The immune response against SARS‐CoV‐2 is comprised of both cellular and humoral arms. Antibody‐based serological tests are being developed to achieve higher sensitivity and specificity. Administration of convalescent plasma containing anti‐viral antibodies was proposed to improve the outcome in severe cases. In this paper, we review some of the aspects associated with the development of antibodies against SARS‐CoV‐2 and their potential use for improved diagnosis and therapy.

Abstract
The immune response against severe acute respiratory syndrome‐corona virus 2 (SARS‐CoV‐2) is comprised of both cellular and humoral arms. While current diagnostic methods are mainly based on polymerase chain reaction, they suffer from insensitivity. Therefore, antibody‐based serologic tests are being developed to achieve higher sensitivity and specificity. Current efforts in treating SARS‐CoV‐2 infection include blocking of viral entry into the host cells, prohibiting viral replication and survival in the host cells, and reducing the exaggerated host immune response. Administration of convalescent plasma containing antiviral antibodies was proposed to improve the outcome in severe cases. In this paper, we review some of the aspects associated with the development of antibodies against SARS‐CoV‐2 and their potential use for improved diagnosis and therapy.
Abbreviations
- ACE
angiotensin converting enzyme
- COVID‐19
corona virus disease 2019
- ELISA
enzyme‐linked immunosorbent assay
- FcR
Fc receptor
- h
human
- Ig
immunoglobulin
- mAb
monoclonal antibody
- MERS
Middle East respiratory syndrome
- N
nucleocapsid
- NAb
neutralizing antibody
- PCR
polymerase chain reaction
- RBD
receptor binding domain
- rN
recombinant nucleocapsid protein
- rS
recombinant spike protein
- S
spike
- SARS‐CoV‐2
severe acute respiratory syndrome‐corona virus 2
- TLR
toll‐like receptor
Severe acute respiratory syndrome‐corona virus 2 (SARS‐CoV‐2) is an infectious RNA virus responsible for causing corona virus disease 2019 (COVID‐19).( 1 ) While current diagnostic methods for COVID‐19 diagnosis are mainly based on polymerase chain reaction (PCR), they suffer from insensitivity. Widespread reports of both false‐positive and false‐negative tests have been reported. Therefore, serologic tests are being developed to identify patients suffering from COVID‐19 and to assist in identifying subjects who have been diseased and may now be immune to reinfection or to severe disease.
The host immune response mounted toward the virus contributes to disease severity. The immune response toward SARS‐CoV‐2 is comprised of both the cellular and humoral arms. Current evidence points to the severe manifestation of COVID‐19 disease as being driven by inappropriate hyperactivation of the immune system, associated cytokine storm, and end‐organ damage.( 2 , 3 ) Current efforts for the treatment of COVID‐19 include blocking viral entry into the host cells, prohibiting viral replication and survival in the host cells, or reducing the exaggerated host immune response. However, these strategies have shown limited efficacy.( 4 ) Administration of convalescent plasma was proposed to improve patient outcomes in severe cases. In this paper, we review some of the aspects associated with the development of antibodies against SARS‐CoV‐2, their biology, potential uses, expected advantage, and disadvantages.
SARS‐CoV‐2 Epitopes as Potential Targets for the Humoral Immune Response
SARS‐CoV‐2 is an enveloped single‐stranded RNA virus. The viral genome encodes four structural proteins, including the spike (S), envelope, membrane, and nucleocapsid (N), as well as other nonstructural proteins. The S protein of SARS‐CoV‐2 consists of two subunits, S1 and S2. Several distinctive elements of SARS‐CoV‐2 are compared with other coronaviruses in Fig. 1.
FIG. 1.
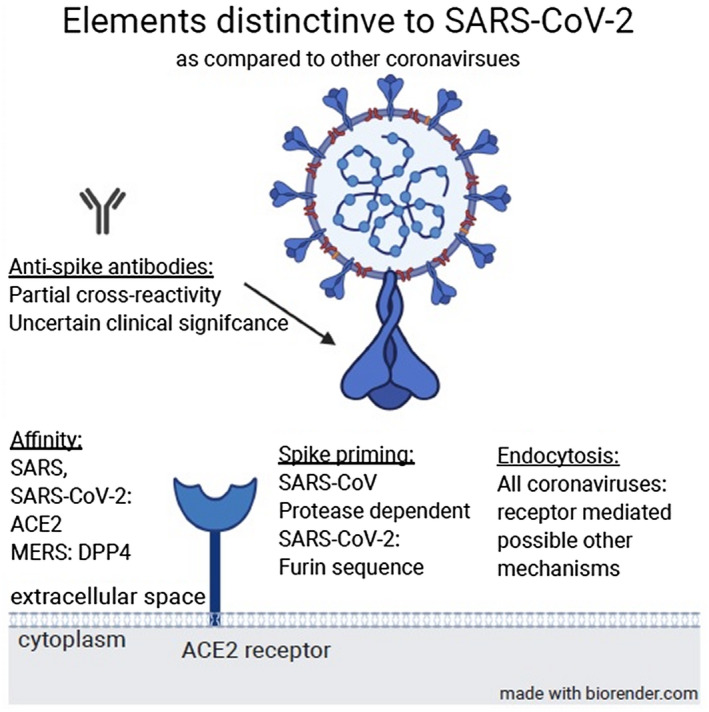
Several distinctive elements of SARS‐CoV‐2 compared with other coronaviruses. Abbreviation: DPP4, dipeptidyl peptidase 4.
Acting as a homotrimer, the heavily glycosylated S protein binds its cellular receptor, angiotensin converting enzyme 2 (ACE2), present on the pneumocytes and enterocytes, by the C‐terminal domain of the S1 subunit in the receptor binding domain (RBD) region.( 5 , 6 ) Extending from the viral membrane, the S protein extends outward from the virion. While the S1 subunit extends furthest from the virus membrane, the inner S2 subunits consist of a mostly helical structure, leading toward the viral membrane. The interaction of the S1–ACE receptor leads to conformational changes in the helical S2 subunit. The next event in viral binding and entry includes cleavage of the S1/S2 protein subunits by cellular proteases. This proteolytic activity may be performed by furin protease, a feature not unique to SARS‐CoV‐2 among the coronaviruses but absent in SARS‐CoV.( 7 ) The cleaving protease, dictating the exact exposed viral amino acid sequence, also determines the pattern of viral–cell fusion.( 8 , 9 ) The release of newly constructed virions and the later activities of these new virions are also dependent on specific protease activity.( 6 )
Among the sites enumerated in this description, several appear as attractive targets for biologically active antibodies. Of note, while new data are continuously and vigorously obtained, specifically regarding SARS‐CoV‐2, much of the functional data regarding coronavirus activity and mechanisms come from research on SARS‐CoV and Middle East respiratory syndrome‐corona virus (MERS‐CoV). This appears particularly poignant where homologies in the structure and function between these viruses are sought. While sequence and biological similarities are common, major differences exist, influencing virus function and antibody biology. These range from matters such as cleavage by similar proteases (although SARS‐CoV‐2 shows unique furin sensitivity) to receptor binding where it shares the affinity toward ACE2 with SARS‐CoV through highly conserved RBD residues.( 10 )
Development of Antibodies Against SARS‐CoV‐2
The final event of protective and effective antibody production is the differentiation of B cells into plasma cells, a change accompanied by robust antibody production. A fraction of these cells will differentiate into memory B cells, allowing for an early antibody response following reinfection; this differentiation has been demonstrated after SARS‐CoV infection.( 11 ) Presumably, the "first contact" of SARS‐CoV‐2 with the immune system occurs following introduction of viable viral particles into the airways. The first responding part of the immune system may be the epithelial cells, both acting as antigen‐presenting cells( 12 ) and internally expressing antiviral proteins, specifically type‐I interferons.( 13 ) Type‐I interferon signaling is usually initiated by toll‐like receptors (TLRs). Variance in the vulnerability to the virus, namely men being more vulnerable, has been attributed partially to superior TLR7 signaling in women, possibly resulting in enhanced antibody production.( 14 ) Notably, TLR7 functions in B cells as well and may contribute to enhanced function and differentiation of plasma cells.( 15 ) Following initial contact with epithelium, innate immune cells come in contact with the virus and with infected cells. Superficial intraepithelial dendritic cells (DCs) in the lungs adjacent to the airways are required for antibody production.( 16 ) After antigen encounter, they move to the regional lymph nodes and help trigger robust antibody production by activation of cluster of differentiation (CD)4 “follicular helper” T cells, supporting B‐cell function.( 17 ) Some DC functions, including type‐I interferon secretion in response to viral stimulation, is also dependent on TLR signaling.( 18 )
While existing research is focused on the endogenic immune response to SARS‐CoV‐2 and its possible beneficial manipulations, isolation of neutralizing antibodies (NAbs) from infected persons or laboratory manufacturing of these antibodies is another subject of intense interest. Monoclonal antibodies (mAbs) with some neutralizing activities were demonstrated to occur in infected human sera.( 19 ) NAbs may be defined in various ways and commonly as the antibody concentration required to prevent or decrease infectivity.( 20 ) The most attractive antibodies are those targeting the S protein, whether in the RBD or other regions, including the S1/S2 proteolytic cleavage site.( 21 ) It is plausible that antibodies targeting these sites will block essential viral functions, including viral antigen binding (expected from S1‐RBD antibodies), and/or interfere with S protein‐mediated viral fusion or cell entry.( 21 , 22 , 23 )
Multiple specific regions in SARS‐CoV‐2 show high homology to the SARS‐CoV virus, suggesting potential B‐ and T‐cell epitopes for SARS‐CoV‐2.( 24 ) A set of B‐cell and T‐cell epitopes were derived from S and N proteins, which (excluding notable differences) are generally conserved between SARS‐CoV and SARS‐CoV‐2. The lack of mutations in these identified epitopes allows assessment of possible SARS‐CoV‐2 immune targets.( 25 ) This study showed that no mutations occurred between SARS‐CoV and SARS‐CoV‐2 in these sequences, confirming the possibility of antibody cross‐reactivity and humoral immunity.
