ABSTRACT
Objectives
To examine the characteristics and distribution of possible severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) target cells in the human trophectoderm (TE) and placenta.
Methods
Bioinformatics analysis was performed based on published single‐cell transcriptomic datasets of early TE and first‐ and second‐trimester human placentae. We conducted the transcriptomic analysis of 4198 early TE cells, 1260 first‐trimester placental cells and 189 extravillous trophoblast cells (EVTs) from 24‐week placentae (EVT_24W) using the SMART‐Seq2 method. In addition, to confirm the bioinformatic results, we performed immunohistochemical staining of three first‐trimester, three second‐trimester and three third‐trimester placentae from nine women recruited prospectively to this study. We evaluated the expression of the SARS‐CoV‐2‐related molecules angiotensin‐converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2).
Results
Via bioinformatic analysis, we identified the existence of ACE2 and TMPRSS2 expression in human TE as well as in first‐ and second‐trimester placentae. In the human TE, 54.4% of TE1 cells, 9.0% of cytotrophoblasts (CTBs), 3.2% of EVTs and 29.5% of syncytiotrophoblasts (STBs) were ACE2‐positive. In addition, 90.7% of TE1 cells, 31.5% of CTBs, 22.1% of EVTs and 70.8% of STBs were TMPRSS2‐positive. In placental cells, 20.4% of CTBs, 44.1% of STBs, 3.4% of EVTs from 8‐week placentae (EVT_8W) and 63% of EVT_24W were ACE2‐positive, while 1.6% of CTBs, 26.5% of STBs, 1.9% of EVT_8W and 20.1% of EVT_24W were TMPRSS2‐positive. Pathway analysis revealed that EVT_24W cells that were positive for both ACE2 and TMPRSS2 (ACE2 + TMPRSS2‐positive) were associated with morphogenesis of branching structure, extracellular matrix interaction, oxygen binding and antioxidant activity. The ACE2 + TMPRSS2‐positive TE1 cells were correlated with an increased capacity for viral invasion, epithelial‐cell proliferation and cell adhesion. Expression of ACE2 and TMPRSS2 was observed on immunohistochemical staining in first‐, second‐ and third‐trimester placentae.
Conclusions
ACE2‐ and TMPRSS2‐positive cells are present in the human TE and placenta in all three trimesters of pregnancy, which indicates the possibility that SARS‐CoV‐2 could spread via the placenta and cause intrauterine fetal infection. © 2020 International Society of Ultrasound in Obstetrics and Gynecology
Keywords: ACE2, COVID‐19, placenta, SARS‐CoV‐2, TMPRSS2, trophectoderm
CONTRIBUTION —
What are the novel findings of this work?
The angiotensin‐converting enzyme 2 and transmembrane protease serine 2, which can facilitate the entry of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) into host cells, are expressed in human trophectoderm (TE) and the placenta throughout the three trimesters of pregnancy.
What are the clinical implications of this work?
Our findings revealed the presence of SARS‐CoV‐2 target cells in human TE and placenta, suggesting that pregnancies complicated by the novel coronavirus disease 2019 are potentially at risk of intrauterine fetal SARS‐CoV‐2 infection or placental insufficiency, which could lead to fetal growth restriction and even fetal loss.
INTRODUCTION
The coronavirus disease 2019 (COVID‐19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), is a global health crisis 1 . At present, there is much controversy relating to the possibility of vertical transmission of SARS‐CoV‐2 in pregnancy. In addition, there are limited data on whether SARS‐CoV‐2 could cause fetal infection, congenital malformation, fetal growth restriction and even fetal loss. High‐quality evidence is needed to address these issues.
The placenta is the interface between the mother and the fetus, mediating protection for the fetus against various infections 2 . Chorionic villi serve as functional units of the placenta and contain three types of trophoblasts: syncytiotrophoblasts (STBs), extravillous trophoblasts (EVTs) and cytotrophoblasts (CTBs). The STBs form the outer layer of the trophoblasts and establish an interface between maternal blood and embryonic extracellular fluid, facilitating exchange of materials between the mother and the fetus. The EVTs form cell columns at the base of the stem villi, and the CTBs line the inside of the STBs and function as trophoblastic stem cells. When maternal infection occurs, the STBs usually serve as a barrier against pathogen invasion. Unlike the STBs, the EVTs represent a possible route through which pathogens can breach the placental barrier and cause vertical transmission 3 , 4 .
Recent studies have indicated that angiotensin‐converting enzyme 2 (ACE2) serves as a putative surface receptor of sensitive cells for SARS‐CoV‐2, which employs downstream serine protease, transmembrane protease serine 2 (TMPRSS2), for S protein priming, enabling viral invasion into host cells 5 , 6 , 7 , 8 , 9 . Cysteine proteases, such as cathepsin B (CTSB) and cathepsin L (CTSL), have also been shown to facilitate entry into certain cell lines of coronaviruses, such as severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus, and filoviruses, such as Ebola virus 10 .
In recent years, the development of single‐cell RNA‐sequencing (scRNA‐seq) technology has offered tremendous transcriptomic information at unprecedented resolution. To better understand the possibility of vertical transmission of SARS‐CoV‐2, we examined the characteristics and distribution of possible SARS‐CoV‐2 target cells in human trophectoderm (TE) and placenta.
