Abstract
Background: microRNAs (miRNAs) are a small, endogenous non-coding RNAs that are involved in post-transcriptional gene regulation of many biological processes, including embryo implantation and placental development. In our previous study, miR-146a-5p was found expressed higher in the serum exosomes of pregnant sows than non-pregnant. The research on miR-146a-5p has been mainly related to human diseases, but there are few studies on its effects on the reproduction of sows in early pregnancy.
Objective: In this article, our motivation is to study the role of miR-146a-5p in the early pregnancy of sows on the cell proliferetion and apoptosis by targeting SMAD3 and SMAD4.
Methods: Bioinformatics software was used to identify the target genes of miR-146a-5p. The wild-type and mutant-type recombinant plasmids of dual-luciferase reporter with 3'-UTR of Smad3 or 3'-UTR of Smad4 were constructed, and co-transfected in porcine kidney cell (PK-15 cell) with miR-146a-5p mimic, mimic-NC(M-NC), inhibitor and inhibitor-NC(IN-NC), then dual-luciferase activity analysis, qRT-PCR and Western blot were performed to verify the target genes. After the transfection of BeWo choriocarcinoma cell (BeWo cell) with miR-146a-5p mimic, M-NC, inhibitor and IN-NC, the mRNA expression of Caspase-3, BAX and Bcl-2 was measured using qRT-PCR, and the cell proliferation was measured using CCK-8 kit.
Results: The luciferase, mRNA and protein expression of Smad3 in PK-15 cells treated by Smad3-3'-UTR-W co-transfected with miR-146a-5p mimic were significantly lower than that with miR-146a-5p M-NC, and the results of Smad4 were similar to Smad3, but the protein expression had a trend to lower in mimic group. The expression level of Bcl-2 in the miR-146a-5p mimic group was significantly lower than that in the miR-146a-5p M-NC group, but the expression pattern of Caspase-3 was just opposite. The mimic of miR-146a-5p reduced the proliferation of BeWo cells, however the inhibitor increased.
Conclusion: Smad3 and Smad4 are the direct target genes of miR-146a-5p. The expression of Smad3 and Smad4 were affected by the mimic and inhibitor of miR-146a-5p. miR-146a-5p affects cell apoptosis and proliferation by regulating their target genes. This study provided new data to understand the regulation mechanism of early pregnancy in sows.
Keywords: Smad3, Smad4, miR-146a-5p, cell proliferation, apoptosis, luciferase
1. INTRODUCTION
microRNAs (miRNAs) are short RNAs that direct messenger RNA degradation or disrupt mRNA translation in a sequence-dependent manner [1]. The primary biological function of miRNAs is to repress the translation of messenger targets, typically RNAs (mRNA), by binding to highly complementary the 3’ untranslated region [2]. In addition, the biological characteristics of microRNAs are extremely stable. For example, microRNAs are well preserved in tissue samples even after formalin fixation and paraffin embedding, and they can be efficiently extracted and evaluated [3]. Therefore, the stability and longevity of microRNAs in body fluids have made them attractive targets as biomarkers for a variety of conditions, including pregnancy disorders [4].
Lawrie et al. reported that miR-21 had the potential as a diagnostic biomarker to diffuse large B-cell lymphoma (DLBCL) and that the levels of serum miR-21 were associated with relapse-free survival in DLBCL patients [5]. Bogunia-Kubik et al. found that miR-146a might be involved in the pathogenesis of rheumatoid arthritis (RA) and suggested that miR-146a-3p polymorphism might be associated with miR-146a-5p levels in serum after anti-TNF-α treatment [6]. Momen-Heravi et al. found that the exosome-based delivery of miRNA-155 mimic or inhibitor had significant biological effects on hepatocytes and macrophages [7]. Certainly, many exosome miRNAs play important roles in human diseases. Vaksman et al. found that the exosomal miRNAs from ovarian carcinoma (OC) effusion supernatants affected cells in both tumors and the surrounding microenvironment, and they could induce more aggressive disease [8]. Wenchao et al. identified and analyzed the miRNAome of endometrial tissues on the 9th, the 12th and the 15th day of the pregnancy and the 12th day of non-pregnancy in Meishan and Yorkshire sows using Illumina sequencing and identified some miRNAs which may be involved in the embryonic implantation [9]. Xipeng Zhang et al. found that microRNA-23b inhibited the proliferation and migration of heat-denatured fibroblasts by targeting Smad3 [10]. Interestingly, in patients with endometriosis, miR-142-5p, miR-146a-5p, miR-1281, miR-940, and miR-4634 were significantly upregulated implantation [11]. At the implantation window phase, miR-142-5p and miR-146a-5p showed significantly enhanced expression in endometrium tissues of endometriosis infertility patients and may affect embryo implantation by acting on a variety of endometrial receptivity marker molecules [12].
Smad3 bound to the promoter region of P450arom and regulated aromatase expression in endometriosis [13]. Smad 2/3 transcripts were present in oocytes, early cleavage stages of embryos, and blastocysts. The presence of TGF-β receptors and their transducer proteins Smad 2/3 in the oocyte indicates a possibility that TGF-β can regulate oocyte processes [14]. During murine embryo development, Smad4 is required for gastrulation and later for anterior development [15]. In addition, the expression levels of Smad2, 3 and 4 in human granulosa cells are correlated with the oocyte quality and may contribute to fertilization rates [16].