In spite of this high homology, cross‐reactivity of SARS‐CoV antibody is limited between two viral S proteins.( 26 , 27 ) Murine polyclonal SARS‐CoV antibodies directed against the S protein inhibited SARS‐CoV‐2 entry into cells, indicating that cross‐NAbs targeting conserved S epitopes can be produced.( 6 ) S1‐targeting mAbs from immunized transgenic mice expressing human immunoglobulin (Ig) variable heavy and light chains can neutralize SARS‐CoV‐2 and SARS‐CoV infections.( 28 , 29 ) In a previously mentioned trial, 206 SARS‐CoV‐2 RBD‐specific mAbs were generated, among which only two clones showed significant blocking of viral entry; this was associated with a high competitive capacity against ACE2 receptor binding.( 19 ) Similar results were observed in studies using sera from patients recovered from SARS and COVID‐19 where limited cross‐neutralization occurred, suggesting that cross‐NAbs are either incompletely reactive or insufficient for disease prevention.( 28 )
Before and concurrently with the isolation of specific antibodies, SARS‐CoV S1‐specific serum from patients convalescing from SARS or from animals was proposed to cross‐neutralize the SARS‐CoV‐2 infection by reducing S protein‐mediated SARS‐CoV‐2 entry.( 8 ) Cross‐reactivity of the antibodies from patients with SARS‐CoV‐2 against the S proteins but not against the RBD of SARS‐CoV and MERS‐CoV has been documented.
The roles played by the RBD in the invasion of SARS‐CoV‐2 into host cells make the RBD a potential target for NAbs. Blocking binding between the RBD and its respective receptor may restrict the conformational change of S or hamper S2‐mediated membrane fusion, thereby inhibiting viral infection of host cells.( 21 ) The human NAbs S230.15 and m396 were isolated from patients infected with SARS‐CoV. They neutralize SARS‐CoV infection by interacting with the RBD and by blocking binding between the viral RBD and ACE2 receptor.( 30 ) The SARS‐CoV RBD‐specific human NAb, CR3022, binds the SARS‐CoV‐2 RBD with high affinity and recognizes an epitope on the RBD that does not overlap with the ACE2‐binding site.( 27 ) The S109.8 and S227.14 mAbs can neutralize the infectious clones of SARS‐CoV and protect mice against four different homologous and heterologous SARS‐CoV strains.( 31 , 32 ) Of note, such mAbs produced in the chimeric mouse cells and originating from patients with SARS‐CoV were shown to neutralize SARS‐CoV‐2 virus particles by an ACE2‐independent mechanism; this probably has to do with S protein fusion or proteolysis and preventing viral fusion.( 28 )
While these studies hold both promise and interest, isolation and analysis of neutralizing antibodies remain a difficult task. A majority of 26 patients recovered from COVID‐19 showed high titers of SARS‐CoV‐2 S1‐specific IgG antibodies when tested by enzyme‐linked immunosorbent assay (ELISA).( 29 ) However, only 3 out of these 26 patients manifested an effective blockade of SARS‐CoV‐2 RBD binding to human (h)ACE2 when tested in vitro.( 29 ) The transient and dynamic conformational states of the S protein have been suggested to provide a narrow window for exposure of the immunogenic epitopes of RBD to B lymphocytes.( 33 ) Early and transient peak levels of anti‐S antibody response were associated with a less favorable outcome for patients compared with a more delayed and sustained response.( 34 )
The phage display method, allowing rapid and wide display of proteins directly correlated to their associated genes, can detect NAbs against SARS‐CoV from both naive and immune antibody libraries that are capable of blocking the binding of the S1 domain, thereby showing virus neutralization and prophylaxis capability either in vitro or in animal models.( 30 , 32 , 35 ) Another method, possibly allowing the production and use of existing NAbs, may include the use of Epstein‐Barr virus transformation of human B cells to improve the isolation of NAbs from the memory B cells harvested from patients infected with SARS‐CoV.( 11 ) Transgenic mice with human Ig genes that are effective for virus prophylaxis in animal models are being developed to produce NAbs against SARS‐CoV by antigen immunization.( 36 , 37 )
Cloning of human mAbs using samples from patients recovered from COVID‐19 whose sera showed hACE2 receptor binding inhibition has been reported.( 38 ) Following antibody cloning, three pairs of IgG variable heavy‐chain and light‐chain inserted expression plasmids were expressed and named as 311mAb‐31B5, 311mAb‐32D4, and 311mAb‐31B9. All three mAbs bind to the RBD protein. While mAb‐31B5 and 311mAb‐32D4 blocked SARS‐CoV‐2 RBD‐hACE2 interaction and neutralized a SARS‐CoV‐2 S pseudotyped lentiviral particle,( 28 ) 311mAb‐31B5 and 311mAb‐32D4 neutralized pseudovirus entry into host cells ectopically expressing hACE2.( 29 ) Several NAbs, such as B1, 1F8, and 5E9, toward epitopes on SARS‐CoV S2 manifested neutralization properties.( 39 , 40 )
N‐specific antibodies have also been demonstrated in the sera of infected patients. Most studies assessing N antibodies have not differentiated these antibodies from other antibodies directed against SARS‐CoV‐2 in studies that seem to show similar kinetics to that of the general antibody response.( 41 ) No studies have shown the occurrence of definitive NAbs directed at the N protein or the nature of the immune response triggered by such antibodies.
