Abstract
Background
To explore the clinical features and CT findings of clinically cured coronavirus disease 2019 (COVID-19) patients with viral RNA positive anal swab results after discharge.
Methods
Forty-two patients with COVID-19 who were admitted to Yongzhou Central Hospital, Hunan, China, between January 20, 2020, and March 2, 2020, were tested for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using anal swab viral RT-PCR. In this report, we present the clinical characteristics and chest CT features of six patients with positive anal swab results and compare the clinical, laboratory, and CT findings between the positive and negative groups.
Results
The anal swab positivity rate for SARS-CoV-2 RNA in discharged patients was 14.3% (6/42). All six patients were male. In the positive group, 40% of the patients (2/5) had a positive stool occult blood test (OBT), but none had diarrhea. The median duration of fever and major symptoms (except fever) in the positive patients was shorter than that of the negative patients (1 day vs. 6 days, 4.5 days vs. 10.5 days, respectively). The incidence of asymptomatic cases in the positive group (33.3%) was also higher than that of the negative group (5.6%). There were no significant differences in the CT manifestation or evolution of the pulmonary lesions between the two groups.
Conclusion
In our case series, patients with viral RNA positive anal swabs did not exhibit gastrointestinal symptoms, and their main symptoms disappeared early. They had similar CT features to the negative patients, which may be easier to be ignored. A positive OBT may indicate gastrointestinal damage caused by SARS-CoV-2 infection.
Keywords: COVID-19, Discharged patients, Anal swab, CT scan
Keypoints
Discharged patients with an anal swab positive for SARS-CoV-2 RNA could be concealed and easily ignored. This positive result may not represent a recurrence of the disease, but rather represents the intermittent excretion of the virus.
Background
COVID-19 is a novel acute respiratory infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1], which is highly contagious. As of August 3, 2020, 88,573 COVID-19 patients have been diagnosed in China. In addition, 17.9 million laboratory-confirmed cases and 686,703 deaths have been reported in 215 countries and regions outside of China, and the number of infected people is still rising rapidly [2–5].
As most patients with COVID-19 show symptoms of respiratory tract infections, the current diagnosis, and epidemic control standards for COVID-19 rely on the collection of throat swabs, sputum, and lower respiratory tract secretions for quantitative reverse transcription polymerase chain reaction (qRT-PCR) based detection of SARS-CoV-2 RNA. Patients can only be discharged after their RNA throat swab tests are negative twice (sampling interval of at least 1 day) [6]. Some researchers have found a small number of patients who present with SARS-CoV-2 RNA positive stool samples [7], and that the respiratory RNA test results may not be consistent with those from stool samples [8, 9]. However, the clinical and imaging findings from discharged patients with negative throat-swab but positive anal-swab RNA test results have not yet been reported.
In this study, we wanted to evaluate the clinical characteristics and radiographic features of patients with SARS-CoV-2 RNA positive anal swabs after discharge, with a view to improving our understanding of the pathology and risk profiles of these patients.
Methods
Study design and participants
This study was approved by the Ethics Commission of Xiangya Hospital Central South University (approval number 202002019), which waived the need for written informed consent due to the nature of COVID-19. All patients with a confirmed COVID-19 diagnosis admitted to our institution who met the national discharge criteria between January 20, 2020, and February 22, 2020, were enrolled in this study. All enrolled patients were diagnosed using the WHO interim guidance [10] and confirmed by a positive SARS-CoV-2 RNA throat swab qRT-PCR result. Patients were characterized as mild, common, severe, and fatal types based on the clinical guidelines for COVID-19 (Trial Version 5) proposed by China National Health Commission [6]. All confirmed cases were subject to clinical observation and treated in the isolation ward of our institution. The development of a treatment plan was based on the COVID-19 guidelines (Trial Version 5) [6]. Discharge criteria of patients with COVID-19 [6] were as follows: 1) body temperature dropped to normal for more than 3 days, 2) significant improvement in respiratory symptoms, 3) obvious inflammatory absorption on pulmonary imaging, and 4) negative RNA test results for two consecutive respiratory samples (sampling interval ≥ 1 day). Data on the clinical characteristics, laboratory findings, treatment, and outcomes were collected from electronic medical records.
Forty-two confirmed COVID-19 patients meeting the discharge criteria were subjected to anal swab sample collection and these samples were then tested for SARS-CoV-2 RNA. Throat and anal swab samples were collected and evaluated using the Chinese Center for Disease Control and Prevention–standardized quantitative polymerase chain reaction assay [11]. All samples were processed by the Department of Laboratory Medicine at Yongzhou Central Hospital. All enrolled patients were divided into positive (6 cases) and negative (36) groups based on their qRT-PCR results. Patients with positive anal-swab results were kept in isolation and placed under medical observation for a further 2 weeks at a secondary site to prevent spread and all the respiratory samples were rechecked to ensure that they were viral RNA negative. Anal swab samples were reevaluated 7 days later, with five patients returning a negative result. The two severe COVID-19 patients had a negative anal swab test 9 days later (Table 1). Patients were released only when we received two consecutive negative results (sampling interval ≥ 1 day) and at the same time, all their close contacts were found and quarantined at home for 14 days in accordance with the guidelines [6]. Thirty-one patients underwent a CT scan at admittance and all enrolled patients underwent a CT scan on discharge, with 25 patients needing another CT scan 7–14 days after discharge.
Table 1.
