Abstract
Increased platelet activation and apoptosis are characteristic of diabetic (DM) platelets, where a Parkin-dependent mitophagy serves a major endogenous protective role. We now demonstrate that Parkin is highly expressed in both healthy platelets and diabetic platelets, compared to other mitochondria-enriched tissues such as the heart, muscle, brain, and liver. Abundance of Parkin in a small, short-lived anucleate cell suggest significance in various key processes. Through proteomics we identified 127 Parkin-interacting proteins in DM platelets and compared them to healthy controls. We assessed the 11 highest covered proteins by individual IPs and confirmed seven proteins that interacted with Parkin; VCP/p97, LAMP1, HADHA, FREMT3, PDIA, ILK, and 14-3-3. Upon further STRING analysis using GO and KEGG, interactions were divided into two broad groups: targeting platelet activation through (1) actions on mitochondria and (2) actions on integrin signaling. Parkin plays an important role in mitochondrial protection through mitophagy (VCP/p97), recruiting phagophores, and targeting lysosomes (with LAMP1). Mitochondrial β-oxidation may also be regulated by the Parkin/HADHA interaction. Parkin may regulate platelet aggregation and activation through integrin signaling through interactions with proteins like FREMT3, PDIA, ILK, and 14-3-3. Thus, platelet Parkin may regulate the protection (mitophagy) and stress response (platelet activation) in DM platelets. This study identified new potential therapeutic targets for platelet mitochondrial dysfunction and hyperactivation in diabetes mellitus.
Keywords: platelet, diabetes, Parkin
1. Introduction
Diabetes mellitus (DM) is a progressive and chronic metabolic disorder characterized by hyperglycemia arising from impaired insulin levels, insulin sensitivity, and/or insulin activity. Currently, over 19.7 million adults in the USA have diagnosed DM, and an estimated 8.2 million have undiagnosed DM [1]. Cardiovascular disease is the major cause of morbidity and mortality among DM patients with approximately 65% of deaths caused by thrombotic events like myocardial and cerebrovascular ischemia and infarction [2]. Platelets play key roles in thrombotic occlusions of major vessels and tissue death.
Platelets are short-lived (7 to 10 days) circulating cells (2 to 4 μm) that contain many critical factors required for the regulation of thrombus formation, vascular homeostasis, and immune responses [3,4,5,6]. Platelets are capable of many fundamental cellular functions despite having no transcriptional capabilities (anucleate) including de novo protein synthesis [4,7,8] and programmed cell death [9,10]. Basal autophagy [11,12], which is distinct from induced autophagy [9,13,14], has only recently been described in platelets. This well-orchestrated process requires considerable energy for prepacking all the relevant mRNAs (no transcription) to maintain normal cellular function (basal autophagy) and for protection (induced autophagy) against severe oxidative stressors as observed with DM [13]. DM platelets have a highly induced protective autophagy processes including Parkin-dependent mitophagy [9,14].
Parkin is a Parkinson’s disease (PD)-related protein with several mutations identified in PD patients [15,16]. Parkin has ubiquitin E3 ligase activity and increased substrate ubiquitination via lysine 27, 29, 48, and 63 of ubiquitin [17,18,19]. Recent reports suggest that Parkin is inactivated by post-translational modifications like oxidation [14] and nitrosylation [16,20]. Additionally, Parkin regulates mitochondrial quality control through a well-orchestrated mitophagy process [21]. During mitochondrial damage, PTEN-induced kinase1 (PINK1) accumulates on damaged mitochondria and recruits Parkin, after which damaged mitochondria is then recognized by LC3-conjugated phagophore structures [21,22]. Parkin expression was increased in DM platelets and readily interacted with MsrB2 to remove damaged mitochondria [9,14].
Parkin is highly expressed in platelets compared to other mitochondrial-enriched tissues. However, the roles of Parkin remain largely unexplored in platelets. To determine alternative functions of Parkin in platelets, interacting proteins in healthy control and DM platelets were documented through literature investigations, we suggest functional associations between platelet aggregation, mitochondrial damage, mitophagy, and Parkin in DM platelets.
2. Results
We recently demonstrated that Parkin plays a key protective role against oxidative stress in platelets by inducing mitophagy [9,14]. The absence of Parkin during such stressors like DM and H2O2 leads to platelet apoptosis [9,14].
2.1. Parkin Interacts with Various Key Platelet Proteins
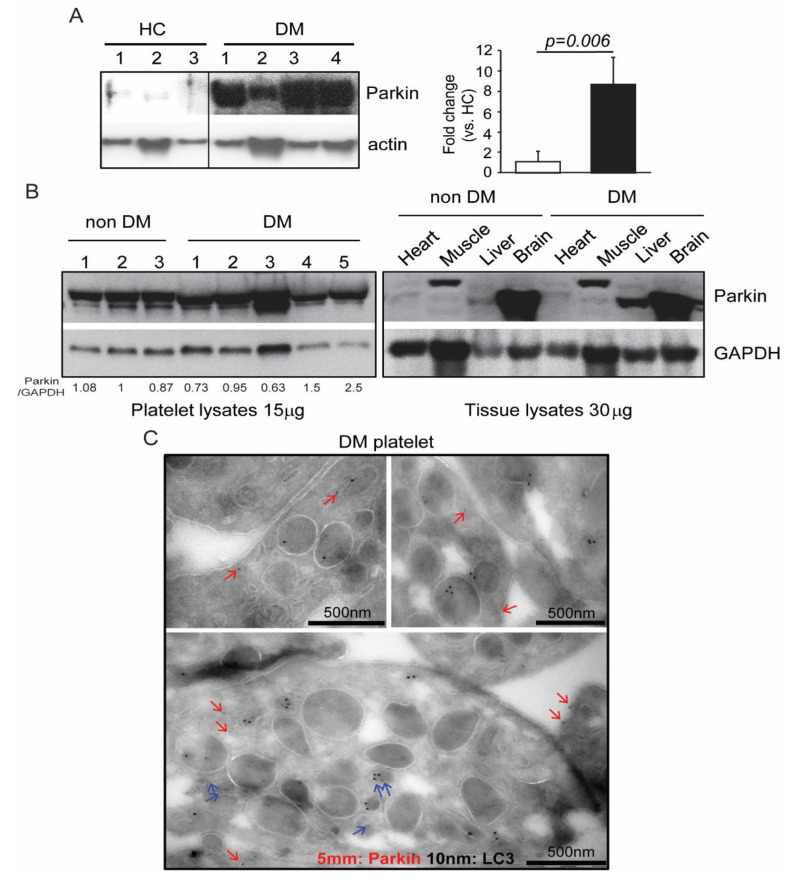
We previously verified that mitophagy induction in DM platelets occurs through a Parkin-dependent mechanism [9]. Surprisingly, Parkin is highly expressed in human DM platelet (Figure 1A) [9,14]. Although Parkin expression was only slightly increased in murine DM platelets, its expression was more than double that of other mitochondria-rich tissues like the heart, muscle, liver, and brain. (Figure 1B) [14]. Electron microscopy demonstrated that Parkin is localized within mitochondria and granules (blue arrows), cytosol, and cell membrane (red arrows) in DM platelets, further elaborating on its potentially diverse functions (Figure 1C). Mitochondrial Parkin also colocalizes with LC3, an autophagy marker (Figure 1C). No other functions beyond mitophagy induction in platelets have been reported for Parkin [14].
Figure 1.
Parkin was highly expressed in platelets. (A) Western blot analysis of Parkin expression in human platelets (platelets isolated from three healthy control and four diabetic patients) (left). Quantification of Parkin in HC and DM (right) (B) Platelets, heart, muscle, liver, and brain tissues isolated from non-DM and DM mice (platelets isolated from three non-DM and five DM mice). Western blot analysis of Parkin in each sample with lanes representing individual mice. (C) Parkin and LC3 immuno-EM analysis of DM platelets where 5 nm dots indicate immunogold-labeled Parkin clusters, and 10 nm dots indicate immunogold-labeled LC3 clusters. Representative areas of clusters of gold labeling in DM patients (A–C) are presented. Blue arrows indicate mitochondrial and granular Parkin. Red arrows indicate parkin in other regions.
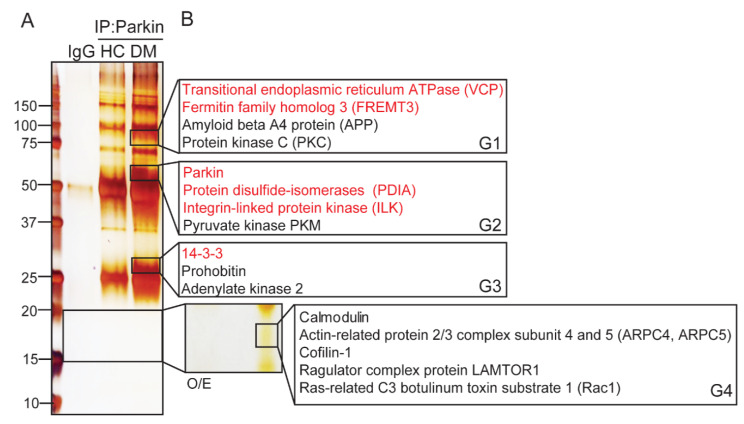
To understand the various functions of Parkin in platelets, we identified Parkin-interacting proteins through immunoprecipitation (IP) with Parkin-specific antibodies and LC-MS/MS (Figure 2). We identified 127 interacting proteins (Table 1, Table 2 and Table 3). Coverage was more than 40% for 33 interacting proteins (Table 1). These proteins include contain platelet aggregating factors such as fibrinogen, coagulation factors, and platelet factors 4. Ubiquitin E2 ligase (UBE2V1), actin related (ARPC5 and cofilin-1), and mitochondria-related proteins (prohibitin, NDUFA2 and ATP5l) were also identified. Moreover, several isoforms of 14-3-3 isoforms were also identified. The 94 proteins with coverage below 40% are listed in Table 2 and Table 3.
Figure 2.
Identification of Parkin-interacting proteins in human DM platelets. (A) Immunoprecipitation of Parkin-interacting proteins in human DM platelets visualized by silver staining. We isolated 10 enriched bands from DM and compared it with HC for LC-MS/MS analysis. (B) Potential interacting proteins were analyzed by LC-MS/MS and using Mascot analysis.