Antibody‐Based Diagnosis of COVID‐19
Serum IgG, IgM, and IgA antibodies against SARS‐CoV appeared in patients after primary SARS infection.( 42 ) Data on the production of IgG and IgM is important for improved diagnosis of COVID‐19.( 43 ) Several studies have described the dynamics of antibody production in these patients. While it is too early to definitively summarize the characteristics of antibody dynamics, certain conclusions seem consistent across these studies. Broadly, antibody titers increase and the prevalence of viral RNA decreases as time progresses from the onset of symptomatic disease.( 44 , 45 )
ELISA‐based diagnostic kits often report a specificity of ~ 90%,( 46 ) with some trials reporting a higher percentage.( 44 ) While this is an impressive figure by itself, it may yield a relatively poor positive predictive value when employed on a large scale to a disease with relatively low prevalence. ELISA tests were argued to be efficient when trying to augment the sensitivity of testing of close contacts( 45 , 47 ) or deciding to allow a person to leave from quarantine. This specificity may be further reduced when testing a person recently exposed to the milder coronaviruses circulating within humans and livestock. However, to our knowledge, this question has not been directly assessed.
IgG and IgM antibodies may appear simultaneously or sequentially, with cases of IgM antibodies appearing last being described in one study.( 47 ) Conversion from seronegativity to seropositivity is likely to occur between 14 and 21 days after the onset of symptoms. Data from some of these studies show that patients with more severe illness were more likely to mount a high‐titer and high‐affinity antibody response, which was not necessarily associated with a reduction in the viral RNA assayed from their blood.( 48 ) This is supported by reports of recurring PCR positivity after IgG seroconversion.( 49 ) If these studies become the prevalent findings, they may stand in stark contrast to well‐established viral disease behaviors where high IgG levels are thought to denote virtual immunity to the disease, allowing at most a mild manifestation following re‐exposure. It seems that in COVID‐19, as our current understanding stands, antibody titers should be thought of as disease markers and not as definitive markers of immunity or disease resolution in the actively ill.
In antibody detection, different ELISA kits show variable results based on recombinant SARS‐CoV‐2 N protein (rN) and recombinant S protein (rS). In a study of 214 patients with confirmed COVID‐19, 68% were diagnosed with rN‐based IgM, 70% with an IgG, 77% with rS‐based IgM, and 74% with IgG tests. The positive rates for rN‐based and rS‐based ELISA detections were 80% for IgM and 82% for IgG. The sensitivity of the rS‐based ELISA for IgM was higher than that of the rN‐based test. Here also, antibody positivity increased as disease time progressed.( 50 )
Another stratum of results expected from antibodies is the identification of immune and recovered persons who may be able to work in critical locations during the times of pandemic. The ability to definitively identify specific NAbs in the serum of recovered patients could also allow identifying potential plasma donors for the development of passive immunization and may assist in evaluating the effectiveness of various treatments in addition to assisting in determining prognosis.( 51 )
Most convalescent plasmas obtained from individuals who recover from COVID‐19 do not contain high levels of NAbs. A recent analysis of plasma from 149 individuals convalescing from COVID‐19 that was collected an average of 39 days after the onset of symptoms showed variable half‐maximal pseudovirus neutralizing titers below 1:50 in 33% and below 1:1,000 in 79%. Only 1% showed titers above 1:5,000. Expanded clones of RBD‐specific memory B cells expressing closely related antibodies in different individuals were identified. The antibodies were directed against three distinct epitopes on the RBD. Rare but recurring RBD‐specific antibodies with potent antiviral activity were identified in all recovered subjects.( 52 ) The relevance of the titers for clinical effect has yet to be determined.
A recent review analyzed the diagnostic accuracy of antibody tests for SARS‐CoV‐2 infection, for assessing past infections, and for use in seroprevalence surveys.( 53 ) A total of 57 publications reporting cohorts with 15,976 samples, of which 8,526 were from cases of SARS‐CoV‐2 infection, were evaluated. These showed substantial heterogeneity in sensitivities of IgA, IgM, and IgG Abs or combinations thereof for results aggregated across different time periods postsymptom onset. Pooled results for IgG, IgM, IgA, total antibodies, and IgG/IgM showed low sensitivity during the first week since onset of symptoms, rising in the second week, and reaching their highest values in the third week. The sensitivity of antibody tests was proposed to be too low in the first week since symptom onset to have a primary role for diagnosis but was suggested to have a role complementing other testing in individuals presenting later, when real‐time PCR tests are negative. Antibody tests are useful for detecting previous SARS‐CoV‐2 infection if used 15 or more days after the onset of symptoms.( 53 )
Several currently available COVID‐19 antibody tests that are used in diagnostics and epidemiology, with a focus on their strengths and weaknesses, are summarized in Table 1.
Table 1.
Several COVID‐19 antibody tests that are used in diagnostics and epidemiology
| Technology Used | Sensitivity | Specificity | Strengths | Weaknesses | |
|---|---|---|---|---|---|
| IgG | CGIA( 96 ) | 29.7%‐88.2% | 98.8%‐99.5% |
|
|
| CLIA( 124 ) | |||||
| ELISA( 125 ) | |||||
| LFA( 126 ) | |||||
| IgM | CGIA( 96 ) | 23.2%‐75.4% | 97.3%‐99.6% |
|
|
| CLIA( 124 ) | |||||
| ELISA( 125 ) | |||||
| LFA( 126 ) | |||||
| IgG/IgM | CGIA( 44 ) | 30.1%‐91.4% | 94.1%‐99.4% |
|
|
| CLIA( 127 ) | |||||
| ELISA( 128 ) | |||||
| LFA( 96 ) |
Abbreviations: CGIA, colloidal gold immunoassay; CLIA, chemiluminescence immunoassay; LFA, lateral flow assay; N/A not applicable.
