Abstract
Stem cell-mediated tissue repair is a promising approach in regenerative medicine. Intestinal epithelium is the most rapidly self-renewing tissue in adult mammals. Recently, using lineage tracing and molecular marker labeling, intestinal stem cells (ISCs) have been identified in Drosophila adult midgut. ISCs reside at the basement membrane and are multipotent as they produce both enterocytes and enteroendocrine cells. The adult Drosophila midgut provides an excellent in vivo model organ to study ISC behavior during aging, stress, regeneration, and infection. It has been demonstrated that Notch, Janus kinase/signal transducer and activator of transcription, epidermal growth factor receptor/mitogen-activated protein kinase, Hippo, and wingless signaling pathways regulate ISCs proliferation and differentiation. There are plenty of genetic tools and markers developed in recent years in Drosophila stem cell studies. These tools and markers are essential in the precise identification of stem cells as well as manipulation of genes in stem cell regulation. Here, we describe the details of genetic tools, markers, and immunolabeling techniques used in identification and characterization of adult midgut stem cells in Drosophila.
Keywords: Drosophila, Adult midgut, Intestinal stem cell, Genetic techniques, Immunolabeling
1. Introduction
Stem cells have been identified in many adult tissues. These unspecialized cells are involved in the maintenance of tissue homeostasis throughout the life of an organism by their ability to self-renew and continuously produce differentiated cells to replenish aged or damaged cells. Stem cells are regulated by both intrinsic and extrinsic (niche) factors. The abnormal balance between stem cell proliferation and differentiation results in organ degeneration, premature aging, and cancer (1–7). Elucidating the mechanisms, which regulate this balance, is critical in adult stem cell biology and regenerative medicine.
The intestinal epithelium is the most vigorously self-renewing tissue in adult mammals. Homeostasis in mammalian intestine epithelium is mediated by intestinal stem cells (ISCs), which are located near the base of each crypt (3, 8). It takes only 3–5 days for ISCs at the bottom of the crypt to proliferate and differentiate toward the lumen to become absorptive and secretory cells types and can replenish cell loss from villi. ISCs are intermixed with paneth cells at the bottom of crypt and shown to contain an overlapping population of Lgr5+ and Bmi1+ cells (3, 8). There are several signaling pathways known to regulate ISCs proliferation (3, 8). However, how ISCs and its progenitor cells respond to pathogens and mediate intestinal regeneration needs further investigation (3, 8–10).
Drosophila has been a very attractive model organism to study stem cell biology. Similar to mammalian intestine, the Drosophila adult midgut also see the rapid turnover by presence of approximately 1,000 ISCs (11–13). The identification of ISC in the adult Drosophila midgut makes it a very useful model organ system to study ISC physiology during aging, stress, regeneration, and infection (14–30). Drosophila ISCs are distributed evenly throughout the gut and localized basically to mature enterocytes (Fig. 1a–c). ISCs upon division produce two daughter cells, with one retaining ISC properties and the other becomes an immature daughter cell, enteroblasts (EBs). ISCs are characterized by expression of high levels of Delta, a ligand for the Notch receptor, which triggers Notch signaling in neighboring EBs (13). Escargot (esg), a transcription factor, is expressed in ISCs and EBs. Su(H)GBE-lacZ, a transcriptional reporter of Notch signaling, has been used as EBs cells marker (11–13). The EBs do not divide further and directly differentiate into absorptive enterocyte (ECs) (90%) and the secretory enteroendocrine (ee) cells (10%) (Fig. 1d). The two differentiated cell types, EC and ee cells, are more apically localized toward the lumen. The ee cells express the homeodomain transcription factor Prospero (Pros) in the nucleus. The mature ECs can be unambiguously distinguished from other cell types by their polyploid nuclei and large cell bodies as well as by expression of PDM1 (25, 31, 32). The young ECs express ferritin-1 heavy chain homologue (FerlHCH) (14). As in mammalian intestinal epithelium, the ECs and ee cells are continually migrated from the basal location toward the gut lumen to replace the lost cells on the surface of the epithelium. It has been demonstrated that all the epithelial cells in the adult midgut are generated by adult midgut progenitors (AMPs) from the larval gut (27). Further, it is shown that signaling through the epidermal growth factor receptor (EGFR)/RAS/mitogen-activated protein kinase (MAPK) pathway is necessary and limiting for AMP proliferation (27). AMPs divide asymmetrically, and Notch pathway directs its first daughter to become a PC, which acts as a niche (33). The AMP and its daughters can remain undifferentiated in response to a Dpp signal from the PC and proliferate to form AMP islands (33).