Table 1.
Identification of Parkin-interacting proteins in DM platelets (above 40% coverage).
| Protein ID | Protein Name | MW (Da) | % Coverage |
|---|---|---|---|
| FIBG_HUMAN | Fibrinogen gamma chain OS = Homo sapiens GN = FGG PE = 1 SV = 3 | 51,479 | 78.8 |
| TERA_HUMAN | Transitional endoplasmic reticulum ATPase OS = Homo sapiens GN = VCP PE = 1 SV = 4 | 89,266 | 78.5 |
| FIBB_HUMAN | Fibrinogen beta chain OS = Homo sapiens GN = FGB PE = 1 SV = 2 | 55,892 | 75.4 |
| 1433Z_HUMAN | 14-3-3 protein zeta/delta OS = Homo sapiens GN = YWHAZ PE = 1 SV = 1 | 27,728 | 74.7 |
| ARPC5_HUMAN | Actin-Related protein 2/3 complex subunit 5 OS = Homo sapiens GN = ARPC5 PE = 1 SV = 3 | 16,310 | 72.2 |
| 1433F_HUMAN | 14-3-3 protein eta OS = Homo sapiens GN=YWHAH PE = 1 SV = 4 | 28,201 | 69.1 |
| DYL1_HUMAN | Dynein light chain 1, cytoplasmic OS = Homo sapiens GN = DYNLL1 PE = 1 SV = 1 | 10,359 | 64 |
| 1433B_HUMAN | 14-3-3 protein beta/alpha OS = Homo sapiens GN = YWHAB PE = 1 SV = 3 | 28,065 | 63 |
| GSTO1_HUMAN | Glutathione S-transferase omega-1 OS=Homo sapiens GN = GSTO1 PE = 1 SV = 2 | 27,548 | 61.4 |
| TPM4_HUMAN | Tropomyosin alpha-4 chain OS=Homo sapiens GN = TPM4 PE = 1 SV = 3 | 28,504 | 60.1 |
| 1433E_HUMAN | 14-3-3 protein epsilon OS = Homo sapiens GN = YWHAE PE = 1 SV = 1 | 29,155 | 59.2 |
| TSP1_HUMAN | Thrombospondin-1 OS = Homo sapiens GN = THBS1 PE = 1 SV = 2 | 129,300 | 58.1 |
| FIBA_HUMAN | Fibrinogen alpha chain OS = Homo sapiens GN = FGA PE = 1 SV = 2 | 94,914 | 56.9 |
| URP2_HUMAN | Fermitin family homolog 3 OS = Homo sapiens GN = FERMT3 PE = 1 SV = 1 | 75,905 | 55.8 |
| PRDX6_HUMAN | Peroxiredoxin-6 OS = Homo sapiens GN = PRDX6 PE = 1 SV = 3 | 25,019 | 54 |
| CAP1_HUMAN | Adenylyl cyclase-associated protein 1 OS=Homo sapiens GN = CAP1 PE = 1 SV = 5 | 51,869 | 53.5 |
| S10A8_HUMAN | Protein S100-A8 OS = Homo sapiens GN = S100A8 PE = 1 SV = 1 | 10,828 | 52.7 |
| 1433T_HUMAN | 14-3-3 protein theta OS = Homo sapiens GN = YWHAQ PE = 1 SV = 1 | 27,747 | 51.4 |
| F13A_HUMAN | Coagulation factor XIII A chain OS = Homo sapiens GN = F13A1 PE = 1 SV = 4 | 83,215 | 51.2 |
| PDIA5_HUMAN | Protein disulfide-isomerase A5 OS=Homo sapiens GN = PDIA5 PE = 1 SV = 1 | 59,556 | 50.5 |
| PHB_HUMAN | Prohibitin OS = Homo sapiens GN = PHB PE = 1 SV = 1 | 29,786 | 50.4 |
| ILK_HUMAN | Integrin-linked protein kinase OS = Homo sapiens GN =I LK PE = 1 SV = 2 | 51,386 | 49.1 |
| MMRN1_HUMAN | Multimerin-1 OS = Homo sapiens GN = MMRN1 PE = 1 SV = 3 | 138,023 | 48.4 |
| COF1_HUMAN | Cofilin-1 OS = Homo sapiens GN = CFL1 PE = 1 SV = 3 | 18,491 | 47 |
| 1433G_HUMAN | 14-3-3 protein gamma OS = Homo sapiens GN = YWHAG PE = 1 SV = 2 | 28,285 | 47 |
| LTOR1_HUMAN | Ragulator complex protein LAMTOR1 OS = Homo sapiens GN = LAMTOR1 PE = 1 SV = 2 | 17,734 | 45.3 |
| UB2V1_HUMAN | Ubiquitin-conjugating enzyme E2 variant 1 OS = Homo sapiens GN = UBE2V1 PE = 1 SV = 2 | 16,484 | 44.2 |
| CALR_HUMAN | Calreticulin OS = Homo sapiens GN = CALR PE = 1 SV = 1 | 48,112 | 43.9 |
| PLF4_HUMAN | Platelet factor 4 OS = Homo sapiens GN = PF4 PE = 1 SV = 2 | 10,838 | 43.6 |
| PDIA6_HUMAN | Protein disulfide-isomerase A6 OS = Homo sapiens GN = PDIA6 PE = 1 SV = 1 | 48,091 | 42 |
| ATP5I_HUMAN | ATP synthase subunit e, mitochondrial OS = Homo sapiens GN = ATP5I PE = 1 SV = 2 | 7928 | 40.6 |
| EMIL1_HUMAN | EMILIN-1 OS = Homo sapiens GN = EMILIN1 PE = 1 SV = 2 | 106,601 | 40.4 |
| NDUA2_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 OS = Homo sapiens GN = NDUFA2 PE = 1 SV = 3 | 10,915 | 40.4 |
Table 2.
Identification of Parkin-interacting proteins in DM platelets (39~20% coverage).
| Protein ID | Protein Name | MW (Da) | % Coverage |
|---|---|---|---|
| PDE5A_HUMAN | cGMP-specific 3′,5′-cyclic phosphodiesterase OS = Homo sapiens GN = PDE5A PE = 1 SV = 2 | 99,921 | 38.7 |
| CXCL7_HUMAN | Platelet basic protein OS = Homo sapiens GN = PPBP PE = 1 SV = 3 | 13,885 | 38.3 |
| ATP5L_HUMAN | ATP synthase subunit g, mitochondrial OS = Homo sapiens GN = ATP5L PE = 1 SV = 3 | 11,421 | 37.9 |
| S10A9_HUMAN | Protein S100-A9 OS = Homo sapiens GN = S100A9 PE = 1 SV = 1 | 13,234 | 37.7 |
| NDUA4_HUMAN | Cytochrome c oxidase subunit NDUFA4 OS = Homo sapiens GN = NDUFA4 PE = 1 SV = 1 | 9,364 | 37 |
| CALM_HUMAN | Calmodulin OS = Homo sapiens GN = CALM1 PE = 1 SV = 2 | 16,827 | 36.9 |
| S10A4_HUMAN | Protein S100-A4 OS = Homo sapiens GN = S100A4 PE = 1 SV = 1 | 11,721 | 36.6 |
| MIC60_HUMAN | MICOS complex subunit MIC60 OS = Homo sapiens GN = IMMT PE = 1 SV = 1 | 83,626 | 35.6 |
| FINC_HUMAN | Fibronectin OS = Homo sapiens GN = FN1 PE = 1 SV = 4 | 262,460 | 34.5 |
| NEXN_HUMAN | Nexilin OS = Homo sapiens GN = NEXN PE = 1 SV = 1 | 80,609 | 34.5 |
| ITB3_HUMAN | Integrin beta-3 OS = Homo sapiens GN = ITGB3 PE = 1 SV = 2 | 87,000 | 33.6 |
| PSA7_HUMAN | Proteasome subunit alpha type-7 OS = Homo sapiens GN = PSMA7 PE = 1 SV = 1 | 27,870 | 33.1 |
| SAMP_HUMAN | Serum amyloid P-component OS = Homo sapiens GN = APCS PE = 1 SV = 2 | 25,371 | 32.7 |
| SKAP2_HUMAN | Src kinase-associated phosphoprotein 2 OS = Homo sapiens GN = SKAP2 PE = 1 SV = 1 | 41,191 | 32.3 |
| NUBP2_HUMAN | Cytosolic Fe-S cluster assembly factor NUBP2 OS = Homo sapiens GN = NUBP2 PE = 1 SV = 1 | 28,807 | 31.4 |
| SSBP_HUMAN | Single-Stranded DNA-binding protein, mitochondrial OS = Homo sapiens GN = SSBP1 PE = 1 SV = 1 | 17,249 | 31.1 |
| GPIX_HUMAN | Platelet glycoprotein IX OS = Homo sapiens GN = GP9 PE = 1 SV = 3 | 19,034 | 30.5 |
| A4_HUMAN | Amyloid beta A4 protein OS = Homo sapiens GN = APP PE = 1 SV = 3 | 86,888 | 30 |
| PSME2_HUMAN | Proteasome activator complex subunit 2 OS = Homo sapiens GN = PSME2 PE = 1 SV = 4 | 27,384 | 29.3 |
| CXCL7_HUMAN | Platelet basic protein OS = Homo sapiens GN = PPBP PE = 1 SV = 3 | 13,885 | 27.3 |
| KAD2_HUMAN | Adenylate kinase 2, mitochondrial OS=Homo sapiens GN = AK2 PE = 1 SV = 2 | 26,461 | 27.2 |
| UB2L3_HUMAN | Ubiquitin-conjugating enzyme E2 L3 OS = Homo sapiens GN = UBE2L3 PE = 1 SV = 1 | 17,850 | 26.6 |
| KPYM_HUMAN | Pyruvate kinase PKM OS = Homo sapiens GN = PKM PE = 1 SV = 4 | 57,900 | 26.2 |
| DHB4_HUMAN | Peroxisomal multifunctional enzyme type 2 OS = Homo sapiens GN = HSD17B4 PE = 1 SV = 3 | 79,636 | 25.7 |
| NDUA7_HUMAN | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex subunit 7 OS =Homo sapiens GN = NDUFA7 PE =1 SV=3 | 12,544 | 25.7 |
| PSA3_HUMAN | Proteasome subunit alpha type-3 OS = Homo sapiens GN = PSMA3 PE = 1 SV = 2 | 28,415 | 24.7 |
| PRDX1_HUMAN | Peroxiredoxin-1 OS = Homo sapiens GN = PRDX1 PE = 1 SV = 1 | 22,096 | 23.6 |
| PLMN_HUMAN | Plasminogen OS = Homo sapiens GN = PLG PE = 1 SV = 2 | 90,510 | 23.5 |
| UBP5_HUMAN | Ubiquitin carboxyl-terminal hydrolase 5 OS = Homo sapiens GN = USP5 PE = 1 SV = 2 | 95,725 | 23.3 |
| HECD3_HUMAN | E3 ubiquitin-protein ligase HECTD3 OS = Homo sapiens GN = HECTD3 PE = 1 SV = 1 | 97,051 | 22.8 |
| CAN1_HUMAN | Calpain-1 catalytic subunit OS = Homo sapiens GN = CAPN1 PE = 1 SV = 1 | 81,838 | 22.5 |
| RGS18_HUMAN | Regulator of G-protein signaling 18 OS = Homo sapiens GN = RGS18 PE = 1 SV = 1 | 27,565 | 22.1 |
| PSME1_HUMAN | Proteasome activator complex subunit 1 OS = Homo sapiens GN = PSME1 PE = 1 SV = 1 | 28,705 | 22.1 |
| PDIA1_HUMAN | Protein disulfide-isomerase OS = Homo sapiens GN = P4HB PE = 1 SV = 3 | 57,081 | 22 |
| GPX1_HUMAN | Glutathione peroxidase 1 OS = Homo sapiens GN = GPX1 PE = 1 SV = 4 | 22,075 | 21.7 |
| PDIA3_HUMAN | Protein disulfide-isomerase A3 OS = Homo sapiens GN = PDIA3 PE = 1 SV = 4 | 56,747 | 21 |
| VWF_HUMAN | von Willebrand factor OS = Homo sapiens GN = VWF PE = 1 SV = 4 | 309,058 | 20.1 |
Table 3.
