Abstract
The rise in human pandemics demands prudent approaches in antiviral material development for disease prevention and treatment by effective protective equipment and therapeutic strategies. However, the current state of the antiviral materials research is predominantly aligned towards drug development and its related areas, catering to the field of pharmaceutical technology. This Review distinguishes the research advances in terms of innovative materials exhibiting antiviral activities that take advantage of fast‐developing nanotechnology and biopolymer technology. Essential concepts of antiviral principles and underlying mechanisms are illustrated, followed by detailed descriptions of novel antiviral materials including inorganic nanomaterials, organic nanomaterials, and biopolymers. The biomedical applications of the antiviral materials are also elaborated based on the specific categorization. Challenges and future prospects are discussed to facilitate the research and development of protective solutions and curative treatments.
Keywords: antiviral materials, biopolymers, drug delivery, nanoparticles, viruses
Biomedical applications: In this Review, the current state of antiviral materials research is summarized, categorized, and discussed. It distinguishes the research advances in the development of drug carriers, drug delivery systems, nanoparticles, biopolymers and protective equipment as therapeutic agents or protective equipment against virus infections.
1. Introduction
Human society is entering a new pandemic age fueled by factors such as the rise in global travel, intensive urbanization, deforestation and changing agricultural practices, all of which increase the possibility of human exposure to animal species that carry potentially deadly viral infections.1 Human enteric viruses pose a serious threat for disease transmission leading to illness and death, as the human species are unlikely to have immunity to the emerging viruses.2 A variety of pathogenic viruses have existed and evolved to cause severe harmful impacts, e. g., severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) epidemics. Besides, H5 N1 may be evolving faster than our ability to understand it, becoming harder to predict the occurrence of a human pandemic.3 The global mortality rate of H5 N1 was 63 %, peaking as high as 82 % in Indonesia.2 The worldwide epidemic of the Hong Kong influenza (H3 N2) in 1968 was reported to be a recombinant virus between human and animal/avian virus, and was even not a mutant virus as previously reported.4 Thus, human pandemics can result from numerous undetected avian influenza strains in existence that may arise out of recombination at any point in time.4
In this highly connected world, new viruses are able to spread rapidly, causing devastating effects that science must retool to encounter the possible pandemic threats.1 Research is vital in developing the technology, systems, and services required to achieve universal health care.5 The conventional methods of prevention involve vaccines and antiviral drugs. However, the development of a vaccine for a new strain could take from few months to several decades; meanwhile, the virus could spread globally and substantially affect the health care system and the global economy.6 To date, there have been numerous reports and publications on antiviral materials. Biomedical applications of antiviral materials dominate the literature over food applications.7 However, antiviral materials have not been widely applied in commercial use for personal protection and safety. Influenza viruses have claimed the lives of millions, causing annual epidemics and occasional pandemics.6a In cases of fast‐moving pathogens such as influenza viruses and coronaviruses, one of the most basic protective strategies is to prevent viral transmission via human contact and aerial discharge. The current state of the protective care products offers surgical or N95 protective masks that, in most instances, are insufficient to provide adequate protection against viral transmission since the virus is small enough to penetrate the microfibers of most masks.8
Viruses are acellular obligate parasites. The life cycle of a virus, or rather, viral replication cycle, consists of six basic stages: attachment, penetration, uncoating, replication, assembly, and virion release (Figure 1).9 Based on the two different virion release methods: lysis and budding, viruses are categorized into cytolytic and cytopathic, respectively. In terms of structures, viruses can be separated into an enveloped and a non‐enveloped structure. The enveloped viruses have primarily lipid envelopes, which are less stable in the environment. On the contrary, the non‐enveloped viruses are more stable in wastewater and surfaces and remain resistant to disinfectants.10 Generally, antiviral behaviors could be divided into two categories. One is by interfering in the viral replication cycle as therapeutic agents. The other is by acting as a protective shield against viral infections.
Figure 1.
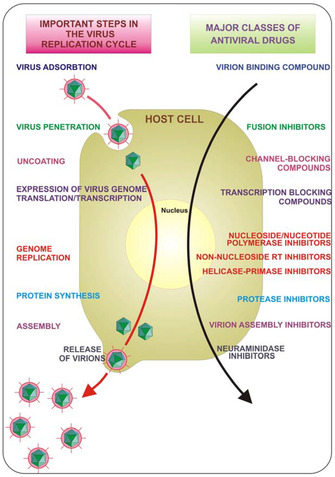
Key steps in the viral replication cycle that provide antiviral targets. Reproduced from reference [17] with permission from the Multidisciplinary Digital Publishing Institute.
Nanotechnology and biopolymer technology have demonstrated to be thriving to deliver essential changes to the development of antiviral therapeutics.11 The research on antiviral technologies such as the use of metal nanoparticles,12 carbon‐based nanomaterials,13 organic nanomaterials,14 nanocomposites,15 and biopolymers16 cast new light onto the development of antiviral protective solutions. For example, the antiviral properties of the nanoparticles are attributed to their charge, size, shape, surface functionality and composition, forcing interference with the viral replication cycle.11a Hence, in this review, we evaluate the latest published literatures on the antiviral materials that exhibit potential as either protective shields or therapeutic agents to prevent or alleviate viral infection. The review also provides information on the current state of the developments in the biomedical applications of the antiviral materials. Challenges and future prospects are discussed in the end to facilitate the research and development of protective solutions and curative treatments.
2. Antiviral principles and mechanisms
2.1. Viral entry inhibition
Inhibition action prior to viral entry is one of the most attractive approaches against viral infection because the extracellular intervention is relatively accessible. The attachment of virus to host cells and viral entry are recognition interactions involving specific components on viruses’ surface and receptors on the host cell membrane,17, 18 which are promising targets for viral entry inhibition.
Inactivation of viruses prior to entry is the most direct antiviral strategy. This antiviral mechanism is proved via indirect evidence, frequently. Yang et al. reported a study about antiviral activities of curcumin (CCM) modified silver nanoparticles, showing that the modified silver nanoparticles (AgNPs) inhibited respiratory syncytial virus (RSV) infection via direct inactivation of viral envelope glycoproteins.19 Also, Speshock et al. reported that the interactions of AgNPs with Tacaribe virus (TCRV) could inhibit arenavirus infection at the early stage of viral replication cycle.20
As a matter of fact, plenty of materials play their antiviral roles via blockage of viral entry.13d, 21 Similar to the virus inactivation mechanism, researchers observed the antiviral activities at the early state prior to viral entry and made the speculation upon different results or behaviors. For example, Barras et al. reported a type of surface‐functionalized carbon dots (CDs) that could interfere with the entry of herpes simplex virus type 1 (HSV‐1).22 Their results indicated that the modified CDs prevented HSV‐1 infection at certain concentrations and by specifically acting at the early stage of viral entry. However, they also claimed that CDs might make a difference in limiting viruses spreading from cell to cell. Gao et al. reported a study on 3,6‐sulfated chitosan (36S) inhibiting human papillomavirus (HPV).21a They claimed that the 36S might directly bind to the viral capsid proteins and thus block HPV adsorption.
Antiviral agents can also take effect via interference with the virus‐cell binding process. Research about polysaccharide‐coated AgNPs against monkeypox virus showed that the modified 10–80 nm particles were capable of blocking virus‐cell binding and penetration.23 As shown in Figure 2, Papp et al. reported a study on the inhibition of influenza virus infection using multivalent sialic‐acid‐functionalized gold nanoparticles (AuNPs). This study demonstrated a relatively clear mechanism: the receptor of the target virus fusion protein is sialic acid receptor while the modified multivalent sialic‐acid‐functionalized AuNPs are expected to compete the binding process and thus inhibit the viral infection.24 Similar mechanisms were reported in multiple studies, where the specific mechanisms were called competition for the binding of virus to the cell, interfaces with viral attachment, preferential binding to the cell proteins, and inhibition of binding to specific receptors.25 Despite their slightly different description, the speculated mechanisms are rather similar: antiviral materials compete or inhibit the binding process between viruses and host cells and thus inhibit viral infections.
Figure 2.
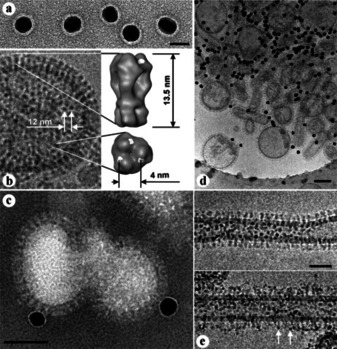
a) AuNP 16 (14 nm), scale bar is 20 nm. b) Influenza A X31 virion. c) Cryo‐TEM preparation of influenza A virions after 60 min incubation with AuNP 16, scale bar is 50 nm. d) Preparation as in (c). e) Cryo‐TEM preparations of influenza A virions before (top) and after (bottom) 60 min incubation with AuNP 14 (2 nm), scale bar is 50 nm. Reproduced from reference [24] with permission from Wiley‐VCH.
Specifically, another paradigm can be introduced for enveloped viruses, targeting viral fusion process. Enveloped viruses are one of the major causes of human viral diseases and the viral entry process of all enveloped viruses involves membrane fusion.26 Vigant et al. summarized this particular aspect in their review article.26 Briefly, the fusion process consists of viral attachment via cell surface receptors as well as conformation changes of protein and components. Both proteins involve in the fusion process and lipids on virus and host cell membranes are potential targets for broad‐spectrum antiviral agents. Relevant antiviral strategies are gaining more attention since the outbreak of COVID‐19, as researchers are looking for similarities among pathogenic viruses instead of only developing traditional point‐to‐point methodology.27 Numerous antiviral agents targeting the viral envelope were reported, including titanium dioxide (TiO2) nanomaterials,28 silica nanoparticles (SiNPs),29 liposomes,30 peptides,31 etc. For example, Jackman et al. reported a therapeutic strategy that introduced potential membrane‐active peptides targeting against mosquito‐borne viruses.32 Badani et al. summarized recent studies of peptide entry inhibitors against enveloped viruses.33 Detailed examples will be introduced in the following sections.
It should be noted that many of those mechanisms were speculation or preliminary conclusion from limited in vivo or in vitro experiments. Various materials show clear antiviral activity against targeted viruses and are speculated that they affect the early stage of viral replication cycle. Although the limited evidence is insufficient to confirm a specific single antiviral mechanism of these materials, the importance of previous studies remains beneficial as the materials exhibited antiviral behavior in the well‐designed experiments. Moreover, under various circumstances, multiple mechanisms are effective at the same time.13d A single antiviral material can also exert inhibitory effects against multiple virus species with different antiviral mechanisms.13c
2.2. Inhibition of other steps from viral replication cycle
Antiviral agents can target at various stages of the viral replication cycle after viral entry. It includes inhibition of viral nucleic acid synthesis; transcription of mRNA via antisense nucleotides; translation and transcription in the nucleus (DNA‐based viruses) or cytoplasm (RNA‐based viruses); protease, integrase, protein synthesis at Golgi and endoplasmic reticulum; uncoating of virus; envelope formation; virus release, and lytic process (cytolytic viruses manipulate apoptosis process and spread from cell to cell). Besides, agents can also target indirect system such as immune system for host defense modification. A large number of antiviral agents taking effects after viral entry are well‐designed upon their behaviors towards specific steps. Some materials serving as drug carriers may have even clearer antiviral mechanisms.34 For example, Zhu et al. investigated the antiviral effect of drug‐loaded single‐walled carbon nanotubes (SWCNTs) against grass carp reovirus (GCRV). The carbon nanotubes served as carriers while the loaded ribavirin functioned as the primary antiviral agent targeting viral RNA‐dependent RNA polymerase.35
Ghaffari et al. reported on the antiviral activity of zinc oxide nanoparticles (ZnO NPs) against H1 N1 influenza virus. The in vitro experiments showed that the modified ZnO NPs exhibited antiviral properties when added for 1 h after viral infection, indicating the inhibitory effect occurred after viral entry.36 Tavakoli et al. reported a study of antiviral materials targeting virus genome. The copper oxide nanoparticles they synthesized exhibited significant antiviral activities against herpes simplex virus type 1 (HSV‐1) via oxidation of viral proteins or degradation of viral genome. Although various antiviral activities were observed at the stages after viral entry, the evidence to identify the exact antiviral mechanisms was still insufficient and remains to be further developed in the future.37
3. Detailed description of various antiviral materials
3.1. Inorganic nanomaterials
Currently, numerous types of inorganic nanomaterials, such as metallic nanoparticles, carbon‐based nanomaterials and silica nanoparticles, have intrigued tremendous interest in biomedical applications because of their attractive physical and chemical characteristics, including superior biocompatibility, good stability, unique structures, large surface‐area‐to‐volume ratios.38 As shown in Table 1, it was found in many studies that inorganic nanomaterials exhibited comparable or even stronger antiviral effect when compared with traditional pharmaceutical ingredient,39 significantly improved the antiviral drug delivery efficiency as nanocarriers,40 or exerted synergistic effect against viruses when in combination with antiviral drugs.41 Although most of the inorganic nanomaterials possessing antiviral activities were reported to be biocompatible, their potential toxic effects on humans and the environment still require additional attention. For example, AuNPs are generally more biocompatible when compared with AgNPs.42 One of the possible toxic mechanisms may be due to the release of silver ions from AgNPs and the exact mechanism is still under debate.43 In this section, we mainly focus on the inhibitory effect of inorganic nanomaterials against various viruses as well as different strategies used to enhance their antiviral activity, stability and biocompatibility.
Table 1.
Antiviral inorganic nanomaterials.
