Abstract
A recent outbreak of coronavirus disease (COVID‐19) caused by the novel severe acute respiratory syndrome coronavirus 2 has driven a global pandemic with catastrophic consequences. The rapid development of promising therapeutic strategies against COVID‐19 is keenly anticipated. Family Coronaviridae comprises positive, single‐stranded RNA viruses that use RNA‐dependent RNA polymerase (RdRP) for viral replication and transcription. As the RdRP of viruses in this family and others plays a pivotal role in infection, it is a promising therapeutic target for developing antiviral agents against them. A critical genetic driver for many cancers is the catalytic subunit of telomerase: human telomerase reverse transcriptase (hTERT), identified initially as an RNA‐dependent DNA polymerase. However, even though hTERT is a DNA polymerase, it has phylogenetic and structural similarities to viral RdRPs. Researchers worldwide, including the authors of this review, are engaged in developing therapeutic strategies targeting hTERT. We have published a series of papers reporting that hTERT has RdRP activity and that this RdRP activity in hTERT is essential for tumor formation. Here, we review the enzymatic function of RdRP in virus proliferation and tumor development, reminding us of how the study of the novel coronavirus has brought us to the unexpected intersection of cancer research and RNA virus research.
Keywords: RdRP inhibitor, RNA virus, RNA‐dependent RNA polymerase, severe acute respiratory syndrome coronavirus 2, telomerase reverse transcriptase
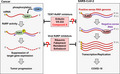
In cancer cells, human telomerase reverse transcriptase (hTERT) is phosphorylated by cyclin‐dependent kinase 1 (CDK1), and the phosphorylated hTERT shows RNA‐dependent RNA polymerase (RdRP) activity. The phosphorylated hTERT regulates the expression of target genes, leading to tumor progression. After the entry of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) into the cell, the RdRP plays a role in transcription and replication of viral RNAs. The inhibition of RdRP activity is a promising molecular target for cancer and SARS‐CoV‐2 infection.
1. INTRODUCTION
In December 2019, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) emerged, causing a severe global outbreak of coronavirus disease 19 (COVID‐19). 1 , 2 As of 10 August 2020, the WHO has amassed reports of more than 19.7 million cases and 720 000 deaths globally. Human coronaviruses, including SARS‐CoV, Middle East Respiratory Syndrome (MERS)‐CoV, and SARS‐CoV‐2, are positive‐sense single‐stranded RNA (ssRNA) viruses that use RNA‐dependent RNA polymerase (RdRP) for their genome replication. 3 Other RNA viruses, such as hepatitis C virus (HCV), poliovirus, dengue virus, influenza virus, Ebola virus (EBOV), and measles virus, also use RdRP for their genome replication. 4 As these RNA viruses are representative organisms in the phylogenetic tree and therefore thought to be some of the oldest, it is not surprising that viral RdRP, an evolutionally primitive polymerase, is required to maintain such ancestral organisms. 5
Original reports describing the cloning of human telomerase reverse transcriptase (hTERT) indicated that hTERT has conserved reverse transcriptase motifs that also contain right‐handed architecture (fingers, thumb, and palm domains) by alignment prediction analysis. 6 , 7 In addition, the authors clearly mentioned in the original reports that hTERT is closely related to viral RdRPs rather than reverse transcriptase by phylogenetic analysis. 6 More recently, 3‐D structural analyses have confirmed that hTERT has the characteristic right‐handed architecture similar to RdRPs found in RNA viruses. 8 , 9 , 10 We have recently reported that hTERT possesses RdRP activity that is essential for tumorigenesis, 11 , 12 suggesting that RNA virus and cancer share a similar survival strategy of RNA‐dependent RNA synthesis. Since the 1970s, cancer research has advanced along with research about oncogenic RNA viruses such as Rous sarcoma virus, resulting in the identification of various cellular oncogenes. Researchers have identified the cellular oncogene c‐src on the Rous sarcoma virus‐encoded oncogene v‐src, which transforms cells and promotes the replication of proviruses integrated into the host genome. 13 , 14 RNA virus and cancer, thus, share a survival strategy of utilizing polynucleotide synthesis that is dependent on the RNA template. The structural similarity of viral RdRPs and hTERT proteins suggests that an RdRP inhibitor would be effective in treating both RNA virus infections and cancer.
Recently, investigators worldwide found a positive, therapeutic effect of RdRP inhibitors in patients with COVID‐19 even though these drugs target other RNA viruses. 15 , 16 , 17 , 18 This response in patients emphasizes the importance of RdRP as a target in antiviral strategies. Here, we review the functional role of RdRP activity in RNA viruses and mammalian cells and discuss the importance of RdRP inhibitors in the treatment of RNA viral diseases and cancer (summarized as Table 1).
TABLE 1.