Identification of Parkin-interacting proteins in DM platelets (below 20% coverage).
| Protein ID | Protein Name | MW (Da) | % Coverage |
|---|---|---|---|
| RAC1_HUMAN | Ras-Related C3 botulinum toxin substrate 1 OS = Homo sapiens GN = RAC1 PE = 1 SV = 1 | 21,436 | 19.8 |
| KAP0_HUMAN | cAMP-dependent protein kinase type I-alpha regulatory subunit OS = Homo sapiens GN = PRKAR1A PE = 1 SV = 1 | 42,955 | 19.4 |
| PSA4_HUMAN | Proteasome subunit alpha type-4 OS = Homo sapiens GN = PSMA4 PE = 1 SV = 1 | 29,465 | 19.2 |
| FA5_HUMAN | Coagulation factor V OS = Homo sapiens GN = F5 PE = 1 SV = 4 | 251,546 | 17.9 |
| NDUAC_HUMAN | NADH dehydrogenase (ubiquinone)1 alpha subcomplex subunit 12 OS =Homo sapiensGN = NDUFA12 PE =1 SV=1 | 17,104 | 17.9 |
| VATG1_HUMAN | V-type proton ATPase subunit G 1 OS = Homo sapiens GN = ATP6V1G1 PE = 1 SV = 3 | 13,749 | 16.9 |
| S10A6_HUMAN | Protein S100-A6 OS = Homo sapiens GN = S100A6 PE = 1 SV = 1 | 10,173 | 16.7 |
| EMRE_HUMAN | Essential MCU regulator, mitochondrial OS = Homo sapiens GN = SMDT1 PE = 1 SV = 1 | 11,434 | 15.9 |
| ACO13_HUMAN | Acyl-Coenzyme A thioesterase 13 OS = Homo sapiens GN = ACOT13 PE = 1 SV = 1 | 14,951 | 15.7 |
| UBL4A_HUMAN | Ubiquitin-Like protein 4A OS = Homo sapiens GN = UBL4A PE = 1 SV = 1 | 17,766 | 15.3 |
| IMB1_HUMAN | Importin subunit beta-1 OS = Homo sapiens GN = KPNB1 PE = 1 SV = 2 | 97,108 | 15.1 |
| STAT3_HUMAN | Signal transducer and activator of transcription 3 OS = Homo sapiens GN = STAT3 PE = 1 SV = 2 | 88,011 | 13.6 |
| ATP8_HUMAN | ATP synthase protein 8 OS = Homo sapiens GN = MT-ATP8 PE = 1 SV = 1 | 7986 | 13.2 |
| RBX1_HUMAN | E3 ubiquitin-protein ligase RBX1 OS = Homo sapiens GN = RBX1 PE = 1 SV = 1 | 12,266 | 13 |
| THIO_HUMAN | Thioredoxin OS = Homo sapiens GN = TXN PE = 1 SV = 3 | 11,730 | 12.4 |
| TGFB1_HUMAN | Transforming growth factor beta-1 OS = Homo sapiens GN = TGFB1 PE = 1 SV = 2 | 44,313 | 12.1 |
| ROCK2_HUMAN | Rho-Associated protein kinase 2 OS = Homo sapiens GN = ROCK2 PE = 1 SV = 4 | 16,0799 | 12 |
| S10A9_HUMAN | Protein S100-A9 OS = Homo sapiens GN = S100A9 PE = 1 SV = 1 | 13,234 | 11.4 |
| QCR9_HUMAN | Cytochrome b-c1 complex subunit 9 OS = Homo sapiens GN = UQCR10 PE = 1 SV = 3 | 7304 | 11.1 |
| PSB7_HUMAN | Proteasome subunit beta type-7 OS = Homo sapiens GN = PSMB7 PE = 1 SV = 1 | 29,946 | 10.8 |
| TGFI1_HUMAN | Transforming growth factor beta-1-induced transcript 1 protein OS = Homo sapiens GN = TGFB1I1 PE = 1 SV = 2 | 49,782 | 10.6 |
| GLRX1_HUMAN | Glutaredoxin-1 OS = Homo sapiens GN = GLRX PE = 1 SV = 2 | 11,768 | 10.4 |
| ANT3_HUMAN | Antithrombin-III OS = Homo sapiens GN = SERPINC1 PE = 1 SV = 1 | 52,569 | 10.1 |
| UB2L6_HUMAN | Ubiquitin/ISG15-Conjugating enzyme E2 L6 OS = Homo sapiens GN = UBE2L6 PE = 1 SV = 4 | 17,757 | 9.8 |
| UFM1_HUMAN | Ubiquitin-Fold modifier 1 OS = Homo sapiens GN = UFM1 PE = 1 SV = 1 | 9112 | 9.4 |
| MGST3_HUMAN | Microsomal glutathione S-transferase 3 OS = Homo sapiens GN = MGST3 PE = 1 SV = 1 | 16,506 | 9.2 |
| ECHA_HUMAN | Trifunctional enzyme subunit alpha, mitochondrial OS = Homo sapiens GN = HADHA PE = 1 SV = 2 | 82,947 | 9 |
| ECHB_HUMAN | Trifunctional enzyme subunit beta, mitochondrial OS = Homo sapiens GN = HADHB PE = 1 SV = 3 | 51,262 | 8.9 |
| PSA5_HUMAN | Proteasome subunit alpha type-5 OS = Homo sapiens GN = PSMA5 PE = 1 SV = 3 | 26,394 | 8.7 |
| PSA2_HUMAN | Proteasome subunit alpha type-2 OS = Homo sapiens GN = PSMA2 PE = 1 SV = 2 | 25,882 | 8.1 |
| DNJA2_HUMAN | DnaJ homolog subfamily A member 2 OS = Homo sapiens GN = DNAJA2 PE = 1 SV = 1 | 45,717 | 8 |
| SODC_HUMAN | Superoxide dismutase (Cu–Zn) OS = Homo sapiens GN = SOD1 PE = 1 SV = 2 | 15,926 | 7.8 |
| UB2D2_HUMAN | Ubiquitin-Conjugating enzyme E2 D2 OS = Homo sapiens GN = UBE2D2 PE = 1 SV = 1indistinguishable | 16,724 | 7.5 |
| UBE2N_HUMAN | Ubiquitin-Conjugating enzyme E2 N OS = Homo sapiens GN = UBE2N PE = 1 SV = 1indistinguishable | 17,127 | 7.2 |
| GSTM3_HUMAN | Glutathione S-transferase Mu 3 OS = Homo sapiens GN = GSTM3 PE = 1 SV = 3 | 26,542 | 7.1 |
| CH10_HUMAN | 10 kDa heat shock protein, mitochondrial OS = Homo sapiens GN = HSPE1 PE = 1 SV = 2 | 10,925 | 6.9 |
| UBE2O_HUMAN | (E3-independent) E2 ubiquitin-conjugating enzyme OS = Homo sapiens GN = UBE2O PE = 1 SV = 3 | 141,205 | 6.9 |
| NDUS5_HUMAN | NADH dehydrogenase (ubiquinone) iron–sulfur protein 5 OS = Homo sapiens GN = NDUFS5 PE = 1 SV = 3 | 12,509 | 6.6 |
| TIM16_HUMAN | Mitochondrial import inner membrane translocase subunit TIM16 OS = Homo sapiens GN = PAM16 PE = 1 SV = 2 | 13,816 | 6.4 |
| COX5B_HUMAN | Cytochrome c oxidase subunit 5B, mitochondrial OS=Homo sapiens GN=COX5B PE=1 SV=2 | 13,687 | 6.2 |
| CD36_HUMAN | Platelet glycoprotein 4 OS = Homo sapiens GN = CD36 PE = 1 SV = 2 | 53,019 | 5.3 |
| COX41_HUMAN | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial OS = Homo sapiens GN = COX4I1 PE = 1 SV = 1 | 19,564 | 4.7 |
| AN32B_HUMAN | Acidic leucine-rich nuclear phosphoprotein 32 family member B OS = Homo sapiens GN = ANP32B PE = 1 SV = 1 | 28,770 | 4.4 |
| TXND9_HUMAN | Thioredoxin domain-containing protein 9 OS = Homo sapiens GN = TXNDC9 PE = 1 SV = 2 | 26,517 | 4 |
| ITB1_HUMAN | Integrin beta-1 OS = Homo sapiens GN = ITGB1 PE = 1 SV = 2 | 88,357 | 3.9 |
| ROCK1_HUMAN | Rho-Associated protein kinase 1 OS = Homo sapiens GN = ROCK1 PE = 1 SV = 1 | 158,076 | 3.8 |
| GRCR2_HUMAN | Glutaredoxin domain-containing cysteine-rich protein 2 OS = Homo sapiens GN = GRXCR2 PE = 3 SV = 1 | 28,266 | 3.6 |
| CATA_HUMAN | Catalase OS = Homo sapiens GN = CAT PE = 1 SV = 3 | 59,719 | 3.6 |
| VPS35_HUMAN | Vacuolar protein sorting-associated protein 35 OS = Homo sapiens GN = VPS35 PE = 1 SV = 2 | 91,649 | 3.6 |
| ITA2B_HUMAN | Integrin alpha-IIb OS = Homo sapiens GN = ITGA2B PE = 1 SV = 3 | 113,306 | 2.9 |
| ANGL5_HUMAN | Angiopoietin-rrelated protein 5 OS = Homo sapiens GN = ANGPTL5 PE = 2 SV = 3 | 44,115 | 2.6 |
| FA10_HUMAN | Coagulation factor X OS = Homo sapiens GN = F10 PE = 1 SV = 2 | 54,697 | 1.4 |
| UBP8_HUMAN | Ubiquitin carboxyl-terminal hydrolase 8 OS = Homo sapiens GN = USP8 PE = 1 SV = 1 | 127,444 | 1 |
