Abstract
Peroxynitrite is a highly reactive oxidant effecting cell signaling and cell death. Here we report a fluorescent protein probe to selectively detect peroxynitrite. A novel unnatural amino acid, thyronine (Thy), was genetically encoded in E. coli and mammalian cells by evolving an orthogonal tRNAPyl/ThyRS pair. Incorporation of Thy into the chromophore of sfGFP or cpsGFP afforded a virtually non-fluorescent reporter. Upon treatment with peroxynitrite, Thy was converted into tyrosine via O-dearylation, regenerating GFP fluorescence in a time- and concentration-dependent manner. Genetically encoded thyronine may also be valuable for other redox applications.
Keywords: Peroxynitrite, Thyronine, Fluorescence, Genetic code expansion, Unnatural amino acid
1. Introduction
Peroxynitrite (ONOO−) is formed from the reaction of nitric oxide and superoxide at a rate of around 1×1010 M−1 s−1.1 In biological systems this short-lived yet highly reactive oxidant generates radicals such as carbonate (CO3•−), nitrogen dioxide (•NO2), and hydroxyl (•OH), which initiate many oxyradical damages in several diseases.1, 2 Specifically, peroxynitrite has been found to nitrate the tyrosine of cytochrome c, exacerbating oxidative damage to mitochondria and signaling downstream cellular events in cell death.3, 4 High peroxynitrite contribute to DNA damage and cellular NAD+ and ATP depletion, resulting in necrotic cell death.5, 6 Peroxynitrite also plays essential roles in inflammation and immune responses through modification of receptors including epidermal growth factor (EGF) receptor, insulin receptor substrate 1 (IRS1), and PPARγ.7 Moreover, peroxinitrite can inhibit viral replication as well as viral RNA entry.8
Due to its biological importance, tools for studying peroxynitrite are desirable. The low steady-state concentrations in vivo (in the range of nanomolar)9 together with the short half-life (~10 ms) of peroxynitrite make it difficult to isolate or directly measure peroxynitrite in specific cells or tissues. Small molecule probes have been developed for monitoring peroxynitrite in living cells with supplied peroxynitrite donors. For instance, a near-IR reversible fluorescent probe containing an organoselenium functional group has been designed to monitor peroxynitrite under physiological conditions.10 A three channel fluorescent probe PN600 is able to differentiate peroxynitrite from other reactive oxygen and nitrogen species.11 However, these probes generally lack cell or tissue specificity. Genetic incorporation of unnatural amino acids (Uaas) into the chromophore of fluorescent proteins (FPs) can modulate the fluorescence properties,12, 13 and the mutant FP may serve as a genetically encodable fluorescent sensor.14, 15 Indeed, incorporation of p-boronate-phenylalanine (pBoPhe) at Tyr66 position of GFPUV enables detection of H2O2, which oxidizes the aryl boronate and converts pBoPhe into Tyr leading to fluorescence increase.16 Incorporation of pBoPhe into the circularly permuted GFP (cpGFP) also leads to fluorescence increase in response to H2O2 and ONOO− via the aryl boronate oxidation mechanism.17 Moreover, further mutation and selection has identified one mutant pnGFP, which responds to ONOO− dramatically more than H2O2.17
Here we report a FP-based ONOO− reporter whose sensor could directly distinguish ONOO− from H2O2 and other less reactive oxygen species using a different O-dearylation mechanism. A novel Uaa thyronine (Thy) was genetically encoded into proteins in E. coli and mammalian cells. When Thy was incorporated into the chromophore of super fold GFP (sfGFP) or cpsGFP, the resultant reporter was virtually nonfluorescent but could be turned on by peroxynitrite only in a time and concentration-dependent manner.
2. Results and Discussion
2.1. Thyronine-based O-dearylation for hROS detection
Aryloxyphenols can be O-dearylated by highly reactive oxygen species (hROS) including hydroxyl radical (•OH), ONOO−, and hypochlorous acid (HOCl), but not by the less reactive oxygen species such as H2O2, superoxide (O2•−), and singlet oxygen (1O2). This reaction has been exploited to design small molecule probes for hROS by installing oxyphenols onto fluorescein, removing of which through O-dearylation regenerates fluorescence.18, 19 As bulky fluorophores may not pass through the ribosome for genetic encoding,20 we reasoned to genetically incorporate the less bulky thyronine (Thy) into the chromophore of FP in place of the chromophore-forming Tyr; O-dearylation of Thy into Tyr may modulate FP fluorescence to afford a new mechanism for sensing hROS (Fig. 1a). Thy is the metabolite of thyroid hormones (T4, T3, and rT3) devoid of the iodine atoms, and thus should be relatively stable and nontoxic to mammalian cells.
Figure 1.