Fig. 1.
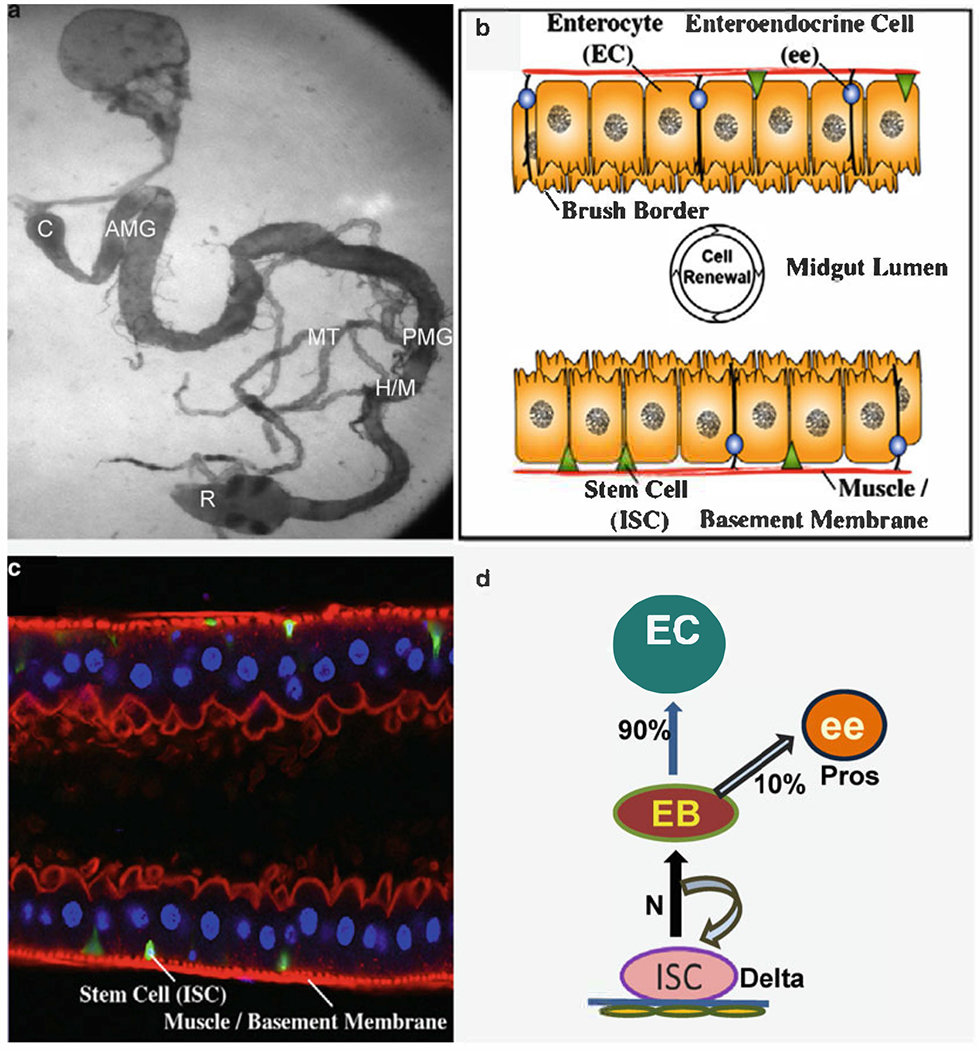
The adult Drosophila midgut is maintained by a population of multipotent intestinal stem cells (ISCs). (a) Wild-type Drosophila gastrointestinal tract visualized by phase-contrast microscopy. (b) Diagram of the adult midgut in cross section. ISCs (green) occupy a basal position in a niche adjacent to the basement membrane and the visceral muscle (red). ISCs give rise to two types of differentiated daughters, enteroendocrine (ee) cells (blue) and enterocytes (ECs, orange). (c) A cross section of the adult midgut showing ISCs marked by esgGal4, UAS-GFP (greed). ECs have large polyploid nuclei (blue, DAPI) and form a polarized cellular monolayer with an actin-rich (red, phalloidin) brush border on their luminal surface; ee cells are not marked, (d) Summary of ISCs lineage that shows that ISCs give rise to daughters (EB), which become 90% enterocytes (EC) and 10% enteroendocrine (ee) cells. C cardia; AMG anterior midgut; PMG posterior midgut; H/M hindgut/midgut junction; MT malpighian tubules; R rectum. (b, c) The figure is adapted with permission from Lee et al. (31).