Types of RNA‐dependent RNA polymerase (RdRP) and agents against them
| RdRP | Structure | Host | Function | Agent | References |
|---|---|---|---|---|---|
| Viral RdRP | Right‐hand | ssRNA viruses | Genome replication/transcription |
Ribavirin Remdesivir Favipiravir Sofosbuvir |
4, 22, 23 |
| Cellular RdRP | Double‐barrel |
Plants Fungi Nematodes Eukaryotes |
RNA silencing | — | 33, 34, 35, 36, 37 |
| Mammalian RdRP (hTERT) | Right‐hand | Cancer cells | Gene expression regulation |
Eribulin VX‐222 (compound X) |
6, 9, 10, 11, 12, 57, 59 |
|
Undifferentiated cells Stem cells |
Unknown | Unknown | 47, 48, 49, 50 |
2. RNA VIRUSES AND RdRPs
2.1. RNA viruses
The genome of many RNA viruses is ssRNA, whereas the genome of others, such as reovirus, is double‐stranded RNA (dsRNA). Furthermore, ssRNA viruses are classified as positive‐ and negative‐sense ssRNA viruses that have positive‐sense (mRNA‐sense) and negative‐sense (antisense) RNA genomes, respectively (Figure 1). 19
FIGURE 1.
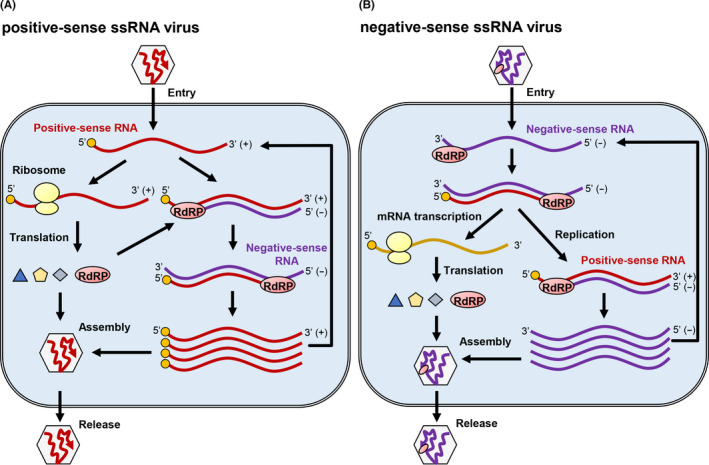
Life cycle of positive‐ and negative‐sense single‐stranded RNA (ssRNA) viruses. A, B, Flowchart of RNA synthesis by RNA‐dependent RNA polymerase (RdRP) of positive‐sense (A) and negative‐sense (B) ssRNA viruses. Small circles at the 5′‐ends of positive‐sense RNA denote m7G‐cap structures, 32 which occur in most of viral mRNAs
The RNA genome of positive‐sense ssRNA viruses can function as mRNA that is directly translated into viral proteins. It is used as a template RNA for the synthesis of complementary negative‐sense RNA that subsequently serves as the template for synthesizing the new positive‐sense RNA (Figure 1A). In contrast, the RNA genome of negative‐sense ssRNA viruses is first transcribed to mRNAs, which are then used for translation to a full‐length positive‐sense RNA, which is subsequently used as a template to yield the full‐size genomic RNA. All these are carried out by viral RdRP packaged in the virus particle (Figure 1B). 20 The family of ssRNA viruses also include retroviruses such as HIV. Retroviruses use reverse transcriptase to generate dsDNA from the positive‐sense viral RNA genome, which is incorporated into the genomic DNA of host cells to form an intermediary stage, provirus. 21
2.2. Viral RdRPs
The RdRP is encoded by all ssRNA viruses (Table 2), except for the retroviruses. Structural analyses of several viruses have shown that their RdRP protein consists of palm, finger, and thumb domains, which are referred to as a closed, right‐handed polymerase (Figure 2A). 22 , 23 In contrast, RdRP proteins of plants, fission yeast, and nematodes have a double‐barrel structure (Figure 2B) (also see Section 3). 24
TABLE 2.
Single‐stranded RNA (ssRNA) virus‐encoded RNA‐dependent RNA polymerases (RdRPs)
| Genome | Species/strain | Family | RdRP |
|---|---|---|---|
| ssRNA(+) | Severe acute respiratory syndrome coronavirus (SARS‐CoV) | Coronaviridae | nsp12 |
| Middle East respiratory syndrome coronavirus (MERS‐CoV) | Coronaviridae | nsp12 | |
| Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) | Coronaviridae | nsp12 | |
| Poliovirus | Picornaviridae | PVgp1 | |
| Hepatitis C virus (HCV) | Flaviviridae | NS5B | |
| Dengue virus (DENV) | Flaviviridae | NS5 | |
| West Nile virus (WNV) | Flaviviridae | NS5 | |
| Zika virus (ZIKV) | Flaviviridae | NS5 | |
| ssRNA(−) | Influenza viruses | Orthomyxoviridae | PB1 |
| Ebola virus (EBOV) | Filoviridae | L | |
| Marburg virus | Filoviridae | L | |
| Measles virus | Paramyxoviridae | L |
FIGURE 2.
