Abstract
The lysosomal membrane protein Niemann‐Pick type C1 (NPC1) and Niemann‐Pick type C2 (NPC2) are main players of cholesterol control in the lysosome and it is known that the mutation on these proteins leads to the cholesterol trafficking‐related neurodegenerative disease, which is called the NPC disease. The mutation R518W or R518Q on the NPC1 is one of the type of disease‐related mutation that causes cholesterol transports to be cut in half, which results in the accumulation of cholesterol and lipids in the late endosomal/lysosomal compartment of the cell. Even though there has been significant progress with understanding the cholesterol transport by NPC1 in combination with NPC2, especially after the structural determination of the full‐length NPC1 in 2016, many details such as the interaction of the full‐length NPC1 with the NPC2, the molecular motions responsible for the cholesterol transport during and after this interaction, and the structure and the function relations of many mutations are still not well understood. In this study, we report the extensive molecular dynamics simulations in order to gain insight into the structure and the dynamics of NPC1 lumenal domain for the cholesterol transport and the disease behind the mutation (R518W). It was found that the mutation induces a structural shift of the N‐terminal domain, toward the loop region in the middle lumenal domain, which is believed to play a central role in the interaction with NPC2 protein, so the interaction with the NPC2 protein might be less favorable compared to the wild NPC1. Also, the simulation indicates the possible re‐orientation of the N‐terminal domain with both the wild and the R518W‐mutated NPC1 after receiving the cholesterol from the NPC2 that align to form an internal tunnel, which is a possible pose for further action in cholesterol trafficking. We believe the current study can provide a better understanding of the cholesterol transport by NPC1 especially the role of NTD of NPC1 in combination with NPC2 interactions.

Keywords: cholesterol trafficking, COVID‐19, molecular dynamics simulation, neurodegenerative Niemann‐Pick C disease, NPC1, NPC2, R518W mutant
We have performed a series of molecular dynamics simulation with lumenal domain Niemann‐Pick type C1 (NPC1) protein, which is one of the key protein that controls the cholesterol transport in lysosome, to investigate the dynamic role of N‐terminal domain (NTD). The simulation shows that the NTD makes proper orientation change for cholesterol trafficking depending on the environment. The same simulation with R518W‐mutated NPC1 shows possible reduced interaction between NPC1 and NPC2, which works in tandem with NPC1. The current modeling study provides insight into the structure and function relation of NPC1, including the disease causing R518W mutation in NPC1.

Abbreviations
- CTD
C‐terminal lumenal domain
- MD
molecular dynamics
- MLD
middle lumenal domain
- NPC
Niemann‐Pick type C
- NPC1
Niemann‐Pick type C1
- NPC2
Niemann‐Pick type C2
- NTD
N‐terminal domain
- TMD
transmembrane domain
1. INTRODUCTION
The cholesterol homeostasis is maintained by many different proteins depending on the tissues in the body (Luo, Yang, & Song, 2019). The blood cholesterol levels are regulated by several processes, which include bio‐synthesis, cholesterol absorption/re‐absorption, and biliary clearance and excretion (Grundy, 1983; Y Litvinov, V. Savushkin, A. Garaeva, & D. Dergunov, 2016). Elevated blood cholesterol levels contribute to atherosclerotic coronary heart disease (Goldstein & Brown, 1990; Horton, Goldstein, & Brown, 2002). Previous studies have shown that lowering the levels of plasma cholesterol significantly reduces the risk of cardiovascular diseases associated with diabetes mellitus even in diabetic patients with normal levels of plasma cholesterol (Malhotra, Gill, Dudeja, & Alrefai, 2016). Therefore, control of the cholesterol for therapeutic purposes regarding diseases such as cardiovascular disease has a fundamental importance. There has been a tremendous amount of effort to understand how different organ systems such as the liver and the intestine coordinate their cellular mechanisms to control the body cholesterol homeostasis even though the same process in the brain is less explored (Luo et al., 2019; Malhotra et al., 2016).
The transmembrane protein Niemann‐Pick type C1 (NPC1) inside the lysosome is one of the key players in the cholesterol transport, which is also mediated by the Niemann‐Pick type C2 (NPC2) (Betters & Yu, 2010; Wang et al., 2010). The NPC1 facilitates the transport of the low‐density lipoprotein (LDL)‐derived cholesterol out of the lysosomes for the subsequent delivery to the endoplasmic reticulum and the plasma membrane. The NPC1 functions mostly in tandem with NPC2, which is a soluble lysosomal protein, in order to move the unesterified cholesterol (Kwon et al., 2009). The abnormal function of NPC1 as result of the mutation and the environment change causes the accumulation of the cholesterol within the lysosome, which leads to a neurodegenerative disease called the NPC disease. Not only the crystal structures of N‐terminal lumenal domain (NTD) of NPC1 with and without cholesterol (PDB id: 3GKI and 3GKH) (Kwon et al., 2009), as well as the cryo‐EM (electron microscope) of the full‐length NPC1 with and without cholesterol in NTD (PDB id: 3JD8 and 5JNX) were reported (Gong et al., 2016). The NPC1 glycoprotein has 13 helixes in the transmembrane domains (TMD) and three relatively large, lumenally oriented domains. The N‐terminal lumenal domain (NTD) contains a cholesterol binding pocket, but the newly determined structure of the cysteine‐rich C‐terminal domain (CTD) contains a loop region that is close to the NTD, which indicates the importance of this region in the cholesterol transport in conjunction with maintaining the possible orientation of the NTD to receive cholesterol from the NPC2 (Li et al., 2017). It is believed that the soluble NPC2 accepts the LDL cholesterol and delivers it to the NTD directly, which is also referred to as the so‐called hands‐off mechanism (Deffieu & Pfeffer, 2011). It is known that the two protruding middle lumenal domain (MLD) loops in the NPC1 interacts with the NPC2. Interestingly, these loops interact with the glycoprotein of the Ebola virus also and both the structure of the MLD with the Ebola glycoprotein and the MLD with the NPC2 has been determined (Gong et al., 2016; Infante, Abi‐Mosleh, et al., 2008; Infante, Radhakrishnan, et al., 2008; Li, Saha, Li, Blobel, & Pfeffer, 2016; Wang et al., 2016). In fact, a study of the NPC1 has importance as a mediator of the cholesterol transport as well as a mediator of coronavirus such as SARS and the Ebola virus. Especially, a lot of attention has been paid to the NPC1 as a one of the possible target proteins to mimic the NPC disease in relation to the recent pandemic outbreak of the COVID‐19 virus because the inhibition of this protein could reduce the replication of a coronavirus, which includes the COVID‐19 virus (Ballout, Sviridov, Bukrinsky, & Remaley, 2020; Sturley et al., 2020).
There have been several computational modeling studies regarding the cholesterol transport through the NPC1 that complement the experimental results. Previously, two working models of the NPC1 (NTD) in complex with the NPC2 were proposed. One is based on the surface residue identification for the interaction between NPC1 (NTD) and NPC2 (Kwon et al., 2009; Wang et al., 2010), and the other is based on the X‐ray structure of NPC2 in complex with the NPC1‐MLD region (Li, Saha, et al., 2016). The NPC1(NTD)–NPC2 interfaces proposed by Brown & Goldstein and Li & Pfeffer, which are coined the “Texas” and the “California” models by Elghobashi‐Meinhardt (Hodošček & Elghobashi‐Meinhardt, 2018), differ with the alignment of the residues. The detailed sliding like hands‐off cholesterol transport especially the conformational change in the cholesterol during the transport from NPC2 to NTD was reported with the QM/MM study within a fixed NPC2/NTD conformational framework (Elghobashi‐Meinhardt, 2014). From the molecular dynamics simulations it was found that the “Texas” model has favorable interface between NPC2 and NTD in NPC2/NTD complex when the cholesterol is in NPC2 side while the same complex dissociates when the cholesterol is in NTD side or there was no cholesterol on both sides (Estiu, Khatri, & Wiest, 2013; Hodošček & Elghobashi‐Meinhardt, 2018). On the other hand, the “California” model showed the opposite behaviors. Based on these observations, it was suggested that the “Texas” model may corresponds to the initial structure for the cholesterol transport from NPC2 to NTD, while the “California” model corresponds to the structure after the cholesterol transport (Hodošček & Elghobashi‐Meinhardt, 2018).
