells were washed with PBS with 2% FCS and permeabilized with 0.1% Triton X in PBS and stained for intracellular markers. HSV-1 was used at the same concentration of virus as used for PBMC to stimulate purified pDCs resuspended at 0.25 X 106 cells/ml.
For cGAMP and ISD stimulation, purified pDCs were stimulated in round-bottomed 96 well plates with different doses of cGAMP and ISD with lipofectamine at 37°C with 5% CO2 concentration. To assess IFN release, the supernatants were collected at different time intervals. For measurement of intracellular cytokines, Brefeldin A (Sigma-Aldrich) was added to each sample two hours before fixation. The next day, the cells were permeabilized and stained for intracellular cytokines.
Flow Cytometry
PBMCs were stained for pDC markers anti-CD123 and anti-BDCA2. Samples were acquired via BD LSR-II or LSR-Fortessa (BD Biosciences) and analysis was done with FlowJo software (BD Informatics). Spade analysis was performed using Cytobank (Santa Clara, CA). For each sample of PBMCs, 300,000 – 500,000 events were acquired while for each sample of purified pDCs, 5,000 events were acquired.
Phospho-flow analyses
Freshly isolated pDCs were stimulated as described in the figure legends, fixed with an equal volume of 4% ice cold PFA in PBS, and kept on the ice. Next, cells were washed, surface stained with HLA-DR–Pacific Blue and CD123-PECY7, followed by washing and another 10 minutes of 4% PFA fixation. Then, the cells were permeabilized either with chilled 100% methanol (for phospho-IRF3) or with chilled 70% ethanol (for phospho-IRF7), which was added dropwise to fixed cells incubated on ice and kept for 45 minutes. Permeabilized cells were washed with PBS containing 2% FCS and stained using anti-phospho antibodies.
ELISA
The supernatants were collected after stimulating purified pDCs with viruses, cGAMP, ISD, or CpG-A. The manufacturer’s protocols were followed to measure the concentration of secreted IFN-α (Mabtech, Cincinnati, OH), TNF-α (BD Biosciences), IFN-λ1 (R&D Systems, Minneapolis, MN), IFN-β (R&D systems, Minneapolis, MN), and IL-10 (R&D systems, Minneapolis, MN).
Imaging Flow Cytometry
Flow imaging experiments were done by Amnis ImageStream X Mark II (EMD Millipore®). PBMCs were stained for pDC markers and fixed in PFA, permeabilized the next day, and stained for different intracellular markers and with the nuclear stain, Draq5, and acquired using the 40X camera. Data were analyzed by IDEAs software (EMD Millipore®) with gating as shown in Figure 3. Cytoplasmic to nuclear translocation of signaling molecules and co-localization of two molecules were calculated according to the method described in (21).
FIGURE 3.
cGAS co-localizes with dsDNA in a time-dependent manner, while STING resides in the ER and dissociates upon stimulation of the upstream pathway. pDCs were isolated from peripheral blood of healthy donors and transfected with FITC-conjugated ISD or ssDNA. Co-localization of DNA and cGAS was determined by Amnis ImageStream. (A) Schemata of identification of pDCs among PBMC population. Single cells are selected out of PBMCs, first by plotting the population with area vs. aspect ratio. Next, focused singlets were determined by plotting the single cells with area vs. side scatter. Finally, pDCs were identified out of focused singlets as CD123+BDCA2+ cells. (B) Sample histograms showing co-localization between cGAS and DNA by bright detail similarity score in time-dependent manner. (C) Representative image of a single cell showing cGAS and dsDNA co-localization. (D) Quantification of co-localization between cGAS and DNA by bright detail similarity score. One way ANOVA was performed between all groups, and then post-hoc T tests were performed. Each dot represents an individual experiment (n = 3). (E, F) pDCs were purified and transfected with unconjugated ISD or ssDNA for 2 hours, fixed overnight, permeabilized, and stained for Calnexin and STING. (E) Histogram of Calnexin and STING co-localization with or without DNA transfection (Data represents one of two independent experiment). (F) Representative images of single cells showing Calnexin and STING co-localization with or without DNA transfection.
siRNA-mediated knock-down
Freshly-isolated purified pDCs were reconstituted in complete RPMI supplemented with rhIL-3 (10 ng/ml) and seeded in 96-well U-bottom plates at a concentration of 0.25 X 106 cells/ml. STING-targeting siRNA was mixed with lipofectamine-RNAiMaX to prepare the transfection mix containing 0.3 μl of lipofectamine per pmol of siRNA. Similar transfection mix with negative control scrambled siRNA was prepared in parallel. Transfection reagents were kept together for 20 minutes in room temperature. The transfection mix was added to the pDCs to achieve a concentration of 10 nM. After 24 hours of lipofection, samples from each group were tested for their survival by staining with Zombie-UV (BioLegend) and gene knockdown efficiency by staining for STING and doing flow cytometric analysis.
Virus uptake
pDC, enriched from PBMCs, were pre-incubated with or without cGAMP or ISD for 3 hours or 6 hours. Next, the cells were washed and resuspended again in enriched RPMI, followed by addition of HSV-GFP at an MOI of 10. Then the cells were cultured for 30 minutes, harvested, washed with PBS with 5 mM EDTA at 4° C to remove viruses attached to the surface of the cell. Next the cells were stained for pDC markers and fixed with 2% PFA before acquiring in Amnis ImageStream.
Statistical Analysis
The statistical significance of differences between two groups of samples was determined by pairwise student’s T-test and differences between multiple groups of samples were determined by ANOVA with Tukey’s post-hoc test. P values <0.05 were considered significant. * indicates that p value is less than or equal to 0.05 and ** indicates that p value is less than or equal to 0.01.
Results
Human primary pDCs constitutively express cGAS and STING
A growing volume of literature indicates that cGAS-STING pathway plays a pivotal role in innate sensing of cytosolic DNA in mammalian cells [reviewed in (22)]. To investigate if a parallel cGAS-STING mediated pathway operates in the human pDCs alongside the TLR9-mediated endosomal pathway, we first tested the expression of cGAS and STING in human pDCs. We purified “untouched” pDCs from peripheral blood of healthy volunteers via negative selection from PBMCs. Freshly-isolated pDCs were lysed and total RNA was extracted. qRT-PCR was performed, using undifferentiated THP1 and HEK293T cells as the positive and negative controls, respectively. We observed both cGAS and STING to be constitutively transcribed in primary human pDCs (Fig. 1A). Next, we employed flow cytometry to examine the protein level of cGAS and STING in pDCs. Both cGAS and STING were observed to be expressed in pDCs derived from peripheral blood (Fig. 1B).
FIGURE 1.
Human pDCs constitutively express cGAS and STING. (A) cGAS and STING transcript level in human pDCs with respect to THP1 cells ( positive control) and HEK293T cells (negative control): Total RNA was extracted from purified pDC, THP1 and HEK293T cells, mRNA was reverse transcribed to cDNA and expression level of cGAS and STING was measured by qRT-PCR Data represent mean ± 1 SD, (n = 3 independent experiments). (B) Protein expression of cGAS and STING in human pDCs. pDCs were distinguished among PBMCs as CD123+BDCA2+ cells. Singlet pDCs were gated from the double-positive population and cGAS and STING expression was measured via intracellular staining. STING expression was measured against the background of specific isotype and cGAS expression was measured against the secondary antibody (Goat anti-rabbit FITC) without the primary anti-cGAS antibody. Spade analysis showing relative expression of (C) cGAS and (D) STING in different subsets of human PBMCs. PBMCs were stained for CD3, CD4, CD8, CD19, CD14, CD123, and BDCA2 to identify CD4+ and CD8+ T cells, B cells, monocytes and pDCs. Cells were permeabilized and stained for cGAS and STING and spade analysis was performed to determine relative expression. The colors represent relative expression of proteins as a heat-map where blue represents low expression, green represents intermediate expression, and red and yellow represent high expression.