Exosome miRNAs play an important role in reproduction, however, there were few reports on the miRNAs in serum exosome of sows during early pregnancy. In our previous study, miR-146a-5p was found differentially expressed in the serum exosomes of pregnant and non-pregnant sows on the 14th day after mating, and the predicted target genes were involved in the regulation of embryonic implantation. Therefore, we conducted this study to confirm it's predicted targeting genes Smad3, Smad4 and their regulation on cell proliferation and apoptosis.
2. MATERIALS AND METHODS
2.1. Prediction of Target Genes of miR-146a-5p and Construction of Dual-Luciferase Reporter Vector
Three online software programs (miRanda, RNAhybrid, and targetscan) were used to predict the target genes of miR-146a-5p, and the intersection of the three results was selected as candidate target genes [17, 18]. The candidate target genes were screened using the sequencing data and by referring to the published references.
The primers of wild type and mutant type vectors were designed according to the binding site predicted in 3'-UTR of target genes (Table 1). The recombinant plasmid of the dual-luciferase reporter with wild type or mutant type 3'-UTR of Smad3 was constructed as per the manual of dual-luciferase reporter system, so did Smad4.
Table 1.
Primers information of candidate target genes.
| Target Name | Sequence (5'→3') | Products Length (bp) |
|---|---|---|
| Ssc-Smad3-W-F | CCCTCGAGAGGCAATCAGGGCAGTTCTCGG | 413 |
| Ssc-Smad3-M-F | CCCTCGAGAGGCAATCAGGGCAGTGACCGG | |
| Ssc-Smad3-W-R | ATTTGCGGCCGCTCGGGGCTTGTCTCAAAATGT | |
| Ssc-Smad4-W-F | CCCTCGAGTTAAAGGCAGAGAAGTTCTCAAAG | 442 |
| Ssc-Smad4-M-F | CCCTCGAGTTAAAGGCAGAGAAGGGATCAAAG | |
| Ssc-Smad4-W-R | ATTTGCGGCCGCTTTAGTAACCAACCTTGTGCCT |
Note: Bold and italic bases are protected bases, and the underlined sequences are recognition sequences of restriction enzyme Not I and Xho I.
2.2. Dual-Luciferase Assay
PK15 cells cultured in a culture flask were digested when the cell density reached 60% to 80% confluence. The cells were starved with Opti-MEMTM medium and transfected 24 hours later. Each group of samples was set up in three parallel experimental groups with three replicates. The miRNA mimics, mimics NC (Ribobio, Guangzhou), psiCHECK-2-3'-UTR-wild-type plasmid and psiCHECK-2-3'-UTR-mutant plasmid were co-transfected into PK15 cells, respectively. Six hours after transfection, the culture medium was replaced, and cells were harvested 24 hours later.
2.3. qRT-PCR and Western Blot of Smad3 and Smad4
The PK-15 cells were resuscitated, passaged, and transfected at 60%-80% confluence. Four treatments were set in this experiment: miR-146a-5p mimic, miR-146a-5p M-NC, miR-146a-5p inhibitor and miR-146a-5p IN-NC. The cells were harvested 24 hours after transfection and then the cellular RNA and protein were extracted. Reverse transcription of cellular RNA was performed for qPCR (Table 2). The extracted cellular proteins were checked and denatured for Western blot using antibody Rabbit mAb #9523 for Smad3 (C67H9), Rabbit mAb #38454 (CST, USA) for Smad4 (D3M6U) and β-actin (I102) polyclonal antibody (Bioworld Technology, USA) as an internal reference.
Table 2.
Real-time PCR primers of SMAD3 and SMAD4.
| Target Name | Sequence (5'→3') | Products Length (bp) | Tm (ºC) |
|---|---|---|---|
| Smad3 | F:AGGAGGAGAAGTGGTGCGAGAA | 196 | 64 |
| R:AGGCGGCAGTAGATGACATGGG | |||
| Smad4 | F:CCATAGACAAGGTGGAGAGAGTG | 225 | 59 |
| R:CTCCAGAGACGAGCATAAATCAC | |||
| GAPDH | F:AACATCATCCCTGCTTCTACCG | 126 | 59 |
| R:GGTCAGATCCACAACCGACAC |
2.4. qRT-PCR of Apoptosis-related Genes
BeWo cells were cultured and transfected with the miR-146a-5p mimic, miR-146a-5p M-NC, miR-146a-5p inhibitor and miR-146a-5p IN-NC. The cells were seeded in 96-well plates at 1 x 104 cells/well. Transfection was conducted when the cells reached 60%-80% confluence. The cells were harvested and total RNA was purified for reverse transcription, then qRT-PCR was performed (Table 3).
Table 3.
Real-time PCR primers of apoptosis-related genes.
| Target Name | Sequence (5'→3') | Products Length (bp) | Tm (ºC) |
|---|---|---|---|
| Caspase-3 | F:GTCGATGCAGCAAACCTCAGGGAA | 156 | 60. 8 |
| R:GCTCAGAAGCACACAAACAAAACT | |||
| Bcl-2 | F:CTGTCGCAGAGGGGCTACGAGTGG | 154 | 60. 8 |
| R:TCTGCAGCGGCGAGGTCCTGGCGA | |||
| BAX | F:CCTTTTGCTTCAGGGTTTCATCCA | 188 | 60. 8 |
| R:AGTCTGTGTCCACGGCGGCAATCA | |||
| GAPDH | F:GCACCGTCAAGGCTGAGAAC | 138 | 60. 8 |
| R:TGGTGAAGACGCCAGTGGA |
2.5. Proliferation of BeWo Cells
BeWo cells were cultured as the proliferation of cells was determined at day 1, 2, 3, 4, and 5 after culturing using CCK-8 solution (Beyotime).
