Abstract
Since the first case of COVID‐19 reported in late December of 2019 in Wuhan, China, the SARS‐CoV‐2 virus has caused approximately 20 million infections and 732 thousand deaths around the world by 11 August 2020. Although the pathogen generally infects the respiratory system, whether it is present in the bloodstream and whether it poses a threat to the blood supply during the period of the outbreak is of serious public concern. In this study, we used enzyme‐linked immunosorbent assay (ELISA) to screen total antibodies against SARS‐CoV‐2 in 2199 blood donors, who had donated blood at the Guangzhou Blood Center during the epidemic. The Ig‐reactive samples were further characterized for IgA, IgG, and IgM subtypes by ELISA and viral nucleic acid by real‐time polymerase chain reaction. Among the 2199 plasma samples, seven were reactive under total antibodies' screening. Further testing revealed that none of them had detectable viral nucleic acid or IgM antibody, but two samples contained IgA and IgG. The IgG antibody titers of both positive samples were 1:16 and 1:4, respectively. Our results indicated a low prevalence of past SARS‐CoV‐2 infection in our blood donors, as none of the tests were positive for viral nucleic acid and only 2 out of 2199 (0.09%) of samples were positive for IgG and IgA. There would be a limited necessity for the implementation of such testing in blood screening in a COVID‐19 low‐risk area.
Keywords: blood, coronavirus, epidemiology, immune responses, immunoglobulin, virus classification
Highlights
The prevalence of past SARS‐CoV‐2 infection was relatively low among voluntary blood donors in Guangzhou, China
Screening of SARS‐CoV‐2 among voluntary blood donors may not be in priority in Guangzhou, China, because of the low risk of transmission via transfusion.
1. INTRODUCTION
Coronavirus disease 2019 (COVID‐19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). The most commonly reported clinical symptoms include fever, cough, and fatigue. Other symptoms such as nasal congestion, rhinorrhea, sore throat, muscle pain, and diarrhea are less common. 1 Although the majority of cases result in mild symptoms, some progress to viral pneumonia and multiorgan failure. 2
SARS‐CoV‐2 is an enveloped virus in the genus of the beta‐coronavirus, but it is significantly different from SARS‐CoV and MERS‐CoV. 3 , 4 Until now, there has been no solid evidence indicating that such respiratory viruses can be transmitted by blood transfusion. 5 , 6 However, considering the incubation period of SARS‐CoV‐2 infections (median, 5.2 days) and the fact that some can be asymptomatic, blood safety remains a general concern. 7 , 8 , 9 , 10 Currently, the diagnosis of COVID‐19 mainly relies on the laboratory detection of SARS‐CoV‐2 RNA in throat swab samples and clinical diagnosis is supported by pulmonary computed tomography. 11 The positive rate of SARS‐CoV‐2 RNA found in blood samples of clinically confirmed patients is still relatively low (15%‐20%). 1 , 12 A recent study recruited blood donors (BDs) in Wuhan city of China between 25 January and 4 March 2020. The results showed 4 out of 2430 BDs (0.16%) to be positive for SARS‐CoV‐2 RNA. 13 This was during the peak of the SARS‐CoV‐2 outbreak in Wuhan, with approximately 50 000 confirmed cases. Although the blood products have not been transfused to clinical patients, the possibility of transfusion‐associated SARS‐CoV‐2 transmission in areas with an intense epidemic cannot be excluded.
The first COVID‐19 case in Guangzhou, a city of South China, was diagnosed on 22 January 2020. A total of 499 cases had been confirmed by 17 April. To study the status of SARS‐CoV‐2 infection among BDs in Guangzhou and evaluate the risk of transfusion transmission, we conducted tests on the antibodies against the virus supported by epidemiological evaluation. Preliminary studies indicated that antibody testing was suitable for serosurvey of blood samples of individuals who had been exposed to SARS‐CoV‐2. 14 In this study, after total antibodies screening, three different antibody Ig subtypes (IgM, IgA, and IgG) and viral nucleic acid were assessed to indicate past and active COVID‐19 infection.