Several groups have characterized two stable functionally different subsets of human pDCs by differential surface expression of CD2 (23–25). Our previous work suggests that these two subsets show dissimilar expression of TLR9 (Mwangi et al., in preparation), with the CD2+ pDC subpopulation expressing higher levels of TLR9. Hence, we investigated how the expression of cGAS and STING compare in these two subsets. The expression of cGAS and STING in CD2high and CD2low subsets of pDCs was found to be comparable (Supplemental Fig. 1, A,B).
Previously, our lab has performed comparative studies between pDCs derived from secondary lymphoid organs such as tonsils and peripheral blood derived pDCs (26). We now tested the relative expression of cGAS and STING in both peripheral blood and tonsils. Our data indicate that the cGAS expression pattern is similar in tonsil and peripheral blood derived pDCs, although STING levels were significantly higher in tonsil samples (Supplemental Fig. 1, C,D).
We also tested how the expression of cGAS and STING compare between major subsets of immune cells. We stained PBMCs for CD4+ and CD8+ T cells, B cells, monocytes, and pDCs and compared the expression of cGAS and STING in different groups of cells by Spade analysis. We observed cGAS and STING expression in pDCs and monocytes to be similar and higher than in T cells and B cells. (Fig. 1C, 1D).
cGAS expression is inducible in pDCs
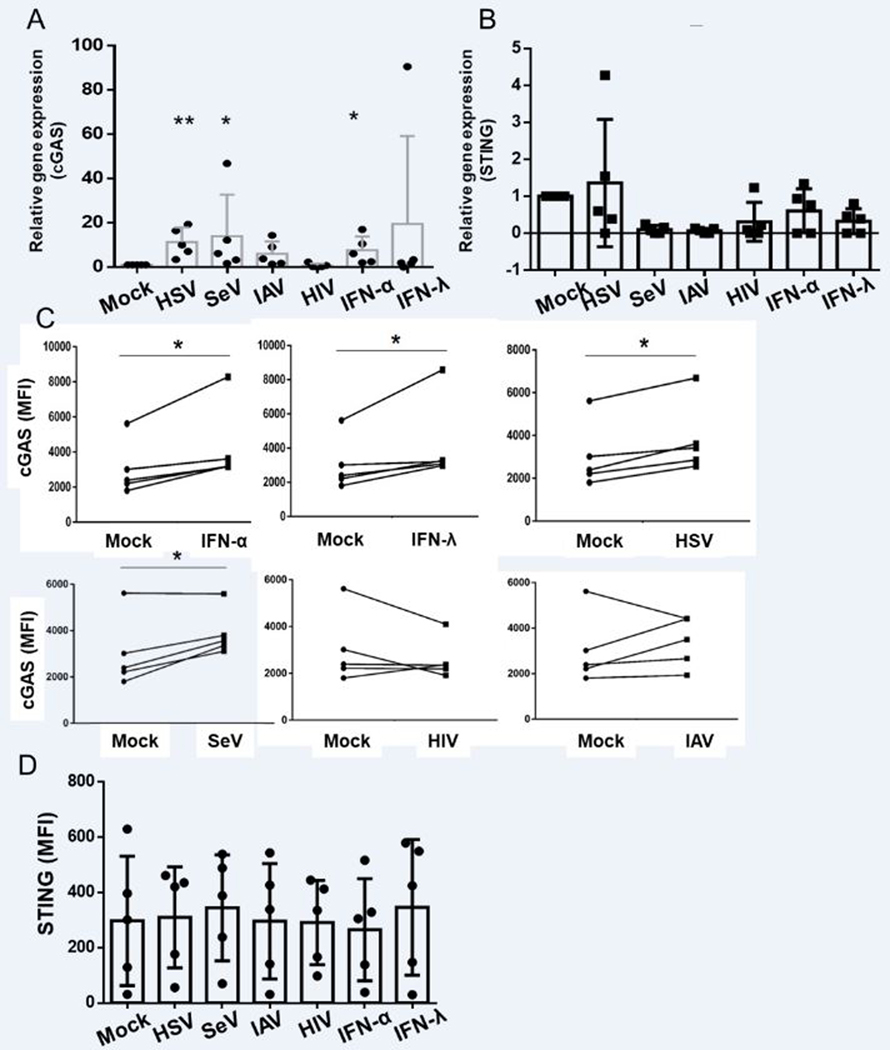
To determine how cGAS and STING expression is modulated in pDCs following different stimuli, we stimulated freshly-isolated pDCs with live viruses including a DNA virus (HSV-1) and RNA viruses including IAV and SeV as well as AT2-inactivated HIV-1 for 4 hours. Since cGAS is a known ISG (27, 28), and pDCs express both type-I and type-III interferon receptors (5), we also stimulated pDCs with rhIFN-α−2b (IFN-α) and rhIFN-λ1 (5). The expression of cGAS mRNA were upregulated following stimulation with type-I and type-III IFNs as well as with HSV, SeV, and IAV, but was unchanged at this timepoint in the HIV-stimulated group (Fig. 2A). In contrast, the expression of STING either remained unaltered or was downregulated with these stimuli (Fig. 2B). When PBMCs were stimulated with the same viruses and interferons for 8 hours and the protein expression of cGAS and STING was measured via flow cytometry, similar patterns of cGAS upregulation but no STING upregulation were observed (Fig 2C, D). We also stimulated PBMCs with rhIFN-β for 8 hrs and observed upregulation of cGAS expression in pDCs (Supplementary Fig. 1E). We did not see any alteration of cGAS or STING protein expression in pDCs with shorter simulation with either IFN-α or IFN-λ or with viral stimuli (data not shown).
FIGURE 2.
Stimulation with viruses and interferons upregulate cGAS but not STING expression in pDCs (A, B) Purified pDCs were stimulated with HSV, SeV, IAV, HIV, IFN-α, or IFN-λ for 4 hours. Total RNA was collected and cDNA was prepared. qPCR was performed to test how the expression of cGAS and STING is altered following each stimulation and are expressed relative to mock (n = 5 independent experiments). (C, D) PBMCs were incubated with IFN-α, IFN-λ, HSV, SeV, IAV, and HIV for 8 hours and expression of (C) cGAS and (D) STING in pDC (CD123+/BDCA2+ cells) were measured by flow cytometry and data expressed as the median fluorescent intensity (MFI) for the mock vs. stimulated pairs, with each donor represented by a line (n=5 independent experiments). Pairwise T tests were performed to test both mRNA and protein expression affected by each stimulation as compared to Mock.
Cytosolic cGAS in pDCs co-localizes with dsDNA
The putative DNA sensor IFI16 is predominantly expressed in the nucleus (29). On the contrary, only the role of cytosolic cGAS in innate sensing has been studied and the subcellular compartmentalization of cGAS has not been fully explored. A recent report suggests that cytosolic cGAS translocates to nucleus upon DNA damage (30). In order to test the subcellular pattern of cGAS expression in human pDCs, we used Amnis ImageStream imaging flow cytometry to see how cGAS is distributed in the cytosol of pDCs. We observed no co-localization of cGAS with the early endosome (EEA1), or late endosome (LAMP1) with or without stimuli (Supplementary Fig. 2). Next, we probed the ability of cytosolic cGAS to bind to non-CpG DNA transfected into the cells. We chose the model of immuno-stimulatory DNA (ISD) introduced by the Medzhitov lab (31), as ISD can be easily tagged with fluorescein and be tracked inside the cell. We lipofected pDCs with either hybridized double-stranded (ds) or single-stranded (ss, only the sense strand of ISD) DNA and stained for cGAS to observe the co-localization of cGAS and the transfected DNA. After 2 hours of transfection, dsDNA co-localized with cGAS while ssDNA remained unengaged, indicating that cGAS is preferentially binding to dsDNA (Figure 3B, 3C).The co-localization measured as bright detail similarity of cGAS and DNA decreased over time, but ssDNA remained unassociated with cGAS (Fig. 3D).