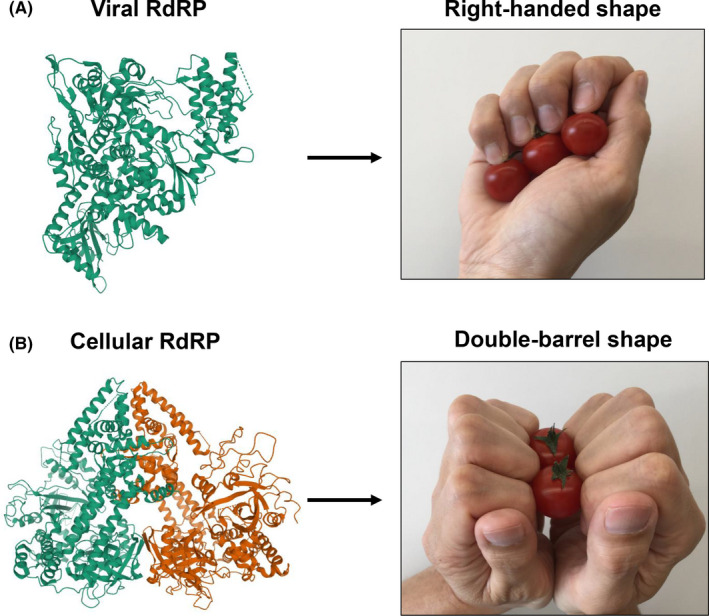
Structure of viral and cellular RNA‐dependent RNA polymerases (RdRPs). A, Viral RdRP has a typical right‐handed structure. The structure of severe acute respiratory syndrome coronavirus 2 RdRP is shown. B, Cellular RdRPs in plants, fungi, and nematodes have a double‐barrel shaped structure. The structure of Neurospora crassa RdRP is shown. Tomatoes held by hands depict RNA chains synthesized by RdRP. Ribbon diagrams are from Protein Data Bank (https://www.rcsb.org/, accession numbers 2J7N and 7BTF)
For positive‐sense ssRNA viruses, such as HCV causing chronic hepatitis, cirrhosis, and hepatocellular carcinoma, the RdRP nonstructural protein 5B (NS5B) carries out the viral replication. 25 As NS5B plays a central role in the replication of HCV in its life cycle, several NS5B inhibitors, such as sofosbuvir, have been developed (see Section 5 for details). 25
Influenza viruses are negative‐sense ssRNA viruses that have eight genomic ssRNAs. 26 After entry of the virus into recipient cells, the virus core containing genomic RNAs and RdRP translocate into the nucleus. Here, the RNA serves as a template: at first, for mRNA synthesis and then for replication by the RdRP complex, consisting of PB1, PB2, and PA. 27 Before viral mRNA synthesis is carried out, host mRNA molecules that were incorporated in the core are cleaved by an “influenza virus‐specific cap snatching reaction,” at 10‐13 bases downstream of the 5′‐end termini by the cap‐dependent endonuclease activity contained in the RdRP complex. The resulting cap‐containing oligonucleotide fragments are used as a primer for the initiation of transcription, which is followed by RNA elongation with RdRP. 28
The positive‐sense ssRNA virus SARS‐CoV‐2 has a large RNA genome (~30 kb) and shares high sequence homology to SARS‐CoV (~80%) and MERS‐CoV (~50%) (Figure 3A). 29 Two polyproteins, polyprotein1a (pp1a) and polyprotein1ab (pp1ab), are encoded in ORF1a and ORF1b regions of the genomic RNA (gRNA). The pp1a and pp1ab polypeptides made from gRNA are, in turn, cleaved into 16 nonstructural proteins (nsps) by nsp3/5 virus‐specific proteases. Among these nsps, nsp12 is an RdRP that later forms an RdRP complex with cofactors nsp7 and nsp8. 16 , 30 This nascent RdRP complex then synthesizes negative‐sense RNAs, perhaps of various sizes, using the positive‐sense gRNA as a template, although the mechanism behind this process remains to be elucidated. These negative‐sense RNA intermediates could serve as templates for the synthesis of subgenomic positive‐sense RNAs (sgRNAs), equivalent to mRNAs, in addition to the full‐size gRNA (Figure 3A). Intriguingly, the gRNA and sgRNAs contains a 5′‐cap structure and 3′‐polyA, in addition to common leader sequences of ~70 bases at their 5′‐ends. 29 Alternatively, each sgRNA (ie, mRNA) is synthesized by discontinuous transcription that is similar to the transcription of vesicular stomatitis virus RNA. 29 , 31 , 32 Although nothing is certain yet about the transcription mechanism of coronaviruses, it is hypothesized that the RdRP jumps to the 3′‐end leader sequence region of the template gRNA from transcription‐regulatory sequences, which are short motifs located in the middle of gRNA (Figure 3A), generating negative‐sense RNA intermediates with the 3′‐end leader sequence. Recently, the protein structure of SARS‐CoV‐2 nsp12 has been analyzed by electron cryomicroscopy, which showed that SARS‐CoV‐2 nsp12 has a right‐handed polymerase structure, like that of SARS‐CoV. 16 , 30 Comparative analysis of structures of nsp12 encoded by SARS‐CoV and SARS‐CoV‐2 has implied that SARS‐CoV‐2 nsp12 has a characteristic β‐hairpin motif in the nidovirus RdRP‐associated nucleotidyltransferase (NiRAN) domain (Figure 3A). 16
FIGURE 3.