Using Convalescent Plasma as a Therapy for COVID‐19
The lack of specific SARS‐CoV‐2‐targeted treatments and vaccines poses great challenges for the management of patients with severe illness. IgG levels against SARS‐CoV, drawn from affected patients, reach peak serum concentration during the convalescent phase and are reduced following recovery.( 54 ) While the capacities of antibodies to neutralize the virus were highly variable in the required concentration, some of them indeed showed such capability and have been shown to provide protection against reinfection in a mouse model.( 11 ) Use of convalescent plasma and development of NAbs are attractive methods for the treatment of viral infections.( 55 , 56 ) Blocking mAbs with high antigen specificity have been proposed as potential candidates for neutralizing infections.( 57 , 58 , 59 )
Convalescent plasma has intermittently emerged during the last few decades as a treatment for various infectious diseases,( 60 , 61 , 62 ) enjoying attention whenever diseases prove resistant to more conventional treatment methods. Plasma‐derived NAbs can provide passive immune responses to viral infections and were effective in patients with severe illnesses caused by other viruses.( 63 , 64 ) A meta‐analysis showed that mortality was reduced after receiving various doses of convalescent plasma in patients with severe acute respiratory infections, with no adverse events or complications after treatment.( 65 ) Antibodies from convalescent plasma were proposed to reduce viremia by enhancing viral clearance, blocking infection of new cells, and contributing in the clearance of infected cells.( 56 , 66 , 67 )
During the 2003 SARS epidemic, severely ill patients who deteriorated despite treatment with methylprednisolone were given convalescent plasma at around the fourteenth day of disease onset. Earlier plasma administration correlated with a better prognosis and higher rate of hospital discharge at day 22.( 63 ) Convalescent plasma or Igs were effective in patients with SARS whose condition continued to deteriorate. Some studies suggested a shorter hospital stay and lower mortality rate following convalescent plasma administration.( 56 , 63 , 68 , 69 , 70 , 71 , 72 ) A similar trend for treatment timing was described for 27 patients with Lassa fever in Nigeria treated with convalescent plasma.( 73 ) The empirical use of convalescent plasma for Ebola virus disease showed some positive results.( 64 , 74 , 75 )
Experimental and clinical data on the use of convalescent plasma products and humanized monoclonal antibodies for H5N1 influenza infection have also shown positive outcomes, and this treatment was proposed as a means for overcoming antiviral drug resistance.( 61 , 76 , 77 ) In a study involving 20 patients with severe pandemic influenza A virus subtype H1N1 2009 virus infection, administration of convalescent plasma reduced respiratory tract viral load, serum cytokine response, and mortality.( 59 ) A prospective cohort study during the 2009 pandemic showed reduction in the relative risk of mortality in patients treated with convalescent plasma, demonstrating reduction of viral loads without any adverse effects.( 59 ) A randomized trial of convalescent plasma failed to achieve its primary endpoint, a reduction of mortality; however, a subgroup multivariate analysis performed on 22 of the 35 patients enrolled in the trial demonstrated that human intravenous Ig treatment was the only factor independently associated with reduced mortality.( 78 )
Development of NAbs against SARS‐CoV‐2 was proposed as a method for developing therapeutic agents for COVID‐19.( 10 , 23 , 35 , 79 ) Several SARS‐CoV‐2 proteins (discussed above) prove attractive targets for NAbs. The SARS‐CoV‐2 S protein is a target for developing NAbs to block its binding and fusion.( 35 ) Currently, no SARS‐CoV‐2‐specific NAbs have been reported.( 21 ) However, polyclonal antibodies from patients recovered from SARS‐CoV‐2 are being used to treat patients with severe infections. While many patients will develop an antibody response following their illness, specific characterization of these antibodies and their properties as NAbs have yet to be determined.( 47 , 48 ) Early administration of convalescent plasma was advised in order to maximize its viral clearance effect.( 80 )
Plasma collection is done by apheresis. In order to qualify for donation, the donor must meet several conditions: diagnosis of prior COVID‐19 infection confirmed by PCR, donation needs to take place 14‐28 days after resolution of the symptoms followed by two consecutive negative PCR results, donors need to be tested for absence of transmissible pathogens, and donation should be done from male or nulliparous female donors, with no previous exposure to blood products in order to minimize the risk of transfusion‐associated acute lung injury. Plasma (200‐600 mL) is donated according to ABO compatibility. Pathogen inactivation measures need to be undertaken.( 81 ) It is advised to administer up to 2 units of plasma, possibly from two different donors.( 81 )
Several studies that described the administration of convalescent plasma to patients critically ill with COVID‐19 suggested posttransfusion viral elimination and clinical improvement. A study of critically ill patients (N = 5) reported clinical improvement in patients’ status and laboratory indication of viral clearance for up to 12 days posttransfusion of two consecutive doses of convalescent plasma (total 400 mL).( 82 ) Three of the patients were on mechanical ventilation and 2 on extracorporeal membrane oxygenation; treatment was provided between 10 and 20 days after hospitalization, following which improvements in fever, the ratio of arterial oxygen partial pressure to fractional inspired oxygen, and viral clearance were noted. Three patients were discharged from the hospital and 2 were in stable condition at the end of the follow‐up period. Although a clinical effect was obtained, the delay of up to 3 weeks in administration and the concurrent use of other therapies make it difficult to assess the effect of plasma.( 83 )
Administration of convalescent plasma in 6 critically ill patients was followed by discontinued viral shedding 3 days after infusion, without reducing mortality.( 84 ) A study of 6 patients with COVID‐19 showed clinical, radiologic, and laboratory improvement following administration of ABO‐compatible convalescent plasma, indicating that this therapy is effective and specific.( 85 ) In a study of 10 patients with severe disease, administration with 200 mL of convalescent plasma showed improved clinical, laboratory, and radiologic status without severe adverse effects.( 86 ) In this study, the antibody titers of donors’ plasma were assessed and found to be elevated in the majority of donors, along with a concurrent increase in NAb titers in the patients' sera following transfusion. Treatment within 2 weeks of initial symptoms improved the response.( 86 ) Differences in outcomes between the studies may reflect temporal variations of administration, including the time lag between plasma donation and administration as well as the time from disease detection to treatment.
Safety evaluation of candidate antibodies must not be overlooked. Although antibodies are generally protective, the antibody‐dependent enhancement (ADE) phenomenon of viral infections is documented for dengue virus and other viruses.( 87 ) In SARS‐ CoV infection, ADE is mediated by the engagement of Fc receptors (FcRs) expressed on various immune cells, including monocytes, macrophages, and B cells.( 88 ) Preexisting SARS‐CoV‐specific antibodies were proposed to promote viral entry into FcR‐expressing cells. Internalization of virus–antibody immune complexes may induce inflammation and tissue injury by activating myeloid cells through FcRs.( 88 )
Several putative and proven NAb interactions in COVID‐19, such as antibody targets and functions, including those associated with disruption and nondisruption binding mechanisms and those targeting the virus itself, are shown in Fig. 2. In addition, non‐neutralizing antibodies, cross‐reactive antibodies, and antibodies with low specificity or low titers, which are unable to act as Nabs, are also generated.
FIG. 2.
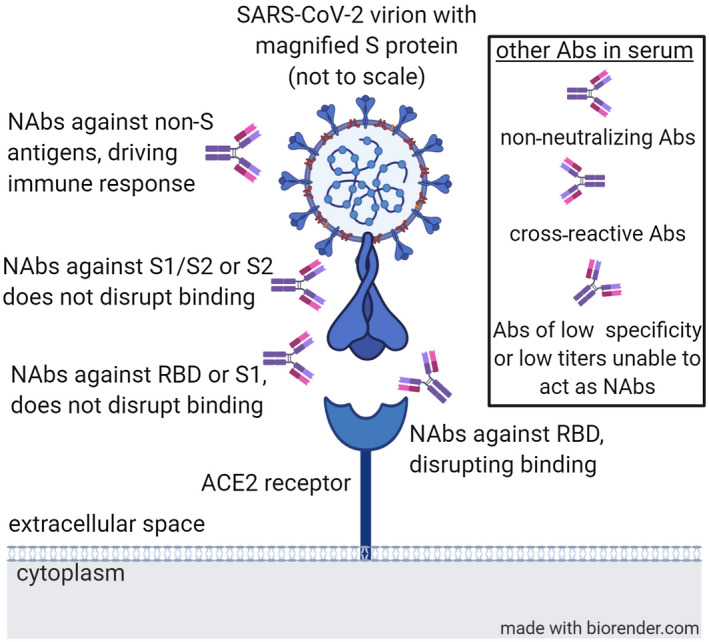
Several putative and proven NAbs in COVID‐19 antibody targets and functions. Abbeviation: Abs, antibodies.