STING is an ER-resident protein, and triggering of the cGAS-STING pathway is characterized by phosphorylation and dissociation of STING from the ER and relocation in perinuclear space where it works as a scaffold for TBK1 and IRF3 (13, 14, 32). To test if the co-localization of dsDNA and cGAS is a functional binding that triggers the downstream pathway in pDC, we tested if STING is dissociated from ER upon transfection of non-CpG, dsDNA. We stained the ER-resident protein Calnexin and measured the co-localization of Calnexin and STING in pDCs after transfection of ISD for 2 hours. In unstimulated pDCs, STING was found to be highly co-localized with Calnexin, supporting its ER-resident status. Upon transfection of dsDNA, STING was dissociated from ER, while a similar effect was not seen following ssDNA transfection (Fig. 3E, 3F).
pDCs produce IFN-α and IFN-λ upon cGAS-STING pathway stimulation
Triggering of the cGAS-STING pathway has been shown to result in type I IFN production in monocytes, macrophages, and dendritic cell populations (33). Given the pathway’s reliance on IRF3-mediated cytokine induction, IFN-β has been the focus of most cGAS-STING pathway-mediated interferon studies (9, 34). Human pDCs are known as the most potent producers of IFN-α and earlier studies have found IFN-α production by pDCs upon cGAS-STING pathway induction (19, 20). As type I and type III interferons are both produced by pDCs upon TLR7 and TLR9-mediated stimulation, we hypothesized that cGAS-STING pathway stimulation will also induce production of these two cytokines. We further explored the production of an NF-κB-dependent cytokine, TNF-α (35), upon cGAS-STING pathway stimulation in pDCs.
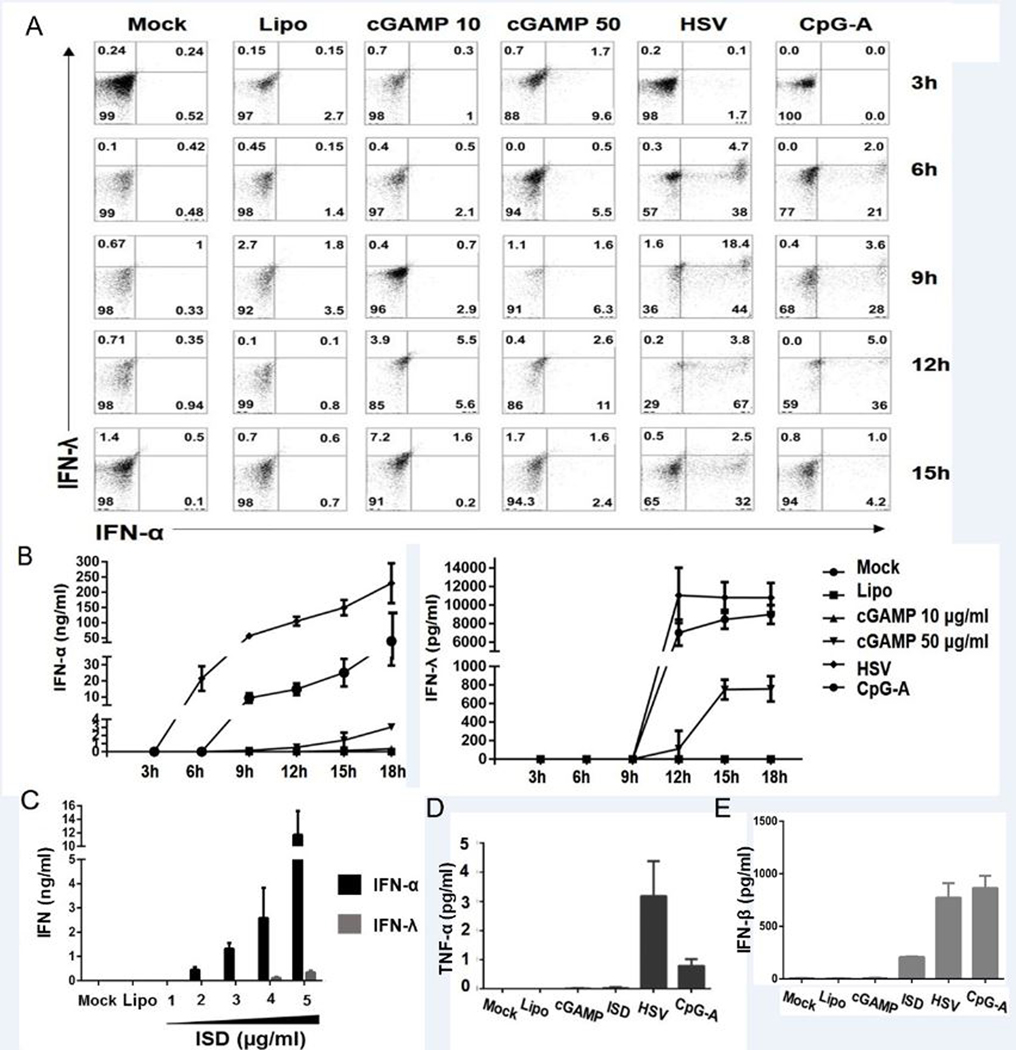
We observed that stimulation with synthetic cGAMP induced both IFN-α and IFN-λ in pDCs in a time-dependent manner (Fig. 4A, 4B). It is notable that the amount of production of both IFN-α and IFN-λ was almost 10-fold and 100-fold less compared to TLR9-agonist CpG-A-mediated or dsDNA virus HSV-1-mediated interferon production, respectively, and was only observed with the 50 μg/ml but not with the 10 μg/ml concentrations of cGAMP. Upon overnight stimulation with non-CpG double-stranded DNA ISD, pDCs produced both type I and type III interferon (Fig. 4C). However, no induction of TNF-α was observable via intracellular flow-cytometry (Supplemental Fig. 3A) or ELISA (Fig. 4 D). We also tested if stimulation with cGAMP can induce maturation of pDCs. We observed minimal-to-no induction of CD40 and CD86 expression at time-points out to 18 hr following stimulation with cGAMP (Supplemental Fig. 3, B,C). We also tested the IFN-β yield by pDCs upon stimulation of cGAS-STING and TLR9-pathway. We observed no IFN-β production upon 6 hour stimulation (data not shown), but we observed IFN-β yield with ISD, HSV, and CpG-A stimulation overnight (Fig. 4E).
FIGURE 4.
pDCs produce type-I and type-III interferon, but not TNF-α, upon stimulation of cGAS-STING pathway. (A)Purified pDCs were stimulated for 3, 6, 9, 12, and 15 hr. with cyclic GMP-AMP (cGAMP), HSV and the TLR9 agonist CpG-A and the expression of IFN-α and IFN-λ were tested via flow cytometry. BFA was added 2 hrs prior to each time point. Cells were stained for pDC markers, fixed, permeabilized, stained for intracellular cytokines and acquired by flow cytometer. Data are from a representative experiment from two individual donors. (B) Purified pDCs (0.25 × 106 cells/ml) were stimulated with HSV, CpG-A or lipofected with cGAMP (10 and 50 μg/ml) and supernatant was collected at each time point to perform ELISA for IFN-α and IFN-λ. Data are shown as mean ± 1 SD (n = 3 independent experiments). (C) pDCs were lipofected with ISD for 18 hours. Supernatants were collected and production of IFN-α and IFN-λ was measured by ELISA. Data are shown as mean ± 1 SD (n = 3 independent experiments). (C) pDCs were lipofected with ISD for 18 hours. Supernatants were collected and production of IFN-α was measured by ELISA. Data are shown as mean ± 1 SD (n = 3 independent experiments). (D, E) pDCs were stimulated with HSV, CpG-A or lipofected with cGAMP (50 μg/ml) or ISD (5 μg/ml) for 18 hours. Supernatants were collected and production of (D) TNF-α and (E) IFN-β was measured by ELISA. Data are shown as mean ± 1 SD (n = 3 independent experiments).
We have used THP1 cells as a positive control to measure the efficiency of cGAMP lipofection and measured IFN-β yield upon overnight stimulation. IFN-β production by THP1 cells was comparable to previous report (Supplementary Fig. 2) (9).