Even though there has been significant progress regarding understanding the dietary cholesterol exchange in a cellular environment in connection with the NPC1 especially after the structural determination of the full‐length NPC1, many details, such as the interaction of the full‐length NPC1 with the NPC2 and the structural change in the full NPC1 as the cholesterol is passed from one place to other places, have still not been directly observed. The location or the orientation of NTD presented in the full‐length NPC1 structure might not be the active form, and there the NTD orientation/location from the full‐length NPC1 structure could have changed during the cholesterol transfer from NPC2 for proper alignment (Gong et al., 2016; Li et al., 2017; Pfeffer, 2019). Eventually, it is believed that once the cholesterol has been transferred to NTD from NPC2, it could be delivered to the sterol sensing domain (SSD) (Li, Wang, et al., 2016; Ohgami et al., 2004; Wheeler, Schmid, & Sillence, 2019), which is in between the helix bundle within the membrane region, and it could possibly trigger a sequence of events once it is transferred there (Pfeffer 2016).
The transfer of the cholesterol from NTD to SSD could proceed possibly through a conduit‐like channel that was observed in the Patched protein by the proton‐driven network (Winkler et al., 2019; Zhang et al., 2018). Note that the modeling study with the disease‐causing L472P mutation indicates a breakdown of this tunnel in NPC1 (Vanharanta et al., 2020), which emphasizes the importance of this tunnel with the cholesterol transport. The recent cryo‐EM structure of NPC1 with NPC1‐blocker itraconazole shows the blocker is located in this tunnel, which supports this scheme (Long et al., 2020). In this context, the possible cholesterol binding sites have been recently suggested through the modeling study (Elghobashi‐Meinhardt, 2019). We note that the channel was observed in a short molecular dynamics simulation with no cholesterol in NTD (Elghobashi‐Meinhardt, 2020). Interestingly, the very recent molecular dynamics study of wild NPC1 with the cholesterol in itraconazole binding site indicates the migration of the cholesterol laterally toward the bilayer direction unlike the mutation P691S, which migrates away from SSD, suggesting that cholesterol might be inserted into bilayer, although this lateral motion for cholesterol transport out of the lysosome (Elghobashi‐Meinhardt, 2020). There is a possibility that the cholesterol is transferred from NTD of neighboring NPC1 such as the inter‐protein transfer (Trinh, Brown, Seemann, Goldstein, & Lu, 2018). In either case, there should be some structural re‐arrangement of the NPC1 protein after the cholesterol is transferred to NTD from NPC2, which could possibly include significant amounts of the NTD being re‐orientated or translocated for further actions (Elghobashi‐Meinhardt, 2019; Li, Saha, et al., 2016).
The mutational study, both theoretically and experimentally, may provide the opportunities to directly or indirectly gain insight into the mechanisms of the NPC1 cholesterol transfer process. Since the first discovery of the mutation on NPC1 protein and its connection to NPC‐related diseases (Yamamoto, Nanba, & Ninomiya, 1999), numerous mutations either on NPC1 or NPC2 have been found additionally (Li et al., 2017; Park et al., 2003). The point mutation R518W (or R518Q) is one example (Millat et al., 2001; Park et al., 2003; Yamamoto et al., 1999) and it has been reported that the cholesterol transfer activity is reduced by 50% by this mutation (Gong et al., 2016). At the same time, it was found that the binding affinity of the NPC2 to the full‐length NPC1 is noticeably reduced (Deffieu & Pfeffer, 2011; Gong et al., 2016). The transfer efficiency was maximized under pH 5.5 (Deffieu & Pfeffer, 2011; Infante, Wang, et al., 2008). From experiments, it has been reported that the interaction between NPC2 and NPC1 has two aspects that include the cholesterol‐independent weak interactions and the cholesterol depending strong interactions (Deffieu & Pfeffer, 2011). After that, it was found that this cholesterol‐dependent binding affinity is as a result of the structural difference in the NPC2 near the MLD loops binding region depending on the presence of cholesterol (Li, Saha, et al., 2016). The computational study of the point mutations I1061T, P1007A, and G992W on the CTD shows the structural instability of the mutated NPC1, especially the instabilities in NTD, which suggest the importance of a correct orientation or the stability of NTD in NPC1 (Martínez‐Archundia, Hernández Mojica, Correa‐Basurto, Montaño, & Camacho‐Molina, 2019). On the other hand, the computer simulation with the NTD protein and its two mutations, which include the Q92R and the Q92S, show the importance of the correct electrostatic distribution near the entrance of the cholesterol pocket as well as the structural stability for suitably bind the cholesterol to the NTD (Petukh & Zhulin, 2018). In combination with the experiment, the molecular dynamics simulation of the L472P mutation indicates a disruption of the tunnel between MLD and CTD, which is believed to play an essential role with the cholesterol transport that was previously mentioned (Vanharanta et al., 2020). Since it was pointed that the R518W mutation decreases the cholesterol transfer activity not because of misfolding of the NPC1 but because of the functional defection of the NPC1 (Deffieu & Pfeffer, 2011), the atomic‐level detailed mechanism behind this defective functionality could provide insights about the structure and the functional relationship of the NPC1, especially with its interaction with the NPC2.
In this study, we present extensive molecular dynamics simulations of the NPC1 in the presence of cholesterol and in the absence of cholesterol in the NTD in order to understand the structural and dynamical characteristics in conjunction with the R518W mutation and its effects on the overall cholesterol transport efficiency in connection with the possible re‐location or orientation of NTD and the interaction of NPC1 with NPC2 (Yamamoto et al., 1999).
2. METHODS
We built the initial full‐length NPC1 structure using an X‐ray structure (PDB ID: 5U74) as a template (Li et al., 2017). The missing NTD structure was introduced by overlapping the overall structure into the cryo‐EM structure presented by Gong et al. (PDB ID 3JD8) (Gong et al., 2016). For the simulation with the R518W mutation, we mutated the Arg518 to the Trp518 with the PyMOL (Schrodinger, 2015) by selecting the lowest steric hindrance conformer. The pH of the simulation was set to 5.5 considering the lumenal environment of the lysosome (Deffieu & Pfeffer, 2011; Elghobashi‐Meinhardt, 2019; Infante, Wang, et al., 2008). For this purpose, considering the intrinsic pKa values of the amino acids, the protonation states of every Asp, Glu, and His was obtained by PROPKA 3.1 (Li, Robertson, & Jensen, 2005). The details of the protonation states obtained from the PROPKA are listed in Table 1. The overall structure was solvated with TIP3P water, and the total charge was balanced with the addition of Na+ ions. In this study, we only considered the lumenal exposed part of the full‐length NPC1 in order to reduce the overall computational cost by imposing the position restraints on the C α atoms connected to the TM. The restraints constant was set to 1000 kJ mol−1 nm2, and its locations were Pro259, Lys392, Glu610, Tyr871, and Tyr1088. Note that we have included the Proline‐rich long strand that is connected to the NTD starting from the membrane surface in our simulation. Of course, this simplified model may not fully reflect the behavior of the full‐length NPC1 including the membrane.
Table 1.