Several signaling pathways and genes are known to regulate the ISCs behavior in Drosophila midgut (5, 11–46). Notch is known to promote the gut homeostasis by initiating ISC differentiation and specifying the fates of EBs (13). It has been shown that active Notch inhibits the proliferation of ISC and blocks its turnover (12, 13). Loss of Notch signaling resulted in large, tumorous mosaic clones that contain both Pros+ ee-like cells as well as Dl+ ISCs, which suggests that in normal condition, Notch signaling blocks ISC proliferation by inhibiting EBs to ee cells differentiation. It has been demonstrated that E-cad is required for interaction between ISC and EB, and adhesion with ECs (34). E-cad is necessary for Notch signaling to achieve proper cell differentiation (34). Loss of E-cad-mediated cell adhesion attenuates migration of EB and its differentiation to EC (34). It has also been shown that ISC tumor by Notch inactivation is assisted by a defect in E-cad down-regulation. The above findings suggest that Notch inhibit tumorigenesis by lowering the E-cad for proper midgut cell turnover (34). Bardin et al. (25) identified the Enhancer of split complex [E(spl)-C] genes as the key Notch targets, which is repressed by Hairless to ensure ISC maintenance. Recently, we reported that the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling promotes ISCs proliferation by activating the ISCs to go through either self-renewal or differentiation (39). Under normal conditions, this function is suppressed by Notch at least through a transcriptional repression of the JAK-STAT signaling ligand, unpaired (upd). Our work suggests that Notch, working as a differentiation signal, has a negative feedback to the ISCs activation process, which results in maintenance of stable cellular architecture of the gut epithelium, important for its proper physiological functions (39). JAK-STAT is also known to regulate ISCs proliferation (37) and/or control self-renewal and lineage differentiation of Drosophila ISCs (40).
Lin et al. (38) demonstrated that Wingless (Wg) is specifically secreted from the underlying smooth muscle cells near the basement membrane and activates the Wg signal transduction pathway in ISCs to regulate its self-renewal (38). Loss of the Wg signaling resulted in slow division of ISCs or differentiation. However, overexpression of wg resulted in ISC overproliferation. They also found that Notch is downstream of wg signaling to regulate the ISCs self-renewal and differentiation. The above study also demonstrated that circular muscle constitutes the ISC niche (38). It has been shown that loss of Adenomatous polyposis coli (APC) results in disruption of midgut homeostasis as evidenced by hyperplasia and multilayering of the midgut epithelium (31). APC is required to regulate ISC proliferation, without affecting the normal differentiation. Further, it has been found that APC hyperplasia is suppressed by reduction in Wnt signaling (31). Taken together, these findings suggest that Wg signaling is required only for ISC proliferation.