METHODS
This study is composed of two parts: scRNA‐seq analysis, which was based on previously published single‐cell transcriptomic profiles of TE and first‐ and second‐trimester placentae, and immunohistochemical staining, for which women were recruited prospectively to the study. A total of nine healthy pregnant women, three from each trimester of pregnancy, were recruited to the study and informed consent was obtained. Placentae of the following gestational ages were collected: 8 weeks (induced abortion (n = 3)), 17 weeks (n = 1), 18 weeks (n = 1), 24 weeks (n = 1), 38 weeks, (n = 1) and 39 weeks (n = 2). After delivery, the placentae were placed on ice and sent to the State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, China. For the second‐ and third‐trimester cases, placentae with decidua around the center (maternal side) were cut into 0.5 × 0.5 × 0.5 cm pieces and fixed with 4% paraformaldehyde overnight at 4°C before paraffin section.
Preprocessing, clustering and visualization of scRNA‐seq data
For the bioinformatics analysis, we explored published scRNA‐seq profiles of first‐ and second‐trimester human placentae and in‐vitro cultured human embryos 11 , 12 . The datasets were acquired from the National Center for Biotechnology Information Gene Expression Omnibus database; the datasets of the human placentae were from GSE89497, and the datasets of in‐vitro cultured human embryos were from GSE109555. All the transcriptomic libraries of scRNA‐seq data analyzed in this study were constructed using the Smart‐seq2 method 13 , 14 . The expression levels of genes were normalized to transcripts‐per‐million mapped reads (TPM). In the human‐placenta datasets, single‐cell transcriptomes of eight placentae were analyzed 11 . We retained cells with at least 3000 genes detected, and extracted 1260 placental cells from the 8‐week placentae (EVT_8W) and 189 EVT cells from the 24‐week placentae (EVT_24W). In total, 16 845 genes with TPMs over five in at least two cells were used for further analysis. In the in‐vitro cultured human‐embryo datasets, single‐cell transcriptomes (5911 cells) of 48 embryos (6, 8, 10, 12 and 14 days postfertilization) were used 12 . We filtered out cells with fewer than 5000 genes detected and genes expressed in fewer than three cells. Eventually, 4198 TE‐derived cells with 22 359 genes were used for clustering analysis and visualization.
Single‐cell data were mainly analyzed using Seurat R package (Satija Lab, New York Genome Center, NY, USA) 15 , 16 . We used the ‘ElbowPlot’ function to determine the appropriate number of dimensions to perform the non‐linear dimensional reduction (using t‐distributed stochastic neighbor embedding (t‐SNE) or uniform manifold approximation and projection (UMAP)). The unsupervised clustering method was used to identify cell clusters with the ‘FindClusters’ function of the Seurat R package. In the human‐placenta datasets, the t‐SNE method was used to visualize the clusters and cell types, using Seurat V3.0 R package 15 , 16 . In the datasets of TE‐derived cells from in‐vitro cultured human embryos, the clusters and cell types were visualized with the UMAP method using Seurat V3.0 R package 15 , 16 . ggplot2 V3.3.0 R package was used to illustrate markers of different cell types 17 .
ACE2 and TMPRSS2 expression and functional analysis
The ‘Featureplot’ function of Seurat V3.0 R package 15 , 16 was used to illustrate the expression of ACE2, TMPRSS2, CTSB and CTSL in all cell types. To show the percentage of cells that were positive for individual molecules in each cell type, we used the ‘geom_bar’ function of the ggplot2 V3.3.0 R package to generate bar plots 17 . Pie charts were used to show the percentages of different cell types in double‐positive (ACE2 + TMPRSS2‐positive) cells (ggplot2 V3.3.0 R package) 17 .
We further conducted functional analysis of the ACE2 + TMPRSS2‐positive cells in different cell types to explore the possible impact of SARS‐CoV‐2 infection on TE or placental function. Differentially expressed genes that were specifically highly regulated in the ACE2 + TMPRSS2‐positive cells were used for further gene ontology (GO) analysis. To compare the functional differences between ACE2 + TMPRSS2‐positive cells and the rest of the cells from EVT_24W and TE1 cells, we used the ‘FindMarkers’ function of the Seurat V3.0 R package to acquire the upregulated genes in ACE2 + TMPRSS2‐positive cells. Then ClusterProfiler V3.14.0 R package was utilized for subsequent GO analysis 18 .
Paraffin sections and immunohistochemical staining
Paraffin sections were prepared as described by Hoffman et al. 19 . Briefly, tissues were collected, fixed, dehydrated and cleared before being embedded in paraffin. Embedded tissues from the first‐, second‐ and third‐trimester placentae were sliced into 5 µm per section, floated on the same glass slide and subjected to further immunohistochemical staining, which was performed as described by Hoffman et al. 19 . In brief, paraffin sections were incubated with antibodies against KRT7 (keratin 7; ZSGB Biotech (Beijing, China), ZA‐0573), HLA‐G (human leukocyte antigen G; Santa Cruz, SC‐21799), VIM (vimentin; ZSGB, ZA0511), TMPRSS2 (Abcam, ab109131) and ACE2 (Abcam, ab246511) after deparaffinization, hydration, endogenous peroxidase activity blocking and antigen retrieval. 3,3′‐diaminobenzidine tetrahydrochloride (DAB) substrate solution (ZSGB, ZLI9018) was used for color staining after section incubation with biotinylated secondary antibody and Sav‐HRP conjugates (ZSGB, SP9001). The sections were counterstained, dehydrated, cleared and coverslipped for long‐term storage at room temperature. Finally, the sections were imaged with a Leica Aperio VESA8 scanner (Leica, Wetzlar, Germany).
