Abstract
The recently discovered 340-cavity in influenza neuraminidase (NA) N6 and N7 subtypes has introduced new possibilities for rational structure-based drug design. However, the plasticity of the 340-loop (residues 342–347) and the role of the 340-loop in NA activity and substrate binding have not been deeply exploited. Here, we investigate the mechanism of 340-cavity formation and demonstrate for the first time that seven of nine NA subtypes are able to adopt an open 340-cavity over 1.8 μs total molecular dynamics simulation time. The finding that the 340-loop plays a role in the sialic acid binding pathway suggests that the 340-cavity can function as a druggable pocket. Comparing the open and closed conformations of the 340-loop, the side chain orientation of residue 344 was found to govern the formation of the 340-cavity. Additionally, the conserved calcium ion was found to substantially influence the stability of the 340-loop. Our study provides dynamical evidence supporting the 340-cavity as a druggable hotspot at the atomic level and offers new structural insight in designing antiviral drugs.
Keywords: influenza, neuraminidase, 340-cavity, molecular dynamics simulations
1. Introduction
Since the beginning of the last century, multiple pandemics caused by influenza viruses have severely impacted public health [1]. Although vaccines and antiviral drugs have been developed to prevent and control upcoming influenza outbreaks, unpredictable novel strains and constantly mutating existing strains still pose a great threat to human life [2]. In 2020, COVID-19, caused by the SARS-CoV-2 coronavirus, has already affected more than 200 countries, taken more than 500,000 lives, and instigated a significant global economic downturn [3]. Influenza, which also spreads readily via respiratory droplets, cannot be neglected for its high morbidity and mortality [4]. New antiviral drugs are a promising and straightforward approach to treating influenza by reducing case severity and transmission, which can be particularly impactful for responding to new outbreaks [5]. Neuraminidase (NA), which releases progeny virions by cleaving the terminal sialic acid (SA) residues on the infected cell surface, is considered the most promising target for antiviral drug development [6]. Therefore, exploring the structural features of NAs and identifying the SA binding path are of great importance in designing anti-influenza drugs [7,8].
Nine NA alleles are classified into two groups according to phylogenic and structural differences (group-1: N1, N4, N5, N8; group-2: N2, N3, N6, N7, N9) [9]. The crystal structures of group-1 NAs contain a unique characteristic, the 150-cavity, which can be exploited to develop new anti-influenza drugs [10]. It is noteworthy that an additional pocket, the 340-cavity, adjacent to the active site of NA capped by the 340-loop (residues 342–347) was observed in the crystal structures of N6 and N7 [11]. Our group discovered that the hydrogen bond between G342 and Q296 and the calcium ion bound in the 340-loop contribute to the special conformation of the N7 340-cavity [12]. However, questions regarding whether the 340-cavity is prevalent in NAs, whether the 340-loop participates in the substrate binding process, and the structural dynamics of the 340-loop in other NA subtypes have never been explored.
To answer the above questions, normal molecular dynamics (MD) simulations were performed on N1 to N9 subtypes in calcium-bound and calcium-free systems. Additionally, the binding mechanism of natural substrate SA was identified by sliding binding-box docking. Our study reveals that seven of the nine NA subtypes exhibit an open 340-cavity, the calcium ion plays an important role in maintaining the conformation of the 340-loop, and the 340-loop is a canonical approach site in the SA binding mechanism. Our results indicate that the 340-loop is a critical motif in NA, and the 340-cavity is a valuable hotspot for anti-influenza drug design.