3. RESULTS
3.1. Target Gene Prediction and Construction of psiCHECK-2 Recombinant Plasmid
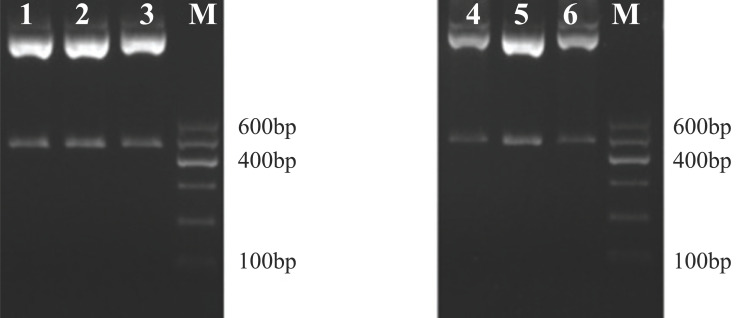
Smad3 and Smad4 were predicted as the target genes of miR-146a-5p. The 3′-UTR regions of the target genes were amplified by PCR. The recombinant psiCHECK plasmids with wildtype and mutation 3'-UTR of Smad3 or Smad4 were constructed using primer site-directed mutation of miRNA binding sites (Figure 1). The psiCHECK-2-3'-UTR-Smad3/4-W and psiCHECK- 2-3'-UTR-Smad3/4-M plasmids were successfully constructed (Figure 2).
Figure 1.
The binding site of miR-146a-5p to the Smad3 / Smad4 3'-UTR region.
Note: The underlined area indicates the target gene mutation site.
Figure 2.
Double enzyme digestion picture of psiCHECK-2-3'-UTR -Smad3/4-M recombinant plasmid.
Note: M is DNA Maker DL 600, 1-3 is the double enzyme digestion picture of psiCHECK-2-Smad3-M; 4-6 is the double enzyme digestion picture of psiCHECK-2-Smad4-M.
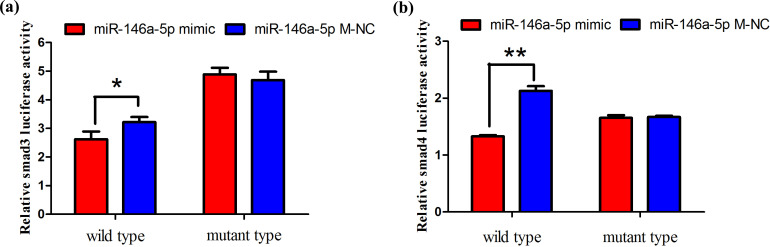
3.2. Dual-Luciferase Assay
Co-transfection of psiCHECK-2-Smad3/4-W or psiCHECK-2-Smad3/4-M dual-luciferase reporter vectors into PK15 cells was performed with the miR-146a-5p mimic or miR-146a-5p M-NC. The dual-fluorescence reporter system showed that luciferase activity of the Smad3-3'-UTR-W recombinant plasmid in the miR-146a-5p co-transformation group was significantly lower than that of the miR-146a-5p M-NC group, whereas, in the co-transformed Smad3-3'-UTR-M group, there was no significant change in luciferase activity between the miR-146a-5p mimic and miR-146a-5p M-NC, this further confirmed that the ssc-miR-146a-5p binds to the 3'-UTR of Smad3 via base comple-mentation (Figure 3a). The miR-146a-5p transfected group significantly reduced luciferase activity, indicating that Smad4 is the direct target gene of miR-146a-5p. When the recombinant plasmid Smad4-3'-UTR-M was co-transfected, there was no significant difference between the two groups, which further confirmed the above results (Figure 3b).
Figure 3.
(a, b) Double luciferase activity analysis of the miR-146a-5p on it’s target genes.
