Abstract
Neoglycoproteins, such as BSA-glycosides, contain carbohydrates covalently attached to a protein carrier via nonnaturally occurring linkages. These conjugates have been used for decades to study carbohydrate–protein interactions and are frequently used as immunogens to raise antibodies to carbohydrate antigens. In fact, neoglycoproteins have been used extensively as vaccine antigens and several have obtained FDA approval. More recently, neoglycoproteins have been used in the construction of glycan arrays to produce “neoglycoprotein microarrays.” In this chapter, two methods for preparing neoglycoproteins are described along with methods to immobilize these conjugates on epoxide-coated glass microscope slides to produce arrays.
Keywords: BSA-glycosides, Neoglycoprotein, Glycan array, Lectin inhibitors, Cancer biomarker
1. Introduction
Carbohydrate–protein interactions play fundamental roles in many biological processes, such as fertilization, inflammation, viral and bacterial infection, and cancer metastasis (1). Therefore, there have been significant efforts to identify and characterize carbohydrate–protein interactions and to develop agonists/antagonists of these interactions. For these objectives, neoglycoproteins have been extremely useful (2, 3). Neoglycoproteins contain one or more copies of a carbohydrate covalently attached to a carrier protein, such as albumin. They are distinct from natural glycoproteins in that the linkage between the carbohydrate and protein is nonnatural. More importantly, neoglycoproteins are normally much more homogeneous than natural glycoproteins, which are usually produced as complex mixtures of glycoforms having different glycan chains attached to the protein core and/or have varying degrees of glycosylation site occupancy. Neoglycoproteins can be used in many common assays developed for proteins/glycoproteins, such as ELISAs and Western blots. They have also been used extensively as multivalent inhibitors of carbohydrate–protein interactions and as vaccine antigens.
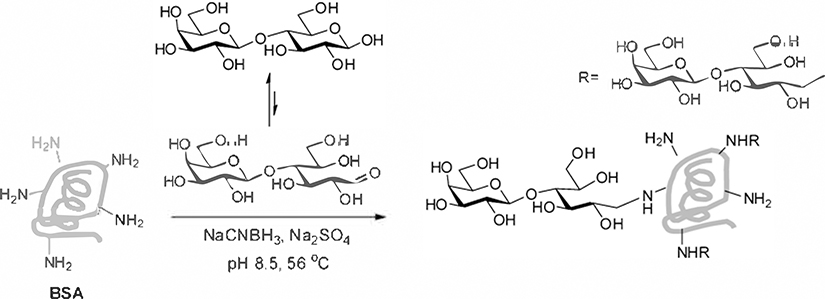
Many methods have been developed for conjugating carbohydrates to proteins to produce neoglycoproteins. Polysaccharides and oligosaccharides that contain a free reducing end can be coupled directly to lysine residues via reductive amination (see Fig. 1) (4, 5). The process results in ring opening of the reducing-end monosaccharide residue, but this is often acceptable, especially for longer glycan chains. Alternatively, carbohydrates can be synthesized or derivatized with a linker to facilitate conjugation. For example, linkers containing a free carboxylic acid can be coupled to free amines on proteins via formation of an activated ester, such as an N-hydroxysuccinimide ester (see Fig. 2) (6) or p-nitrophenyl esters (7). Linkers containing a free thiol can be conjugated to maleimide-derivatized proteins or dehydroalanine-containing proteins (8). Other conjugation methods include copper(I)-catalyzed azide-alkyne cycloaddition (9) and conjugation of two amines with a squaric decyl ester ( 10 ). Typically, the resulting product contains a distribution of conjugates, with varying attachment sites and conjugation ratios. For example, coupling to lysines might produce a product with an average conjugation ratio of 20 carbohydrates per molecule of protein. However, there is a significant proportion of conjugates with 18, 19, 21, or 22 carbohydrates per molecule of protein, and the exact lysines that are modified vary from one molecule of protein to another. Nonetheless, they each have only one type of carbohydrate structure attached. Methods for site-specific attachment of glycan chains to proteins have also been developed (11–14).
Fig. 1.
Coupling via reductive amination.
Fig. 2.