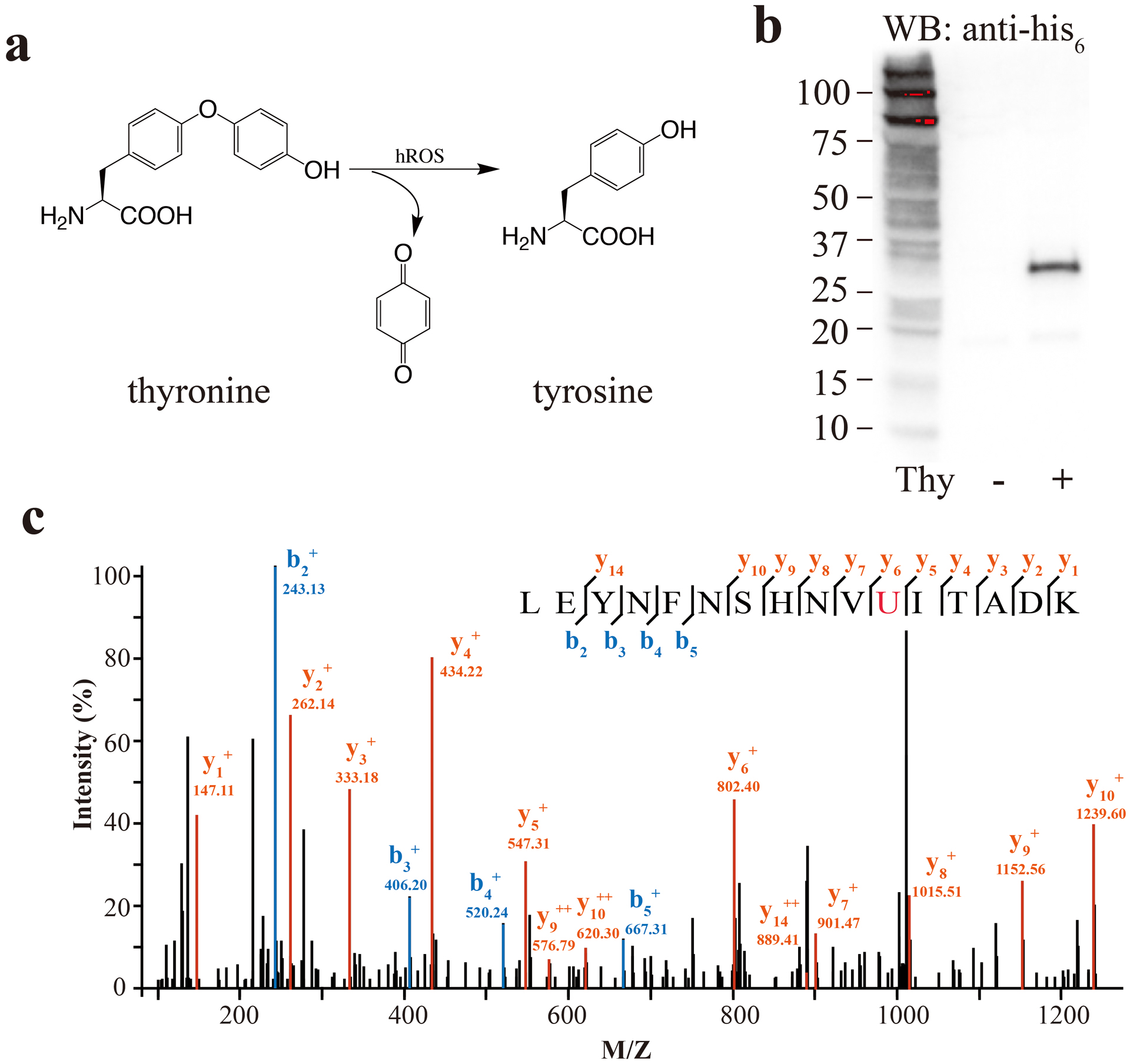
Genetically encode Thy into sfGFP in E. coli. (a) Chemical structure of Thy and its O-dearylation into Tyr upon exposure to hROS such as ONOO−. (b) Western blot analysis of Thy incorporation into sfGFP at position 151. Whole-cell lysate from the same amount of cells were separated on SDS-PAGE, and an anti-His6 antibody was used to detect the Hisx6 tag appended at the C-terminus of sfGFP. (c) Tandem MS spectrum of trypsin-digested sfGFP(151Thy). U in red represents Thy (calculated molecular weight 273.29).
To test if ONOO− could O-dearylated Thy, we incubated Thy with ONOO− and analyzed the reaction with HPLC (Fig. S1). After incubation, the peak for Thy disappeared with concomitant increase of the Tyr peak, confirming the conversion of Thy to Tyr by ONOO−.
2.2. Genetically encode Thy in E.coli
To genetically encode Thy, a Methanosarcina mazeia pyrrolysyl-tRNA synthetase (MmPylRS) mutant library was generated by mutating residues Ala302, Leu305, Tyr306, Leu309, Ile322, Asn346, Cys348, Tyr384, Val401, and Trp417 using the small-intelligent mutagenesis approach, which allows more residues to be mutated simultaneously than the conventional NNK saturation method.21 Selection was performed as described previously.22 Colonies showing Thy-dependent phenotype converged on the same PylRS mutant containing the following mutations: L309G, N346A, C348I, V401K, and W417I, which was named as ThyRS. To confirm the incorporation specificity of ThyRS, we co-expressed the sfGFP gene containing a TAG codon at position 151 with the MmtRNAPyl/ThyRS pair in E. coli. In the absence of Thy, no full-length sfGFP was detected on Western blot; when 0.5 mM Thy was added to the growth media, full-length sfGFP(151Thy) was expressed (Fig. 1b). The purified sfGFP(151Thy) was digested with trypsin and analyzed with tandem MS. The identified b and y ions confirmed the successful incorporation of Thy at the TAG codon-specified position 151 (Fig. 1c).