The intrinsic pKa values of Asp, Glu, and His obtained from PROPKA for four systems we have used in this work. The pKa values less than or equal to 5.5 are shown as NA
| Amino acids | Domain | Wild | R518W | Wild w/. chol. | R518W w/. chol |
|---|---|---|---|---|---|
| Asp524 | MLD | NA | 5.58 | NA | 5.58 |
| Asp944 | CTD | 5.78 | 5.78 | 5.78 | 5.78 |
| Glu30 | NTD | NA | NA | 5.73 | 5.76 |
| Glu109 | NTD | 6.90 | 6.89 | 7.03 | 7.02 |
| Glu406 | MLD | 11.47 | 11.61 | 11.47 | 11.61 |
| Glu451 | MLD | 6.39 | 6.39 | 6.39 | 6.39 |
| Glu606 | MLD | 7.50 | 7.59 | 7.50 | 7.59 |
| His215 | NTD | 6.41 | 6.41 | 6.41 | 6.41 |
| His441 | MLD | 6.60 | 6.60 | 6.60 | 6.60 |
| His492 | MLD | 6.04 | 6.04 | 6.04 | 6.04 |
| His497 | MLD | 6.75 | 6.75 | 6.75 | 6.75 |
| His510 | MLD | 6.46 | 6.42 | 6.46 | 6.42 |
| His530 | MLD | 6.21 | 6.21 | 6.21 | 6.21 |
| His884 | CTD | 6.04 | 5.96 | 6.04 | 5.96 |
| His1016 | CTD | 5.83 | 5.85 | 5.83 | 5.85 |
| His1029 | CTD | 6.45 | 6.45 | 6.45 | 6.45 |
MLD, middle lumenal domain
CTD, C‐terminal lumenal domain
The recent molecular dynamics simulation (Elghobashi‐Meinhardt, 2020) with full NPC1 when cholesterol is present in the NPC1 inhibiting itraconazole binding site (Long et al., 2020), which is at the interface between the membrane and the lumenal region, shows there is a non‐negligible distance correlation coefficient between the TMD and the rest of the domains. Therefore, the model presented here need to be understood within a simplified framework. The initial structure we used in this study is shown in Figure 1. All the figures of the protein structure were drawn with PyMOL (Schrodinger, 2015). We also used CHARMM36 force field (Huang et al., 2017) for both the protein and the cholesterol when needed. The simulation box was constructed by setting the minimum distance from the protein edge to the simulation box as 10.0 Å. The final box size was 102.58, 110.22, and 83.82 Å along the x, y, and z direction, respectively. The periodic boundary condition was imposed along the three different directions. The non‐bonded cutoff distance was set to 12.0 Å with pair list updated every 25 time steps. The long‐range electrostatic interaction was treated with the particle‐mesh‐Ewald (PME) method with a PME order of 4 and a Fourier spacing of 1.2 Å. The initially prepared system was energy minimized with the steepest descent method, which was followed by the molecular dynamics simulation with constraints to all‐bond in protein using the LINCS algorithm for 1 ns before the production run. For the final production, the simulation was run under the constant temperature and constant pressure condition, i.e., NPT simulation. The temperature was set to 310K using the velocity rescale method with a coupling constant of 0.1 ps and the pressure was set to 1 atm using the Berendsen pressure coupling method with a coupling constant of 2.0 ps with a compressibility of 4.5x10‐5 bar‐1.
Figure 1.
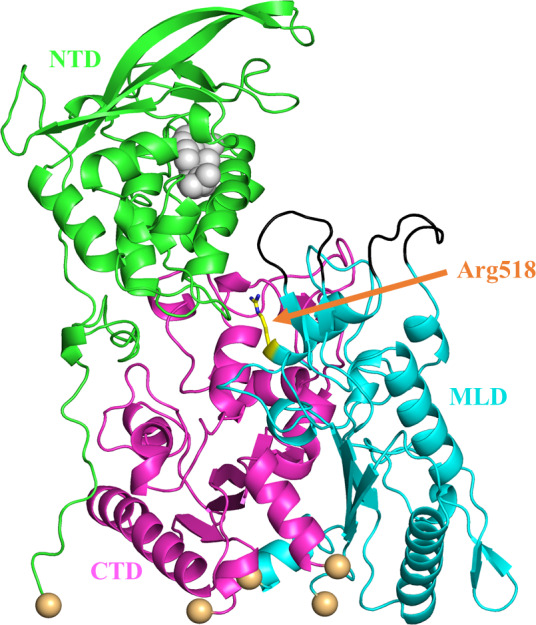
The initial structure we have prepared for simulations, including the cholesterol in the NTD. The cholesterol is shown as a gray sphere. The NTD, C‐terminal lumenal domain (CTD), and MLD domains are shown as green, magenta, and cyan, respectively. The yellow stick is the Arg518 residue which is subject to the mutation of the Trp in our simulation. The Niemann‐Pick type C2 binding loops are shown in black and the five restrained C α atoms are shown as spheres in a wheat color
The whole simulation was run using Gromacs‐2018.6 (Van Der Spoel et al., 2005). We used the leap‐frog time integrator with a time step of 4.0 fs to speed up the simulations by increasing the hydrogen atom mass by factors of 4 while keeping the total atomic mass unchanged by subtracting the same mass from the H‐bonded heavy atoms as implemented in Gromacs (Feenstra, Hess, & Berendsen, 1999; Yang, Kulkarni, Lim, & Pak, 2017). We have performed the molecular dynamics for four different systems, which included the wild NPC1, the R518W mutated NPC1, the wild NPC1 with cholesterol on the NTD, and the R518W‐mutated NPC1 with cholesterol on NTD. The reason we included a system with cholesterol in the NTD for mutated NPC1 in our simulation is we would like to trace the effects of the mutation after a successful cholesterol transfer from NPC2 to NTD because as much as half of the cholesterol is transferred to NPC1 from NPC2 according to an experiment in mutation (Gong et al., 2016). The simulation time and the total number of independent simulations are described in detail in the Results and Discussion section.
3. RESULTS
Initially, we ran eight independent wild NPC1 simulations with no cholesterol in the NTD for 200 ns. One of the simulation trajectories showed a noticeable change with the NTD tilting toward the outside of the NPC1 with a backbone RMSD of 8.7 Å compared to the initial structure. Only the heavy atoms are considered in backbone RMSD calculations. Therefore, we further proceeded increasing the simulation up to 1.0 μs with this trajectory. The rest of the trajectories essentially showed no noticeable changes from the starting structure in terms of the NTD tilting, with the RMSD of the whole system being around 6.0 Å from the starting structure. Among those remaining seven trajectories, we randomly selected two more trajectories and proceeded with the simulation up to 1.0 μs, but these two trajectories remained essentially steady in terms of the NTD angle changes relative to the other domains. The results presented here were obtained from the trajectory with the large NTD displacement mentioned above. To monitor the angle change in the NTD, we obtained the relative angles between each of the domains as a function of the simulation time, which is illustrated in Figure 2. The angles shown here are the relative angles between the long helices from each domain. It is worth noting that the RMSD of each domain obtained from the alignment of the corresponding domain only is relatively small during the simulations (less than 3.5 Å), which is shown in Figure S1. Therefore, monitoring the angles between the well‐preserved helices from each domain could be a measure of the relative change in the orientation between them. For this purpose, we defined a single vector from each helix and calculated the angles between them. For a given helix, the single vector from a helix can be obtained based on the three consecutive C α coordinates at the beginning of the helix and the three consecutive C α coordinates at the end of the helix (Kahn, 1989). The corresponding residues we have used are Ser99 ~ Thr112 in NTD, Glu575 ~ Asn593 in MLD, and Phe1051 ~ Met1069 in CTD. The direction of the axis is set from the low residue number to the high residue number. The helices we defined as well as their vectors are shown in Figure S2. Panel A shows the sudden change in angles, especially the angle between the CTD and the NTD, which started from around 100 ns and remained steady with some fluctuations after 300 ns. Compared to panel A, the same plot on panels B and C, which corresponded to the angles in those two randomly selected trajectories, showed minor angle changes. Figure 3a–c show the superposition of the simulation structures from our 1.0 μs simulation trajectory over the initial structure at 0.2, 0.5, and 1.0 μs, respectively. One can see the displacement of the NTD away from the MLD domain starting from the initial structure and the maximization of the displacement/tilting near 0.5 μs, followed by somewhat decrease displacement/tilting again near 1.0 μs. Therefore, there is a possibility that the NTD might move back and forth between the structure (A) and the structure (B), even though the current simulation is too short to observe such a behavior repeatedly. Note the direction of NTD displacement after 1.0 μs is still away from the MLD. The movie file corresponding to this trajectory can be found in the Video S1. The detailed structural information on the interaction or the complex formation of NPC2 with NTD in the presence of the full‐length NPC1 could significantly enhance our understanding of the cholesterol transfer from NPC2 to NTD. We note that the structure of putative NTD–NPC2 complex “Texas” model when there is cholesterol in the NPC2 side (Estiu et al., 2013; Hodošček & Elghobashi‐Meinhardt, 2018), generates significant structural crash when superimposed on the cryo‐EM structure, indicating that possible involvement of some structural re‐orientation or displacement of NTD to adapt the NPC2 if the “Texas” model is right. The fitting of “the Texas interface” and the “California” model into the current simulation structure after 1.0 μs in Figure 4 shows no more crashes like the crash that was previously mentioned, and a favorable interaction is possible between the full‐length NPC1 and the NPC2. Both the “Texas” interface and the “California” interface have no crash with the current simulation structure. The “California” model has no additional interactions with the full NPC1 other than the direct interface with NTD. Therefore, we presume the dissociative interaction of NPC2 with NTD when the cholesterol is in NPC2 that was observed in previous molecular dynamics simulations with NTD/NPC2 complex only remains valid (Hodošček & Elghobashi‐Meinhardt, 2018). Certainly, it would be interesting to keep track of this “Texas” interface‐based putative full‐length NPC1 + NPC2 structure with the long‐time molecular dynamics in the presence of the cholesterol. More importantly, the modeling of the full‐length NPC1 in complex with the cholesterol bound NPC2 with the addition of the NPC2 to current simulation structure by aligning the MLD of the MLD–NPC2 complex experimental structure (Li, Saha, et al., 2016) could be a good starting point to understand the detailed interaction between the NPC2 and the NTD in the full‐length NPC1. This is because there might be an initial binding of NPC2 to NPC1 MLD loops and the stable complex formation of the NPC2 with the NTD when there is cholesterol on the NPC2. The modeling study can provide information about how the NTD acts as a possible anchoring player, which was pointed out by Gong et al. (Gong et al., 2016).