|
Type of Nanomaterials |
Nanomaterials Characteristics |
Virus |
Antiviral mechanism |
References |
|---|---|---|---|---|
|
AgNPs |
Amantadine‐modified silver nanoparticles |
H1 N1 |
Prevent viral attachment to the host cell; inhibit caspase‐3 mediated apoptosis via ROS generation |
Li et al., 201667 |
|
Silver nanorods conjugated with sodium 2‐mercaptoethane sulfonate |
HIV,HSV‐1 |
Inhibit viral replication |
Etemadzade et al., 201668 |
|
|
AgNPs decorated by oseltamivir |
H1 N1 |
Block viral entry and inhibit ROS‐mediated signaling pathways |
Li et al., 201641 |
|
|
Curcumin modified silver nanoparticles |
RSV |
Direct virus inactivation |
Yang et al., 201647 |
|
|
AgNPs functionalized with zanamivir |
H1 N1 |
Inhibit the neuraminidase activity of the H1 N1 virus, resist virus‐ induced apoptosis of the host cells |
Lin et al., 201769 |
|
|
Electrochemical‐synthesized AgNPs |
Poliovirus |
Inhibition of viral binding to RD cells |
Huy et al., 201770 |
|
|
AgNPs with a size of 20–25 nm |
Bovine herpesvirus‐1 (BoHV‐1) |
Inhibit viral replication |
El‐Mohamady et al., 201871 |
|
|
Tannic acid modified AgNPs |
HSV‐2 |
Enhance anti‐HSV‐2 immune response |
Orłowski et al., 201872 |
|
|
Green synthesis of AgNPs from medicinal plants |
Chikungunya virus (CHIKV) |
– |
Sharma et al., 201973 |
|
|
Poly‐vinylpyrrolidone (PVP) coated biopure AgNPs |
RSV |
Attach to viral glycoproteins and block viral entry |
Morris et al., 201974 |
|
|
Ag2S NCs |
Glutathione‐capped Ag2S NCs |
PEDV |
Blockage of viral RNA synthesis and budding; activate immune system |
Du et al., 201845 |
|
AuNPs |
HA‐AuNP /interferon α complex |
HCV |
Enhance innate immune response |
Lee et al., 201250 |
|
AuNPs conjugated with thiolated raltegravir molecules |
HIV‐1 |
Inhibit HIV integrase |
Garrido et al., 201540 |
|
|
Pectin‐reduced AuNPs carry antiretroviral drug zidovudine |
HIV |
Target HIV reservoir sites |
Borker et al., 201775 |
|
|
Gallic acid stabilized monodispersed AuNPs |
HSV‐1, HSV‐2 |
Interfered with virus attachment and proliferation |
Halder et al., 201876 |
|
|
Delivery of antisense peptide nucleic acids by AuNPs |
Bovine viral diarrhea virus (BVDV) |
Inhibit the translation and replication of the virus |
Ghaffari et al., 201977 |
|
|
AuNPs synthesized by using garlic extract |
Measles virus (MeV) |
Block virus directly and inhibit virus replication |
Meléndez‐Villanueva et al., 2019229 |
|
|
AuNCs |
Glutathione‐stabilized fluorescent AuNCs |
PRRSV |
Direct PRRSV inactivation and blockage of viral absorption |
Bai et al., 201851 |
|
CuO |
Nearly spherical CuO NPs with an average size of 40 nm |
HSV‐1 |
Oxidation of viral proteins or degradation of the viral genome |
Tavakoli et al., 202078 |
|
Cu2O |
Spherical Cu2O NPs with an average size of 45 nm |
HCV |
Interaction with virion surface |
Hang et al., 201579 |
|
Cu2O |
Solid‐state copper(I) compounds |
H1 N1, bacteriophage Qβ |
Denature protein structures on viral surfaces |
Minoshima et al., 201680 |
|
CuI |
CuI NPs ranging 100–400 nm |
FCV |
Capsid protein oxidation |
Shionoiri et al., 201281 |
|
CuI NPs with an average size of 160 nm |
H1 N1 |
Degradation of viral proteins |
Fujimori et al., 201282 |
|
|
TiO2 |
TiO2 nano‐colloids synthesized by sonochemical method |
NDV |
Destroy lipid in viral envelope |
Akhtar et al., 2019 |
|
SiNPs |
Silicon nanoparticles prepared by grinding of porous silicon |
HIV, RSV |
Virions blockage and inactivation |
Osminkina et al., 201483 |
|
SiNPs |
Surface‐modified SiNPs |
HIV |
Interaction with specific virus envelope |
Silva et al., 201684 |
|
Mesoporous SiNPs |
Acyclovir‐Loaded and glycosaminoglycan functionalized mesoporous SiNPs |
HSV‐1, HSV‐2 |
Inhibition of viral entry and DNA replication. |
Lee et al., 201885 |
|
Mesoporous SiNPs |
Lipid‐coated and ML336‐ loaded mesoporous SiNPs |
Venezuelan equine encephalitis virus (VEEV) |
Virus inactivation by chemical inhibitor |
LaBauve et al., 201866 |
|
ZnO |
Micro–nano filopodia‐like ZnO structures |
HSV‐1 |
Trap the virions and block viral entry |
Mishra et al., 201186 |
|
ZnO tetrapod micro‐nanostructures |
HSV‐2 |
Block viral entry and neutralize HSV‐2 virions |
Antoine et al., 201287 |
|
|
PEGylated ZnO NPs |
HSV‐1 |
Direct interaction with virus, trap the virions and subsequently block viral entry |
Tavakoli et al., 201821c |
|
|
PEGylated ZnO NPs |
H1 N1 |
Inactivate virus after viral entry |
Ghaffari et al., 201936 |
|
|
Fe3O4 NPs |
Polymer coated superparamagnetic Fe3O4 nanoparticles |
H1 N1 |
Inhibition of viral RNA synthesis |
Kumar et al., 201488 |
|
Fe3O4 NPs |
Glycine coated Fe3O4 NPs |
H1 N1 |
Preferential binding to the protein knobs of influenza virus |
Kumar et al., 201989 |
|
Magnetic NPs |
Aptamer‐conjugated iron oxide nanoparticles |
HCV |
Bind to E1E2 glycoprotein of HCV and lower the viral load |
Delaviz et al., 201590 |
|
SeNPs |
Selenium nanoparticles (SeNPs) functionalized with oseltamivir |
H1 N1 |
Inhibition of caspase 3‐mediated apoptosis via ROS generation |
Li et al., 201791 |
|
SeNPs loaded with zanamivir |
H1 N1 |
Suppress the activation of caspase‐3 and cleavage of PARP; down‐regulate p38 and JNK signaling pathways |
Lin et al., 201792 |
|
|
SeNPs decorated by ribavirin |
H1 N1 |
Resist caspase‐3 apoptotic pathway |
Lin et al., 201893 |
|
|
SeNPs functionalized with amantadine |
H1 N1 |
Depress cell apoptosis via ROS‐mediated AKT signaling pathways |
Li et al., 201894 |
|
|
Fullerene |
Glycodendrofullerenes with 36 mannoses |
EBOV |
Blockage of DC‐SIGN mediated viral entry |
Luczkowiak et al., 201395 |
|
Pyridine/pyridinium‐type fullerene derivatives |
HIV |
Inhibit HIV reverse transcriptase activity |
Yasuno et al., 201596 |
|
|
Fullerene |
C70 fullerene derivatives 2a‐c |
HIV‐1 |
Inhibit viral maturation by interrupting Gag and Gag‐Pol processing |
Castro et al., 201697 |
|
Fullerene derivatives 1,2,3 |
HIV‐1 |
Inhibit HIV‐1 replication |
Martinez et al., 201698 |
|
|
Fullerene derivatives 1a‐e |
HCV |
Inhibit HCV NS3/4 A protease and NS5B polymerase activity |
Kataoka et al., 201699 |
|
|
Mannosylated 3D fullerenes C60 |
EBOV |
Interfere with lectin‐mediated EBOV infection through multivalent interaction |
Illescas et al., 201754 |
|
|
Multivalent disaccharide/[60] fullerene nanoballs |
Zika virus (ZIKV), dengue virus (DENV) |
Blockage of DC‐SIGN mediated viral entry |
Ramos‐Soriano et al., 2019100 |
|
|
Chlorofullerenes C60Cl6 and C70Cl8 |
HIV‐1, HIV‐2 |
Inhibit HIV‐1 attachment and entry to host cells |
Kraevaya et al., 2020101 |
|
|
SWCNTs |
SWCNTs loading HIV‐1 IN inhibitor, 5ClTEP |
HIV‐1 |
Disrupt the DNA binding channel of HIV‐1 integrase (IN) |
Zhang et al., 201357 |
|
MWCNTs |
Highly hydrophilic carboxylated MWCNTs loading antiretroviral drugs CHI360 and CHI415 |
HIV‐1 |
Strong interaction with viral enzyme |
Iannazzo et al., 201513b |
|
SWCNTs |
SWCNTs loaded with ribavirin |
GCRV |
Direct inhibition of the viral RNA polymerase |
Zhu et al., 201535 |
|
MWCNTs |
Tyrosinase immobilized MWCNTs |
HSV‐1, HSV‐2, coxsackievirus type B3 (Cox B3), cytomegalovirus (CMV) |
Inhibition of virus replication |
Botta et al., 2015102 |
|
SWCNTs |
Isoprinosine delivery with SWCNTs |
Nervous necrosis virus (NNV) |
Activation of immune response |
Zhu et al., 2019103 |
|
GO |
GO and partially reduced sulfonated GO (rGO–SO3) |
HSV‐1 |
Inhibition of viral entry |
Sametband et al., 2014104 |
|
GO |
GO conjugated with PVP |
PRV, PEDV |
Inactivate virus via structural destruction |
Ye et al., 201513c |
|
GO |
Reduced GO modified with sulfonated magnetic nanoparticles |
HSV‐1 |
Virion capture and photothermal therapy |
Deokar et al., 2017105 |
|
GO |
β‐cyclodextrin (CD) functionalized GO loading curcumin |
RSV |
Directly inactivate the virus and inhibit viral attachment |
Yang et al., 201758 |
|
Graphene |
Graphene derivatives covered with polyglycerol sulfate and alkyl chains |
HSV‐1 |
Interact with virus and destroy viral membrane |
Donskyi et al., 2019106 |
|
CDs |
CDs modified with boronic acid or amine |
HSV‐1 |
Blockage of viral entry |
Barras et al., 2016107 |
|
Cationic CDs by using ascorbic acid as carbon precursor |
PRV, PRRSV |
Induce immune response to inhibit viral replication |
Du et al., 2016108 |
|
|
Graphene‐like CDs derived from citric acid and modified with boronic acid |
HIV‐1 |
Blockage of viral entry |
Fahmi et al., 2016109 |
|
|
CDs modified by surface passivation molecules |
Human norovirus virus‐like‐particles (VLPs) |
Inhibition of VLPs’ binding to HBGA receptors |
Dong et al., 2017110 |
|
|
CDs |
Cationic CDs by using curcumin as carbon precursor |
PEDV |
Blockage of viral entry, viral RNA synthesis and virus budding; suppression of ROS accumulation; inhibition of virus replication by activating immune system |
Du et al., 201861 |
|
Cationic CDs by using benzoxazine monomers as carbon precursor |
Japanese encephalitis virus (JEV), ZIKV, DENV, porcine parvovirus (PPV), adenovirus‐associated virus (AAV) |
Directly interact with virions; might limit the transmission of the virus |
Huang et al., 201960 |
|
|
CDs derived from curcumin |
EV71 |
Block the attachment of EV71 virus to the cell; inhibit virus replication |
Lin et al., 2019111 |
|
|
CDs derived from ethylenediamine/citric acid and postmodified with boronic acid ligands; CDs derived from 4‐ aminophenylboronic acid |
Human Coronavirus (HCoV) |
Inhibit viral entry; interfere with virus replication |
Łoczechin et al., 2019112 |
3.1.1. Silver nanoparticles
AgNPs are one of the most extensively researched classes of metallic nanomaterials used for antiviral applications. Numerous studies have reported that AgNPs exhibit broad‐spectrum antiviral efficacy on different stages of the viral replication cycle.17, 44 Recently, Du et al. developed biocompatible glutathione‐capped Ag2S nanoclusters (NCs), serving as a highly efficient antiviral agent against porcine epidemic diarrhea virus (PEDV). Mechanism analysis (Figure 3) showed that Ag2S NCs inhibited the synthesis of viral negative‐strand RNA and viral budding. In the meantime, Ag2S NCs also induced the production of IFN‐stimulating genes and upregulated proinflammation cytokines expression. Their findings suggested that Ag2S NCs can become a promising antiviral agent in the treatment of other types of coronavirus, such as SARS and MERS coronavirus.45
Figure 3.
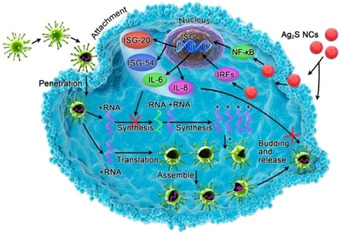
Possible antiviral activity of glutathione‐capped Ag2S NCs. Reproduced from reference [45] with permission from the American Chemical Society.
Apart from exhibiting direct antiviral activities, AgNPs were also reported to exert enhanced antiviral effect when modified with pharmaceutical ingredients. For instance, curcumin is an active component of turmeric, which was known as a type of antiviral and antimicrobial agent. However, its poor bioavailability and low solubility in water become a major limitation for further biomedical applications.46 Yang et al. reported that CCM could act as a reducing and capping agent to prepare uniform and stable AgNPs without using organic solvents. The experiments revealed that CCM modified AgNPs could exert synergistic antiviral effect against the RSV via direct virus inactivation.47 Other studies also explored the combinational antiviral potential of the AgNPs and common antiviral drugs. Li et al. decorated AgNPs with Oseltamivir (Ag@OTV), which exhibited significant antiviral efficacy against H1 N1 infection. The underlying mechanism indicated that Ag@OTV could block viral entry and, more importantly, inhibit reactive oxygen species (ROS)‐mediated signaling pathways to reduce cell apoptosis. The above findings exhibited the great potential of utilizing antiviral drug‐modified AgNPs in the treatment of various types of viruses.41
3.1.2. Gold nanoparticles
AuNPs usually possess large surface‐area‐to‐volume ratios with a diameter ranging from 1 to 100 nm, allowing conjugation of various drugs or ligands.48 In comparison with other types of metallic nanomaterials that may release heavy metals during the treatment, AuNPs possess superior biocompatibility and facile preparation approaches.49 The above advantages make AuNPs become attractive materials for antiviral applications. Lee et al. designed a hyaluronic acid‐gold nanoparticle/interferon α (HA‐AuNP/IFNα) complex, serving as an efficient IFNα delivery platform with long circulation time. They discovered that the complex remained in murine liver tissue for more than 7 days postinjection, therefore enhancing innate immune response to HCV infection in liver tissue.50 Bai et al. reported that the glutathione‐stabilized fluorescent gold nanocluster (AuNCs) showed different antiviral influences on pseudorabies virus (PRV) and porcine reproductive and respiratory syndrome virus (PRRSV). The results showed that AuNCs could selectively inhibit PRRSV (RNA virus) propagation by the direct viral inactivation and blockage of viral absorption but not that of PRV (DNA virus), indicating the possibility of applying AuNCs in the treatment of RNA virus infection.51
3.1.3. Carbon‐based nanomaterials
Carbon‐based nanomaterials, such as fullerenes, carbon nanotubes (CNTs), graphene oxide (GO), or carbon dots, are among the most widely investigated nanomaterials possessing antiviral properties.
3.1.3.1. Fullerenes
Fullerenes are allotropic forms of carbon with hollow spherical, ellipsoid, or tubular structures.52 The fullerene family has raised considerable interest in antiviral applications due to its distinctive physical and chemical properties. Illescas et al. prepared 3D fullerene C60 with a spherical shape, which could act as a biocompatible scaffold for the multivalent presentation of functional carbohydrates.53 They demonstrated that the mannosylated fullerenes could interfere with lectin‐mediated Ebola virus (EBOV) infection via a multivalent manner.54 Fullerenes and their derivatives were also reported as potential human immunodeficiency virus type 1 (HIV‐1) inhibitors. Castro et al. synthesized novel cationic C70 derivatives that exhibited over 99 % HIV‐1 inhibition rate. Further, it was the first time to prove that the fullerene derivatives could interfere with viral maturation by impairing Gag and Gag‐Pol processing.55
3.1.3.2. Carbon nanotubes
Carbon nanotubes as novel nanomaterials have shown intriguing applications in the biomedical field. They usually serve as drug carriers due to their functionalized surface with multiple drug binding sites and their capacity to penetrate cellular membranes.56 Iannazzo et al. prepared highly hydrophilic carboxylated multiwalled‐carbon nanotubes (ox‐MWCNTs) and the same materials loading antiretroviral drugs CHI360 and CHI415. They proved that both ox‐MWCNT and MWCNT−C‐CHI360 could have strong interaction with viral enzyme and act as potential HIV‐1 inhibitor.13b Zhang et al. also explored the antiviral activity of CNTs against HIV‐1. They discovered that CNTs could not only disrupt the DNA binding channel of HIV‐1 integrase (IN) but also stably carry HIV‐1 IN inhibitor molecules, exhibiting enhanced inhibitory effect as a dual‐functional agent against HIV infection.57
3.1.3.3. Graphene oxide
GO and its derivatives have been broadly investigated for biomedical applications owing to their unique physicochemical properties. For example, Ye et al.13c reported that the GO nanosheets showed strong antiviral effect against PRV and PEDV, due to their extraordinary single‐layer structure and negative surface charge. Based on the antiviral effect of GO, Yang et al. loaded CCM on the surface of β‐cyclodextrin functionalized GO and explored their synergistic antiviral effect against RSV infection. The results showed that CCM loaded GO could effectively suppress RSV infection through direct virus inactivation and viral attachment inhibition.58
3.1.3.4. Carbon dots
CDs are an intriguing type of fluorescent carbon nanomaterial with a size below 10 nm.59 The surface functionalization of CDs significantly enables them to interact closely with the interface in various biological systems.60 Though the antiviral research of CDs is still in the initial stage, studies carried so far have already shown the promising antiviral activity of CDs that derived from various carbon precursors. Du et al. synthesized cationic CDs by using CCM as a single‐layer carbon precursor and observed that the CCM–CDs exhibited outstanding antiviral activity against PEDV infection. The mechanism analysis revealed that CDs could inhibit virus entry, suppress the synthesis of negative‐strand RNA within the virus, hinder the budding of the virus and prevent the accumulation of ROS caused by PEDV infection. In addition, this material could also inhibit viral replication by stimulating the host cells to produce proinflammatory cytokines genes and interferon‐stimulating genes (Figure 4). The results suggested that CCM–CDs carry the great potential to be developed into a multi‐target antiviral agent in the future.61
Figure 4.
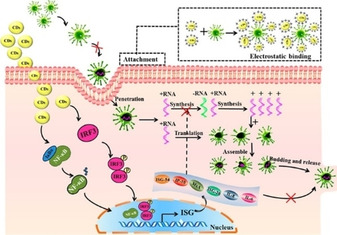
Possible antiviral mechanism of CCM‐CDs against PEDV infection. Reproduced from reference [61] with permission from the American Chemical Society.
3.1.4. Other types of inorganic nanomaterials
Other inorganic nanomaterials made of copper, zinc, titanium, silica, iron and selenium were also reported with various antiviral activities, mainly including viral entry inhibition, intrinsic virucidal effect and delivery of antiviral drugs. For instance, SiNPs with hydrophobic/hydrophilic surface properties can interact with specific virus envelope that has similar surface properties. It prevented the contact between the host cell receptors and viral envelope, and significantly reduced viral transduction ability.62 Similarly, Hang et al. proved that cuprous oxide nanoparticles (CO‐NPs) could inhibit HCV infection via interaction with virion surface, therefore interfere with viral attachment and entry.63 Also, Tavakoli et al. reported that PEGlated ZnO NPs showed higher antiviral effect against the H1 N1 influenza virus and HSV‐1 than that of bare ZnO NPs, indicating PEGlation is an effective approach to increase antiviral ability and reduce cytotoxicity.21c, 36 Moreover, Akhtar et al. synthesized titanium dioxide nano‐colloids (TiO2‐NCs) via sonochemical method. The antiviral experiments of TiO2‐NCs against Newcastle disease virus (NDV) showed virucidal efficacy at a minimum dose of 6.25 μg/ml, and the possible mechanism of virus inactivation was by lipid damage in the viral envelope.64 Mesoporous silica nanoparticles (MSNs) have been widely used as drug delivery systems owing to their large surface area, tunable pore structure, size and shape, as well as ease of synthesis and modification.65 Recently, LaBauve et al. developed lipid‐coated MSNs loading antiviral molecule ML336 against the Venezuelan equine encephalitis virus (VEEV). The MSNs core significantly increased hydrophobic drug loading while the lipid coating retained the loaded drug, achieving sustained drug release and enhanced material biocompatibility.66
3.2. Organic nanomaterials
Organic nanomaterials are equipped with favorable properties such as good biocompatibility, biodegradability, colloidal stability and easy modification, owing to their size, morphology as well as surface characteristics.113 The organic nanomaterials, including polymeric nanoparticles, lipid‐based nanomaterials, dendrimers and micelles, are also extensively evaluated for their antiviral properties. Their inherent virucidal characteristics and capabilities to load therapeutic agents make them suitable candidates for effective virus treatment.
3.2.1. Polymeric nanoparticles
Polymeric nanoparticles are usually made of natural or synthetic polymers (Table 2) with a size ranging from 10 to 1000 nm.114 Antiviral drugs can be either encapsulated in the polymeric matrix or absorbed on the surface to increase their bioavailability, prolong blood circulation time and reduce the possible side effect. Li et al. reported a unique cocktail therapeutic strategy for antiviral treatment. They developed biodegradable polymeric nanoparticles to trap reverse transcriptase inhibitor and conjugate with HIV‐1 fusion inhibitor, revealing combinational virucidal effects against HIV‐1. Besides, the results also showed improved intracellular uptake, lengthened blood circulation time and controlled release.14a Polymeric nanoparticles can also inhibit virus replication via ROS regulation. Kim et al. synthesized poly(aniline‐co‐pyrrole) polymerized nanoregulators (PASomes) which can control ROS levels in vitro. After the downregulation of intracellular ROS, virus propagation was significantly suppressed in Madin‐Darby Canine Kidney (MDCK) cells.115 Besides, Jamali et al. reported that siRNA‐loaded chitosan (CS) nanoparticles effectively targeted virus nucleoprotein to reduce virus infections. Moreover, they also indicated that the intranasal administration of CS/siRNA nanoparticles showed therapeutic effect on mice attacked with a lethal dose of influenza virus, revealing the antiviral activity in vivo.116
Table 2.