Stimulation of pDCs by cGAMP and ISD is dependent upon STING
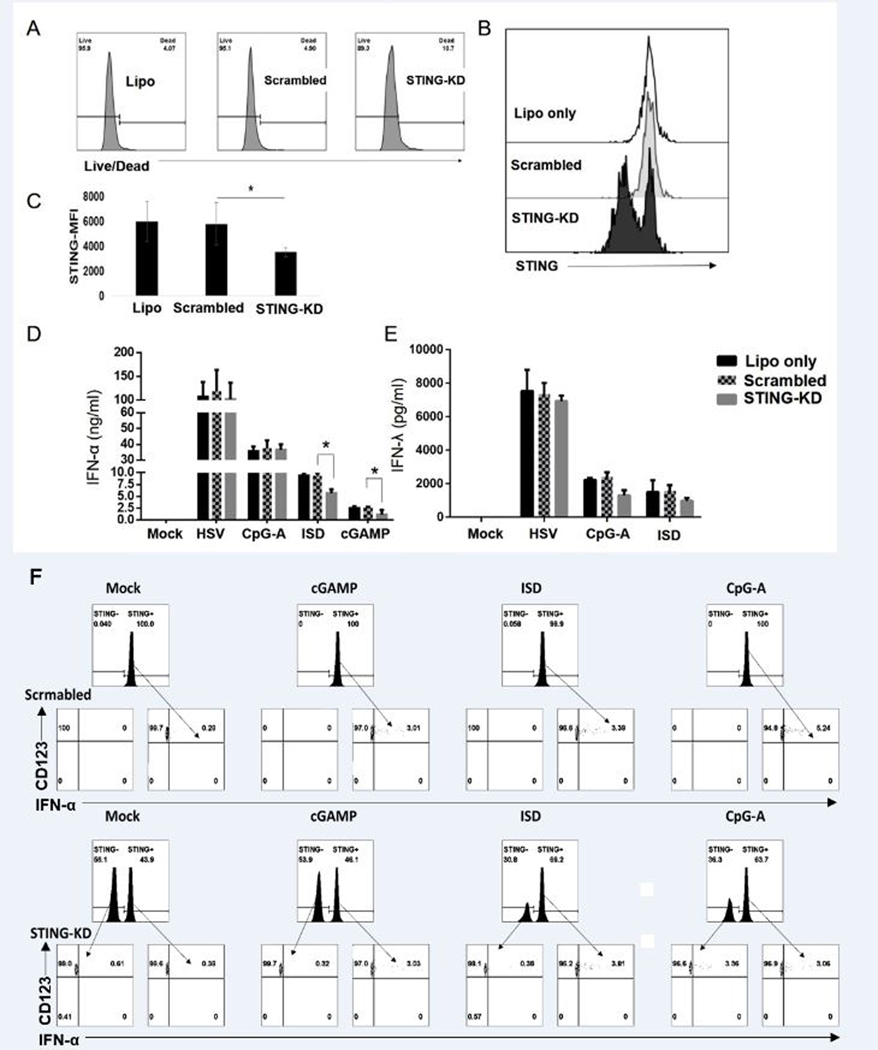
Apart from the cGAS-STING pathway, other cytosolic DNA sensing pathways have been reported involving the innate sensors including DAI, IFI16, and AIM2 (17, 36). To confirm that our observed immunostimulatory effect of cGAMP and ISD on pDCs is mediated by STING, we performed siRNA-mediated knockdown of STING in primary pDCs. We lipofected the purified pDCs from healthy donors with siRNA specific for STING or a scrambled version of the same siRNA using a silencer select siRNA that in itself does not have any immunostimulatory effect. As evidenced by live/dead staining (Zombie dye, BioLegend), siRNA-mediated knockdown did not affect the survival of pDCs (Fig. 5A) and significantly knocked down the expression of STING (Fig. 5B, 5C).
FIGURE 5.
STING knock-down of pDCs down-regulates interferon production upon ISD or cGAMP stimulation. pDCs were lipofected with siRNA targeting STING or scrambled siRNA for 24 hours. (A) The survival of pDCs was measured via live/dead staining by flow cytometry. (B) Sample histograms showing the efficacy of knock-down of STING. (C) Summary data for STING expression (MFI) in pDCs lipofected with or without siRNA targeting STING or scrambled siRNA (n=3 independent experiments). (D, E) pDCs incubated with siRNA for 24 hours were washed and resuspended in fresh media and stimulated with HSV, CpG-A, ISD, and cGAMP for another 24 hours. Supernatants were collected and ELISA was performed to measure IFN-α and IFN-λ (n = 3 indepdendent experiments). (F) pDCs lipofected with scrambled siRNA or siRNA targeting STING for 48 hours were washed, resuspended in media, and stimulated with cGAMP or ISD for 3 hours, or CpG-A for 6 hours. IFN-α production by pDCs were measured via flow cytometry (Data represents of one of 2 independent experiments).
Next, we stimulated the pDCs with or without STING knockdown with HSV, CpG-A, ISD, and cGAMP for another 24 hours, collected the supernatant, and performed ELISA to measure IFN-α and IFN-λ1. STING-KD significantly downregulated the production of IFN-α induced by both ISD and cGAMP (Fig. 5D) but had less effect on IFN-λ1 production (Fig. 5E). We also performed flow-cytometric analysis to test the effect of STING-KD on cGAS-STING-mediated and TLR9-mediated IFN production. We observed that in the pDC-subset where STING-KD was successful, IFN-α production via cGAMP and ISD stimulation was elimininated. Yet, STING-KD did not affect CpG-A-mediated IFN-α production (Fig. 5F).
cGAS-STING pathway induces type I and type III IFN production in pDCs via the TBK1-IRF3 axis
Interferon production in pDCs in response to viral stimuli mostly relies on phosphorylation and nuclear translocation of IRF7 (37). On the other hand, cGAS-STING pathway has been reported to be operating via phosphorylation, dimerization, and nuclear translocation of IRF3 facilitated by TBK1 (38). To investigate the role of TBK1 in the cGAS-STING pathway in pDCs, we pre-treated purified pDCs with the TBK1 inhibitor BX795 or an inhibitory ODN (iODN) that blocks TLR9 signaling. After pre-treating with either of the inhibitors for 6 hours, we stimulated pDCs with cGAMP, ISD, HSV, or CpG-A overnight, and performed ELISAs to measure IFN-α, IFN-β, and IFN-λ production. Pre-treatment with the TBK1 inhibitor downregulated IFN-α, and IFN-β production in pDCs upon stimulation with ISD or cGAMP but not HSV or CpG-A, while pre-treatment with iODN downregulated IFN-α production by CpG-A and HSV but not ISD or cGAMP-induced IFN-α (Fig. 6A). We have also seen inhibitory effect of BX795 on pDCs upon 1 hour pretreatment following overnight stimulation (Supplementary Figure 3E). Our results support that TBK1 is involved in cGAS-STING pathway in pDCs and not in TLR9-mediated IFN production.
FIGURE 6.