2.3. Genetically encode Thy in mammalian cells
To incorporate Thy into proteins in mammalian cells, we tested and optimized Thy incorporation in a HeLa cell line with the EGFP reporter stably integrated in the genome and in the HEK293 cell line with the EGFP reporter transiently transfected. The ThyRS gene was cloned into plasmid pMP-3xtRNAPyl, which contains three copies of tRNAPyl driven by a type-3 pol III promoter to increase the Uaa incorporation efficiency in mammalian cells.22–24 The resultant pMP-3xtRNAPyl-ThyRS plasmid was transfected into the HeLa-GFP(182TAG) reporter cells, in which the EGFP gene containing a TAG stop codon at position 182 is stably integrated into the genome.23 Position 182 of EGFP is a permissive site tolerating various mutations. FACS analysis of the HeLa-GFP (182TAG) reporter cells showed that strong green fluorescence was detected at either 24 h or 48 h only from cells fed with 0.5 mM Thy (Fig. 2a). Consistent with FACS analysis, fluorescence images of the HeLa-GFP (182TAG) reporter cells confirmed that GFP fluorescence was observed only in cells grown in the presence of Thy (Fig. 2b). Both experiments support that Thy was incorporated into GFP in HeLa cells. In addition, we also transiently transfected HEK293 cells with plasmid pMP-3xtRNAPyl-ThyRS and a reporter plasmid pcDNA3.1-EGFP (182TAG) at 2:1 ratio, followed by culturing with or without 0.5 mM Thy for 48 h. Again, strong EGFP fluorescence was observed only when Thy was supplied to the media (Fig. 2c). These data indicate efficient incorporation of Thy into proteins in mammalian cell lines.
Figure 2.
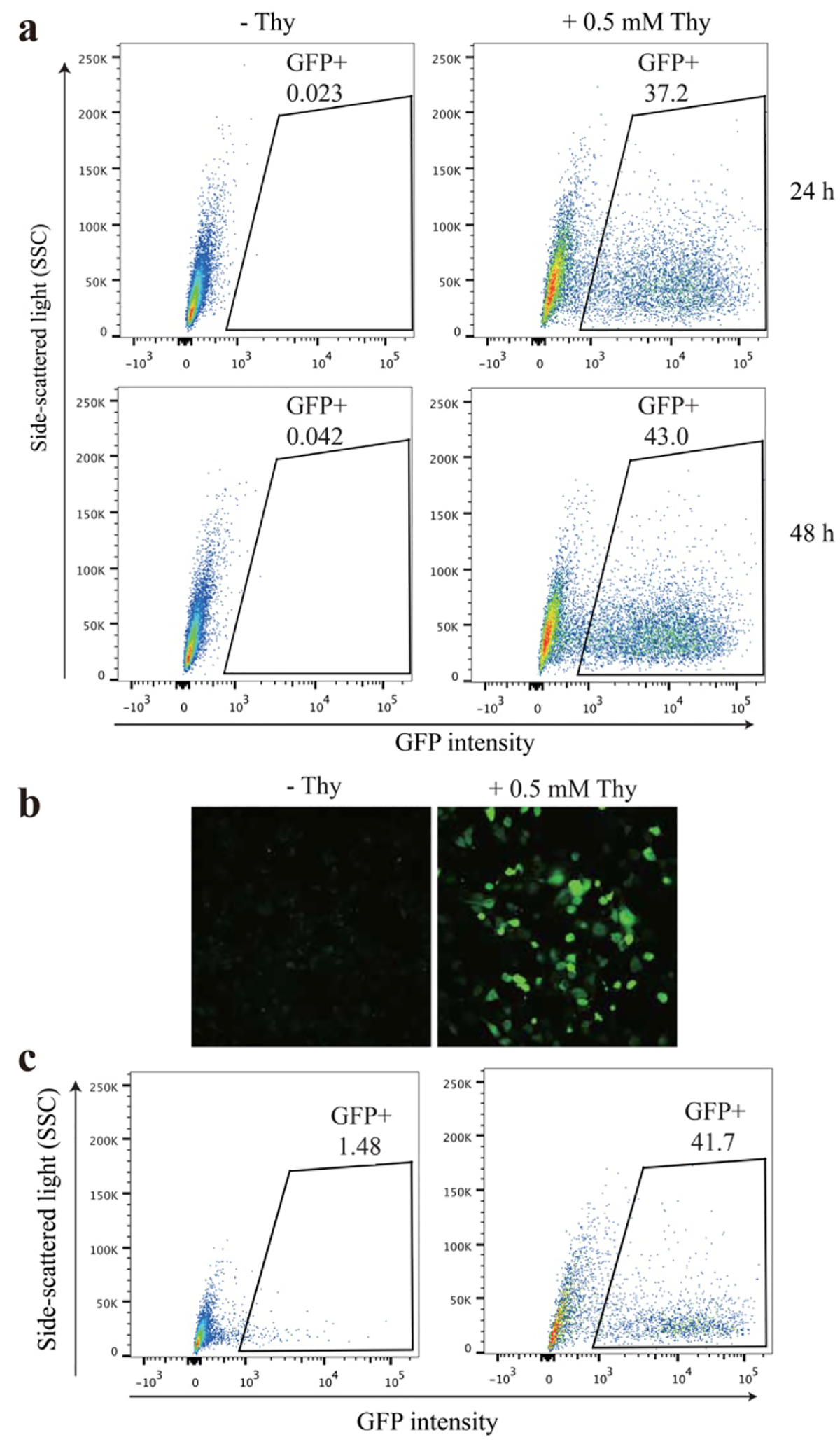
Genetic incorporation of Thy into proteins in mammalian cells. (a) FACS analysis of Thy incorporation into EGFP in the HeLa-EGFP(182TAG) stable reporter cells. HeLa-EGFP(182TAG) stable cell line was transfected with plasmid pMP-3xtRNAPyl-ThyRS, cultured with or without 0.5 mM Thy, and analyzed at indicated time. (b) Fluorescence images of HeLa-EGFP-182TAG cells that were transfected with pMP-3xtRNAPyl-ThyRS, with or without 0.5 mM Thy in the media, and incubated for 24 h. (c) FACS analysis of Thy incorporation into EGFP in HEK293 cells. HEK293 cells were cotransfected with pMP-3xtRNAPyl-ThyRS and pcDNA3.1-EGFP(182TAG), treated with or without 0.5 mM Thy, and analyzed at 48 h.