Clinical Characteristics and CT findings of patients with positive anal swab viral RNA
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | |
|---|---|---|---|---|---|---|
| Sex | Male | Male | Male | Male | Male | Male |
| Age-ranges, y | 15–20 | 5–10 | 60–70 | 40–50 | 20–30 | 30–40 |
| Clinical type | Common | Common | Severe | Severe | Mild | Common |
| Maximum temperature, °C | 36.9 | 36.0 | 36.1 | 39.5 | 37.5 | 37.8 |
| Diarrhea | No | No | No | No | No | No |
| OBT | Negative | Positive | Positive | Negative | Negative | |
| Duration of fever, d | 0 | 0 | 0 | 5 | 2 | 10 |
| Duration of major symptoms (except fever), d | 0 | 5 | 7 | 4 | 0 | 5 |
| Duration of throat swab positive, d | 11 | 9 | 13 | 12 | 2 | 9 |
| Duration of anal swab positive, d | 7 | 7 | 9 | 9 | 7 | 7 |
| Hospital stays, d | 13 | 15 | 20 | 21 | 5 | 14 |
| CT score on admission | 0 | 1 | 17 | 13 | ||
| CT score on discharge | 0 | 0 | 14 | 10 | 0 | 0 |
| CT score after discharge | 0 | 10 | 7 |
CT scan and image interpretation
All the study participants were scanned using a Siemens SOMATOM Emotion CT scanner in a supine position, and breath was held after inhalation. Images spanned from the apex of the lungs to the top of the diaphragm and the acquisition parameters were as follows: 120 kV; adaptive tube current (CARE Dose 4D); layer thickness, 5 mm; matrix, 512 × 512; high-resolution algorithm reconstruction; layer interval, 2 mm; and reconstruction layer thickness 1.5 mm.
All chest CT scans were subject to blind review by two radiologists (with 5 and 18 years of experience in reading chest CT) in consensus. If the results were inconsistent, a consensus was reached after negotiation. All images were viewed on both the lung (width, 1100–1300 HU; level, − 800 HU) and mediastinal (width, 300–400 HU; level, 40–50 HU) settings on the picture archiving and communication system. CT features including lesion location (two lungs, single lung, lung lobe, and lung segment), degree of lung involvement; focal (single lesion), multiple (two or more lesions, limited to 2 lung lobes), diffuse (multiple lesions, distributed in more than 2 lung lobes), lesion distribution characteristics (in the pleura, along the bronchial vascular bundle, both in pleura and along the bronchial vascular bundle), lesion morphological characteristics (ground-glass opacity, ground-glass nodules, crazy-paving pattern, air bronchogram) were recorded. These images were then evaluated using the semi-quantitative scoring method proposed by Chang et al. [12, 13] with lung involvement being determined using the separate score from each of the five lung lobes. Each lung lobe was 0–5 points, with a total possible score of 0–25 (0 point represented no involvement, 1 point less than 5% affected, 2 points 6–25% affected, 3 points 26–49% affected, 4 points 50–75% affected, 5 points more than 75% affected). The CT findings on admission, discharge, and after discharge were compared between the two groups.
Statistical analysis
Statistical analysis was performed using SPSS software (version 23.0). Categorical variables are represented as number and frequency, while continuous variables are described using the mean, standard deviation, median, and interquartile range (IQR). The clinical and CT characteristics of the positive and negative viral RNA groups were compared using the χ 2 test or Fisher’s exact test. One-Way ANOVA or independent-sample t test was performed on the continuous variables based on their distribution. A P value of < 0.05 was considered statistically significant. Multiple comparisons were not analyzed, and given the possibility of Type I errors, these findings should be interpreted as exploratory and descriptive.
Results
Clinical characteristics
This case study included 42 patients with confirmed COVID-19 who were clinically cured and discharged based on the COVID-19 guidelines (Trial Version 5) [6]. The positivity rate of the anal swab samples from discharged patients was 14.3% (6/42), all of which were male (100%) (Table 1). There was no recurrence in the throat swab samples after anal swab specimens tested positive, and no close contacts were found to be infected. In the six patients with positive anal swab results two patients (33.3%) were identified through contact tracing, and four (66.7%) were identified via a family cluster. The six cases spanned all of the clinical presentations and included one mild type, three common types, and two severe types of infections. There was no significant difference in clinical type, the use of antibiotics or glucocorticoids during hospitalization between the two groups. The incidence of GI symptoms was 16.7% (7/42), 6 patients (85.7%) presenting with diarrhea were classified as common type, and one patient (14.3%) presenting with nausea and vomiting was classified as severe type. None of the six patients, positive for viral RNA on the anal swab, presented with GI symptoms. The most common symptom at onset is fever (69.0%); with other symptoms such as cough, dyspnea, myalgia, and diarrhea frequently persisting after the patient’s temperature has returned to normal. The median duration of fever in the positive group was 1 day (IQR, 0–6.3 days) which was shorter than that of the negative group (6 days [IQR, 2–9.8 days]), but the difference between the two groups was not statistically significant. The median duration of other major symptoms (except fever) in the positive patients was also shorter than that of patients with negative results (4.5 days [IQR 0–5.5 days] vs. 10.5 days [IQR 4.3–14 days], P = 0.022). The incidence of asymptomatic cases in the positive group was higher than that in the negative group (33.3% vs. 5.6%, P = 0.034) and there were no significant differences in the other clinical characteristics between the two groups (Table 2).
Table 2.