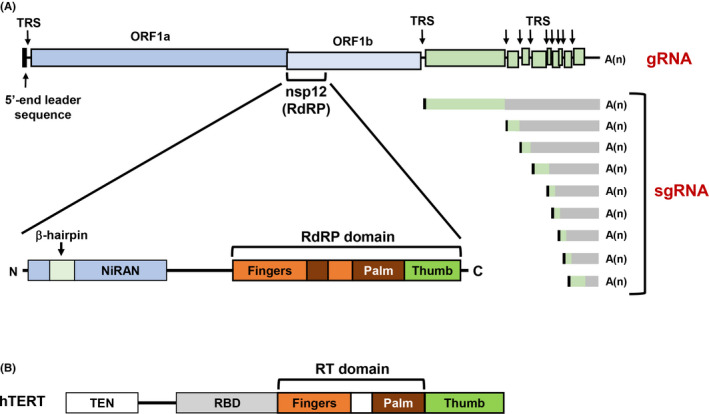
Structure of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) and human telomerase reverse transcriptase (hTERT). A, Genomic structure of SARS‐CoV‐2. The RNA‐dependent RNA polymerase (RdRP) nsp12 comprises the nidovirus RdRP‐associated nucleotidyltransferase (NiRAN) and RdRP domains. In addition to genomic RNA (gRNA), subgenomic RNAs (sgRNAs) are produced by discontinuous transcription. 32 The gRNA and sgRNAs contain a common leader sequence at their 5′‐end. TRS, transcription‐regulatory sequence. B, Structure of hTERT. The right‐handed structure, composed of fingers, palm, and thumb domains, is shared with the viral RdRPs such as nsp12 of SARS‐CoV‐2. RBD, RNA‐binding domain; RT, reverse transcriptase; TEN, telomerase essential N‐terminal domain
3. CELLULAR RdRP
Viral RdRP plays a pivotal role in viral replication and transcription, whereas RdRP in plant, fungi, nematodes, and eukaryotic cells is a key molecule in RNA silencing. 33 , 34 , 35 Especially in insects and plants, RNA silencing functions as a defense mechanism for eliminating exogenous nucleic acids. Here, the viral dsRNAs are processed by the dsRNA‐specific endonuclease Dicer into primary siRNAs. 36 , 37 Using one strand of viral dsRNA as a template, cellular RdRP amplifies dsRNAs that are subject to the Dicer‐mediated processing afterwards. The secondary siRNAs thus generated by these processes are incorporated into an RNA‐induced silencing complex (RISC) that cleaves target viral RNAs complementary to the siRNA. 37
In fission yeast Schizosaccharomyces pombe, RdRP participating in heterochromatin formation in the centromere region produces dsRNAs using transcripts from the region as a template and subsequently processes the dsRNAs into siRNAs. The resulting siRNAs recruit a RISC‐like RNA‐induced transcriptional silencing (RITS) complex to the centromere region, promoting heterochromatin formation. 38 Inhibition of the expression of RdRPs and RITS complex components leads to derepression of transcription from the centromere region, causing abnormalities in the heterochromatin structure. 39 Thus, RdRP is essential for proper chromosome segregation and mitosis progression in these model organisms.
4. RNA‐DEPENDENT RNA POLYMERASE IN MAMMALIAN CELLS
4.1. RNA‐dependent RNA polymerase activity in hTERT in human cancer cells
Telomeres are located at the terminus of linear chromosomes and protect the chromosome ends. The telomere is elongated by telomerase that was initially identified as an RNA‐dependent DNA polymerase (reverse transcriptase). 6 , 7 , 40 The minimum, essential components of telomerase are a catalytic component of hTERT and a template RNA (hTERC: human telomerase RNA component). 41 Human telomerase reverse transcriptase is upregulated in the majority of human cancers and contributes directly to cell transformation. 7 , 42 , 43 The role of telomerase is the maintenance of telomere structure by its reverse transcriptase activity; however, recent studies have shown that hTERT has other important functions beyond telomere maintenance. 44 , 45
Phylogenetic analysis indicates that hTERT is closely related to viral RdRPs rather than reverse transcriptase. 6 Also, structural analyses have long hypothesized that hTERT also has the characteristic right‐handed architecture similar to RdRPs found in RNA viruses (Figures 2A and 3B). 6 , 8 , 9 We previously reported that hTERT has RdRP activity, which generates dsRNAs that are processed to siRNAs for the purpose of downregulating gene expression. 11 In addition, we know that the amount of hTERT protein and its RdRP activity is higher during the mitotic phase of the cell cycle, supporting their involvement in transcriptional upregulation and heterochromatin formation. 46 More recently, we found that the hTERT molecule is phosphorylated at threonine 249 by the serine/threonine kinase cyclin‐dependent kinase 1 (CDK1) and that this phosphorylation enhances the RdRP activity in hTERT without affecting telomerase activity or telomere maintenance. 12 Remarkably, the hTERT RdRP synthesizes an antisense RNA against target genes, such as tumor suppressor gene transcript, and negatively regulates the expression of the suppressor, eventually leading to cancer progression (Figure 4, left).
FIGURE 4.