Antiviral organic nanomaterials.
|
Type of Nanomaterials |
Nanomaterials Characteristics |
Therapeutic Agents |
Virus |
Antiviral mechanism |
References |
|---|---|---|---|---|---|
|
Polymeric nanoparticles |
PLGA nanoparticles encapsulating combination of antiretroviral drugs |
Maraviroc, etravirine, raltegravir |
HIV‐1 |
Inhibit reverse transcriptase; block viral entry; inhibit integrase |
Jiang et al., 2015129 |
|
PEG‐PLA‐NPs encapsulate HIV‐1 entry inhibitor and conjugate with reverse transcriptase inhibitor |
DAAN‐14 f, T1144 |
HIV‐1 |
Inhibit HIV‐1 entry and transcription |
Li et al., 201614a |
|
|
Polymeric nanoparticles |
Poly(aniline‐co‐pyrrole) polymerized nanoregulators |
– |
H1 N1, H3 N2, H9 N2 |
ROS regulated inhibition of viral propagation |
Kim et al., 2017115 |
|
Chitosan nanoparticles loaded with siRNA |
siRNA‐1496 |
H1 N1 |
Inhibit influenza virus replication |
Jamali et al., 2018116 |
|
|
L‐HSA conjugated PLGA NPs encapsulating antiviral drug |
Lamivudine |
HBV |
Inhibit HBV polymerase and terminate viral DNA chain synthesis |
Dhoke et al., 2018130 |
|
|
Efavirenz‐loaded PLGA NPs modified by transferrin receptor‐ binding peptide |
Efavirenz |
HIV |
Target at the blood‐brain barrier and release antiviral drug |
Martins et al., 2019131 |
|
|
Prodrug‐loaded PLGA NPs |
Raltegravir prodrug |
HIV |
Antiretroviral drug delivery |
Creighton et al., 2019132 |
|
|
Nanodecoy with polymeric core and host cell membrane coating |
– |
ZIKV |
Prevent viral entry by trapping ZIKV |
Rao et al., 2019133 |
|
|
Liposomes |
Liposome‐mediated RNA sensor delivery and expression |
Retinoic acid‐inducible gene‐I |
HBV |
Induce immune response and inhibit HBV replication |
Sato et al., 2015134 |
|
Engineered liposomes as nanocarriers for antiviral agent |
Ivermectin |
DENV |
Inhibit DENV replication |
Croci et al., 2016135 |
|
|
Antibody fragments grafted liposomes encapsulating dapivirine |
Dapivirine |
HIV‐1 |
Neutralization of HIV‐1 by binding to virus envelope glycoprotein |
Wang et al., 2016136 |
|
|
Lipid raft‐like liposomes loading chimeric entry‐ inhibitor peptide |
Chimeric peptides |
HIV‐1 |
Target HIV‐1 gp41 and block viral entry |
Gómara et al., 2017137 |
|
|
Single‐layer and multi‐layer liposomes conjugated with S‐NeuAc‐α (2‐6)‐di‐ LacNAc |
– |
H1 N1 |
Block viral entry into MDCK cells |
Cheng et al., 2018138 |
|
|
Anti‐RSV peptide‐loaded liposomes |
RF‐482 |
RSV |
Inhibit the RSV fusion and block viral entry |
Joshi et al., 2018139 |
|
|
Cationic liposomes incorporated with stearylamine |
– |
Baculovirus, HSV‐1 |
Interact with the lipid envelope of viruses |
Tahara et al., 2018118 |
|
|
Transferrin (Tf)‐conjugated liposomes |
Ganciclovir |
Cytomegalo‐virus (CMV) |
Inhibit the expression of CMV glycoprotein B |
Asasutjarit et al., 202014b |
|
|
Solid lipid nanoparticles |
Solid lipid nanoparticles bearing short hairpin RNA |
shRNA74 |
HCV |
Silencing of HCV replicon |
Torrecilla et al., 2016140 |
|
Solid lipid nanoparticles encapsulating HIV‐1 protease inhibitor |
Ritonavir |
HIV‐1 |
Inhibit virus production by using HIV‐1 protease inhibitor |
Javan et al., 2017141 |
|
|
Solid lipid nanoparticles |
Solid lipid nanoparticles with a drug loading of 67.44 % |
Acyclovir |
HSV‐1 |
Sustained‐release of antiviral drug |
Kondel et al., 2019120 |
|
Planar lipid bilayers |
Discoidal phospholipid bilayers wrapped by two copies of amphipathic membrane scaffold protein |
– |
H1 N1 |
Perforate the viral envelope and cause virus inactivation |
Kong et al., 2019121 |
|
Dendrimers |
Multivalent glycodendrimers bearing different carbohydrates or glycomimetic DC‐SIGN ligands |
bis‐benzylamide 4 |
HIV, DENV |
Block DC‐SIGN mediated uptake of DENV; inhibit HIV trans‐infection |
Varga et al., 2014142 |
|
Dendrimers with polyphenolic core and 24 sulfonate surface groups |
– |
HCV |
Prevent virions absorption to the target cell |
Sepúlveda‐Crespo et al., 2017143 |
|
|
Anionic PEG‐citrate G2 dendrimer conjugated with multi‐epitopic HIV‐1 vaccine candidate |
Multi‐epitopic rHIVtop4 |
HIV‐1 |
Induce Th1 immune responses |
Abdoli et al., 2017144 |
|
|
6’‐sialyllactose‐polyamidoamine (6SL‐ PAMAM) conjugates |
– |
H1 N1 |
Inhibit virus attachment and viral entry |
Kwon et al., 2017124 |
|
|
Ammonium‐terminated amphiphilic Janus dendrimers |
Camptothecin |
HCV |
Inhibit NS3 protease of HCV and restrict virus replication |
Lancelot et al., 2017145 |
|
|
Biocompatible G1 and G2 anionic citrate‐ PEG‐citrate dendrimer |
– |
HIV |
Blockage of viral attachment |
Kandi et al., 2019146 |
|
|
Polyanionic carbosilane dendrimers with a polyphenolic core and sulfonate or carboxylate end‐groups |
– |
HIV‐1 |
Virions inactivation and gp120 shedding |
Sepúlveda‐Crespo et al., 201814c |
|
|
Anionic poly(alkylideneamine) dendrimers with carboxylate and sulfonate terminal groups |
– |
HIV‐1 |
Blockage of viral entry by interacting with target proteins on the virus |
Maciel et al., 2019147 |
|
|
3′‐sialyllactose‐ and 6′‐ sialyllactose‐conjugated PAMAM dendrimers |
– |
IAV |
Host‐specific inhibition of IAV infection |
Günther et al., 2020148 |
|
|
Micelle |
Stearic acid‐g‐chitosan oligosaccharide micelle conjugated with antiviral drug |
Acyclovir |
HBV |
Inhibit the expressions of hepatitis B surface antigen (HBsAg), hepatitis B e antigen (HBeAg) and HBV DNA |
Huang et al., 2011149 |
|
Cross‐linked polymeric micelles loading antiviral agent |
Camptothecin |
HCV |
Inhibition of HCV replicon |
Jiménez‐Pardo et al., 2015150 |
|
|
Camptothecin‐loaded amphiphilic polymeric micelles |
Camptothecin |
HCV |
Inhibit HCV replication |
Concellón et al., 2016151 |
|
|
Chitosan‐g‐stearic acid micelles delivering DNAzyme |
DrzBS |
HBV |
Suppress HBV S gene expression |
Hong et al., 2019128 |
3.2.2. Lipid‐based nanomaterials
Liposomes are vesicular systems consisting of unilamellar or multilamellar phospholipid bilayers.117 They have received much attention in the biomedical area owning to their outstanding biocompatibility, biodegradability, drug loading capacity and low toxicity. For example, ganciclovir (GCV) is applied for the treatment of retinitis caused by cytomegalovirus (CMV). Asasutjarit et al. developed transferrin (Tf)‐conjugated liposomes to deliver GCV via intravitreal injection and topical instillation. After Tf receptors‐mediated endocytosis, the drug‐loaded liposomes effectively inhibited the expression of CMV glycoprotein B, serving as a promising antiviral agent in the CMV retinitis therapy.14b Tahara et al. prepared a kind of cationic liposomes incorporated with stearylamine (SA) and proved their antiviral activity against recombinant baculovirus without loading active pharmaceutical ingredients. They demonstrated that the binding of SA liposomes to host cell membranes prevented viral entry and the antiviral activity of SA liposomes on HSV‐1 is comparable to that of antiviral drug acyclovir.118
Solid lipid nanoparticles (SLN) consist of lipids that are solid at body temperature, such as fatty acids and triglycerides. They have advantages including lower toxicity, lower cost, higher stability, broader available source, higher drug loading capacity compared with that of synthetic polymeric nanoparticles and liposomes.119 Kondel et al. prepared acyclovir SLN to treat HSV infection and demonstrated that a single dose of this formulation had comparative antiviral efficacy to the multiple‐dose regiment of traditional acyclovir.120
Another interesting study reported a novel virucidal nanodisc which was a self‐assembled discoidal planar lipid bilayer wrapped by two copies of amphipathic membrane scaffold protein (MSP) and modified with sialic acid on the surface. Mechanistically, the nanodiscs could bind to influenza virions via sialic acid interaction and then co‐endocytosed into host cells. Under low pH endosomal environment, the nanodiscs effectively perforated the viral envelope and lead to virus inactivation.121
3.2.3. Dendrimers
Dendrimers are spherical macromolecules consisting of a central core and three‐dimensional branched architecture with abundant end groups.122 The interior cavity is suitable for drug encapsulation while the exterior surface can be easily conjugated with drugs and targeting ligands.123 The use of functionalized dendrimers as antiviral agents has been widely explored. For instance, a study has shown that sulfonate‐ended carbosilane dendrimers presented virucidal activity against HIV‐1 infection through virions inactivation and gp120 shedding.14c Kwon et al. investigated the virucidal effect of multivalent 6’‐sialyllactose‐polyamidoamine (6SL‐PAMAM) conjugates against influenza A virus (IAV). These conjugates resisted H1 N1 induced hydrolysis and protected 75 % of mice from fatal attack with H1 N1, exhibiting the potential to be further developed as IAV inhibitors in virus treatment. Apart from the inherent antiviral activity of functionalized dendrimers, they also act as efficient nanocarriers for drug delivery.124 Lancelot et al. reported a type of amphiphilic Janus dendrimers consisting of two dendritic blocks with different end groups. The result showed that these dendrimers could encapsulate camptothecin while maintaining the activity of the drug. From the antiviral studies, the Janus dendrimers exerted effective anti‐HCV activity at low camptothecin concentration.125
3.2.4. Micelles
Micelles are colloidal systems synthesized from amphiphilic copolymers with particle size at 5–100 nm range.126 The inner core formed by hydrophobic blocks can encapsulate drugs with poor water solubility, while the outer shell formed by hydrophilic blocks can be readily functionalized with various chemical groups.127 Over the past years, micelles have attracted considerable attention as a drug delivery system. Hong et al. synthesized a DNAzyme called DrzBS which has the potential to inhibit HBV S gene expression via sequence‐specific mRNA cleavage. However, the application of this enzyme was limited due to the lack of an exogenous delivery system. To overcome this challenge, they constructed chitosan‐g‐stearic acid (CSO‐SA) micelles to effectively deliver DrzBS for HBV gene therapy. The results indicated that, compared with common transfection reagent LipofectamineTM, the DNAzyme delivered by micelles exhibited higher HBV inhibition rate and even prolonged therapeutic time to 96 h.128
3.3. Nanocomposites
Nanocomposites can be defined as heterogeneous materials with at least one component that has one, two or three dimensions of nanoscale size.152 Using combinational nanomaterials for the antiviral applications can effectively integrate the advantages of each component in the nanocomposites, which may exhibit fascinating and potent antiviral activity (Table 3). For instance, Du et al. prepared silver nanoparticle‐modified graphene oxide (GO‐AgNPs) nanocomposites and investigated the antiviral effect of the material on the replication of PRRSV. The results indicated that GO‐AgNPs effectively impeded PRRSV infection and exerted stronger antiviral effect than single GO and AgNPs, suggesting great potential of developing hybrid nanomaterials for antiviral applications.153 Besides, nanoscale TiO2 is one of the most popular photocatalysts possessing strong oxidizing ability under UV light.154 Zan et al. first investigated the photocatalysis effect of TiO2 nanoparticles and TiO2‐coated ceramic plate on HBsAg, which is involved in the attachment of HBV to hepatocytes. Both materials showed a destructive impact on HBsAg, suggesting their potential use as HBV inhibitor and providing feasible ideas to develop TiO2 based nanocomposites for antiviral applications.15a Recently, Monmaturapoj et al. modified TiO2 on the surface of hydroxyapatite and significant antiviral activity of HA/TiO2 composite against H1 N1 was observed. They demonstrated that the virus was firstly absorbed onto the surface of HA, followed with ROS generation from TiO2 under UV radiation that directly inactivated the virus.155 In another study, Ishiguro et al. deposited copper ion on their previously developed TiO2‐coated cordierite foam for air cleaner, which exhibited stronger antiviral and antibacterial activities than that of TiO2‐coated cordierite foam.156 Copper ions were also incorporated in zeolite‐textile materials and showed high and rapid inactivation of the avian influenza virus (AIV) H5 subtypes.15b
Table 3.
Antiviral nanocomposites.
|
Nanocomposites |
Virus |
Antiviral mechanism |
References |
|---|---|---|---|
|
TiO2‐coated ceramic plate |
HBV |
Inactivate HBV by photocatalysis effect |
Zan et al., 200715a |
|
TiO2 and DNA nanocomposites via polylysine (PL) linker |
H1 N1, H5 N1, H3 N2 |
Target conservative regions of viral RNA; inhibit virus reproduction |
Levina et al., 2016160 |
|
TiO2‐modified hydroxyapatite |
H1 N1 |
ROS‐induced virus inactivation |
Monmaturapoj et al., 2018155 |
|
Silver‐doped TiO2 nanocomposites |
H1 N1 |
Inactivate virus by enhanced photocatalytic reaction |
Moongraksathum et al., 2019161 |
|
Cu2+/ TiO2‐coated cordierite foam |
Qβ bacteriophage T4 bacteriophage |
Inactivate virus by enhanced photocatalytic reaction |
Ishiguro et al., 2013156 |
|
Cu2+ incorporated in zeolite‐textile materials |
AIV H5 subtypes |
Destruction of virions by Cu2+ |
Imai et al., 201215b |
|
AgNPs/CS composites |
H1 N1 |
Interact with virions and block viral entry |
Yasutaka et al., 2013162 |
|
Polyquaternary polyphosphonium‐ oligochitosans (PQPOC) decorated with AgNPs |
Hepatitis A virus (HAV), Noroviruses (NoV), and CoxB4. |
AgNPs bind to virions active sites; interaction between PQPOC and viruses; chitosan‐induced viral RNA degradation inhibits virus replication |
Sofy et al., 2019163 |
|
AgNPs‐modified GO |
Infectious bursal disease virus (IBDV), feline coronavirus(FcoV) |
Block viral entry and interfere with viral membrane fusion |
Chen et al., 2016164 |
|
AgNPs‐modified GO |
PRRSV |
Inhibition of viral entry and proliferation |
Du et al., 2018153 |
|
Nanosilver based anionic linear globular dendrimer |
HIV‐1 |
Inhibition of virus replication |
Ardestani et al., 2015165 |
|
Nanohybrids with SWCNTs, MWCNTs, and carbon nanohorns (SWCNHs) as scaffolds connecting to glycofullerenes |
EBOV |
Block DC‐SIGN mediated viral entry |
Rodríguez‐Pérez et al., 2018166 |
|
Nanofibrous membranes consist of electrospun nanofibers and the daylight‐active photosensitizers |
T7 bacteriophage |
ROS‐induced virus inactivation |
Si et al., 2018159 |
|
Quaternized chitosan nanofibers containing graphene |
PPV |
Bind and remove virus in solution |
Bai et al., 2013167 |
|
Chitosan‐chondroitin sulfate nanocomplex encapsulating tenofovir |
HIV‐1 |
Inhibit viral reverse transcriptase |
Wu et al., 2016168 |
|
PLGA‐CS coated nanoparticles loading two antiretrovirals |
HIV‐1 |
– |
Makita‐Chingombe et al., 2016169 |
|
Zidovudine loaded PVP/SA‐PEG nanoparticles |
HIV |
– |
K. S. et al., 2018170 |
|
S‐Linked sialyloligosaccharides bearing liposomes and micelles |
H1 N1 |
Block viral entry into the MDCK cells |
Yeh et al., 2015171 |
Nanofibers are a type of nanoscale fibrous material with high surface‐to‐volume ratio, facile surface functionalization and excellent mechanical properties, which have gathered great interest for applications in different biomedical fields.157 When mixed in nanocomposites, nanofibers usually exhibit desired synergistic effect in combination with other components. Bai et al. prepared quaternized electrospun chitosan nanofibers containing graphene, which exerted excellent virus removal efficacy by binding up to 95 % of porcine parvovirus. The addition of graphene can increase nanofiber formation and also the reduction of virus.158 Lately, Si et al. also reported a fascinating study of antiviral nanocomposites. They fabricated a novel daylight‐driven rechargeable nanofibrous membranes (RNMs) with virucidal activities through the integration of electrospun nanofibers and the photosensitizers. The photoactive RNMs can rapidly produce and accumulate ROS under daylight and release them under dim light or dark environment. With their high ROS production rate, durable activity and high biocidal efficacy, the RNMs possess great potential to serve as a biocidal layer on protective equipment.159
3.4. Antiviral biopolymers
3.4.1. Saccharides
Monosaccharides,172 oligosaccharides173 and polysaccharides174 and their derivatives175 are found to possess attractive properties, such as low toxicity, biocompatibility as well as antiviral efficacy. There exist numerous review papers discussing the structural properties and antiviral activities of saccharides, especially the widely studied oligosaccharides and polysaccharides.175, 176 Some latest research findings are illustrated in this review, and further details can be found in relevant references.