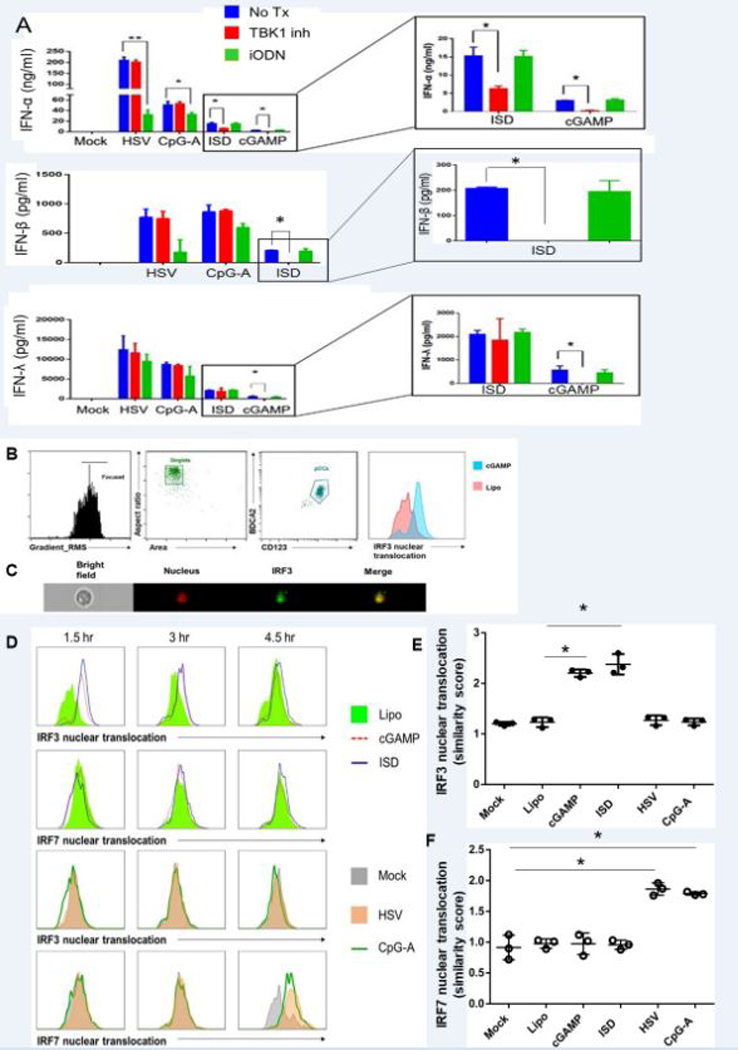
cGAS-STING pathway mediated interferon production in pDCs is dependent on the TBK1-IRF3 axis. (A) Purified pDCs were pre-treated for 6 hours with TBK1 inhibitor (BX795) or TLR9 inhibitor (iODN) and stimulated with HSV, CpG-A or ISD for 18 hours. Supernatants were collected and ELISAs for IFN-α and IFN-λ were performed. (B, C, D) pDCs transfected with cGAMP or ISD were stained for surface markers, fixed, permeabilized, and stained for nucleus (DRAQ5) and IRF3 or IRF7 and acquired via Amnis ImageStream. (B) Gating strategy showing how pDCs were identified among single-cells as CD123+BDCA2+ cells. Nuclear co-localization was seen in pDCs expressing IRF3. (C) Single cell image of an interferon producing pDC showing nuclear co-localization of IRF3 upon cGAMP stimulation. (D) Nuclear translocation of IRF3 and IRF7 upon cGAMP, ISD, HSV, and CpG-A stimulation was measured at 1.5, 3, and 4.5 hrs. (E,F) Quantification of nuclear translocation of IRF3 and IRF7 upon different stimuli. Each dot represents one independent experiment.
Next, we investigated if the cGAS-STING pathway functions via TBK1-mediated IRF3 phosphorylation and translocation. We stimulated purified pDCs with cGAMP, ISD, HSV, or CpG-A and observed phosphorylation and nuclear translocation of IRF3 and IRF7 respetively by flow cytometry and Amnis ImageStream. Upon stimulation with cGAMP and ISD for 2 hours we did see phosphorylation of IRF3, which was not seen in HSV and CpG-A mediated stimulation of 4 hour (Supplementary Fig 4A). On the contrary, we observed IRF7 phosphorylation with HSV and CpG-A stimulation but not by cGAMP and ISD stimulation (Supplementary Fig.4B). We found a time-dependent nuclear translocation of IRF3 upon stimulation by either cGAMP or ISD, while we did not observe any translocation of IRF7 (Fig. 6B, C). In contrast, when we stimulated pDCs with CpG-A and HSV, we observed IRF7 translocation as expected with no IRF3 translocation (Fig. 6D, 6E). Our data indicate that, unlike TLR9-mediated activation, the cGAS-STING pathway in human pDCs relies on IRF3 and not IRF7.
The cGAS-STING pathway has been reported to be able to induce IKK-β-mediated nuclear translocation of NF-κB. Although we did not see the induction of NF-κB-dependent cytokines or co-stimulatory molecules in pDCs upon cGAS-STING stimulation (Fig. 4), we investigated if there is any observable nuclear translocation of NF-κB, or another IKK-β-dependent transcription factor, IRF5. We did not observe any nuclear translocation of NF-κB or IRF5 upon cGAMP or ISD stimulation (data not shown), suggesting that this arm of the cGAS-STING pathway is not functioning in human pDC.
cGAS-STING pathway stimulation inhibits TLR9-mediated IFN production, possibly by inducing SOCS1 and SOCS3 expression
Our observations suggested that the cGAS-STING pathway and TLR9-mediated pathway in human pDCs do not share mediators to induce IFN production and, instead, work parallel to each other. Hence we hypothesized that triggering of both pathways will have an additive effect in terms of total yield of cytokines. To our surprise, when pDCs were pre-stimulated either with cGAMP or ISD for 6 hours and challenged by the synthetic TLR9 agonists CpG-A or dsDNA virus HSV-1 overnight, we observed a marked inhibition of total interferon production compared to stimulation with CpG-A or HSV-1. In contrast, pre-treatment with CpG-A, which signals through TLR9, had not effect on subsequent response to HSV-1 (Fig. 7A). Instead of pre-treatment, if we simultaneously stimulated pDCs with cGAMP and HSV or ISD and HSV, we also observed some inhibition, especially with ISD. However, the inhibitory effect is more moderate than with pre-treatment (Supplementary Figure 4D).
FIGURE 7.
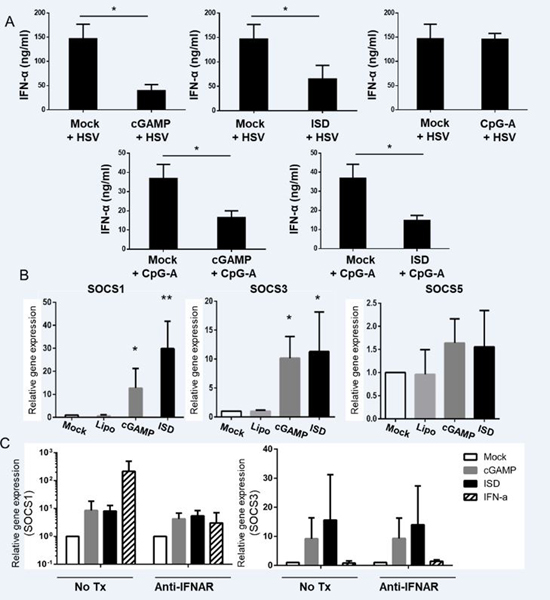
Pre-stimulation of cGAS-STING pathway inhibits TLR9-mediated cytokine production in pDCs probably by inducing SOCS1/3 but not SOCS5. (A) pDCs were stimulated for 6 hours for cGAS-STING pathway (via cGAMP or ISD) or TLR9 pathway (via CpG-A) followed by stimulation with HSV or CpG-A for 18 hours. Supernatants were collected and ELISAs were performed to measure IFN-α (n =3 independent experiments). (B) pDCs stimulated with cGAMP or ISD for 3 hours and qRT-PCR was performed to measure the induction of SOCS1, SOCS3, and SOCS5. (n =3 independent experiments) (C) pDCs pre-treated with or without anti IFNAR antibody for 1 hour were stimulated with cGAMP, ISD, or IFN-α for 3 hours. qRT-PCR was performed to measure the induction of SOCS1 and SOCS3 (n = 3 independent experiments).
Next, we investigated how the cGAS-STING pathway effects its inhibitory function on pDCs. SOCS molecules have been shown to play a key role in limiting the immune activation in cell types including pDCs (39). Recently, it was reported that plasmodium infection in a murine models exerts an inhibitory effect on pDCs by inducing SOCS molecules via the cGAS-STING pathway (40). Hence, we hypothesized that a similar mechanism might be in place in human pDCs. We stimulated primary human pDCs with cGAMP, ISD, or CpG-A for 3 hours and performed qRT-PCR to test the gene transcription of SOCS1, SOCS3, and SOCS5. We observed that SOCS1 and SOCS3, but not SOCS5 were upregulated with cGAS-STING stimulation (Fig.7B). To test if the SOCS1 and SOCS3 induction by cGAMP and ISD is direct or via an IFN-α-mediated autocrine effect, we pre-treated pDCs with anti-IFNAR antibody for 1 hour and stimulated pDCs with cGAMP and ISD for 3 hours. Treatment with anti-IFNAR antibody did not inhibit the expression of SOCS1 and SOCS3, but did inhibit their upregulation by exogenous IFN-α, indicating that their induction by cGAMP and ISD was directly mediated by the cGAS-STING pathway (Fig.7C).