RESULTS
ACE2‐ and TMPRSS2‐positive cells in human trophectoderm
Using publicly available scRNA‐seq datasets from human TE 12 , we divided the cells into four clusters according to the expression levels of characteristic markers, comprising TE1, CTB, EVT and STB (Figure 1). Unsupervised clustering analysis presented an unbiased distribution of 4198 cells using the UMAP method. The cells in the TE1 cluster exhibited a high expression level of the TE‐related molecule CDX2 (caudal‐type homeobox 2), and these cells were mainly from embryonic cells obtained on day 6 after fertilization. Therefore, we considered TE1 cells to be TE cells before implantation. High expression of HLA‐G was observed in the EVT cluster. The cluster with high expression of CYP19A1 (encoding the enzyme cytochrome P450 family 19 subfamily A member 1) was STB. The fourth cluster that showed high expression of PARP1 (encoding poly ADP‐ribose polymerase‐1) was considered to be CTB (Figure 1a,b). With respect to the expression of SARS‐CoV‐2‐related molecules, 54.4% of TE1 cells, 9.0% of CTBs, 3.2% of EVTs and 29.5% of STBs were ACE2‐positive (Figure 1c,g), while 90.7% of TE1 cells, 31.5% of CTBs, 22.1% of EVTs and 70.8% of STBs were TMPRSS2‐positive (Figure 1d,h). At the same time, 100% of TE1 cells, 95.9% of CTBs, 96.8% of EVTs and 97.4% of STBs were CTSB‐positive (Figure 1e,i), and 100% of TE1 cells, 99.9% of CTBs, 99.7% of EVTs and 100% of STBs were CTSL‐positive (Figure 1f,j).
Figure 1.
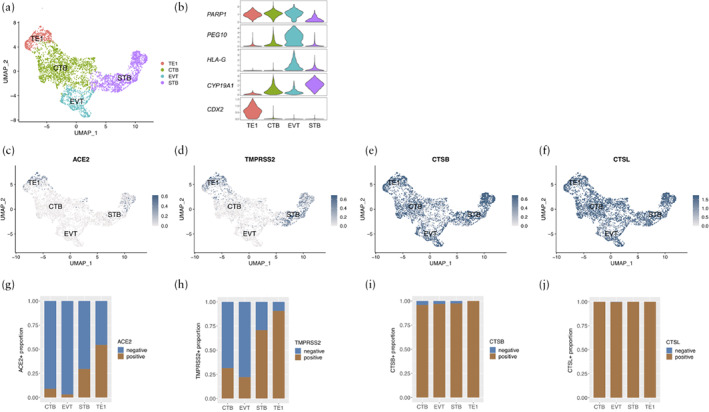
(a,b) Uniform manifold approximation and projection (UMAP) distributions showing cell clusters (a) and violin plots displaying expression levels of representative markers in each cell type (b) of single‐cell RNA sequencing data from embryonic trophectoderm (TE) cells obtained at 6–14 days after fertilization. Cells were classified into four clusters: trophectoderm 1 (TE1), syncytiotrophoblasts (STB), extravillous trophoblasts (EVT) and cytotrophoblasts (CTB) (c–f). Expression in early TE cells of SARS‐CoV‐2‐related molecules: angiotensin‐converting enzyme 2 (ACE2) (c), transmembrane protease serine 2 (TMPRSS2) (d), cathepsin B (CTSB) (e) and cathepsin L (CTSL) (f). (g–j) Proportion of cells that were positive for ACE2 (g), TMPRSS2 (h), CTSB (i) and CTSL (j) according to cell type.
ACE2‐ and TMPRSS2‐positive cells in first‐ and second‐trimester placentae
Based on online scRNA‐seq data from first‐ and second‐trimester placentae 11 , we categorized the cells into seven different clusters. Unsupervised clustering analysis presented the unbiased distribution of 1449 placental cells using the t‐SNE method. From the first‐trimester placenta, the cluster that exhibited high expression of HLA‐G was EVT_8W. The CTB cluster showed high expression of PARP1. The cluster macrophage showed CD68 (encoding the glycoprotein Cluster of Differentiation 68) expression. Mesenchymal cells showed high expression of VIM, which is a classical marker for stromal cells. Cells with high expression of CYP19A1 were identified as STBs. Blood cells showed high expression of ALAS2 (encoding 5′‐aminolevulinate synthase 2) and HBB (encoding beta hemoglobin). EVT_24W cells showed high expression of HLA‐G (Figure 2a,b). In first‐trimester placental cells, 20.4% of CTBs, 44.1% of STBs and 3.4% of EVT_8W were ACE2‐positive, while 1.6% of CTBs, 26.5% of STBs and 1.9% of EVT_8W were TMPRSS2‐positive (Figure 2g,h). Compared with trophoblasts, macrophages, blood cells and mesenchymal stromal cells showed lower expression levels of ACE2 and TMPRSS2 (Figure 2c,d). With respect to the EVT_24W cluster, 63% and 20.1% of cells were ACE2‐positive and TMPRSS2‐positive, respectively (Fig 2c,d,g,h). All cells in all clusters were CTSB‐positive (Figure 2e,i). In addition, 99.6% of CTBs, 98.1% of blood cells, 98.0% of mesenchymal cells and all (100%) cells from other cell types were CTSL‐positive (Figure 2f,j).