2. Results
2.1. N1, N3, N4, N5, N6, N7, and N8 Exhibit an Open 340-Cavity
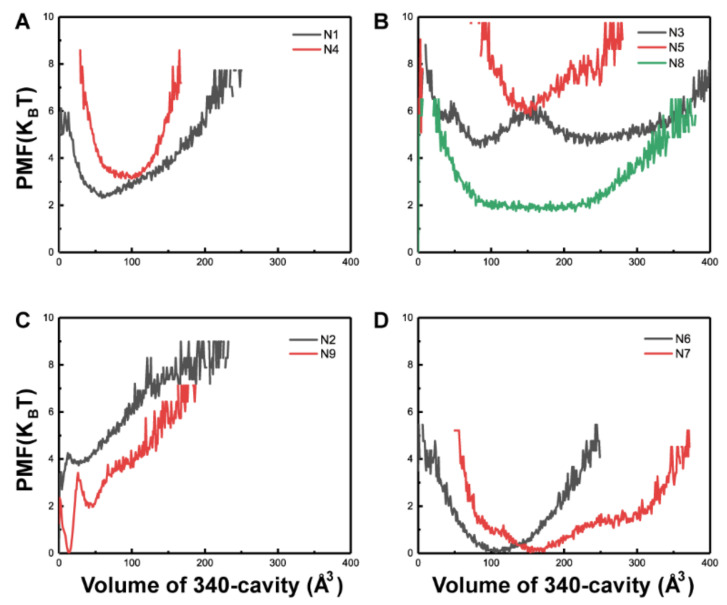
To probe 340-cavity formation in NAs, the volumes of the 340-cavity for all NA subtypes were calculated using POVME. Maps of the potential of mean force (PMF) from N1 to N9 as a function of 340-cavity volume were constructed based on the simulation data (Figure 1). To compare the size of the 340-cavity, the volume of the N7 crystal structure 340-cavity, 102 Å3, was given as a reference. It revealed that most of the NA subtypes, except N2 and N9, are able to adopt open 340-cavity conformations in normal MD simulations. Moreover, the NA subtypes can be divided into three types based on their 340-cavity volume.
Figure 1.
Potential of mean force (PMF) of nine neuraminidase (NA) subtypes based on 340-cavity volume calculation. Potential of mean force (PMF) based on the volume of the 340-cavity in N1 and N4 (A), N3, N5, and N8 (B), N2 and N9 (C), N6 and N7 (D).
Type-1 NAs comprise N1, N3, N4, N5, and N8. Although no 340-cavity was observed in the crystal structures of type-1 NAs (Figure 2), their 340-loops transitioned to open conformations during simulation. In particular, one local minimum indicating the open conformation of the 340-cavity in N1 and N4 was discovered, and the local minimum is centered around 75 and 100 Å3 in N1 and N4, respectively (Figure 1A). For N3, N5, and N8, local minima indicating a larger 340-cavity with volumes 232, 154, and 175 Å3 are shown in Figure 1B. Type-2 NAs comprise N2 and N9, wherein almost no 340-cavity forms during simulation (Figure 1C). Type-3 NAs comprise N6 and N7, of which the crystal structures already exhibit an open 340-cavity (Figure 2). Based on the PMF maps, local minima with volumes of 107 and 154 Å3 are associated with the lowest free energy in N6 and N7, respectively (Figure 1D). PMF maps illustrate that the 340-cavity of N1, N3, N4, N5, N6, N7, and N8 are able to exhibit the open configuration, while N2 and N9 prefer the closed conformation of the 340-cavity.
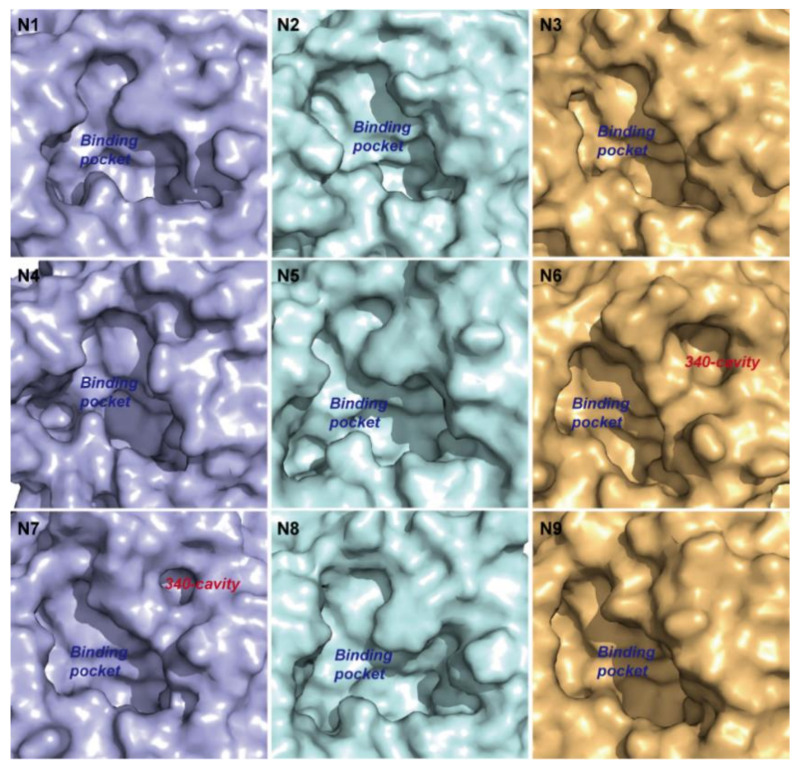
Figure 2.