Several large trials using convalescent plasma are being conducted.( 89 ) Identifying and cloning mAbs that target viral proteins to block entry into host cells is being explored for preventing and treating COVID‐19.( 29 , 35 ) Computational simulation of antibody–antigen complexes can improve the design of these therapies. Key residues between RBD and NAbs can be identified, and models are being used to assess the interaction between S protein and human ACE2 or antibodies.( 27 , 35 , 90 , 91 , 92 , 93 )
Methods for Improving Potential Use of Antibodies for COVID‐19 Treatment
Several methods for improving the effectiveness of convalescent plasma or NAbs are being considered. The outcomes of passive convalescent plasma therapy from recovered donors are unpredictable due to variability among donors in both the levels and types of antibodies.( 94 ) Appropriate selection of donors is required for improving the quality of collected plasma. Assessment of antibody titers needs to be performed before harvesting due to marked variability in titers among donors. Titers correlate with disease severity, timing of donation, use of steroids during acute illness,( 95 , 96 ) and quality of antibodies (i.e., whether they are NAbs or not). Timing of plasmapheresis is a major factor as lower levels of antibodies are detected within the first 2 weeks following recovery.( 47 ) More data are required on the amount of virus neutralization by antibodies following exposure to convalescent plasma. In vitro testing for neutralizing and/or cross‐neutralizing activity and in vivo evaluation in available COVID‐19 animal models for protective efficacy along with preclinical studies and clinical trials testing safety and efficacy are needed for optimizing this therapeutic option.( 21 )
The sex of the donor also plays a role in mounting a significant response. The degree of activation of immune cells is higher in women than in men, which correlates with triggering of TLR7 and production of proinflammatory cytokines. TLR7 is expressed in innate immune cells, which recognize single‐strand RNA virus by promoting production of viral antibodies and the generation of interleukin (IL)‐6 and IL‐1 inflammatory cytokines. TLR7 is higher in women than in men, and its expression may lead to better immune responses and increased resistance to viral infections.( 14 ) Pairing human leukocyte antigen typing with COVID‐19 was proposed to improve the assessment of disease severity and assist in preferred donor selection.( 97 )
The use of hyperimmune globulin rather than whole plasma was proposed for improving the efficacy and validity of the therapy. The main advantages are associated with an ability to provide patients with controlled quantities of antibodies in lower volumes.( 82 ) Similar techniques for concentrating antibodies are being used for the treatment and prevention of other diseases.( 98 ) This is similar to the concept of using hyperimmune globulin for various indications, including viral diseases in immunocompetent and immunocompromised hosts.( 99 , 100 , 101 ) A “cocktail antibody approach” for SARS‐CoV‐2 was proposed based on studies suggesting that the combination of antibodies from diverse donors may exert a synergistic neutralization effect.( 35 ) A mixture of two antibodies showed a synergistic neutralization effect due to recognition of different epitopes on the RBD.( 102 )
Use of immune adjuvants may also improve the response to the antibodies.( 103 ) Sphingolipid‐based adjuvants, when administered with antibodies, augmented the antiviral response( 104 ) and improved the systemic anti‐inflammatory effects of antibodies.( 105 ) Use of hyperimmune bovine colostrum comprised of antibodies and sphingolipids was effective in reducing systemic inflammation.( 106 , 107 , 109 ) Mode of antibody administration may also have an impact on the effect of antibody‐based therapy. Oral administration of antibodies ameliorated viral‐mediated chronic inflammation by promotion of regulatory T cells,( 109 ) and oral administration of viral antigens augmented an antiviral immunity while reducing inflammation.( 110 , 111 )
Data on the possible harmful effects of antibody‐mediated immune response in the development of pulmonary complications of SARS‐CoV is controversial. Several patients who died of SARS manifested strong NAb responses and pulmonary inflammation, suggesting that the NAbs could be associated with deterioration of the lung disease.( 35 , 112 ) Similar notions have been proposed for explaining the more severe phenotype of COVID‐19 prevalent in China. This may be related to the higher degree of exposure to milder coronaviruses and a "priming" of the immune system by preexisting antibodies, leading to immune dysfunction and overfunction.( 113 ) This notion is supported by mild disease manifestations in patients with agammaglobulinemia.( 114 ) Previous exposure to coronaviruses may also explain a relatively high prevalence of S protein‐reactive CD4 cells in healthy donors in a study.( 115 )
A major obstacle for implementing immune‐based therapies for the viruses, including the administration of mAbs, is associated with development of viral resistance due to immune evasion mechanisms, which the virus generates in response to the immune pressure imposed on it by immunomodulatory agents.( 116 ) Prolonged exposure to antiviral drugs is associated with drug resistance, leading to persistent viremia or severe disease. In cases where antiviral treatment is highly effective, leading to viral elimination, resistance is less likely to occur. However, immunotherapy, including administration of antibodies, is associated with selective pressure that may result in rapid viral and host adaptations leading to resistance to the therapy.( 117 ) Both host and viral factors are associated with development of resistance. Viral‐related tools include mechanisms of viral replication, genomic inference, and high rates of viral mutations.( 117 , 118 , 119 )
An immune adaptation process toward antibody‐induced pressure on the virus or on antiviral humoral and cellular responses may limit the efficacy and longevity of these therapies. A combination of several potent NAbs could improve the sensitivity to neutralization.( 35 ) Methods for overcoming resistance by implementing host‐tailored variability are being developed based on data generated from the use of these methods for improving the effects of other immunomodulatory drugs.( 120 , 121 , 122 , 123 ) These include implementing artificial intelligence methods for overcoming host compensatory responses in sepsis and its sequel( 120 ) and for improving the effects of adjuvants.( 122 ) Algorithm‐controlled treatment regimens are now being used in several clinical trials for overcoming drug resistance (NCT03843697; NCT03747705).
Concluding Remarks
The lack of accurate diagnostic and effective therapeutic methods for patients infected with SARS‐CoV‐2 led to the need to develop humoral‐based approaches. While this approach holds promise, more data are needed for optimizing antibody‐based diagnosis and for improving the implementation of convalescent plasma and other antibody‐based therapies. The potential development of effective vaccines will benefit from the results achieved from these diagnostic and therapeutic attempts. Immunotherapeutic methods are expected to require targeting the cellular arm of the immune system either in addition to or as part of the design of antibody‐based approaches, mainly for alleviating immune‐mediated target organ damage in COVID‐19.
Potential conflict of interest: Dr. Ilan is the founder of Oberon Sciences and is a consultant for Teva, ENZO, Protalix, Betalin Therapeutics, Immuron, SciM, Natural Shield, Tiziana Pharma, Plantylight, and Exalenz Bioscience. The other authors have nothing to report.
References
- 1. Conti P, Gallenga CE, Tete G, Caraffa A, Ronconi G, Younes A, et al. How to reduce the likelihood of coronavirus‐19 (CoV‐19 or SARS‐CoV‐2) infection and lung inflammation mediated by IL‐1. J Biol Regul Homeost Agents 2020;34:333‐338. [DOI] [PubMed] [Google Scholar]