Figure 2.
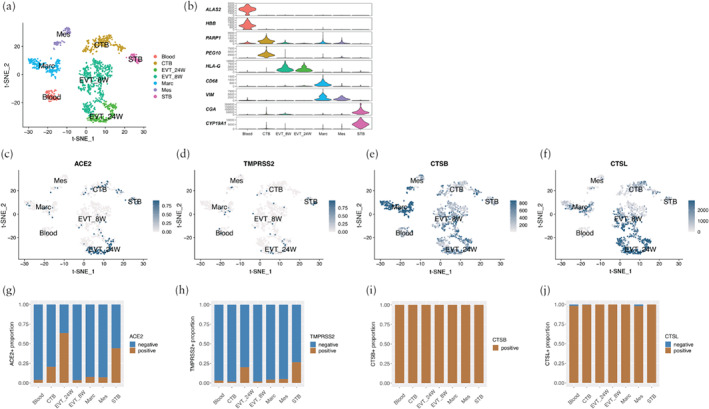
(a,b) t‐distributed stochastic neighbor embedding (t‐SNE) distributions showing cell clusters (a) and violin plots displaying expression levels of representative markers in each cell type (b) of single‐cell RNA sequencing data from 8‐week and 24‐week placentae. Cells were classified into seven clusters: blood, cytotrophoblasts (CTB), extravillous trophoblasts from 8‐week (EVT_8W) and 24‐week (EVT_24W) placentae, macrophages (Marc), mesenchymal cells (Mes) and syncytiotrophoblasts (STB) (c–f). Expression in first‐ and second‐trimester placentae of SARS‐CoV‐2‐related molecules angiotensin‐converting enzyme 2 (ACE2) (c), transmembrane protease serine 2 (TMPRSS2) (d), cathepsin B (CTSB) (e) and cathepsin L (CTSL) (f). (g–j) Proportion of cells that were positive for ACE2 (g), TMPRSS2 (h), CTSB (i) and CTSL (j) according to cell type.
Immunohistochemical analysis of ACE2 and TMPRSS2 in human placentae
Immunohistochemical staining of ACE2 and TMPRSS2 on serial sections of first‐, second‐ and third‐trimester human placentae confirmed the expression of ACE2 and TMPRSS2 in the placenta in all trimesters of pregnancy. Markers including KRT7, HLA‐G and VIM were also stained to show the distribution of trophoblasts, EVTs and stromal cells, respectively. In the first‐trimester placentae, ACE2 was expressed mainly in the STBs, and TMPRSS2 was positive in STBs and EVTs as well as CTBs (Figure 3a). In the second‐trimester placentae, the main sites of ACE2 expression were EVTs and STBs, and TMPRSS2 was expressed in all trophoblasts (Figure 3b). In the third‐trimester placentae, ACE2 was expressed mainly in EVTs and STBs, and TMPRSS2 was expressed in all trophoblasts (Figure 3c).
Figure 3.
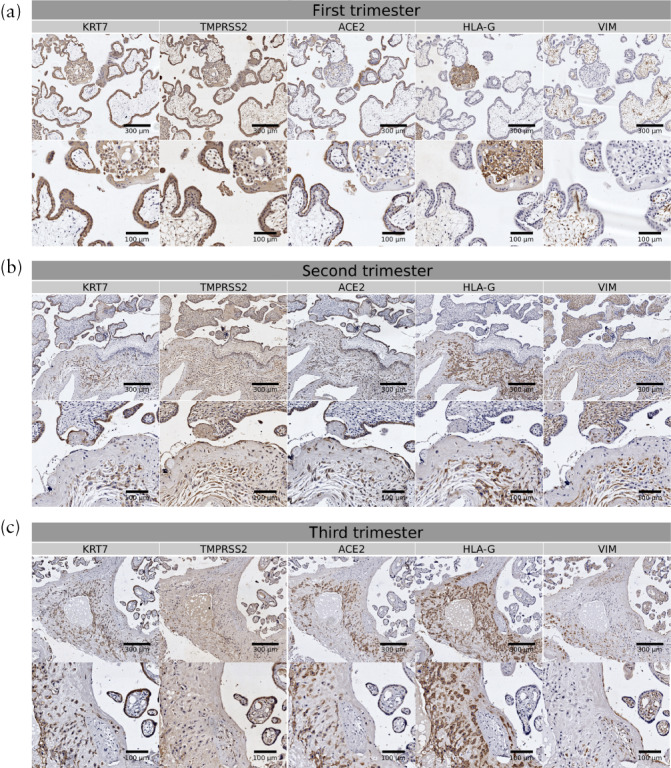
Immunohistochemical staining of angiotensin‐converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) in first‐ (a), second‐ (b) and third‐ (c) trimester human placentae. Staining of keratin 7 (KRT7; marker of trophoblasts), human leukocyte antigen G (HLA‐G; marker of extravillous trophoblasts) and vimentin (VIM; marker of stromal cells) was also conducted to show different cell types. Scale bars in upper rows of figure parts represent 300 µm and those in lower rows represent 100 µm.