Demographics and baseline characteristics of patients with COVID-19
| Total (N = 42) | Anal swab test positive (n = 6) | Anal swab test negative (n = 36) | P Valuea | |
|---|---|---|---|---|
| Sex | ||||
| Female | 25(59.5%) | 6(100.0%) | 19(52.8%) | 0.031 |
| Male | 17(40.5%) | 0(0.0%) | 17(47.2%) | |
| Age, median (IQR), y | 40(30.8 ~ 46.0) | 28.5(13.3 ~ 50.5) | 42(32 ~ 46) | 0.250 |
| >50 | 9(21.4%) | 1(16.7%) | 8(22.2%) | 0.762 |
| ≤ 50 | 33(78.6%) | 5(83.3%) | 28(77.8%) | |
| BMI, median (IQR) | 24.4(22.2 ~ 25.7) | 24.6(16.0 ~ 27.9) | 24.1(22.2 ~ 25.6) | 1.000 |
| Epidemiology | ||||
| Direct exposure history | 22(52.4%) | 2(33.3%) | 20(55.6%) | 0.148 |
| Indirect exposure history | 6(14.3%) | 0(0.0%) | 6(16.7%) | |
| Family outbreak | 14(33.3%) | 4(66.7%) | 10(27.8%) | |
| Incubation period, median (IQR), d | 7(3 ~ 10) | 9(4.5 ~ 11.3) | 6.5(3 ~ 10) | 0.320 |
| Onset of symptom to, median (IQR), d | ||||
| Hospital admission | 3.5(2 ~ 6) | 3.0(2 ~ 5.5) | 3.5(2 ~ 6) | 0.697 |
| Anal swab test | 27(25 ~ 32.3) | 26.5(21.5 ~ 35.3) | 27(25 ~ 32.3) | 0.836 |
| Signs and symptoms | ||||
| Fever | 29(69.0%) | 3(50.0%) | 26(72.2%) | 0.281 |
| Maximum temperature, median (IQR), °C | 37.7(36.8 ~ 38.5) | 37.2(36.1 ~ 38.2) | 37.9(37.0 ~ 38.7) | 0.235 |
| <37·3 | 13(31.0%) | 3(50.0%) | 10(27.8%) | 0.541 |
| 37·3–38 | 14(33.3%) | 2(33.3%) | 12(33.3%) | |
| 38·1–39 | 8(19.0%)) | 0(0.0%) | 8(22.2%) | |
| >39 | 7(16.7%) | 1(16.7%) | 6(16.7%) | |
| Cough | 35(83.3%) | 4(66.7%) | 31(86.1%) | 0.242 |
| Dyspnea | 3(7.1%) | 1(16.7%) | 2(5.6%) | 0.333 |
| Fatigue | 3(7.1%) | 1(16.7%) | 2(5.6%) | 0.333 |
| Myalgia | 5(11.9%) | 0(0.0%) | 5(13.9%) | 0.336 |
| Headache | 4(9.5%) | 0(0.0%) | 4(11.1%) | 0.396 |
| Nausea and vomiting | 1(2.4%) | 0(0.0%) | 1(2.8%) | 0.683 |
| Diarrhea | 6(14.3%) | 0(0.0%) | 6(16.7%) | 0.285 |
| No symptoms | 4(9.5%) | 2(33.3%) | 2(5.6%) | 0.034 |
| Comorbidities | ||||
| Diabetes | 2(4.8%) | 0(0.0%) | 2(5.6%) | 0.558 |
| Hypertension | 2(4.8%) | 0(0.0%) | 2(5.6%) | 0.558 |
| Hepatitis or liver cirrhosis | 2(4.8%) | 0(0.0%) | 2(5.6%) | 0.558 |
| Chronic renal failure | 1(2.4%) | 0(0.0%) | 1(2.8%) | 0.683 |
| Malignancy | 1(2.4%) | 0(0.0%) | 1(2.8%) | 0.683 |
| Digestive diseases | 3(7.1%) | 1(16.7%) | 2(5.6%) | 0.333 |
| Clinical type | ||||
| Mild type | 2(4.8%) | 1(16.7%) | 1(2.8%) | 0.701 |
| Common type | 30(71.4%) | 3(50.0%) | 27(75.0%) | |
| Severe type | 7(16.7%) | 2(33.3%) | 5(13.9%) | |
| Critical type | 3(7.1%) | 0(0.0%) | 3(8.3%) | |
| Mild + Common type | 32(76.2%) | 4(66.7%) | 28(77.8%) | 0.558 |
| Severe + Critical type | 10(23.8) | 2(33.3%) | 8(22.2%) | |
| Treatment | ||||
| Antibiotic | 29(69.0%) | 3(50.0%) | 26 (72.2%) | 0.281 |
| Duration of antibiotic | 6.5(0 ~ 8.3) | 2.5(0 ~ 10.5) | 7.0(0 ~ 8) | 0.562 |
| Glucocorticoid | 29(69.0%) | 4(66.7%) | 25(69.4%) | 0.892 |
| Duration of glucocorticoid | 4(0 ~ 5) | 4.5(0 ~ 6.5) | 4(0 ~ 5) | 0.739 |
| Non-Invasive Ventilation | 3(7.1%) | 1(16.7%) | 2(5.6%) | 0.333 |
| Duration of throat swab positive, median (IQR), d | 7.5(5 ~ 10.3) | 10.0(7.3 ~ 12.3) | 7.0(3 ~ 10) | 0.208 |
| Duration of fever, median (IQR), d | 5(0.8 ~ 9.3) | 1(0 ~ 6.3) | 6(2 ~ 9.8) | 0.120 |
| Duration of major symptoms (except fever), median (IQR), d | 8.5(3.8 ~ 13.3) | 4.5(0 ~ 5.5) | 10.5(4.3 ~ 14) | 0.022 |
| Hospital stays, median (IQR), d | 12.5(9 ~ 18) | 14.5(11 ~ 20.3) | 11.5(9 ~ 17.5) | 0.471 |
a P values indicate differences between anal swab test positive and negtive patients. P < .05 was considered statistically significant
Laboratory findings
The positive rate of the stool occult blood test (OBT) during hospitalization was 40% (2/5) in patients with positive anal swabs, and 8.7% (2/23) in the negative group (Table 3). Common causes of gastrointestinal (GI) damage in patients with positive anal swab tests were ruled out by the careful evaluation of the patients prior medical history and related examinations. The median oxygenation index of the positive anal swab group was higher than that of the negative group (486.2 mmHg [IQR 409.5–680.1 mmHg] vs. 381.0 mmHg [IQR 318.6–467.8 mmHg], P = 0.040). One severe-type patient had five positive throat swab samples but was negative on the anal swab test. There was no significant difference between the two groups in any of the other laboratory findings (Table 3).