2. MATERIALS AND METHODS
2.1. Study participants
The study participants consisted of 2199 voluntary BDs, who were randomly selected from 23 March through 2 April 2020. Among them, 1489 were male and 710 were female. The median age was 34 years old (ranging from 18 to 59 years old). Before donation, the BDs were required to meet the defined criteria of whole blood and platelet apheresis donation. During the COVID‐19 outbreak, BDs were required to answer an additional questionnaire on whether in the past 28 days they (a) had close contact with confirmed or suspected cases of COVID‐19; (b) had traveled in areas with active COVID‐19 epidemic; (c) had such symptoms as fever, cough, fatigue, sore throat, muscle pain or diarrhea. In addition, the BDs were tested for body temperature. For BDs who were positive for SARS‐CoV‐2 nucleic acid or antibody testing, an epidemiological survey was conducted, and their family members and close contacts were given the same tests.
For each participant, 10 mL of whole blood sample was drawn for the tests.
2.2. Testing of antibodies by enzyme‐linked immunosorbent assays
Total antibodies against SARS‐CoV‐2 were screened using an enzyme‐linked immunosorbent assay (ELISA) developed by Wantai Biological Pharmacy Enterprise (WT; Beijing, China). Total antibody‐reactive samples were further tested for IgA and IgM by individual assay (WT), and IgG by two independent reagents by WT and Lizhu Diagnostics (LZ; Zhuhai, China), respectively.
2.3. Titration of anti‐SARS‐CoV‐2 IgG antibody
Titration of anti‐SARS‐CoV‐2 IgG antibody was performed using the ELISA reagent (WT) based on a doubling dilution method.
2.4. Nucleic acid testing of SARS‐CoV‐2
The nucleic acid of SARS‐CoV‐2 was tested using the assay from Sansure Biotechnology (Hunan, China). Viral RNA was extracted from 200μL plasma using magnetic beads, followed by reverse transcription into complementary DNA (cDNA) at 50°C for 30 minutes. The cDNA was then used for real‐time polymerase chain reaction (PCR) amplification under the following conditions: 95°C for 1 minute, followed by 45 cycles of 95°C for 15 seconds and then 60°C for 30 seconds. A C t value lower than 40 was considered to be positive. The analytical sensitivity of the assay was 200 copies/mL.
3. RESULTS
3.1. Frequency of nucleic acid and antibodies against SARS‐CoV‐2 in BDs
Among the total cohort, seven donors were reactive for total antibodies against SARS‐CoV‐2. Further tests revealed that all seven individuals were negative for viral nucleic acid and IgM antibody (Table 1). However, only two samples (BD2 and BD5) developed IgG together with IgA antibody subtypes (0.09%; 2 out of 2199). Notably, the presence of the IgG antibody was confirmed by two assays using the receptor‐binding domain (RBD) of the spike protein (S‐protein) and the recombinant nucleocapsid protein (N‐protein) as antigens, respectively. The other five total antibody‐reactive samples that were negative for IgA and IgG tests were likely to be false positive.
Table 1.
Epidemiological information and testing results of the donors reactive for total antibodies against SARS‐CoV‐2
| ID | Sex | Age | Ethnicity | Occupation | Total antibodies (S/CO) | IgM | IgA | IgG (WT) | IgG (LZ) | Nucleic acid |
|---|---|---|---|---|---|---|---|---|---|---|
| BD1 | Male | 45 | Han | Public officer | +(8.15) | − | − | − | − | − |
| BD2 | Male | 29 | Han | Catering worker | +(18.0) | − | + | + | + | − |
| BD3 | Male | 31 | Han | Clerk | +(2.52) | − | − | − | − | − |
| BD4 | Female | 26 | Han | Clerk | +(1.92) | − | − | − | − | − |
| BD5 | Female | 40 | Han | Nurse | +(18.0) | − | + | + | + | − |
| BD6 | Male | 43 | Han | Public officer | +(1.21) | − | − | − | − | − |
| BD7 | Male | 45 | Han | Public officer | +(9.06) | − | − | − | − | − |
Note: The donors who were positive for IgA and IgG antibody are indicated in bold.
Abbreviations: Ig, immunoglobulin; LZ, Lizhu Diagnostics; WT, Wantai Biological Pharmacy Enterprise.
3.2. Titer of the IgG antibody against SARS‐CoV‐2
As there is currently no quantitative assay available for the SARS‐CoV‐2 antibodies, the IgG antibody was titrated using the IgG ELISA assay (WT) through a doubling dilution procedure. The S/CO of the two IgG‐positive samples (BD2 and BD5) declined in a dose‐dependent manner along with titration. The titers for BD2 and BD5 were 1:16 and 1:4, respectively (Figure 1).