- 2. Pedersen SF, Ho YC. SARS‐CoV‐2: a storm is raging. J Clin Invest 2020;130:2202‐2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rokni M, Ghasemi V, Tavakoli Z. Immune responses and pathogenesis of SARS‐CoV‐2 during an outbreak in Iran: comparison with SARS and MERS. Rev Med Virol 2020;30:e2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shetty R, Ghosh A, Honavar SG, Khamar P, Sethu S. Therapeutic opportunities to manage COVID‐19/SARS‐CoV‐2 infection: present and future. Indian J Ophthalmol 2020;68:693‐702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang T, Bidon M, Jaimes JA, Whittaker GR, Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res 2020;178:104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell 2020;181:281‐292.e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, et al. Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci U S A 2020;117:11727‐11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hoffmann M, Kleine‐Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271‐280.e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, et al. Crystal structure of SARS‐CoV‐2 main protease provides a basis for design of improved α‐ketoamide inhibitors. Science 2020;368:409‐412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou P, Yang X‐L, Wang X‐G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Traggiai E, Becker S, Subbarao K, Kolesnikova L, Uematsu Y, Gismondo MR, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med 2004;10:871‐875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wosen JE, Mukhopadhyay D, Macaubas C, Mellins ED. Epithelial MHC class II expression and its role in antigen presentation in the gastrointestinal and respiratory tracts. Front Immunol 2018;9:2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Makris S, Paulsen M, Johansson C. Type I interferons as regulators of lung inflammation. Front Immunol 2017;8:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conti P, Younes A. Coronavirus COV‐19/SARS‐CoV‐2 affects women less than men: clinical response to viral infection. J Biol Regul Homeost Agents 2020;34:339‐343. [DOI] [PubMed] [Google Scholar]
- 15. Simchoni N, Cunningham‐Rundles C. TLR7‐ and TLR9‐responsive human B cells share phenotypic and genetic characteristics. J Immunol 2015;194:3035‐3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambrecht BN, Hammad H. Lung dendritic cells in respiratory viral infection and asthma: from protection to immunopathology. Annu Rev Immunol 2012;30:243‐270. [DOI] [PubMed] [Google Scholar]
- 17. Ueno H, Schmitt N, Palucka AK, Banchereau J. Dendritic cells and humoral immunity in humans. Immunol Cell Biol 2010;88:376‐380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen X, Yang X, Zheng Y, Yang Y, Xing Y, Chen Z. SARS coronavirus papain‐like protease inhibits the type I interferon signaling pathway through interaction with the STING‐TRAF3‐TBK1 complex. Protein Cell 2014;5:369‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Catalan‐Dibene J. Human antibodies can neutralize SARS‐CoV‐2. Nat Rev Immunol 2020;20:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klasse PJ. Neutralization of virus infectivity by antibodies: old problems in new perspectives. Adv Biol 2014;2014:157895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS‐CoV‐2 and other human coronaviruses. Trends Immunol 2020;41:355‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS‐CoV spike protein: a key target for antivirals. Expert Opin Ther Targets 2017;21:131‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang S, Du L, Shi Z. An emerging coronavirus causing pneumonia outbreak in Wuhan, China: calling for developing therapeutic and prophylactic strategies. Emerg Microbes Infect 2020;9:275‐277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS‐CoV‐2. Cell Host Microbe 2020;27:671‐680.e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID‐19 coronavirus (SARS‐CoV‐2) based on SARS‐CoV immunological studies. Viruses 2020;12:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science 2020;367:1260‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tian X, Li C, Huang A, Xia S, Lu S, Shi Z, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerg Microbes Infect 2020;9:382‐385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS‐CoV‐2 on virus entry and its immune cross‐reactivity with SARS‐CoV. Nat Commun 2020;11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen X, Li R, Pan Z, Qian C, Yang Y, You R, et al. Human monoclonal antibodies block the binding of SARS‐CoV‐2 spike protein to angiotensin converting enzyme 2 receptor. Cell Mol Immunol 2020;17:647‐649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu Z, Chakraborti S, He Y, Roberts A, Sheahan T, Xiao X, et al. Potent cross‐reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci U S A 2007;104:12123‐12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rockx B, Corti D, Donaldson E, Sheahan T, Stadler K, Lanzavecchia A, et al. Structural basis for potent cross‐neutralizing human monoclonal antibody protection against lethal human and zoonotic severe acute respiratory syndrome coronavirus challenge. J Virol 2008;82:3220‐3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sui J, Li W, Murakami A, Tamin A, Matthews LJ, Wong SK, et al. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc Natl Acad Sci U S A 2004;101:2536‐2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, et al. Immunogenicity and structures of a rationally designed prefusion MERS‐CoV spike antigen. Proc Natl Acad Sci U S A 2017;114:E7348‐E7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Infantino M, Damiani A, Gobbi FL, Grossi V, Lari B, Macchia D, et al. Serological assays for SARS‐CoV‐2 infectious disease: benefits, limitations and perspectives. Isr Med Assoc J 2020;22:203‐210. [PubMed] [Google Scholar]
- 35. Zhou G, Zhao Q. Perspectives on therapeutic neutralizing antibodies against the novel coronavirus SARS‐CoV‐2. Int J Biol Sci 2020;16:1718‐1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coughlin M, Lou G, Martinez O, Masterman SK, Olsen OA, Moksa AA, et al. Generation and characterization of human monoclonal neutralizing antibodies with distinct binding and sequence features against SARS coronavirus using XenoMouse. Virology 2007;361:93‐102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Greenough TC, Babcock GJ, Roberts A, Hernandez HJ, Thomas WD Jr, Coccia JA, et al. Development and characterization of a severe acute respiratory syndrome‐associated coronavirus‐neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J Infect Dis 2005;191:507‐514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc 2009;4:372‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duan J, Yan X, Guo X, Cao W, Han W, Qi C, et al. A human SARS‐CoV neutralizing antibody against epitope on S2 protein. Biochem Biophys Res Commun 2005;333:186‐1 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elshabrawy HA, Coughlin MM, Baker SC, Prabhakar BS. Human monoclonal antibodies against highly conserved HR1 and HR2 domains of the SARS‐CoV spike protein are more broadly neutralizing. PLoS One 2012;7:e50366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2−specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis 2020;26:1478‐1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Woo PC, Lau SK, Wong BH, Chan KH, Chu CM, Tsoi HW, et al. Longitudinal profile of immunoglobulin G (IgG), IgM, and IgA antibodies against the severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in patients with pneumonia due to the SARS coronavirus. Clin Diagn Lab Immunol 2004;11:665‐668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. di Mauro G, Cristina S, Concetta R, Francesco R, Annalisa C. SARS‐Cov‐2 infection: response of human immune system and possible implications for the rapid test and treatment. Int Immunopharmacol 2020;84:106519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, et al. Antibody detection and dynamic characteristics in patients with COVID‐19. Clin Infect Dis 2020; 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID‐19). Clin Infect Dis 2020;71:778‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jin Y, Wang M, Zuo Z, Fan C, Ye F, Cai Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis 2020;94:49‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Long Q‐X, Liu B‐Z, Deng H‐J, Wu G‐C, Deng K, Chen Y‐K, et al. Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med 2020;26:845‐848. [DOI] [PubMed] [Google Scholar]
- 48. Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019. Clin Infect Dis 2020; 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fu W, Chen Q, Wang T. Letter to the Editor: three cases of re‐detectable positive SARS‐CoV‐2 RNA in recovered COVID‐19 patients with antibodies. J Med Virol 2020; 10.1002/jmv.25968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein‐based enzyme‐linked immunosorbent assays for detecting antibodies against SARS‐CoV‐2. J Clin Microbiol 2020;58:e00461‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winter AK, Hegde ST. The important role of serology for COVID‐19 control. Lancet Infect Dis 2020;20:758‐759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, et al. Convergent antibody responses to SARS‐CoV‐2 in convalescent individuals. Nature 2020;584:437‐442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor‐Phillips S, et al.; Cochrane COVID‐19 Diagnostic Test Accuracy Group . Antibody tests for identification of current and past infection with SARS‐CoV‐2. Cochrane Database Syst Rev 2020;6:CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS‐associated coronavirus after recovery. N Engl J Med 2007;357:1162‐1163. [DOI] [PubMed] [Google Scholar]