| UBR4_HUMAN | E3 ubiquitin-protein ligase UBR4 OS = Homo sapiens GN = UBR4 PE = 1 SV = 1 | 573,476 | 0.9 |
| HD_HUMAN | Huntingtin OS = Homo sapiens GN = HTT PE = 1 SV = 2 | 347,383 | 0.7 |
| MYCB2_HUMAN | E3 ubiquitin-protein ligase MYCBP2 OS = Homo sapiens GN = MYCBP2 PE = 1 SV = 3 | 509,759 | 0.6 |
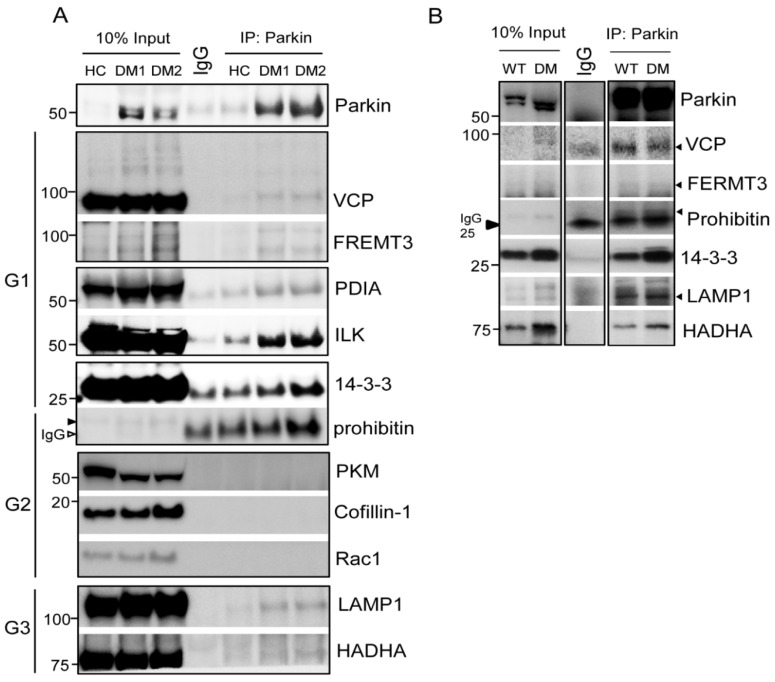
We confirmed interactions between Parkin and the identified proteins in human and murine platelets (Figure 3). Initially, we selected 11 candidates for Parkin interaction verification. VCP/p97, FREMT3, PDIA, ILK, and 14-3-3 (G1) interacted with Parkin in human platelets but prohibitin, PKM, cofillin-1, and Rac1 did not (G2) (Figure 3A). Additionally, the mitochondrial β-oxidation related protein Hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA thiolase/enoyl-CoA hydratase alpha subunits (HADHA), and LAMP1 (mitophagy related) (G3) also interacted with Parkin in platelets (Figure 3A). VCP/p97, FERMT3, LAMP1, HADHA, 14-3-3, and prohibitin interacted with Parkin in murine platelets (Figure 3B).
Figure 3.
Confirmation of interactions between Parkin and selected target proteins in human and murine platelets. (A) Immunoprecipitation of each specific antibody in pooled human DM platelets (4 HC, 5DM1, and 6DM2). We incubated 500 μg protein lysates incubated with specific antibodies overnight at 4 °C with 10% input as control. G1 represents interacting protein groups from LC-MS/MS results. G2 represents the non-interacting proteins group in LC-MS/MS results. G3 represents protein that interacted with Parkin and that were not found in the LC-MS/MS results. (B) Immunoprecipitation of each specific antibody in pooled murine platelets (3 WT and 5DM). We incubated 500 μg protein lysates incubated with specific antibodies overnight at 4 °C with 10% input as control.
2.2. Determining the Potential Roles of Platelet Parkin-Interacting Proteins through Literature Analysis
2.2.1. Parkin Plays an Important Role in Mitochondrial Protection through Mitophagy
Transitional Endoplasmic Reticulum ATPase (VCP/p97)
VCP/p97 is a hexameric protein of the AAA (ATPases associated with diverse cellular activities) family which generally utilizes energy from ATP hydrolysis [23]. VCP/p97 has been linked to various membrane trafficking processes, including Golgi reassembly post-mitosis [24] and control of lipid droplet biogenesis [25]. Emerging evidence has connected VCP/p97 to lysosomal protein degradation through its ability to facilitate cargo sorting via the endosomal pathway and autophagy [26,27,28]. One VCP/p97 mutation causes a rare multisystem disease, IBMPFD (inclusion body myopathy with Paget’s disease and frontotemporal dementia) [28]. Several recent studies reported the involvement of VCP/p79 in mitophagy [29,30]. Here, we identified that VCP/p97 interacts with Parkin in DM platelets, possibly regulating the mitophagy process (VCP/p97: 78.5% coverage, Table 1).
Lysosomal-Associated Membrane Protein 1 (LAMP1)
LAMP1 is a well-known lysosomal protein that we previously confirmed to colocalize with Parkin and LC3 in DM platelets [9,14] and, again, in this study through IP (Figure 3). Although not identified in the LC-MS/MS, LAMP1 was used to confirm Parkin’s role in autophagy activation (Table 1, Table 2 and Table 3). This highlights the potential deficiencies and false negatives of the LC-MS/MS, potentially due to the complex processing.
Mitochondrial Three Functional Protein A (TFPα, HADHA)
TFPα is a multienzyme mitochondrial complex harboring three major enzymes from the β-oxidation cycle of long-chain fatty acids. TFPα deficiencies present in neonates as a severe cardiac phenotype, often with death in the first weeks. Deficiency is related to maternal HELLP (hydrolysis, elevated liver enzyme and low platelets) syndrome and reduced birth weight [31]. HADHA is involved in long-chain fatty acid-induced autophagy of intestinal epithelial cells and is therefore proposed as a new therapeutic target for inflammatory bowel disease (IBD) [32]. A functional relationship may exist between Parkin and HADHA.
Prohibitin (PHB, Murine Only)
Two members of the prohibitin family, PHB1 and PHB2, are highly homologous proteins localized to the mitochondrial inner membrane [33,34]. The PHB complexes perform diverse functions in mitochondria, including regulation of membrane protein degradation, chaperones, regulation of oxidative phosphorylation, maintenance of mitochondrial genetic stability, and regulation of mitochondrial morphology [33,34]. PHB1 and 2 also function as autophagy receptors [35,36,37,38]. PHB2 directly interacts with LC3 potentially regulating the mitophagy process [37]. In platelets, prohibitin is expressed in membranes and is involved in PAR1-mediated platelet aggregation [39]. Here, we confirmed that prohibitin can interact with Parkin in murine platelets but not in human DM platelets (Figure 3B).
2.2.2. Parkin Regulates DM Platelets through Protein Interactions with Integrin Complex Proteins
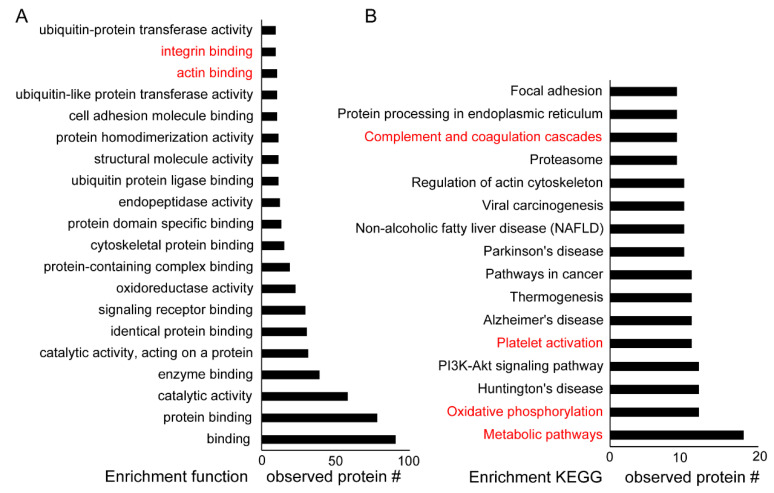
We analyzed the enriched function (GO) and KEGG pathways using the identified proteins (Figure 4). Parkin-interacting proteins were associated with integrin binding, signaling, and ubiquitin proteasome system in DM platelets (Figure 4A). Parkin may participate in the metabolic pathway and oxidative phosphorylation of DM platelets as well as platelet activation and the coagulation cascade (Figure 4B). Parkin is localized to the mitochondrial outer membrane during mitophagy activation. Furthermore, Parkin may be associated with integrin signaling through 14-3-3, Amyloid beta A4 protein [1], Calreticulin (CALR), EMILIN1, fermitin family homolog3 (FERMT3), integrin-linked protein kinase [1], integrin beta 1 and 3 (ITGB1 and ITGB3), protein disulfide isomerase (PDI/P4HB), thrombospondin-1 (THBS1), and von Willebrand factor (VWF) in platelets (Table 1, Table 2 and Table 3). Our study confirmed the interaction between Parkin and 14-3-3, FREMT3, PDI, and ILK (Figure 3). Parkin may integrate outside-in signaling in platelets and may induce platelet activation through these interactions.