Figure 2.
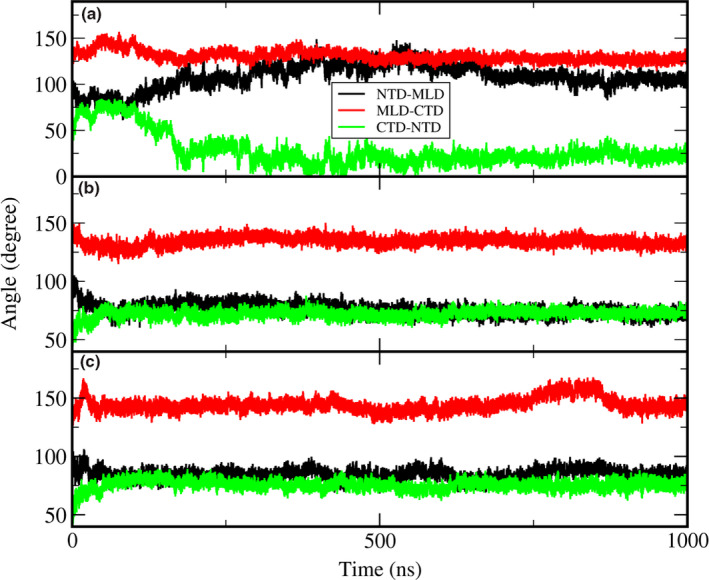
The time evolution of the angles among the NTD, MLD, and C‐terminal lumenal domain (CTD) for three trajectories. Panel a corresponds to the trajectory with the titled NTD. We selected a helix from each domain and calculated the angles between each helix axis. Please refer to Figure S2 for the selected helices in each domain. The black, red, and green lines correspond to the angle between NTD and MLD, angle between MLD and CTD, and angle between CTD and NTD, respectively. The angles between each domain are obtained based on a specific helix as an axis from each domain
Figure 3.
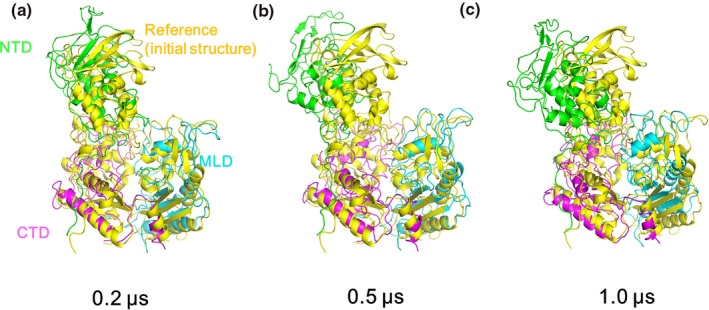
The structure obtained from the current simulation with a wild type after 0.2 μs (a), 0.5 μs (b), and 1.0 μs (c) that is in overlapped with the initial structure (yellow). The color of each domain is same as the color used in Figure 1
Figure 4.
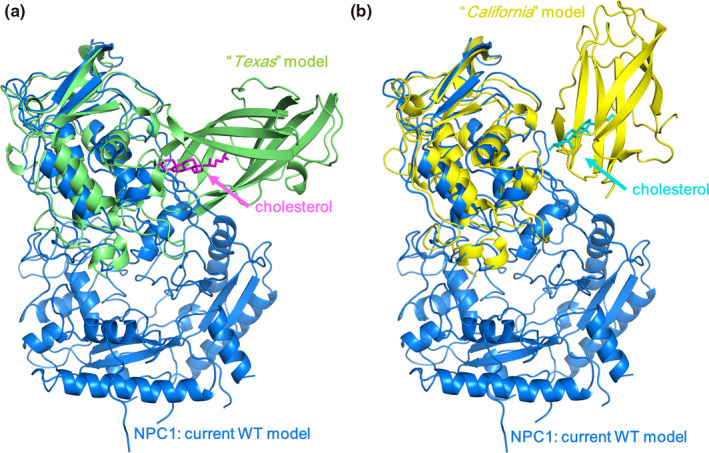
Fitting of (a) the “Texas” model (lime) and (b) the “California” model (yellow) onto the structure of the WT Niemann‐Pick type C1 (NPC1) obtained from the current simulation without cholesterol (blue). The coordinate of “Texas” model is from Estiu et al. (Estiu et al., 2013). We obtained the “California” model structure following the procedure in the reference (Li, Saha, et al., 2016). Namely, we overlapped Niemann‐Pick type C2 (NPC2) binding X‐ray structure (PDB ID: 5KWY) onto cryo‐EM structure (PDB ID: 3JD8). Then, we aligned the two NPC2 binding loops on MLD in 5KWY to the same loops of 3JD8 along with the NPC2 in 5KWY. The structure in figure (a) is a result from combination of the NTD/NPC2 complex having the “Texas” interface with full‐length NPC1 from current simulation. If the “Texas” model is reasonable, this is a putative structure of full‐length NPC1 in complex with cholesterol bound NPC2 for the cholesterol transport from NPC2 to NTD. This model may serve as a template for further study such as monitoring the relative orientation/distance change right after NPC2 binding in MLD loops
As for the R518W mutation simulation, we generated six independent trajectories that were 200 ns each. Basically, no noticeable changes were observed except in one trajectory where the NTD was displaced toward the MLD side, which is the opposite direction of the wild‐type simulation. To monitor the possible swing between different states, we selected this trajectory and keep running the simulation up to 1.5 μs, but we were not able to see the corresponding behavior and the NTD remains steady with displacement toward the MLD. The relative angles between each domain and the RMSDs corresponding to this trajectory are shown in Figure S3. Note the significant decrease in the angle between the NTD and the MLD (black line), which is an indication of NTD tilting/displacement toward MLD side. Possibly, this causes a less favorable interaction with the NPC2 as a result of steric hindrance, and it could be the signature of the experimentally observed cholesterol‐independent weak interaction of the NPC2 with the mutated NPC1 (Deffieu & Pfeffer, 2011; Gong et al., 2016). In fact, the overlap of the MLD from the last frame of this trajectory over the MLD of the X‐ray structure (PDB ID: 5KWY)(Li, Saha, et al., 2016) generates a structural crash in the interface between the NPC2 and the NTD, more specifically the residue A34 ~ K38 of the NTD (magenta) and the residue G80 ~ P84 of the NPC2 (yellow) (Figure 5). Another possible cause of the weakened interaction is the structural change in the NPC2 binding of the MLD loops upon the mutation. It is reported that the seven residues, which include Q421, Y423, P424, D502, F503, F504, and Y506, in the two protruding MLD loops are involved in the interaction with the NPC2 (Li, Saha, et al., 2016). To understand this possibility, we tried to compare the binding affinity of the NPC2 binding loops of the MLD with the NPC2 before and after the mutation indirectly by calculating the backbone RMSD of the loops from the mutated trajectory using the experimentally determined NPC2 binding MLD loops as a reference (Li, Saha, et al., 2016). For this purpose, we obtained the RMSD by aligning the two protruding loops only from the last 1.0 μs from the trajectory to the reference loops structure. As can be seen in the resulting Figure S4, the RMSD shows narrow distribution centered at 1.8 Å, which indicates the loops structure from the simulation trajectory is not much different from the loop structure in wild NPC1. Based on this observation, we think that the difference in the NPC2 binding affinity between the wild and the mutated NPC1 is small. Note that the residues D502 are E502 in our simulation since we have used a human NPC1 that is unlike an NPC1 X‐ray structure, which is bovine. Therefore, we believe the reduced binding affinity of the mutated R518W is as a result of the displacement of the NTD toward the MLD, which leads to less favorable NTD–NPC2 interaction compared to a state where the interface is water exposed. This can be understood by noting that the interactions in this region are mostly between the hydrophilic residues or the charged residues from the NTD and the hydrophobic residues from the NPC2. Even though the current mutation simulation explains the reduced binding affinity of the NPC2 to the NPC1 in the R518W mutation that was observed in the in vitro experiments (Deffieu & Pfeffer, 2011; Gong et al., 2016), it is not clear at this moments if this reduced binding is the main culprit behind the disease in cellular environment because the pH of lysosome lumenal side with the NPC1‐deficient lysosome is increased from 5.5 according to the experiments with live imaging (Leung, Chakraborty, Saminathan, & Krishnan, 2019; Wheeler, Haberkant, et al., 2019).