Surface of the NA binding pocket in different subtypes. Surface of the binding pocket in the crystal structures of different NA subtypes. N6 and N7 have an open 340-cavity in the crystal structures.
2.2. 340-Cavity Formation Is Governed by the Side-Chain Orientation of Residue 344
To identify the conformational alterations that induce 340-cavity opening, root mean squared fluctuation (RMSF) calculation and clustering analysis of each NA subtype were carried out. The 340-loop residues of each NA subtype are listed in Table 1. RMSF values indicate that residues 342, 343, and 344 were associated with larger flexibility than that of residue 345, 346, and 347 in the 340-loop of type-1 NAs (N1, N3, N4, N5, and N8), while RMSF values of the 340-loop residues in type-2 NAs (N2 and N9) showed little discrepancies (Table 2). The RMSF results suggest that the open conformation of the 340-cavity in type-1 NAs may be controlled by residues 342–344 in the 340-loop.
Table 1.
Residues of the 340-loop in each NA subtype.
| NA Subtype | 340-Loop Residues |
|---|---|
| N1 | 342-SSNGAY-347 |
| N2 | 342-NERGTQ-347 |
| N3 | 342-NVNGGP-347 |
| N4 | 342-NGKGRY-347 |
| N5 | 342-GGSGTY-347 |
| N6 | 342-GGSPDP-347 |
| N7 | 342-GSPGAP-347 |
| N8 | 342-G-NQGY-347 |
| N9 | 342-YPGNNN-347 |
N8 (PDB 2HT5) lacks residue 343.
Table 2.
Root mean squared fluctuation (RMSF) for 340-loop residues over the whole simulation in type-1 and type-2 NAs.
| Residue No. | RMSF of Type-1 NAs (nm) | Type-2 NAs (nm) | |||||
|---|---|---|---|---|---|---|---|
| N1 | N3 | N4 | N5 | N8 | N2 | N9 | |
| 342 | 0.2524 | 0.3295 | 0.1536 | 0.1119 | 0.1072 | 0.1234 | 0.1072 |
| 343 | 0.1571 | 0.3867 | 0.2273 | 0.1326 | N.A. | 0.0775 | 0.1057 |
| 344 | 0.2590 | 0.3599 | 0.1575 | 0.2121 | 0.2154 | 0.0969 | 0.1396 |
| 345 | 0.0973 | 0.0982 | 0.0759 | 0.1113 | 0.1384 | 0.0612 | 0.1474 |
| 346 | 0.0742 | 0.0905 | 0.0973 | 0.0902 | 0.0839 | 0.0598 | 0.1623 |
| 347 | 0.1240 | 0.0861 | 0.1321 | 0.1659 | 0.1365 | 0.0709 | 0.1496 |
The 340-loop of the representative structures in open configuration displays a great deviation from its locations in the crystal structures for type-1 NAs. Moreover, the side-chain of residue 344 was discovered to be oriented to the opposite direction, which left a space for the formation of 340-cavity and induced the transition state of the 340-loop from the closed to the open conformation in type-1 NAs (Figure 3A–E).
Figure 3.
Mechanism for 340-cavity formation. Cluster analysis based on the 340-loop root mean squared fluctuation was performed. Cluster centers representing the open conformation of the 340-cavity were superimposed on the crystal structures of type-1 and type-2 NAs. Panels (A–E) show a structural comparison between the cluster center and crystal structure of the 340-loop in type-1 NAs N1, N3, N4, N5, and N8. Panels (G,H) show a structural comparison between the cluster center and crystal structure of the 340-loop in type-2 NAs N2 and N9. The 340-loops of cluster centers and crystal structures are colored in blue and orange, respectively. Panels (F,I) illustrate the 340-cavity and binding pocket of type-3 NA N6 and N7 with surface representation.
In contrast, ARG-344 in type-2 NA N2 never changed orientation from its initial position. The strong salt bridge interaction between ARG-344 and ASP-369 in N2 locked the side-chain of ARG-344 and intensified the rigidity of the 340-loop, leading to an always-closed conformation of the 340-loop (Figure 3G). In N9, the 344th position was occupied by the amino acid glycine, which has no side-chain, and the 340-loop maintained its starting crystal structure conformation during the entire simulation (Figure 3H). In N6 and N7, the comparison between the crystal structure and representative structure is not displayed because the 340-cavity in the crystal structures already exhibit an open conformation (Figure 3F,I).