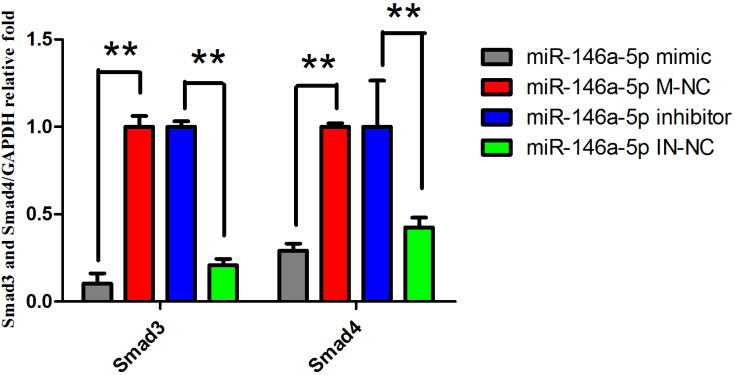
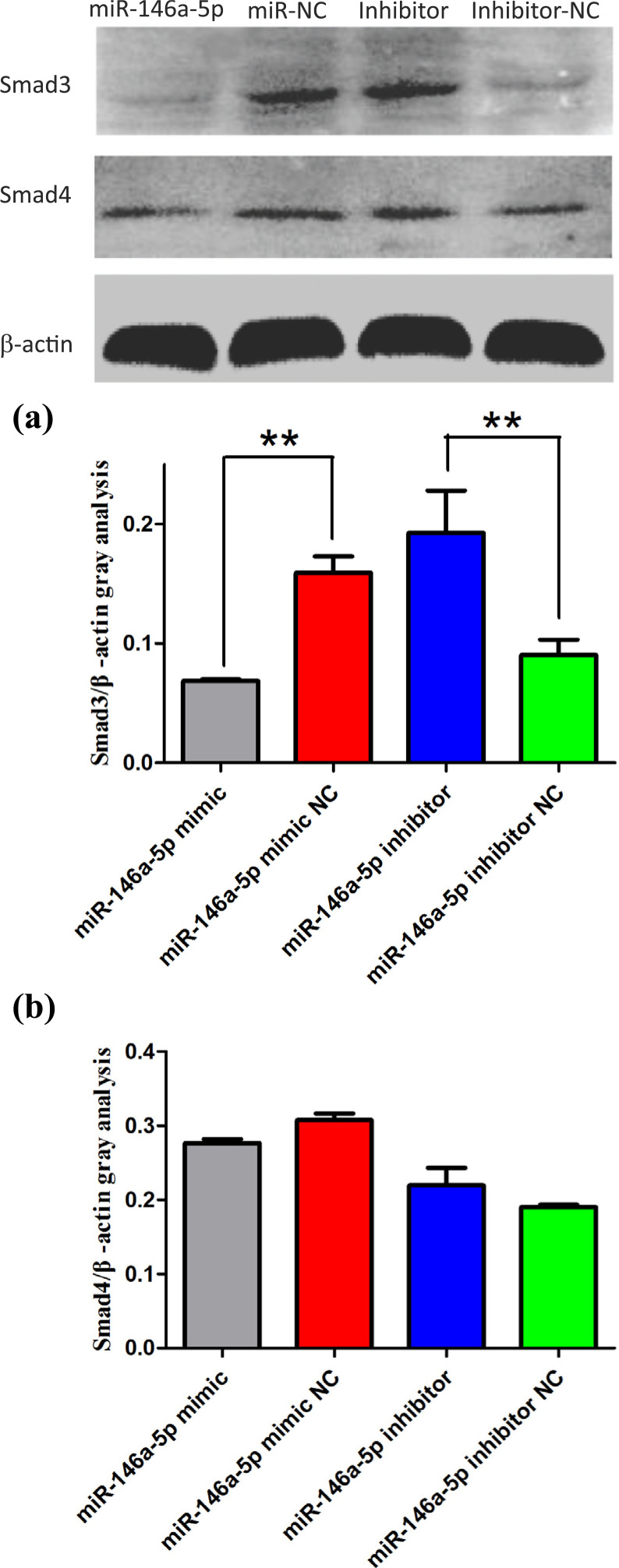
3.3. Mimic and Inhibitor of miR-146a-5p Affect the Expression of Smad3 and Smad4
To further study the role of miRNAs in the regulation of the target genes, the expression levels of Smad3 and Smad4 in the miR-146a-5p mimic, miR-146a-5p M-NC, miR-146a-5p inhibitor and miR-146a-5p IN-NC groups were measured using qRT-PCR and Western blotting (Figures 4 and 5). Compared with the miR-146a-5p M-NC group, the expression of Smad3 mRNA and protein was extremely significantly decreased in the miR-146a-5p mimic group (P<0. 01). Compared with the IN-NC group, both mRNA and protein expression of the Smad3 gene were significantly increased in the inhibitor group (P<0. 01). Smad4 was also proved as target gene of miR-146a-5p, because transfection of mimic reduced the mRNA expression of Smad4 significantly (P<0.01) and trend to reduce the protein expression of Smad4 compared with the M-NC group, the miRNA inhibitor group increased the mRNA expression of Smad4 extremely significantly (P<0.01) and had a trend to reduce the protein expression of Smad4 compared with the M-NC group. miR-146a-5p showed inhibitory effects on both mRNA and protein expression of SMAD3/4.
Figure 4.
mRNA expression of Smad3 and Smad4 in different treatment groups
Note: **P<0.01, The difference is highly significant. *P<0.05, The difference is significant, the same below.
Figure 5.
(a, b) Protein expression of Smad3 and Smad4 in different treatment groups.