Figure 5.
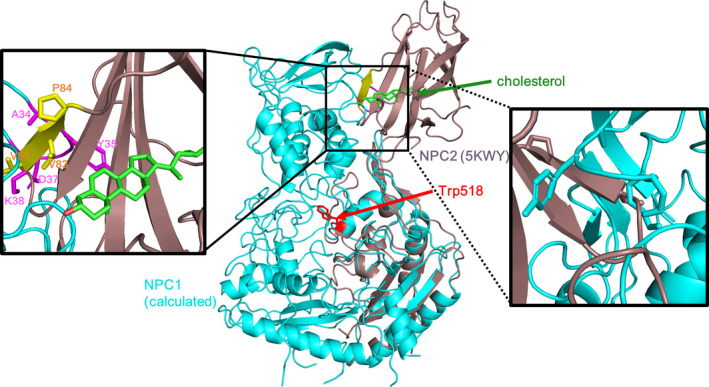
The overlap of the structure obtained from the mutation simulation over the X‐ray structure of the Niemann‐Pick type C2 (NPC2) that is in binding with the MLD. It generates some structural crashes near the interface between NPC2 and NTD. The detailed structure of the interface is shown along with the residues. The mutated residue Trp518 was presented as a red stick for convenience. For clarity, the same structure from the rear side is shown as the magnified figure on the right hand side
We also performed the molecular dynamics simulation with the cholesterol loaded in the NTD for the wild NPC1. The seven independent simulations to 200 ns showed no distinctive titling motions or distance changes between the domains. There were no noticeable structural differences between any two different trajectories as well. Therefore, we selected one of the trajectories and proceeded the simulation to 2.0 μs. We then observed a change with the NTD orientation. The backbone RMSD of the whole system as well as each individual domains using the starting structure as a reference were plotted in Figure 6 as a function of time. We observed that the trajectory quickly makes a conduit‐like channel as a result of this re‐orientation, which includes the cholesterol that contains the NTD as a part of this channel that was mentioned above. From Figure 7, which is the same angle profile as Figure 2, its relative orientation between the domains is different from the structure with no cholesterol at least within our simulation time scale. We have selected a structure near 1.8 μs, which has an RMSD of 9.0 Å, and presented the identified tunnel in Figure 8. The tunnel was constructed using MoleOnline (Berka et al., 2012) and imported into PyMOL (Schrodinger, 2015). For this tunnel identification, we have used the default MoleOnline parameters. It is somewhat surprising that this long tunnel is kept surviving even with the large structural fluctuation. This tunnel might act as a channel for the cholesterol transport from NTD to SSD, which is a lot like the experimentally observed channel in the NCR1 (Winkler et al., 2019). Of course, the simulation has not reached the fully equilibrated state. Hence, the tunnel we observed should be understood as a snapshot of a dynamically fluctuating system and its shape might be not ideal. However, this simulation shows that the trajectory can form at tunnel, which was not observed in the trajectory with no cholesterol on NTD, and it can facilitate the transport of the cholesterol. Also, there is an onset of the NTD alignment to this channel when there is cholesterol on the NTD, which is possibly for the proper transport of the cholesterol from the NTD all the way to the SSD.
Figure 6.
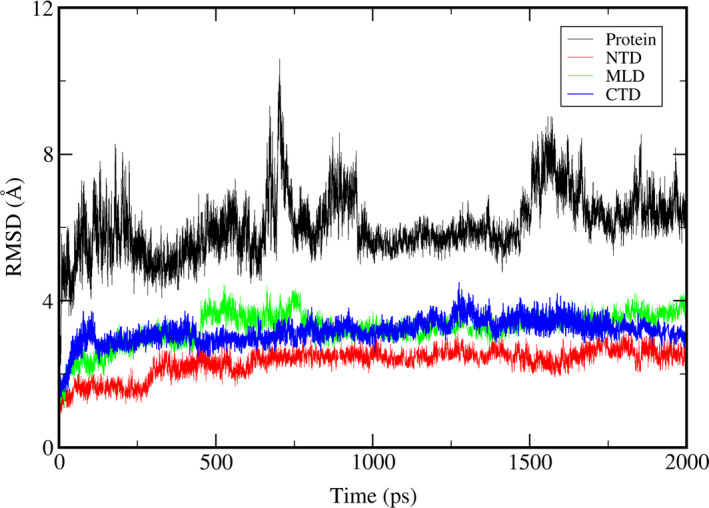
The RMSD time profile of the wild‐type Niemann‐Pick type C1 (NPC1) simulation with cholesterol in the NTD. The RMSD of the whole system is shown as black line. The RMSD of NTD, MLD, and C‐terminal lumenal domain (CTD) are shown as red, green, and blue lines, respectively
Figure 7.
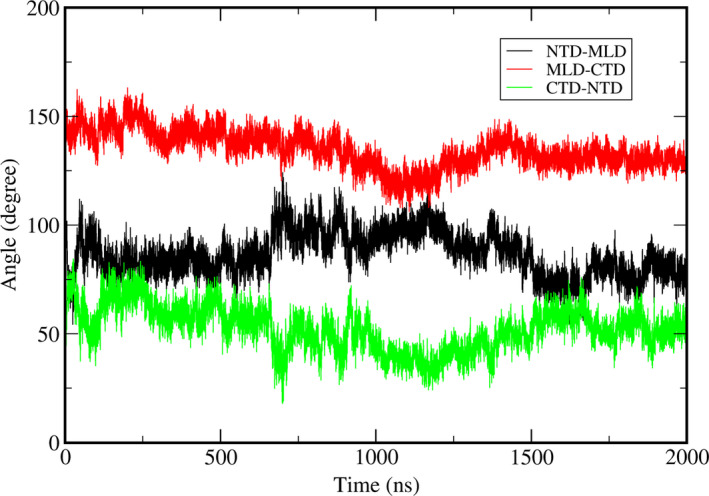
The change in the angles as a function of time for a wild‐type trajectory with cholesterol in the NTD. The angles here are obtained using the same method as in Figure 2
Figure 8.
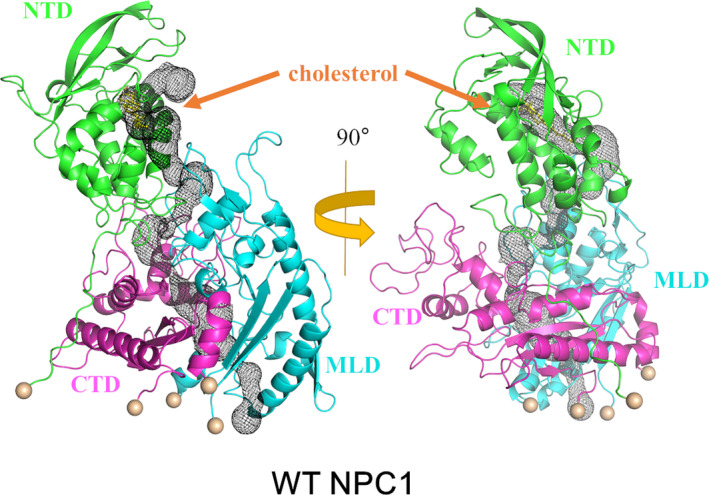
The tunnel obtained from the wild‐type cholesterol containing an NTD simulation. The tunnel is identified with MoleOnline (Berka et al., 2012) and is represented as black mesh. The NTD, C‐terminal lumenal domain (CTD), and MLD domains are shown as green, magenta, and cyan, respectively
The introduction of the disulfide bond between the MLD and the CTD, which modifies the tunnel, fails to rescue the cholesterol accumulation in the NPC1−/− HeLa cells (Saha et al., 2020). This demonstrates the importance of the relative movement between the domains especially between the MLD and the CTD. Also, the restriction of the NTD movement away from the rest of the domain with the formation of the disulfide bond between the P251 and the L929 does not affect its functionality, which suggests that NTD movement away from the rest of the domain is not necessary for the cholesterol transport (Saha et al., 2020). However, this does not preclude the possibility of the NTD movement within the domain, such as the tilting we have observed, which possibly accumulates the cholesterol from the NPC2 and delivers it to the internal tunnel in present study.