Among various marine organisms, seaweeds, also known as algae, are the most abundant source of polysaccharide, especially sulfated polysaccharides.177 Carrageenans are linear sulfated polysaccharides extracted from red seaweeds, which have been widely investigated as an antiviral agent.178 Boulho et al. applied both conventional and microwave‐assisted extraction (MAE) methods to harvest carrageenans from Solieria chordalis and explored their antiviral ability. Results indicated that by using MAE methods, the extracted carrageenans exhibited better antiviral ability against HSV‐1, equal to that of antiviral drug acyclovir.179 Guo et al. reported that iota‐carrageenan (CG) displayed potent antiviral activity against PRRSV by blocking viral entry and inhibiting virus‐induced NF‐κB activation, which is essential for viral gene transcription and replication.21b Another type of well‐known sulfated polysaccharides are fucoidans obtained from brown seaweeds. Recent studies have shown that fucoidans can exhibit antiviral properties. For example, Ponce et al. extracted fucoidans from brown seaweed Scytosiphon lomentaria and then isolated heavily sulfated galactofucans and uronofucoidans by cetrimide fractionation. They demonstrated that galactofucans exhibited significant antiviral activity against HSV‐1 and HSV‐2 infection while uronofucoidans showed no effect.180 Sun et al. also reported that fucoidans possessed anti‐HSV‐2 activity by interfering with the absorption of virions to the host cells.174 Generally, as shown in Figure 5, the possible antiviral mechanism of seaweed polysaccharides could be the prevention of virus adsorption into the host cells and/or inhibition of the new virion production inside the host cells.175
Figure 5.
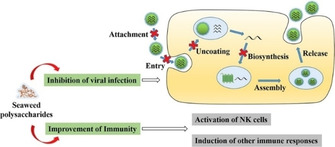
The antiviral activities and mechanisms of seaweed polysaccharides. Reproduced from reference [175] with permission from Elsevier.
Chitosan is a natural biocompatible and biodegradable biopolymer derived from chitin deacetylation.181 Sulfated chitosan has shown intriguing biological properties including antiviral effect.182 Gao et al. developed 36S derived from marine polysaccharides and demonstrated its broad anti‐HPV activities. 36S may directly block HPV entry by interacting with viral capsid protein. In addition, it may also interfere with the cellular PI3 K/Akt/mTOR pathway and therefore suppress cell autophagy.21a Besides, sulfated chitosan extracted from the cuttlebone of Sepia pharaonic was confirmed with antiviral effect against NDV by binding to virus receptors to prevent viral proliferation in the avian bloodstream.183
Cyclodextrins are natural cyclic oligosaccharides mainly composed of six to eight glucopyranoside units, with unique ring structure, internal hydrophobic cavity and hydrophilic external surface.184 The above properties make cyclodextrins become potent candidates in the treatment of viral infection via drug delivery, direct virucidal action or synergistic therapy. Recently, Jones et al. developed a nontoxic cyclodextrin modified with sulfonic acid, exhibiting irreversible virucidal activity against a wide range of heparan sulfate‐dependent viruses and posing a high barrier for the emergence of drug resistance.185
In summary, the structural diversity and complexity of saccharides and their derivatives could contribute their antiviral activities at different stages of viral infection processes, revealing considerable potential in future clinical transformation.
3.4.2. Peptides
Antiviral peptides have received much attention over the last few years because of their increasing discoveries of antiviral properties. Such peptides can derive from natural sources, such as bacteria, plants and animals, or can be rationally designed and synthesized.186 They have been reported to target different stages of the virus replication cycle, and the primary mechanism can be summarized as follows: First, some peptides can interact with the virus or host cells, block viral attachment and prevent viral fusion. Second, several types of peptides present direct biocidal activity by disrupting virus envelope. Third, specific peptides can interact with viral polymerase complex or stimulate immune response to inhibit viral replication.187 Lately, several reviews have comprehensively introduced the studies of antiviral peptides,186, 187, 188 readers may refer to those reviews for detailed information. In this section, we mainly focus on the latest development of antiviral peptides.
Recently, Mechlia et al. investigated the anti‐rabies activity of dermaseptins S3, S4 and their derivatives, which are excreted from amphibian skin glands. The results have shown that dermaseptins not only disrupted the viral envelope before viral entry but also affected downstream stages of the virus replication cycle after infection. Data also showed that S4 M4 K exhibited the highest therapeutic effect that protected 50–60 % infected mice from lethal challenge with Rabies virus (RABV).16b In another study, poly‐gamma‐glutamic acid (γ‐PGA), an anionic polypeptide generated from Bacillus species, showed its antiviral efficacy against norovirus infection. After oral administration, γ‐PGA can interfere with viral entry and increase IFN‐β production, which can effectively suppress virus replication in host cells.189 Also, β‐defensins, a family of endogenous cysteine‐rich and cationic peptides, demonstrated broad‐spectrum antiviral properties against various viruses, especially influenza virus.187b, 190 Zhao et al. synthesized different peptides derived from mouse β‐defensin‐4 and discovered that peptide P9 effectively inhibited influenza A virus (H1 N1, H3 N2, H5 N1, H7 N7 and H7 N9) and coronavirus (SARS‐CoV and MERS‐CoV). Mechanism analysis revealed that P9 bound to viral envelope glycoproteins, entered into the cells with the virus and prevented endosomal acidification, which impeded membrane fusion and viral RNA release.191
In addition, rationally synthesized peptides have also been extensively explored for their antiviral effect. Zhao et al. developed a type of dual‐functional peptide TAT P1, loading defective interfering genes (DIG‐3) of influenza virus. On the one hand, the delivered DIG‐3 significantly suppressed virus replication through cell transfection. On the other hand, the vector TAT P1 showed intrinsic antiviral activity by preventing endosomal acidification.192 This study has paved the way for developing transfection vectors as promising therapeutic agents in virus treatment.
Besides, membrane‐active peptide has received much attention over the past years in terms of developing antiviral agents. Many medically important viruses are equipped with lipid envelopes derived from host cell membranes, which play a crucial role in viral structural integrity and become attractive targets for membrane‐active peptide.193 For example, α‐helical (AH) peptide is known as a broad‐spectrum antiviral agent against a wide range of enveloped viruses via lipid membrane destabilization.194 Recently, Jackman et al. demonstrated a promising antiviral strategy named ‘lipid envelope antiviral disruption’ (LEAD), using AH peptide as the template (Figure 6). The engineered peptide could penetrate through the blood‐brain barrier and preferentially target at Zika virus, resulting in significantly reduced viral infection in mice model.31 Previous studies indicated that AH peptide possesses unique size‐selective disruptive behavior,194, 195 which can form pores in highly curved membranes (e. g., small vesicles, viral envelopes) and subsequently contribute to membrane lysis after reaching a critical pore intensity.196 It was further identified that the flexible conformation of AH peptide enables it to exhibit higher membrane targeting selectivity compared with C5 A peptide.197 The above findings suggest that LEAD strategy opens the door to the treatment of mosquito‐borne or other types of enveloped virus infection in a feasible approach.
Figure 6.
Scheme illustration of LEAD antiviral strategy. Reproduced from reference [197] with permission from the American Chemical Society.
4. State of the art in current antiviral biomedical applications
Infectious diseases caused by viruses are still a major threat to public health and associated with significant economic losses throughout the world.198 As of 20 August 2020, there have been over 22 million confirmed cases of COVID‐19 with 3.5 % mortality rate.199 The successful synthesis (including biosynthesis) of those fascinating antiviral materials may provide new insights into the development of antiviral protection solutions, potential antiviral agents, antiviral drug carriers and antiviral drug delivery systems.
4.1. Protective equipment against aerosol and droplet based viral entry
Proper antiviral protection can effectively prevent infectious disease, reduce economic losses and save lives.200 Viruses can spread among humans through direct or indirect contact to blood and other body fluids and/or exposure to respiratory aerosols or droplets from infectious individuals.201 If there is no vaccine for the specific virus, the best way to prevent spread and infection is to avoid being exposed to the viruses.202 Non‐pharmaceutical interventions, such as facemasks, goggles, gloves, and protective suits have been used to protect against virus infection during a pandemic.8b, 203
As most viruses range in size from 5 to 300 nm, the pore sizes of materials are critical for antiviral protection.204 The moisture‐proof plastic and rubber are commonly used for the fabrication of goggles and gloves, respectively. Compared with goggles and gloves, protective suits require more soft and comfortable permeable materials.205 However, porous materials are insufficient for antiviral protection. An antiviral material layer is required for protective suits by providing contact killing against virus either in aerosol or in liquid forms.206 The antiviral nanoparticles have been mechanically infused into textiles for protective suits and other protective products.159, 207
Facemasks and respirators are the key pieces of personal protective equipment (PPE) that are generally applied for the protection from the viruses transmitted by respiratory aerosols and droplets.208 Reusable cloth masks have been widely used by general public and even health care workers globally, particularly in Asia.209 The efficacy of cloth masks against specific virus infectious threats such as influenza and coronavirus could be extremely limited. And thus, the common practices such as reuse of masks are discouraged on the basis of public health judgment. Therefore, cost‐effective antiviral materials are preferred as the raw materials for disposable facemasks to lower the risk of severe illness from virus infection.
Currently, medical masks are recommended for the public to prevent respiratory virus infections.210 The medical masks, also known as surgical masks, are typically three layers that include outer, barrier and inner layers in sequence. The outer and inner layers are made of non‐woven fabric materials, and the barrier layer is made of a melt‐blown material. The barrier layer acts as the filter that prevents the virus as well as other microbes and particles from entering or exiting the mask. To increase the antiviral effectiveness, antiviral nanoparticles, such as silver, copper and zinc nanoparticles have been coated as a film on the outer layer of masks to rapidly destroy viruses before entering the barrier layer.159, 207, 211 Furthermore, an additional antiviral layer has also been developed using antiviral materials to co‐support with other conventional layers. The antiviral layer, which placed between outer layer and barrier layer could be made from hybrid materials, such as silver nanoparticle‐containing fibers, silver‐containing polymer nanocomposites.212 The design of an antiviral medical mask is shown in Figure 7. The main functions of outer layer, antiviral layer, barrier layer and inner layer are trapping viruses and penetration of air, inactivating or killing viruses, filtration of air, and final barrier, respectively.
Figure 7.
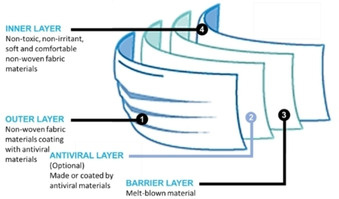
The design of medical masks against viruses
Besides the medical masks, protective respirators have also been used for antiviral applications.213 The respirators are tight‐fitting protective devices with superior antiviral properties when compared to the medical masks.214 The N95 respirators have been certified to filter at least 95 % of particles that are recommended to the healthcare workers to wear for prevention from contracting the virus.215 Antiviral materials, such as copper oxide nanoparticles could be impregnated into N95 respirators or coated on the surfaces of N95 respirators with an antiviral layer to further enhance the antiviral performance.216 Furthermore, elastomeric respirators are reusable devices with exchangeable cartridge filters that offer a viable protection option to healthcare workers for updating respiratory protection programs.217 Obviously, the application of nanomaterials, nanocomposites or biopolymers with antiviral properties to the structure of elastomeric respirators could offer many potential advantages, such as enhanced antiviral efficiency, reduced disinfection frequency, and extended wear time.218 For instance, the graphene containing chitosan nanofibers and daylight‐driven rechargeable nanofibrous membranes mentioned in the previous section would become suitable candidates in the filter design.158, 159
4.2. Potential antiviral agents
The emergence of various viruses has increased the demand for potential antiviral agents, which can be used to treat viral infections.219 The antiviral agents are different from viricides and should be relatively harmless to the hosts.220 Nowadays, researchers are working on expanding the scope of antiviral agents to different viruses, such as coronavirus, HIV, HBV, HCV, HSV, and influenza A and B viruses.220b, 221 However, it is difficult to design safe and effective antiviral agents without damaging host biological cells. Furthermore, the emergence of widespread drug resistance and viral mutation makes developing antiviral agents even more difficult. Research on potential antiviral drugs must therefore be continued, and all possible strategies should be challenging.
Some antiviral materials, especially biopolymers are promising candidates as antiviral agents.7, 222 Most biopolymers are isolated from natural sources, which possess low toxicity, less side‐effects, renewable supply and biodiversity. These biopolymers could offer complementary and overlapping mechanisms of antiviral action by inhibiting viral replication and/or viral genome synthesis. Compared with standard combinatorial chemistry, these biopolymers have higher chemical diversity and biochemical specificity, providing an important opportunity to find new lead structures that are bioactive against a wide range of viruses. In addition, the structures of the biopolymers can lead to chemical modification with improved antiviral activities.223
In recent years, researchers have confirmed that some polysaccharides,,[222b,222c][224] proteins,225 peptides,226 polyphenols,227 poly‐and oligonucleotides228 and some natural products derived materials229 possess antiviral activities and wide‐ranging beneficial therapeutic effects. Specifically, the polysaccharides have emerged as an essential antiviral agent, both in vitro and in vivo. Due to unparalleled chemical diversity, polysaccharides offer unlimited opportunities for new antiviral agents.222b, 222c, 230 Recently, the polysaccharides were reported as a novel strategy for inhibiting or slowing down the viral infection without cytotoxic effects. The polysaccharides, particularly sulfated polysaccharides, demonstrated to have antiviral bioactivities against multiple viruses, such as coronavirus, DENV, influenza virus, poliovirus, HSV and varicella‐zoster viruses.230
Nevertheless, more studies on structure‐activity relationships, mechanism of action, drug metabolism, molecular simulation, and combinatorial chemistry are required for better utilization of those biopolymers for biomedical applications. Besides, some biopolymers, such as sulfated polysaccharides, polyphenols and peptides, can be used as synergistic enhancers to boost the effect of other antiviral drugs.231 The synergistic effect can reduce the therapeutic dose and toxicity of antiviral drugs, and minimize or delay the induction of antiviral resistance.
4.3. Antiviral drug carriers
Currently, antiviral drugs suffer from low antiviral efficacy, low compound solubility, low bioavailability when administered in conventional dosage forms. Short half‐lives of active compounds, undesired systemic toxic and side effects hinder the development of antiviral drugs.232 A variety of carrier systems have been developed for antiviral drugs to improve the effectiveness and specificity of them. By far, the most widely studied antiviral drug carriers are biodegradable materials and nanomaterials. Besides physical immobilization and encapsulation, the conjugation of antiviral drugs to biodegradable polymeric carriers is usually designed by the presence of covalent bond between the water‐soluble polymer and the antiviral drug molecule. The concept of polymer‐drug conjugates opens up a new perspective for drug carriers in modern pharmacy.233 Compared with conventional antiviral drugs, the antiviral effectiveness of polymer‐drug conjugates can be achieved by either part of the polymeric backbone or the side chains, which offer several attractive advantages, such as improved water solubility and stability, controlled administration, and improved pharmacokinetics and biodistribution. To develop antiviral polymer conjugates, several biodegradable polymers are currently being studied, such as poly(N‐(2‐hydroxypropyl)methacrylamide), lignins, (glycol)proteins, deoxynucleotide and biocompatible dendrimers for reduced cytotoxicity and enhanced activity of antiviral drugs.234
Advances in nanotechnology have a profound impact on drug carriers, leading to the development of nanomaterials with larger loading capacity and higher targeting accuracy for the treatment of viral diseases.235 Nanomaterials with antiviral intrinsic activity can be considered drug carriers to enhance the effectiveness of antiviral drugs by synergistic effects. However, they are often related to solubility and bioavailability issues. To overcome these limitations, biodegradable nanoparticles, such as lipid‐based nanoparticles and biopolymeric nanoparticles have been commonly used as carriers for antiviral drug delivery in the treatment of various viruses. By using these nanocarriers, it is possible to overcome the problems of many antiviral drugs in conventional dosage forms, which may help to address solubility and dissolution issues, increase the bioavailability of drugs, protect sensitive drugs from degradation, reduce adverse side effects, improve tissue tolerance to drugs and reduce treatment costs.34b, 236
4.4. Antiviral drug delivery systems
As viruses are characterized by rapid replication within host cells and carry the ability to attack any part of the host cell, the clinical efficacy of antiviral drugs and their bioavailability becomes more critical considerations in the treatment of viral infections.237 This makes it difficult to find targets for antiviral drugs that can interfere with the viruses without harming the host cells. In recent years, more and more new antiviral drugs have entered the pharmaceutical market. Therefore, various antiviral drug delivery systems have been used for drug site‐specific targeting to enhance the effectiveness of the treatment. The targets are not only specific cells, but also specific organelles for antiviral and antiretroviral therapy. Figure 8 outlines the drug delivery systems most commonly studied for antiviral and antiretroviral therapy.238
Figure 8.
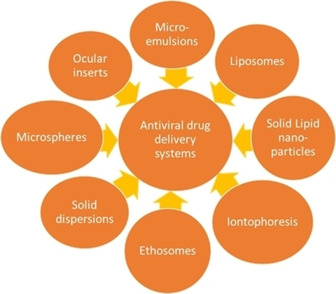
Schematic diagram of antiviral drug delivery systems. Reproduced from reference [238] with permission from Medknow.
Among the different materials of drug delivery systems being currently investigated by pharmaceutical scientists, “smart” polymer biomaterials hold a great potential for antiviral drug delivery.239 As an essential feature of human body systems is their ability to change important properties in response to tiny environmental signals, the development of “smart” polymer biomaterials with biomimetic properties could be applied as intelligent antiviral drug delivery systems. “Smart” polymers, or stimuli‐responsive polymers in a more scientific term, are capable of altering their chemical and/or physical properties upon exposure to external stimuli.239, 240 These materials have been intensively studied over the years for on‐demand drug deliveries. The schematic representation of “smart” polymers as a drug delivery system for the transport of active antiviral drugs is shown in Figure 9. Inspired by viruses trafficking from endo‐lysosomes, “smart” polymers have been used as an effective drug delivery system. The stimuli‐responsive degradation properties of “smart” polymers have shown great possibilities in exhibiting enhanced release of antiviral therapeutics into targeted cells, even the specific organelles.241
Figure 9.