Coupling via EDC activation. (a) Examples of sugar acids used for coupling to albumin and (b) reaction sequence for coupling.
Glycan arrays have recently been developed as high-throughput tools for studying carbohydrate–protein interactions (15–17). Glycan arrays contain many different carbohydrates immobilized on a solid support in a spatially defined arrangement. They can be constructed in many ways, such as immobilization of linker-modified carbohydrates, lipid-modified carbohydrates, and neoglycoproteins (15–18). Our group has focused on printing neoglycoproteins on epoxide-coated glass microscope slides to produce neoglycoprotein microarrays (6, 19, 20). Since many neoglycoproteins are commercially available and others are readily prepared from oligosaccharides, this approach provides a convenient method to produce glycan arrays. In addition, the neoglycoproteins used on the array surface can be used in solution as agonists/antagonists of carbohydrate–protein interactions and can be used in many other assays, enabling one to readily transition from array results to other applications.
In this chapter, we describe two methods for preparing and characterizing neoglycoproteins. The first method involves reductive amination of oligosaccharides containing a free reducing end. The second involves activation of glycans that contain a linker with a free carboxylic acid. In addition, we describe methods to immobilize neoglycoproteins on epoxide-coated glass microscope slides to prepare glycan arrays, along with quality control checks to verify effective printing.
2. Materials
2.1. Solutions for Reductive Amination Method
BSA solution (10×): 150 mg/mL bovine serum albumin (BSA, essentially protease free, ≥98%, Sigma, St. Louis, MO).
Borate buffer (10×): 400 mM sodium tetraborate (Sigma), adjusted with 1 M NaOH to pH 8.5. Store at room temperature. If a precipitate forms, heat at 70°C until the solution is clear.
Sodium sulfate solution (6×): Approximately 3 M sodium sulfate (Sigma) was prepared at a temperature of 50 °C to produce a saturated solution. Solutions are stored at room temperature but are reheated to 50 ° C prior to use.
Oligosaccharide solutions: 20 mM oligosaccharides in water. Solutions were stored frozen at −20°C until used.
Sodium cyanoborohydride solution (10×): 3 M sodium cyanoborohydride (Sigma). Prepare a fresh solution prior to reactions.
Dialysis buffer: 6 mM NaCl (Sigma) solution.
Dialysis units (MWCO 10 000, Slide-A-Lyzer MINI Dialysis Unit, Pierce, Rockford, IL).
Applied Biosystem Voyager-DE Pro MALDI-time of flight (TOF) mass spectrometry (International Equipment Trading Ltd., Vernon Hills, IL) for evaluating the content of glycans on BSA.
MALDI matrix solution: Sinapinic acid (3,5-dimethoxy-4-hydroxycinnamic acid, Sigma) as matrix for all experiments. Matrix was prepared as a saturated solution in 50% acetonitrile/H2O (0.1% TFA) (v/v).
2.2. Solutions for Active Ester Method
1-Ethyl-3-(3-dimethyllaminopropyl)carbodiimide hydrochloride (EDC; Anaspec, Fremont, CA) was dissolved in water and then diluted with an equal volume of DMF to give a final concentration of 300 mM. Prepare a fresh solution prior to reactions.
300 mM N-Hydroxysuccinimide (NHS; Aldrich, St. Louis, MO). Prepare a fresh solution prior to reactions.
4 mg/mL BSA in sodium borate buffer (10 mM sodium borate, 90 mM NaCl, pH 8.0).
Glycan solutions: Glycans containing a linker with a free carboxylic acid (sugar acids) were dissolved in water at a final concentration of 150 mM. Solutions were stored frozen at −20 °C until used. Sugar acids are typically obtained by chemical synthesis. For example, glycopeptides that contain a free C-terminal carboxylic acid can be prepared via solid-phase peptide synthesis, deprotection, and purification.
2.3. Carbohydrate Microarray Fabrication
Printing buffer: l× PBS, 2.5% glycerol, 0.006% Triton-X 100.
PBS buffer (20×): 2.74 M NaCl, 60 mM KCl, 200 mM Na2HPO4, 36 mM KH2PO4, pH 7.4. Store at room temperature.