To observe the behavior of the mutated NPC1 once the cholesterol is transferred to the NTD from the NPC2, which might be relatively easy after the binding of the NPC2 to the MLD loops, we performed the same simulations with the cholesterol in the NTD for the R518W mutation. The number of the total independent simulation trajectories was nine, and the simulation was run up to 500 ns each for each of the eight trajectories and 1.6 μs for one trajectory. Among these nine trajectories, two of them showed similar behaviors, i.e., the formation of a tunnel or a tunnel‐like structure, which is shown in Figure 9. We also examined the possibility of blocking of the tunnel as a result of the R518W mutation since it is located near the tunnel. However, tunnel blocking was not observed in all the simulations. The RMSD time profile of the whole system as well as each individual domain of the trajectory corresponding to 1.6 μs run are shown in Figure S5. These observations are in agreement with the experimental findings that the reduced cholesterol trafficking of the R518W NPC1 is mainly caused by the functional inability instead of misfolding as stated above.
Figure 9.
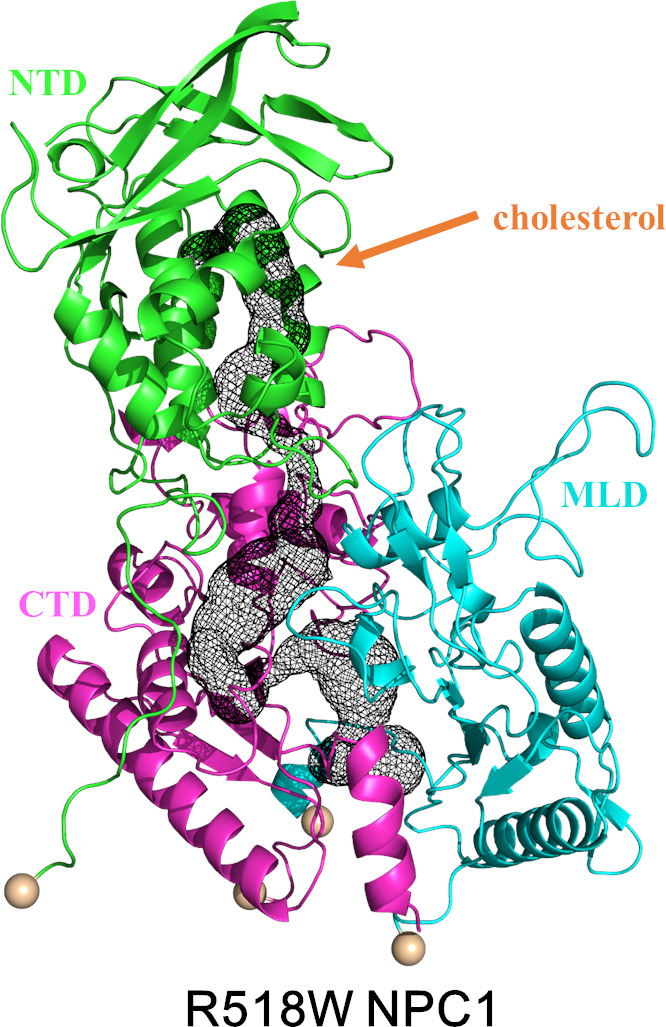
The same Figure as Figure 9 with an R518W mutation with cholesterol in the NTD. The structure shown is extracted at 200 ns. The NTD, C‐terminal lumenal domain (CTD), and MLD domains are shown as green, magenta, and cyan, respectively
Strictly speaking, the re‐orientation of the NTD is not an independent event. It is a result of the concerted motion between each of the domains. The introduction of disulfide bond between the MLD and the CTD, which modifies the tunnel, fails to rescue the cholesterol accumulation in the NPC1−/− HeLa cells (Saha et al., 2020). This demonstrates the importance of the relative movement between the domains especially between the MLD and the CTD. Also, the restriction of the NTD movement away from the rest of the domain with the formation of a disulfide bond between the P251 and the L929 does not affect its functionality, which suggests that the NTD movement away from the rest of the domain is not necessary for the cholesterol transport (Saha et al., 2020). However, this does not preclude the possibility of the NTD movement within the domain, such as the tilting we have observed, which facilitates the accommodation of the cholesterol from the NPC2 and its delivery to the internal tunnel in present study.
The relatively large structural movement between each of the domains observed in the current simulations and the long‐time simulation of the full‐length NPC1 suggest that the conformational stability of the TMD is higher, and there was not a lot of structural change starting from the initial structure with C α RMSD below 2 Å, unlike the lumenal exposed three domains (Vanharanta et al., 2020).
4. DISCUSSION
Based on the mutational study here, it seems that the reduced binding affinity NPC2 to the R518W‐mutated NPC1 is caused by the unfavorable orientation of the NTD for the NPC2 binding, which thereby decreased the cholesterol transfer activity from NPC2 to NTD. On the other hand, it seems there is no difficulty for a proper alignment of the NTD to the cholesterol transporting channel after the cholesterol is transferred to the NTD from the NPC2 with same mutation, which supports the stepwise process for the cholesterol transport from the NPC2 to the SSD in the lysosomal membrane. Before the transfer of the cholesterol from NPC2 to NTD, the NTD has to have a proper pose for a favorable interaction with the NPC2, which is indicated by the titling of the NTD from the current simulation. The structure of the full‐length NPC1 + NPC2 (with cholesterol) is not observed and it would be interesting to model this complex starting from the putative structure presented in this work. The “Texas” model is based on the NTD and the NPC2 interface proposed by Brown and Goldstein (Kwon et al., 2009; Wang et al., 2010) and there is the possibility that their interaction structure might be different under the presence of the full‐length NPC1. Right after the cholesterol transfer from the NPC2 to the NTD, the alignment of the cholesterol in the NTD must be directed more or less to the NPC2 cholesterol leaving the region. It is believed that the next step is the re‐orientation of the NTD starting from this conformation toward the long channel that may lead to the SSD. This could happen within a single NPC1 or between the neighboring NPC1 proteins. This process of leaving the NPC2 from the NTD and the NTD re‐orientation could happen independently, or they could be connected. In either case, it seems reasonable to assume that from our simulations, there is change in the NTD orientation toward the favorable formation of a tunnel that can facilitate the release of the cholesterol from the NTD to the tunnel for further process. This behavior was not observed in the previous computational studies. The very recent experimental cryo‐EM study (Qian et al., 2020) identified a tunnel between the MLD and the CTD and observed two structurally distinct NTD orientations at pH 5.5. One is the structure with a cholesterol entrance NTD side properly oriented toward the channel (PDB ID: 6W5U) and the other structure has an NTD orientation away from it (PDB ID: 6W5T). Combined with the observation of the cholesterol both in the tunnel and the NTD, they mentioned that the cholesterol from the NTD “eventually enters the central tunnel”, which agrees with the current simulation results.
We note that the cholesterol transfer efficiency from the NPC2 to the NPC1 is reduced by 90% with the deletion of the NTD (Gong et al., 2016). Therefore, the role of the NTD must be crucial for the proper cholesterol transport through the NPC1. In the current study, we demonstrated the importance of the NTD as a dynamic domain depending on the existence of cholesterol on it along with the mutation R518W.