Figure 4.
Enriched function (GO) and KEGG analysis of identified proteins. STRING (functional protein association networks) used in the grouping of identified proteins. (A) Top 20 enriched functions of Parkin-interacting proteins in human DM platelets. (B) Top 16 enriched KEGG pathways of Parkin-interacting proteins in human DM platelets.
14-3-3
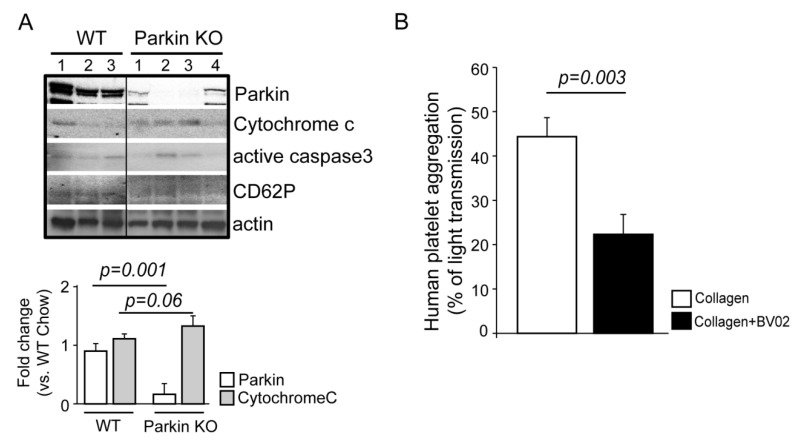
Hyperactivation and hyperaggregation are well-known phenotypes of DM platelets [5,6]. Additionally, 14-3-3 is involved in platelet aggregation with GPIB-IX-V [40,41]. The 14-3-3 proteins family are highly conserved intracellular proteins with several isoforms: β, ε, ζ, γ, η, τ, and σ. These isoforms associate as homo- and heterodimers interact with over 200 different proteins including serine and threonine phosphorylated intracellular proteins. The 14-3-3 proteins interact with diverse intracellular molecules, signaling proteins, metabolic enzymes, cytoskeletal proteins, transcription factors, and apoptosis-related proteins and can regulate platelet mitochondrial respiratory [42,43,44]. It has also been associated with neuronal diseases [36,45,46]; 14-3-3 η is a well-known negative regulator of Parkin E3 activity through direct interactions in mice brains [46]. Six 14-3-3 isoforms have been detected in human platelets including β, ε, ζ, γ, η and τ, with ζ and γ expressed at high levels [43,47]. The first identified 14-3-3 ζ binds to the cytoplasmic tails of GPIbα and GPIbβ [40,48] while other reports have suggested that the GPIb-14-3-3ζ interactions can promote VWF-dependent integrin αIIββ3-activation and cell spreading [48,49]. Mice that are 14-3-3ζ-deficient exhibit defective pro-coagulant function reduced arterial thrombosis, reduced thrombin generation, and pulmonary embolism in vivo. Platelet bioenergetics has revealed enhanced mitochondrial respiratory reserve capacities in 14-3-3ζ-deficient platelets that correlated with sustained levels of metabolic ATP levels. Moreover, 14-3-3ζ serves as an important regulator of platelet bioenergetic functioning and, therefore, also pro-coagulant and thrombosis in vivo [47]. The LC-MS/MS results indicated that Parkin interacted with 14-3-3 ε, λ, θ, η, δ, ζ, α, and β in DM platelets (Table 1). Therefore, Parkin may interact indirectly with GPIbα-V-IX, integrin beta3, and cellular signaling through interactions with diverse 14-3-3 isoforms. The 14-3-3 inhibitor, BVO2, inhibited the GPIb-IX-V complex and collagen induced platelet aggregation (Figure 5B). Parkin may regulate integrin-14-3-3-induced platelet activation. Collectively, our results and previous reports, predict that Parkin may regulate platelet aggregation through 14-3-3 and GPIb-IX-V complexes. Furthermore, Parkin participates in platelet activation through integrin signaling.
Figure 5.
Parkin associated with apoptosis and platelet aggregation. (A) Western blot analysis of Parkin KO mice platelets. Parkin, Cytochrome C, active caspase3, and CD62P antibodies were used for this experiment. The numbers indicate individual mice. (B) Quantification of each group using ImageJ software. (B) Collagen induced platelet aggregation in human platelets (n = 3) after BVO2 (14-3-3 inhibitor) treatment.
Fermitin Family Homolog 3 (FERMT3: URP2/Kindlin-3)
The kindlin family members, including kindlin-1, kindlin-2, and kindlin-3, have high sequence homology but display different tissue expression patterns [50]. These proteins strongly associate with human diseases, as a lack of kindlin-1 in humans cause Kindler syndrome. Kindlin-2 deficiencies have not been reported with lethal consequences [51,52,53]. Kindlin-3 deficiencies display severe bleeding and recurrent infections due to the dysfunctional integrin in platelet and leukocytes [54,55,56]. Kindlin-3 deficiencies are key to supporting integrin activation and platelet thrombus formation [50]. Parkin interacts with kindlin-3 in human and murine DM platelets (kindlin-3/FERMT3: 55.8% coverage, Figure 3). Based on this and Parkin’s role in mitophagy, we suggest that FERMT3 is involved in platelet mitophagy and thrombus formation through integrin signaling with Parkin.
Protein Disulfide-Isomerase (PDI, P4HB)
Protein disulfide isomerase (PDI) was identified 20 years ago as an endoplasmic reticulum protein that facilitates the formation of correct disulfide bonds in nascent proteins [57]. There are more than 20 members of the PDI, seven containing a CGHC-active site [57]. Among CGHC-active site members, four are associated with platelet function and thrombosis (PDI, ERp57, ERp72, and ERp5) [57]. PDI was the first from this protein family identified in integrin-mediated platelet aggregation, adhesion, and thrombosis [58,59]. Here, in our LC-MS/MS results, we identified PDIA 1, 3, 5, and 6 as Parkin-interacting proteins (Table 1, Table 2 and Table 3). Parkin-associated integrin-mediated platelet function, homeostasis, and thrombosis are dependent on PDIs to form appropriate disulfide bonds.
Integrin Linked Kinase
Integrin-linked kinase was reported to interact with the cytoplasmic tail of β-integrin subunits and its serine/threonine kinase activity is upregulated through platelet stimulation [60,61,62]. ILKs have functions as adaptor proteins, interacting and regulating β1 and β3 integrin subunits [63]. ILK-deficient mice exhibit reduced platelet activation and aggregation and increased bleeding [60,62]. ILK regulates the rate of platelet activation rates and is essential for the formation of stable thrombi by controlling platelet response rates to collagen via GPVI [62]. From this, Parkin may regulate platelet functioning by binding with integrin and its related proteins like ILK (49.1% coverage, Table 1).
2.3. New Functions of Parkin and Parkin-Interacting Proteins in Platelets
Through GO and KEGG analysis in conjunction with literature reviews, we present and support additional Parkin functions through confirmed Parkin-interacting proteins. Key platelet functions including activation, aggregation, and mitochondrial functions appear to be regulated, at least partially, by Parkin. We previously verified Parkin-dependent mitophagy activation in a T2DM mouse model [9,14]. To confirm the importance of Parkin in platelets, we incorporated Parkin KO mice platelets. In Parkin KO mice platelets, there was increased cytochrome C and active caspase3 indicative of increased platelet apoptosis (Figure 5A). The platelet activation maker, CD62P (pSelectin), was slightly decreased but not significantly. The LC-MS/MS analysis verified the functional relationship between platelet activation-associated proteins and Parkin (Table 1, Table 2 and Table 3), suggesting that Parkin regulates platelet activation. We then used a 14-3-3 inhibitor to interrupt the interaction with Parkin (Figure 5B) which decreased platelet aggregation induced by collagen through 14-3-3 inhibitor treatment.
3. Discussion
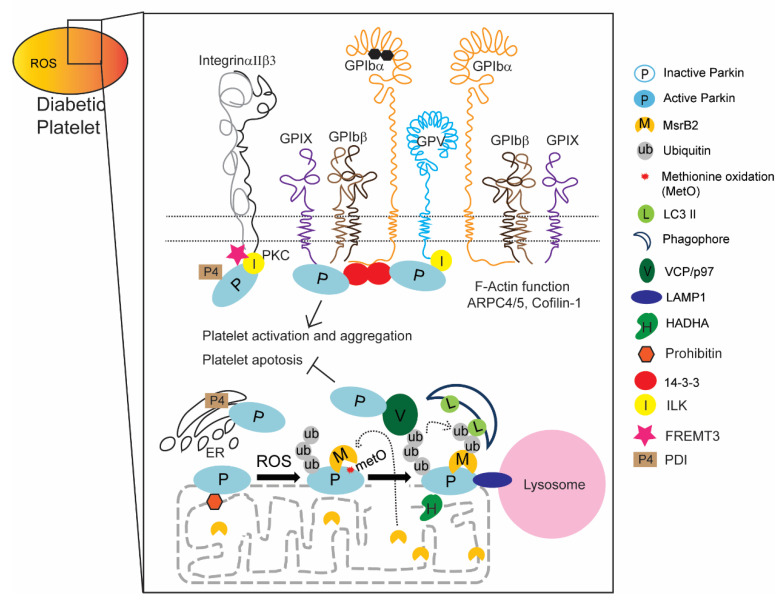
We previously verified that Parkin expression levels were high in HC platelets, even more so in DM [9,14] and underwent post-translational modification (MetO) under oxidative stress [14]. Cysteine oxidation is significantly increased in PD and includes methionine oxidation [64]. We confirmed that the ubiquitylation and methionine oxidation that occurs in Parkin lead to mitophagy. In this study, we aimed to uncover and verify other key platelet functions regulated by Parkin through the identification of Parkin-interacting proteins using IP and LC-MS/MS. Through this proteomic approach, we provided experimental and literature evidence that Parkin may regulate various processes beyond mitophagy, including integrin-dependent signaling, mitochondrial energy metabolism, platelet activation/aggregation, and ER-mitochondrial cross-talk (Figure 6). Furthermore, surface membrane receptor interactions with Parkin provide a possible link between external signaling and internal cellular processes.