2.3. The 340-Loop Is Involved in the SA Binding Pathway
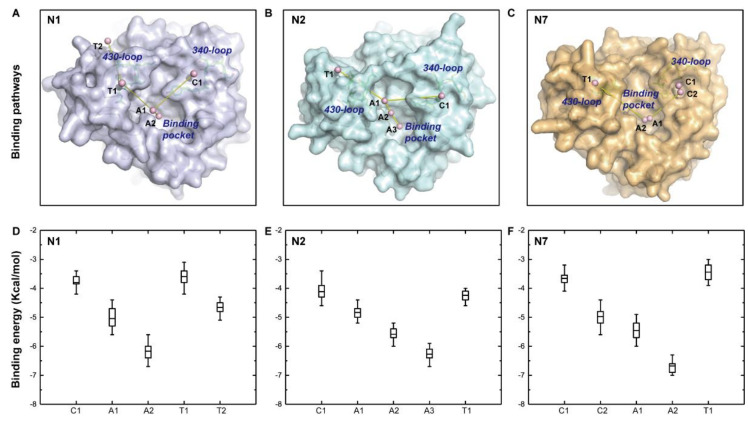
The discovery of the 340-cavity in seven of nine NA subtypes via simulation indicates that small molecule compounds that target this cavity could have pan-NA activity. The resultant compounds would interfere with NA function if the 340-loop or 340-cavity is involved in natural SA substrate binding. To figure out the function of the 340-loop in SA binding, sliding binding-box docking was performed on N1, N2, and N7, representing the three types of NAs. The docking procedure can be observed in Figure S1. Two lowest binding energy pathways of SA were proposed in the three NA subtypes, designated the climbing path and the tunneling path. This method was inspired by a previous sliding binding-box docking study which revealed the binding mechanism of a NA inhibitor [13]. The binding path of SA and the averaged binding energy of cluster components are illustrated in Figure 4.
Figure 4.
Binding pathway of sialic acid (SA) and distribution of binding energies along the pathway. The two lowest binding energy pathways of sialic acid (SA), the climbing path and tunneling path, are illustrated in N1 (A), N2 (B), and N7 (C). The harbors of the climbing path, active site, and tunneling path were abbreviated as C, A, and T, respectively. Panels (D–F) represent binding energies in accordance with the harbor sites in N1, N2, and N7, respectively. Boxes represent binding energies between 25% and 75% of the cluster components, lines within the boxes represent mean values, and vertical bars represent binding energies within 1.5 interquartile range (IQR).
In the climbing path, the natural substrate SA did a brief layover in the 340-loop region in the three NAs. Although the 340-loop in the N1 and N2 crystal structures displayed a closed conformation, SA was still held in the 340-loop region for a time while approaching the final binding pocket in both NAs (Figure 4A,B). There were two harbors in the climbing path of SA in N7 since the 340-loop exhibited an open conformation in the N7 crystal structure (Figure 4C). After a brief layover at the 340-loop region, the SA continued to move to the active site via two (A1 and A2 in N1 and N7) or three steps (A1, A2, and A3 in N2). The large decrease in binding energy made this movement energetically favorable (Figure 4D–F).
In the tunneling path, the 430-loop (residues 430–433) adjacent to the binding pocket was discovered to act as a harbor that facilitated SA landing in N1, N2, and N7. The average binding energies of the cluster centers in the tunneling path were larger than that of cluster centers in the climbing path (Figure 4D–F). Thus, the probability of SA binding via the tunneling path was lower than that of the climbing path, a finding in good agreement with a previous study [13]. The SA binding mechanisms determined here suggest that the 340-loop is an important motif in shepherding SA to the final binding pocket, and this finding indicates that the 340-cavity is a potential target pocket for drug discovery.