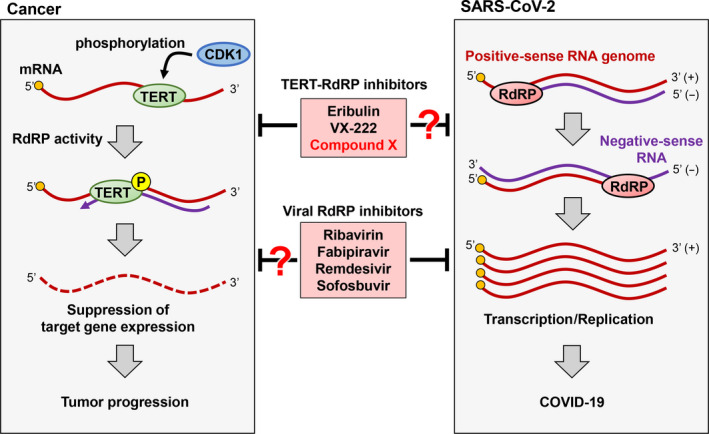
Roles of RNA‐dependent RNA polymerase (RdRP) in cancer development and severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection. In cancer cells, human telomerase reverse transcriptase (hTERT) is phosphorylated by cyclin‐dependent kinase 1 (CDK1), and phosphorylated hTERT shows RdRP activity. 12 Phosphorylated hTERT regulates the expression of target genes, leading to tumor progression. After the entry of SARS‐CoV‐2 into the cell, RdRP plays a role in transcription and replication of viral RNAs. The inhibition of RdRP activity is a promising molecular target for cancer and SARS‐CoV‐2 infection. COVID‐19, coronavirus disease
4.2. RNA‐dependent RNA polymerase in normal cells
In most somatic cells, hTERT is not expressed, and if so, it is expressed at extremely low levels, 47 whereas germline cells and somatic stem cells, as well as cancer cells, show high levels of hTERT expression. 48 For example, induced pluripotent stem (iPS) cells show a high level of mouse TERT (mTERT) 49 and hTERT expression 50 to prevent concurrent telomere shortening, which occurs as a result of successive rounds of cell division. However, the specific expression of hTERT in undifferentiated cells might suggest that the RdRP activity in hTERT plays some other role or roles in the regulation of gene expression. In other words, RdRP might be involved in differentiation, development, or both. Intriguingly, the reprogramming efficiency of mTERT‐KO fibroblasts to iPS cells was lower than that of WT fibroblasts, and this phenotype was recovered by the expression of an mTERT mutant without telomerase activity, suggesting that the activity of mTERT beyond telomere maintenance could have some effects on the reprogramming activity of iPS cells. 51 It is, thus, necessary to examine whether hTERT shows RdRP activity in undifferentiated cells, such as iPS cells.
Recently, it has also been suggested that RNA polymerase II (RNAPII) possesses RdRP activity. In one study, RdRP of purified human RNAPII extends short interspersed nuclear element (SINE)‐encoded mouse B2 RNA by using B2 RNA as a template. However, the extension of mouse B2 RNA by human RNAPII destabilizes the B2 RNA. 52 It remains unclear whether only the mouse B2 RNA serves as a template for the human RNAPII/RdRP activity in vitro or whether human and mouse RNAPII indeed show the RdRP activity in cells.
5. RNA‐DEPENDENT RNA POLYMERASE INHIBITORS
5.1. Inhibitors targeting viral RdRPs
Single‐stranded RNA viruses utilize RdRPs for transcription of viral genes and replication of the viral genome. Therefore, RdRP is a plausible target for the development of antiviral drugs, and many pharmaceutical companies have engaged in developing inhibitors against RdRPs of RNA viruses (Table 1).
Sofosbuvir, indicated to treat chronic hepatitis caused by HCV infection, is a nucleoside analog that shows anti‐HCV activity by interacting with HCV RdRP coded by the viral NS5B gene. Sofosbuvir is metabolized in hepatocytes to the active metabolite, sofosbuvir triphosphate, which is incorporated into the RNA chain by NS5B and acts as a chain terminator. 53 Sofosbuvir has high therapeutic efficacy in combination with other HCV therapeutic agents, as do the NS5B inhibitor ribavirin and the HCV NS5A inhibitor ledipasvir. 25
Favipiravir (T‐705), approved in Japan to treat emerging influenza viruses resistant to current drugs, blocks RdRP of influenza viruses. 54 , 55 After entering cells, favipiravir is metabolized to favipiravir triphosphate; it is then incorporated into the viral RNA chain where it acts as a chain terminator.
There is another RdRP inhibitor that is used to treat deadly filovirus infections such as EBOV disease and Marburg virus disease. The adenosine analog antiviral remdesivir (GS‐5734) inhibits EBOV RdRP as a chain terminator and shows a broad spectrum of antiviral activity against various pathogenic RNA viruses, including multiple variants of EBOV, other filoviruses and human coronaviruses. 56
Now, the development of therapeutic agents for COVID‐19 is ongoing. Among several SARS‐CoV‐2 proteins, RdRP nsp12 has attracted attention because it is involved in the essential reaction required for viral RNA replication and transcription. As the structure of viral RdRPs is highly conserved among various RNA viruses, a viral RdRP inhibitor is likely to inhibit RdRPs of other RNA viruses. Indeed, recent studies confirmed that SARS‐CoV‐2 nsp12 has a right‐hand structure, similar to other viral RdRPs. 16 , 30 These data are encouraging, promoting an investigation of whether existing RdRP inhibitors such as favipiravir for influenza virus or remdesivir for EBOV could be repositioned to treat COVID‐19. Wang et al 15 examined the efficiency of several RdRP inhibitors on SARS‐CoV‐2 infection. They found that favipiravir showed a moderate inhibitory effect (EC50, 61.88 μM), whereas remdesivir showed the most prominent inhibitory effect on viral proliferation (EC50, 0.77 μM). Moreover, remdesivir has been reported to show an antiviral effect against other species of coronaviruses besides EBOV and Marburg virus. 56 Therefore, a phase III clinical study of remdesivir was begun for COVID‐19 patients in the US, and preliminary results reported that remdesivir accelerated recovery by 31% (11 from 15 days) and improved the mortality rate (8.0% from 11.6%), 17 indicating that the use of RdRP inhibitors against RNA viruses is highly promising.