2.4. Incorporation of Thy into GFP chromophore to detect peroxynitrite
To sense hROS via Thy O-dearylation, we incorporated Thy into sfGFP at residue Tyr66, which is part of the chromophore and is known to modulate the fluorescence of sfGFP.13, 25 E. coli cells were co-transformed with plasmids pTAK-sfGFP(66TAG) and pBK-ThyRS, grown in the presence of 0.5 mM Thy, and induced overnight at 18 °C. SDS-PAGE analysis of purified proteins indicated that full-length sfGFP(66Thy) was produced, which ran at the similar position as the wildtype (WT) sfGFP protein (Fig. 3a). The purified sfGFP(66Thy) was further analyzed with electrospray ionization time-of-flight mass spectrometry (ESITOF MS) (Fig. 3b). A peak observed at 27777 Da corresponds to intact sfGFP(66Thy) with Thy incorporation and chromophore maturation (expected 27777 Da). The other peak measured at 27645 Da corresponds to the sfGFP(66Thy) with a mature chromophore but lacking the initiating Met (expected 27646 Da). These data indicated that the sfGFP(66Thy) protein was successfully produced and Thy incorporation at site 66 did not prevent chromophore formation. We then measured the fluorescence emission using the excitation wavelength of 485 nm. As expected, WT sfGFP showed strong fluorescence with emission maximum at 510 nm, whereas sfGFP (66Thy) had negligible fluorescence intensity (Fig. 3c). Therefore, Thy incorporation at position 66 resulted in a non-fluorescent sfGFP(66Thy) protein, which is desirable for detection through fluorescence intensity increase.
Figure 3.
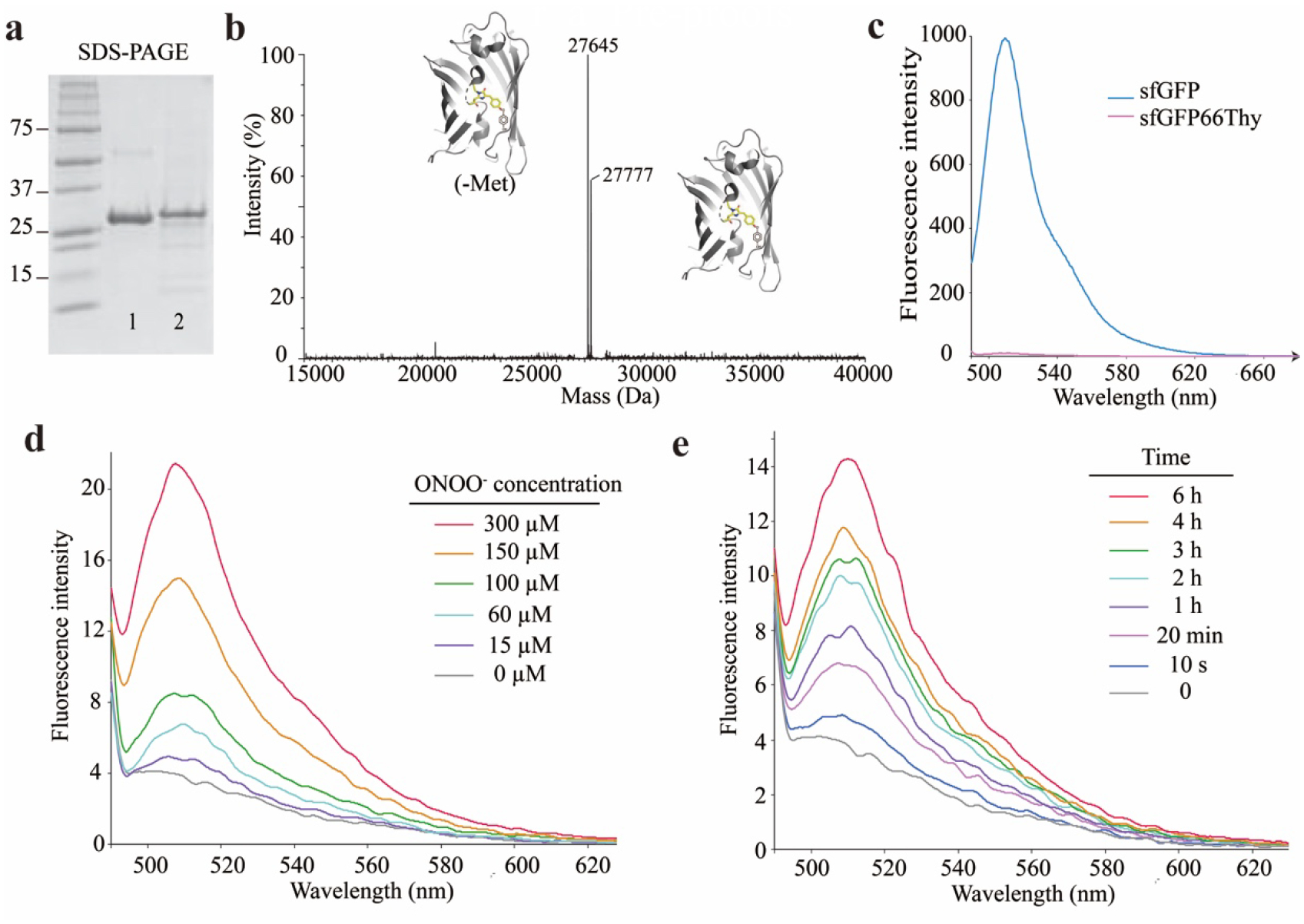
Construction of sfGFP(66Thy) reporter to detect peroxynitrite. (a) SDS-PAGE analysis of purified sfGFP and sfGFP(66Thy) protein. 1, sfGFP; 2, sfGFP(66Thy). (b) ESI-TOF MS spectrum of intact sfGFP(66Thy) protein confirmed Thy incorporation and chromophore formation. (c) Fluorescence intensity of sfGFP and sfGFP(66Thy) under the excitation wavelength of 485 nm. (d) Fluorescence emission spectra of sfGFP(66Thy) protein with the treatment of indicated concentrations of ONOO− for 6 h. Excitation wavelength = 485 nm. (e) Fluorescence emission spectra of sfGFP(66Thy) protein after treatment with 150 μM ONOO− for different time period. Excitation wavelength = 485 nm.