Functional analysis of ACE2‐ and TMPRSS2‐positive cells
We further conducted functional analysis of the cells that were both ACE2‐ and TMPRSS2‐positive (ACE2 + TMPRSS2‐positive) in different cell types. First, distributions of the ACE2 + TMPRSS2‐positive cells from each cell type were described in the dataset from the first‐ and second‐trimester placentae and the TE (Figure 4). Among all cell types, the EVT_24W and TE1 cells contributed to the ACE2 + TMPRSS2‐positive cells with the highest proportion. GO analysis identified the functional differences between the ACE2 + TMPRSS2‐positive cells and the rest of the cells from these two cell types (EVT_24W and TE1) 20 , 21 . The ACE2 + TMPRSS2‐positive EVT_24W cells were shown to be associated with morphogenesis of branching structure, extracellular matrix interaction, oxygen binding and antioxidant activity (Figure 5). The ACE2 + TMPRSS2‐positive TE1 cells showed a strong association with increased capacity for viral invasion, epithelial‐cell proliferation and cell‐adhesion‐molecule binding (Figure 6).
Figure 4.
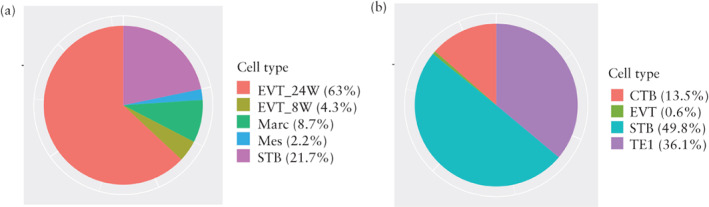
Distribution of cells that were positive for both angiotensin‐converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) in 8‐week (8W) and 24‐week (24W) human placentae (a) and in early trophectoderm (b), across cell types. CTB, cytotrophoblasts; EVT, extravillous trophoblasts; Marc, macrophages; Mes, mesenchymal cells; STB, syncytiotrophoblasts; TE1, trophectoderm 1.
Figure 5.
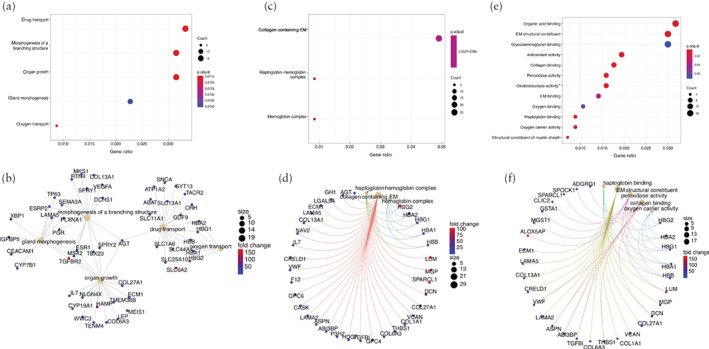
Gene ontology (GO) analysis showing enriched terms in extravillous trophoblasts from 24‐week human placentae (EVT_24W) that were positive for both angiotensin‐converting enzyme 2 and transmembrane protease serine 2 in comparison to rest of EVT_24W cells that were not, including GO biological process analysis (a,b), GO cellular component analysis (c,d) and GO molecular function analysis (e,f). Colors in dot plots (a,c,e) are coded according to significance of enrichment scores and sizes are on basis of counts of overlapping genes. In network plots (b,d,f), size of each dot represents count of overlapping genes and color of each gene dot represents significance of enrichment scores. *Acting on peroxide as acceptor. EM, extracellular matrix.
Figure 6.
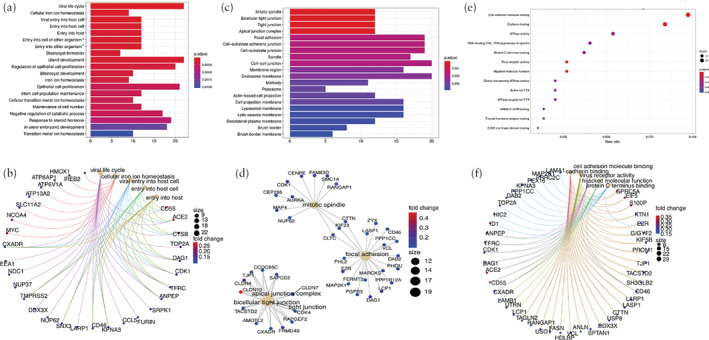
Gene ontology (GO) analysis of upregulated genes in trophectoderm 1 (TE1) cells from early embryo that were positive for both angiotensin‐converting enzyme 2 and transmembrane protease serine 2 in comparison to rest of TE1 cells that were not, including GO biological process analysis (a,b), GO cellular component analysis (c,d) and GO molecular function analysis (e,f). Colors in bar plots (a,c) are coded according to significance of enrichment scores and heights are on basis of overlapping genes. Colors in dot plot (e) are coded according to significance of enrichment scores and sizes are on basis of counts of overlapping genes. In network plots (b,d,f), size of each dot represents count of overlapping genes and color of each gene dot represents significance of enrichment scores. *Involved in symbiotic interaction. TRA, transcription repressor activity; TTA, transmembrane transporter activity.
DISCUSSION
Based on scRNA‐seq analysis and immunohistochemical staining, the main findings of this study are: (1) ACE2 and TMPRSS2 are expressed in human TE and placenta throughout pregnancy; (2) ACE2 + TMPRSS2‐positive EVTs from second‐trimester placentae show association with morphogenesis of branching structure, extracellular matrix interaction, oxygen binding and antioxidant activity; and (3) ACE2 + TMPRSS2‐positive early TE cells show a strong association with increased capacity for viral invasion, epithelial‐cell proliferation and cell‐adhesion‐molecule binding.
