Abstract
Background
Long noncoding RNAs (lncRNAs) are emerging as crucial contributors to the development of breast cancer and are involved in the stemness regulation of breast cancer stem cells (BCSCs). LncRNAs are closely associated with the prognosis of breast cancer patients. It is critical to identify BCSC-related lncRNAs with prognostic value in breast cancer.
Methods
A co-expression network of BCSC-related mRNAs-lncRNAs from The Cancer Genome Atlas (TCGA) was constructed. Univariate and multivariate Cox proportional hazards analyses were used to identify a stemness risk model with prognostic value. Kaplan–Meier analysis, univariate and multivariate Cox regression analyses and receiver operating characteristic (ROC) curve analysis were performed to validate the risk model. Principal component analysis (PCA) and Gene Set Enrichment Analysis (GSEA) functional annotation were conducted to analyze the risk model.
Results
In this study, BCSC-related lncRNAs in breast cancer were identified. We evaluated the prognostic value of these BCSC-related lncRNAs and eventually obtained a prognostic risk model consisting of 12 BCSC-related lncRNAs (Z68871.1, LINC00578, AC097639.1, AP003119.3, AP001207.3, LINC00668, AL122010.1, AC245297.3, LINC01871, AP000851.2, AC022509.2 and SEMA3B-AS1). The risk model was further verified as a novel independent prognostic factor for breast cancer patients based on the calculated risk score. Moreover, based on the risk model, the low- risk and high-risk groups displayed different stemness statuses.
Conclusions
These findings suggested that the 12 BCSC-related lncRNA signature might be a promising prognostic factor for breast cancer and can promote the management of BCSC-related therapy in clinical practice.
Background
Breast cancer is the most commonly diagnosed malignancy and is the leading cause of cancer-associated mortality among women worldwide [1, 2]. In the field of clinical treatment, increasing attention has been focused on individual and precise therapeutic strategies. Thus, the identification of novel prognostic biomarkers and promising targets is considered to be an effective way to achieve this goal.
Heterogeneity is a hallmark of solid tumors, including breast cancer, which results from the enrichment of cancer stem cells (CSCs) [3, 4]. CSCs represent a dynamic subpopulation of tumor cells characterized by self-renewal, pluripotency and limitless proliferative properties [5]. Breast cancer stem cells (BCSCs) are considered the source of tumor aggression, metastasis, worse prognosis, chemoresistance and recurrence in breast cancer [6]. Therefore, identifying key stemness regulators of BCSCs is of great importance for both theoretical studies and clinical practice.
Long noncoding RNAs (lncRNAs) are a class of transcript RNAs longer than 200 nucleotides that are not translated into proteins [7]. LncRNAs are involved in the development and progression of various cancers at different levels, including epigenetic, transcriptional and posttranscriptional regulation, and are considered one of the most sensitive and specific cancer biomarkers [8–11]. Recently, lncRNAs have become a hot topic in stemness regulation of CSCs and prediction of prognosis in numerous cancers. Therefore, it is essential to identify key lncRNAs closely related to the stemness of BCSCs and prognosis in breast cancer.
In this study, we analyzed a dataset of lncRNA expression in breast cancers from The Cancer Genome Atlas (TCGA) and screened prognostic lncRNAs related to the stemness of BCSCs. We identified a 12 BCSC-related lncRNA signature with the ability to predict the survival prognosis of breast cancer patients.
Methods
Patient data sets
Clinical information and pathology records of patients with breast cancer were taken from the TCGA (https://cancergenome.nih.gov/). The edgeR package was used to normalize gene expression. A total of 1053 TCGA female breast cancer patients with lncRNA expression profiles were utilized in the present study.
Among them, 986 patients with complete follow-up information and survival time ≥ 30 days and 539 patients with complete clinicopathological data were applied to subsequent analyses. The clinical characteristics are detailed in Table 1.
Table 1.
Clinical pathological parameters of patients with breast cancer
| Feature | N (539) | % |
|---|---|---|
| Age (years) | ||
| > 60 | 227 | 42.1 |
| ≤ 60 | 312 | 57.9 |
| T classification | ||
| T1 (< 2 cm) | 147 | 27.3 |
| T2 (2-5 cm) | 323 | 59.9 |
| T3 (≥ 5 cm) | 55 | 10.2 |
| T4 (chest wall and/or skin invasion) | 14 | 2.6 |
| N classification (pN) | ||
| N0 (no metastasis) | 259 | 48.1 |
| N1 (1–3 metastasis) | 178 | 33 |
| N2 (4–9 metastasis) | 64 | 11.9 |
| N3 (≥ 10 metastasis) | 38 | 7 |
| M classification | ||
| M0 (no distantmetastasis) | 528 | 98 |
| M1 (distant metastasis) | 11 | 2 |
| TNM stage | ||
| I | 96 | 17.8 |
| II | 318 | 59 |
| III | 114 | 21.2 |
| IV | 11 | 2 |
| ER | ||
| Negative | 127 | 23.6 |
| Positive | 412 | 76.4 |
| PR | ||
| Negative | 175 | 32.5 |
| Positive | 364 | 67.5 |
| HER2 | ||
| Negative | 440 | 81.6 |
| Positive | 99 | 18.4 |
| Molecular subtypes | ||
| HER2 amplification | 92 | 17.1 |
| Luminal A/B | 419 | 77.7 |
| TNBC | 28 | 5.2 |
T tumor size, N lymph node, M distant metastasis, TNM stage according to AJCC 8th classification, TNBC triple-negative breast cancer
Identification of BCSC-related lncRNAs in breast cancer
A total of 213 BCSC-related encoding genes (mRNAs) were extracted from the Molecular Signatures Database of Gene Set Enrichment Analysis (GSEA: M14079, M4740, M9246, and M13135). Finally, 1198 BCSC-related lncRNAs were identified by constructing BCSC-related mRNAs-lncRNAs co-expression network according to the criteria of |Correlation Coefficient|> 0.3 and p < 0.001 by Pearson correlation analysis using the limma R package [12].