Figure 6.
Summary of Parkin functions in DM platelets.
Parkin transfers the changes in the external environment, like diabetes, to the internal environment by interacting with membrane receptor proteins, like integrin, inducing mitophagy. Parkin also indirectly interacts with cytosolic proteins, F-actin functional ARP4/5, and directly with VCP/p97 associated with mitophagy activation. PINK1 is well-known to interact with Parkin and VCP/p97, after which this complex regulates dendritic arborization [65]. Based on these results, we hypothesize that Parkin regulates organelle cross-talk potentially transferring external signals to internal platelet environments and cell–cell cross-talk among other cells. Proteomic analyses also suggest that Parkin participates in diverse processes and mitophagy induction in DM platelets, like phagophore recruitment to damaged mitochondria through interactions with MsrB2 [14] and regulation of autolysosome formation through interactions with LAMP1. Parkin-deficient mice exhibit increased apoptotic platelets compared with healthy control mice (Figure 5A) [9]. Parkin interacts with 14-3-3, PDIA, FREMT3, ILK, and F-actin-related proteins (well-known proteins associated with platelet activation and aggregation), and so we suggest that Parkin is also involved with platelet aggregation and activation (Figure 6). In DM platelets, mitochondrial outer membrane is disrupted with mitochondrial protein release and exposed inner membrane proteins [14]. Consequently, the interaction between Parkin and HADHA and prohibitin may occur in human and murine DM platelets (Table 2), suggesting that Parkin is likely to participate in platelet energy metabolism in DM (Figure 6).
In conclusion, our results suggest that Parkin with its many actions on platelet mitochondria, activation, and aggregation may be a therapeutic target for antiplatelet treatment. However, further detailed molecular studies are required that focus on the individual binding partners of Parkin in DM platelets.
4. Material and Methods
4.1. Preparation of Human Platelet
Venous blood was drawn from healthy and patients at Yale University School of Medicine (HIC#1005006865) from multiple outpatient clinics including the cardiovascular, diabetes, and neurology clinics. Informed consent was obtained from all subjects, and the experiments conformed to the principles set out in the WMA Declaration of Helsinki and the Department of Health and Human Services Belmont Report (IRB#1006006865, 5/11/2017). All healthy subjects were free from medication or diseases known to interfere with platelet function [5,6,9]. Upon informed consent, a venous blood sample (approximately 20 cc) was drawn by standard venipuncture and collected into tubes containing 3.8% trisodium citrate (w/v). Blood samples were prepared as previously described [66]. Platelet-rich plasma (PRP) was obtained by differential centrifugation. Purity of platelet preparation was determined by Western blot analysis using platelet markers (CD41), monocyte markers (CD14), and red blood cell markers (CD235a) [9].
4.2. Preparation of Mice Platelets
Blood (0.7–1 mL) was directly aspirated from the right cardiac ventricle into 1.8% sodium citrate (pH 7.4) in WT (C57Bl/6) and diabetic mice (mice were 8 weeks of age; STZ injected for 5 days followed by high-fat diet for 12 weeks). Citrated blood from several mice with identical genotype was pooled and diluted with equal volume of HEPES/Tyrode’s buffer. PRP was prepared by centrifugation at 100 g for 10 min and then used for Immunoprecipitation and Western blotting. All mice were of C57Bl/6 background (WT and Parkin whole-body knockout). The experiment of Parkin KO mice experiment was performed at the Yale Animal Facility and 300 George St. New Haven, CT, USA, under the supervision of YARC (Yale Animal Resources Center) and Rita Weber (Animal facility manager, YARC). All experiments were performed in accordance with guidelines and regulations as outlined by IACUC (the Yale Institutional Animal Care and Use Committee) under the approved protocol (IACUC# 2017-11413 (3/31/2017)). Diabetic mice were generated in a Korea CDC animal facility. All experiments were performed in accordance with guidelines and regulations under the approved protocol (#KCDC-109-18-2A (2019), #KCDC-116-19-2A (2020), and #KCDC-115-19-2A (2020)).
4.3. Western Blotting
Standard Western blot analysis protocols were used. A 10% Input of IP lysates was loaded in each well as loading control. We used specific individual antibodies and dilutions (Parkin: abcam #ab15954 (1:1000), VCP: Invitrogen #MA3-004 (1:1000), FREMT3: abcam #ab68040 (1:1000), PDIA: abcam #ab2792 (1:1000), ILK: abcam #ab52480 (1:1000), 14-3-3: Thermo Fisher scientific #51-0700 (1:1000), prohibitin: abcam #ab28172 (1:1000), LAMP1: cell signaling #9091 (1:1000), HADHA: abcam #ab203114 (1:1000), PKM: abcam #ab137791 (1:1000), cofillin-1: GeneTex #GTX102156 (1:1000), Rac1: Sigma–Aldrich #SAB4502560 (1:1000), GAPDH: cell signaling #3683 (1:1000), Cytochrome C: abcam #ab90529 (1:2000), CD62P: abcam #182135 (1:1000), Actin: Santa Cruz #SC47778 (1:1000)).
4.4. Immunoprecipitation
The 500 μg pulled healthy/DM platelet lysates and cell lysates (after transient transfection) were mixed with 1 μg specific target antibodies and the same species IgG control with HC) and incubated overnight at 4 °C. Then 50% slurry protein A sepharose beads and 50% slurry protein G sepharose beads were mixed 1:1. Next, 30 μL of the 50% slurry washed A/G beads with lysates/antibodies mixture was incubated for 1 h at 4 °C. After 3 more washes with lysis buffer, we used 1–10% lysates were used for the input. We pooled 4 healthy subjects and 11 human DM platelets (5 pooled in DM1 and 6 pooled in DM2) for Parkin immunoprecipitation (Figure 3A) and pooled 3 WT and 5 DM mice platelets (Figure 3B).
4.5. Silver Staining and LC-Mass Analysis
Mass spectrometry was performed according to the manufacturer’s protocols by Pierce silver stain, and individual bands were excised for LC-MS/MS after silver staining. In-gel digestion, LC-MS/MS, and peptide identification were performed by Yale MS & Proteomics Resource.
In-Gel Digestion: Silver-stained gel bands were treated with 5% acetic acid for 10 min with rocking. The acid was removed, and the bands were covered with freshly prepared destaining solution (made fresh by mixing in a 1:1 ratio stock solutions of 30 mM potassium ferricyanide in water and 100 mM sodium thiosulfate in water) until the brownish color disappeared. The bands were then rinsed three times with 0.5 mL of water for 5 min to remove the acid and chemical reducing agents. The gel bands were cut into small pieces and washed for 30 min on a tilt-table with 450 µL 50% acetonitrile/100 mM NH4HCO3 (ammonium bicarbonate) followed by a 30 min wash with 50% acetonitrile/12.5 mM NH4HCO3. The gel bands were shrunk by the brief addition then removal of acetonitrile, and then dried by speed vacuum. Each sample was resuspended in 100 µL of 25 mM NH4HCO3 containing 0.5 µg of digestion grade trypsin (Promega, V5111) and incubated at 37 °C for 16 h. Supernatants containing tryptic peptides were transferred to new Eppendorf tubes and the gel bands were extracted with 300 µL of 80% acetonitrile/0.1% trifluoroacetic acid for 15 min. Supernatants were combined and dried by speed vacuum. Peptides were dissolved in 24 µL MS loading buffer (2% acetonitrile, 0.2% trifluoroacetic acid) with 5 µL injected for LC-MS/MS analysis.
LC-MS/MS on the Thermo Scientific Q Exactive Plus: LC-MS/MS analysis was performed on a Thermo Scientific Q Exactive Plus equipped with a Waters nanoAcquity UPLC system utilizing a binary solvent system (A: 100% water, 0.1% formic acid; B: 100% acetonitrile, 0.1% formic acid). Trapping was performed at 5 µL/min, 97% Buffer A for 3 min using a Waters Symmetry® C18 180 µm and 20 mm trap column (Waters, USA). Peptides were separated using an ACQUITY UPLC PST [67] C18 nanoACQUITY Column 1.7 µm, 75 µm × 250 mm (37 °C) and eluted at 300 nL/min with the following gradient: 3% buffer B at initial conditions; 5% B at 1 min; 35% B at 50 min; 50% B at 60 min; 90% B at 65 min; 90% B at 70 min; return to initial conditions at 71 min. MS was acquired in profile mode over the 300–1700 m/z range using 1 microscan, 70,000 resolution, AGC target of 3E6, and a maximum injection time of 45 ms. Data dependent MS/MS were acquired in centroid mode on the top 20 precursors per MS scan using 1 microscan, 17,500 resolution, AGC target of 1E5, maximum injection time of 100 ms, and an isolation window of 1.7 m/z. Precursors were fragmented by HCD activation with collision energy of 28%. MS/MS were collected on species with an intensity threshold of 2E4, charge states 2–6, and peptide match preferred. Dynamic exclusion was set to 20 s.
Peptide Identification: Tandem mass spectra were extracted by Proteome Discoverer software (version 1.3, Thermo Scientific) and searched in-house using the Mascot algorithm (version 2.6.0, Matrix Science). The data were searched against the SwissProt database (version 2017_01) with taxonomy restricted to Homo sapiens (20,172 sequences). Search parameters included trypsin digestion up to 2 missed cleavages, peptide mass tolerance of 10 ppm, MS/MS fragment tolerance of 0.02 Da, and methionine oxidation and propionamide adduct to cysteine as variable modifications. Normal and decoy database searches were run with the confidence level set to 95% (p < 0.05).