5.2. Inhibitors targeting RdRP activity in hTERT
The structure of hTERT is similar to the characteristic right‐handed architecture of RNA virus RdRPs. 6 , 8 , 9 In addition, the inhibition of RdRP activity in hTERT by a genetic approach suppressed the growth of cancer cell lines, 12 suggesting that efforts to search for inhibitors against RdRP activity in hTERT are, in our opinion, a promising strategy for finding candidate anticancer drugs. Thus, we hypothesized that inhibitors against viral RdRPs would also inhibit the RdRP activity in hTERT. We reported that the HCV RdRP inhibitor VX‐222 showed a significant inhibitory effect on RdRP activity in human hTERT. 57 VX‐222 is an allosteric inhibitor that binds to the thumb domain of NS5B. 58 Although the binding site between VX‐222 and hTERT remains unknown, further studies, such as assessment by a 3‐D docking model analysis between VX‐222 and hTERT, would be required to determine the molecular mechanism of how this molecule inhibits the RdRP activity in hTERT. These data indicate that inhibitors against viral RdRPs could also inhibit the RdRP activity in hTERT (Figure 4). Using similar approaches, we are searching for other inhibitors that would have anti‐RdRP activity in hTERT and could potentially be used as an anticancer drug.
During our analysis of the effects of high expression of hTERT in ovarian cancer cells, we had discovered another compound that suppresses RdRP activity in hTERT: eribulin mesylate (eribulin). Eribulin is a nontaxane inhibitor of microtubule dynamics that induces irreversible mitotic blockade, leading to apoptosis. Although the underlying mechanism of how eribulin inhibits the RdRP activity in hTERT is unknown, we confirmed that ovarian cancer cells with high RdRP activity are more sensitive to eribulin. 59 , 60 Based on the promising preclinical results obtained with tumor‐bearing mice, a phase II trial of eribulin for patients with recurrent glioblastoma has begun in Japan (UMIN000030359, https://upload.umin.ac.jp/cgi‐open‐bin/ctr_e/ctr_view.cgi?recptno=R000034631).
5.3. Side‐effects of RdRP inhibitors
It had long been considered that RdRP is specific to RNA viruses and conventional model organisms, such as yeast Schizosaccharomyces pombe, and mammalian cells do not express RdRP. Recently, the anti‐HCV RdRP inhibitor ribavirin and the antiinfluenza agent favipiravir were found to be teratogenic in animal models. 55 , 61 Looking at these observations from a different perspective, these results imply that RdRP is expressed in mammalian cells in early developmental stages. Therefore, fetal development might be disturbed by inhibitors against viral RdRPs. Also, previous studies have shown that hTERT and mTERT are specifically expressed in stem cells and embryonic cells as well as in cancer cells, 7 , 47 , 48 and that mTERT plays an extratelomeric role in normal undifferentiated cells (see the Section 4.2). 48 , 51 , 62 While we do not know whether viral RdRP inhibitors inhibit RdRP in these cells, we should carefully assess whether the RdRP inhibitors affect normal cells at the stage of differentiation and development by the inhibition of RdRP activity in hTERT. Moreover, as the telomere biology, including functional roles of TERT protein, significantly differs between mouse and human, we should carefully evaluate whether mouse models could accurately reflect functions of TERT in humans.
6. CONCLUSION
As viral RdRPs are conserved across RNA virus species and are essential enzymes in the life cycle of viruses, RdRP is the ideal target of an anti‐RNA virus discovery strategy. The RdRP of RNA viruses have a characteristic right‐handed shape. Thus, each RdRP inhibitor could share structural features that relate to a common inhibitory effect, even if the RNA viruses are different. The RdRP inhibitors favipiravir (against influenza virus) and remdesivir (against EBOV) showed inhibitory effects on the COVID‐19 virus, although their efficacies and the methods of inhibition are different. 15 , 17 Notably, hTERT also has a right‐handed structure, which is shared by RdRP of RNA viruses 6 , 9 and shows the RdRP activity that is essential for enhanced tumorigenicity. 12 We have already identified several compounds that show inhibitory activity against the RdRP activity in hTERT, which, thus far, has never been targeted in the anticancer drug‐hunting portfolio, 57 , 59 , 60 and we are now searching for and developing new inhibitors against the RdRP of hTERT. Obviously, RNA viruses and cancers operate differently in their quest for survival; however, they seem to share a survival strategy in their utilization of RdRPs (Table 1). Several inhibitors to specific viral RdRPs have been used to treat RNA virus infections, other than the intended target. Thus, inhibitors against the RdRP activity in hTERT might be used as an antiviral or anticancer drug that would be highly promising and versatile (Figure 4) because inhibitors against the RdRP activity in hTERT might have therapeutic effects on both RNA viruses and cancers. The recent outbreak of SARS‐CoV‐2 has caused a global pandemic and catastrophic consequences. Such RdRP inhibitors could be used as a countermeasure against these threats to humans.