We also tested if pre-treatment with cGAMP or ISD inhibits virus uptake by pDCs and, thus, if the inhibitory effect we observed is merely a mechanistic one. We pretreated purified pDCs with or without cGAMP or ISD for 3 hours and 6 hours and incubated them with HSV constitutively expressing GFP for 30 minutes. Next, we washed the cells, stained for surface markers of pDCs, and acquired them via imaging flow to measure uptake of GFP-tagged HSV by them. We did not observe any moderation of virus uptake following cGAMP and ISD pretreatment (Supplementary Fig. 4C).
Inhibitory cytokine IL-10 is known for imparting an inhibitory effect on innate immune cells by inducing SOCS3 (but not SOCS1) expression (41, 42). Hence we wanted to make sure that the inhibitory effect of cGAS-STING was not via induction and autocrine effect of IL-10 on pDCs. We did not observe any detectable IL-10 being produced by purified pDCs upon stimulation with cGAMP or ISD for 6 hours or 18 hours, suggesting that the SOCS induction via cGAS-STING pathway stimulation is a direct effect of the pathway, rather than mediated through IL-10. Yu and colleagues recently suggested a mechanism of SOCS1 and SOCS3-mediated inhibition of IFN production. They showed SOCS1/3 can induce degradation of IRF7 when transfected into HEK239 cells (54). To test whether a similar mechanism is at play in primary pDCs, we measured the expression of IRF7 in pDCs stimulated by cGAMP and ISD (Supplementary fig. 4E). We did not observe any downregulation of IRF7 suggesting that our observed inhibition possibly follows an alternate mechanism. Thus, our data suggest that that triggering of the cGAS-STING pathway in pDCs can induce expression of SOCS molecules which, in turn, is correlated with the inhibition of the TLR9 pathway-mediated IFN production.
Discussion
In the present study, we have investigated the role of two major innate DNA-sensing pathways in primary human pDCs and probed how they intersect with each other. We confirmed that both the cGAS-STING and TLR9 pathway are operative in pDCs and are capable of inducing type I and type III IFN, but use separate cellular machinery. We further demonstrated that activating the cGAS-STING pathway in pDCs induces SOCS1 and SOCS3, which may contribute to inhibition of TLR9-mediated interferon production.
The innate sensing of DNA in the cytosolic compartment of cells has been a major field of investigation. Early studies have pointed towards the existence of cytosolic DNA sensors in different cell types including murine pDCs (31, 43). The role of STING was established in the cytosolic sensing of DNA, and multiple segments in the transmembrane portion of its C-terminal domain were found to be able to bind to the bacterial cyclic di-nucleotide cyclic di-GMP (11). The crystal structure of STING showed that the C-terminal segments form V-shaped dimers to form the groove for binding of cyclic di-nucleotides (44). However, the lack of DNA-binding domain on STING made a separate DNA sensor necessary for its function. The discovery of cGAS provided the upstream arm of the pathway. The recent upsurge in the publications related to this pathway has cemented its central role in innate sensing of cytosolic DNA sensing.
The exclusivity of the STING-TBK1-IRF3 axis is not universal in all cell types. Earlier studies suggested that there might be a possible interaction between STING and both IRF3 and IRF7, but not IRF9 (45). Later, it was shown that STING-mediated cytokine production, induced by a DNA vaccine, was IRF3 as well as IRF7 dependent in a murine model. In fact, antigen presentation of a DNA vaccine was reported to be IRF7-dependent but independent of IRF3 as well as cGAS (46). Given the high expression of IRF7 in pDCs (3), a role for this transcription factor in cGAS-STING-mediated IFN production was possible. However, we did not see any nuclear translocation of IRF7 in pDC upon cGAS-STING pathway stimulation.
The role of pDCs as the principal type I IFN producers places them at a key position of host-microbe interaction as well as autoimmune disorders. The equilibrium between induction and inhibition of IFN production is a delicate balance, the disruption of which causes either pathology or inadequate immune response (47). Several mechanisms of inhibitory immune modulation have been proposed in pDCs. While molecules like indoleamine 2,3-dioxygenase (IDO) (48, 49), T cell Ig and mucin-domain containing molecule-3 (Tim-3) (50) have been shown to exert an inhibitory effect on pDCs, other molecules such as CCR9 and Granzyme-B (51) render pDCs more tolerogenic. Whether SOCS molecules play a role to subdue the immune response in human pDCs is a relatively open question. Earlier studies indicated that SOCS molecules impart a feedback regulatory effect on activated dendritic cells (52). Hepatatis B virus surface antigen was reported to inhibit interferon expression in pDCs by induction of SOCS1 (53). A recent study has also reported SOCS1 and SOCS3-mediated inhibition of IFN-α production in pDCs following polyubiquitination and degradation of IRF7 (54).
We have previously published the inhibitory role of IL-10 on IFN production by PBMCs and pDCs (55, 56). Hence, we tested the possibility of a cGAS-STING-mediated induction of IL-10 in pDCs imparting an autocrine autoinhibitory effect. We did not find any detectable IL-10 production by pDCs with stimulation with either cGAMP or ISD, indicating that the inhibitory effect downstream to cGAS-STING pathway is not mediated by an IL-10 autocrine loop.
SOCS molecules have been used by different pathogens to evade the IFN-mediated immune response (39, 57). The TLR-mediated pathway has been indicated in the induction by SOCS molecules downstream to RSV infection, but the data suggests that the effect is majorly orchestrated by an autocrine-loop of type I IFN (58). The interaction between SOCS molecules with the SH2 domain of STAT molecules to modulate the JAK-STAT pathway is well-documented (59). The surface TLRs such as TLR2 and TLR4 are modulated by SOCS via the poly-ubiquitination and degradation of the adaptor protein Mal (60). How the SOCS molecule may downregulate TLR9-mediated IFN production is yet to be explored. It is possible that SOCS molecules are capable of ubiquitinating and degrading any of the molecules downstream to TLR9 pathway, namely IRAKs, components of the IKK complex or IRFs, especially IRF7. In fact, the targeting of IRF by SOCS was recently documented as an immune evasive mechanism employed by HTLV1 (61).
In spite of its central role in cytosolic DNA-mediated IFN production, the immunosuppressive role of STING has also been reported. DNA-sensing involving STING seemed to impart a tolerogenic effect on murine myeloid dendritic cells (62). In a lupus mouse model, STING deficiency, but not IRF3 deficiency, resulted in marked immune activation. STING-deficient murine macrophages demonstrated an exacerbated reaction to both TLR7 and TLR9 stimuli. Immune activation in STING null mice was similar to that of SOCS1 and SOCS3 null mice, indicating a possible IRF3-independent immunosuppressive role via STING-SOCS1/3 (63). In a mouse model of malarial infection, cGAS-STING pathway has been shown to induce SOCS1/3 to downregulate TLR9 signaling (64). On the contrary, a synergistic role of TLR9 and STING has also been reported (65).
Our study has shown that the role of cGAS-STING and TLR9-mediated pathways in pDCs are more antagonistic than synergistic. We have established that cGAS-STING pathway is functional in primary human pDCs and are capable of producing type I and type III IFNs upon triggering. Unlike TLR9-mediated interferon production, cGAS-STING pathway uses IRF3 instead of IRF7 to induce IFN production. The cGAS-STING pathway activation inhibits TLR9-mediated IFN production, possibly by inducing the inhibitory molecules SOCS1 and SOCS3. These results indicate that the cGAS-STING pathway in human pDCs might be harnessed to inhibit TLR9-mediated aberrant and untrammeled IFN production leading to autoimmune disorders.