Fluorophore-labeled BSA (Invitrogen Corporation, Carlsbad, CA) and BSA-glycosides were diluted with printing buffer to a concentration of 125 μg/mL.
Arraylt SuperEpoxy 2 glass slides (TeleChem International, Inc., Sunnyvale, CA).
PBST0.05 washing buffer: 1× PBS, 0.05% tween 20.
Robotic microarrayer: MicroGrid II 600/610 (Genomic Solutions, BioRobotics, Ann Arbor, MI).
Arraylt stealth pins: SMP2 pins (uptake volume 0.25 μL, delivery volume 0.5 nL) were purchased from TeleChem International, Inc. (Sunnyvale, CA).
Sample plates: 384-well V-bottom sample plates (Genetix, San Jose, CA).
16-well slide modules (Grace Bio-Labs, Bend, OR).
Genepix 4000A microarray scanner (Molecular Devices Corporation, Union City, CA).
3. Methods
3.1. Preparation of BSA-Glycosides by Reductive Amination Method
Mix BSA (2 μL, 150 mg/mL) with sodium borate (5.5 μL, 400 mM, pH 8.5), sodium sulfate (3.7 μL, saturated at 50 °C), oligosaccharide (3.3 μL, 20 mM solution for 15 equiv), and water (5.3 μL) in a 200-μL PCR tube. Add sodium cyanoborohydride (2.2 μL, 3M) and cap the tube (see Notes 1 and 2).
Incubate the solution in a PCR thermal cycler at 56°C for 96 h with a heated lid (see Note 3).
Dilute the reaction with H2O (100 μL), transfer to a 500-μL dialysis tube, and dialyze against H2O (2.5 L) containing 6 mM NaCl three times.
Dilute the dialyzed BSA conjugates to a final concentration of 1.0 mg/mL and store at −20 ° C.
3.2. Preparation of BSA-Glycosides by Active Ester Method
Mix sugar acid, NHS (300 mM), and then EDC (300 mM) in an Eppendorf tube at a volume ratio of 2:1:1 and incubate at room temperature with occasional gentle mixing to form the NHS ester in situ (see Note 4).
After 1 h, add the solution containing the NHS ester to a solution of BSA (4 mg/mL in sodium borate buffer, pH 8.0). Precool the solution to 4°C.
After 15 min, warm the solution to room temperature and incubate for another 1 h.
Dialyze BSA-conjugates against H2O (2.5 L) containing 6 mM NaCl three times using dialysis units.
3.3. MALDI MS Analysis of BSA-Glycoside
BSA conjugates are analyzed by MALDI-TOF MS and the extent of conjugation is determined by subtracting the average mass of BSA (66431) from the average mass of the conjugate and then dividing by the molecular weight of the sugar acid minus 18 (see Notes 5 and 6).
The instrument is operated in linear mode under positive condition with the accelerating voltage of 25 kV, guide wire 0.15%, and grid voltage 91.5%.
A nitrogen laser should be used at 337 nm with 250 laser shots averaged per spectrum.
Sample preparation is a modified “dried droplet” procedure, whereby 0.3 μL of samples are spotted on a ^MALDI sample plate followed by O.3 μL of matrix. The mixture is then allowed to air dry prior to analysis.
Data analysis is carried out using Data Explorer software resident on Voyager mass spectrometer and used BSA as a standard for external calibration.
3.4. Carbohydrate Microarray Fabrication
Transfer 15 μL of fluorophore-labeled BSA and neoglycoprotein solution (125 μg/mL) to wells of a 384-well V-bottom sample plates (see Notes 7 and 8).
Print carbohydrate microarrays on epoxide-modified glass slides using a robotic microarrayer fitted with SMP2 Microspotting Pins (see Note 9).
The humidity level should be maintained at −50% with an ultrasonic humidifier.
Each BSA-glycoside component should be printed in duplicate (see Figs. 3 and 4). The diameter of each spot is around 90 μm and the pitch is 200 μm (see Note 10).
Printed slides are allowed to dry at room temperature for an additional few hours (keeping the total exposure time less than 36 h) and then stored at −20 °C until use (see Notes 11 and 12).
Fig. 3.