4.6. Platelet Aggregation Test
Platelet suspensions were incubated with BV02 (2-(2,3-Dihydro-1,5-dimethyl-3-oxo-2-phenyl-1H-pyrazol-4-yl)-2,3-dihydro-1,3-dioxo-1H-isoindole-5-carboxylic acid, 14-3-3 inhibitor, 10 nM) for 1 h. Platelet aggregation was monitored at 37 °C with constant stirring (1200 rpm) in a dual-channel lumi-aggregometer (model 700; Chrono-Log). Platelet aggregation was measured as the increase in light transmission for 10 min, starting with the addition of 2 μL of 1 mg/mL collagen (Chrono-Log) to a 500 μL reaction as a pro-aggregatory stimulus; the final concentration was 4 μg/mL. The maximum aggregation was expressed as a percentage of maximum light transmission with non-stimulated PRP of 0% and PPP of 100%.
Acknowledgments
Specially thanks to Jean Kanyo at Yale MS & Proteomics Resource.
Author Contributions
S.H.L. wrote the manuscript, and planned and performed experiments, data analysis, and figure development; J.D. recruited patients for the studies and helped prepare samples; W.-H.K. provided critical input for the manuscript; J.H. oversaw the study development and data analysis and wrote and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grants from NIH NHLBI (RO1 HL122815, HL115247, and HL117798). J.D. was supported by an NIH T32 Vascular Biology Training grant, and S.H.L. and W.-H.K. were supported by Korea National Institute of Health intramural research grant 4800-4845-302 (2018-NI008-00 and 2018-NI007-00).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Benjamin E.J., Blaha M.J., Chiuve S.E., Cushman M., Das S.R., Deo R., de Ferranti S.D., Floyd J., Fornage M., Gillespie C., et al. Heart disease and stroke statistics-2017 update: A report from the american heart association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreiro J.L., Angiolillo D.J. Diabetes and antiplatelet therapy in acute coronary syndrome. Circulation. 2011;123:798–813. doi: 10.1161/CIRCULATIONAHA.109.913376. [DOI] [PubMed] [Google Scholar]
- 3.Leytin V. Apoptosis in the anucleate platelet. Blood Rev. 2012;26:51–63. doi: 10.1016/j.blre.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 4.Lindemann S., Tolley N.D., Dixon D.A., McIntyre T.M., Prescott S.M., Zimmerman G.A., Weyrich A.S. Activated platelets mediate inflammatory signaling by regulated interleukin 1beta synthesis. J. Cell Biol. 2001;154:485–490. doi: 10.1083/jcb.200105058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang W.H., Stitham J., Gleim S., Di Febbo C., Porreca E., Fava C., Tacconelli S., Capone M., Evangelista V., Levantesi G., et al. Glucose and collagen regulate human platelet activity through aldose reductase induction of thromboxane. J. Clin. Investig. 2011;121:4462–4476. doi: 10.1172/JCI59291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang W.H., Stitham J., Jin Y., Liu R., Lee S.H., Du J., Atteya G., Gleim S., Spollett G., Martin K., et al. Aldose reductase-mediated phosphorylation of p53 leads to mitochondrial dysfunction and damage in diabetic platelets. Circulation. 2014;129:1598–1609. doi: 10.1161/CIRCULATIONAHA.113.005224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mason K.D., Carpinelli M.R., Fletcher J.I., Collinge J.E., Hilton A.A., Ellis S., Kelly P.N., Ekert P.G., Metcalf D., Roberts A.W., et al. Programmed anuclear cell death delimits platelet life span. Cell. 2007;128:1173–1186. doi: 10.1016/j.cell.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 8.Weyrich A.S., Dixon D.A., Pabla R., Elstad M.R., McIntyre T.M., Prescott S.M., Zimmerman G.A. Signal-dependent translation of a regulatory protein, Bcl-3, in activated human platelets. Proc. Natl. Acad. Sci. USA. 1998;95:5556–5561. doi: 10.1073/pnas.95.10.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S.H., Du J., Stitham J., Atteya G., Lee S., Xiang Y., Wang D., Jin Y., Leslie K.L., Spollett G., et al. Inducing mitophagy in diabetic platelets protects against severe oxidative stress. EMBO Mol. Med. 2016;8:779–795. doi: 10.15252/emmm.201506046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouseph M.M., Huang Y., Banerjee M., Joshi S., MacDonald L., Zhong Y., Liu H., Li X., Xiang B., Zhang G., et al. Autophagy is induced upon platelet activation and is essential for hemostasis and thrombosis. Blood. 2015;126:1224–1233. doi: 10.1182/blood-2014-09-598722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng W., Chang C., Luo D., Su H., Yu S., Hua W., Chen Z., Hu H., Liu W. Dissection of autophagy in human platelets. Autophagy. 2014;10:642–651. doi: 10.4161/auto.27832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Bermudez M., Lopez-Mejias R., Gonzalez-Juanatey C., Castaneda S., Miranda-Filloy J.A., Blanco R., Fernandez-Gutierrez B., Balsa A., Gonzalez-Alvaro I., Gomez-Vaquero C., et al. Association of the methionine sulfoxide reductase A rs10903323 gene polymorphism with cardiovascular disease in patients with rheumatoid arthritis. Scand. J. Rheumatol. 2012;41:350–353. doi: 10.3109/03009742.2012.677063. [DOI] [PubMed] [Google Scholar]
- 13.Domenech E., Maestre C., Esteban-Martinez L., Partida D., Pascual R., Fernandez-Miranda G., Seco E., Campos-Olivas R., Perez M., Megias D., et al. AMPK and PFKFB3 mediate glycolysis and survival in response to mitophagy during mitotic arrest. Nat. Cell Biol. 2015;17:1304–1316. doi: 10.1038/ncb3231. [DOI] [PubMed] [Google Scholar]
- 14.Lee S.H., Lee S., Du J., Jain K., Ding M., Kadado A.J., Atteya G., Jaji Z., Tyagi T., Kim W.H., et al. Mitochondrial MsrB2 serves as a switch and transducer for mitophagy. EMBO Mol. Med. 2019;11:e10409. doi: 10.15252/emmm.201910409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foroud T., Uniacke S.K., Liu L., Pankratz N., Rudolph A., Halter C., Shults C., Marder K., Conneally P.M., Nichols W.C. Heterozygosity for a mutation in the parkin gene leads to later onset Parkinson disease. Neurology. 2003;60:796–801. doi: 10.1212/01.WNL.0000049470.00180.07. [DOI] [PubMed] [Google Scholar]
- 16.Sunico C.R., Nakamura T., Rockenstein E., Mante M., Adame A., Chan S.F., Newmeyer T.F., Masliah E., Nakanishi N., Lipton S.A. S-Nitrosylation of parkin as a novel regulator of p53-mediated neuronal cell death in sporadic Parkinson’s disease. Mol. Neurodegener. 2013;8:29. doi: 10.1186/1750-1326-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty J., Basso V., Ziviani E. Post translational modification of Parkin. Biol. Direct. 2017;12:6. doi: 10.1186/s13062-017-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding D., Ao X., Liu Y., Wang Y.Y., Fa H.G., Wang M.Y., He Y.Q., Wang J.X. Post-translational modification of Parkin and its research progress in cancer. Cancer Commun. 2019;39:77. doi: 10.1186/s40880-019-0421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Durcan T.M., Fon E.A. The three ‘P’s of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2019;29:989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozawa K., Komatsubara A.T., Nishimura Y., Sawada T., Kawafune H., Tsumoto H., Tsuji Y., Zhao J., Kyotani Y., Tanaka T., et al. S-nitrosylation regulates mitochondrial quality control via activation of parkin. Sci. Rep. 2013;3:2202. doi: 10.1038/srep02202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Truban D., Hou X., Caulfield T.R., Fiesel F.C., Springer W. PINK1, Parkin, and Mitochondrial Quality Control: What can we Learn about Parkinson’s Disease Pathobiology? J. Parkinsons Dis. 2017;7:13–29. doi: 10.3233/JPD-160989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen G.F., Toth R., James J., Ganley I.G. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer H., Weihl C.C. The VCP/p97 system at a glance: Connecting cellular function to disease pathogenesis. Pt 18J. Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer H.H. Golgi reassembly after mitosis: The AAA family meets the ubiquitin family. Biochim. Biophys. Acta. 2005;1744:108–119. doi: 10.1016/j.bbamcr.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 25.Olzmann J.A., Richter C.M., Kopito R.R. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. USA. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritz D., Vuk M., Kirchner P., Bug M., Schutz S., Hayer A., Bremer S., Lusk C., Baloh R.H., Lee H., et al. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat. Cell Biol. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bug M., Meyer H. Expanding into new markets—VCP/p97 in endocytosis and autophagy. J. Struct. Biol. 2012;179:78–82. doi: 10.1016/j.jsb.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Bayraktar O., Oral O., Kocaturk N.M., Akkoc Y., Eberhart K., Kosar A., Gozuacik D. IBMPFD Disease-Causing Mutant VCP/p97 Proteins Are Targets of Autophagic-Lysosomal Degradation. PLoS ONE. 2016;11:e0164864. doi: 10.1371/journal.pone.0164864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X., Sun X., Hu D., Wang Y.J., Fujioka H., Vyas R., Chakrapani S., Joshi A.U., Luo Y., Mochly-Rosen D., et al. VCP recruitment to mitochondria causes mitophagy impairment and neurodegeneration in models of Huntington’s disease. Nat. Commun. 2016;7:12646. doi: 10.1038/ncomms12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z.H., Clark C., Geisbrecht E.R. Drosophila clueless is involved in Parkin-dependent mitophagy by promoting VCP-mediated Marf degradation. Hum. Mol. Genet. 2016;25:1946–1964. doi: 10.1093/hmg/ddw067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiekerkoetter U., Sun B., Khuchua Z., Bennett M.J., Strauss A.W. Molecular and phenotypic heterogeneity in mitochondrial trifunctional protein deficiency due to beta-subunit mutations. Hum. Mutat. 2003;21:598–607. doi: 10.1002/humu.10211. [DOI] [PubMed] [Google Scholar]