Identification of BCSC-related lncRNA prognostic signatures for breast cancer
To identify BCSC-related lncRNAs associated with survival, univariate Cox proportional hazards analysis was performed with p < 0.01 as the criteria, and multivariate Cox analysis was subsequently performed to establish the optimal prognostic risk model based on the Akaike information criterion (AIC = 1422.28) using the survival R package. Then, the risk score for each patient was calculated based on the following formula:
Risk score = coef (lncRNA1) × expr (lncRNA1) + coef (lncRNA2) × expr (lncRNA2) + …… + coef (lncRNAn) × expr (lncRNAn).
coef (lncRNAn) was defined as the coefficient of lncRNAs correlated with survival.
expr (lncRNAn) was defined as the expression of lncRNAs.
Breast cancer patients in the TCGA were divided into a high-risk group and a low-risk group according to the median risk score. Kaplan–Meier survival analysis was conducted to evaluate the survival difference between the two groups using the survival and survminer R packages.
Independent prognostic analysis and ROC curve plotting
Univariate and multivariate Cox regression analyses were performed to assess the relationship between survival prognosis and age; estrogen receptor (ER) expression; progesterone receptor (PR) expression; human epidermal growth factor receptor 2 (HER2) expression; Tumor, Node, Metastasis (TNM) stage; tumor size (T); lymph node (N) metastasis; distant metastasis (M); and risk score using the survival R package. Time-dependent receiver operating characteristic (ROC) curves were plotted to evaluate the predictive accuracy for survival time through different clinical pathological factors and risk scores using the survival ROC R package.
Statistical analysis
All statistical analyses were performed using R software (version 3.6.2). A co-expression network of BCSC-related lncRNAs-mRNAs with prognostic value in breast cancer was constructed visualized by Cytoscape and Sankey diagram. The correlation between the expression of BCSC-related lncRNAs and clinicopathological factors was analyzed by ggpubr R package. Principal component analysis (PCA) was performed for effective dimension reduction, pattern recognition, and exploratory visualization of high-dimensional data of the whole genome, 213 BCSC-related encoding genes and the risk model of BCSC-related lncRNAs expression profiles, respectively [13, 14]. GSEA was used for functional annotation. Two-tailed p < 0.05 was considered statistically significant.
Results