Schematic representation of a “smart” polymer‐based delivery system for the transport of active antiviral drugs.239, 240
5. Challenges and future prospects
In general, nanomaterials have been used as cost‐effective biomedical materials for developing antiviral protective solutions. Antiviral biopolymers exhibit equivalent potential for antiviral applications due to their inherent low toxicity, broad‐spectrum antiviral activity, high specificity, effectiveness, wide acceptability, biodegradability, and relatively low production costs.11c, 242 All the efforts dedicated to the synthesis and characterization of these antiviral materials have paved the way for various biomedical applications. However, it was highlighted that dosage regulation and effectivity need to be addressed before industrial‐scale applications.7 Most studies were in the preliminary stages of testing and implementation. For challenging biomedical applications, such as using antiviral drug carriers and antiviral drug delivery systems, it has been found that the current antiviral materials are far from meeting all the clinical requirements. Nevertheless, antiviral biodegradable nanomaterials and “smart” polymer biomaterials are still expected to solve the related biomedical problems and accelerate the transition to clinical applications.
On the one hand, the most important limitation observed is that most of the conclusions derived were based on the observations from the in vitro antiviral studies. Further exploration into in vivo studies with animal and clinical testing are essential to progress towards real biomedical applications. On the other hand, the complexity of interactions during the antiviral activity limits the ability to understand the antiviral mechanism. Currently, most of the reported mechanisms are based on speculations. Although the results from the antiviral studies were conclusive, the underlying mechanism of action remains unresolved due to the lack of sufficient evidence. Profound understandings of the antiviral mechanisms would facilitate precise antiviral material development.
The reviewed candidates for the antiviral applications demonstrate significant potential in the fight against future viral pandemics. The use of different virus strains and types further complicates the correlation among studies. The most common viral strains used are HIV and HSV, which restrict the application scope of antiviral materials. It would be ideal to develop and test the antiviral materials on certain viruses that represent each viral category. A universal protocol for such studies would facilitate the applications to a broader spectrum of viral pathogens. It was emphasized that there is an urgent need to develop more effective antiviral therapies to reduce morbidity and mortality.243 A concerted effort is essential to focus the research towards specific goals of achieving the antiviral materials with desired performance.
Amidst the viral infections, there are numerous unexplained factors that determine interspecies transmission, reassortment, human‐to‐human transmission, and exposure‐to‐infection ratio of the novel viruses.3, 6a The opportunity to observe real‐time virus evolution would provide us with invaluable information on the factors that determine pathogenicity and/or transmissibility.6a The availability of immense amount of data from the previous pandemics can aid along with the help of automation, artificial intelligence and bioinformatics to direct the development of the research area. In the case of H5 N1, Yong reported that 99.9 % of the exposed population with antibodies carried in their blood failed to develop the disease. However, rapid viral replication was observed in infected people.3 The cases were clustered within the genetically susceptible blood relatives while others possessed genetic variants that protected them.3 The complications in understanding the pattern of viral infection need resolution.
The economic cost of the pandemics is high with the loss of lives, loss in productivity, social disruption, and incurred government and medical expenses. For example, the economic cost of the 1957 and 1968 pandemics combined was $32 billion (in 1995 dollars).244 Hence, it is essential for governments and interested institutions to venture and invest in active research addressing the gap in the field. In principle, the H5 N1 can become airborne and unpredictable.3 Preparedness for such an airborne pandemic could prevent large‐scale implications. The development and/or evolution of novel viruses are predicted to be imminent that would increase the number of pandemics in the future. The probability of pandemic occurrence increases over time.244, 245 Moreover, the higher the frequency of pandemics the greater the failure of containment efforts due to the shortage of workforce and resources.245 Hanvoravongchai et al. studied the pandemic preparedness and health system challenges of six Asian countries including Thailand, Indonesia, Taiwan, Lao PDR, Cambodia and Viet Nam.246 The shortage of highly skilled workers, insufficient stockpiles of the antivirals and PPE were some of the resource challenges attributed to the developing countries. Unfortunately, it is also suggested that the human understanding of the emergence of pandemics is incomplete and may not be as frequent as indicated by scientists.245
Further, a WHO report stated that creativity and imagination are essential to the research enterprise with new ideas emerging to resolve health problems.5 It emphasized that the constraints are in the means to implement these ideas and highest quality research into dependable and practical applications to facilitate better health. Research investments are attained by demonstrating results from translatable scientific investigations into accessible and affordable health services. It will be ever more arduous and convoluted to develop antiviral materials with the emergence of new virus strains. The development of novel methods of treatment or inhibition would eventually be overcome by nature (in this case, viruses) over time. Hence, this fight against the viruses will be never ending.
Conflict of interest
The authors declare no conflict of interest.
Biographical Information
Lili Liang obtained her B.S. in pharmaceutics from Zhejiang University, China in 2019. From 2019, she is a PhD student in the School of Civil and Environmental Engineering and Nanyang Environment and Water Research Institute at Nanyang Technological University (NTU), Singapore. Her current research interests mainly focus on conversion of plastic waste to carbon‐based nanomaterials and their applications in biomedical, analytical areas.

Biographical Information
Ashiq Ahamed is a researcher at the Nanyang Technological University (NTU), Singapore and a doctoral candidate at the Abo Akademi University, Finland. His current research interests include Environmental Impact Assessment, Toxicity, Waste Management, and Electrochemical applications. He has worked in various government‐funded and consulting projects at NTU. He is trained in Life Cycle Assessment tools such as GaBi and SimaPro, Nanotools such as AFM, Ellipsometry, ATR‐FTIR and QCMD, USEtox® 2.0 – Characterization fact.

Biographical Information
Liya Ge obtained her B.Eng. in chemical engineering from Zhejiang University, China in 1999. From 1999–2002, she worked as an Engineer in State Oceanic Administration, China. She subsequently completed her PhD study in analytical chemistry from Nanyang Technological University (NTU), Singapore in 2005. Since then, Dr. Ge has been working as a researcher and later a senior researcher in NTU. Her current research interests cover analytical chemistry including the development of sensors, biosensors and micro total analysis systems for rapid on‐site detection and point‐of‐care diagnosis.

Biographical Information
Xiaoxu Fu is currently a PhD student at the School of Civil and Environmental Engineering at Nanyang Technological University (NTU), Singapore. He got his master degree in the same school in NTU and his bachelor degree in Shanghai Jiao Tong university in China. His scienfic research and interest is focused on conducting polymers and their application on biomedical, analytical and environmental applications.

Biographical Information
Grzegorz Lisak obtained his M.Sc. (Chem. Eng.) in physical chemistry in 2007 from Poznań University of Technology, Poland. He obtained his D.Sc. (Tech.) in analytical chemistry in 2012 from Åbo Akademi University, Finland. Between 2013 and 2015 he was a research fellow at University of Wollongong, Australia and Malmö University, Sweden. In 2016, he was made assistant professor at Nanyang Technological University, Singapore. Since 2017 he is a Director of the Residues and Resources Reclamation Center at Nanyang Environment and Water Research Institute, Singapore.

Acknowledgements
The authors would like to acknowledge the Nanyang Environment and Water Research Institute, Nanyang Technological University (Singapore), and Economic Development Board (Singapore) for the financial support of this research
L. Liang, A. Ahamed, L. Ge, X. Fu, G. Lisak, ChemPlusChem 2020, 85, 2105.
References
- 1. Holmes E. C., Nature 2011, 478, 319–320. [Google Scholar]
- 2. Butler D., Nature 2009, 458, 1082–1083. [DOI] [PubMed] [Google Scholar]
- 3. Yong E., Nature 2012, 486, 456–459. [DOI] [PubMed] [Google Scholar]
- 4. Correspondent O. C. B., Nature 1973, 243, 187–188. [Google Scholar]
- 5.W. H. Organization, 2014.
- 6.
- 6a. Neumann G., Noda T., Kawaoka Y., Nature 2009, 459, 931–939; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6b. Mahmoud A., Microb. Biotechnol. 2016, 9, 549–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Randazzo W., Fabra M. J., Falcó I., López-Rubio A., Sánchez G., Compr. Rev. Food Sci. Food Saf. 2018, 17, 754–768. [DOI] [PubMed] [Google Scholar]
- 8.
- 8a. MacIntyre C. R., Cauchemez S., Dwyer D. E., Seale H., Cheung P., Browne G., Fasher M., Wood J., Gao Z., Booy R., Emerging Infect. Dis. 2009, 15, 233; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Bischoff W. E., Reid T., Russell G. B., Peters T. R., J. Infect. Dis. 2011, 204, 193–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. J. Flint, L. W. Enquist, V. R. Racaniello, A. M. Skalka, Principles of Virology : Pathogenesis and Control, ASM Press, Washington, UNITED STATES, 2008.
- 10.
- 10a. Chan Y. F., Abu Bakar S., Med J Malaysia 2005, 60, 246–248; [PubMed] [Google Scholar]
- 10b. Wutzler P., Sauerbrei A., Lett. Appl. Microbiol. 2004, 39, 194–198. [DOI] [PubMed] [Google Scholar]
- 11.
- 11a. Szunerits S., Barras A., Khanal M., Pagneux Q., Boukherroub R., Molecules 2015, 20, 14051–14081; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b. Chen L., Huang G., Int. J. Biol. Macromol. 2018, 115, 77–82; [DOI] [PubMed] [Google Scholar]
- 11c. Boas L. C. P. V., Campos M. L., Berlanda R. L. A., de Carvalho Neves N., Franco O. L., Cell. Mol. Life Sci. 2019, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Joe Y. H., Park D. H., Hwang J., J. Hazard. Mater. 2016, 301, 547–553; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b. Elechiguerra J. L., Burt J. L., Morones J. R., Camacho-Bragado A., Gao X., Lara H. H., Yacaman M. J., J. Nanobiotechnol. 2005, 3, 6; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12c. Kosiol P., Hansmann B., Ulbricht M., Thom V., J. Membr. Sci. 2017, 533, 289–301; [Google Scholar]
- 12d. Bowman M.-C., Ballard T. E., Ackerson C. J., Feldheim D. L., Margolis D. M., Melander C., J. Am. Chem. Soc. 2008, 130, 6896–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.
- 13a. Park K.-T., Hwang J., Carbon 2014, 75, 401–410; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b. Iannazzo D., Pistone A., Galvagno S., Ferro S., De Luca L., Monforte A. M., Da Ros T., Hadad C., Prato M., Pannecouque C., Carbon 2015, 82, 548–561; [Google Scholar]
- 13c. Ye S., Shao K., Li Z., Guo N., Zuo Y., Li Q., Lu Z., Chen L., He Q., Han H., ACS Appl. Mater. Interfaces 2015, 7, 21571–21579; [DOI] [PubMed] [Google Scholar]
- 13d. Du T., Lu J., Liu L., Dong N., Fang L., Xiao S., Han H., ACS Applied Bio Materials 2018, 1, 1286–1293. [DOI] [PubMed] [Google Scholar]
- 14.
- 14a. Li W., Yu F., Wang Q., Qi Q., Su S., Xie L., Lu L., Jiang S., AIDS 2016, 30, 827–838; [DOI] [PubMed] [Google Scholar]
- 14b. Asasutjarit R., Managit C., Phanaksri T., Treesuppharat W., Fuongfuchat A., Int. J. Pharm. 2020, 119084; [DOI] [PubMed] [Google Scholar]
- 14c. Sepulveda-Crespo D., de la Mata F. J., Gomez R., Munoz-Fernandez M. A., Nanoscale 2018, 10, 8998–9011. [DOI] [PubMed] [Google Scholar]
- 15.
- 15a. Zan L., Fa W., Peng T., Gong Z.-k., J. Photochem. Photobiol. B, Biol. 2007, 86, 165–169; [DOI] [PubMed] [Google Scholar]
- 15b. Imai K., Ogawa H., Bui V. N., Inoue H., Fukuda J., Ohba M., Yamamoto Y., Nakamura K., Antiviral Res. 2012, 93, 225–233. [DOI] [PubMed] [Google Scholar]
- 16.
- 16a. Gao Y., Liu W., Wang W., Zhang X., Zhao X., Carbohydr. Polym. 2018, 198, 329–338; [DOI] [PubMed] [Google Scholar]
- 16b. Mechlia M. B., Belaid A., Castel G., Jallet C., Mansfield K. L., Fooks A. R., Hani K., Tordo N., Vaccine 2019, 37, 4694–4700. [DOI] [PubMed] [Google Scholar]
- 17. Galdiero S., Falanga A., Vitiello M., Cantisani M., Marra V., Galdiero M., Molecules 2011, 16, 8894–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.
- 18a. Dimitrov D. S., Nat. Rev. Microbiol. 2004, 2, 109–122; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18b. Vitiello M., Galdiero M., Galdiero M., Protein Pept. Lett. 2009, 16, 786–793. [DOI] [PubMed] [Google Scholar]
- 19. Yang X. X., Li C. M., Huang C. Z., Nanoscale 2016, 8, 3040–3048. [DOI] [PubMed] [Google Scholar]
- 20. Speshock J. L., Murdock R. C., Braydich-Stolle L. K., Schrand A. M., Hussain S. M., J. Nanobiotechnol. 2010, 8, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.
- 21a. Gao Y., Liu W., Wang W., Zhang X., Zhao X., Carbohydr. Polym. 2018, 198, 329–338; [DOI] [PubMed] [Google Scholar]
- 21b. Guo C., Zhu Z., Yu P., Zhang X., Dong W., Wang X., Chen Y., Liu X., Antivir. Ther. 2019, 24, 261–270; [DOI] [PubMed] [Google Scholar]
- 21c. Tavakoli A., Ataei-Pirkooh A., Mm Sadeghi G., Bokharaei-Salim F., Sahrapour P., Kiani S. J., Moghoofei M., Farahmand M., Javanmard D., Monavari S. H., Nanomedicine 2018, 13, 2675–2690. [DOI] [PubMed] [Google Scholar]
- 22. Barras A., Pagneux Q., Sane F., Wang Q., Boukherroub R., Hober D., Szunerits S., ACS Appl. Mater. Interfaces 2016, 8, 9004–9013. [DOI] [PubMed] [Google Scholar]
- 23. Rogers J. V., Parkinson C. V., Choi Y. W., Speshock J. L., Hussain S. M., Nanoscale Res. Lett. 2008, 3, 129–133. [Google Scholar]
- 24. Papp I., Sieben C., Ludwig K., Roskamp M., Bottcher C., Schlecht S., Herrmann A., Haag R., Small 2010, 6, 2900–2906. [DOI] [PubMed] [Google Scholar]
- 25.
- 25a. Baram-Pinto D., Shukla S., Gedanken A., Sarid R., Small 2010, 6, 1044–1050; [DOI] [PubMed] [Google Scholar]
- 25b. Baram-Pinto D., Shukla S., Perkas N., Gedanken A., Sarid R., Bioconjugate Chem. 2009, 20, 1497–1502; [DOI] [PubMed] [Google Scholar]
- 25c. Kumar R., Pandey K., Sahoo G. C., Das S., Das V., Topno R. K., Das P., Mater. Sci. Eng. C 2017, 75, 1465–1471; [DOI] [PubMed] [Google Scholar]
- 25d. Lu L., Sun R. W., Chen R., Hui C. K., Ho C. M., Luk J. M., Lau G. K., Che C. M., Antivir Ther 2008, 13, 253–262. [PubMed] [Google Scholar]
- 26. Vigant F., Santos N. C., Lee B., Nat. Rev. Microbiol. 2015, 13, 426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cho N. J., Glenn J. S., Nat. Mater. 2020, 19, 813–816. [DOI] [PubMed] [Google Scholar]
- 28. Akhtar S., Shahzad K., Mushtaq S., Ali I., Rafe M. H., Fazal-ul-Karim S. M., Mater. Res. Express 2019, 6. [Google Scholar]
- 29. de Souza e Silva J. M., Hanchuk T. D. M., Santos M. I., Kobarg J., Bajgelman M. C., Cardoso M. B., ACS Appl. Mater. Interfaces 2016, 8, 16564–16572. [DOI] [PubMed] [Google Scholar]
- 30. Tahara K., Kobayashi M., Yoshida S., Onodera R., Inoue N., Takeuchi H., Int. J. Pharm. 2018, 543, 311–317. [DOI] [PubMed] [Google Scholar]
- 31. Jackman J. A., Costa V. V., Park S., Real A. L. C. V., Park J. H., Cardozo P. L., Ferhan A. R., Olmo I. G., Moreira T. P., Bambirra J. L., Queiroz V. F., Queiroz-Junior C. M., Foureaux G., Souza D. G., Ribeiro F. M., Yoon B. K., Wynendaele E., De Spiegeleer B., Teixeira M. M., Cho N.-J., Nat. Mater. 2018, 17, 971–977. [DOI] [PubMed] [Google Scholar]
- 32. Jackman J. A., Shi P.-Y., Cho N.-J., ACS Infect. Dis. 2019, 5, 4–8. [DOI] [PubMed] [Google Scholar]
- 33. Badani H., Garry R. F., Wimley W. C., Biochim. Biophys. Acta 2014, 1838, 2180–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.