2.4. A Calcium Ion Controls the Stability of the 340-Loop
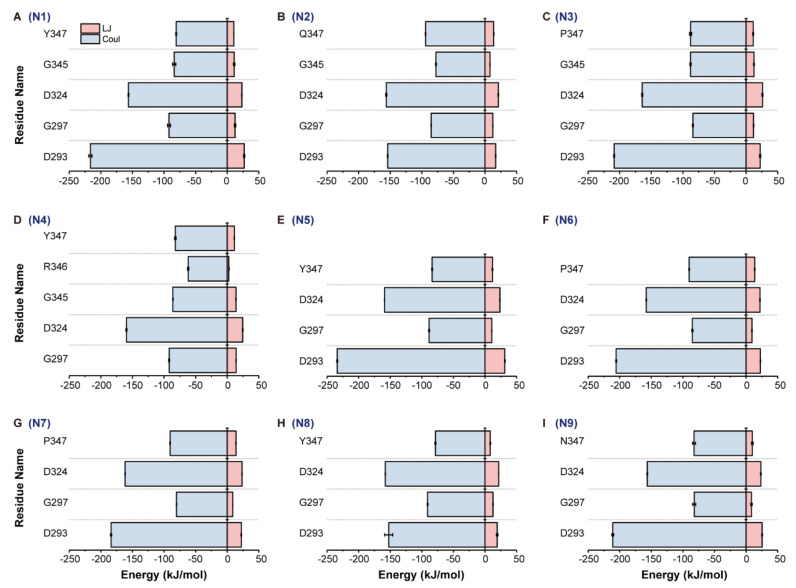
The 340-loop was found to be involved in the SA binding mechanism. Therefore, the conformation and stability of the 340-loop need to be carefully characterized. A conserved calcium ion found to influence the activity and thermostability of NA is located near the 340-loop [14,15]. At least one residue on the 340-loop interacts with this cation ion (Table 3). The interaction energy between the calcium ion and residues in Table 3 contains two parts, the Lennard-Jones (LJ) and Coulomb (Coul) interactions. The LJ and Coul interactions between the calcium ion and interacted residues of the nine NA subtypes were well-preserved during simulations, since error values of block averages are relatively small (Figure 5).
Table 3.
Residues interact with the conserved calcium ion in each NA subtype.
| NA Subtype | Residues Interact with the Calcium Ion |
|---|---|
| N1 | D293, G297, D324, G345, Y347 |
| N2 | D293, G297, D324, G345, Q347 |
| N3 | D293, G297, D324, G345, P347 |
| N4 | G297, D324, G345, R346, Y347 |
| N5 | D293, G297, D324, Y347 |
| N6 | D293, G297, D324, P347 |
| N7 | D293, G297, D324, P347 |
| N8 | D293, G297, D324, Y347 |
| N9 | D293, G297, D324, N347 |
Residues in bold font are 340-loop residues.
Figure 5.
Interaction energy between the conserved calcium ion and interacted residues in all NA subtypes. Interaction energy, Lennard-Jones (LJ) and Coulomb (Coul), between the calcium ion and interacted residues was calculated based on the whole trajectory of each NA subtype. Panels (A) to (I) represent the LJ and Coul interactions of N1 to N9, respectively.
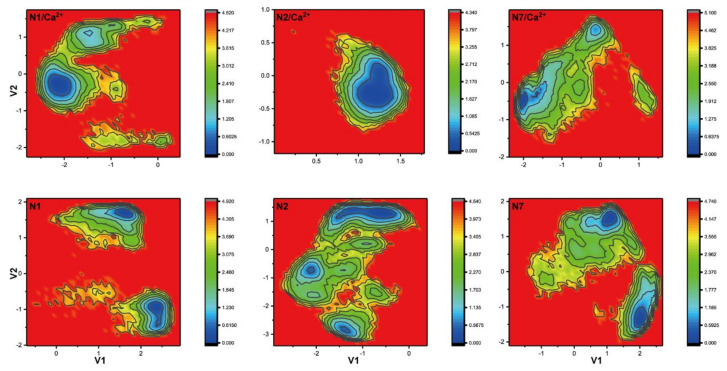
To further characterize the conformational changes of the 340-loop due to influence of the calcium ion, dihedral principal components analysis (dPCA) of the 340-loop in the calcium-bound and calcium-free systems was performed. Free energy landscapes of N1, N2, and N7, representing the three 340-cavity types of NA, are shown in Figure 6. It is evident that the free energy landscapes of the 340-loop in the calcium-bound and calcium-free systems of all NA subtypes are distinct from each other, indicating disparate configurations of the 340-loop (Figure S2). Specifically, only one local minimum was observed in the calcium-bound NA systems, while two or more local minima were observed in the calcium-free systems. Interestingly, comparing free energy landscapes of the 340-loop in the calcium-bound systems, configurations of the 340-loops in N1 and N7 experienced a wider space than that in N2 system, which is directly related to the 340-cavity volume calculation. In the volume calculation, the N2 340-loop maintained its closed conformation during the whole trajectory, while the N1 340-loop transitioned from closed to open state, and the N7 340-cavity opened to a larger extent during the simulations. The dPCA analysis revealed that the conserved calcium ion plays an important role in maintaining the stability of the 340-loop, and removal or disruption of this cation ion could severely influence the 340-loop conformation.