Table 3.
Laboratory Findings of Patients Infected With COVID-19 on Admission to Hospital
| Normal range | Median (IQR) | P Valuea | |||
|---|---|---|---|---|---|
| Total (N = 42) | Anal swab test positive (n = 6) | Anal swab test positive (n = 36) | |||
| Leukocyte count, × 109/L | 3.5 ~ 9.5 | 5.2(3.9 ~ 6.2) | 5.0(3.6 ~ 6.6) | 5.3(4.1 ~ 6.3) | 0.820 |
| Lymphocyte count, ×109/L | 0.8 ~ 4 | 1.3(0.9 ~ 1.8) | 1.8(0.6 ~ 2.3) | 1.2(1.0 ~ 1.6) | 0.539 |
| lymphocyte percentage, % | 20 ~ 40 | 26.9(20.4 ~ 33.1) | 33.4(17.6 ~ 36.0) | 26.2(20.1 ~ 32.7) | 0.408 |
| Haemoglobin, g/L | 130 ~ 175 | 142.5(131.0 ~ 155.5) | 147(134.3 ~ 160.0) | 139(131.0 ~ 154.8) | 0.516 |
| Platelet count, ×109/L | 125 ~ 350 | 181(145.8 ~ 231.0) | 234.5(127.5 ~ 305.3) | 179.5(146.3 ~ 223.8) | 0.493 |
| ESR, mm/1 h | 0 ~ 15 | 25(11.3 ~ 44.8) | 18(7.1 ~ 36.0) | 26(12.8 ~ 45.3) | 0.210 |
| C-reactive protein, mg/L | 0 ~ 6 | 11.3(2.4 ~ 35.2) | 9.3(1.7 ~ 33.3) | 11.3(2.4 ~ 36.6) | 0.471 |
| Procalcitonin, μg/L | 0.0 ~ 0.5 | 0.05(0.03 ~ 0.05) | 0.05(0.03 ~ 0.05) | 0.05(0.03 ~ 0.05) | 0.661 |
| Total bilirubin, μmol/L | 3.42 ~ 8.2 | 7.2(4.7 ~ 8.7) | 6.5(2.8 ~ 7.5) | 7.5(4.8 ~ 8.9) | 0.159 |
| Alanine aminotransferase, U/L | 0 ~ 40 | 25.5(14.0 ~ 40.0) | 30(7.3 ~ 57.5) | 24(15.0 ~ 37.3) | 0.793 |
| Aspartate aminotransferase, U/L | 5 ~ 49 | 27.5(22.0 ~ 35.5) | 27.5(22.0 ~ 43.8) | 27.5(22.0 ~ 35.0) | 0.739 |
| Albumin, g/L | 60 ~ 85 | 43.7(40.6 ~ 45.9) | 46.1(43.2 ~ 48.2) | 42.7(40.5 ~ 45.5) | 0.075 |
| Creatine kinase, U/L | 26 ~ 196 | 91.5(68.8 ~ 133.3) | 108.5(85.5 ~ 195.5) | 90(61.3 ~ 126.3) | 0.281 |
| Lactate dehydrogenase, U/L | 109 ~ 245 | 244.5(202.5 ~ 308.5) | 240(193 ~ 290) | 244.5(202.8 ~ 309.5) | 0.586 |
| Creatinine, μmol/L | 59 ~ 104 | 61.0(51.5 ~ 70.9) | 60.9(52.0 ~ 64.5) | 62.7(51.2 ~ 71.2) | 0.713 |
| Blood urea nitrogen, mmol/L | 1.43 ~ 8.2 | 3.3(2.5 ~ 5.0) | 4.7(4.0 ~ 5.3) | 3.1(1.5 ~ 7.7) | 0.040 |
| Glucose, mmol/L | 3.89 ~ 6.11 | 5.6(5.2 ~ 6.8) | 5.0(4.7 ~ 6.6) | 5.7(5.3 ~ 6.9) | 0.123 |
| D-dimers, mg/L | 0 ~ 0.55 | 0.3(0.2 ~ 0.4) | 0.3(0.2 ~ 0.5) | 0.3(0.2 ~ 0.4) | 1.000 |
| Prothrombin time, s | 9 ~ 14 | 11.4(11.0 ~ 11.9) | 11.0(10.8 ~ 12.9) | 11.4(11.0 ~ 11.9) | 0.586 |
| APTT, s | 20 ~ 40 | 26.1(23.7 ~ 28.6) | 26.2(23.1 ~ 30.6) | 26.1(24.5 ~ 27.6) | 0.958 |
| Thrombin time, s | 14 ~ 21 | 16.4(15.8 ~ 17.6) | 16.9(15.8 ~ 18.0) | 16.3(15.8 ~ 17.5) | 0.562 |
| Fibrinogen, g/L | 2 ~ 4 | 3.4(2.9 ~ 4.5) | 3.1(2.4 ~ 4.5) | 3.5(3.1 ~ 4.5) | 0.314 |
| Endotoxin, pg/mL | <10 | 90.4(8.3 ~ 176.0) | 31.4(4.4 ~ 388.9) | 100.7(8.5 ~ 176.0) | 0.576 |
| SpO2, % | 91.9 ~ 99.9 | 98(97 ~ 99) | 98(98 ~ 100) | 98(97 ~ 99) | 0.265 |