- 34a. Kinman L., Brodie S. J., Tsai C. C., Bui T., Larsen K., Schmidt A., Anderson D., Morton W. R., Hu S.-L., Ho R. J., J. Acquired Immune Defic. Syndr. 2003, 34, 387–397; [DOI] [PubMed] [Google Scholar]
- 34b. Vyas T. K., Shah L., Amiji M. M., Expert Opin. Drug Delivery 2006, 3, 613–628. [DOI] [PubMed] [Google Scholar]
- 35. Zhu B., Liu G. L., Ling F., Wang G. X., Antiviral Res. 2015, 118, 29–38. [DOI] [PubMed] [Google Scholar]
- 36. Ghaffari H., Tavakoli A., Moradi A., Tabarraei A., Bokharaei-Salim F., Zahmatkeshan M., Farahmand M., Javanmard D., Kiani S. J., Esghaei M., Pirhajati-Mahabadi V., Monavari S. H., Ataei-Pirkooh A., J. Biomed. Sci. 2019, 26, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tavakoli A., Hashemzadeh M. S., J. Virol. Methods 2020, 275, 113688. [DOI] [PubMed] [Google Scholar]
- 38. Khalid K., Tan X., Mohd Zaid H. F., Tao Y., Lye Chew C., Chu D.-T., Lam M. K., Ho Y.-C., Lim J. W., Chin Wei L., BioEngineering 2020, 11, 328–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saravanan M., Asmalash T., Gebrekidan A., Gebreegziabiher D., Araya T., Hilekiros H., Barabadi H., Ramanathan K., Pharmaceutical nanotechnology 2018, 6, 17–27. [DOI] [PubMed] [Google Scholar]
- 40. Garrido C., Simpson C. A., Dahl N. P., Bresee J., Whitehead D. C., Lindsey E. A., Harris T. L., Smith C. A., Carter C. J., Feldheim D. L., Future Med. Chem. 2015, 7, 1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y., Lin Z., Zhao M., Xu T., Wang C., Hua L., Wang H., Xia H., Zhu B., ACS Appl. Mater. Interfaces 2016, 8, 24385–24393. [DOI] [PubMed] [Google Scholar]
- 42. Shankar S., Jaiswal L., Aparna R. S. L., Prasad R. G. S. V., Mater. Lett. 2014, 137, 75–78. [Google Scholar]
- 43. Dos Santos C. A., Seckler M. M., Ingle A. P., Gupta I., Galdiero S., Galdiero M., Gade A., Rai M., J. Pharm. Sci. 2014, 103, 1931–1944. [DOI] [PubMed] [Google Scholar]
- 44. Akbarzadeh A., Kafshdooz L., Razban Z., Dastranj Tbrizi A., Rasoulpour S., Khalilov R., Kavetskyy T., Saghfi S., Nasibova A. N., Kaamyabi S., Kafshdooz T., Artificial Cells, Nanomedicine, and Biotechnology 2018, 46, 263–267. [DOI] [PubMed] [Google Scholar]
- 45. Du T., Liang J., Dong N., Lu J., Fu Y., Fang L., Xiao S., Han H., ACS Appl. Mater. Interfaces 2018, 10, 4369–4378. [DOI] [PubMed] [Google Scholar]
- 46.
- 46a. Zorofchian Moghadamtousi S., Abdul Kadir H., Hassandarvish P., Tajik H., Abubakar S., Zandi K., BioMed Res. Int. 2014, 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46b. Singh D. K., Jagannathan R., Khandelwal P., Abraham P. M., Poddar P., Nanoscale 2013, 5, 1882–1893. [DOI] [PubMed] [Google Scholar]
- 47. Yang X. X., Li C. M., Huang C. Z., Nanoscale 2016, 8, 3040–3048. [DOI] [PubMed] [Google Scholar]
- 48. Singh P., Pandit S., Mokkapati V., Garg A., Ravikumar V., Mijakovic I., Int. J. Mol. Sci. 2018, 19, 1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yadavalli T., Shukla D., Nanomed. Nanotech. Biol. Med. 2017, 13, 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee M.-Y., Yang J.-A., Jung H. S., Beack S., Choi J. E., Hur W., Koo H., Kim K., Yoon S. K., Hahn S. K., ACS Nano 2012, 6, 9522–9531. [DOI] [PubMed] [Google Scholar]
- 51. Bai Y., Zhou Y., Liu H., Fang L., Liang J., Xiao S., ACS Appl. Nano Mater. 2018, 1, 969–976. [Google Scholar]
- 52. Malhotra B. D., Ali M. A., Nanomaterials for Biosensors (Eds.: B. D. Malhotra, M. A. Ali), William Andrew Publishing, 2018, pp. 75–103. [Google Scholar]
- 53. Muñoz A., Sigwalt D., Illescas B. M., Luczkowiak J., Rodríguez-Pérez L., Nierengarten I., Holler M., Remy J.-S., Buffet K., Vincent S. P., Rojo J., Delgado R., Nierengarten J.-F., Martín N., Nat. Chem. 2016, 8, 50–57. [DOI] [PubMed] [Google Scholar]
- 54. Illescas B. M., Rojo J., Delgado R., Martín N., J. Am. Chem. Soc. 2017, 139, 6018–6025. [DOI] [PubMed] [Google Scholar]
- 55. Castro E., Martinez Z. S., Seong C. S., Cabrera-Espinoza A., Ruiz M., Hernandez Garcia A., Valdez F., Llano M., Echegoyen L., J. Med. Chem. 2016, 59, 10963–10973. [DOI] [PubMed] [Google Scholar]
- 56.
- 56a. Höfinger S., Melle-Franco M., Gallo T., Cantelli A., Calvaresi M., Gomes J. A. N. F., Zerbetto F., Biomaterials 2011, 32, 7079–7085; [DOI] [PubMed] [Google Scholar]
- 56b. Sahoo N. G., Bao H., Pan Y., Pal M., Kakran M., Cheng H. K. F., Li L., Tan L. P., Chem. Commun. 2011, 47, 5235–5237. [DOI] [PubMed] [Google Scholar]
- 57. Zhang Z., Wang B., Wan B., Yu L., Huang Q., Biochem. Biophys. Res. Commun. 2013, 436, 650–654. [DOI] [PubMed] [Google Scholar]
- 58. Yang X. X., Li C. M., Li Y. F., Wang J., Huang C. Z., Nanoscale 2017, 9, 16086–16092. [DOI] [PubMed] [Google Scholar]
- 59. Xu X., Ray R., Gu Y., Ploehn H. J., Gearheart L., Raker K., Scrivens W. A., J. Am. Chem. Soc. 2004, 126, 12736–12737. [DOI] [PubMed] [Google Scholar]
- 60. Huang S., Gu J., Ye J., Fang B., Wan S., Wang C., Ashraf U., Li Q., Wang X., Shao L., Song Y., Zheng X., Cao F., Cao S., J. Colloid Interface Sci. 2019, 542, 198–206. [DOI] [PubMed] [Google Scholar]
- 61. Ting D., Dong N., Fang L., Lu J., Bi J., Xiao S., Han H., ACS Appl. Nano Mater. 2018, 1, 5451–5459. [DOI] [PubMed] [Google Scholar]
- 62. de Souza e Silva J. M., Hanchuk T. D. M., Santos M. I., Kobarg J., Bajgelman M. C., Cardoso M. B., ACS Appl. Mater. Interfaces 2016, 8, 16564–16572. [DOI] [PubMed] [Google Scholar]
- 63. Hang X., Peng H., Song H., Qi Z., Miao X., Xu W., J. Virol. Methods 2015, 222, 150–157. [DOI] [PubMed] [Google Scholar]
- 64. Akhtar S., Shahzad K., Mushtaq S., Ali I., Rafe M. H., Fazal-ul-Karim S. M., Mater. Res. Express 2019, 6. [Google Scholar]
- 65. Bamrungsap S., Zhao Z., Chen T., Wang L., Li C., Fu T., Tan W., Nanomedicine 2012, 7, 1253–1271. [DOI] [PubMed] [Google Scholar]
- 66. LaBauve A. E., Rinker T. E., Noureddine A., Serda R. E., Howe J. Y., Sherman M. B., Rasley A., Brinker C. J., Sasaki D. Y., Negrete O. A., Sci. Rep. 2018, 8, 13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Li Y., Lin Z., Zhao M., Guo M., Xu T., Wang C., Xia H., Zhu B., RSC Adv. 2016, 6, 89679–89686. [Google Scholar]
- 68. Etemadzade M., Ghamarypour A., Zabihollahi R., shabbak G., Shirazi M., Sahebjamee H., Vaziri A. Z., Assadi A., Ardestani M. S., Shandiz S. A. S., Aghasadeghi M. R., Asian Pac. J. Trop. Dis. 2016, 6, 854–858. [Google Scholar]
- 69. Lin Z., Li Y., Guo M., Xu T., Wang C., Zhao M., Wang H., Chen T., Zhu B., RSC Adv. 2017, 7, 742–750. [Google Scholar]
- 70. Huy T. Q., Hien Thanh N. T., Thuy N. T., Chung P. V., Hung P. N., Le A.-T., Hong Hanh N. T., J. Virol. Methods 2017, 241, 52–57. [DOI] [PubMed] [Google Scholar]
- 71. El-Mohamady R. S., Ghattas T., Zawrah M., Abd El-Hafeiz Y., Int. J. Veterinary Sci. Med. 2018, 6, 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Orłowski P., Kowalczyk A., Tomaszewska E., Ranoszek-Soliwoda K., Węgrzyn A., Grzesiak J., Celichowski G., Grobelny J., Eriksson K., Krzyzowska M., Viruses 2018, 10, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sharma V., Kaushik S., Pandit P., Dhull D., Yadav J. P., Kaushik S., Appl. Microbiol. Biotechnol. 2019, 103, 881–891. [DOI] [PubMed] [Google Scholar]
- 74. Morris D., Ansar M., Speshock J., Ivanciuc T., Qu Y., Casola A., Garofalo R. P., Viruses 2019, 11, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Borker S., Patole M., Moghe A., Pokharkar V., Gold Bull. 2017, 50, 235–246. [Google Scholar]
- 76. Halder A., Das S., Ojha D., Chattopadhyay D., Mukherjee A., Mater. Sci. Eng. C 2018, 89, 413–421. [DOI] [PubMed] [Google Scholar]
- 77. Ghaffari E., Rezatofighi S. E., Ardakani M. R., Rastegarzadeh S., Nanomedicine 2019, 14, 1827–1840. [DOI] [PubMed] [Google Scholar]
- 78. Tavakoli A., Hashemzadeh M. S., J. Virol. Methods 2020, 275, 113688. [DOI] [PubMed] [Google Scholar]
- 79. Hang X., Peng H., Song H., Qi Z., Miao X., Xu W., J. Virol. Methods 2015, 222, 150–157. [DOI] [PubMed] [Google Scholar]
- 80. Minoshima M., Lu Y., Kimura T., Nakano R., Ishiguro H., Kubota Y., Hashimoto K., Sunada K., J. Hazard. Mater. 2016, 312, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shionoiri N., Sato T., Fujimori Y., Nakayama T., Nemoto M., Matsunaga T., Tanaka T., J. Biosci. Bioeng. 2012, 113, 580–586. [DOI] [PubMed] [Google Scholar]
- 82. Fujimori Y., Sato T., Hayata T., Nagao T., Nakayama M., Nakayama T., Sugamata R., Suzuki K., Appl. Environ. Microbiol. 2012, 78, 951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Osminkina L. A., Timoshenko V. Y., Shilovsky I. P., Kornilaeva G. V., Shevchenko S. N., Gongalsky M. B., Tamarov K. P., Abramchuk S. S., Nikiforov V. N., Khaitov M. R., Karamov E. V., J. Nanopart. Res. 2014, 16. [Google Scholar]
- 84. de Souza E. S. J. M., Hanchuk T. D., Santos M. I., Kobarg J., Bajgelman M. C., Cardoso M. B., ACS Appl. Mater. Interfaces 2016, 8, 16564–16572. [DOI] [PubMed] [Google Scholar]
- 85. Lee E. C., Nguyen C. T. H., Strounina E., Davis-Poynter N., Ross B. P., ACS Omega 2018, 3, 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mishra Y. K., Adelung R., Röhl C., Shukla D., Spors F., Tiwari V., Antiviral Res. 2011, 92, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Antoine T. E., Mishra Y. K., Trigilio J., Tiwari V., Adelung R., Shukla D., Antiviral Res. 2012, 96, 363–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kumar S. R., Paulpandi M., ManivelRaja M., Mangalaraj D., Viswanathan C., Kannan S., Ponpandian N., RSC Adv. 2014, 4, 13409–13418. [Google Scholar]
- 89. Kumar R., Nayak M., Sahoo G. C., Pandey K., Sarkar M. C., Ansari Y., Das V. N. R., Topno R. K., Bhawna, Madhukar M., Das P., J. Infect. Chemother. 2019, 25, 325–329. [DOI] [PubMed] [Google Scholar]
- 90. Delaviz N., Gill P., Ajami A., Aarabi M., RSC Adv. 2015, 5, 79433–79439. [Google Scholar]
- 91. Li Y., Lin Z., Guo M., Xia Y., Zhao M., Wang C., Xu T., Chen T., Zhu B., Int. J. Nanomed. 2017, 12, 5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lin Z., Li Y., Guo M., Xiao M., Wang C., Zhao M., Xu T., Xia Y., Zhu B., RSC Adv. 2017, 7, 35290–35296. [Google Scholar]
- 93. Lin Z., Li Y., Gong G., Xia Y., Wang C., Chen Y., Hua L., Zhong J., Tang Y., Liu X., Int. J. Nanomed. 2018, 13, 5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Li Y., Lin Z., Guo M., Zhao M., Xia Y., Wang C., Xu T., Zhu B., Int. J. Nanomed. 2018, 13, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Luczkowiak J., Muñoz A., Sánchez-Navarro M., Ribeiro-Viana R., Ginieis A., Illescas B. M., Martín N., Delgado R., Rojo J., Biomacromolecules 2013, 14, 431–437. [DOI] [PubMed] [Google Scholar]
- 96. Yasuno T., Ohe T., Takahashi K., Nakamura S., Mashino T., Bioorg. Med. Chem. Lett. 2015, 25, 3226–3229. [DOI] [PubMed] [Google Scholar]
- 97. Castro E., Martinez Z. S., Seong C.-S., Cabrera-Espinoza A., Ruiz M., Hernandez Garcia A., Valdez F., Llano M., Echegoyen L., J. Med. Chem. 2016, 59, 10963–10973. [DOI] [PubMed] [Google Scholar]
- 98. Martinez Z. S., Castro E., Seong C.-S., Cerón M. R., Echegoyen L., Llano M., Antimicrob. Agents Chemother. 2016, 60, 5731–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kataoka H., Ohe T., Takahashi K., Nakamura S., Mashino T., Bioorg. Med. Chem. Lett. 2016, 26, 4565–4567. [DOI] [PubMed] [Google Scholar]
- 100. Ramos-Soriano J., Reina J. J., Illescas B. M., de la Cruz N., Rodriguez-Perez L., Lasala F., Rojo J., Delgado R., Martin N., J. Am. Chem. Soc. 2019, 141, 15403–15412. [DOI] [PubMed] [Google Scholar]
- 101. Kraevaya O. A., Peregudov A. S., Godovikov I. A., Shchurik E. V., Martynenko V. M., Shestakov A. F., Balzarini J., Schols D., Troshin P. A., Chem. Commun. 2020. [DOI] [PubMed] [Google Scholar]
- 102. Botta G., Bizzarri B. M., Garozzo A., Timpanaro R., Bisignano B., Amatore D., Palamara A. T., Nencioni L., Saladino R., Bioorg. Med. Chem. 2015, 23, 5345–5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhu S., Li J., Huang A.-G., Huang J.-Q., Huang Y.-Q., Wang G.-X., Aquaculture 2019, 512, 734377. [Google Scholar]
- 104. Sametband M., Kalt I., Gedanken A., Sarid R., ACS Appl. Mater. Interfaces 2014, 6, 1228–1235. [DOI] [PubMed] [Google Scholar]
- 105. Deokar A. R., Nagvenkar A. P., Kalt I., Shani L., Yeshurun Y., Gedanken A., Sarid R., Bioconjugate Chem. 2017, 28, 1115–1122. [DOI] [PubMed] [Google Scholar]
- 106. Donskyi I. S., Azab W., Cuellar-Camacho J. L., Guday G., Lippitz A., Unger W. E., Osterrieder K., Adeli M., Haag R., Nanoscale 2019, 11, 15804–15809. [DOI] [PubMed] [Google Scholar]
- 107. Barras A., Pagneux Q., Sane F., Wang Q., Boukherroub R., Hober D., Szunerits S., ACS Appl. Mater. Interfaces 2016, 8, 9004–9013. [DOI] [PubMed] [Google Scholar]
- 108. Du T., Liang J., Dong N., Liu L., Fang L., Xiao S., Han H., Carbon 2016, 110, 278–285. [Google Scholar]
- 109. Fahmi M. Z., Sukmayani W., Khairunisa S. Q., Witaningrum A. M., Indriati D. W., Matondang M. Q. Y., Chang J. Y., Kotaki T., Kameoka M., RSC Adv. 2016, 6, 92996–93002. [Google Scholar]
- 110. Dong X., Moyer M. M., Yang F., Sun Y. P., Yang L., Sci. Rep. 2017, 7, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lin C. J., Chang L., Chu H. W., Lin H. J., Chang P. C., Wang R. Y. L., Unnikrishnan B., Mao J. Y., Chen S. Y., Huang C. C., Small 2019, 15, e1902641. [DOI] [PubMed] [Google Scholar]
- 112. Loczechin A., Seron K., Barras A., Giovanelli E., Belouzard S., Chen Y. T., Metzler-Nolte N., Boukherroub R., Dubuisson J., Szunerits S., ACS Appl. Mater. Interfaces 2019, 11, 42964–42974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Naahidi S., Jafari M., Edalat F., Raymond K., Khademhosseini A., Chen P., J. Controlled Release 2013, 166, 182–194. [DOI] [PubMed] [Google Scholar]
- 114. Yadav H. K. S., Almokdad A. A., shaluf S. I. M., Debe M. S., Nanocarriers for Drug Delivery (Eds.: S. S. Mohapatra, S. Ranjan, N. Dasgupta, R. K. Mishra, S. Thomas), Elsevier, 2019, pp. 531–556. [Google Scholar]
- 115. Kim H. O., Yeom M., Kim J., Kukreja A., Na W., Choi J., Kang A., Yun D., Lim J. W., Song D., Small 2017, 13, 1700818. [DOI] [PubMed] [Google Scholar]
- 116. Jamali A., Mottaghitalab F., Abdoli A., Dinarvand M., Esmailie A., Kheiri M. T., Atyabi F., Drug Delivery Transl. Res. 2018, 8, 12–20. [DOI] [PubMed] [Google Scholar]
- 117. Daraee H., Etemadi A., Kouhi M., Alimirzalu S., Akbarzadeh A., Artificial cells, nanomedicine, and biotechnology 2016, 44, 381–391. [DOI] [PubMed] [Google Scholar]
- 118. Tahara K., Kobayashi M., Yoshida S., Onodera R., Inoue N., Takeuchi H., Int. J. Pharm. 2018, 543, 311–317. [DOI] [PubMed] [Google Scholar]
- 119.