Figure 6.
Free energy landscape of the 340-loop from dihedral principle component analysis. Free energy landscapes of the 340-loop in N1, N2, and N7 calcium-bound and calcium-free systems were obtained using dihedral principle component analysis (dPCA).
3. Discussion
The 340-loop was discovered to be a canonical accommodating pocket along SA’s binding mechanism to NA across multiple NA subtypes. This implicates that the SA binding process may be influenced or terminated if the 340-loop is reconfigured or blocked from its conventional configuration. Our results are consistent with a previous study in which the NA inhibitor binding pathway was explored and oseltamivir was found to loosely localize between the two loops 294-NWH-296 and 345-GAY-347 [13]. Although the authors stated that this finding may not be prevalent in other NAs, we have found that the 340-loop participates in SA binding of N1, N2, and N7, which represent three types of NAs. The 340-cavity was found in seven of nine NA subtypes, which indicated that this cavity could guide the design of robust novel ligands which function as NA inhibitors by interfering with the SA binding mechanism. Our study provides more evidence that supports the 340-cavity as a potential drug target.
Additionally, the conserved calcium ion was found to maintain the stability of the 340-loop due to its intimate contact with residues on the loop. Specifically, the backbone carbonyl group of residue Y/P/Q/N347 stably interacts with the calcium ion in all NA subtypes (Table 3 and Figure 5). Y347 in N1 was identified as the key framework residue that can “clamp” the ligand into a favorable binding pose in one study [16]. Two other studies found that residue 347 could mediate oseltamivir on the NA surface via a hydrogen bond [13,17]. All these findings, including our own, indicate that the conserved calcium ion plays an important role in stabilizing the conformation of the 340-loop, thus facilitating SA/drug binding. Therefore, disruption of the calcium ion binding can also influence NA function and thus stop viral release.
Moreover, the 430-loop was discovered to be a common stop in the SA tunneling pathway of the three NA subtypes. The displacement of the 430-loop was found to induce a larger opening of the binding pocket based on crystal structural analysis and MD simulations [18,19]. The 430-loop was also predicted to be a novel druggable hot spot by a computational solvent mapping study [20]. Our study provides new evidence that supports the 430-loop as a potential drug-targeting region.
In conclusion, this study demonstrates the plasticity of the 340-loop in different NA subtypes and the importance of the conserved calcium ion in influencing the stability of the 340-loop. The open 340-cavity discovered in the seven NA subtypes and the mechanism of 340-cavity formation offer new structural insights for antiviral drug development. Moreover, the 340-loop’s role as a canonical stop in the SA climbing path suggests it can function as a novel drug-binding hotspot for combating influenza viruses.
4. Methods
4.1. MD Simulation Details
Normal MD simulations of nine subtypes of NA were performed, and the X-ray crystal structures of N1 to N9 were taken from PDB 2HTY, 1NN2, 4HZV, 3HTV, 3SAL, 4QN4, 4QN3, 2HT5, and 4MWJ, respectively [10,11,21,22,23,24]. All the NA structures were crystallized with the conserved calcium ion, and the calcium ion coordinated with interacted residues (Table 3) was preserved in the calcium-bound system and deleted in the calcium-free system. The simulation time for each NA subtype with or without calcium ion was 100 ns, with total simulation of 1.8 μs in this study. All the systems were solvated with TIP3P waters in an octahedral box with periodic boundary conditions [25]. Sodium and chloride ions were added to a concentration of 100 mM. The GROMACS and Amber99SB force field were used in all simulations [26]. All systems were first equilibrated for 5 ns by restraining all the heavy atoms, then normal MD simulations were performed in an isothermal, isobaric ensemble (300 K, 1 bar) for 100 ns. Bond length constraints were applied to all bonds that contained hydrogen atoms based on the LINCS protocol [27]. An integration step of 0.002 ps was used in all the simulations. Electrostatic interactions were treated with Particle Mesh Ewald with a cutoff of 0.9 nm with grid spacing for the FFT grid < 0.12 nm [28].