| PaCO2, mmHg | 35 ~ 45 | 37.0(34 ~ 41) | 36.0(33.4 ~ 42.7) | 37.0(34.1 ~ 41.0) | 0.933 |
| PaO2:FIO2, mmHg | 400 ~ 500 | 388.1(330.2 ~ 487.1) | 486.2(409.5 ~ 680.1) | 381.0(318.6 ~ 467.8) | 0.040 |
| BNP, ng/L | 0 ~ 125 | 50(50 ~ 58.3) | 50(50 ~ 50) | 50(50 ~ 82.3) | 0.600 |
| Troponin I, μg/L | 0 ~ 1.68 | 0.8(0.6 ~ 0.9) | 0.8(0.6 ~ 0.9) | 0.8(0.6 ~ 0.9) | 0.741 |
| Fecal occult blood test, positive | 4(14.3%) | 2(40%) | 2(8.7%) | 0.135 | |
aP values indicate differences between anal swab test positive and negtive patients. P < .05 was considered statistically significant. ESR erythrocyte sedimentation rate, APTT Activated partial thromboplastin time
CT findings
For the patients in the positive group: Patient 1 did not show any abnormalities on CT. Abnormal CT findings were found in patients 2, 3, and 4 on admission (Figs. 1, 2 and 3). Patient 2 was a child with a focal GGO in the left lower lobe and a CT score of 1. Two severe-type patients (patients 3 and 4) presented with multiple GGO and consolidations in both lungs. CT scores on admission, discharge, and after discharge for Patient 3 were 17, 14, and 10, respectively, and the corresponding CT scores for Patient 4 were 13, 10, and 7, respectively. There were no significant differences in the number of segments involved, distribution of lesions, imaging features, or CT score on admission between the two groups. The absorption on the CT scans of patients 3 and 4 was slow (Figs. 1, 2, 3 and 4), but there were no significant difference in the CT scores at discharge between the two groups. The CT scores of the positive group were generally higher than those of the negative group after discharge (8.5 ± 2.1 vs. 3.8 ± 2.2, P = 0.038) (Table 4).
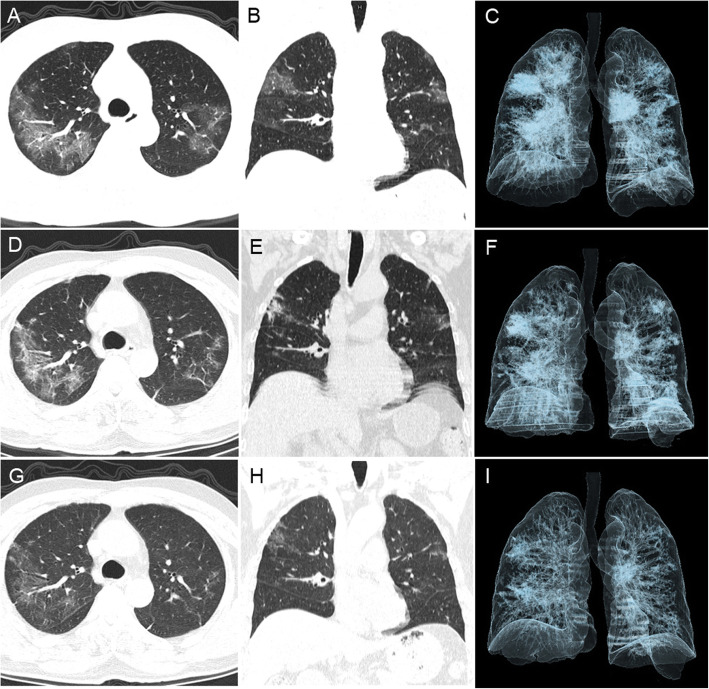
Fig. 1.
Patient 3 had a positive anal swab sample after discharge. This aged male, who was admitted to the hospital because of a cough and dyspnea, presented without fever (body temperature 36.1 °C) or diarrhea. His OBT was positive and his throat swab RNA tests were positive for 13 days. His chest CT, taken on admission (February 10, 2020), showed bilateral multiple GGO and intralobular interstitial thickening and peripheral distribution (CT score = 17) (A-C). His CT on discharge (February 20, 2020) showed absorption of multiple GGO, but consolidation and aggravated linear opacities, the lesions absorption was slow (CT score = 14) (D-E). CT after discharge (March 8, 2020) showed continuous absorption of multiple lesions, with a small amount of remaining GGO and linear opacities (CT score = 10) (G-I)
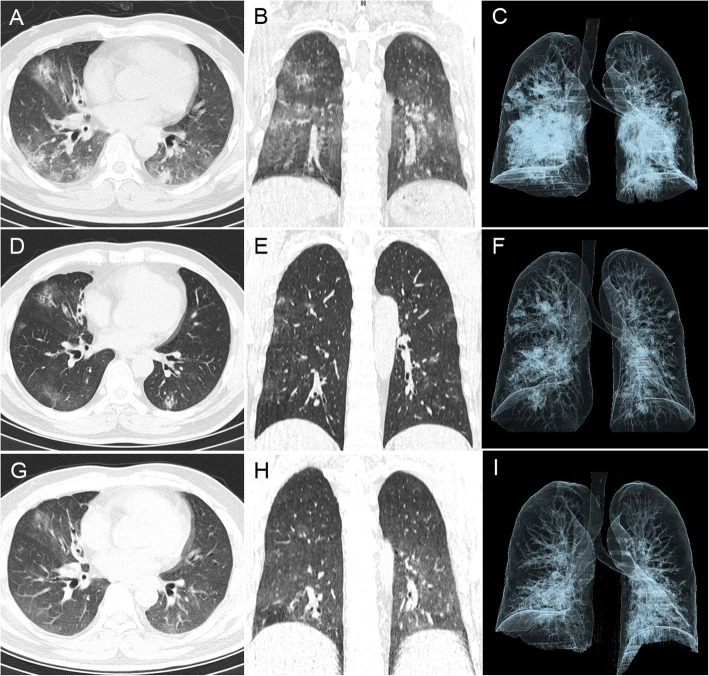
Fig. 2.