The non-fluorescent sfGFP(66Thy) was treated with ONOO− in a range of concentrations (0 to 300 μM), and the fluorescence emission was recorded (Fig. 3d). The fluorescence intensity increased upon ONOO− treatment in a concentration dependent manner; over 50% increase was measured with 60 μM ONOO− and four-fold increase with 300 μM ONOO−. The fluorescence intensity increase of sfGFP(66Thy) also showed a time-dependent response to ONOO− treatment from 0 to 6 h (Fig. 3e). In addition, the fluorescence emission peak of sfGFP(66Thy) after ONOO− treatment had the same wavelength as the WT sfGFP, suggesting that Thy66 was converted into Tyr as designed. Nonetheless, the fluorescence intensity of sfGFP(66Thy) after peroxynitrite treatment was relatively low in comparison to WT sfGFP even after 6 h. Since the WT sfGFP fluorophore could recover more than 95% of the starting fluorescence within 4 min through renaturation26 and ONOO− has a short lifetime in neutral aqueous solution,27 we speculated that the relatively low fluorescence intensity of this reporter was due to the inefficient conversion of Thy to Tyr. The limit of detection (LOD) was determined to be 66 μM of ONOO− under these experimental conditions. Further optimization through mutagenesis and selection may improve the performance.
The circularly permutated sfGFP (cpsGFP) has also been used to detect certain analytes.15, 17 To determine if Thy would work in the chromophore of cpsGFP, we incorporated Thy at site Tyr66 of the cpsGFP, which has the original N- and C- termini of sfGFP connected through a peptide linker and a new terminal break introduced between residue 145N and 146F.15 The resultant cpsGFP(66Thy) protein was purified and found to have extremely low fluorescence. Upon treating with ONOO−, cpsGFP(66Thy) also showed fluorescence intensity increase in a time- and ONOO− concentration-dependent manner (Fig. S2). We also treated the cpsGFP(66Thy) with H2O2 and HClO, respectively. No fluorescence change was detected for either oxidant (Fig. S3 and Fig. S4), suggesting good selectivity of the cpsGFP(66Thy) reporter for ONOO−.
3. Conclusion
We evolved an orthogonal tRNAPyl/ThyRS pair for genetic incorporation of a novel Uaa Thy into proteins in E. coli and mammalian cells. Incorporation of Thy in the chromophore of sfGFP and cpsGFP afforded fluorescent reporters to detect ONOO− in vitro via the O-dearylation mechanism. Incorporation of pBoPhe into GFP and cpsGPF has been previously reported to sense H2O2 and ONOO− via aryl boronate oxidation, and an evolved mutant pnGFP shows high specificity towards ONOO−.16,17 The Thy-based reporters were highly selective for ONOO− through the chemical mechanism, yet their detection limits need further optimization for cellular applications. Given the relatively small number of redox amino acids available, genetically encoding the redox sensitive thyronine in E. coli and mammalian cells may inspire other interesting applications.