3.4. The effect of miRNA on mRNA Expression of Apoptosis-related Genes in BeWo cell
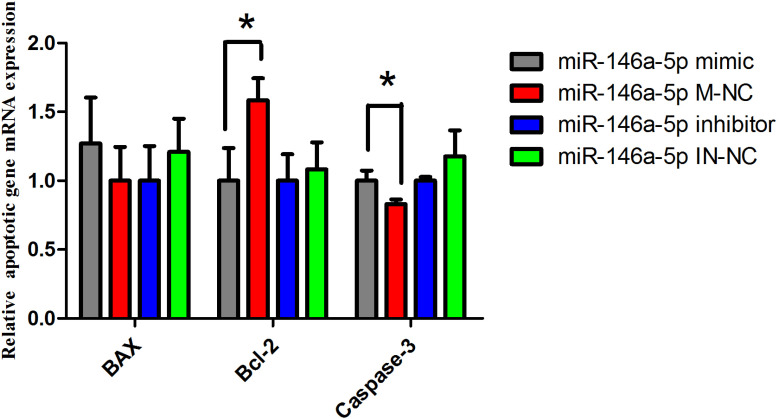
We examined the effects of miR-146a-5p via over-expression and inhibition of the miRNA in cultured BeWo cells. We quantified the mRNA expression levels of apoptosis-related genes by qRT-PCR. After 24 hours, we evaluated the expression level of the apoptosis genes Bcl-2, Caspase-3 and BAX. The expression level of the apoptosis suppressor gene Bcl-2 in the miR-146a-5p mimic group was significantly lower than that in the miR-146a-5p M-NC group (P<0.05), but there was no significant difference between the treatment of miR-146a-5p inhibitor and IN-NC (Figure 6). The expression of Caspase-3 mRNA was significantly increased after overexpression of the miRNA (P<0. 05), but there was no significant difference between the miR-146a-5p inhibitor group and IN-NC group; another apoptotic gene, BAX, had no difference between the miR-146a-5p mimic and M-NC group or the miR-146a-5p inhibitor and IN-NC group. But after the overexpression of miR-146a-5p, the expression of the apoptotic gene BAX appeared to be higher than that in the M-NC group. Adversely, after inhibition of miR-146a-5p, the expression level of BAX was lower than that in the IN-NC group (Figure 6). The above results demonstrated that the cells may undergo apoptosis after transfection with miR146a-5p mimic.
Figure 6.
mRNA expression of apoptotic genes.
3.5. Regulation of Cell Proliferation by miR-146a-5p
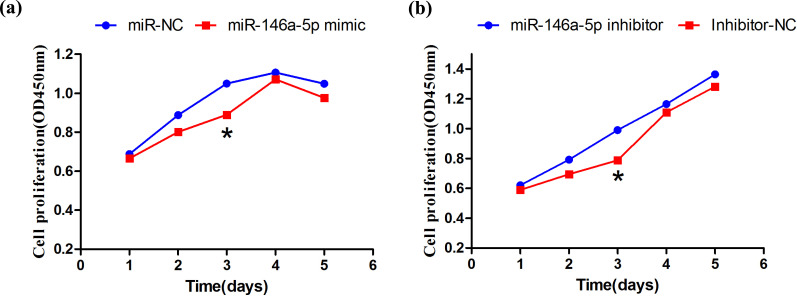
After miR-146a-5p mimic, miR-146a-5p M-NC, miR-146a-5p inhibitor and miR-146a-5p IN-NC were transfected into BeWo cells. The cell proliferation was measured at day 1, 2, 3, 4, and 5 after culturing using CCK-8 solution. On the third day, cell proliferation in the miRNA mimic group was significantly lower that in the M-NC group (Figure 7a). On the fifth day, the overexpressed miRNA and negative control cells showed a decreased trend. The cell proliferation under inhibition of miR-146a-5p was significantly higher than that of the NC group on the third day (Figure 7b), and the cell proliferation showed a gradually increased trend. The results demonstrated that the overexpression and inhibition of miR146a-5p affected cell proliferation.
Figure 7.
(a, b) Effect of miR-146a-5p on cell proliferation.
4. DISCUSSION
The stability and specificity of miRNAs as biomarkers have made them a topic of interest in recent years [19]. Related studies have shown that miRNAs play important roles in many processes involved in embryonic development [20], organogenesis [21], fetal development [22] and pluripotency of embryonic stem cells, cell differentiation, proliferation, apoptosis, organ development, and growth control [23].
This study showed that miR-146a-5p regulates cell proliferation and apoptosis by targeting Smad3/4. Zhang et al. used high-throughput sequencing to identify differentially expressed miRNAs in swine lung tissues infected with PCV2, and found that four miRNAs were upregulated. They suggested that the target genes of those miRNAs may play an important role in disease resistance [24]. It had been reported that miR-146a-5p targeted IR and inhibited protein expression, thereby inhibited adipogenesis [25]. The expression of miR-146a-5p can be used as a biomarker for the prognosis of patients with Ias [26]. It can also be used as a novel biomarker of cisplatin in the treatment of non-small cell lung cancer patients and in monitoring drug resistance [27]. The above studies demonstrate that miRNAs may play important roles in mediating the occurrence, diagnosis, and treatment of diseases.
TGF-β exerts its biological effects mainly through the TGF-β/Smad pathway. Smad3/4 play important roles in mammalian physiological processes. Previous studies have showed in a mouse model that Smad4 is the target gene of miR-146a-5p, and overexpression of Smad4 inhibits miR-146a-5p expression in muscle cells [28]. In addition, miR-146a may affect the proliferation of APL cells through the TGF-β1/Smad signaling pathway [29]. In this study, a dual luciferase assay demonstrated that Smad3/4 are the target genes of miR-146a-5p, the same results were obtained by qRT-PCR and Western Blot. The above results showed that miR-146a-5p significantly inhibited Smad3/4 at the mRNA level, and also had an inhibitory effect on the post-transcriptional levels of Smad3/4, which is consistent with the hypothesis of miRNA regulation of target genes.