Previous scRNA‐seq findings concerning SARS‐CoV‐2 target cells in placentae
The recent development of scRNA‐seq technologies has enabled us to examine the expression of certain SARS‐CoV‐2‐related genes at the single‐cell level 22 . Recently, the National Institutes of Health Human Cell Atlas (NIH‐HCA) released the analysis of ACE2 and CTSL expression in first‐ and third‐trimester placental scRNA‐seq transcriptomes 23 , 24 , 25 , 26 . Their research also demonstrated that only a small fraction of the maternal decidua and fetal trophoblasts in first‐trimester placentae and some of the EVTs and STBs in third‐trimester placentae express both ACE2 and CTSL. In comparison to their findings, our findings indicate a higher proportion of SARS‐CoV‐2 target cells in the TE and the first‐ and second‐trimester placenta. The main difference between our study and that of the NIH‐HCA with regard to the sequencing methodology was that the NIH‐HCA research employed 10 × Genomics and Drop‐seq platforms for all scRNA‐seq while, in this study, we utilized transcriptomes with the SMART‐Seq2 method, which has been shown to have higher sensitivity of gene detection and lower dropout rates 27 .
Clinical implications
It is well accepted that SARS‐CoV‐2 is transmitted via respiratory droplets and direct contact 28 , 29 . During pregnancy, there is the additional concern that the virus can be transmitted to the fetus, however, existing evidence on the possibility of vertical transmission remains highly controversial 30 , 31 , 32 . Previous studies have evaluated the risk of intrauterine infection by testing for SARS‐CoV‐2 RNA in amniotic fluid, cord blood, placental tissue and vaginal secretion samples collected from pregnant women with confirmed COVID‐19 and pharyngeal swab samples collected from neonates born to these women 33 , 34 , 35 . In all three studies, all biological samples tested negative 33 , 34 , 35 . Zeng et al. 36 reported that three of 33 (9%) infants were diagnosed with neonatal early‐onset COVID‐19 based on positive SARS‐CoV‐2 RNA results in two consecutive nasopharyngeal and anal swabs obtained on day 2 and day 4 of age. All three infants tested negative for SARS‐CoV‐2 RNA on day 6 or 7 of age. Though strict infection control and prevention measures were implemented during delivery, the possibility of postpartum neonatal infection cannot be completely excluded because of the delay in testing 36 . For some pathogens, such as Rubella virus, the risk of vertical transmission varies at different stages of pregnancy 37 . With respect to SARS‐CoV‐2, we observed a lower level of ACE2 and TMPRSS2 expression in EVTs in second‐trimester placentae than in first‐trimester placentae based on scRNA‐seq analysis. More recently, Vivanti et al. 38 reported a case of maternal and neonatal SARS‐CoV‐2 infection based on positive SARS‐CoV‐2 RNA results in the maternal nasopharyngeal swab, vaginal swab, blood, placenta and amniotic fluid, as well as neonatal blood, nasopharyngeal swab and rectal swab.
Our findings confirm the presence of the target cells of SARS‐CoV‐2 in the human TE and placenta, highlighting the possibility of a transplacental approach for vertical transmission. A case of miscarriage at 19 weeks' gestation in a pregnant woman with COVID‐19 has been reported recently 39 . In this case, a reverse transcription polymerase chain reaction test for SARS‐CoV‐2 RNA was positive in the placenta but negative in the abortus. Such findings have led us to speculate that SARS‐CoV‐2 can cause miscarriage, especially when there is a long duration of viral exposure and heavy viral load. More data are needed to answer the question of whether COVID‐19 in pregnancy is associated with fetal growth restriction and structural malformation.
Previous findings have shown that ACE2 + TMPRSS2‐positive cells are considered to be associated with viral invasion 9 . Considering that SARS‐CoV‐2 invasion might cause functional impairment to its target cells, we conducted functional analysis in the two cell types that showed the highest percentage of ACE2 + TMPRSS2‐positive cells. Compared with the rest of the EVT_24W cells, ACE2 + TMPRSS2‐positive EVT_24W cells were associated with morphogenesis of branching structure, extracellular matrix interaction, oxygen binding and antioxidant activity. Compared with the rest of the early TE cells, ACE2 + TMPRSS2‐positive early TE cells showed correlation with increased capacity for viral invasion, epithelial‐cell proliferation and cell‐adhesion‐molecule binding. These findings suggest that pregnancies complicated by COVID‐19 are potentially at risk of placental insufficiency, which might lead to fetal growth restriction and even fetal loss.
Strengths and limitations
This study has several strengths. First, scRNA‐seq transcriptomic analysis was used to identify SARS‐CoV‐2 target cells at the single‐cell level. Second, differences in SARS‐CoV‐2 target cell numbers in placentae from the three trimesters of pregnancy were explored. Third, the SMART‐Seq2 sequencing platform was used to achieve better accuracy of the analysis. Fourth, immunohistochemical staining analysis was performed to confirm the bioinformatic results. Finally, functional analysis of SARS‐CoV‐2 target cells was conducted to illustrate the potential impact of SARS‐CoV‐2 infection on placental function.
A major limitation of the study is that for third‐trimester placentae we only performed immunohistochemical staining without scRNA‐seq analysis, for the reason that, as far as we know, the scRNA‐seq transcriptome of the third‐trimester placenta has not been analyzed by the SMART‐Seq2 platform until now. In addition, since our bioinformatic analysis and immunohistochemical experiments were based on a limited number of samples, considering the impact of individual heterogeneity, whether the expression patterns of ACE2 and TMPRSS2 can be extended to the general population requires further exploration.