Clearly, there is limitation of the current simulations since our model includes lysosomal lumenal domains only instead of full‐length NPC1, which include the membrane and the cytoplasmic loops. Therefore, the possible concerted motion between domains via the TMD and cytoplasmic loops is not fully reflected in the current study. Moreover, the current simulation is too short to access the equilibrium state, even though we employed the 4.0 fs time step. The increased time step of 4.0 fs in current simulation might generate somewhat different trajectory from the trajectory obtained from the usual 1.0 ~ 2.0 fs time step. To check this possibility, we performed another set of simulations using 2.0 fs time step with wild NPC1 with no cholesterol on NTD. We have not observed the tilting behavior that was shown above in total 10 independent trajectories up to 200 ns each. Therefore, we extended one of the simulations up to 400 ns and we were able to observe the similar tilting behavior even though the degree of tilting was less than the results obtained from simulation trajectory with 4.0 fs time step. The resulting figure is shown in Figure S6. These results might partially support the validity of the time step we have used here. Therefore, the simulation results presented here should be understood as ‘a dynamical signature’ of the corresponding behaviors instead of the final equilibrium property. Perhaps the engaging and disengaging of the NTD orientation is in dynamic equilibrium, and the real system. Nevertheless, the current study provides insights into the structure function relation of NPC1, especially the shift of NTD orientation depending on the stage of cholesterol transport in connection to the NPC2 and effect of these motions or NPC1 lumenal domain conformations by R518W mutation. We believe the current study can enhance our understanding of cholesterol absorption/re‐absorption process via NPC1 with atomic detail.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
AUTHOR CONTRIBUTIONS
H.‐J.Y., H.H.L., and S.J. conceived and designed the experiments. H.J performed computations. All authors analyzed the data, and wrote and edited the manuscript.
Supporting information
Fig S1‐S6
Video S1
ACKNOWLEDGEMENTS
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (NRF‐2018R1D1A1B07040808) to HJY. SJ acknowledges National Research Foundation of Korea (NRF) 2017M3D9A1073784. This article was previously posted as a pre‐print on BioRxiv (www.biorxiv.org/content/10.1101/2020.06.09.141630v1).
All experiments were conducted in compliance with the ARRIVE guidelines.
Yoon H‐J, Jeong H, Lee HH, Jang S. Molecular dynamics study with mutation shows that N‐terminal domain structural re‐orientation in Niemann‐Pick type C1 is required for proper alignment of cholesterol transport. J. Neurochem.2021;156:967–978. 10.1111/jnc.15150
Hye‐Jin Yoon and Hyunah Jeong have equal contribution.
Contributor Information
Hyung Ho Lee, Email: hyungholee@snu.ac.kr.
Soonmin Jang, Email: sjang@sejong.ac.kr.
REFERENCES
- Ballout R. A., Sviridov D., Bukrinsky M. I., Remaley A. T. (2020). The lysosome: A potential juncture between SARS‐CoV‐2 infectivity and Niemann‐Pick disease type C, with therapeutic implications. The FASEB Journal, 34(6), 7253–7264. 10.1096/fj.202000654r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka, K. , Hanák, O. , Sehnal, D. , Banáš, P. , Navratilova, V. , Jaiswal, D. , … Otyepka, M. (2012). MOLE online 2.0: Interactive web‐based analysis of biomacromolecular channels. Nucleic Acids Research, 40, W222–W227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betters, J. , & Yu, L. (2010). Transporters as drug targets: Discovery and development of NPC1L1 inhibitors. Clinical Pharmacology and Therapeutics, 87, 117–121. 10.1038/clpt.2009.209 [DOI] [PubMed] [Google Scholar]
- Deffieu, M. S. , & Pfeffer, S. R. (2011). Niemann‐Pick type C 1 function requires lumenal domain residues that mediate cholesterol‐dependent NPC2 binding. Proceedings of the National Academy of Sciences, 108, 18932–18936. 10.1073/pnas.1110439108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elghobashi‐Meinhardt, N. (2014). Niemann‐Pick type C disease: A QM/MM study of conformational changes in cholesterol in the NPC1 (NTD) and NPC2 binding pockets. Biochemistry, 53, 6603–6614. 10.1021/bi500548f [DOI] [PubMed] [Google Scholar]
- Elghobashi‐Meinhardt, N. (2019). Computational tools unravel putative sterol binding sites in the lysosomal NPC1 protein. Journal of Chemical Information and Modeling, 59, 2432–2441. 10.1021/acs.jcim.9b00186 [DOI] [PubMed] [Google Scholar]
- Elghobashi‐Meinhardt, N. (2020). Cholesterol transport in wild‐type NPC1 and P691S: Molecular dynamics simulations reveal changes in dynamical behavior. International Journal of Molecular Sciences, 21, 2962. 10.3390/ijms21082962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estiu, G. , Khatri, N. , & Wiest, O. (2013). Computational studies of the cholesterol transport between NPC2 and the N‐terminal domain of NPC1 (NPC1 (NTD)). Biochemistry, 52, 6879–6891. 10.1021/bi4005478 [DOI] [PubMed] [Google Scholar]
- Feenstra, K. A. , Hess, B. , & Berendsen, H. J. (1999). Improving efficiency of large time‐scale molecular dynamics simulations of hydrogen‐rich systems. Journal of Computational Chemistry, 20, 786–798. [DOI] [PubMed] [Google Scholar]
- Goldstein, J. L. , & Brown, M. S. (1990). Regulation of the mevalonate pathway. Nature, 343, 425. 10.1038/343425a0 [DOI] [PubMed] [Google Scholar]
- Gong, X. , Qian, H. , Zhou, X. , Wu, J. , Wan, T. , Cao, P. , … Yan, N. (2016). Structural insights into the Niemann‐Pick C1 (NPC1)‐mediated cholesterol transfer and Ebola infection. Cell, 165, 1467–1478. 10.1016/j.cell.2016.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, S. M. (1983). Absorption and metabolism of dietary cholesterol. Annual Review of Nutrition, 3, 71–96. 10.1146/annurev.nu.03.070183.000443 [DOI] [PubMed] [Google Scholar]
- Hodošček, M. , & Elghobashi‐Meinhardt, N. (2018). Simulations of NPC1 (NTD): NPC2 protein complex reveal cholesterol transfer pathways. International Journal of Molecular Sciences, 19, 2623. 10.3390/ijms19092623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, J. , Goldstein, J. , & Brown, M. (2002). SREBPs: Transcriptional mediators of lipid homeostasis. Cold Spring Harbor symposia on quantitative biology, Vol. 67 (pp. 491–498). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Rauscher, S. , Nawrocki, G. , Ran, T. , Feig, M. , de Groot, B. L. , … MacKerell, A. D. (2017). CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nature Methods, 14, 71–73. 10.1038/nmeth.4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante, R. E. , Abi‐Mosleh, L. , Radhakrishnan, A. , Dale, J. D. , Brown, M. S. , & Goldstein, J. L. (2008). Purified NPC1 protein I. Binding of cholesterol and oxysterols to a 1278‐amino acid membrane protein. Journal of Biological Chemistry, 283, 1052–1063. 10.1074/jbc.M707943200 [DOI] [PubMed] [Google Scholar]
- Infante, R. E. , Radhakrishnan, A. , Abi‐Mosleh, L. , Kinch, L. N. , Wang, M. L. , Grishin, N. V. , … Brown, M. S. (2008). Purified NPC1 Protein II. Localization of sterol binding to a 240‐amino acid soluble luminal loop. Journal of Biological Chemistry, 283, 1064–1075. 10.1074/jbc.M707944200 [DOI] [PubMed] [Google Scholar]
- Infante, R. E. , Wang, M. L. , Radhakrishnan, A. , Kwon, H. J. , Brown, M. S. , & Goldstein, J. L. (2008). NPC2 facilitates bidirectional transfer of cholesterol between NPC1 and lipid bilayers, a step in cholesterol egress from lysosomes. Proceedings of the National Academy of Sciences, 105, 15287–15292. 10.1073/pnas.0807328105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn, P. C. (1989). Defining the axis of a helix. Computers and Chemistry, 13, 185–189. 10.1016/0097-8485(89)85005-3 [DOI] [Google Scholar]