4.2. Sliding Binding-Box Docking
Sliding binding-box docking was performed using Autodock Vina software [13,29]. The N1, N2, and N7 crystal structures were treated as receptors, and the direction from the binding pocket to the exposed solvent was defined as the z-axis. Thirteen overlapping grid boxes along the z-direction were generated, wherein the z plane covers the binding pocket and the xy planes cover the entire protein area. Each grid box was set to size 48 × 48 × 10.5 Å, with an overlap of 2 Å in the z-direction. In detail, the x and y coordinates of the overlapped grid boxes were kept the same as those of the thirteen grid boxes in the Autodock setup procedure, while the z coordinate was increased by 2 Å for each grid box. The representative image of sliding binding-box docking is shown in Figure S1. Sialic acid (SA) was prepared as the ligand. An exhaustiveness value of 20 was set in Autodock Vina. The possible binding mechanism of SA was revealed by k-means clustering: the x, y, and z coordinates of the center of mass of the ligand and the docking score were treated as coordinates in four dimensions, and the docking score was weighted by a factor of 0.7. The number of cluster centers was set at 5.
4.3. 340-Volume Calculation
The volume of the 340-cavity was calculated using POVME [30]. NA structure snapshots were extracted from the trajectory of each NA subtype with the calcium ion every 5 ps and superimposed onto the reference structure (N7) with a pregenerated 3D-grid representing the 340-cavity. The volume was calculated by counting the grid points located in the 340-cavity.
4.4. Dihedral Principal Components Analysis (dPCA)
Before dPCA, all backbone dihedral angles of the 340-loop in N1 to N9 were collected from trajectories every 1 ps [31]. dPCA was performed on the angles collected from each NA in the calcium-bound and calcium-free systems. All calculated snapshots were projected to the first two eigenvectors to get the first two principal components to make a two-dimensional free energy landscape.
Acknowledgments
We would like to thank Lihua Meng for providing data analysis help and Kevin Shi for providing language help.
Abbreviations
| dPCA | dihedral principle component analysis |
| MD | Molecular dynamics |
| NA | Neuraminidase |
| PMFRMSF | Potential of mean forceRoot mean squared fluctuation |
Supplementary Materials
Supplementary Materials can be found at https://www.mdpi.com/1422-0067/21/16/5655/s1.
Author Contributions
N.H. conceived and designed the experiments. N.H., J.T.Y.N., and Y.L. performed the experiments and analyzed the data. N.H. prepared the original draft. Y.M. and Z.H. reviewed and edited the manuscript. All authors have read and agree to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 31660240) and Xijiang Innovation and Entrepreneurship Team of Zhaoqing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Fauci A.S. Pandemic influenza threat and preparedness. Emerg. Infect. Dis. 2006;12:73–77. doi: 10.3201/eid1201.050983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann G., Noda T., Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabutti G., d’Anchera E., Sandri F., Savio M., Stefanati A. Coronavirus: Update Related to the Current Outbreak of COVID-19. Infect. Dis. Ther. 2020:1–13. doi: 10.1007/s40121-020-00295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eccles R. Understanding the symptoms of the common cold and influenza. Lancet Infect. Dis. 2005;5:718–725. doi: 10.1016/S1473-3099(05)70270-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiraki K., Daikoku T. Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharmacol. Ther. 2020;209:107512. doi: 10.1016/j.pharmthera.2020.107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das K., Aramini J.M., Ma L.C., Krug R.M., Arnold E. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol. 2010;17:530–538. doi: 10.1038/nsmb.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perot S., Sperandio O., Miteva M.A., Camproux A.C., Villoutreix B.O. Druggable pockets and binding site centric chemical space: A paradigm shift in drug discovery. Drug Discov. Today. 2010;15:656–667. doi: 10.1016/j.drudis.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y., Wu Y., Zhang W., Qi J., Gao G.F. Enabling the ’host jump’: Structural determinants of receptor-binding specificity in influenza A viruses. Nat. Rev. Microbiol. 2014;12:822–831. doi: 10.1038/nrmicro3362. [DOI] [PubMed] [Google Scholar]