Conclusions
Our findings have shown the presence of ACE2‐ and TMPRSS2‐positive cells in the human placenta at different stages of pregnancy as well as in the human TE, which indicates the possibility that SARS‐CoV‐2 could spread via the placenta and cause intrauterine fetal infection. Functional analysis revealed that ACE2 + TMPRSS2‐positive EVT_24W cells are associated with morphogenesis of branching structure, extracellular matrix interaction, oxygen binding and antioxidant activity, and that ACE2 + TMPRSS2‐positive early TE cells are correlated with an increased capacity for viral invasion, epithelial cell proliferation and cell adhesion.
ACKNOWLEDGMENTS
This study was supported by grants 31900602 and 31900664 from the National Natural Science Foundation of China and 2018YFC1004101 from the Ministry of Science and Technology of the People's Republic of China. The funding sources had no involvement in the study. The authors declare no conflict of interest.
Contributor Information
L. C. Poon, Email: liona.poon@cuhk.edu.hk.
H. Wang, Email: wanghm@ioz.ac.cn.
H. Yang, Email: yanghuixia@bjmu.edu.cn.
REFERENCES
- 1. Wang C, Chen D, Yang H. Updates on COVID‐19 Infection. Matern Med 2020; 2: 65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maltepe E, Fisher SJ. Placenta: the forgotten organ. Annu Rev Cell Dev Biol 2015; 31: 523–552. [DOI] [PubMed] [Google Scholar]
- 3. Pereira L. Congenital Viral Infection: Traversing the Uterine‐Placental Interface. Annu Rev Virol 2018; 5: 273–299. [DOI] [PubMed] [Google Scholar]
- 4. Arora N, Sadovsky Y, Dermody TS, Coyne CB. Microbial Vertical Transmission during Human Pregnancy. Cell Host Microbe 2017; 21: 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature 2020; 581: 215–220. [DOI] [PubMed] [Google Scholar]
- 6. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS‐CoV‐2 by full‐length human ACE2. Science 2020; 367: 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J. Structural and Functional Basis of SARS‐CoV‐2 Entry by Using Human ACE2. Cell 2020; 181: 894–904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS‐CoV‐2. Nature 2020; 581: 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann M, Kleine‐Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N‐H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. SARS‐CoV‐2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020; 181: 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion RJ, Nunneley JW, Barnard D, Pöhlmann S, McKerrow JH, Renslo AR, Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res 2015; 116: 76–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Fan X, Wang R, Lu X, Dang YL, Wang H, Lin HY, Zhu C, Ge H, Cross JC, Wang H. Single‐cell RNA‐seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta. Cell Res 2018; 28: 819–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou F, Wang R, Yuan P, Ren Y, Mao Y, Li R, Lian Y, Li J, Wen L, Yan L, Qiao J, Tang F. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 2019; 572: 660–664. [DOI] [PubMed] [Google Scholar]
- 13. Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart‐seq2 for sensitive full‐length transcriptome profiling in single cells. Nat Methods 2013; 10: 1096–1098. [DOI] [PubMed] [Google Scholar]
- 14. Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full‐length RNA‐seq from single cells using Smart‐seq2. Nat Protoc 2014; 9: 171–181. [DOI] [PubMed] [Google Scholar]
- 15. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018; 36: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive Integration of Single‐Cell Data. Cell 2019; 177: 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer International Publishing: Houston, TX, USA, 2016. [Google Scholar]
- 18. Yu G, Wang L‐G, Han Y, He Q‐Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16: 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoffmann P, Feige J‐J, Alfaidy N. Expression and oxygen regulation of endocrine gland‐derived vascular endothelial growth factor/prokineticin‐1 and its receptors in human placenta during early pregnancy. Endocrinology 2006; 147: 1675–1684. [DOI] [PubMed] [Google Scholar]
- 20. Carbon S, Douglass E, Dunn N, Good B, Harris NL, Lewis SE, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, Hartline E, Fey P, Thomas PD, Albou LP, Ebert D, Kesling MJ, Mi H, Muruganujan A, Huang X, Poudel S, Mushayahama T, Hu JC, LaBonte SA, Siegele DA, Antonazzo G, Attrill H, Brown NH, Fexova S, Garapati P, Jones TEM, Marygold SJ, Millburn GH, Rey AJ, Trovisco V, Dos Santos G, Emmert DB, Falls K, Zhou P, Goodman JL, Strelets VB, Thurmond J, Courtot M, Osumi DS, Parkinson H, Roncaglia P, Acencio ML, Kuiper M, Lreid A, Logie C, Lovering RC, Huntley RP, Denny P, Campbell NH, Kramarz B, Acquaah V, Ahmad SH, Chen H, Rawson JH, Chibucos MC, Giglio M, Nadendla S, Tauber R, Duesbury MJ, Del NT, Meldal BHM, Perfetto L, Porras P, Orchard S, Shrivastava A, Xie Z, Chang HY, Finn RD, Mitchell AL, Rawlings ND, Richardson L, Sangrador‐Vegas A, Blake JA, Christie KR, Dolan ME, Drabkin HJ, Hill DP, Ni L, Sitnikov D, Harris MA, Oliver SG, Rutherford K, Wood V, Hayles J, Bahler J, Lock A, Bolton ER, De Pons J, Dwinell M, Hayman GT, Laulederkind SJF, Shimoyama M, Tutaj M, Wang SJ et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019; 47(D1): D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel‐Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS, Bals J, Hauser BM, Feldman J, Muus C, Wadsworth MH 2nd, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi L‐E, Barbry P, Leslie A, Kiem H‐P, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK, Ordovas‐Montanes J. SARS‐CoV‐2 Receptor ACE2 Is an Interferon‐Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020; 181: 1016–1035.