Recent studies from several group suggests that ISCs in the Drosophila midgut are activated in response to tissue stress, damage, aging, or feeding chemicals or microbial pathogens (17–42). It has been demonstrated that platelet-derived growth factor (PDGF), JNK, insulin receptor, and other signaling pathways are involved in these events (14–17). Park et al. (42) reported that p38b MAPK pathway plays a central role in the balance between ISC proliferation and differentiation in the midgut. Further, they showed that D-p38b acts downstream of PVF2/PVR signaling in this age-related ISC behavior. Furthermore, they reported that D-p38b is regulated by the DREF in ISC and EBs and that DREF is involved in the regulation of proliferation and differentiation of ISCs and EBs (42). Recently, Hochmuth et al. (43) demonstrated that Keapl and Nrf2 act as a critical redox management system to regulate ISC and intestinal homeostasis.
Several groups have demonstrated that damaged or stressed ECs produce the cytokines such as Upd, Upd2, and Upd3, which activate the JAK-STAT signaling in ISCs and EBs and promote their division and differentiation in normal and regenerating midgut (20–22, 26). JNK pathway also required for bacteria-induced stem cell proliferation (20). Hippo (Hpo) signaling has been shown to be a mediator of regenerative response in Drosophila midgut (30, 32, 44, 45). Loss of Hpo signaling or overactivation Yorki (YKi) in midgut induces regenerative response by producing cytokines of the Upd family and multiple EGFR ligands (Spitz, Keren, and Vein), which activate JAK-STAT and EGFR signaling pathways in ISCs to stimulate their proliferation (20, 25, 32, 44). Further, it has been shown that Hpo target, Yki, plays a critical and cell-autonomous role in ISCs, whose activity is suppressed by the upstream Hpo pathway members, Fat and Dachsous (Ds) (44). Moreover, Warts and Yorkie mediate a transition from low- to high-level ISC proliferation to facilitate gut regeneration (45).
Jiang et al. (28) reported that ISC proliferation induced by JAK-STAT signaling is dependent upon EGFR signaling. Their results suggest that EGFR/Ras/MAPK signaling pathway plays a central role in intestinal homeostasis. Xu et al. (5) recently reported that EGFR together with Wg and JAK-STAT signaling regulates ISC maintenance and division. They found that simultaneous disruption of all three signaling leads to rapid and complete ISC elimination, and visceral muscle is a critical component of the ISC niche. Biteau and Jasper (46) have shown that EGF receptor signaling is required to maintain the ISCs proliferation as its ligand Vein is expressed in the visceral muscle, which provides a permissive signal for ISC proliferation. They further found that AP-1 transcription factor FOS acts as a mediator for EGF and JNK to promote ISCs proliferation in response to stress. They emphasize that visceral muscle serves as a functional “niche” for ISCs (46). Amcheslavsky et al. (4) demonstrated that tuberous sclerosis complex (TSC) plays a critical role in balancing ISC growth and division as loss of TSC in the results in large ISCs formation, which show defect in cell cycle. Stem cells have also been reported to present in the other regions of the Drosophila gut and are regulated by some of the similar pathways reported in Drosophila posterior midgut (47–51).
Drosophila serves as an excellent genetic model system to study adult stem cells. There are several sophisticated techniques that have been used in lineage analysis to identify stem cells as well as to generate mosaic mutant cells in living flies. Most of the techniques used to identify the adult stem cells are based on mitotic recombination. Further, function of specific genes in the stem cells can be manipulated by using the genetic tools such as flipase/flipase recombination target (FLP/FRT) system-based tubulin-lacZ positive-labeling system, (FLP/FRT) and GAL4/UAS system-based positively marked mosaic lineage (PMML), and GAL80-based mosaic analysis with a repressible cell marker (MARCM) (Figs. 2–4). The lineage tracing is the key in the stem cell identification. It has been suggested that most dividing cells marked in the lineage tracing are not stem cells (52). In the marked clones, non-stem cells can generate transient clones, which are short-lived and produce only differentiated cells. However, stem cells clones grow larger, maintain longer, and contain undifferentiated cells, trans-amplifying cells, and terminally differentiated cells (52). Following the stem cell lineage over time is critical in understanding the stem cell potency, replacement, and aging as well as helpful in characterization of the stem cell niches (52). Clonal analysis of stem cells, molecular markers, available phenotypes, sophisticated genetic tools, and immunohistological-staining methods, together with functional genomics and proteomics approach, will be helpful in identifying the genes, and signaling pathways regulate ISCs behavior in adult midgut (53–56). In this chapter, we have presented some of the standard techniques used in the identification and characterization of ISCs in adult Drosophila. The details of the molecular markers and transgenes expressed in the different cell types in midgut are presented in Tables 1 and 2, respectively.