4. Experimental
4.1. Primers used for cloning
| Primers name | Sequence |
|---|---|
| sfGFP_Spel_forward | GTTGTTACTAGTAAGGGCGAGGAGCTGTTCACCGGGGTGGTG |
| sfGFP_BlplHis_reverse | GGTGGTGCTCAGCTTAGTGATGGTGATGGTGATGCTTGTACAGCTCGTCC |
| sfGFP66TAG_forward | TGACCACCCTGACCTAgGGCGTGCAGTGCTTCAG |
| sfGFP66TAG_reverse | CTGAAGCACTGCACGCCcTAGGTCAGGGTGGTCA |
| sfGFP151TAG_forward | CAACAGCCACAACGTCTAGATCACCGCCG |
| sfGFP151TAG_reverse | CGGCGGTGATCTAGACGTTGTGGCTGTTG |
| MmThyRS_Nde1_forward | GTTGTTCATATGGATAAAAAGTCTCTGAACACTCTGATT |
| MmThyRS_Pst1_reverse | GTTGTTCTGCAGCCACCCGCCTGGTTATAGATTGGTTGAAATCCCATT |
| MmThyRS_Nco1_forward | GGTGGTCCATGGATAAAAAGTCTCTGAACACTCTGATTTCTGCGA |
| MmThyRS_Nhe1_reverse | GGTGGTGCTAGCTTATAGATTGGTTGAAATCCCATTGTAATAGGACTCGG |
| cpsGFP_F1 | GTTGTTCATATGGGCAGCAGTCCGTATAACAGCCACAAGGTCTATATCACCGCCGACAAG |
| cpsGFP_R2 | ACCGGTGCCACCGCTGCCACCATCCACCTTGTACAGCTCGTCCATGCCCAGAGTGAT |
| cpsGFP_F3 | GTGGATGGTGGCAGCGGTGGCACCGGTGTGAGTAAGGGCGAGGAGCTGTTCACCGGG |
| cpsGFP_R4 | GTTGTTGAGCTCCCAGTTGTACTCCAGCTTGTGCCCCAGGATGTTGCCGTCCTCCTT |
| cpsGFP66TAG_forward | GTGCCCTGGCCCACCCTCGTGACCACCCTGACCTAGGGCGTGCAGTGCTTCAGCC |
| cpsGFP66TAG_reverse | GGCTGAAGCACTGCACGCCCTAGGTCAGGGTGGTCACGAGGGTGGGCCAGGGCAC |
4.2. Molecular cloning
Primers were synthesized and purified by Integrated DNA Technologies (IDT). Plasmids were created through overlap PCR cloning and sequenced by GENEWIZ. pBAD-sfGFP was purchased from Addgene.
pTAK-sfGFP: primers sfGFP_Spe1_forward and sfGFP_Blp1 His_reverse were used to generate pTAK-sfGFP using pBAD-sfGFP as template. pTAK-sfGFP (151TAG): primers sfGFP151TAG_forward and sfGFP151TAG_reverse were used to generate the 151TAG amber codon using pTAK-sfGFP as template. pTAK-sfGFP (66TAG): primers sfGFP66TAG_forward and sfGFP66TAG_reverse were used to generate the 66TAG amber codon using pTAK-sfGFP as template. Amino acid sequence of sfGFP with Tyr66 highlighted in red and Tyr151 highlighted in blue is shown below.
MTSKGEELFTGVVPILVELDGDVNGHKFSVRGEGEGD ATNGKLTLKFICTTGKLPVPWPTLVTTLTYGVQCFSRYPD HMKRHDFFKSAMPEGYVQERTISFKDDGTYKTRAEVKFE GDTLVNRIELKGIDFKEDGNILGHKLEYNFNSHNVYITAD KQKNGIKANFKIRHNVEDGSVQLADHYQQNTPIGDGPVL LPDNHYLSTQSVLSKDPNEKRDHMVLLEFVTAAGITHGM DELYKHHHHHH
pTAK-cpsGFP: primers cpsGFP_F1 and cpsGFP_R2, or cpsGFP_F3 and cpsGFP_R4 were used to amplify sfGFP fragments in two separate PCR reactions using pTAK-sfGFP as template. The resulting two fragments were joined in an overlap PCR with two primers cpsGFP_F1 and cpsGFP_R4 to create the cpsGFP. pTAK-cpsGFP (66TAG): primers cpsGFP 66TAG_forward and cpsGFP66TAG_reverse were used to generate the 66TAG amber codon using pTAK-cpsGFP as template. Amino acid sequence of cpsGFP with Tyr66 highlighted in red is shown below.
MGSSPYNSHKVYITADKQKNGIKANFKIRHNVEDGSV QLADHYQQNTPIGDGPVLLPDNHYLSTQSVLSKDPNEKR DHMVLLEFVTAAGITLGMDELYKVDGGSGGTGVSKGEEL FTGVVPILVELDGDVNGHKFSVRGEGEGDATNGKLTLKFI CTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKRHDFFKS AMPEGYVQERTISFKDDGTYKTRAEVKFEGDTLVNRIELK GIDFKEDGNILGHKLEYNWELHHHHHH
pBK-ThyRS: primers MmThyRS_Nde1_forward and MmThyRS_Pst1_reverse were used to amplify the ThyRS from the identified hits. pMP-3xtRNAPyl-ThyRS: primers MmThyRS_Nco1_forward and MmThyRS_Nhe1_reverse were used to generate pMP-3xtRNAPyl-ThyRS using pMP-3xtRNAPyl-FnbYRS as template.28
4.3. Library construction and ThyRS selection
To evolve a synthetase specific for Thy, a PylRS library was constructed by mutating residues Ala302, Leu305, Tyr306, Leu309, Ile322, Asn346, Cys348, Tyr384, Val401, and Trp417 using the small-intelligent mutagenesis approach.21 The library was subjected to selection using procedures described previously.22 All hits conferring Thy-dependent phenotype contained the same amino acid mutations in the synthetase gene, converging into L309G/N346A/C348I/V401K/W417I, which was named as ThyRS.