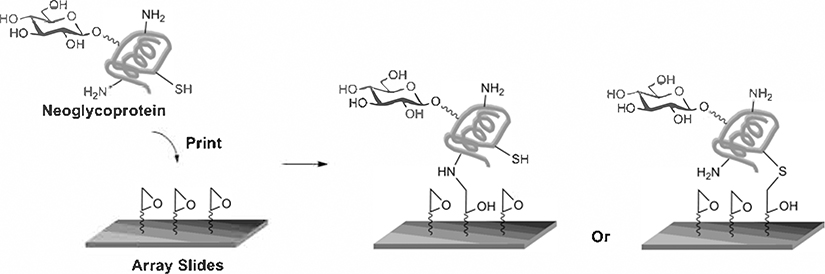
Microarray printing. Hollow pins are dipped into wells of a 384-well plate containing print solutions. The filled pins are then moved to the slides and lightly tapped on the glass to print array spots.
Fig. 4.
Immobilization of neoglycoproteins on epoxide-coated slides.
3.5. Quality Assessment of Microarray Slides
Most slides have a complete array grid. Toward the end of a print run, pins may occasionally run out of liquid resulting in missing spots. In addition, pins may occasionally get clogged, resulting in missing spots. Tolerance for missing spots depends on the particular application. For highest quality data, only slides containing a complete grid should be used. For less-critical experiments, such as optimizing reaction parameters, missing spots are acceptable. Below, we describe two procedures for quality assessment of microarray slides.
3.5.1. Physical Inspection of Printed Arrays
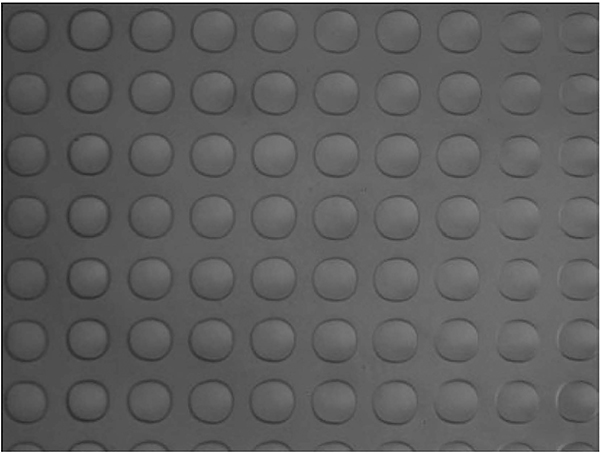
Inspect each printed slide under a microscope at 10× magnification (see Fig. 5).
Note the slide, block, column, and row number for any missing spots.
Evaporation can complicate the analysis. Inspect slides immediately after printing.
Fig. 5.
View of microarray spots after printing. A magnified view of liquid spots after printing showing uniform spots with consistent spacing and no missing spots.
3.5.2. Evaluate Binding with One or More Lectins
Fit slides with 16-well slide modules.
Block slides by addition of 3% BSA/PBS (200 μL per well) and incubate at room temperature for 2.0 h.
After removing block solution, incubate slides with one or more lectins (diluted in 1% BSA/PBS; 100 μL per well) at room temperature for 2 h. Lectins are typically assayed at a concentration of 1–10 μg/mL (see Note 13).
After washing 3× with PBST0.05 (200 μL per well), incubate slides with Cy3–streptavidin (2 μg/mL in 1% BSA/PBS; 100 μL per well) at room temperature for 2 h.
After washing 3× with PBST0.05 (200 μL per well), remove slides from the well module and immerse in PBST0.05 for 15 min.
Place the slide in a 50-mL conical tube and centrifuge for 5 min at 200 ×g to dry the slide.
Scan slides using a Genepix 4000A microarray scanner at PMT voltage settings, where no saturated pixels are obtained. Image analysis is carried out with Genepix Pro 6.0 analysis software from the same company. Spots are defined as circular features with maximum diameter of 100 μm. The background-corrected median feature intensity, F532median-B532, should be used for data processing.
Binding data is compared with results from previous batches of slides and with published binding data for each lectin.