Figure 1.
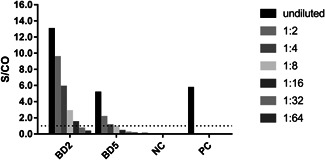
Titration of IgG antibody against SARS‐CoV‐2. The plasma samples of the two IgG‐positive donors (BD2 and BD5) were doubling diluted and then tested with anti‐SARS‐CoV‐2 IgG ELISA assay (WT). NC and PC served as the negative control and positive control sera for the assay and were tested without dilution, respectively. The dotted line indicates a value of 1.0 for the signal to cut‐off (S/CO)
3.3. Epidemiological survey
Neither BD2 and BD5 had any history of close contact with COVID‐19 patients or travel in the epidemic regions. However, BD5 worked as a nurse, and she reported a history of SARS infection in 2003. Nevertheless, her four family members tested negative for all the SARS‐CoV‐2 tests.
4. DISCUSSION
Currently, little is known about the prevalence of SARS‐CoV‐2 in populations other than COVID‐19 patients. The prevalence of antibodies against the virus in healthcare workers, who have been recognized as a high‐risk group, has been reported to be as high as 3.8% in China 15 and 1.6% in Germany. 16 Low‐risk populations such as voluntary BDs have been tested for SARS‐CoV‐2 antibodies in France (3%), 17 Italy (5.1%), 18 Denmark (1.8%), 19 and Jordan (0%). 20 In this study, although seven samples were reactive to the initial total antibodies test, no asymptomatic active COVID‐19 infection was found because all seven BDs reactive for total antibodies were negative for the viral nucleic acid and IgM antibody against anti‐SARS‐CoV‐2. In addition, only two samples (0.09%) were positive for IgA and IgG antibody tests. We speculated that the presence of antibodies in both donors (BD2 and BD5) were reliable because they were confirmed by one IgA assay and two IgG assays that coated the RBD of the S‐protein and N‐protein, respectively. 21 The other five total antibody‐reactive samples were likely to be false positive, even if the sensitivity and specificity of the ELISA assays were not clearly elucidated. The sampling time was also an influencing factor responsible for the low prevalence because the antibody against SARS‐CoV‐2 was hardly produced in the early stage of the disease and become detectable later around day 10.
The sampling time may also be responsible for the low antibody titer (1:4 and 1:16) in our BDs. It has been reported that the antibody level was relatively low (<1:100) in patients with mild clinical signs but high (>1:800) in patients with severe SARS. 22 Theoretically, the low antibody titer (1:4 and 1:16) in our BDs may relate to the stage of infection. In the early stage of the infection, the studied individuals might have been asymptomatic and generated a low level of IgG. Naturally, the level of antibody could peak at around 10 days after infection and then maintain but gradually decrease over time. Previous studies have established that level of IgG antibody against SARS‐CoV remain relatively high after infection, and they do not decline significantly for 4 years. 23 , 24 , 25 Although the exact time of infection for the BDs was unknown, the low level of antibody titers found here cannot be attributed to the fading over time. However, long‐term study regarding the retention time of anti‐SARS‐CoV‐2 antibodies is still lacking. Furthermore, the same ELISA tests were performed with plasma samples from convalescent COVID‐19 patients recruited in the same institution. The IgG antibody against SARS‐CoV‐2 was detected in only 10 out of 15 samples (data not shown). Further study is required to find out whether the low detection rate was due to the limitation of sensitivity of the assays or whether the antibodies were actually absent from some of these patients.
SARS‐CoV‐2 RNA can be detected by real‐time PCR as early as 1 to 3 days before the clinical presentation of the disease. In contrast, antibody production typically occurs between 7 and 11 days after exposure, which varies with different infected individuals. Recently, Zhao et al 26 reported the median seroconversion time of IgM and IgG against SARS‐CoV‐2 to be 12 and 14 days, respectively. The IgM level declined significantly in 35 days after the onset of disease, and less than 50% of the infected cases can be detected. 15 In this study, IgG but not IgM or viral RNA was found in two donors, indicating past infection of SARS‐CoV‐2.