CONFLICT OF INTEREST
Kenkichi Masutomi received patent transfer gains from Carna Biosciences. No potential conflicts of interest were disclosed by the remaining authors.
ACKNOWLEDGMENTS
We thank Mary Y. Nishikawa, MA, ELS of Lexaly Communications for her editorial review of the manuscript. The Japan Agency for Medical Research and Development (AMED) supported this work through the P‐CREATE grant 20cm0106115h0005 and the Emerging/Re‐emerging Infectious Diseases Project grant 20fk0108274h0001 (K.M.).
Machitani M, Yasukawa M, Nakashima J, Furuichi Y, Masutomi K. RNA‐dependent RNA polymerase, RdRP, a promising therapeutic target for cancer and potentially COVID‐19. Cancer Sci. 2020;111:3976–3984. 10.1111/cas.14618
Funding information
Japan Agency for Medical Research and Development, P‐CREATE grant 20cm0106115h0005; Emerging/Re‐emerging Infectious Diseases Project grant 20fk0108274h0001.
REFERENCES
- 1. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shi M, Lin XD, Chen X, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556:197‐202. [DOI] [PubMed] [Google Scholar]
- 5. Koonin EV, Senkevich TG, Dolja VV. The ancient Virus World and evolution of cells. Biol Direct. 2006;1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakamura TM, Morin GB, Chapman KB, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955‐959. [DOI] [PubMed] [Google Scholar]
- 7. Meyerson M, Counter CM, Eaton EN, et al. hEST2, the putative human telomerase catalytic subunit gene, is up‐regulated in tumor cells and during immortalization. Cell. 1997;90:785‐795. [DOI] [PubMed] [Google Scholar]
- 8. Gillis AJ, Schuller AP, Skordalakes E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature. 2008;455:633‐637. [DOI] [PubMed] [Google Scholar]
- 9. Jiang J, Wang Y, Susac L, et al. Structure of telomerase with telomeric DNA. Cell. 2018;173:1179‐1190.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nguyen THD, Tam J, Wu RA, et al. Cryo‐EM structure of substrate‐bound human telomerase holoenzyme. Nature. 2018;557:190‐195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maida Y, Yasukawa M, Furuuchi M, et al. An RNA‐dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230‐235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yasukawa M, Ando Y, Yamashita T, et al. CDK1 dependent phosphorylation of hTERT contributes to cancer progression. Nat Commun. 2020;11:1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gibbs CP, Tanaka A, Anderson SK, et al. Isolation and structural mapping of a human c‐src gene homologous to the transforming gene (v‐src) of Rous sarcoma virus. J Virol. 1985;53:19‐24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vogt PK. Retroviral oncogenes: a historical primer. Nat Rev Cancer. 2012;12:639‐648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019‐nCoV) in vitro. Cell Res. 2020;30:269‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao Y, Yan L, Huang Y, et al. Structure of the RNA‐dependent RNA polymerase from COVID‐19 virus. Science. 2020;368:779‐782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid‐19 ‐ preliminary report. N Engl J Med. 2020. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 18. Cai Q, Yang M, Liu D, et al. Experimental treatment with favipiravir for COVID‐19: an open‐label control study. Engineering (Beijing). 2020. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ahlquist P. Parallels among positive‐strand RNA viruses, reverse‐transcribing viruses and double‐stranded RNA viruses. Nat Rev Microbiol. 2006;4:371‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA‐binding protein pivotal to virus replication. J Gen Virol. 2002;83:723‐734. [DOI] [PubMed] [Google Scholar]
- 21. Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395‐425. [DOI] [PubMed] [Google Scholar]
- 22. Hansen JL, Long AM, Schultz SC. Structure of the RNA‐dependent RNA polymerase of poliovirus. Structure. 1997;5:1109‐1122. [DOI] [PubMed] [Google Scholar]
- 23. Bressanelli S, Tomei L, Roussel A, et al. Crystal structure of the RNA‐dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA. 1999;96:13034‐13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salgado PS, Koivunen MR, Makeyev EV, Bamford DH, Stuart DI, Grimes JM. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006;4:e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Afdhal N, Zeuzem S, Kwo P, et al. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med. 2014;370:1889‐1898. [DOI] [PubMed] [Google Scholar]
- 26. McGeoch D, Fellner P, Newton C. Influenza virus genome consists of eight distinct RNA species. Proc Natl Acad Sci USA. 1976;73:3045‐3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang TS, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990;64:5669‐5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Plotch SJ, Bouloy M, Ulmanen I, Krug RM. A unique cap(m7G pppXm)‐dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847‐858. [DOI] [PubMed] [Google Scholar]
- 29. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS‐CoV‐2 transcriptome. Cell. 2020;181:914‐921.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hillen HS, Kokic G, Farnung L, Dienemann C, Tegunov D, Cramer P. Structure of replicating SARS‐CoV‐2 polymerase. Nature. 2020;584:154–156. [DOI] [PubMed] [Google Scholar]