Identification of significant prognostic value of BCSC-related lncRNAs in breast cancer
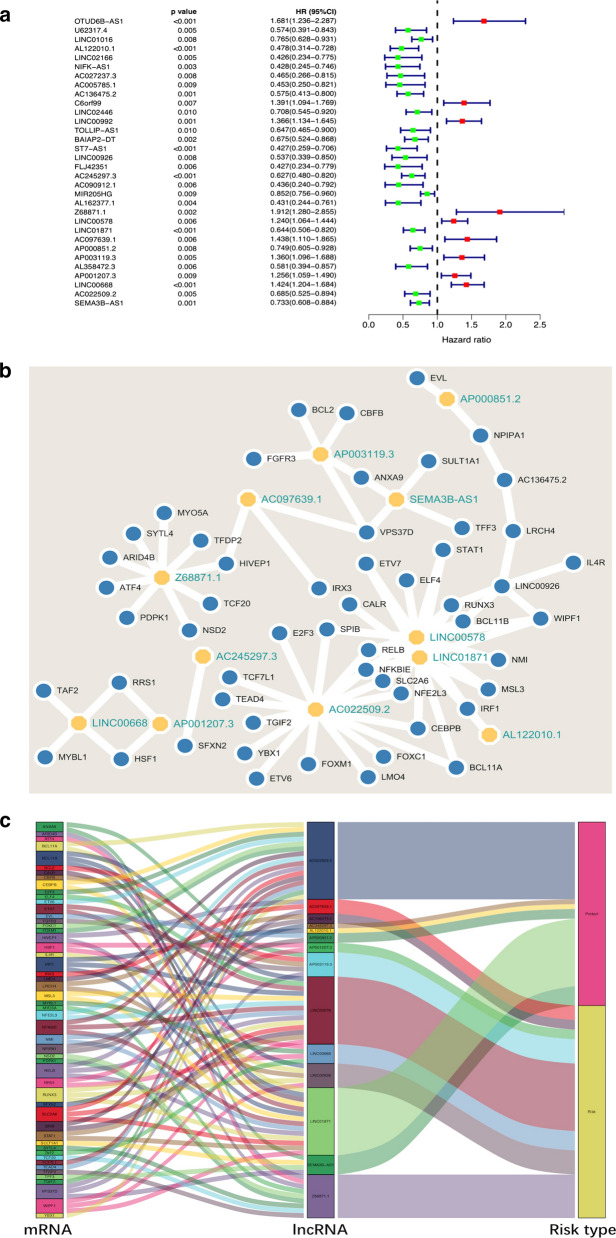
A total of 1198 BCSC-related lncRNAs were obtained by constructing co-expression networks with 213 BCSC-related encoding genes (mRNAs) (Additional files 1, 2 and 3). Among them, 32 BCSC-related lncRNAs were significantly associated with the survival of breast cancer patients from the TCGA (p < 0.01) by Cox proportional hazards analysis, including 23 lncRNAs with low risk (hazard ratio (HR) < 1) and 9 lncRNAs with high risk (HR > 1) (Fig. 1). Subsequently, multivariate Cox analysis further screened 12 lncRNAs from the above 23 BCSC-related lncRNAs with prognostic significance, namely, Z68871.1, LINC00578, AC097639.1, AP003119.3, AP001207.3, LINC00668, AL122010.1,
Fig. 1.
Identification of BCSC-related lncRNAs with significant prognostic value in breast cancer. a The forest showed the HR (95%CI) and p-value of selected lncRNAs by univariate Cox proportional-hazards analysis. b, c A co-expression network of the 12 BCSC-related lncRNAs-mRNAs with prognostic value was constructed and visualized using Cytoscape and Sankey diagram
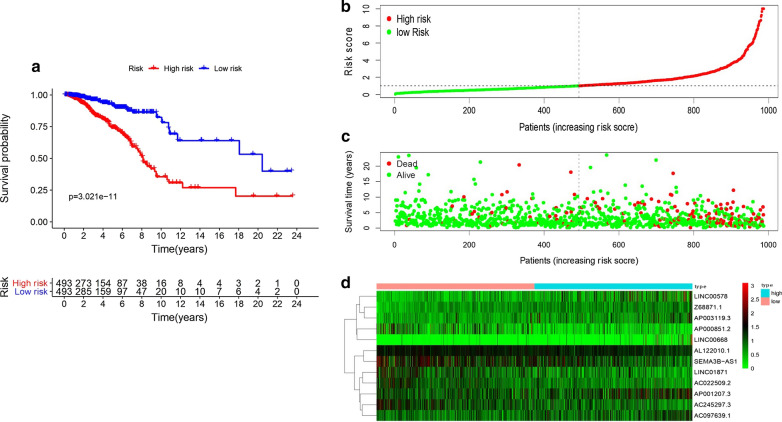
AC245297.3, LINC01871, AP000851.2, AC022509.2 and SEMA3B-AS1 (Table 2). These 12 lncRNAs were constructed into the optimal prognostic risk model of BCSC-related lncRNAs. As shown in Fig. 1a–c visualization co-expression network of BCSC-related lncRNAs-mRNAs with prognostic value was established. According to the risk score formula and the calculated median risk score, breast cancer patients were divided into a high-risk group and a low-risk group. Kaplan–Meier survival analysis showed that the high-risk group presented worse overall survival (OS) than the low risk group (p = 3.021E−11) (Fig. 2a), suggesting that the risk score had prognostic value. The risk curve and scatterplot were made to illustrate the risk score and the corresponding survival status of breast cancer patients. The results showed that the higher the risk score was, the more mortality occurred (Fig. 2b, c). The heatmap of the expression of these 12 BCSC-related lncRNAs in breast cancer samples showed that Z68871.1, LINC00578, AC097639.1, AP003119.3, AP001207.3 and LINC00668 were upregulated in the high risk group, while AL122010.1, AC245297.3, LINC01871, AP000851.2, AC022509.2 and SEMA3B-AS1 were highly expressed in the low risk group (Fig. 2d). Therefore, these studies identified 12 BCSC-related lncRNAs with prognostic significance for breast cancer.
Table 2.