In rat primary follicle granulosa cells and follicular cells, Smads 1, 2, 3, 4, 5 and 7 are present at all stages of follicular development [30]. The regulatory factors of Smad2/3 in the TGF-β signaling pathway play an important role in the regulation of trophoblast cell function and placental preeclampsia [31, 32]. Smad 2/3 and Smad4 are located in the cytoplasm of oocytes, ovarian granulosa cells and fetal ovarian granulosa cells [33].
BeWo cell, an immortalized human placental chorionic trophoblast cell line, is similar in morphology and function to trophoblast cells. BeWo cell is often used as cell models to study cell invasion, secretion, proliferation and apoptosis. Zhang et al. showed that under hypoxic conditions, HIF-1α and miR-146a promoted the autophagy of chondrocytes by inhibiting the expression of Bcl-2 [34]. Zhou et al. found that the apoptosis genes Caspase7 and Bcl-2 were target genes of miR-146a through bioinformatic analysis and dual-luciferase assays [35]. The overexpression of miR-146a can effectively reduce the mRNA and protein expression of Caspase-7 and Bcl-2, thereby promoting apoptosis. Zhou et al. showed that knockdown of miR-146a suppressed cell proliferation and colony formation in vitro and in vivo [36]. Zou et al. found that miR-146a-5p could regulate the TGF-β1/Smad and LPS/NF-κB/Bambi pathways to attenuate liver fibrosis, and the overexpression of miR-146a-5p also suppressed the proliferation and the activation of HSCs [37]. Inactivating mutations in the TGF-β Type-II receptors and SMAD4 have been described in tumors even though cancer cells can lose the cytostatic responsiveness due to defects in downstream to SMAD factors [38]. Li et al. found that the overexpression of Smad3 inhibited the expression of both MyoD and myogenin in chicken, and suppressed the cell proliferation and differentiation [39]. Other miRNAs such as miR-224 and miR-26b also targeted the regulation of Smad4 gene and affected the proliferation of granulosa cells [40]. Tomic et al. showed that knocking out the Smad3 gene caused female mice to aggravate the apoptosis of ovarian granulosa cells, hindering the development of follicles, leading to an increase in follicular atresia [41]. In our study, the expression of Bcl-2 gene in cells treated by miR-146a-5p mimic was significantly lower than that by miR-NC, which was similar to the previous researchers. Therefore, we speculate that miR-146a-5p may regulate the target gene Smad3/4, and thus regulate the expression of Bcl-2 and Caspase-3 genes. In addition, on the third day, the proliferation of cells treated by miR-146a-5p mimic was significantly lower than that by M-NC, while the proliferation of cells treated by miR-146a-5p inhibitor was significantly higher than that by IN-NC. These results suggested that miR-146a-5p may affect cell proliferation by regulating the Smad3/4 genes.
CONCLUSION
This study verified that Smad3 and Smad4 are the direct target genes of miR-146a-5p, and miR-146a-5p can regulate cell proliferation and apoptosis by targeting Smad3 and Smad4.
ACKNOWLEDGEMENTS
This work was supported by Shihezi University and the Manasi Tiankang animal husbandry of China.
AUTHORS CONTRIBUTION
T. H. conceived and designed the experiments. T. L. and M. Q. performed the experiments. T. L., M. Q., and B. W. analyzed the data. T. L., M. Q., and H. G. contributed reagents/materials/analysis tools. M. Q. and T. L. wrote the paper.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study is approved by the Medical Ethics Committee, First Affiliated Hospital, Medical College, Shihezi University, China (Approval no. 2014-073-01).
HUMAN AND ANIMAL RIGHTS
No humans were used in the study. All reported experiments on animals were performed in accordance to the protocol approved by the Animal care and use committee of Shihezi University.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data supporting the findings of the article cannot be shared as it is design for the patent application.