- 55. Tiberghien P, de Lambalerie X, Morel P, Gallian P, Lacombe K, Yazdanpanah Y. Collecting and evaluating convalescent plasma for COVID‐19 treatment: why and how. Vox Sang 2020; 10.1111/vox.12926. [DOI] [PubMed] [Google Scholar]
- 56. Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID‐19. Lancet Infect Dis 2020;20:398‐400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Marston HD, Paules CI, Fauci AS. Monoclonal antibodies for emerging infectious diseases ‐ borrowing from history. N Engl J Med 2018;378:1469‐1472. [DOI] [PubMed] [Google Scholar]
- 58. Saylor C, Dadachova E, Casadevall A. Monoclonal antibody‐based therapies for microbial diseases. Vaccine 2009;27(Suppl. 6):G38‐G46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hung IF, To KK, Lee CK, Lee KL, Chan K, Yan WW, et al. Convalescent plasma treatment reduced mortality in patients with severe pandemic influenza A (H1N1) 2009 virus infection. Clin Infect Dis 2011;52:447‐456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dunmire GB. Some observations on treating cases of diphtheria. Read in the section on Diseases of Children, at the Forty‐Fourth Annual Meeting of the American Medical Association. JAMA 1893;XXI:853‐857. [Google Scholar]
- 61. Luke TC, Kilbane EM, Jackson JL, Hoffman SL. Meta‐analysis: convalescent blood products for Spanish influenza pneumonia: a future H5N1 treatment? Ann Intern Med 2006;145:599‐609. [DOI] [PubMed] [Google Scholar]
- 62. Arabi Y, Balkhy H, Hajeer AH, Bouchama A, Hayden FG, Al‐Omari A, et al. Feasibility, safety, clinical, and laboratory effects of convalescent plasma therapy for patients with Middle East respiratory syndrome coronavirus infection: a study protocol. Springerplus 2015;4:709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheng Y, Wong R, Soo YO, Wong WS, Lee CK, Ng MH, et al. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur J Clin Microbiol Infect Dis 2005;24:44‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kraft CS, Hewlett AL, Koepsell S, Winkler AM, Kratochvil CJ, Larson L, et al.; Nebraska Biocontainment Unit and the Emory Serious Communicable Diseases Unit . The use of TKM‐100802 and convalescent plasma in 2 patients with Ebola virus disease in the United States. Clin Infect Dis 2015;61:496‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mair‐Jenkins J, Saavedra‐Campos M, Baillie JK, Cleary P, Khaw FM, Lim WS, et al.; Convalescent Plasma Study Group . The effectiveness of convalescent plasma and hyperimmune immunoglobulin for the treatment of severe acute respiratory infections of viral etiology: a systematic review and exploratory meta‐analysis. J Infect Dis 2015;211:80‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schoofs T, Klein F, Braunschweig M, Kreider EF, Feldmann A, Nogueira L, et al. HIV‐1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV‐1. Science 2016;352:997‐1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lu CL, Murakowski DK, Bournazos S, Schoofs T, Sarkar D, Halper‐Stromberg A, et al. Enhanced clearance of HIV‐1‐infected cells by broadly neutralizing antibodies against HIV‐1 in vivo. Science 2016;352:1001‐1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lai ST. Treatment of severe acute respiratory syndrome. Eur J Clin Microbiol Infect Dis 2005;24:583‐591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Soo YO, Cheng Y, Wong R, Hui DS, Lee CK, Tsang KK, et al. Retrospective comparison of convalescent plasma with continuing high‐dose methylprednisolone treatment in SARS patients. Clin Microbiol Infect 2004;10:676‐678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li G, Chen X, Xu A. Profile of specific antibodies to the SARS‐associated coronavirus. N Engl J Med 2003;349:508‐509. [DOI] [PubMed] [Google Scholar]
- 71. Corman VM, Albarrak AM, Omrani AS, Albarrak MM, Farah ME, Almasri M, et al. Viral shedding and antibody response in 37 patients with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis 2016;62:477‐483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hsueh PR, Huang LM, Chen PJ, Kao CL, Yang PC. Chronological evolution of IgM, IgA, IgG and neutralisation antibodies after infection with SARS‐associated coronavirus. Clin Microbiol Infect 2004;10:1062‐1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Frame JD, Verbrugge GP, Gill RG, Pinneo L. The use of Lassa fever convalescent plasma in Nigeria. Trans R Soc Trop Med Hyg 1984;78:319‐324. [DOI] [PubMed] [Google Scholar]
- 74. World Health Organization . Use of convalescent whole blood or plasma collected from patients recovered from Ebola virus disease for transfusion, as an empirical treatment during outbreaks: interim guidance for national health authorities and blood transfusion services. https://www.who.int/csr/resources/publications/ebola/convalescent‐treatment/en/. Published September 2014. Accessed May 2020. [Google Scholar]
- 75. Florescu DF, Kalil AC, Hewlett AL, Schuh AJ, Stroher U, Uyeki TM, et al. Administration of brincidofovir and convalescent plasma in a patient with Ebola virus disease. Clin Infect Dis 2015;61:969‐973. [DOI] [PubMed] [Google Scholar]
- 76. Hanson BJ, Boon ACM, Lim APC, Webb A, Ooi EE, Webby RJ. Passive immunoprophylaxis and therapy with humanized monoclonal antibody specific for influenza A H5 hemagglutinin in mice. Respir Res 2006;7:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhou B, Zhong N, Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N Engl J Med 2007;357:1450‐1451. [DOI] [PubMed] [Google Scholar]
- 78. Hung IFN, To KKW, Lee CK, Lee KL, Yan WW, Chan K, et al. Hyperimmune IV immunoglobulin treatment: a multicenter double‐blind randomized controlled trial for patients with severe 2009 influenza A(H1N1) infection. Chest 2013;144:464‐473. [DOI] [PubMed] [Google Scholar]
- 79. Kruse RL. Therapeutic strategies in an outbreak scenario to treat the novel coronavirus originating in Wuhan, China. F1000Res 2020;9:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhao Q, He Y. Challenges of convalescent plasma therapy on COVID‐19. J Clin Virol 2020;127:104358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Epstein J, Burnouf T. Points to consider in the preparation and transfusion of COVID‐19 convalescent plasma. Vox Sang 2020; 10.1111/vox.12939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, et al. Treatment of 5 critically ill patients with COVID‐19 with convalescent plasma. JAMA 2020;323:1582‐1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Roback JD, Guarner J. Convalescent plasma to treat COVID‐19: possibilities and challenges. JAMA 2020; 10.1001/jama.2020.4940. [DOI] [PubMed] [Google Scholar]
- 84. Zeng Q‐L, Yu Z‐J, Gou J‐J, Li G‐M, Ma S‐H, Zhang G‐F, et al. Effect of convalescent plasma therapy on viral shedding and survival in patients with coronavirus disease 2019. J Infect Dis 2020;222:38‐43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ye M, Fu D, Ren Y, Wang F, Wang D, Zhang F, Xia X, et al. Treatment with convalescent plasma for COVID‐19 patients in Wuhan, China. J Med Virol 2020; 10.1002/jmv.25882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, et al. Effectiveness of convalescent plasma therapy in severe COVID‐19 patients. Proc Natl Acad Sci 2020;117:9490‐9496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID‐19. Nat Rev Immunol 2020;20:339‐341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang SF, Tseng SP, Yen CH, Yang JY, Tsao CH, Shen CW, et al. Antibody‐dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun 2014;451:208‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sheridan C. Convalescent serum lines up as first‐choice treatment for coronavirus. Nat Biotechnol 2020;38:655‐658. [DOI] [PubMed] [Google Scholar]