The risk model of 12 BCSC-related lncRNAs with prognostic value in breast cancer
| LncRNA | Coef | HR | HR.95L | HR.95H | p-value | Risk |
|---|---|---|---|---|---|---|
| AL122010.1 | − 0.488472 | 0.61356332 | 0.40085805 | 0.9391353 | 0.02450628 | Low |
| AC245297.3 | − 0.39251 | 0.6753596 | 0.51409558 | 0.88720971 | 0.00480743 | Low |
| Z68871.1 | 0.675654 | 1.96531726 | 1.25113209 | 3.08718155 | 0.00336423 | High |
| LINC00578 | 0.189555 | 1.20871114 | 1.02961181 | 1.41896451 | 0.0205253 | High |
| LINC01871 | − 0.397982 | 0.67167386 | 0.51597499 | 0.87435591 | 0.00309782 | Low |
| AC097639.1 | 0.207522 | 1.23062483 | 0.94799722 | 1.59751257 | 0.11903949 | High |
| AP000851.2 | − 0.271595 | 0.7621631 | 0.59823533 | 0.97101016 | 0.02794552 | Low |
| AP003119.3 | 0.190492 | 1.20984515 | 0.95504527 | 1.53262399 | 0.11439212 | High |
| AP001207.3 | 0.274701 | 1.31613764 | 1.12443772 | 1.54051956 | 0.00062571 | High |
| LINC00668 | 0.26132 | 1.29864313 | 1.08803894 | 1.55001251 | 0.00379653 | High |
| AC022509.2 | − 0.211675 | 0.80922784 | 0.64043779 | 1.02250321 | 0.07614398 | Low |
| SEMA3B-AS1 | − 0.466346 | 0.62729049 | 0.50223943 | 0.78347763 | 3.94E−05 | Low |
coef the coefficient of lncRNAs correlated with survival, HR hazard ratio, HR.95L low 95%CI of HR, HR.95H high 95%CI of HR
Fig. 2.
The prognostic value of the risk model of the 12 BCSC-related lncRNAs in the TCGA cohort. a Kaplan–Meier survival analysis of the high-risk and low-risk groups based on the risk model and median risk score. b The risk curve based on the risk score of each sample. c The scatterplot based on the survival status of each sample. The green and red dots represent survival and death, respectively. d The heatmap displayed the expression levels of BCSC-related lncRNAs in the high-risk and low-risk groups
Evaluation of the risk model of the 12 BCSC-related lncRNAs as independent prognostic factor for breast cancer patients
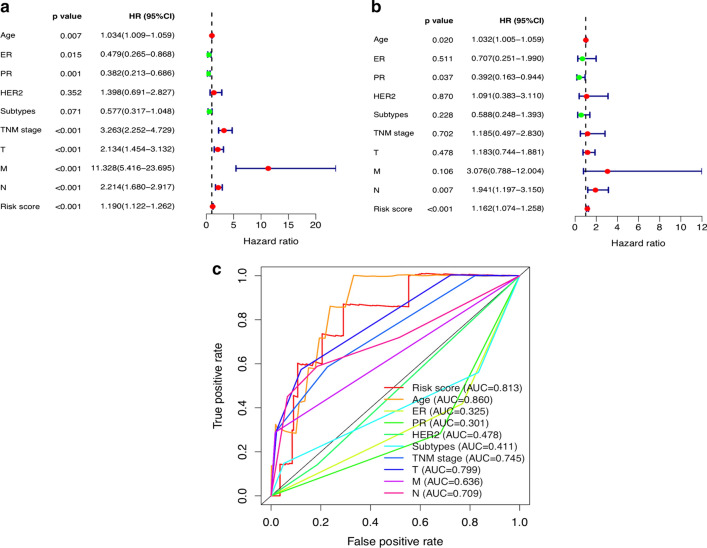
Univariate and multivariate Cox regression analyses were performed to assess whether the risk model of the above 12 BCSC-related lncRNAs was an independent prognostic factor for breast cancer. The HR of the risk score and 95% CI were 1.190 and 1.122–1.262 (p < 0.001) in univariate Cox regression analysis (Fig. 3a) and 1.162 and 1.074–1.258 (p < 0.001) in multivariate Cox regression analysis (Fig. 3b), respectively, indicating that the risk model of the 12 BCSC-related lncRNAs was the most significant prognostic factor for breast cancer, independent of clinicopathological parameters such as age, ER expression, PR expression, HER2 expression, molecular subtypes, TNM stage, tumor size, lymph node metastasis and distant metastasis. To evaluate the predictive specificity and sensitivity of the risk score on the prognosis of breast cancer patients, the area under the ROC curve (AUC) of the risk score was estimated. The AUC of the risk score was 0.813, followed by the AUC of age and more than the AUCs of other clinicopathological factors (Fig. 3c), suggesting that the prognostic risk model of the 12 BCSC-related lncRNAs for breast cancer was considerably reliable. All of the above results indicated that the 12 BCSC-related lncRNAs were significant independent prognostic factors for breast cancer patients.
Fig. 3.
Assessment of the prognostic risk model of the 12 BCSC-related lncRNAs in breast cancer. a The univariate and b multivariate Cox regression analysis of risk model score and clinical features regarding prognostic value. c The AUC for risk model score and clinical features according to the ROC curves. Clinical features: Age, ER, PR, HER2, Subtypes (molecular subtypes), TNM stage, T (tumor size), N (lymph node metastasis) and M (distant metastasis)
Correlation of the expression of the 12 BCSC-related lncRNAs with clinicopathological factors
To further investigate whether the 12 BCSC-related lncRNAs were involved in the development of breast cancer, we assessed the association of the expression of the 12 BCSC-related lncRNAs with clinicopathological factors. There were significant correlations between most of the 12 BCSC-related lncRNAs and ER expression, PR expression and molecular subtypes, as shown in Fig. 4.