4. Notes
Reductive amination
-
1
The reaction conditions have been optimized for BSA. Other proteins may be more or less tolerant of the high salt conditions and high temperature.
-
2
Reactions occasionally result in precipitation, especially at higher temperatures.
-
3
Evaporation and condensation of liquid on the lid of the reaction tube can be a significant problem, especially for small scale reactions. The use of a PCR tube and a PCR thermal cycler with a heated lid minimizes this problem.
EDC/NHS coupling
-
4
EDC should be added last to the reaction mixture.
MALDI MS analysis
-
5
The ratio of sugar acid to protein can be modulated to produce conjugates with varying numbers of carbohydrates. For the active ester method, at a ratio of 30:1, we typically get about 15 oligosaccharides per molecule of BSA. At 7:1, the final ratio is typically around 4/BSA.
-
6
For the reductive amination method, at a starting ratio of 9:1, the product typically has about 4–5 oligosaccharides per molecule of BSA. At a ratio of 35:1, the final conjugate should have around 15 oligosaccharides per molecule of BSA.
Printing
-
7
We typically print Cy5–BSA as the first component and Cy3–BSA toward the end of the grid. These fluorophore–BSA conjugates simplify alignment of the array grid when analyzing experiments and serve as quality control checks for the print procedure. For example, carryover due to insufficient washing of pins is easily detected.
-
8
Print solutions in the source plate evaporate over time. Water should be added to compensate for evaporation. For example, when a source plate is in use for 4–5 h, we would add about 2 mL of water to each well. The exact amount depends on the total time, humidity, temperature, and arrayer.
-
9
Clean pins thoroughly to avoid carryover from one print component to the next. The optimal conditions should be determined experimentally. For our recommended print solutions and a MicroGrid II microarrayer, we recommend at least two complete wash cycles with a 5-mm submission height, a 3-s dwell time in each of two wash baths, and an 8-s drain time (where the water in the pins is removed by vacuum).
-
10
The optimal printing conditions are dependent on the type of microarrayer. For a MicroGridII fitted with SMP2 pins, our preferred conditions are listed in Tables 1 and 2.
-
11
Keep the total time for the printing process below 36 h to avoid protein degradation.
-
12
Cy5 is sensitive to degradation and photobleaching. Slides containing Cy5–BSA or Cy5-labeled proteins should be stored in the dark at −20°C.
Table 1.
Prespotting settings (to remove excess liquid on the pins)
| Parameter | Description | Recommended value |
|---|---|---|
| Number of prespots | Spotting times of a freshly loaded pin | 6 |
| Delay before spotting | Time of suspended pins above slide | 0.5 s |
| Target height | Distance of pin into contact with slide | 15 mm |
| Speed | Speed of pin into contact with slide | 4 mm/s |
| Surface height | Thickness of slide | 1 mm |
| Dwell time | Time tool is held in place at its target height | 0.250 s |
| Multiple strikes | Number of tapping for each spot | 1 |
| Pitch | Distance between the centers of adjacent spots | 0.45 mm |
Table 2.
Printing parameters
| Parameter | Description | Recommended value |
|---|---|---|
| Number of slides | High-print-quality slides | 12–15 |
| Delay before spotting | Time of suspended pins above slide | 0 s |
| Target height | Distance of pin into contact with slide | 1 mm |
| Speed | Speed of pin into contact with slide | 4 mm/s |
| Surface height | Thickness of slide | 1 mm |
| Dwell time | Time tool is held in place at its target height | 0 s |
| Pitch | Distance between the center of spots | 0.2 mm |
Quality control checks
-
13
The choice of lectin(s) for quality control assessment depends on the composition of the array. We frequently use wheat germ agglutinin (WGA), ricinus communis agglutinin (RCA120), bauhinia purpurea agglutinin (BPA), and concanavalin A (ConA).
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NCI.