- 90. Zhao Q, Ahmed M, Tassev DV, Hasan A, Kuo TY, Guo HF, et al. Affinity maturation of T‐cell receptor‐like antibodies for Wilms tumor 1 peptide greatly enhances therapeutic potential. Leukemia 2015;29:2238‐2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li D, Gong R, Zheng J, Chen X, Dimitrov DS, Zhao Q. Engineered antibody CH2 domains binding to nucleolin: isolation, characterization and improvement of aggregation. Biochem Biophys Res Commun 2017;485:446‐453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhao Q, Ahmed M, Guo HF, Cheung IY, Cheung NK. Alteration of electrostatic surface potential enhances affinity and tumor killing properties of anti‐ganglioside GD2 monoclonal antibody hu3F8. J Biol Chem 2015;290:13017‐13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Barderas R, Desmet J, Timmerman P, Meloen R, Casal JI. Affinity maturation of antibodies assisted by in silico modeling. Proc Natl Acad Sci U S A 2008;105:9029‐9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jawhara S. Could intravenous immunoglobulin collected from recovered coronavirus patients protect against COVID‐19 and strengthen the immune system of new patients? Int J Mol Sci 2020;21:2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang L, Pang R, Xue X, Bao J, Ye S, Dai Y, Zheng Y, et al. Anti‐SARS‐CoV‐2 virus antibody levels in convalescent plasma of six donors who have recovered from COVID‐19. Aging (Albany NY) 2020;12:6536‐6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yongchen Z, Shen H, Wang X, Shi X, Li Y, Yan J, et al. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID‐19 patients. Emerg Microbes Infect 2020;9:833‐836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nguyen A, David JK, Maden SK, Wood MA, Weeder BR, Nellore A, et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J Virol 2020;94:e00510‐e00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Matson MA, Schenker E, Stein M, Zamfirova V, Nguyen HB, Bergman GE. Safety and efficacy results of simulated post‐exposure prophylaxis with human immune globulin (HRIG; KEDRAB) co‐administered with active vaccine in healthy subjects: a comparative phase 2/3 trial. Hum Vaccin Immunother 2020;16:452‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Nigro G. Hyperimmune globulin in pregnancy for the prevention of congenital cytomegalovirus disease. Expert Rev Anti Infect Ther 2017;15:977‐986. [DOI] [PubMed] [Google Scholar]
- 100. Wasserman RL, Greener BN, Mond J. RI‐002, an intravenous immunoglobulin containing high titer neutralizing antibody to RSV and other respiratory viruses for use in primary immunodeficiency disease and other immune compromised populations. Expert Rev Clin Immunol 2017;13:1107‐1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Carbone J. The immunology of posttransplant CMV infection: potential effect of CMV immunoglobulins on distinct components of the immune response to CMV. Transplantation 2016;100(Suppl. 3):S11‐S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. ter Meulen J, van den Brink EN, Poon LL, Marissen WE, Leung CS, Cox F, et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med 2006;3:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Vogel FR. Improving vaccine performance with adjuvants. Clin Infect Dis 2000;30(Suppl. 3):S266‐S270. [DOI] [PubMed] [Google Scholar]
- 104. Mizrahi M, Lalazar G, Ben Ya'acov A, Livovsky DM, Horowitz Y, Zolotarov L, et al. Beta‐glycoglycosphingolipid‐induced augmentation of the anti‐HBV immune response is associated with altered CD8 and NKT lymphocyte distribution: a novel adjuvant for HBV vaccination. Vaccine 2008;26:2589‐2595. [DOI] [PubMed] [Google Scholar]
- 105. Ilan Y, Maron R, Tukpah AM, Maioli TU, Murugaiyan G, Yang K, et al. Induction of regulatory T cells decreases adipose inflammation and alleviates insulin resistance in ob/ob mice. Proc Natl Acad Sci U S A 2010;107:9765‐9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Adar T, Ben Ya'acov A, Lalazar G, Lichtenstein Y, Nahman D, Mizrahi M, et al. Oral administration of immunoglobulin G‐enhanced colostrum alleviates insulin resistance and liver injury and is associated with alterations in natural killer T cells. Clin Exp Immunol 2012;167:252‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Mizrahi M, Shabat Y, Ben Ya'acov A, Lalazar G, Adar T, Wong V, et al. Alleviation of insulin resistance and liver damage by oral administration of Imm124‐E is mediated by increased Tregs and associated with increased serum GLP‐1 and adiponectin: results of a phase I/II clinical trial in NASH. J Inflamm Res 2012;5:141‐1 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Abdelmalek MF, Freilich BL, Harrison SA, Powell EE, Rinella MEM, Tobis N, et al. Imm‐124E improves metabolic endotoxemia and markers of liver injury in nonalcoholic steatohepatitis [Abstract]. Hepatology, 2018;68:108A. [Google Scholar]
- 109. Halota W, Ferenci P, Kozielewicz D, Dybowska D, Lisovoder N, Samira S, et al. Oral anti‐CD3 immunotherapy for HCV‐nonresponders is safe, promotes regulatory T cells and decreases viral load and liver enzyme levels: results of a phase‐2a placebo‐controlled trial. J Viral Hepat 2015;22:651‐657. [DOI] [PubMed] [Google Scholar]
- 110. Israeli E, Safadi R, Melhem A, Pappo O, Shibolet O, Klein A, et al. Induction of oral immune regulation towards liver‐extracted proteins for treatment of chronic HBV and HCV hepatitis: results of a phase I clinical trial. Liver Int 2004;24:295‐307. [DOI] [PubMed] [Google Scholar]
- 111. Safadi R, Israeli E, Papo O, Shibolet O, Melhem A, Bloch A, et al. Treatment of chronic hepatitis B virus infection via oral immune regulation toward hepatitis B virus proteins. Am J Gastroenterol 2003;98:2505‐2515. [DOI] [PubMed] [Google Scholar]
- 112. Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, et al. Anti‐spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS‐CoV infection. JCI Insight 2019;4:e123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Tetro JA. Is COVID‐19 receiving ADE from other coronaviruses? Microbes Infect 2020;22:72‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Quinti I, Lougaris V, Milito C, Cinetto F, Pecoraro A, Mezzaroma I, et al. A possible role for B cells in COVID‐19? Lesson from patients with agammaglobulinemia. J Allergy Clin Immunol 2020;146:211‐213.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Pia L. SARS‐CoV‐2‐reactive T cells in patients and healthy donors. Nat Rev Immunol 2020;20:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Gelman R, Bayatra A, Kessler A, Schwartz A, Ilan Y. Targeting SARS‐CoV‐2 receptors as a means for reducing infectivity and improving antiviral and immune response: an algorithm‐based method for overcoming resistance to antiviral agents. Emerg Microbes Infect 2020;9:1397‐1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Irwin KK, Renzette N, Kowalik TF, Jensen JD. Antiviral drug resistance as an adaptive process. Virus Evol 2016;2:vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Vilas Boas LCP, Campos ML, Berlanda RLA, de Carvalho NN, Franco OL. Antiviral peptides as promising therapeutic drugs. Cell Mol Life Sci 2019;76:3525‐3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mazzon M, Marsh M. Targeting viral entry as a strategy for broad‐spectrum antivirals. F1000Res 2019;8:F1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kenig A, Ilan Y. A personalized signature and chronotherapy‐based platform for improving the efficacy of sepsis treatment. Front Physiol 2019;10:1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Khoury T, Ilan Y. Introducing patterns of variability for overcoming compensatory adaptation of the immune system to immunomodulatory agents: a novel method for improving clinical response to anti‐TNF therapies. Front Immunol 2019;10:2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Ilan Y. beta‐Glycosphingolipids as mediators of both inflammation and immune tolerance: a manifestation of randomness in biological systems. Front Immunol 2019;10:1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Ilan Y. Generating randomness: making the most out of disordering a false order into a real one. J Transl Med 2019;17:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Perez Garcia F, Perez Tanoira R, Romanyk Cabrera JP, Arroyo Serrano T, Gomez Herruz P, Cuadros Gonzalez J. Rapid diagnosis of SARS‐CoV‐2 infection by detecting IgG and IgM antibodies with an immunochromatographic device: a prospective single‐center study. medRxiv 2020; 10.1101/2020.04.11.20062158. [DOI] [Google Scholar]
- 125. Lippi G, Salvagno GL, Pegoraro M, Militello V, Caloi C, Peretti A, et al. Assessment of immune response to SARS‐CoV‐2 with fully automated MAGLUMI 2019‐nCoV IgG and IgM chemiluminescence immunoassays. Clin Chem Lab Med 2020;58:1156‐1159. [DOI] [PubMed] [Google Scholar]
- 126. Lou B, Li TD, Zheng SF, Su YY, Li ZY, Liu W, et al. Serology characteristics of SARS‐CoV‐2 infection after exposure and post‐symptom onset. Eur Respir J 2020;56:2000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zeng Z, Chen L, Pan Y, Deng Q, Ye G, Li Y, et al. Re: profile of specific antibodies to SARS‐CoV‐2: the first report. J Infect 2020;81:e80‐e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Xiang J, Yan M, Li H, Liu T, Lin C, Huang S, et al. Evaluation of enzyme‐linked immunoassay and colloidal gold‐immunochromatographic assay kit for detection of novel coronavirus (SARS‐Cov‐2) causing an outbreak of pneumonia (COVID‐19). medRxiv 2020; 10.1101/2020.02.27.20028787. [DOI] [Google Scholar]