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vento‐Tormo R, Efremova M, Botting RA, Turco MY, Vento‐Tormo M, Meyer KB, Park JE, Stephenson E, Polański K, Goncalves A, Gardner L, Holmqvist S, Henriksson J, Zou A, Sharkey AM, Millar B, Innes B, Wood L, Wilbrey‐Clark A, Payne RP, Ivarsson MA, Lisgo S, Filby A, Rowitch DH, Bulmer JN, Wright GJ, Stubbington MJT, Haniffa M, Moffett A, Teichmann SA. Single‐cell reconstruction of the early maternal–fetal interface in humans. Nature 2018; 563: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsang JCH, Vong JSL, Ji L, Poon LCY, Jiang P, Lui KO, Ni YB, To KF , Cheng YKY, Chiu RWK, Lo YMD. Integrative single‐cell and cell‐free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci U S A 2017; 114: E7786–E7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Suryawanshi H, Morozov P, Straus A, Sahasrabudhe N, Max KEA, Garzia A, Kustagi M, Tuschl T, Williams Z. A single‐cell survey of the human first‐trimester placenta and decidua. Sci Adv 2018; 4: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Costigan M, Scholz J, Woolf CJ. Neuropathic pain: a maladaptive response of the nervous system to damage. Annu Rev Neurosci 2009; 32: 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet‐Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative Analysis of Single‐Cell RNA Sequencing Methods. Mol Cell 2017; 65: 631–643.e4. [DOI] [PubMed] [Google Scholar]
- 28. Poon LC, Yang H, Dumont S, Lee JCS, Copel JA, Danneels L, Wright A, Costa FDS, Leung TY, Zhang Y, Chen D, Prefumo F. ISUOG Interim Guidance on coronavirus disease 2019 (COVID‐19) during pregnancy and puerperium: information for healthcare professionals – an update. Ultrasound Obstet Gynaecol 2020; 55: 848–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Juan J, Gil MM, Rong Z, Zhang Y, Yang H, Poon LC. Effects of coronavirus disease 2019 (COVID‐19) on maternal, perinatal and neonatal outcome: systematic review. Ultrasound Obstet Gynecol 2020; 56: 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang C, Zhou Y‐H, Yang H‐X, Poon LC. Intrauterine vertical transmission of SARS‐CoV‐2: what we know so far. Ultrasound Obstet Gynecol 2020; 55: 724–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. World Health Organization . Modes of transmission of virus causing COVID‐19: implications for IPC precaution recommendations: scientific brief, 29 March 2020. World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- 32. Dashraath P, Wong JLJ, Lim MXK, Lim LM, Li S, Biswas A, Choolani M, Mattar C, Su LL. Coronavirus Disease 2019 (COVID‐19) pandemic and pregnancy. Am J Obstet Gynecol 2020; 222: 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen H, Guo J, Wang C, Luo F, Yu X, Zhang W, Li J, Zhao D, Xu D, Gong Q, Liao J, Yang H, Hou W, Zhang Y. Clinical characteristics and intrauterine vertical transmission potential of COVID‐19 infection in nine pregnant women: a retrospective review of medical records. Lancet 2020; 395: 809–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lei D, Wang C, Li C, Fang C, Yang W, Chen B, Wei M, Xu X, Yang H, Wang S, Fan C. Clinical characteristics of COVID‐19 in pregnancy: analysis of nine cases. Chin J Perinat Med 2020; 23: 222–228. [Google Scholar]
- 35. Chen S, Huang B, Luo DJ, Li X, Yang F, Zhao Y, Nie X, Huang BX. [Pregnancy with new coronavirus infection: clinical characteristics and placental pathological analysis of three cases]. Zhonghua Bing Li Xue Za Zhi = Chinese J Pathol 2020; 49: 418–423. [DOI] [PubMed] [Google Scholar]
- 36. Zeng L, Xia S, Yuan W, Yan K, Xiao F, Shao J, Zhou W. Neonatal Early‐Onset Infection With SARS‐CoV‐2 in 33 Neonates Born to Mothers With COVID‐19 in Wuhan, China. JAMA Pediatr 2020; 174: 722–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bouthry E, Picone O, Hamdi G, Grangeot‐Keros L, Ayoubi J‐M, Vauloup‐Fellous C. Rubella and pregnancy: diagnosis, management and outcomes. Prenat Diagn 2014; 34: 1246–1253. [DOI] [PubMed] [Google Scholar]
- 38. Vivanti AJ, Vauloup‐Fellous C, Prevot S, Zupan V, Suffee C, Do Cao J, Benachi A, De Luca D. Transplacental transmission of SARS‐CoV‐2 infection. Nat Commun 2020; 11: 3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baud D, Greub G, Favre G, Gengler C, Jaton K, Dubruc E, Pomar L. Second‐Trimester Miscarriage in a Pregnant Woman With SARS‐CoV‐2 Infection. JAMA 2020; 323: 2198–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]