- 31. Spaan W, Delius H, Skinner M, et al. Coronavirus mRNA synthesis involves fusion of non‐contiguous sequences. EMBO J. 1983;2:1839‐1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Furuichi Y. Caps on Eukaryotic mRNAs. Chichester: eLS: John Wiley & Sons, Ltd.; 2014. [Google Scholar]
- 33. Kumagai MH, Donson J, della‐Cioppa G, Harvey D, Hanley K, Grill LK. Cytoplasmic inhibition of carotenoid biosynthesis with virus‐derived RNA. Proc Natl Acad Sci USA. 1995;92:1679‐1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806‐811. [DOI] [PubMed] [Google Scholar]
- 35. Romano N, Macino G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343‐3353. [DOI] [PubMed] [Google Scholar]
- 36. Sijen T, Fleenor J, Simmer F, et al. On the role of RNA amplification in dsRNA‐triggered gene silencing. Cell. 2001;107:465‐476. [DOI] [PubMed] [Google Scholar]
- 37. Meister G, Tuschl T. Mechanisms of gene silencing by double‐stranded RNA. Nature. 2004;431:343‐349. [DOI] [PubMed] [Google Scholar]
- 38. Verdel A, Jia S, Gerber S, et al. RNAi‐mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672‐676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SI. RNA‐dependent RNA polymerase is an essential component of a self‐enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005;102:152‐157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521‐529. [DOI] [PubMed] [Google Scholar]
- 41. Weinrich SL, Pruzan R, Ma L, et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat Genet. 1997;17:498‐502. [DOI] [PubMed] [Google Scholar]
- 42. Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464‐468. [DOI] [PubMed] [Google Scholar]
- 43. Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev. 2015;29:2219‐2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cong Y, Shay JW. Actions of human telomerase beyond telomeres. Cell Res. 2008;18:725‐732. [DOI] [PubMed] [Google Scholar]
- 45. Martinez P, Blasco MA. Telomeric and extra‐telomeric roles for telomerase and the telomere‐binding proteins. Nat Rev Cancer. 2011;11:161‐176. [DOI] [PubMed] [Google Scholar]
- 46. Maida Y, Yasukawa M, Okamoto N, et al. Involvement of telomerase reverse transcriptase in heterochromatin maintenance. Mol Cell Biol. 2014;34:1576‐1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173‐179. [DOI] [PubMed] [Google Scholar]
- 48. Roake CM, Artandi SE. Regulation of human telomerase in homeostasis and disease. Nat Rev Mol Cell Biol. 2020;21:384‐397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663‐676. [DOI] [PubMed] [Google Scholar]
- 50. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861‐872. [DOI] [PubMed] [Google Scholar]
- 51. Kinoshita T, Nagamatsu G, Saito S, Takubo K, Horimoto K, Suda T. Telomerase reverse transcriptase has an extratelomeric function in somatic cell reprogramming. J Biol Chem. 2014;289:15776‐15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wagner SD, Yakovchuk P, Gilman B, et al. RNA polymerase II acts as an RNA‐dependent RNA polymerase to extend and destabilize a non‐coding RNA. EMBO J. 2013;32:781‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sofia MJ, Bao D, Chang W, et al. Discovery of a beta‐d‐2'‐deoxy‐2'‐alpha‐fluoro‐2'‐beta‐C‐methyluridine nucleotide prodrug (PSI‐7977) for the treatment of hepatitis C virus. J Med Chem. 2010;53:7202‐7218. [DOI] [PubMed] [Google Scholar]
- 54. Furuta Y, Gowen BB, Takahashi K, Shiraki K, Smee DF, Barnard DL. Favipiravir (T‐705), a novel viral RNA polymerase inhibitor. Antiviral Res. 2013;100:446‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shiraki K, Daikoku T. Favipiravir, an anti‐influenza drug against life‐threatening RNA virus infections. Pharmacol Ther. 2020;209:107512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sheahan TP, Sims AC, Graham RL, et al. Broad‐spectrum antiviral GS‐5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9:eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Maida Y, Yasukawa M, Masutomi K. De novo RNA synthesis by RNA‐dependent RNA polymerase activity of telomerase reverse transcriptase. Mol Cell Biol. 2016;36:1248‐1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yi G, Deval J, Fan B, et al. Biochemical study of the comparative inhibition of hepatitis C virus RNA polymerase by VX‐222 and filibuvir. Antimicrob Agents Chemother. 2012;56:830‐837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yamaguchi S, Maida Y, Yasukawa M, Kato T, Yoshida M, Masutomi K. Eribulin mesylate targets human telomerase reverse transcriptase in ovarian cancer cells. PLoS One. 2014;9:e112438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Takahashi M, Miki S, Fujimoto K, et al. Eribulin penetrates brain tumor tissue and prolongs survival of mice harboring intracerebral glioblastoma xenografts. Cancer Sci. 2019;110:2247‐2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kochhar DM, Penner JD, Knudsen TB. Embryotoxic, teratogenic, and metabolic effects of ribavirin in mice. Toxicol Appl Pharmacol. 1980;52:99‐112. [DOI] [PubMed] [Google Scholar]
- 62. Sarin KY, Cheung P, Gilison D, et al. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048‐1052. [DOI] [PMC free article] [PubMed] [Google Scholar]