FUNDING
This work was supported by National Natural Science Foundation of China (Grant no. 31460586 and 31960645).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 2.Loux S.C., Scoggin K.E., Bruemmer J.E., Canisso I.F., Troedsson M.H., Squires E.L., Ball B.A. Evaluation of circulating miRNAs during late pregnancy in the mare. PLoS One. 2017;12(4):e0175045. doi: 10.1371/journal.pone.0175045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J., Smyth P., Flavin R., Cahill S., Denning K., Aherne S., Guenther S.M., O’Leary J.J., Sheils O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of Formalin-Fixed Paraffin-Embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7(1):36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang C., Zhu D.X., Dong H.J., Zhou Z.J., Wang Y.H., Liu L., Fan L., Miao K.R., Liu P., Xu W., Li J.Y. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann. Hematol. 2012;91(4):553–559. doi: 10.1007/s00277-011-1350-9. [DOI] [PubMed] [Google Scholar]
- 5.Lawrie C.H., Gal S., Dunlop H.M., Pushkaran B., Liggins A.P., Pulford K., Banham A.H., Pezzella F., Boultwood J., Wainscoat J.S., Hatton C.S., Harris A.L. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008;141(5):672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 6.Bogunia-Kubik K., Wysoczańska B., Piątek D., Iwaszko M., Ciechomska M., Świerkot J. Significance of polymorphism and expression of miR-146a and NFkB1 genetic variants in patients with rheumatoid arthritis. Arch. Immunol. Ther. Exp. (Warsz.) 2016;64(1) Suppl. 1:131–136. doi: 10.1007/s00005-016-0443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Momen-Heravi F., Bala S., Bukong T., Szabo G. Exosome-mediated delivery of functionally active miRNA-155 inhibitor to macrophages. Nanomedicine (Lond.) 2014;10(7):1517–1527. doi: 10.1016/j.nano.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vaksman O., Tropé C., Davidson B., Reich R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis. 2014;35(9):2113–2120. doi: 10.1093/carcin/bgu130. [DOI] [PubMed] [Google Scholar]
- 9.Li W., Xi Y., Xue S., Wang Y., Wu L., Liu H., Lei M. Sequence analysis of microRNAs during pre-implantation between Meishan and Yorkshire pigs. Gene. 2018;646:20–27. doi: 10.1016/j.gene.2017.12.046. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X., Yang J., Zhao J., Zhang P., Huang X. MicroRNA-23b inhibits the proliferation and migration of heat-denatured fibroblasts by targeting Smad3. PLoS One. 2015;10(7):, e0131867. doi: 10.1371/journal.pone.0131867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang P., Wu Z., Ma C., Pan N., Wang Y., Yan L. Endometrial miR-543 is downregulated during the implantation window in women with endometriosis-related infertility. Reprod. Sci. 2018;26(7):900–908. doi: 10.1177/1933719118799199. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Ma C.H., Qiao J. Differential expression of microRNA in eutopic endometrium tissue during implantation window for patients with endometriosis related infertility. Zhonghua Fu Chan Ke Za Zhi. 2016;51(6):436–441. doi: 10.3760/cma.j.issn.0529-567X.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Qu J., Zhu Y., Wu X., Zheng J., Hou Z., Cui Y., Mao Y., Liu J. Smad3/4 binding to promoter II of P450arom so as to regulate aromatase expression in endometriosis. Reprod. Sci. 2017;24(8):1187–1194. doi: 10.1177/1933719116681517. [DOI] [PubMed] [Google Scholar]
- 14.Osterlund C., Fried G. TGFbeta receptor types I and II and the substrate proteins Smad 2 and 3 are present in human oocytes. Mol. Hum. Reprod. 2000;6(6):498–503. doi: 10.1093/molehr/6.6.498. [DOI] [PubMed] [Google Scholar]
- 15.Sirard C., de la Pompa J.L., Elia A., Itie A., Mirtsos C., Cheung A., Hahn S., Wakeham A., Schwartz L., Kern S.E., Rossant J., Mak T.W. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 1998;12(1):107–119. doi: 10.1101/gad.12.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo F.T., Fan K., Ambartsumyan G., Menon P., Ketefian A., Bentsi-Barnes I.K., Pisarska M.D. Relative expression of genes encoding SMAD signal transduction factors in human granulosa cells is correlated with oocyte quality. J. Assist. Reprod. Genet. 2011;28(10):931–938. doi: 10.1007/s10815-011-9609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H., Liu L., Liu X., Zhang M., Li X. Correlation between miRNAs and target genes in response to Campylobacter jejuni inoculation in chicken. Poult. Sci. 2018;97(2):485–493. doi: 10.3382/ps/pex343. [DOI] [PubMed] [Google Scholar]
- 18.Zou L., Xiong X., Yang H., Wang K., Zhou J., Lv D., Yin Y. Identification of microRNA transcriptome reveals that miR-100 is involved in the renewal of porcine intestinal epithelial cells. Sci. China Life Sci. 2019;62(6):816–828. doi: 10.1007/s11427-018-9338-9. [DOI] [PubMed] [Google Scholar]
- 19.Gilchrist G.C., Tscherner A., Nalpathamkalam T., Merico D., LaMarre J. MicroRNA expression during bovine oocyte maturation and fertilization. Int. J. Mol. Sci. 2016;17(3):396. doi: 10.3390/ijms17030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amanai M., Brahmajosyula M., Perry A.C. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol. Reprod. 2006;75(6):877–884. doi: 10.1095/biolreprod.106.056499. [DOI] [PubMed] [Google Scholar]