Supplementary Material
Key points:
A functional cGAS-STING pathway exists in human plasmacytoid dendritic cells.
Activation of cGAS-STING in pDCs inhibits TLR9-mediated interferon production.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases Grants R01AI026806 and R01AI106125 to PFB.
Abbreviations used in this article:
- cGAMP
Cyclic guanosine monophosphate–adenosine monophosphate
- cGAS
Cyclic GMP-AMP synthase
- STING
Stimulator of interferon genes
- IAV
Influenza virus
- SeV
Sendai virus
- IRF
IFN response factor
- MOI
multiplicity of infection
- pDC
plasmacytoid dendritic cell
- TBK
tank-binding protein
- ISD
Immunostimulatory DNA
- PFA
paraformaldehyde
- SOCS
Suppressor of cytokine signaling
References:
- 1.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, and Liu YJ. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science 284: 1835–1837. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald-Bocarsly P, Dai J, and Singh S. 2008. Plasmacytoid dendritic cells and type I IFN: 50 years of convergent history. Cytokine Growth Factor Rev 19: 3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izaguirre A, Barnes BJ, Amrute S, Yeow WS, Megjugorac N, Dai J, Feng D, Chung E, Pitha PM, and Fitzgerald-Bocarsly P. 2003. Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J Leukoc Biol 74: 1125–1138. [DOI] [PubMed] [Google Scholar]
- 4.Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, and Uze G. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur J Immunol 34: 796–805. [DOI] [PubMed] [Google Scholar]
- 5.Yin Z, Dai J, Deng J, Sheikh F, Natalia M, Shih T, Lewis-Antes A, Amrute SB, Garrigues U, Doyle S, Donnelly RP, Kotenko SV, and Fitzgerald-Bocarsly P. 2012. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J Immunol 189: 2735–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desmet CJ, and Ishii KJ. 2012. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat Rev Immunol 12: 479–491. [DOI] [PubMed] [Google Scholar]
- 7.Gilliet M, Cao W, and Liu YJ. 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol 8: 594–606. [DOI] [PubMed] [Google Scholar]
- 8.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, and Vance RE. 2013. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Reports 3: 1355–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, and Chen ZJ. 2013. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339: 826–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, Fitzgerald KA, Hayakawa Y, and Vance RE. 2009. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med 206: 1899–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, and Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478: 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa H, and Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka Y, and Chen ZJ. 2012. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal 5: ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa H, Ma Z, and Barber GN. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito T, Kanzler H, Duramad O, Cao W, and Liu YJ. 2006. Specialization, kinetics, and repertoire of type 1 interferon responses by human plasmacytoid predendritic cells. Blood 107: 2423–2431. [DOI] [PubMed] [Google Scholar]
- 16.Barchet W, Cella M, Odermatt B, Asselin-Paturel C, Colonna M, and Kalinke U. 2002. Virus-induced interferon alpha production by a dendritic cell subset in the absence of feedback signaling in vivo. J Exp Med 195: 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Yuan B, Bao M, Lu N, Kim T, and Liu YJ. 2011. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol 12: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li XD, Wu J, Gao D, Wang H, Sun L, and Chen ZJ. 2013. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341: 1390–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paijo J, Doring M, Spanier J, Grabski E, Nooruzzaman M, Schmidt T, Witte G, Messerle M, Hornung V, Kaever V, and Kalinke U. 2016. cGAS Senses Human Cytomegalovirus and Induces Type I Interferon Responses in Human Monocyte-Derived Cells. PLoS Pathog 12: e1005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bode C, Fox M, Tewary P, Steinhagen A, Ellerkmann RK, Klinman D, Baumgarten G, Hornung V, and Steinhagen F. 2016. Human plasmacytoid dentritic cells elicit a Type I Interferon response by sensing DNA via the cGAS-STING signaling pathway. Eur J Immunol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.George TC, Morrissey PJ, Cui C, Singh S, and Fitzgerald Bocarsly P. 2009. Measurement of cytoplasmic to nuclear translocation. Curr Protoc Cytom Chapter 9: Unit 9.28. [DOI] [PubMed] [Google Scholar]
- 22.Marinho FV, Benmerzoug S, Oliveira SC, Ryffel B, and Quesniaux VFJ. 2017. The Emerging Roles of STING in Bacterial Infections. Trends Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsui T, Connolly JE, Michnevitz M, Chaussabel D, Yu CI, Glaser C, Tindle S, Pypaert M, Freitas H, Piqueras B, Banchereau J, and Palucka AK. 2009. CD2 distinguishes two subsets of human plasmacytoid dendritic cells with distinct phenotype and functions. J Immunol 182: 6815–6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryant C, Fromm PD, Kupresanin F, Clark G, Lee K, Clarke C, Silveira PA, Suen H, Brown R, Newman E, Cunningham I, Ho PJ, Gibson J, Bradstock K, Joshua D, and Hart DN. 2016. A CD2 high-expressing stress-resistant human plasmacytoid dendritic-cell subset. Immunol Cell Biol. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Lepelley A, Azria E, Lebon P, Roguet G, Schwartz O, Launay O, Leclerc C, and Lo-Man R. 2013. Neonatal plasmacytoid dendritic cells (pDCs) display subset variation but can elicit potent anti-viral innate responses. PLoS One 8: e52003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Megjugorac NJ, Jacobs ES, Izaguirre AG, George TC, Gupta G, and Fitzgerald-Bocarsly P. 2007. Image-based study of interferongenic interactions between plasmacytoid dendritic cells and HSV-infected monocyte-derived dendritic cells. Immunological investigations 36: 739–761. [DOI] [PubMed] [Google Scholar]
- 27.Schoggins JW, MacDuff DA, Imanaka N, Gainey MD, Shrestha B, Eitson JL, Mar KB, Richardson RB, Ratushny AV, Litvak V, Dabelic R, Manicassamy B, Aitchison JD, Aderem A, Elliott RM, Garcia-Sastre A, Racaniello V, Snijder EJ, Yokoyama WM, Diamond MS, Virgin HW, and Rice CM. 2014. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505: 691–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, and Bieniasz P. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, and Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11: 997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Zhang H, Wu X, Ma D, Wu J, Wang L, Jiang Y, Fei Y, Zhu C, Tan R, Jungblut P, Pei G, Dorhoi A, Yan Q, Zhang F, Zheng R, Liu S, Liang H, Liu Z, Yang H, Chen J, Wang P, Tang T, Peng W, Hu Z, Xu Z, Huang X, Wang J, Li H, Zhou Y, Liu F, Yan D, Kaufmann SHE, Chen C, Mao Z, and Ge B. 2018. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563: 131–136. [DOI] [PubMed] [Google Scholar]
- 31.Stetson DB, and Medzhitov R. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24: 93–103. [DOI] [PubMed] [Google Scholar]
- 32.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, and Akira S. 2009. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 106: 20842–20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q, Sun L, and Chen ZJ. 2016. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol 17: 1142–1149. [DOI] [PubMed] [Google Scholar]