Fig. 4.
The correlation of the expression of the 12 BCSC-related lncRNAs with clinicopathological factors. a ER expression. b PR expression. c HER2 expression. d molecular subtypes (LuminalA/B; HER2 amplification; TNBC: triple-negative breast cancer). e TNM stage. f Tumor size (T1: < 2 cm; T2: ≥ 2 cm and < 5 cm; T3: ≥ 5 cm; T4: invasion of chest wall and/or skin). g N classification (N0: no lymph node metastasis; N1: 1–3 lymph node metastasis; N2: 4–9 lymph node metastasis; N3: ≥ 10 lymph node metastasis). h M classification (M0: no distant metastasis; M1: distant metastasis). ns: no statistical significance, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
Different stemness statuses in the low-risk and high-risk groups
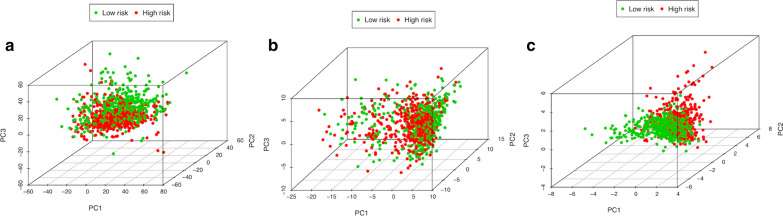
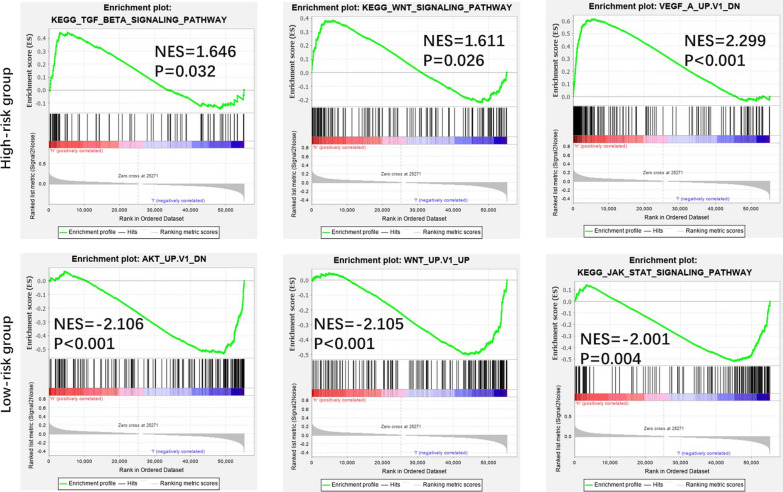
PCA was performed to compare the difference between the low-risk and high-risk groups based on the risk model of the 12 BCSC-related lncRNAs, 224 BCSC-related encoding genes and whole-genome expression profiles, respectively (Fig. 5). The results showed that the low-risk and high-risk groups based on the risk model were distributed in distinct directions, more obvious than the others, suggesting that the risk model could divide breast cancer patients into two parts and that the stemness status of breast cancer patients in the high-risk group differed from those in the low-risk group. Functional annotation was further conducted using GSEA, and the results showed that the differentially expressed genes between the high-risk and low-risk groups based on the risk model of the 12 BCSC-related genes were enriched in stemness-related processes and CSC-related pathways (Fig. 6). These results indicated that the low-risk and high-risk groups showed different stemness statuses.
Fig. 5.
The Low-risk and high-risk groups displayed different stemness statuses. a–c Principal components analysis (PCA) between the low-risk and high-risk groups based on the whole-genome, BCSC-related encoding genes and the risk model of the 12 BCSC-related lncRNAs expression profiles
Fig. 6.