Fig. 2.
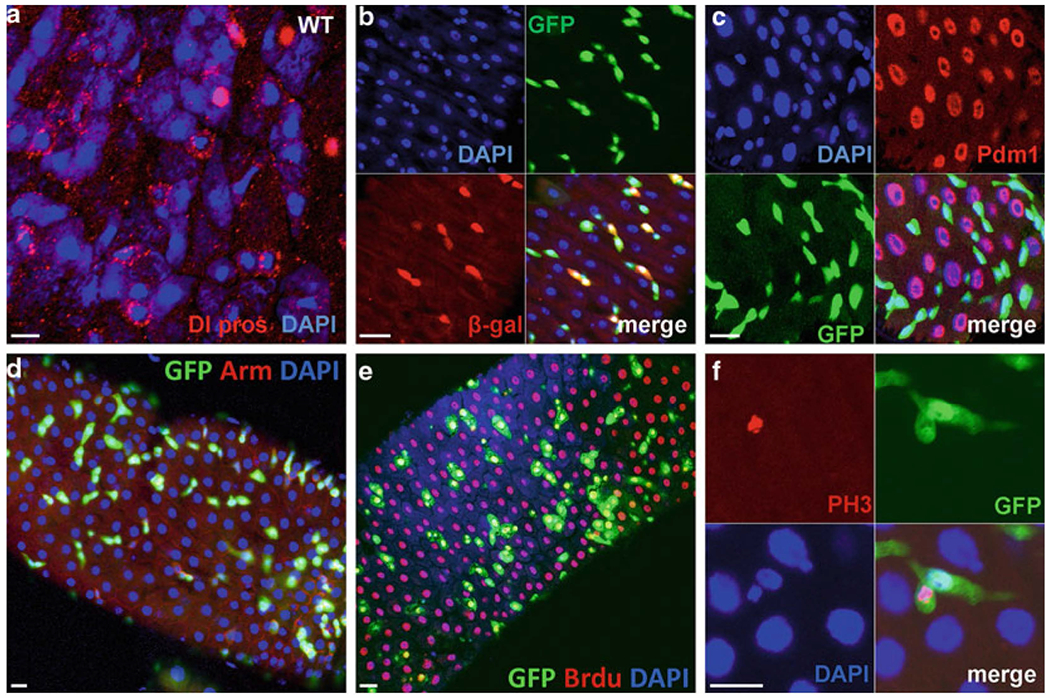
ISCs markers and proliferative cell marker expression in the midgut. (a) Wild-type gut with expression of anti-DI (red-punctate structures resembling endocytic vesicles for ISCs), anti-prospero (red-nuclear staining, ee cells); Dapi staining, blue. (b) Gut stained for 10X STAT-GFP (green-labeled the ISCs and EBs) and Su(H)GBE-LacZ (red-labeled EBs only), Dapi (blue). (c) Gut staining of esg-GAl4-UAS-GFP flies with anti-Pdm1. GFP mark the ISCs and EBs (green), Pdm1-mark the EC cells (Red), Dapi (blue). (d) kr-Gal4-UAS-GFP gut stained for GFP (green-labeled the ISCs and EBs), anti-Arm (red), and Dapi (blue). (e) BrdU pulse to detect the DNA synthesis in the gut using 10× STAT-GFP flies, anti-BrdU (red), GFP (green), Dapi (blue). (f) 10× STAT-GFP (green) gut stained with phospho-histone H3 (red) to detect the mitosis. Scale bars: 10 μm.
Fig. 4.