The prevalence of SARS‐CoV‐2 RNA in our BDs may have been underestimated for the following reasons. First, donors with COVID‐19‐like symptoms or a history of travel or contact with COVID‐19 patients were deferred by a pre‐donation questionnaire. Second, total antibodies were initially screened, and reactive samples were then used for the nucleic acid testing (NAT), which may result in missing window period samples. These samples were positive for viral RNA but negative for antibody (Ab) because the Ab had not yet been generated due to the early stage of the infection. Lastly, the analytical sensitivity of NAT assay used in this study (Sansure) was 200 copies/mL, which was less than that of cobas SARS‐CoV‐2 (Roche; 46 copies/mL). However, in a very recent multicenter report, Chang et al 27 used a NAT assay with high sensitivity (minipool: 62.94 and 33.14 copies/mL for N and ORF1ab regions; individual test: 3.87 and 4.85 copies/mL for two regions using 1600 μL of plasma), but no SARS‐CoV‐2 RNA was found among 98 342 BDs in Hubei, China, where there was an intense epidemic. Taken together, despite some limitations of the assay and strategy, almost no SARS‐COVID‐19 RNA was found in BDs in Guangzhou. A much larger epidemiological study is required to evaluate the risk of transfusion transmission of COVID‐19.
The epidemiological survey revealed neither of the IgG‐positive donors had a history of close contact with COVID‐19 patients or traveling to a highly epidemic region. BD2 was a migrant and lives alone in Guangzhou, and was healthy and had not contracted SARS in the past, but may have had contact with wild animals. The other donor, BD5, was a nurse who had contracted SARS in 2003, but none of her four family members was positive for all the SARS‐CoV‐2 tests (data not shown). Accordingly, the antibodies found in BD5 may have been produced in response to a past SARS infection. As no archived plasma samples were available for these donors, this possibility could not be ruled out. Nevertheless, in many of these cases, the antibodies against SARS were undetectable 5 years after infection. 18
In summary, a low prevalence of SARS‐CoV‐2 past infection was found among BDs in Guangzhou, China, indicating that the risk of transmission of SARS‐CoV‐2 by blood transfusion is relatively low in the city and thus there would be a limited necessity for the implementation of such testing in blood screening.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
RX and JH drafted the main manuscript. RX, CD, QL, ZS, MW, and XR contributed to donor recruitment, tests, and data collection. CL revised the manuscript. HW and YF conceived the study and approved the manuscript. All authors reviewed and approved the final version of the manuscript.
ETHICS STATEMENT
This study protocol was reviewed and approved by the Medical Ethics Committee of Guangzhou Blood Center, Guangzhou, Guangdong, China. The participants have provided written informed consent before enrollment. The current study conformed to the ethical guidelines of the 1975 Declaration of Helsinki.
ACKNOWLEDGMENTS
The authors would like to acknowledge Dr Ling Lu (Third Affiliated Hospital of Sun Yat‐Sen University, Guangzhou, China) and Dr Sentot Santoso (Justus‐Liebig University, Giessen, Germany) for the suggestions on manuscript writing and English language editing, and also Dr Shengxiang Ge (Xiamen University, Xiamen, China) and Yourong Zheng (Guangzhou Blood Center, Guangzhou, China) for the assistance with antibody testing. This study was supported by the grants from the National Science and Technology Major Project of China (No. 2018ZX10302205‐001). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Xu R, Huang J, Duan C, et al. Low prevalence of antibodies against SARS‐CoV‐2 among voluntary blood donors in Guangzhou, China. J Med Virol. 2021;93:1743–1747. 10.1002/jmv.26445
Ru Xu, Jieting Huang, and Chaohui Duan contributed equally to this study.
Contributor Information
Yongshui Fu, Email: fuyongshui1969@yahoo.com.
Hao Wang, Email: 479053321@qq.com.
REFERENCES
- 1. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xie M, Chen Q. Insight into 2019 novel coronavirus—an updated interim review and lessons from SARS‐CoV and MERS‐CoV. Int J Infect Dis. 2020;94:119‐124. 10.1016/j.ijid.2020.03.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. World Health Organization . WHO Recommendations on SARS and Blood Safety; 2003. https://www.who.int/csr/sars/guidelines/bloodsafety/en/. Accessed 4 May 2020.