Functional enrichment analysis based on the risk model of the 12 BCSC-related lncRNAs by GSEA. Significantly enriched KEGG pathways and oncogenic signatures in the high-risk and low-risk groups
Discussion
In clinical practice, although the OS of breast cancer patients has made promising improvements, the occurrence of drug resistance to breast cancer has constantly increased. The increased frequency of chemoresistance and endocrine resistance might result from enrichment with BCSCs. Because large-scale current therapeutic strategies focus on eliminating the majority of non-CSCs, the residual population of chemo-resistant cancer cells that contributes to relapse and metastasis is thought to result from the existence of minimal residual CSCs [15, 16]. Thus, effective treatment aimed at BCSCs has promising prospects. This led us to find potential specific prognostic biomarkers and therapeutic targets for BCSCs. Recently, an increasing number of studies have suggested the crucial role of lncRNAs in regulating the stemness of CSCs and predicting prognosis in numerous cancers such as intestinal cancer, lung cancer, and hepatocellular cancer [17–19]. Similarly, an increasing number of lncRNAs have been implicated in the regulation of stemness in breast cancer. LncRNA-Hh strengthened CSC generation in twist-positive breast cancer by activating the Hedgehog signaling pathway [20]. LncRNA FEZF1-AS1 promoted the stemness of BCSC tumorigenesis by targeting the miR-30a/Nanog axis [21]. LncRNA HOTAIR contributed to the epithelial–mesenchymal transition (EMT) of BCSCs by activating the STAT3 signaling pathway [22]. In this study, we identified the risk model of the 12 BCSC-related lncRNAs as an independent prognostic factor for breast cancer. To date, among these 12 BCSC-related lncRNAs, only LINC00578, LINC00668 and SEMA3B-AS1 have been studied in breast cancer or other cancers. It has been reported that LINC00578 is associated with worse OS in pancreatic cancer and lung adenocarcinoma [23, 24]. LINC00668 promoted tumorigenesis and progression and indicated poor prognosis in not only breast cancer but also other cancers, such as colorectal cancer, hepatocellular carcinoma and non-small-cell lung cancer [25–28]. SEMA3B-AS1 might serve as a new tumor suppressor to inhibit the development of hepatocellular carcinoma, esophageal squamous cell carcinoma and gastric cardia adenocarcinoma [29–31]. Consistent with our results, LINC00578 and LINC00668, as high-risk BCSC-related lncRNAs, were correlated with worse prognosis in breast cancer patients, whereas SEMA3B-AS1, as a low-risk BCSC-related lncRNA, was associated with better prognosis in breast cancer patients. Breast cancer is a complex disease and highly heterogeneous tumor [32]. To assess survival prognosis and guide individual therapeutic decisions, breast cancer has been divided into distinct molecular subtypes based primarily on the expression status of hormonal receptors such as ER, PR, HER2 and Ki67 (tumor proliferation index) as follows: luminal A/B (ER and/or PR positive), HER2 enriched (HER2 positive) and triple-negative breast cancer (ER, PR and HER2 negative) [33]. It is also well known that ER and/or PR positive indicates an effective endocrine therapy outcome and better survival prognosis; HER2 positive represents a more aggressive phenotype but is sensitive to HER2-targeted therapy; triple negative is enriched with BCSCs and associated with a worse prognosis due to the lack of effective therapeutic targets [34, 35]. Moreover, it was demonstrated that distinct molecular subtypes of breast cancer were enriched with different amounts of BCSCs [4]. Thus, distinct ER, PR and HER2 statuses indicated different biological processes of breast cancer and survival outcomes. In line with the abovementioned findings, most of the 12 BCSC-related lncRNAs were remarkably associated with ER expression, PR expression and molecular subtypes, which further suggested that the 12 BCSC-related lncRNAs might be involved in the development and progression of breast cancer, and the risk model was also based on the intrinsic properties of breast cancer. More interestingly, there were no significant relationship between most of the 12 BCSCs-related lncRNAs and tumor size (T), lymph node (N) status, distant metastasis (M) and TNM stage, indicating that the risk model of the 12 BCSCs-related lncRNAs has no close correlation with the sooner or later for finding and diagnosis of breast cancer, but is only strongly linked to the intrinsic biological characteristics of distinct subtypes of breast cancer. Furthermore, the risk model was the second most statistically significant prognostic signature compared with other clinicopathological factors, and the ROC result (AUC = 0.813) confirmed that the risk model is reliable. Combined with the AUC of the risk model score, these results all indicated that the risk model of the 12 BCSC-related lncRNAs had superior prognostic value to other clinicopathological factors. The results of PCA and GSEA functional annotation illustrated that the high-risk and low- risk groups showed different distribution directions and aggregation centers based on the risk model, rather than the whole-genome expression profiles and BCSC-related genes expression profiles, indicating that the significant differences in OS between the high-risk and low-risk groups might result from different stemness and oncogenic statuses induced by the risk model of the 12 BCSC-related lncRNAs. Taken together, these results indicated that the prognostic signature of the 12 BCSC-related lncRNAs might be a feasible independent prognostic factor for breast cancer in clinical practice. To date, a key challenge of precision genomic medicine is to make reliable and accurate predictions of clinical outcomes from high-dimensional molecular data [36]. To solve this problem, there have been some advances in Cox regression with prognostic value in recent years. A Cox elastic net has been used in objective and data-driven feature selection with time-to-event data [37]. Cox-nnet is an artificial neural network approach that has been utilized in predicting low-dimensional survival prognosis [38]. Bayesian-optimized deep survival models (SurvivalNet models) have successfully improved the accuracy of prognostic prediction for high-dimensional cancer genomic profiles [39]. In addition, Cox-nnet has a better performance than SurvivalNet models, and SurvivalNet models provide comparable performance to the Cox elastic net [39, 40]. Moreover, Cox-PASNet, which is a novel pathway-based sparse deep neural network for survival analysis that integrates high-dimensional genomic data and clinical data, has been applied to identify significant prognostic factors [40]. However, our study has some limitations. We applied traditional univariate and multivariate Cox proportional hazards analyses to establish and estimate the prognostic value of the risk model of the 12 BCSC-related lncRNAs. Although the method has been approved and employed in many researches, it is necessary to improve our further study with more advanced methodologies and technologies in the future. To further validate our bioinformatics prediction results, in-depth studies on the 12 BCSC-related lncRNAs, including functional experiments and molecular mechanisms, are needed.