Patient 4 had a positive anal swab after discharge. This middle-aged male was admitted to the hospital because of fever (maximum body temperature 39.5 °C), cough, and dyspnea. His OBT was positive and he had positive throat swab samples for 12 days. The chest CT on admission (January 20, 2020) showed markedly reduced lung translucency, bilateral multiple GGO and consolidations and peripheral distribution (CT score = 13) (A-C). CT on discharge (February 10, 2020) showed lung transparency returned to normal and absorption of multiple GGO, but some irregular consolidations remained (CT score = 10) (D-E). CT after discharge (February 24, 2020) showed continuous absorption of multiple lesions, with only a small amount of GGO remaining (CT score = 7) (G-I)
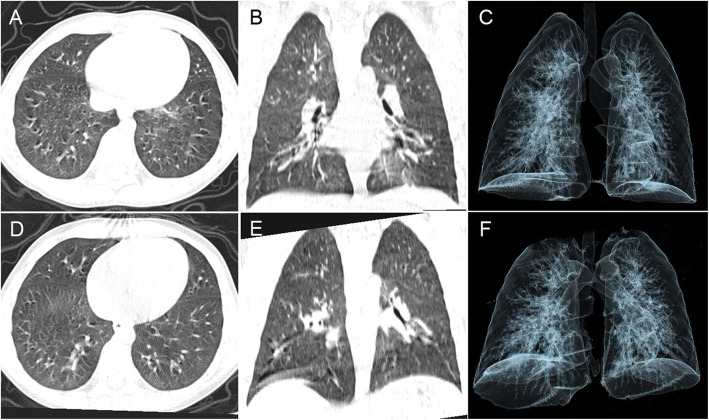
Fig. 3.
Patient 2 had a positive anal swab for SARS-CoV-2 after discharge. The young male, who was admitted to the hospital because of persistent cough (5 days) and presented without fever (body temperature 36.0 °C) or diarrhea. His OBT was negative, and his throat swabs were positive for only 9 days. The chest CT administered on admission (February 10, 2020) showed a focal GGO in the left lower lobe and peripheral distribution (CT score = 1) (A-C). The CT on discharge (February 20, 2020) showed complete absorption of this lesion (CT score = 0) (D-E)
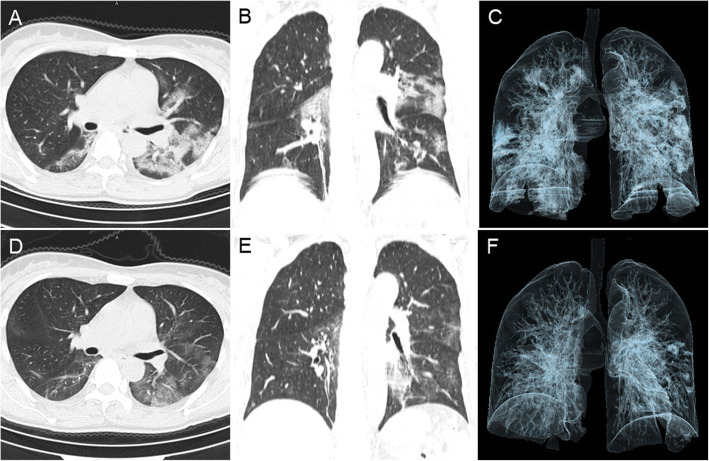
Fig. 4.
A patient with an anal swab negative for SARS-CoV-2 after discharge. This middle-aged female was admitted to the hospital because of a persistent cough (6 days) and fever (maximum body temperature of 39.6 °C), without diarrhea. Her OBT was negative, and she had positive throat swabs for only 6 days. The chest CT on admission (February 6, 2020) showed bilateral multiple GGO and consolidation, central and peripheral distribution (CT score = 11) (A-C). CT on discharge (February 12, 2020) showed significant absorption of these bilateral lung lesions (CT score = 5) (D-E)
Table 4.