- 119a. Gupta U., Jain N. K., Adv. Drug Delivery Rev. 2010, 62, 478–490; [DOI] [PubMed] [Google Scholar]
- 119b. Zazo H., Colino C. I., Lanao J. M., J. Controlled Release 2016, 224, 86–102. [DOI] [PubMed] [Google Scholar]
- 120. Kondel R., Shafiq N., Kaur I. P., Singh M. P., Pandey A. K., Ratho R. K., Malhotra S., Pharm. Nanotechnol. 2019, 7, 389–403. [DOI] [PubMed] [Google Scholar]
- 121. Kong B., Moon S., Kim Y., Heo P., Jung Y., Yu S. H., Chung J., Ban C., Kim Y. H., Kim P., Hwang B. J., Chung W. J., Shin Y. K., Seong B. L., Kweon D. H., Nat. Commun. 2019, 10, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Bosman A. W., Janssen H. M., Meijer E. W., Chem. Rev. 1999, 99, 1665–1688. [DOI] [PubMed] [Google Scholar]
- 123.
- 123a. Cheng Y., Xu Z., Ma M., Xu T., J. Pharm. Sci. 2008, 97, 123–143; [DOI] [PubMed] [Google Scholar]
- 123b. Kim Y., Park E. J., Na D. H., Arch. Pharmacal Res. 2018, 41, 571–582. [DOI] [PubMed] [Google Scholar]
- 124. Kwon S.-J., Na D. H., Kwak J. H., Douaisi M., Zhang F., Park E. J., Park J.-H., Youn H., Song C.-S., Kane R. S., Nat. Nanotechnol. 2017, 12, 48. [DOI] [PubMed] [Google Scholar]
- 125. Lancelot A., Clavería-Gimeno R., Velázquez-Campoy A., Abian O., Serrano J. L., Sierra T., Eur. Polym. J. 2017, 90, 136–149. [Google Scholar]
- 126. Milovanovic M., Arsenijevic A., Milovanovic J., Kanjevac T., Arsenijevic N., Antimicrobial Nanoarchitectonics (Ed.: A. M. Grumezescu), Elsevier, 2017, pp. 383–410. [Google Scholar]
- 127. Lembo D., Donalisio M., Civra A., Argenziano M., Cavalli R., Expert Opin. Drug Delivery 2018, 15, 93–114. [DOI] [PubMed] [Google Scholar]
- 128. Hong Y., Mao D., Wu R., Gao Z., Meng T., Wang R., Liu L., Miao J., RSC Adv. 2019, 9, 15196–15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Jiang Y., Cao S., Bright D. K., Bever A. M., Blakney A. K., Suydam I. T., Woodrow K. A., Mol. Pharm. 2015, 12, 4363–4374. [DOI] [PubMed] [Google Scholar]
- 130. Dhoke D. M., Basaiyye S. S., Khedekar P. B., Int. J. Drug Deliv. Technol. 2018, 47, 77–94. [Google Scholar]
- 131. Martins C., Araújo F., Gomes M. J., Fernandes C., Nunes R., Li W., Santos H. A., Borges F., Sarmento B., Eur. J. Pharm. Biopharm. 2019, 138, 111–124. [DOI] [PubMed] [Google Scholar]
- 132. Creighton R. L., Suydam I. T., Ebner M. E., Afunugo W. E., Bever A. M., Cao S., Jiang Y., Woodrow K. A., ACS Biomater. Sci. Eng. 2019, 5, 4013–4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Rao L., Wang W., Meng Q.-F., Tian M., Cai B., Wang Y., Li A., Zan M., Xiao F., Bu L.-L., Li G., Li A., Liu Y., Guo S.-S., Zhao X.-Z., Wang T.-H., Liu W., Wu J., Nano Lett. 2019, 19, 2215–2222. [DOI] [PubMed] [Google Scholar]
- 134. Sato S., Li K., Kameyama T., Hayashi T., Ishida Y., Murakami S., Watanabe T., Iijima S., Sakurai Y., Watashi K., Tsutsumi S., Sato Y., Akita H., Wakita T., Rice C. M., Harashima H., Kohara M., Tanaka Y., Takaoka A., Immunity 2015, 42, 123–132. [DOI] [PubMed] [Google Scholar]
- 135. Croci R., Bottaro E., Chan K. W. K., Watanabe S., Pezzullo M., Mastrangelo E., Nastruzzi C., Int. J Biomater, 2016, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Wang S. X., Michiels J., Ariën K. K., New R., Vanham G., Roitt I., Nanoscale Res. Lett. 2016, 11, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Gómara M. J., Pérez-Pomeda I., Gatell J. M., Sánchez-Merino V., Yuste E., Haro I., Nanomed. Nanotech. Biol. Med. 2017, 13, 601–609. [DOI] [PubMed] [Google Scholar]
- 138. Cheng H.-W., Wang H.-W., Wong T.-Y., Yeh H.-W., Chen Y.-C., Liu D.-Z., Liang P.-H., Bioorg. Med. Chem. 2018, 26, 2262–2270. [DOI] [PubMed] [Google Scholar]
- 139. Joshi S., Chaudhari A. A., Dennis V., Kirby D. J., Perrie Y., Singh S. R., BioEngineering 2018, 5, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Torrecilla J., del Pozo-Rodríguez A., Solinís M. ÿ., Apaolaza P. S., Berzal-Herranz B., Romero-López C., Berzal-Herranz A., Rodríguez-Gascón A., Colloids and Surf. B Biointerfaces 2016, 146, 808–817. [DOI] [PubMed] [Google Scholar]
- 141. Javan F., Vatanara A., Azadmanesh K., Nabi-Meibodi M., shakouri M., J. Pharm. Pharmacol. 2017, 69, 1002–1009. [DOI] [PubMed] [Google Scholar]
- 142. Varga N., Sutkeviciute I., Ribeiro-Viana R., Berzi A., Ramdasi R., Daghetti A., Vettoretti G., Amara A., Clerici M., Rojo J., Fieschi F., Bernardi A., Biomaterials 2014, 35, 4175–4184. [DOI] [PubMed] [Google Scholar]
- 143. Sepúlveda-Crespo D., Jiménez J. L., Gómez R., De La Mata F. J., Majano P. L., Muñoz-Fernández M. Á., Gastaminza P., Nanomed. Nanotech. Biol. Med. 2017, 13, 49–58. [DOI] [PubMed] [Google Scholar]
- 144. Abdoli A., Radmehr N., Bolhassani A., Eidi A., Mehrbod P., Motevalli F., Kianmehr Z., Chiani M., Mahdavi M., Yazdani S., Artif. Cells, Nanomed., Biotechnol. 2017, 45, 1762–1768. [DOI] [PubMed] [Google Scholar]
- 145. Lancelot A., Clavería-Gimeno R., Velázquez-Campoy A., Abian O., Serrano J. L., Sierra T., Eur. Polym. J. 2017, 90, 136–149. [Google Scholar]
- 146. Kandi M. R., Mohammadnejad J., Shafiee Ardestani M., Zabihollahi R., Soleymani S., Aghasadeghi M. R., Baesi K., Mol. Biol. Rep. 2019, 46, 143–149. [DOI] [PubMed] [Google Scholar]
- 147. Maciel D., Guerrero-Beltrán C., Ceña-Diez R., Tomás H., Muñoz-Fernández M. Á., Rodrigues J., Nanoscale 2019, 11, 9679–9690. [DOI] [PubMed] [Google Scholar]
- 148. Günther S. C., Maier J. D., Vetter J., Podvalnyy N., Khanzhin N., Hennet T., Stertz S., Sci. Rep. 2020, 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Huang S.-T., Du Y.-Z., Yuan H., Zhang X.-G., Miao J., Cui F.-D., Hu F.-Q., Carbohydr. Polym. 2011, 83, 1715–1722. [Google Scholar]
- 150. Jiménez-Pardo I., González-Pastor R., Lancelot A., Claveria-Gimeno R., Velázquez-Campoy A., Abian O., Ros M. B., Sierra T., Macromol. Biosci. 2015, 15, 1381–1391. [DOI] [PubMed] [Google Scholar]
- 151. Concellón A., Clavería-Gimeno R., Velázquez-Campoy A., Abian O., Piñol M., Oriol L., RSC Adv. 2016, 6, 24066–24075. [Google Scholar]
- 152. Chowdhury A. H., Debnath R., Islam S. M., Saha T., Sustainable Polymer Composites and Nanocomposites, Springer, 2019, pp. 1067–1091. [Google Scholar]
- 153. Du T., Lu J., Liu L., Dong N., Fang L., Xiao S., Han H., ACS Appl. Bio Mater. 2018, 1, 1286–1293. [DOI] [PubMed] [Google Scholar]
- 154.
- 154a. Pant H. R., Pandeya D. R., Nam K. T., Baek W.-i., Hong S. T., Kim H. Y., J. Hazard. Mater. 2011, 189, 465–471; [DOI] [PubMed] [Google Scholar]
- 154b. Lee S.-Y., Park S.-J., J. Ind. Eng. Chem. 2013, 19, 1761–1769. [Google Scholar]
- 155. Monmaturapoj N., Sri-on A., Klinsukhon W., Boonnak K., Prahsarn C., Mater. Sci. Eng. C 2018, 92, 96–102. [DOI] [PubMed] [Google Scholar]
- 156. Ishiguro H., Yao Y., Nakano R., Hara M., Sunada K., Hashimoto K., Kajioka J., Fujishima A., Kubota Y., Appl. Catal. B 2013, 129, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.
- 157a.J. Zhang, T. Lin, X. Wang, in Functional Nanofibers and their Applications (Ed.: Q. Wei), Woodhead Publishing, 2012, pp. 55–70;
- 157b.M. Ramalingam, S. Ramakrishna, in Nanofiber Composites for Biomedical Applications (Eds.: M. Ramalingam, S. Ramakrishna), Woodhead Publishing, 2017, pp. 3–29
- 158. Bai B., Mi X., Xiang X., Heiden P. A., Heldt C. L., Carbohydr. Res. 2013, 380, 137–142. [DOI] [PubMed] [Google Scholar]
- 159. Si Y., Zhang Z., Wu W., Fu Q., Huang K., Nitin N., Ding B., Sun G., Sci. Adv. 2018, 4, eaar5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Levina A. S., Repkova M. N., Bessudnova E. V., Filippova E. I., Mazurkova N. A., Zarytova V. F., Beilstein J. Nanotechnol. 2016, 7, 1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Moongraksathum B., Chien M.-Y., Chen Y.-W., J. Nanosci. Nanotechnol. 2019, 19, 7356–7362. [DOI] [PubMed] [Google Scholar]
- 162. Mori Y., Ono T., Miyahira Y., Nguyen V. Q., Matsui T., Ishihara M., Nanoscale Res. Lett. 2013, 8, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Sofy A. R., Hmed A. A., Abd El Haliem N. F., Zein M. A. E., Elshaarawy R. F. M., Carbohydr. Polym. 2019, 226, 115261. [DOI] [PubMed] [Google Scholar]
- 164. Chen Y. N., Hsueh Y. H., Hsieh C. T., Tzou D. Y., Chang P. L., Int. J. Environ. Res. Public Health 2016, 13, 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Ardestani M. S., Fordoei A. S., Abdoli A., Ahangari Cohan R., Bahramali G., Sadat S. M., Siadat S. D., Moloudian H., Nassiri Koopaei N., Bolhasani A., Rahimi P., Hekmat S., Davari M., Aghasadeghi M. R., J. Mater. Sci. Mater. Med. 2015, 26, 179. [DOI] [PubMed] [Google Scholar]
- 166. Rodríguez-Pérez L., Ramos-Soriano J., Pérez-Sánchez A., Illescas B. M., Muñoz A., Luczkowiak J., Lasala F., Rojo J., Delgado R., Martín N., J. Am. Chem. Soc. 2018, 140, 9891–9898. [DOI] [PubMed] [Google Scholar]
- 167. Bai B., Mi X., Xiang X., Heiden P. A., Heldt C. L., Carbohydr. Res. 2013, 380, 137–142. [DOI] [PubMed] [Google Scholar]
- 168. Wu D., Ensinas A., Verrier B., Primard C., Cuvillier A., Champier G., Paul S., Delair T., Mol. Pharm. 2016, 13, 3279–3291. [DOI] [PubMed] [Google Scholar]
- 169. Makita-Chingombe F., Kutscher H. L., DiTursi S. L., Morse G. D., Maponga C. C., J. Drug Deliv. 2016, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Joshy K. S., Snigdha S., Anne G., Nandakumar K., Laly P., Sabu A. T., Chem. Phys. Lipids 2018, 210, 82–89. [DOI] [PubMed] [Google Scholar]
- 171. Yeh H.-W., Lin T.-S., Wang H.-W., Cheng H.-W., Liu D.-Z., Liang P.-H., Org. Biomol. Chem. 2015, 13, 11518–11528. [DOI] [PubMed] [Google Scholar]
- 172. Su Y., Meng L., Sun J., Li W., Shao L., Chen K., Zhou D., Yang F., Yu F., Eur. J. Med. Chem. 2019, 182, 111622. [DOI] [PubMed] [Google Scholar]
- 173. Gao X., Wu D., Wen Y., Gao L., Liu D., Zhong R., Yang C., Zhao C., Int. Dairy J. 2020, 110, 104784. [Google Scholar]
- 174. Sun Q.-L., Li Y., Ni L.-Q., Li Y.-X., Cui Y.-S., Jiang S.-L., Xie E.-Y., Du J., Deng F., Dong C.-X., Carbohydr. Polym. 2020, 229, 115487. [DOI] [PubMed] [Google Scholar]
- 175. Shi Q., Wang A., Lu Z., Qin C., Hu J., Yin J., Carbohydr. Res. 2017, 453–454, 1–9. [DOI] [PubMed] [Google Scholar]
- 176.
- 176a. Chen L., Huang G., Int. J. Biol. Macromol. 2018, 115, 77–82; [DOI] [PubMed] [Google Scholar]
- 176b. Grice I., Mariottini G., Mar. Org. Model Syst. Biol. Med., Springer, 2018, pp. 439–475; [Google Scholar]
- 176c. Valcarcel J., Novoa-Carballal R., Pérez-Martín R. I., Reis R. L., Vázquez J. A., Biotechnol. Adv. 2017, 35, 711–725; [DOI] [PubMed] [Google Scholar]
- 176d. Tanna B., Mishra A., Compr. Rev. Food Sci. Food Saf. 2019, 18, 817–831; [DOI] [PubMed] [Google Scholar]
- 176e. Pomin V. H., Bezerra F. F., Soares P. A. G., Curr. Pharm. Des. 2017, 23, 3405–3414; [DOI] [PubMed] [Google Scholar]
- 176f. Hao C., Yu G., He Y., Xu C., Zhang L., Wang W., Rev. Med. Virol. 2019, 29, e2043; [DOI] [PubMed] [Google Scholar]
- 176g. Wang W., Wang S.-X., Guan H.-S., Mar. Drugs 2012, 10, 2795–2816; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176h. Garrido P. F., Calvelo M., Blanco-González A., Veleiro U., Suárez F., Conde D., Cabezón A., Piñeiro Á., Garcia-Fandino R., Int. J. Pharm. 2020, 119689; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176i. Morozov V., Hansman G., Hanisch F. G., Schroten H., Kunz C., Mol. Nutr. Food Res. 2018, 62, 1700679. [DOI] [PubMed] [Google Scholar]
- 177. Venkatesan J., Lowe B., Anil S., Manivasagan P., Kheraif A. A. A., Kang K.-H., Kim S.-K., Starch/Staerke 2015, 67, 381–390. [Google Scholar]
- 178.
- 178a. Diogo J. V., Novo S. G., González M. J., Ciancia M., Bratanich A. C., Res. Vet. Sci. 2015, 98, 142–144; [DOI] [PubMed] [Google Scholar]
- 178b. Qureshi D., Nayak S. K., Maji S., Kim D., Banerjee I., Pal K., Curr. Pharm. Des. 2019, 25, 1172–1186. [DOI] [PubMed] [Google Scholar]
- 179. Boulho R., Marty C., Freile-Pelegrín Y., Robledo D., Bourgougnon N., Bedoux G., J. Appl. Phycol. 2017, 29, 2219–2228. [Google Scholar]
- 180. Ponce N. M. A., Flores M. L., Pujol C. A., Becerra M. B., Navarro D. A., Córdoba O., Damonte E. B., Stortz C. A., Carbohydr. Res. 2019, 478, 18–24. [DOI] [PubMed] [Google Scholar]
- 181. Jayakumar R., Prabaharan M., Nair S. V., Tamura H., Biotechnol. Adv. 2010, 28, 142–150. [DOI] [PubMed] [Google Scholar]
- 182. Dimassi S., Tabary N., Chai F., Blanchemain N., Martel B., Carbohydr. Polym. 2018, 202, 382–396. [DOI] [PubMed] [Google Scholar]
- 183. Karthik R., Manigandan V., Saravanan R., Rajesh R. P., Chandrika B., Int. J. Biol. Macromol. 2016, 84, 319–328. [DOI] [PubMed] [Google Scholar]
- 184.
- 184a. Kurkov S. V., Loftsson T., Int. J. Pharm. 2013, 453, 167–180; [DOI] [PubMed] [Google Scholar]
- 184b. Jansook P., Ogawa N., Loftsson T., Int. J. Pharm. 2018, 535, 272–284. [DOI] [PubMed] [Google Scholar]
- 185. Jones S. T., Cagno V., Janeček M., Ortiz D., Gasilova N., Piret J., Gasbarri M., Constant D. A., Han Y., Vuković L., Král P., Kaiser L., Huang S., Constant S., Kirkegaard K., Boivin G., Stellacci F., Tapparel C., Sci. Adv. 2020, 6, eaax9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Vilas Boas L. C. P., Campos M. L., Berlanda R. L. A., de Carvalho Neves N., Franco O. L., Cell. Mol. Life Sci. 2019, 76, 3525–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.
- 187a. Skalickova S., Heger Z., Krejcova L., Pekarik V., Bastl K., Janda J., Kostolansky F., Vareckova E., Zitka O., Adam V., Kizek R., Viruses 2015, 7, 5428–5442; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187b. Shartouny J. R., Jacob J., Semin. Cell Dev. Biol. 2019, 88, 147–155. [DOI] [PubMed] [Google Scholar]
- 188.