Conclusion
In conclusion, we identified a BCSC-related lncRNA signature consisting of 12 lncRNAs (Z68871.1, LINC00578, AC097639.1, AP003119.3, AP001207.3, LINC00668, AL122010.1, AC245297.3, LINC01871, AP000851.2, AC022509.2 and SEMA3B-AS1), which can act as a novel independent prognostic factor for breast cancer. In the future, with prospective validation, the 12 BCSC-related lncRNA signature may improve the predictive accuracy and guide individual specific therapy for patients with breast cancer.
Supplementary information
Additional file 1. The list of 1198 BCSC-related lncRNAs.
Additional file 2. The list of 213 BCSC-related mRNAs.
Additional file 3: Fig. S1. Protein-protein interaction network of the 213 BCSC-related encoding genes (mRNAs).
Acknowledgments
The data of this study was download from The Cancer Genome Atlas (TCGA), we gratefully acknowledge the patients and operations.
Abbreviations
- LncRNAs
Long noncoding RNAs
- TCGA
The Cancer Genome Atlas
- ROC
Receiver operating characteristics
- PCA
Principal components analysis
- GSEA
Gene Set Enrichment Analysis
- OS
Overall survival
- HR
Hazard ratio
- ER
Estrogen receptor
- PR
Progesterone receptor
- HER2
Human epidermal growth factor receptor 2
Authors’ contributions
XL, YL, XY and FJ designed the study and conceived the project. XL acquired the data and analyzed the data. XL drafted the manuscript. YL, XY and FJ revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (Nos. 81773163, 81702881).
Data availability
All data utilized in this study are included in this article and all data supporting the findings of this study are available on reasonable request from the corresponding author.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xinmiao Yu and Feng Jin contributed equally to this work
Contributor Information
Xiaoying Li, Email: lixiaoying@cmu.edu.cn.
Yang Li, Email: yangli@cmu.edu.cn.
Xinmiao Yu, Email: xmyu@cmu.edu.cn.
Feng Jin, Email: jinfeng@cmu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12967-020-02497-4.
References
- 1.DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, et al. Breast cancer statistics, 2019. CA Cancer J Clin. 2019;69(6):438–451. doi: 10.3322/caac.21583. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Wicha MS. Cancer stem cell heterogeneity in hereditary breast cancer. Breast Cancer Res. 2008;10(2):105. doi: 10.1186/bcr1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13(11):674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 6.Peitzsch C, Tyutyunnykova A, Pantel K, Dubrovska A. Cancer stem cells: the root of tumor recurrence and metastases. Semin Cancer Biol. 2017;44:10–24. doi: 10.1016/j.semcancer.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Fazal FM, Chang HY. lncRNA structure: message to the heart. Mol Cell. 2016;64(1):1–2. doi: 10.1016/j.molcel.2016.09.030. [DOI] [PubMed] [Google Scholar]
- 8.Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet. 2018;50(12):1705–1715. doi: 10.1038/s41588-018-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang F, Fang E, Mei H, Chen Y, Li H, Li D, et al. Cis-Acting circ-CTNNB1 promotes beta-catenin signaling and cancer progression via DDX3-mediated transactivation of YY1. Cancer Res. 2019;79(3):557–571. doi: 10.1158/0008-5472.CAN-18-1559. [DOI] [PubMed] [Google Scholar]
- 10.Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang Y, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33(4):706–20. doi: 10.1016/j.ccell.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179(5):1033–1055. doi: 10.1016/j.cell.2019.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schober P, Boer C, Schwarte LA. Correlation coefficients: appropriate use and interpretation. Anesth Analg. 2018;126:1763–1768. doi: 10.1213/ANE.0000000000002864. [DOI] [PubMed] [Google Scholar]
- 13.Kim S, Kang D, Huo Z, Park Y, Tseng GC. Meta-analytic principal component analysis in integrative omics application. Bioinformatics. 2018;34:1321–1328. doi: 10.1093/bioinformatics/btx765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Safo SE, Long Q. Incorporating biological information in sparse principal component analysis with application to genomic data. BMC Bioinform. 2017;18:332. doi: 10.1186/s12859-017-1740-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 17.Wang R, Dong HX, Zeng J, Pan J, Jin XY. LncRNA DGCR5 contributes to CSC-like properties via modulating miR-330-5p/CD44 in NSCLC. J Cell Physiol. 2018;233(9):7447–7456. doi: 10.1002/jcp.26590. [DOI] [PubMed] [Google Scholar]
- 18.Zhu P, Wu J, Wang Y, Zhu X, Lu T, Liu B, et al. LncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat Cell Biol. 2018;20(10):1134–1144. doi: 10.1038/s41556-018-0194-0. [DOI] [PubMed] [Google Scholar]