- 34.Abe T, Harashima A, Xia T, Konno H, Konno K, Morales A, Ahn J, Gutman D, and Barber GN. 2013. STING recognition of cytoplasmic DNA instigates cellular defense. Mol Cell 50: 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Falvo JV, Tsytsykova AV, and Goldfeld AE. 2010. Transcriptional Control of the TNF Gene. Current directions in autoimmunity 11: 27–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, and Cheng G. 2012. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol 13: 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai J, Megjugorac NJ, Amrute SB, and Fitzgerald-Bocarsly P. 2004. Regulation of IFN regulatory factor-7 and IFN-alpha production by enveloped virus and lipopolysaccharide in human plasmacytoid dendritic cells. J Immunol 173: 1535–1548. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH 3rd, Becker KG, and Ko MS. 2000. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci U S A 97: 9127–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bode JG, Ludwig S, Ehrhardt C, Albrecht U, Erhardt A, Schaper F, Heinrich PC, and Haussinger D. 2003. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J 17: 488–490. [DOI] [PubMed] [Google Scholar]
- 40.Yu X, Cai B, Wang M, Tan P, Ding X, Wu J, Li J, Li Q, Liu P, Xing C, Wang Helen Y., Su X.-z., and Wang R-F. Cross-Regulation of Two Type I Interferon Signaling Pathways in Plasmacytoid Dendritic Cells Controls Anti-malaria Immunity and Host Mortality. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niemand C, Nimmesgern A, Haan S, Fischer P, Schaper F, Rossaint R, Heinrich PC, and Muller-Newen G. 2003. Activation of STAT3 by IL-6 and IL-10 in primary human macrophages is differentially modulated by suppressor of cytokine signaling 3. J Immunol 170: 3263–3272. [DOI] [PubMed] [Google Scholar]
- 42.Ding Y, Chen D, Tarcsafalvi A, Su R, Qin L, and Bromberg JS. 2003. Suppressor of Cytokine Signaling 1 Inhibits IL-10-Mediated Immune Responses. The Journal of Immunology 170: 1383. [DOI] [PubMed] [Google Scholar]
- 43.Hochrein H, Schlatter B, O’Keeffe M, Wagner C, Schmitz F, Schiemann M, Bauer S, Suter M, and Wagner H. 2004. Herpes simplex virus type-1 induces IFN-alpha production via Toll-like receptor 9-dependent and -independent pathways. Proc Natl Acad Sci U S A 101: 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shu C, Yi G, Watts T, Kao CC, and Li P. 2012. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol 19: 722–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, and Shu HB. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29: 538–550. [DOI] [PubMed] [Google Scholar]
- 46.Suschak JJ, Wang S, Fitzgerald KA, and Lu S. 2016. A cGAS-Independent STING/IRF7 Pathway Mediates the Immunogenicity of DNA Vaccines. J Immunol 196: 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fitzgerald-Bocarsly P, and Jacobs ES. 2010. Plasmacytoid dendritic cells in HIV infection: striking a delicate balance. Journal of Leukocyte Biology 87: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fallarino F, Asselin-Paturel C, Vacca C, Bianchi R, Gizzi S, Fioretti MC, Trinchieri G, Grohmann U, and Puccetti P. 2004. Murine plasmacytoid dendritic cells initiate the immunosuppressive pathway of tryptophan catabolism in response to CD200 receptor engagement. J Immunol 173: 3748–3754. [DOI] [PubMed] [Google Scholar]
- 49.Boasso A, Herbeuval JP, Hardy AW, Anderson SA, Dolan MJ, Fuchs D, and Shearer GM. 2007. HIV inhibits CD4+ T-cell proliferation by inducing indoleamine 2,3-dioxygenase in plasmacytoid dendritic cells. Blood 109: 3351–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz JA, Clayton KL, Mujib S, Zhang H, Rahman AK, Liu J, Yue FY, Benko E, Kovacs C, and Ostrowski MA. 2017. Tim-3 is a Marker of Plasmacytoid Dendritic Cell Dysfunction during HIV Infection and Is Associated with the Recruitment of IRF7 and p85 into Lysosomes and with the Submembrane Displacement of TLR9. J Immunol 198: 3181–3194. [DOI] [PubMed] [Google Scholar]
- 51.Jahrsdorfer B, Vollmer A, Blackwell SE, Maier J, Sontheimer K, Beyer T, Mandel B, Lunov O, Tron K, Nienhaus GU, Simmet T, Debatin KM, Weiner GJ, and Fabricius D. 2010. Granzyme B produced by human plasmacytoid dendritic cells suppresses T-cell expansion. Blood 115: 1156–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jackson SH, Yu CR, Mahdi RM, Ebong S, and Egwuagu CE. 2004. Dendritic cell maturation requires STAT1 and is under feedback regulation by suppressors of cytokine signaling. J Immunol 172: 2307–2315. [DOI] [PubMed] [Google Scholar]
- 53.Xu Y, Hu Y, Shi B, Zhang X, Wang J, Zhang Z, Shen F, Zhang Q, Sun S, and Yuan Z. 2009. HBsAg inhibits TLR9-mediated activation and IFN-alpha production in plasmacytoid dendritic cells. Mol Immunol 46: 2640–2646. [DOI] [PubMed] [Google Scholar]
- 54.Yu C-F, Peng W-M, Schlee M, Barchet W, Eis-Hübinger AM, Kolanus W, Geyer M, Schmitt S, Steinhagen F, and Oldenburg J. 2018. SOCS1 and SOCS3 Target IRF7 Degradation To Suppress TLR7-Mediated Type I IFN Production of Human Plasmacytoid Dendritic Cells. The Journal of Immunology: ji1700510. [DOI] [PubMed] [Google Scholar]
- 55.Payvandi F, Amrute S, and Fitzgerald-Bocarsly P. 1998. Exogenous and Endogenous IL-10 Regulate IFN-α Production by Peripheral Blood Mononuclear Cells in Response to Viral Stimulation. The Journal of Immunology 160: 5861. [PubMed] [Google Scholar]
- 56.Pierog PL, Zhao Y, Singh S, Dai J, Yap GS, and Fitzgerald-Bocarsly P. 2018. Toxoplasma gondii Inactivates Human Plasmacytoid Dendritic Cells by Functional Mimicry of IL-10. J Immunol 200: 186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yokota S, Yokosawa N, Okabayashi T, Suzutani T, Miura S, Jimbow K, and Fujii N. 2004. Induction of suppressor of cytokine signaling-3 by herpes simplex virus type 1 contributes to inhibition of the interferon signaling pathway. J Virol 78: 6282–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oshansky CM, Krunkosky TM, Barber J, Jones LP, and Tripp RA. 2009. Respiratory syncytial virus proteins modulate suppressors of cytokine signaling 1 and 3 and the type I interferon response to infection by a toll-like receptor pathway. Viral Immunol 22: 147–161. [DOI] [PubMed] [Google Scholar]
- 59.Croker BA, Kiu H, and Nicholson SE. 2008. SOCS regulation of the JAK/STAT signalling pathway. Semin Cell Dev Biol 19: 414–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, Nicholson SE, Hilton DJ, O’Neill LA, and Hertzog PJ. 2006. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol 7: 148–155. [DOI] [PubMed] [Google Scholar]
- 61.Oliere S, Hernandez E, Lezin A, Arguello M, Douville R, Nguyen TL, Olindo S, Panelatti G, Kazanji M, Wilkinson P, Sekaly RP, Cesaire R, and Hiscott J. 2010. HTLV-1 evades type I interferon antiviral signaling by inducing the suppressor of cytokine signaling 1 (SOCS1). PLoS Pathog 6: e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang L, Li L, Lemos H, Chandler PR, Pacholczyk G, Baban B, Barber GN, Hayakawa Y, McGaha TL, Ravishankar B, Munn DH, and Mellor AL. 2013. Cutting edge: DNA sensing via the STING adaptor in myeloid dendritic cells induces potent tolerogenic responses. J Immunol 191: 3509–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharma S, Campbell AM, Chan J, Schattgen SA, Orlowski GM, Nayar R, Huyler AH, Nundel K, Mohan C, Berg LJ, Shlomchik MJ, Marshak-Rothstein A, and Fitzgerald KA. 2015. Suppression of systemic autoimmunity by the innate immune adaptor STING. Proc Natl Acad Sci U S A 112: E710–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu X, Cai B, Wang M, Tan P, Ding X, Wu J, Li J, Li Q, Liu P, Xing C, Wang HY, Su XZ, and Wang RF. 2016. Cross-Regulation of Two Type I Interferon Signaling Pathways in Plasmacytoid Dendritic Cells Controls Anti-malaria Immunity and Host Mortality. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Temizoz B, Kuroda E, Ohata K, Jounai N, Ozasa K, Kobiyama K, Aoshi T, and Ishii KJ. 2015. TLR9 and STING agonists synergistically induce innate and adaptive type-II IFN. Eur J Immunol 45: 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.