Full amino acid sequence of ThyRS is the following: MDKKSLNTLISATGLWMSRTGTIHKIKHHEVSRSKIYIEM ACGDHLVVNNSRSSRTARALRHHKYRKTCKRCRVSDEDL NKFLTKANEDQTSVKVKVVSAPTRTKKAMPKSVARAPKP LENTEAAQAQPSGSKFSPAIPVSTQESVSVPASVSTSISSIST GATASALVKGNTNPITSMSAPVQASAPALTKSQTDRLEVL LNPKDEISLNSGKPFRELESELLSRRKKDLQQIYAEERENY LGKLEREITRFFVDRGFLEIKSPILIPLEYIERMGIDNDTELS KQIFRVDKNFCLRPMLAPNLYNYGRKLDRALPDPIKIFEIG PCYRKESDGKEHLEEFTMLAFIQMGSGCTRENLESIITDFL NHLGIDFKIVGDSCMVYGDTLDVMHGDLELSSAKVGPIPL DREWGIDKPIIGAGFGLERLLKVKHDFKNIKRAARSESYY NGISTNL
4.4. Genetic incorporation of Thy into sfGFP (66TAG), cpsGFP(66TAG), and sfGFP (151TAG) in E. coli
Plasmid pTAK-sfGFP (66TAG), pTAK-cpsGFP (66TAG), or pTAK-sfGFP (151TAG) was co-transformed with pBK-ThyRS into E. coli BL21 (DE3), and plated on LB agar plate supplemented with 50 μg/mL kanamycin and 34 μg/mL chloramphenicol. Colonies were picked up and inoculated in 100 mL 2x YT (5 g/L NaCI, 16 g/L Tryptone, 10 g/L Yeast extract). The cells were grown at 37 °C, 200 rpm until OD reaches 0.6. Then the medium was added with either 0.2 % L-arabinose only or 0.2 % L-arabinose plus 0.5 mM Thy, and the expression were carried out at 18 °C, 200 rpm for 18 h. Cells were harvested and the pellet was resuspended in 10 mL lysis buffer (50 mM Tris-HCI, 20 mM imidazole, 500 mM NaCI, pH 7.5, EDTA free protease inhibitor cocktail and 1 μg/mL DNase). The cell suspension was opened by sonication, after which the cell lysis solution was centrifuged at 25,000 g at 4 °C for 15 min. The supernatant was collected and incubated with 200 μL Ni-NTA resin. After 2 times wash, the protein was eluted with elution buffer (50 mM Tris-HCI, 300 mM imidazole, 500 mM NaCI, pH 7.5).
4.5. Genetic incorporation of Thy in mammalian cells
Plasmid pMP-3xtRNAPyl-ThyRS (2 μg) was transfected into Hela-EGFP (182TAG) reporter cells with 6 μL lipofectamine-2000 in 1 mL opti-MEM media when the cells reached 80% confluence. The media was changed to DMEM with 10% FBS after 15 h. The cells were treated with or without 0.5 mM Thy, and were cultured at 37 °C for additional 24 h or 48 h, after which the cells were harvested for fluorescence microscopy imaging and fluorescence-activated cell sorting (FACS) analysis. For HEK293 cells, 2 μg of plasmid pMP-3xtRNAPyl-ThyRS and 2 μg of pcDNA3.1-EGFP (182TAG) were co-transfected.
4.6. Mass spectrometry analysis of Thy incorporation
Mass spectrometric analyses of proteins were preformed as described previously.29 In brief, for intact protein analysis, 10 μg sfGFP (66Thy) protein was analyzed with ESI-TOF. For tandem MS analysis of digested peptide, 10 μg sfGFP (151Thy) was digested with trypsin at 37 °C overnight and terminated with 5 % (v/v) formic acid. The digested peptides were desalted with C18 StageTip, eluted, and dried with SpeedVac.
4.7. Peroxynitrite concentration measurement and fluorescence detection
Peroxynitrite was purchased from Cayman Chemical. The concentration of peroxynitrite was determined by measuring the absorption at 302 nm. The extinction coefficient of peroxynitrite solution in 0.1 M NaOH is 1670 M−1 cm−1 at 302 nm.19 sfGFP (66Thy) (5 μg) or cpsGFP(66Thy) (10 μg) was incubated with indicated concentrations of ONOO− for different time length. The fluorescence emission spectrum was measured with excitation wavelength at 485 nm on a Fluorolog-3 (Horiba). LOD was determined as 3.3(SD/S), wherein SD is the standard deviation of the response and S is the slope of the calibration curve.
Supplementary Material
Acknowledgments
The authors would like to dedicate this paper to Dr. Peter G. Schultz for being awarded the Tetrahedron Prize 2019 for Creativity. This work was supported by the National Institutes of Health (R01GM118384 to L.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplementary Figures S1–S4. DNA sequences of sfGFP(66TAG), cpsGFP(66TAG), and ThyRS.