- 19.Zhao X, Sun B, Liu T, Shao B, Sun R, Zhu D, et al. Long noncoding RNA n339260 promotes vasculogenic mimicry and cancer stem cell development in hepatocellular carcinoma. Cancer Sci. 2018;109(10):3197–3208. doi: 10.1111/cas.13740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou M, Hou Y, Yang G, Zhang H, Tu G, Du YE, et al. LncRNA-Hh strengthen cancer stem cells generation in twist-positive breast cancer via activation of hedgehog signaling pathway. Stem Cells. 2016;34(1):55–66. doi: 10.1002/stem.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Sun L, Zhang Y, Lu G, Li Y, Wei Z. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J Cell Physiol. 2018;233(11):8630–8. doi: 10.1002/jcp.26611. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32(11):2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Li C, Sun Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am J Transl Res. 2018;10(8):2648–2658. [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Zhao H, Xu Y, Li J, Deng C, Deng Y, et al. Systematic identification of lincRNA-based prognostic biomarkers by integrating lincRNA expression and copy number variation in lung adenocarcinoma. Int J Cancer. 2019;144(7):1723–1734. doi: 10.1002/ijc.31865. [DOI] [PubMed] [Google Scholar]
- 25.Qiu X, Dong J, Zhao Z, Li J, Cai X. LncRNA LINC00668 promotes the progression of breast cancer by inhibiting apoptosis and accelerating cell cycle. Onco Targets Ther. 2019;12:5615–5625. doi: 10.2147/OTT.S188933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan S, Yue Y, Wang J, Li W, Sun M, Gu C, et al. LINC00668 promotes tumorigenesis and progression through sponging miR-188-5p and regulating USP47 in colorectal cancer. Eur J Pharmacol. 2019;858:172464. doi: 10.1016/j.ejphar.2019.172464. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Zhou X, Liu J, Liu Z, Zhang L, Gong Y, et al. Genomewide investigation of the clinical implications and molecular mechanism of long noncoding RNA LINC00668 and proteincoding genes in hepatocellular carcinoma. Int J Oncol. 2019;55(4):860–878. doi: 10.3892/ijo.2019.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang R, Hu C, Li Q, Cheng Z, Gu L, Li H, et al. Sodium new houttuyfonate suppresses metastasis in NSCLC cells through the Linc00668/miR-147a/slug axis. J Exp Clin Cancer Res. 2019;38(1):155. doi: 10.1186/s13046-019-1152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong Y, Li Y, Song T, Zhang D. MiR-718 mediates the indirect interaction between lncRNA SEMA3B-AS1 and PTEN to regulate the proliferation of hepatocellular carcinoma cells. Physiol Genomics. 2019;51(10):500–505. doi: 10.1152/physiolgenomics.00019.2019. [DOI] [PubMed] [Google Scholar]
- 30.Dong Z, Liang X, Wu X, Kang X, Guo Y, Shen S, et al. Promoter hypermethylation-mediated downregulation of tumor suppressor gene SEMA3B and lncRNA SEMA3B-AS1 correlates with progression and prognosis of esophageal squamous cell carcinoma. Clin Exp Metastasis. 2019;36(3):225–241. doi: 10.1007/s10585-019-09964-3. [DOI] [PubMed] [Google Scholar]
- 31.Guo W, Liang X, Liu L, Guo Y, Shen S, Liang J, et al. MiR-6872 host gene SEMA3B and its antisense lncRNA SEMA3B-AS1 function synergistically to suppress gastric cardia adenocarcinoma progression. Gastric Cancer. 2019;22(4):705–722. doi: 10.1007/s10120-019-00924-0. [DOI] [PubMed] [Google Scholar]
- 32.Yeo SK, Guan JL. Breast Cancer: Multiple Subtypes within a Tumor? Trends Cancer. 2017;3(11):753–760. doi: 10.1016/j.trecan.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 34.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24(9):2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, et al. Tailoring therapies–improving the management of early breast cancer: St Gallen international expert consensus on the primary therapy of early breast cancer 2015. Ann Oncol. 2015;26(8):1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for Cox's proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ching T, Zhu X, Garmire LX. Cox-nnet: An artificial neural network method for prognosis prediction of high-throughput omics data. PLoS Comput Biol. 2018;14:e1006076. doi: 10.1371/journal.pcbi.1006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yousefi S, Amrollahi F, Amgad M, Dong C, Lewis JE, Song C, et al. Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Sci Rep. 2017;7:11707. doi: 10.1038/s41598-017-11817-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hao J, Kim Y, Mallavarapu T, Oh JH, Kang M. Interpretable deep neural network for cancer survival analysis by integrating genomic and clinical data. BMC Med Genomics. 2019;12:189. doi: 10.1186/s12920-019-0624-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. The list of 1198 BCSC-related lncRNAs.
Additional file 2. The list of 213 BCSC-related mRNAs.
Additional file 3: Fig. S1. Protein-protein interaction network of the 213 BCSC-related encoding genes (mRNAs).
Data Availability Statement
All data utilized in this study are included in this article and all data supporting the findings of this study are available on reasonable request from the corresponding author.