- 9.Air G.M. Influenza neuraminidase. Influenza Other Respir. Viruses. 2012;6:245–256. doi: 10.1111/j.1750-2659.2011.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell R.J., Haire L.F., Stevens D.J., Collins P.J., Lin Y.P., Blackburn G.M., Hay A.J., Gamblin S.J., Skehel J.J. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443:45–49. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 11.Sun X., Li Q., Wu Y., Wang M., Liu Y., Qi J., Vavricka C.J., Gao G.F. Structure of influenza virus N7: The last piece of the neuraminidase "jigsaw" puzzle. J. Virol. 2014;88:9197–9207. doi: 10.1128/JVI.00805-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han N., Mu Y., Miao H., Yang Y., Wu Q., Li J., Ding J., Xu B., Huang Z. The 340-cavity in neuraminidase provides new opportunities for influenza drug development: A molecular dynamics simulation study. Biochem. Biophys. Res. Commun. 2016;470:130–136. doi: 10.1016/j.bbrc.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Tran D.T., Le L.T., Truong T.N. Discover binding pathways using the sliding binding-box docking approach: Application to binding pathways of oseltamivir to avian influenza H5N1 neuraminidase. J. Comput. Aided Mol. Des. 2013;27:689–695. doi: 10.1007/s10822-013-9675-1. [DOI] [PubMed] [Google Scholar]
- 14.Burmeister W.P., Cusack S., Ruigrok R.W. Calcium is needed for the thermostability of influenza B virus neuraminidase. J. Gen. Virol. 1994;75:381–388. doi: 10.1099/0022-1317-75-2-381. [DOI] [PubMed] [Google Scholar]
- 15.Wang H., Dou D., Ostbye H., Revol R., Daniels R. Structural restrictions for influenza neuraminidase activity promote adaptation and diversification. Nat. Microbiol. 2019;4:2565–2577. doi: 10.1038/s41564-019-0537-z. [DOI] [PubMed] [Google Scholar]
- 16.Lawrenz M., Wereszczynski J., Amaro R., Walker R., Roitberg A., McCammon J.A. Impact of calcium on N1 influenza neuraminidase dynamics and binding free energy. Proteins. 2010;78:2523–2532. doi: 10.1002/prot.22761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le L., Lee E.H., Hardy D.J., Truong T.N., Schulten K. Molecular dynamics simulations suggest that electrostatic funnel directs binding of Tamiflu to influenza N1 neuraminidases. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaro R.E., Minh D.D.L., Cheng L.S., Lindstrom W.M., Olson A.J., Lin J.H., Li W.W., McCammon J.A. Remarkable loop flexibility in avian influenza N1 and its implications for antiviral drug design. J. Am. Chem. Soc. 2007;129:7764–7765. doi: 10.1021/ja0723535. [DOI] [PubMed] [Google Scholar]
- 19.Zhu X., Yang H., Guo Z., Yu W., Carney P.J., Li Y., Chen L.M., Paulson J.C., Donis R.O., Tong S., et al. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc. Natl. Acad. Sci. USA. 2012;109:18903–18908. doi: 10.1073/pnas.1212579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landon M.R., Amaro R.E., Baron R., Ngan C.H., Ozonoff D., McCammon J.A., Vajda S. Novel druggable hot spots in avian influenza neuraminidase H5N1 revealed by computational solvent mapping of a reduced and representative receptor ensemble. Chem. Biol. Drug Des. 2008;71:106–116. doi: 10.1111/j.1747-0285.2007.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varghese J.N., Colman P.M. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 A resolution. J. Mol. Biol. 1991;221:473–486. doi: 10.1016/0022-2836(91)80068-6. [DOI] [PubMed] [Google Scholar]
- 22.Li Q., Qi J., Wu Y., Kiyota H., Tanaka K., Suhara Y., Ohrui H., Suzuki Y., Vavricka C.J., Gao G.F. Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance. J. Virol. 2013;87:10016–10024. doi: 10.1128/JVI.01129-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M., Qi J., Liu Y., Vavricka C.J., Wu Y., Li Q., Gao G.F. Influenza A virus N5 neuraminidase has an extended 150-cavity. J. Virol. 2011;85:8431–8435. doi: 10.1128/JVI.00638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y., Bi Y., Vavricka C.J., Sun X., Zhang Y., Gao F., Zhao M., Xiao H., Qin C., He J., et al. Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses. Cell Res. 2013;23:1347–1355. doi: 10.1038/cr.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 26.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory. Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 27.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 28.Darden T., York D., Pedersen L. Particle Mesh Ewald—an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 29.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durrant J.D., Votapka L., Sorensen J., Amaro R.E. POVME 2.0: An Enhanced Tool for Determining Pocket Shape and Volume Characteristics. J. Chem. Theory Comput. 2014;10:5047–5056. doi: 10.1021/ct500381c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mu Y.G., Nguyen P.H., Stock G. Energy landscape of a small peptide revealed by dihedral angle principal component analysis. Proteins Struct. Funct. Bioinform. 2005;58:45–52. doi: 10.1002/prot.20310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.