References and notes
- 1.Radi R; Peluffo G; Alvarez MN; Naviliat M; Cayota A, Unraveling peroxynitrite formation in biological systems. Free Radic Biol Med 2001, 30 (5), 463–88. [DOI] [PubMed] [Google Scholar]
- 2.Radi R; Beckman JS; Bush KM; Freeman BA, Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem 1991, 266 (7), 4244–50. [PubMed] [Google Scholar]
- 3.Jang B; Han S, Biochemical properties of cytochrome c nitrated by peroxynitrite. Biochimie 2006, 88 (1), 53–8. [DOI] [PubMed] [Google Scholar]
- 4.Cassina AM; Hodara R; Souza JM; Thomson L; Castro L; Ischiropoulos H; Freeman BA; Radi R, Cytochrome c nitration by peroxynitrite. J Biol Chem 2000, 275 (28), 21409–15. [DOI] [PubMed] [Google Scholar]
- 5.Szabo C, Multiple pathways of peroxynitrite cytotoxicity. Toxicol Lett 2003, 140–141, 105–12. [DOI] [PubMed] [Google Scholar]
- 6.Virag L; Szabo E; Gergely P; Szabo C, Peroxynitrite-induced cytotoxicity: mechanism and opportunities for intervention. Toxicol Lett 2003, 140–141, 113–24. [DOI] [PubMed] [Google Scholar]
- 7.Szabo C; Ischiropoulos H; Radi R, Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov 2007, 6 (8), 662–80. [DOI] [PubMed] [Google Scholar]
- 8.Padalko E; Ohnishi T; Matsushita K; Sun H; Fox-Talbot K; Bao C; Baldwin WM 3rd; Lowenstein CJ, Peroxynitrite inhibition of Coxsackievirus infection by prevention of viral RNA entry. Proc Natl Acad Sci U S A 2004, 101 (32), 11731–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nalwaya N; Deen WM, Nitric oxide, oxygen, and superoxide formation and consumption in macrophage cultures. Chem Res Toxicol 2005, 18 (3), 486–93. [DOI] [PubMed] [Google Scholar]
- 10.Yu F; Li P; Li G; Zhao G; Chu T; Han K, A near-IR reversible fluorescent probe modulated by selenium for monitoring peroxynitrite and imaging in living cells. J Am Chem Soc 2011, 133 (29), 11030–3. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Q; Zhu Z; Zheng Y; Cheng J; Zhang N; Long YT; Zheng J; Qian X; Yang Y, A three-channel fluorescent probe that distinguishes peroxynitrite from hypochlorite. J Am Chem Soc 2012, 134 (45), 18479–82. [DOI] [PubMed] [Google Scholar]
- 12.Wang L; Brock A; Herberich B; Schultz PG, Expanding the genetic code of Escherichia coli. Science 2001, 292 (5516), 498–500. [DOI] [PubMed] [Google Scholar]
- 13.Wang L; Xie J; Deniz AA; Schultz PG, Unnatural amino acid mutagenesis of green fluorescent protein. J Org Chem 2003, 68 (1), 174–6. [DOI] [PubMed] [Google Scholar]
- 14.Fu C; Kobayashi T; Wang N; Hoppmann C; Yang B; Irannejad R; Wang L, Genetically encoding quinoline reverses chromophore charge and enables fluorescent protein brightening in acidic vesicles. J Am Chem Soc 2018, 140 (35), 11058–11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang X; Wang N; Cerny R; Niu W; Guo J, Fluorescent protein-based turn-on probe through a general protection-deprotection design strategy. ACS Sens 2017, 2 (7), 961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F; Niu W; Guo J; Schultz PG, Unnatural amino acid mutagenesis of fluorescent proteins. Angew Chem Int Ed Engl 2012, 51 (40), 10132–5. [DOI] [PubMed] [Google Scholar]
- 17.Chen ZJ; Ren W; Wright QE; Ai HW, Genetically encoded fluorescent probe for the selective detection of peroxynitrite. J Am Chem Soc 2013, 135 (40), 14940–3. [DOI] [PubMed] [Google Scholar]
- 18.Setsukinai K; Urano Y; Kakinuma K; Majima HJ; Nagano T, Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J Biol Chem 2003, 278 (5), 3170–5. [DOI] [PubMed] [Google Scholar]
- 19.Bai X; Huang Y; Lu M; Yang D, HKOH-1: A highly sensitive and selective fluorescent probe for detecting endogenous hydroxyl radicals in living cells. Angew Chem Int Ed Engl 2017, 56 (42), 12873–12877. [DOI] [PubMed] [Google Scholar]
- 20.Wang L; Schultz PG, Expanding the genetic code. Angew Chem Int Ed Engl 2004, 44 (1), 34–66. [DOI] [PubMed] [Google Scholar]
- 21.Lacey VK; Louie GV; Noel JP; Wang L, Expanding the library and substrate diversity of the pyrrolysyl-tRNA synthetase to incorporate unnatural amino acids containing conjugated rings. Chembiochem 2013, 14 (16), 2100–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takimoto JK; Dellas N; Noel JP; Wang L, Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem Biol 2011, 6 (7), 733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W; Takimoto JK; Louie GV; Baiga TJ; Noel JP; Lee KF; Slesinger PA; Wang L, Genetically encoding unnatural amino acids for cellular and neuronal studies. Nat Neurosci 2007, 10 (8), 1063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coin I; Perrin MH; Vale WW; Wang L, Photo-cross-linkers incorporated into G-protein-coupled receptors in mammalian cells: a ligand comparison. Angew Chem Int Ed Engl 2011, 50 (35), 8077–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heim R; Prasher DC; Tsien RY, Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A 1994, 91 (26), 12501–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pedelacq JD; Cabantous S; Tran T; Terwilliger TC; Waldo GS, Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 2006, 24 (1), 79–88. [DOI] [PubMed] [Google Scholar]
- 27.Ferrer-Sueta G; Radi R, Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chem Biol 2009, 4 (3), 161–77. [DOI] [PubMed] [Google Scholar]
- 28.Liu J; Li S; Aslam NA; Zheng F; Yang B; Cheng R; Wang N; Rozovsky S; Wang PG; Wang Q; Wang L, Genetically encoding photocaged quinone methide to multitarget protein residues covalently in vivo. J Am Chem Soc 2019, 141 (24), 9458–9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N; Yang B; Fu C; Zhu H; Zheng F; Kobayashi T; Liu J; Li S; Ma C; Wang PG; Wang Q; Wang L, Genetically encoding fluorosulfate-l-tyrosine to react with lysine, histidine, and tyrosine via sufex in proteins in vivo. J Am Chem Soc 2018, 140 (15), 4995–4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