- 21.Byrne M.J., Warner C.M. MicroRNA expression in preimplantation mouse embryos from Ped gene positive compared to Ped gene negative mice. J. Assist. Reprod. Genet. 2008;25(5):205–214. doi: 10.1007/s10815-008-9211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui X.S., Shen X.H., Kim N.H. Dicer1 expression in preimplantation mouse embryos: Involvement of Oct3/4 transcription at the blastocyst stage. Biochem. Biophys. Res. Commun. 2007;352(1):231–236. doi: 10.1016/j.bbrc.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Tzur G., Levy A., Meiri E., Barad O., Spector Y., Bentwich Z., Mizrahi L., Katzenellenbogen M., Ben-Shushan E., Reubinoff B.E., Galun E. MicroRNA expression patterns and function in endodermal differentiation of human embryonic stem cells. PLoS One. 2008;3(11):, e3726. doi: 10.1371/journal.pone.0003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang P., Wang L., Li Y., Jiang P., Wang Y., Wang P., Kang L., Wang Y., Sun Y., Jiang Y. Identification and characterization of microRNA in the lung tissue of pigs with different susceptibilities to PCV2 infection. Vet. Res. (Faisalabad) 2018;49(1):18. doi: 10.1186/s13567-018-0512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao Y., Huang B., Chen Y., Hong G., Xu J., Hu C., Wang C. Integrated analyses reveal overexpressed Notch1 promoting porcine satellite cells’ proliferation through regulating the cell cycle. Int. J. Mol. Sci. 2018;19(1):271. doi: 10.3390/ijms19010271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H.L., Li L., Cheng C.J., Sun X.C. Expression of miR-146a-5p in patients with intracranial aneurysms and its association with prognosis. Eur. Rev. Med. Pharmacol. Sci. 2018;22(3):726–730. doi: 10.26355/eurrev_201802_14300. [DOI] [PubMed] [Google Scholar]
- 27.Yuwen D.L., Sheng B.B., Liu J., Wenyu W., Shu Y.Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017;21(11):2650–2658. [PubMed] [Google Scholar]
- 28.Sun Y., Li Y., Wang H., Li H., Liu S., Chen J., Ying H. miR-146a-5p acts as a negative regulator of TGF-β signaling in skeletal muscle after acute contusion. Acta Biochim. Biophys. Sin. (Shanghai) 2017;49(7):628–634. doi: 10.1093/abbs/gmx052. [DOI] [PubMed] [Google Scholar]
- 29.Zhong H., Wang H.R., Yang S., Zhong J.H., Wang T., Wang C., Chen F.Y. Targeting Smad4 links microRNA-146a to the TGF-β pathway during retinoid acid induction in acute promyelocytic leukemia cell line. Int. J. Hematol. 2010;92(1):129–135. doi: 10.1007/s12185-010-0626-5. [DOI] [PubMed] [Google Scholar]
- 30.Drummond A.E. TGFbeta signalling in the development of ovarian function. Cell Tissue Res. 2005;322(1):107–115. doi: 10.1007/s00441-005-1153-1. [DOI] [PubMed] [Google Scholar]
- 31.Stoikos C.J., Salamonsen L.A., Hannan N.J., O’Connor A.E., Rombauts L., Dimitriadis E. Activin A regulates trophoblast cell adhesive properties: Implications for implantation failure in women with endometriosis-associated infertility. Hum. Reprod. 2010;25(7):1767–1774. doi: 10.1093/humrep/deq097. [DOI] [PubMed] [Google Scholar]
- 32.Reddy A., Suri S., Sargent I.L., Redman C.W., Muttukrishna S. Maternal circulating levels of activin A, inhibin A, sFlt-1 and endoglin at parturition in normal pregnancy and pre-eclampsia. PLoS One. 2009;4(2):, e4453. doi: 10.1371/journal.pone.0004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Billiar R.B., St Clair J.B., Zachos N.C., Burch M.G., Albrecht E.D., Pepe G.J. Localization and developmental expression of the activin signal transduction proteins Smads 2, 3, and 4 in the baboon fetal ovary. Biol. Reprod. 2004;70(3):586–592. doi: 10.1095/biolreprod.103.018598. [DOI] [PubMed] [Google Scholar]
- 34.Zhang F., Wang J., Chu J., Yang C., Xiao H., Zhao C., Sun Z., Gao X., Chen G., Han Z., Zou W., Liu T. MicroRNA-146a induced by hypoxia promotes chondrocyte autophagy through Bcl-2. Cell. Physiol. Biochem. 2015;37(4):1442–1453. doi: 10.1159/000438513. [DOI] [PubMed] [Google Scholar]
- 35.Zhou X., Su S., Li S., Pang X., Chen C., Li J., Liu J. MicroRNA-146a down-regulation correlates with neuroprotection and targets pro-apoptotic genes in cerebral ischemic injury in vitro. Brain Res., . 2016;1648(Pt A):136–143. doi: 10.1016/j.brainres.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 36.Zhou C., Jiang C.Q., Zong Z., Lin J.C., Lao L.F. miR-146a promotes growth of osteosarcoma cells by targeting ZNRF3/GSK-3β/β-catenin signaling pathway. Oncotarget. 2017;8(43):74276–74286. doi: 10.18632/oncotarget.19395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou Y., Cai Y., Lu D., Zhou Y., Yao Q., Zhang S. MicroRNA-146a-5p attenuates liver fibrosis by suppressing profibrogenic effects of TGFβ1 and lipopolysaccharide. Cell. Signal. 2017;39:1–8. doi: 10.1016/j.cellsig.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 38.Morikawa M., Derynck R., Miyazono K. TGF-β and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 2016;8(5):a021873. doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X., McFarland D.C., Velleman S.G. Effect of Smad3-mediated transforming growth factor-β1 signaling on satellite cell proliferation and differentiation in chickens. Poult. Sci. 2008;87(9):1823–1833. doi: 10.3382/ps.2008-00133. [DOI] [PubMed] [Google Scholar]
- 40.Yao G., Yin M., Lian J., Tian H., Liu L., Li X., Sun F. MicroRNA-224 is involved in transforming growth factor-β-mediated mouse granulosa cell proliferation and granulosa cell function by targeting Smad4. Mol. Endocrinol. 2010;24(3):540–551. doi: 10.1210/me.2009-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomic D., Miller K.P., Kenny H.A., Woodruff T.K., Hoyer P., Flaws J.A. Ovarian follicle development requires Smad3. Mol. Endocrinol. 2004;18(9):2224–2240. doi: 10.1210/me.2003-0414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of the article cannot be shared as it is design for the patent application.