References
- 1.Dwek R A. (1996) Glycobiology: Toward Understanding the Function of Sugars, Chem. Rev.96(2), 683–720. [DOI] [PubMed] [Google Scholar]
- 2.Roy R (1996) Syntheses and some applications of chemically defined multivalent glycoconjugates, Curr. Opin. Struc. Biol. 6, 692–702. [DOI] [PubMed] [Google Scholar]
- 3.Stowell CP, and Lee YC (1980) Neoglycoproteins: the preparation and application of synthetic glycoproteins, Adv. Carbohyd. Chem. Bi. 37, 225–281. [DOI] [PubMed] [Google Scholar]
- 4.Gildersleeve JC, Oyelaran O, Simpson JT, and Allred B (2008) Improved Procedure for Direct Coupling of Carbohydrates to Proteins via Reductive Amination, Bioconjugate Chem. 19(2), 1485–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clausen M, and Madsen R (2004) Synthesis of oligogalacturonates conjugated to BSA, Carbohyd. Res. 339, 2159–2169. [DOI] [PubMed] [Google Scholar]
- 6.Manimala JC, Roach TA, Li Z, and Gildersleeve JC (2006) High-throughput carbohydrate microarray analysis of 24 lectins, Angew. Chem. Int. Edit. 45, 3607–3610. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Ling C-C, and Bundle DR (2004) A New Homobifunctional p-Nitro Phenyl Ester Coupling Reagent for the Preparation of Neoglycoproteins, Org. Lett. 6, 4407–4410. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Schiller SM, and Schultz PG (2007) A biosynthetic route to dehydroalanine-containing proteins, Angew. Chem. Int. Edit. 46, 6849–6851. [DOI] [PubMed] [Google Scholar]
- 9.van Kasteren SI., Kramer HB, Jensen HH, Campbell SJ, Kirkpatrick J, Oldham NJ, Anthony DC, and Davis BG (2007) Expanding the diversity of chemical protein modification allows post-translational mimicry, Nature 446, 1105–1109. [DOI] [PubMed] [Google Scholar]
- 10.Karelin AA, Tsvetkov YE, Paulovicova L, Bystricky S, Paulovicova E, and Nifantiev NE (2009) Synthesis of a heptasaccharide fragment of the mannan from Candida guilliermondii cell wall and its conjugate with BSA, Carbohydr. Res. 344, 29–35. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Wang L, Brock A, Wong C-H, and Schultz PG (2003) A method for the generation of glycoprotein mimetics, J Am. Chem. Soc. 125, 1702–1703. [DOI] [PubMed] [Google Scholar]
- 12.van Kasteren SI, Kramer HB, Gamblin DP, and Davis BG (2007) Site-selective glycosylation of proteins: creating synthetic glycoproteins, Nat. Protoc. 2, 3185–3194. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Bhatt VS, Sun G, Wang PG, and Palmer AF (2008) Site-Selective Glycosylation of Hemoglobin on Cys β93, Bioconjugate Chem. 19, 2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, and Tolbert TJ (2010) Study of On-Resin Convergent Synthesis of N-Linked Glycopeptides Containing a Large High Mannose N-Linked Oligosaccharide, J Am. Chem. Soc. 132, 3211–3216. [DOI] [PubMed] [Google Scholar]
- 15.Bernardes GJL, Castagner B, and Seeberger PH (2009) Combined Approaches to the Synthesis and Study of Glycoproteins, ACS Chem.Biol. 4, 703–713. [DOI] [PubMed] [Google Scholar]
- 16.Liang C-H, and Wu C-Y (2009) Glycan array: a powerful tool for glycomics studies, Expert Rev. Proteomics 6, 631–645. [DOI] [PubMed] [Google Scholar]
- 17.Oyelaran O, and Gildersleeve JC (2009) Glycan arrays: recent advances and future challenges, Curr. Opin. Chem. Biol. 13, 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao J, Patwa TH, Lubman DM, and Simeone DM (2008) Protein biomarkers in cancer: natural glycoprotein microarray approaches, Curr. Opin. Mol. Ther. 10, 602–610. [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Li Q, Rodriguez LG, and Gildersleeve JC (2010) An Array-Based Method To Identify Multivalent Inhibitors, J Am. Chem. Soc. 132, 9653–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oyelaran O, Li Q, Farnsworth D, and Gildersleeve JC (2009) Microarrays with Varying Carbohydrate Density Reveal Distinct Subpopulations of Serum Antibodies, J Proteome Res. 8, 3529–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]