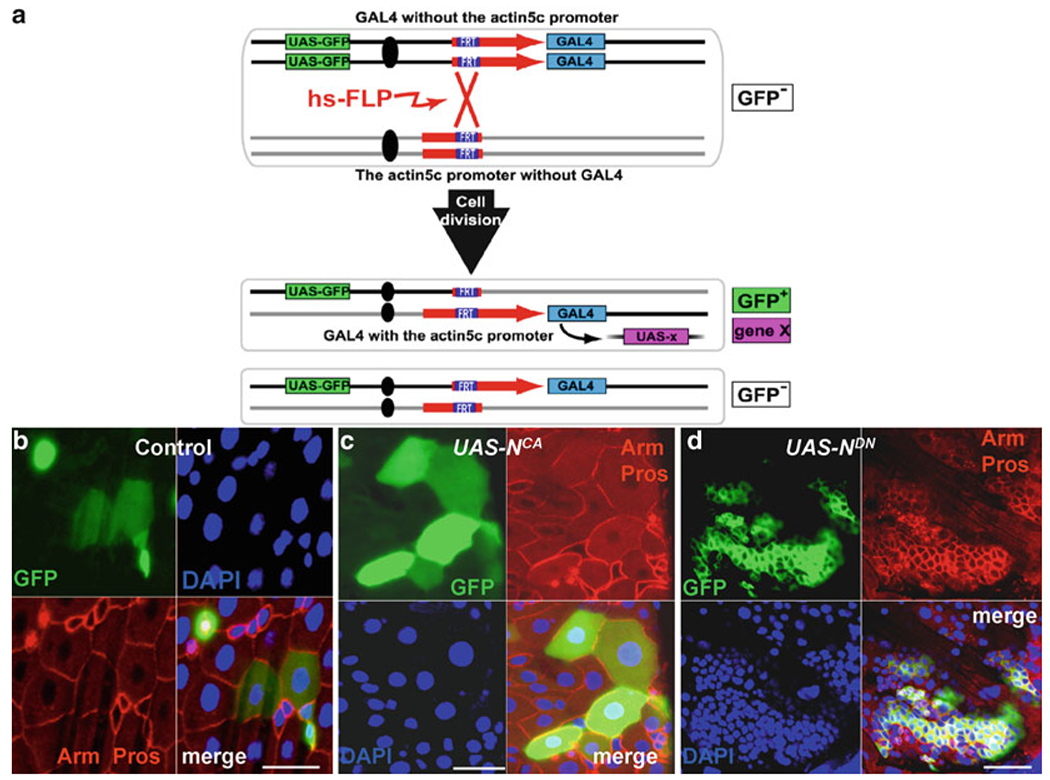
PMML labeling system to generate ISCs clones in the midgut. (a) A schematic diagram showing how to generate a functional actin5C-gal4 gene by using the FLP-mediated FRT recombination technique. A functional actin5C-gal4 gene is reconstituted by heat shock-induced FLP-mediated recombination between inactive but complimentary alleles, actin5C FRT and FRT gal4. The daughter cell that inherits the actin5C-gal4 gene expresses UAS-GFP or any other Iransgene constructs. (b) Gut with GFP-marked wild-type PMML clones. Anti-GFP (green), anti-Arm (red-diploid cell nest), anti-Pros (red-nuclei of ee cells), and Dapi (blue). (c) Gut with GFP-marked UAS-NCA PMML clones. GFP (green), Arm (red-diploid cell nest), anti-Pros (red-nuclei of ee cells), and Dapi (blue). (d) Gut with GFP-marked UAS-NDN PMML clones. Anti-GFP (green), anti-Arm (red-diploid cell nest), anti-Pros (red-nuclei of ee cells), and Dapi (blue). (a) The figure is adapted with permission from Kirilly et al. (58). Scale bars: 10 μm.
Table 1.