Imaging characteristics of patients with COVID-19
| Total (N = 31) | Positive (n = 4) | Negative (n = 27) | P Value a | |
|---|---|---|---|---|
| First CT, negative, (%) | 3(9.7%) | 1(25.0%) | 2(7.4%) | 0.349 |
| First CT, positive, (%) | 28(90.3%) | 3(75.0%) | 25(92.6%) | |
| Number of involved segments | 9.3 ± 6.1 | 11 ± 8.9 | 9.1 ± 5.9 | 0.726 |
| Involvement, (%) | ||||
| Single lobe | 5(17.9%) | 1(33.3%) | 4(16.0%) | 0.459 |
| Bilateral lobe | 23(82.1%) | 2(66.7%) | 21(84.0%) | |
| Distribution, (%) | ||||
| Central | 0 | 0 | 0 | – |
| Peripheral | 15(53.6%) | 1(33.3%) | 14(56.0%) | 0.583 |
| Central + Peripheral | 13(46.4%) | 2(66.7%) | 11(44.0%) | |
| Degree, (%) | ||||
| Single lesion | 4(14.3%) | 1(33.3%) | 3(12.0%) | 0.217 |
| Multiple lesions | 13(46.4%) | 0(0.0%) | 13(52.0%) | |
| Diffuse lesions | 11(39.3%) | 2(66.7%) | 9(36.0%) | |
| Imaging Features, (%) | ||||
| GGO | 27(96.4%) | 1(66.7%) | 25(100.0%) | 0.107 |
| Consolidation | 19(67.9%) | 3(100.0%) | 16(64.0%) | 0.530 |
| Linear opacities | 19(67.9%) | 2(66.7%) | 17(68.0%) | 1.000 |
| Mixed type | 22(78.6%) | 2(66.7%) | 20(80.0%) | 0.530 |
| Interstitial change | ||||
| Interlobular septal thickening | 3(10.7%) | 1(3.3%) | 2(8.0%) | 0.298 |
| Intralobular interstitial thickening | 21(75.0%) | 2(66.7%) | 19(76.0%) | 1.000 |
| Other signs, (%) | ||||
| Nodule | 8(28.6%) | 1(33.3%) | 7(28.0%) | 1.000 |
| Dilatation of vessels | 26(92.9%) | 2(66.7%) | 24(96.0%) | 0.206 |
| Bronchial wall thickening | 13(46.4%) | 2(66.7%) | 11(44.0%) | 0.583 |
| Air bronchogram | 5(17.9%) | 0(0.0%) | 5(20.0%) | 1.000 |
| Crazy-paving pattern | 3(10.7%) | 0(0.0%) | 3(12.0%) | 1.000 |
| Halo sign | 7(25.0%) | 1(33.3%) | 6(24.0%) | 1.000 |
| Reversed halo sign | 0(0.0%) | 0(0.0%) | 0(0.0%) | – |
| Pleural thickening | 4(14.3%) | 0(0.0%) | 4(16.0%) | 1.000 |
| Hydrothorax | 0(0.0%) | 0(0.0%) | 0(0.0%) | – |
| Lymphadenectasis | 0(0.0%) | 0(0.0%) | 0(0.0%) | – |
| CT scores | ||||
| On admission | 7.7 ± 4.6 | 10.3 ± 8.3 | 7.4 ± 4.1 | 0.532 |
| On discharge | 5.6 ± 3.5 | 8.0 ± 7.2 | 5.3 ± 2.9 | 0.433 |
| After discharge | 4.4 ± 2.7 | 8.5 ± 2.1 | 3.8 ± 2.2 | 0.038 |
aP values indicate differences between anal swab test positive and negative patients. P < .05 was considered statistically significant
Discussion
Since COVID-19 mainly affects the respiratory system [14, 15], anal swab and stool sample testing for SARS-CoV-2 RNA has only recently gained attention. Some patients without GI symptoms may present with positive stool samples [3, 7], and a positive SARS-CoV-2 RNA result from an anal swab or stool specimen does not necessarily indicate that there must be live virus in the patient’s stool since the viral particles trapped in the respiratory secretions could be swallowed and processed through the digestive tract. Several case reports have shown that live virus can be detected in stool specimens from COVID-19 patients [16–20]. However, it is still uncertain whether SARS-CoV-2 in feces might be an additional source of transmission. No previous studies have investigated the clinical or imaging characteristics of patients with positive anal swab or stool specimens.
In the current case series, six patients (14.3%) were found to have positive anal swab samples after discharge. The positivity rate for SARS-CoV-2 RNA in feces samples from hospitalized patients with COVID-19 was reported to be between 29 and 53% [19, 20]. The reason why our positive rate was lower than theirs may be that they tested stool specimens from hospitalized patients at earlier time points, while we tested anal swabs from patients who met the discharge criteria and thus represent a later time point. Xiao et al. [18] tested serial stool specimens from a critically ill patient and found that there were only RNA fragments and not infectious viral particles in the feces collected at later time points. In the anal swab positive group, 40% of patients (2/5) tested positive for OBT, while only 8.7% (2/23) were OBT positive in the negative group. There was no diarrhea in the anal swab positive group, but six patients in the negative group reported GI symptoms. Diarrhea does not necessarily correlate with OBT, so we speculate that diarrhea may be only a premonitory symptom of SARS-CoV-2 infection of the GI tract. The higher positive rate of OBT during hospitalization in patients with positive anal swab samples suggests that a positive OBT may be a sign that SARS-CoV-2 is damaging the GI tract. OBT-positive patients were positive for viral RNA on the anal swab samples for longer. This suggests that OBT may be a better indicator of GI SARS-CoV-2 infection and damage than symptomology. The mechanism by which SARS-CoV-2 causes GI damage is still unclear. In humans and other mammals, coronaviruses mainly target the upper respiratory tract, GI tract, and central nervous system [16, 21]. SARS-CoV-2 belongs to the same family as the SARS-CoV that caused the SARS outbreak in 2003. It is currently known that in addition to damaging the respiratory tract, SARS-CoV also damages the intestine. The main driver for this damage is the fact that cell receptor-angiotensin-converting enzyme II (ACE2) is expressed in both the human respiratory tract and the esophagus, small intestine, and colon [21–23]. Some studies indicate that 3–10% of patients with COVID-19 develop GI symptoms [14, 15]. We can thus hypothesize that SARS-CoV-2 is transmitted through ACE2 receptors in the GI tract damaging the intestine, leading to hemorrhage, which could be detected by OBT.