- 32.Maeyashiki C., Oshima S., Otsubo K., Kobayashi M., Nibe Y., Matsuzawa Y., Onizawa M., Nemoto Y., Nagaishi T., Okamoto R., et al. HADHA, the alpha subunit of the mitochondrial trifunctional protein, is involved in long-chain fatty acid-induced autophagy in intestinal epithelial cells. Biochem. Biophys. Res. Commun. 2017;484:636–641. doi: 10.1016/j.bbrc.2017.01.159. [DOI] [PubMed] [Google Scholar]
- 33.Artal-Sanz M., Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol. Metab. 2009;20:394–401. doi: 10.1016/j.tem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Merkwirth C., Langer T. Prohibitin function within mitochondria: Essential roles for cell proliferation and cristae morphogenesis. Biochim. Biophys. Acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Kathiria A.S., Butcher L.D., Feagins L.A., Souza R.F., Boland C.R., Theiss A.L. Prohibitin 1 modulates mitochondrial stress-related autophagy in human colonic epithelial cells. PLoS ONE. 2012;7:e31231. doi: 10.1371/journal.pone.0031231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Triplett J.C., Zhang Z., Sultana R., Cai J., Klein J.B., Bueler H., Butterfield D.A. Quantitative expression proteomics and phosphoproteomics profile of brain from PINK1 knockout mice: Insights into mechanisms of familial Parkinson’s disease. J. Neurochem. 2015;133:750–765. doi: 10.1111/jnc.13039. [DOI] [PubMed] [Google Scholar]
- 37.Wei Y., Chiang W.C., Sumpter R., Jr., Mishra P., Levine B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017;168:224–238.e10. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan C., Gong L., Chen L., Xu M., Abou-Hamdan H., Tang M., Desaubry L., Song Z. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy. 2020;16:419–434. doi: 10.1080/15548627.2019.1628520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Wang Y., Xiang Y., Lee W. Prohibitins are involved in protease-activated receptor 1-mediated platelet aggregation. J. Thromb. Haemost. 2012;10:411–418. doi: 10.1111/j.1538-7836.2011.04607.x. [DOI] [PubMed] [Google Scholar]
- 40.Andrews R.K., Harris S.J., McNally T., Berndt M.C. Binding of purified 14-3-3 zeta signaling protein to discrete amino acid sequences within the cytoplasmic domain of the platelet membrane glycoprotein Ib-IX-V complex. Biochemistry. 1998;37:638–647. doi: 10.1021/bi970893g. [DOI] [PubMed] [Google Scholar]
- 41.Dai K., Bodnar R., Berndt M.C., Du X. A critical role for 14-3-3zeta protein in regulating the VWF binding function of platelet glycoprotein Ib-IX and its therapeutic implications. Blood. 2005;106:1975–1981. doi: 10.1182/blood-2005-01-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrison D. 14-3-3: Modulators of signaling proteins? Science. 1994;266:56–57. doi: 10.1126/science.7939645. [DOI] [PubMed] [Google Scholar]
- 43.MacKintosh C., Meek S.E. Regulation of plant NR activity by reversible phosphorylation, 14-3-3 proteins and proteolysis. Cell. Mol. Life Sci. 2001;58:205–214. doi: 10.1007/PL00000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleppe R., Martinez A., Doskeland S.O., Haavik J. The 14-3-3 proteins in regulation of cellular metabolism. Semin. Cell Dev. Biol. 2011;22:713–719. doi: 10.1016/j.semcdb.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Sata S. Regulation of parkin and Parkinson’s disease. Tanpakushitsu Kakusan Koso. 2006;51(Suppl. 10):1418–1422. [PubMed] [Google Scholar]
- 46.Sato S., Chiba T., Sakata E., Kato K., Mizuno Y., Hattori N., Tanaka K. 14-3-3eta is a novel regulator of parkin ubiquitin ligase. EMBO J. 2006;25:211–221. doi: 10.1038/sj.emboj.7600774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoenwaelder S.M., Darbousset R., Cranmer S.L., Ramshaw H.S., Orive S.L., Sturgeon S., Yuan Y., Yao Y., Krycer J.R., Woodcock J., et al. 14-3-3zeta regulates the mitochondrial respiratory reserve linked to platelet phosphatidylserine exposure and procoagulant function. Nat. Commun. 2016;7:12862. doi: 10.1038/ncomms12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du X., Harris S.J., Tetaz T.J., Ginsberg M.H., Berndt M.C. Association of a phospholipase A2 (14-3-3 protein) with the platelet glycoprotein Ib-IX complex. J. Biol. Chem. 1994;269:18287–18290. [PubMed] [Google Scholar]
- 49.Mangin P., David T., Lavaud V., Cranmer S.L., Pikovski I., Jackson S.P., Berndt M.C., Cazenave J.P., Gachet C., Lanza F. Identification of a novel 14-3-3zeta binding site within the cytoplasmic tail of platelet glycoprotein Ibalpha. Blood. 2004;104:420–427. doi: 10.1182/blood-2003-08-2881. [DOI] [PubMed] [Google Scholar]
- 50.Gao J., Huang M., Lai J., Mao K., Sun P., Cao Z., Hu Y., Zhang Y., Schulte M.L., Jin C., et al. Kindlin supports platelet integrin alphaIIbbeta3 activation by interacting with paxillin. J. Cell Sci. 2017;130:3764–3775. doi: 10.1242/jcs.205641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dowling J.J., Gibbs E., Russell M., Goldman D., Minarcik J., Golden J.A., Feldman E.L. Kindlin-2 is an essential component of intercalated discs and is required for vertebrate cardiac structure and function. Circ. Res. 2008;102:423–431. doi: 10.1161/CIRCRESAHA.107.161489. [DOI] [PubMed] [Google Scholar]
- 52.Dowling J.J., Vreede A.P., Kim S., Golden J., Feldman E.L. Kindlin-2 is required for myocyte elongation and is essential for myogenesis. BMC Cell Biol. 2008;9:36. doi: 10.1186/1471-2121-9-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montanez E., Ussar S., Schifferer M., Bosl M., Zent R., Moser M., Fassler R. Kindlin-2 controls bidirectional signaling of integrins. Genes Dev. 2008;22:1325–1330. doi: 10.1101/gad.469408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabnis H., Kirpalani A., Horan J., McDowall A., Svensson L., Cooley A., Merck T., Jobe S., Hogg N., Briones M. Leukocyte adhesion deficiency-III in an African-American patient. Pediatr. Blood Cancer. 2010;55:180–182. doi: 10.1002/pbc.22386. [DOI] [PubMed] [Google Scholar]
- 55.Robert P., Canault M., Farnarier C., Nurden A., Grosdidier C., Barlogis V., Bongrand P., Pierres A., Chambost H., Alessi M.C. A novel leukocyte adhesion deficiency III variant: Kindlin-3 deficiency results in integrin- and nonintegrin-related defects in different steps of leukocyte adhesion. J. Immunol. 2011;186:5273–5283. doi: 10.4049/jimmunol.1003141. [DOI] [PubMed] [Google Scholar]
- 56.Morrison V.L., MacPherson M., Savinko T., Lek H.S., Prescott A., Fagerholm S.C. The beta2 integrin-kindlin-3 interaction is essential for T-cell homing but dispensable for T-cell activation in vivo. Blood. 2013;122:1428–1436. doi: 10.1182/blood-2013-02-484998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Essex D.W., Wu Y. Multiple protein disulfide isomerases support thrombosis. Curr. Opin. Hematol. 2018;25:395–402. doi: 10.1097/MOH.0000000000000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Essex D.W., Li M. Protein disulphide isomerase mediates platelet aggregation and secretion. Br. J. Haematol. 1999;104:448–454. doi: 10.1046/j.1365-2141.1999.01197.x. [DOI] [PubMed] [Google Scholar]
- 59.Manickam N., Sun X., Li M., Gazitt Y., Essex D.W. Protein disulphide isomerase in platelet function. Br. J. Haematol. 2008;140:223–229. doi: 10.1111/j.1365-2141.2007.06898.x. [DOI] [PubMed] [Google Scholar]
- 60.Tucker K.L., Sage T., Stevens J.M., Jordan P.A., Jones S., Barrett N.E., St-Arnaud R., Frampton J., Dedhar S., Gibbins J.M. A dual role for integrin-linked kinase in platelets: Regulating integrin function and alpha-granule secretion. Blood. 2008;112:4523–4531. doi: 10.1182/blood-2008-03-148502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esfandiarei M., Yazdi S.A., Gray V., Dedhar S., van Breemen C. Integrin-linked kinase functions as a downstream signal of platelet-derived growth factor to regulate actin polymerization and vascular smooth muscle cell migration. BMC Cell Biol. 2010;11:16. doi: 10.1186/1471-2121-11-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones C.I., Tucker K.L., Sasikumar P., Sage T., Kaiser W.J., Moore C., Emerson M., Gibbins J.M. Integrin-linked kinase regulates the rate of platelet activation and is essential for the formation of stable thrombi. J. Thromb. Haemost. 2013;12:1342–1352. doi: 10.1111/jth.12620. [DOI] [PubMed] [Google Scholar]
- 63.Brodehl A., Rezazadeh S., Williams T., Munsie N.M., Liedtke D., Oh T., Ferrier R., Shen Y., Jones S.J.M., Stiegler A.L., et al. Mutations in ILK, encoding integrin-linked kinase, are associated with arrhythmogenic cardiomyopathy. Transl. Res. 2020;208:15–29. doi: 10.1016/j.trsl.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meng F., Yao D., Shi Y., Kabakoff J., Wu W., Reicher J., Ma Y., Moosmann B., Masliah E., Lipton S.A., et al. Oxidation of the cysteine-rich regions of parkin perturbs its E3 ligase activity and contributes to protein aggregation. Mol. Neurodegener. 2011;6:34. doi: 10.1186/1750-1326-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K.Z.Q., Steer E., Otero P.A., Bateman N.W., Cheng M.H., Scott A.L., Wu C., Bahar I., Shih Y.T., Hsueh Y.P., et al. PINK1 Interacts with VCP/p97 and Activates PKA to Promote NSFL1C/p47 Phosphorylation and Dendritic Arborization in Neurons. eNeuro. 2019;5 doi: 10.1523/ENEURO.0466-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saxena S.P., Brandes L.J., Becker A.B., Simons K.J., LaBella F.S., Gerrard J.M. Histamine is an intracellular messenger mediating platelet aggregation. Science. 1989;243:1596–1599. doi: 10.1126/science.2928797. [DOI] [PubMed] [Google Scholar]
- 67.Kravic B., Behrends C., Meyer H. Regulation of lysosome integrity and lysophagy by the ubiquitin-conjugating enzyme UBE2QL1. Autophagy. 2020;16:179–180. doi: 10.1080/15548627.2019.1687217. [DOI] [PMC free article] [PubMed] [Google Scholar]