- Kwon, H. J. , Abi‐Mosleh, L. , Wang, M. L. , Deisenhofer, J. , Goldstein, J. L. , Brown, M. S. , & Infante, R. E. (2009). Structure of N‐terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell, 137, 1213–1224. 10.1016/j.cell.2009.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, K. , Chakraborty, K. , Saminathan, A. , & Krishnan, Y. (2019). A DNA nanomachine chemically resolves lysosomes in live cells. Nature Nanotechnology, 14, 176–183. 10.1038/s41565-018-0318-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Robertson, A. D. , & Jensen, J. H. (2005). Very fast empirical prediction and rationalization of protein pKa values. Proteins: Structure, Function, and Bioinformatics, 61, 704–721. 10.1002/prot.20660 [DOI] [PubMed] [Google Scholar]
- Li, X. , Lu, F. , Trinh, M. N. , Schmiege, P. , Seemann, J. , Wang, J. , & Blobel, G. (2017). 3.3 Å structure of Niemann‐Pick C1 protein reveals insights into the function of the C‐terminal luminal domain in cholesterol transport. Proceedings of the National Academy of Sciences, 114, 9116–9121. 10.1073/pnas.1711716114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Saha, P. , Li, J. , Blobel, G. , & Pfeffer, S. R. (2016). Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proceedings of the National Academy of Sciences, 113(36), 10079–10084. 10.1073/pnas.1611956113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Wang, J. , Coutavas, E. , Shi, H. , Hao, Q. , & Blobel, G. (2016). Structure of human Niemann‐Pick C1 protein. Proceedings of the National Academy of Sciences, 113, 8212–8217. 10.1073/pnas.1607795113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, T. , Qi, X. , Hassan, A. , Liang, Q. , De Brabander, J. K. , & Li, X. (2020). Structural basis for itraconazole‐mediated NPC1 inhibition. Nature Communications, 11, 1–11. 10.1038/s41467-019-13917-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, J. , Yang, H. , & Song, B.‐L. (2019). Mechanisms and regulation of cholesterol homeostasis. Nature Reviews Molecular Cell Biology, 1–21. [DOI] [PubMed] [Google Scholar]
- Malhotra, P. , Gill, R. K. , Dudeja, P. K. , & Alrefai, W. A. (2016). Mellitus and Intestinal Niemann‐Pick C1–Like 1 Gene Expression. In Mauricio D. (ed.), Molecular Nutrition and Diabetes, A Volume in the Molecular Nutrition Series, 277–290. Amsterdam, Netherlands: Elsevier. . [Google Scholar]
- Martínez‐Archundia, M. , Hernández Mojica, T. , Correa‐Basurto, J. , Montaño, S. , & Camacho‐Molina, A. (2019). Molecular dynamics simulations reveal structural differences among wild‐type NPC1 protein and its mutant forms. Journal of Biomolecular Structure and Dynamics, 38, 3527–3532. [DOI] [PubMed] [Google Scholar]
- Millat, G. , Marçais, C. , Tomasetto, C. , Chikh, K. , Fensom, A. H. , Harzer, K. , … Vanier, M. T. (2001). Niemann‐Pick C1 disease: Correlations between NPC1 mutations, levels of NPC1 protein, and phenotypes emphasize the functional significance of the putative sterol‐sensing domain and of the cysteine‐rich luminal loop. The American Journal of Human Genetics, 68, 1373–1385. 10.1086/320606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohgami, N. , Ko, D. C. , Thomas, M. , Scott, M. P. , Chang, C. C. , & Chang, T.‐Y. (2004). Binding between the Niemann‐Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol‐sensing domain. Proceedings of the National Academy of Sciences, 101, 12473–12478. 10.1073/pnas.0405255101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, W. D. , O'Brien, J. F. , Lundquist, P. A. , Kraft, D. L. , Vockley, C. W. , Karnes, P. S. , … Snow, K. (2003). Identification of 58 novel mutations in Niemann‐Pick disease type C: Correlation with biochemical phenotype and importance of PTC1‐like domains in NPC1. Human Mutation, 22, 313–325. [DOI] [PubMed] [Google Scholar]
- Petukh, M. , & Zhulin, I. B. (2018). Comparative study of the effect of disease causing and benign mutations in position Q92 on cholesterol binding by the NPC1 n‐terminal domain. Proteins: Structure, Function, and Bioinformatics, 86, 1165–1175. 10.1002/prot.25597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S. R. (2016). Clues to NPC1‐mediated cholesterol export from lysosomes. Proceedings of the National Academy of Sciences of the United States of America. 113(29), 7941–7943. 10.1073/pnas.1608530113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S. R. (2019). NPC intracellular cholesterol transporter 1 (NPC1)‐mediated cholesterol export from lysosomes. Journal of Biological Chemistry, 294, 1706–1709. 10.1074/jbc.TM118.004165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, H. , Wu, X. , Du, X. , Yao, X. , Zhao, X. , Lee, J. , … Yan, N. (2020). Structural basis of low‐pH‐dependent lysosomal cholesterol egress by NPC1 and NPC2. Cell, 182(1), 98–111.e18. 10.1016/j.cell.2020.05.020 [DOI] [PubMed] [Google Scholar]
- Saha, P. , Shumate, J. L. , Caldwell, J. G. , Elghobashi‐Meinhardt, N. , Lu, A. , Zhang, L. , … Pfeffer, S. R. (2020). Inter‐domain dynamics drive cholesterol transport by NPC1 and NPC1L1 proteins. Elife, 9, e57089. 10.7554/eLife.57089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, L. L. C. (2015). The PyMOL molecular graphics system. Version 1.8.
- Sturley, S. , Márka, Z. , Márka, S. , Hammond, N. , Rajakumar, T. , & Munkacsi, A. (2020) Insights into the COVID‐19 pandemic from a rare neurodegenerative disease.
- Trinh, M. N. , Brown, M. S. , Seemann, J. , Goldstein, J. L. , & Lu, F. (2018). Lysosomal cholesterol export reconstituted from fragments of Niemann‐Pick C1. eLife, 7, e38564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel, D. , Lindahl, E. , Hess, B. , Groenhof, G. , Mark, A. E. , & Berendsen, H. J. C. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Vanharanta, L. , Peränen, J. , Pfisterer, S. G. , Enkavi, G. , Vattulainen, I. , & Ikonen, E. (2020). High‐content imaging and structure‐based predictions reveal functional differences between Niemann‐Pick C1 variants. Traffic, 21, 386–397. 10.1111/tra.12727 [DOI] [PubMed] [Google Scholar]
- Wang, H. , Shi, Y. , Song, J. , Qi, J. , Lu, G. , Yan, J. , & Gao, G. F. (2016). Ebola viral glycoprotein bound to its endosomal receptor Niemann‐Pick C1. Cell, 164, 258–268. 10.1016/j.cell.2015.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. L. , Motamed, M. , Infante, R. E. , Abi‐Mosleh, L. , Kwon, H. J. , Brown, M. S. , & Goldstein, J. L. (2010). Identification of surface residues on Niemann‐Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metabolism, 12, 166–173. 10.1016/j.cmet.2010.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, S. , Haberkant, P. , Bhardwaj, M. , Tongue, P. , Ferraz, M. J. , Halter, D. , … Sillence, D. J. (2019). Cytosolic glucosylceramide regulates endolysosomal function in Niemann‐Pick type C disease. Neurobiology of Disease, 127, 242–252. 10.1016/j.nbd.2019.03.005 [DOI] [PubMed] [Google Scholar]
- Wheeler, S. , Schmid, R. , & Sillence, D. J. (2019). Lipid–protein interactions in Niemann–pick type C disease: Insights from molecular modeling. International Journal of Molecular Sciences, 20, 717. 10.3390/ijms20030717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, M. B. , Kidmose, R. T. , Szomek, M. , Thaysen, K. , Rawson, S. , Muench, S. P. , … Pedersen, B. P. (2019). Structural insight into eukaryotic sterol transport through Niemann‐Pick type C proteins. Cell, 179(2), 485–497.e18. 10.1016/j.cell.2019.08.038 [DOI] [PubMed] [Google Scholar]
- Y. Litvinov, D. , V. Savushkin, E. , A. Garaeva, E. , & D. Dergunov, A. (2016). Cholesterol efflux and reverse cholesterol transport: Experimental approaches. Current Medicinal Chemistry, 23, 3883–3908. 10.2174/0929867323666160809093009 [DOI] [PubMed] [Google Scholar]
- Yamamoto, T. , Nanba, E. , Ninomiya, H. et al (1999). NPC1 gene mutations in Japanese patients with Niemann‐Pick disease typeáC. Human Genetics, 105, 10–16. [DOI] [PubMed] [Google Scholar]
- Yang, C. , Kulkarni, M. , Lim, M. , & Pak, Y. (2017). In silico direct folding of thrombin‐binding aptamer G‐quadruplex at all‐atom level. Nucleic Acids Research, 45, 12648–12656. 10.1093/nar/gkx1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Bulkley, D. P. , Xin, Y. , Roberts, K. J. , Asarnow, D. E. , Sharma, A. , … Beachy, P. A. (2018). Structural basis for cholesterol transport‐like activity of the hedgehog receptor patched. Cell, 175(5), 1352–1364.e14. 10.1016/j.cell.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S6
Video S1