Our study found that the median duration of the major symptoms was shorter in those patients with positive anal swabs than in those without. The reason for the less severe symptoms and faster recovery in the positive group remains unclear, but may indicate that these patients reached the recovery phase of the disease faster. We also found that the anal swabs tests started to come back negative for viral RNA at around 7 days, with this being extended to 9 days in more severe cases. This observation suggests that the virus may exist in severe patients for longer. Live SARS-CoV-2 was isolated from feces [16–20], indicating that fecal-oral or fecal-respiratory transmission is possible via aerosolized feces. During the SARS epidemic, a large-scale community outbreak in Hong Kong, China, was caused by patient excreta eventually resulting in 321 infections and 42 deaths [24]. This indicates that the fecal excreta of patients with SARS can be infectious. Although the infectivity of patients with positive anal swab viral RNA is still uncertain, we recommend that anal swab or fecal samples are tested for viral RNA and that OBT is evaluated at diagnosis and over the course of treatment in cases of COVID-19 to minimize the risk of fecal transmission. Given this discharge and hospital cleaning practices should pay more attention to those who have a positive OBT result. However, this recommendation still needs to be validated in a larger clinical cohort.
This study has several limitations. First, this was a retrospective study. Second, our cohort size was small since only 42 discharged patients had any anal samples collected and the positive rate of the anal swab tests was also very low, thus the findings of this study should be interpreted as exploratory and descriptive. Third, we lack more detailed clinical information, including the absence of patient sera to evaluate viremia. We could not compare the viral loads of respiratory and digestive tract specimens during hospitalization. Fourth, false negatives could potentially interfere with our results. Lastly, the infectivity of patients with positive stool samples cannot be established. We hope that the preliminary results of this study will stimulate further research in this area.
Conclusion
In conclusion, 14.3% of the discharged patients in this study were found to be positive for SARS-CoV-2 RNA in anal swab samples. They did not have GI symptoms and had similar CT features as the negative patients, and their main symptoms disappeared early, which may make these patients easier to ignore. A positive OBT may indicate GI damage caused by SARS-CoV-2. Further follow-up and research are needed.
Acknowledgements
We greatly appreciate the kind assistance of all our colleagues who helped us during the current study.
Authors’ contributions
W.L. and L.S. conceived and designed the research. H.Z., L.Z., L.S. and D.W. acquired the data. H.Z., W.L. and L.Z. analyzed and interpreted the data. H.Z., L.Z. and Z.X. performed statistical analysis. W.L. handled funding and supervision. H.Z., Y.L. drafted the manuscript. W.L. and H.X.B. made critical revision of the manuscript for important intellectual content. All authors have read and approved the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 81671676) and the Scientific Research Project of Hunan Health and Health Commission (Grant No. B2019194).
Availability of data and materials
The datasets used and/or analyzed during this study are available from the corresponding author (Weihua Liao, owenliao@csu.edu.cn) upon reasonable request.
Ethics approval and consent to participate
The study was approved by the Ethics Commission of Xiangya Hospital Central South University (approval number 202002019), which waived the written informed consent for emerging infectious diseases.
Consent for publication
This manuscript is approved by all authors for publication.
Competing interests
We declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Linbo Shi, Email: 692906369@qq.com.
Weihua Liao, Email: owenliao@csu.edu.cn.
References
- 1.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.China National Health Commission. Update on the novel coronavirus pneumonia outbreak (Mar 2, 2020). Beijing: China National Health Commission. http://www.nhc.gov.cn/xcs/s3574/202003/0f6de907780541fc940e847afccf3b64.shtml (Accessed 3 Mar 2020).
- 3.Holshue ML, DeBolt C, Lindquist S, et al. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382(10):929. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phan LT, Nguyen TV, Luong QC, et al. Importation and human-to-human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382:872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization . Coronavirus Disease 2019(COVID-19): situation report-196. 2020. [Google Scholar]
- 6.General Office of National Health Committee. Office of state administration of traditional Chinese medicine. Notice on the issuance of a programme for the diagnosis and treatment of novel coronavirus (2019-nCoV) infected pneumonia (trial fifth edition) (2020-02-26) (EB/OL). http://bgs.satcm.gov.cn/zhengcewenjian/2020-02-06/12847.html).
- 7.Yang Z, Li Q, Dai X, et al. Three cases of novel coronavirus pneumonia with viral nucleic acids still positive in stool after throat swab detection turned negative. Chin J Dig. 2020;40:E002. doi: 10.3760/cma.j.issn.0254-1432.2020.0002. [DOI] [Google Scholar]
- 8.Pan Y, Zhang D, Yang P, Poon L, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Y, SHEN L, XU Z, Zhou J, Zhou H. SARS-CoV-2 May Persist in Digestive Tract Longer than Respiratory Tract. 2020. [Google Scholar]
- 10.World Health Organization . Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected: interim guidance. 2020. [Google Scholar]
- 11.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang YC, Yu CJ, Chang SC, et al. Pulmonary sequelae in convalescent patients after severe acute respiratory syndrome: evaluation with thin-section CT. Radiology. 2005;236:1067–1075. doi: 10.1148/radiol.2363040958. [DOI] [PubMed] [Google Scholar]
- 13.Pan F, Ye T, Sun P, et al. Time Course of Lung Changes On Chest CT During Recovery From 2019 Novel coronavirus (COVID-19) pneumonia. Radiology. 2020;1:200370. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan WJ, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Kang ZI, Gong HY, et al. The digestive system is a potential route of 2019 nCoV infection: a bioinformatics analysis based on single-cell transcriptomes. 2020. [Google Scholar]
- 17.Zhang Y, Chen C, Zhu S, et al. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Weekly. 2020;2(8):123–124. doi: 10.46234/ccdcw2020.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao F, Sun J, Xu Y, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. 2020;26(8):1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Moore MJ, Vasilieva N, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R, Qiao S, Zhang G. Analysis of angiotensin-converting enzyme 2 (ACE2) from different species sheds some light on cross-species receptor usage of a novel coronavirus 2019-nCoV. J Inf Secur. 2020;80(4):469. doi: 10.1016/j.jinf.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during this study are available from the corresponding author (Weihua Liao, owenliao@csu.edu.cn) upon reasonable request.