- 6. Eickmann M, Gravemann U, Handke W, et al. Inactivation of Ebola virus and Middle East respiratory syndrome coronavirus in platelet concentrates and plasma by ultraviolet C light and methylene blue plus visible light, respectively. Transfusion. 2018;58(9):2202‐2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu X, Yang R. COVID‐19 transmission through asymptomatic carriers is a challenge to containment. Influenza Other Respir Viruses. 2020;14(4):474‐475. 10.1111/irv.12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019‐nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382(10):970‐971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020;382(13):1199‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang L, Yan Y, Wang L. Coronavirus disease 2019: coronaviruses and blood safety. Transfus Med Rev. 2020;34(2):75‐80. 10.1016/j.tmrv.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adhikari SP, Meng S, Wu YJ, et al. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID‐19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chang L, Zhao L, Gong H, Wang L, Wang L. Severe acute respiratory syndrome coronavirus 2 RNA detected in blood donations. Emerg Infect Dis. 2020. 2020;26(7):1631‐1633. 10.3201/eid2607.200839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jin Y, Wang M, Zuo Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. 2020;94:49‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu X, Sun J, Nie S, et al. Seroprevalence of immunoglobulin M and G antibodies against SARS‐CoV‐2 in China. Nat Med. 2020;26(8):1193‐1195. 10.1038/s41591-020-0949-6 [DOI] [PubMed] [Google Scholar]
- 16. Korth J, Wilde B, Dolff S, et al. SARS‐CoV‐2‐specific antibody detection in healthcare workers in Germany with direct contact to COVID‐19 patients. J Clin Virol. 2020;128:104437. 10.1016/j.jcv.2020.104437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grzelak L, Temmam S, Planchais C, et al. SARS‐CoV‐2 serological analysis of COVID‐19 hospitalized patients, pauci‐symptomatic individuals and blood donors [preprint]. medRxiv. 2020. 10.1101/2020.04.21.20068858 [DOI] [Google Scholar]
- 18. Valenti L, Bergna A, Pelusi S, et al. SARS‐CoV‐2 seroprevalence trends in healthy blood donors during the COVID‐19 Milan outbreak [preprint]. medRxiv. 2020; 10.1101/2020.05.11.20098442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Erikstrup C, Hother CE, Pedersen OBV, et al. Estimation of SARS‐CoV‐2 infection fatality rate by real‐time antibody screening of blood donors [published online ahead of print, 25 June 2020]. Clin Infect Dis. 2020:ciaa849. 10.1093/cid/ciaa849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sughayer MA, Mansour A, Al Nuirat A, Souan L, Ghanem M, Siag M. Covid‐19 seroprevalence rate in healthy blood donors from a community under strict lockdown measures [preprint]. medRxiv. 2020. 10.1101/2020.06.06.20123919 [DOI] [Google Scholar]
- 21. Liu W, Liu L, Kou G, et al. Evaluation of nucleocapsid and spike protein‐based enzyme‐linked immunosorbent assays for detecting antibodies against SARS‐CoV‐2. J Clin Microbiol. 2020;58(6):e00461‐20. 10.1128/JCM.00461-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang WT, Kao CL, Chung MY, et al. SARS exposure and emergency department workers. Emerg Infect Dis. 2004;10(6):1117‐1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie SY, Zeng G, Xia SC, et al. A three‐year follow‐up study on sera specific antibody in severe acute respiratory syndrome cases after the onset of illness. Zhonghua Liu Xing Bing Xue Za Zhi. 2007;28(4):343‐345. [PubMed] [Google Scholar]
- 24. Wang HJ, Zhang LL, Tan WJ, et al. Consecutive five‐year follow‐up analysis of specific IgG antibody of 22 cases of SARS patients after recovery. Bing Du Xue Bao. 2010;26(4):295‐297. [PubMed] [Google Scholar]
- 25. Li T, Xie J, He Y, et al. Long‐term persistence of robust antibody and cytotoxic T cell responses in recovered patients infected with SARS coronavirus. PLOS One. 2006;1(1):e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019 [published online ahead of print, 28 March 2020]. Clin Infect Dis. 2020:ciaa344. 10.1093/cid/ciaa344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang L, Yan Y, Zhao L, et al. No evidence of SARS‐CoV‐2 RNA among blood donors: a multicenter study in Hubei, China [published online ahead of print, 14 July 2020]. Transfusion. 2020. 10.1111/trf.15943 [DOI] [PMC free article] [PubMed] [Google Scholar]