- 188a. Ahmed A., Siman-Tov G., Hall G., Bhalla N., Narayanan A., Viruses 2019, 11, 704–704; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188b. Voss S., Nitsche C., Bioorg. Med. Chem. Lett. 2020, 30, 126965. [DOI] [PubMed] [Google Scholar]
- 189. Lee W., Kim M., Lee S.-H., Jung H.-G., Oh J.-W., Sci. Rep. 2018, 8, 8667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.
- 190a. Mattar E. H., Almehdar H. A., Yacoub H. A., Uversky V. N., Redwan E. M., Cytokine Growth Factor Rev. 2016, 28, 95–111; [DOI] [PubMed] [Google Scholar]
- 190b. Kalenik B. M., Góra-Sochacka A., Sirko A., Adv. Virus Res. 2018, 247, 10–14; [DOI] [PubMed] [Google Scholar]
- 190c. Hsieh I.-N., Hartshorn K. L., Pharmaceuticals 2016, 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V. K.-M., Chan C. C.-S., Leung H.-C., Fai N., Lin Y.-P., Zhang A. J.-X., Jin D.-Y., Yuen K.-Y., Zheng B.-J., Sci. Rep. 2016, 6, 22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Zhao H., To K. K. W., Chu H., Ding Q., Zhao X., Li C., Shuai H., Yuan S., Zhou J., Kok K.-H., Jiang S., Yuen K.-Y., Nat. Commun. 2018, 9, 2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Jackman J. A., Linardy E., Yoo D., Seo J., Ng W. B., Klemme D. J., Wittenberg N. J., Oh S.-H., Cho N.-J., Small 2016, 12, 1159–1166. [DOI] [PubMed] [Google Scholar]
- 194. Cho N.-J., Dvory-Sobol H., Xiong A., Cho S.-J., Frank C. W., Glenn J. S., ACS Chem. Biol. 2009, 4, 1061–1067. [DOI] [PubMed] [Google Scholar]
- 195.
- 195a. Jackman J. A., Zan G. H., Zhdanov V. P., Cho N.-J., J. Phys. Chem. B 2013, 117, 16117–16128; [DOI] [PubMed] [Google Scholar]
- 195b. Jackman J. A., Saravanan R., Zhang Y., Tabaei S. R., Cho N.-J., Small 2015, 11, 2372–2379. [DOI] [PubMed] [Google Scholar]
- 196. Jackman J. A., Goh H. Z., Zhdanov V. P., Knoll W., Cho N.-J., J. Am. Chem. Soc. 2016, 138, 1406–1413. [DOI] [PubMed] [Google Scholar]
- 197. Park S., Jackman J. A., Cho N.-J., Langmuir 2019, 35, 9934–9943. [DOI] [PubMed] [Google Scholar]
- 198.
- 198a. Jones K. E., Patel N. G., Levy M. A., Storeygard A., Balk D., Gittleman J. L., Daszak P., Nature 2008, 451, 990–993; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198b. Keesing F., Belden L. K., Daszak P., Dobson A., Harvell C. D., Holt R. D., Hudson P., Jolles A., Jones K. E., Mitchell C. E., Nature 2010, 468, 647–652; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198c. Coker R. J., Hunter B. M., Rudge J. W., Liverani M., Hanvoravongchai P., Lancet 2011, 377, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.World Health Organization, “WHO Coronavirus Disease (COVID-19) Dashboard”, can be found under https://covid19.who.int/?gclid=EAIaIQobChMIiuCjh-D46QIVyCMrCh0peQz1EAAYASAAEgIaBfD_BwE, 2020
- 200.
- 200a.C. f. D. Control, Prevention, N. C. f. I. Diseases, Addressing emerging infectious disease threats: a prevention strategy for the United States, Centers for Disease Control and Prevention, 1994; [PubMed]
- 200b. Committee G. I. S. A., CMAJ 2004, 171, 1203–1208.15534314 [Google Scholar]
- 201.
- 201a. Tang J., Li Y., Eames I., Chan P., Ridgway G., J. Hosp. Infect. 2006, 64, 100–114; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201b. Taylor L. H., Latham S. M., Woolhouse M. E., Philos. Trans. R. Soc. London B Biol. Sci. 2001, 356, 983–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.J. D. Siegel, E. Rhinehart, M. Jackson, L. Chiarello, 2007.
- 203.
- 203a. Coia J., Ritchie L., Adisesh A., Booth C. M., Bradley C., Bunyan D., Carson G., Fry C., Hoffman P., Jenkins D., J. Hosp. Infect. 2013, 85, 170–182; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203b. Jefferson T., Del Mar C. B., Dooley L., Ferroni E., Al-Ansary L. A., Bawazeer G. A., Van Driel M. L., Nair S., Jones M. A., Thorning S., Cochrane Database Syst. Rev. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Singh P., Gonzalez M. J., Manchester M., Drug Dev. Res. 2006, 67, 23–41. [Google Scholar]
- 205. Shishoo R., Int. J. Cloth. Sci. Technol. 2002. [Google Scholar]
- 206.
- 206a. Smith A., Taylor J., Google Patents, 2011; [Google Scholar]
- 206b. Haldar J., An D., De Cienguegos L. Á., Chen J., Klibanov A. M., Google Patents, 2010. [Google Scholar]
- 207. Tran Q. H., Le A.-T., Advances in Natural Sciences: Nanoscience and Nanotechnology 2013, 4, 033001. [Google Scholar]
- 208.
- 208a. bin-Reza F., Lopez Chavarrias V., Nicoll A., Chamberland M. E., Influenza Other Respi. Viruses 2012, 6, 257–267; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208b. Cowling B., Zhou Y., Ip D., Leung G., Aiello A., Epidemiol. Infect. 2010, 138, 449–456. [DOI] [PubMed] [Google Scholar]
- 209.
- 209a. MacIntyre C. R., Seale H., Dung T. C., Hien N. T., Nga P. T., Chughtai A. A., Rahman B., Dwyer D. E., Wang Q., BMJ open 2015, 5, e006577; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209b. MacIntyre C. R., Chughtai A. A., BMJ 2015, 350, h694. [DOI] [PubMed] [Google Scholar]
- 210.
- 210a. Loeb M., Dafoe N., Mahony J., John M., Sarabia A., Glavin V., Webby R., Smieja M., Earn D. J., Chong S., JAMA 2009, 302, 1865–1871; [DOI] [PubMed] [Google Scholar]
- 210b. MacIntyre C. R., Wang Q., Cauchemez S., Seale H., Dwyer D. E., Yang P., Shi W., Gao Z., Pang X., Zhang Y., Influenza Other Respi. Viruses 2011, 5, 170–179; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210c. Gralton J., McLaws M.-L., Crit. Care Med. 2010, 38, 657–667. [DOI] [PubMed] [Google Scholar]
- 211.
- 211a. Ren G., Oxford J. S., Reip P. W., Lambkin-Williams R., Mann A., Google Patents, 2013; [Google Scholar]
- 211b. Borkow G., Zhou S. S., Page T., Gabbay J., PLoS One 2010, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.
- 212a. Nonomura A., Google Patents, 2007; [Google Scholar]
- 212b. Wen S. H., Google Patents, 2004; [Google Scholar]
- 212c. Munzarová M., Brno, Nanocon 2013, 16–19. [Google Scholar]
- 213. Ippolito M., Vitale F., Accurso G., Iozzo P., Gregoretti C., Giarratano A., Cortegiani A., Pulmonology 2020, 26, 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.
- 214a. Grinshpun S. A., Haruta H., Eninger R. M., Reponen T., McKay R. T., Lee S.-A., J. Occup. Environ. Hyg. 2009, 6, 593–603; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214b. Lee S.-A., Grinshpun S. A., Reponen T., Ann. Occup. Hyg. 2008, 52, 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Qian Y., Willeke K., Grinshpun S. A., Donnelly J., Coffey C. C., Am. Ind. Hyg. Assoc. J. 1998, 59, 128–132. [DOI] [PubMed] [Google Scholar]
- 216. Borkow G., Zhou S. S., Page T., Gabbay J., PLoS One 2010, 5, e11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.
- 217a. Patolia H. H., Pan J., Harb C., Marr L. C., Baffoe-Bonnie A. W., Infect. Control Hosp. Epidemiol. 2020, 1–2; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217b. Radonovich L., Published, 2017. [Google Scholar]
- 218. Quan F.-S., Rubino I., Lee S.-H., Koch B., Choi H.-J., Sci. Rep. 2017, 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.
- 219a. De Clercq E., Nat. Rev. Microbiol. 2004, 2, 704–720; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219b. De Clercq E., J. Clin. Virol. 2004, 30, 115–133. [DOI] [PubMed] [Google Scholar]
- 220.
- 220a. Al-Jabri A. A., Alenzi F. Q., The open AIDS J. 2009, 3, 1–3; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220b. Müller B., Kräusslich H.-G., Antiviral Strategies, Springer, 2009, pp. 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.
- 221a. De Clercq E., Biochem. Pharmacol. 2013, 85, 727–744; [DOI] [PubMed] [Google Scholar]
- 221b. De Clercq E., Li G., Clin. Microbiol. Rev. 2016, 29, 695–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.
- 222a. Lieleg O., Lieleg C., Bloom J., Buck C. B., Ribbeck K., Biomacromolecules 2012, 13, 1724–1732; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222b. Ahmadi A., Zorofchian Moghadamtousi S., Abubakar S., Zandi K., BioMed Res. Int. 2015, 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222c. Shi Q., Wang A., Lu Z., Qin C., Hu J., Yin J., Carbohydr. Res. 2017, 453, 1–9. [DOI] [PubMed] [Google Scholar]
- 223.
- 223a. Matsuda M., Shigeta S., Okutani K., Mar. Biotechnol. 1999, 1, 68–73; [DOI] [PubMed] [Google Scholar]
- 223b. Yoshida O., Nakashima H., Yoshida T., Kaneko Y., Yamamoto I., Matsuzaki K., Uryu T., Yamamoto N., Biochem. Pharmacol. 1988, 37, 2887–2891; [DOI] [PubMed] [Google Scholar]
- 223c. Lee J.-B., Srisomporn P., Hayashi K., Tanaka T., Sankawa U., Hayashi T., Chem. Pharm. Bull. 2001, 49, 108–110. [DOI] [PubMed] [Google Scholar]
- 224.
- 224a. Chirkov S., Appl. Biochem. Microbiol. 2002, 38, 1–8; [Google Scholar]
- 224b. Davydova V., Nagorskaya V., Gorbach V., Kalitnik A., Reunov A., Solov'eva T., Ermak I., Appl. Biochem. Microbiol. 2011, 47, 103–108. [Google Scholar]
- 225.
- 225a. Diamond M. S., Farzan M., Nat. Rev. Immunol. 2013, 13, 46–57; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225b. Parikh B. A., Tumer N. E., Mini-Rev. Med. Chem. 2004, 4, 523–543; [DOI] [PubMed] [Google Scholar]
- 225c. Eo S.-K., Kim Y.-S., Lee C.-K., Han S.-S., J. Ethnopharmacol. 2000, 72, 475–481. [DOI] [PubMed] [Google Scholar]
- 226.
- 226a. Kliger Y., Gallo S. A., Peisajovich S. G., Muñoz-Barroso I., Avkin S., Blumenthal R., Shai Y., J. Biol. Chem. 2001, 276, 1391–1397; [DOI] [PubMed] [Google Scholar]
- 226b. Gordon Y. J., Huang L. C., Romanowski E. G., Yates K. A., Proske R. J., McDermott A. M., Curr. Eye Res. 2005, 30, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.
- 227a. Droebner K., Ehrhardt C., Poetter A., Ludwig S., Planz O., Antiviral Res. 2007, 76, 1–10; [DOI] [PubMed] [Google Scholar]
- 227b. Sokmen M., Angelova M., Krumova E., Pashova S., Ivancheva S., Sokmen A., Serkedjieva J., Life Sci. 2005, 76, 2981–2993. [DOI] [PubMed] [Google Scholar]
- 228. Carmichael E., Google Patents, 1998. [Google Scholar]
- 229. Meléndez-Villanueva M. A., Morán-Santibañez K., Martínez-Sanmiguel J. J., Rangel-López R., Garza-Navarro M. A., Rodríguez-Padilla C., Zarate-Triviño D. G., Trejo-Ávila L. M., Viruses 2019, 11, 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.
- 230a. Comper W., Google Patents, 2005; [Google Scholar]
- 230b. Talarico L., Pujol C., Zibetti R., Faria P., Noseda M., Duarte M., Damonte E., Antiviral Res. 2005, 66, 103–110; [DOI] [PubMed] [Google Scholar]
- 230c. Pujol C. A., Ray S., Ray B., Damonte E. B., Int. J. Biol. Macromol. 2012, 51, 412–416; [DOI] [PubMed] [Google Scholar]
- 230d. Makarenkova I., Deriabin P., L′vov D., Zviagintseva T., Besednova N., Vopr. Virusol. 2010, 55, 41–45; [PubMed] [Google Scholar]
- 230e. Faccin L., Benati F., Rincão V., Mantovani M., Soares S., Gonzaga M., Nozawa C., Carvalho Linhares R., Lett. Appl. Microbiol. 2007, 45, 24–28; [DOI] [PubMed] [Google Scholar]
- 230f. Huleihel M., Ishanu V., Tal J., Arad S. M., J. Appl. Phycol. 2001, 13, 127–134. [Google Scholar]
- 231.
- 231a. Morán-Santibañez K., Cruz-Suárez L. E., Ricque-Marie D., Robledo D., Freile-Pelegrín Y., Peña-Hernández M. A., Rodríguez-Padilla C., Trejo-Avila L. M., BioMed Res. Int. 2016, 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231b. Morán-Santibañez K., Peña-Hernández M. A., Cruz-Suárez L. E., Ricque-Marie D., Skouta R., Vasquez A. H., Rodríguez-Padilla C., Trejo-Avila L. M., Viruses 2018, 10, 465; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231c. Lampis G., Deidda D., Pinza M., Pompei R., Antiviral Chem. Chemother. 2001, 12, 125–131. [DOI] [PubMed] [Google Scholar]
- 232. Sezer A. D., Recent advances in novel drug carrier systems, BoD-Books on Demand, 2012. [Google Scholar]
- 233. Greco F., Vicent M. J., Front. Biosci. 2008, 13, 2744–2756. [DOI] [PubMed] [Google Scholar]
- 234.
- 234a. Ulbrich K., Šubr V., Adv. Drug Delivery Rev. 2010, 62, 150–166; [DOI] [PubMed] [Google Scholar]
- 234b. Witzler M., Alzagameem A., Bergs M., Khaldi-Hansen B. E., Klein S. E., Hielscher D., Kamm B., Kreyenschmidt J., Tobiasch E., Schulze M., Molecules 2018, 23, 1885; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234c. Fiume L., Busi C., Di Stefano G., Mattioli A., Adv. Drug Delivery Rev. 1994, 14, 51–65; [Google Scholar]
- 234d. Molema G., Jansen R. W., Visser J., Herdewijn P., Moolenaar F., Meijer D. K., J. Med. Chem. 1991, 34, 1137–1141; [DOI] [PubMed] [Google Scholar]
- 234e. Dolce V., Fiermonte G., Runswick M. J., Palmieri F., Walker J. E., Proc. Mont. Acad. Sci. 2001, 98, 2284–2288; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234f. Gillies E. R., Frechet J. M., Drug Discovery Today 2005, 10, 35–43. [DOI] [PubMed] [Google Scholar]
- 235.
- 235a. Lisziewicz J., Tőke E. R., Nanomed. Nanotech. Biol. Med. 2013, 9, 28–38; [Google Scholar]
- 235b. Lembo D., Cavalli R., Antiviral Chem. Chemother. 2010, 21, 53–70. [DOI] [PubMed] [Google Scholar]
- 236.
- 236a. Verrecchia T., Spenlehauer G., Bazile D., Murry-Brelier A., Archimbaud Y., Veillard M., J. Controlled Release 1995, 36, 49–61; [Google Scholar]
- 236b. Kohli E., Han H.-Y., Zeman A. D., Vinogradov S. V., J. Controlled Release 2007, 121, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.
- 237a. Patel S. K., Sandeep K., Singh M., Singh G. P., Lee J.-K., Bhatia S. K., Kalia V. C., Biotechnological applications of polyhydroxyalkanoates, Springer, 2019, pp. 207–225; [Google Scholar]
- 237b. Kabilan S., Ayyasamy M., Jayavel S., Paramasamy G., Int. J. Microbiol. 2012, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Sharma P., Chawla A., Arora S., Pawar P., J. Adv. Pharm. Technol. Res. 2012, 3, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.
- 239a. Mahajan A., Aggarwal G., Int. J. Drug Dev. Res 2011, 3, 16–30; [Google Scholar]
- 239b. Hoffman A. S., Stayton P. S., Press O., Murthy N., Lackey C. A., Cheung C., Black F., Campbell J., Fausto N., Kyriakides T. R., Polym. Adv. Technol. 2002, 13, 992–999. [Google Scholar]
- 240. Grainger S. J., El-Sayed M. E., Biologically-responsive hybrid biomaterials 2010, 171–190. [Google Scholar]
- 241.
- 241a. Song N., Zhou L., Li J., Pan Z., He X., Tan H., Wan X., Li J., Ran R., Fu Q., Nanoscale 2016, 8, 7711–7722; [DOI] [PubMed] [Google Scholar]
- 241b. Duro-Castano A., Movellan J., Vicent M., Biomater. Sci. 2015, 3, 1321–1334. [DOI] [PubMed] [Google Scholar]
- 242. Grice I., Mariottini G., Marine Organisms as Model Systems in Biology and Medicine, Springer, 2018, pp. 439–475. [Google Scholar]
- 243. Hui D. S., Lee N., Chan P. K., Respirology 2017, 22, 1300–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Pennisi E., Science 1995, 270, 1916–1917. [DOI] [PubMed] [Google Scholar]
- 245.M. Enserink, American Association for the Advancement of Science, 2006
- 246. Hanvoravongchai P., Adisasmito W., Chau P. N., Conseil A., De Sa J., Krumkamp R., Mounier-Jack S., Phommasack B., Putthasri W., Shih C.-S., BMC Public Health 2010, 10, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]