Antibodies used to mark Drosophila intestinal stem cell and differentiated cells
| Antibody | Cell type expression | Species | Dilution | Source | References |
|---|---|---|---|---|---|
| Notch | ISC, EB | Mouse | 1:10 | DSHB | (11) |
| Armadillo | Outer layer of ISC, EB, and EC | Mouse | 1:20 | DSHB | (11) |
| Prospero | ee cells | Mouse | 1:100 | DSHB | (13) |
| Allatostatin | ee cells | Mouse | 1:10 | DSHB | (11) |
| Tachykinin | ee cells | Rabbit | 1:2,000 | D. Nässel | (11) |
| Delta | ISC | Mouse | 1:100 | DSHB | (13) |
| Anilin | Mitotic ISC | Rabbit | 1:1,000 | Chris field | (13) |
| Alpha-tubulin | Microtubule and spindle | Rat | 1:50 | Immunologicals direct | (13) |
| Gamma-tubulin | Centrosomes | Mouse | 1:500 | Sigma | (13) |
| Daughterless | ISC, EB, ee, EC | Rabbit | 1:1,000 | Y.-N. Jan | (25) |
| Cdc2 | ISC, EB | Rabbit | 1:500 | Santa Cruz | (4) |
| AcH3 | ISC, EB | Rabbit | 1:500 | Millipore | (4) |
| Pdml | Mature EC | Rabbit | 1:100 | Xiaohang Yang | (31) |
| pH3 | Dividing ISC | Rabbit | 1:1,000 | Upstate | (12) |
| Hairless | ISC and EB, some differentiated progeny | Guinea pig | 1:500 | A. Preiss | (25) |
| EGFR | ISC and/or EB | Goat | 1:200 | Santa Cruz | (5) |
| Yki | ISC/EB | Rat Rabbit |
1:1,000 1:400 |
Helen McNeill Kenneth Irvine |
(44, 45) |
| Fat | ISC, EB | Rat | 1:2,000 | Michael Simon | (44) |
| Ds | Borders of EC cells | Rat | 1:5,000 | Michael Simon | (44) |
| Asense | ee cells | Rabbit | 1:5,000 | Y.-N. Jan | (25) |
| scute RNA | ISC, EB and/or ee | – | – | – | (25) |
| krn RNA | ISC | – | – | – | (28) |
| dpERK | ISC, EB | Rabbit | 1:1,000 | Cell signaling | (32) |
| STAT92E | ISC, EB | Rabbit | 1:500 | S. Hou | (39) |
| pMoe | Interface membranes between the ISC and EB cells and their corners |
Rabbit | 1:500 | Cell signaling | (34) |
| E-cad | Interface membranes between the ISC and EB cells | Rat | 1:20 | DSHB | (34) |
| aPKC | Apical areas of the ISC/EB | Rabbit | 1:100 | Santa Cruz | (34) |
| wg | Circular muscles, basement membrane, and ISCs | Mouse | 1:200 | DSHB | (38) |
| Caudal | All epithelial cells of gut | Guinea Pig |
1:400 | East Asian Distribution Centre, JAPAN | (15,16) |
| Spectrin | Apical side of ECs | Mouse | 1:100 | DSHB | (17) |
| Dapi | Mark the nuclei | – | 1 μg/mL | Sigma | (13) |
| Brdu | S-phase cell marker | Mouse | 1:100 | Invitrogen | (39) |
| β-galactosidase | lacZ expression | Rabbit | 1:2,000 | Cappel | (13) |
| GFP | For GFP-fusion protein lines | Mouse/rabbit | 1:100/1:500 | Invitrogen | (39) |
Table 2.
Transgenes expressed in Drosophila midgut stem cells and differentiated cells
| Transgenes | Expression in midgut | References |
|---|---|---|
| esg-Gal4-UAS-GFP | ISC, EB | (12) |
| Kr-Gal4-UAS-GFP | ISC, EB | Present study |
| Su(H)Gbe-lacZ | EB | (12) |
| Pvf2-lacZ | ISC/EB | (15) |
| Dl-LacZ | ISCs | (26, 37) |
| delta-Gal4 | ISC | (55) |
| Su(H)Gbe-Gal4 | EB | (55) |
| polo-GFP | ISCs metaphase plate | (4) |
| vn-lacZ | Midgut visceral muscle cells | (28) |
| spi (spi-Gal4NF0261) | ISC, EB and low in EC | (28) |
| rhoX81-lacZ | Midgut visceral muscle cells | (28) |
| upd-lacZ | ISC and EB | (39) |
| upd-3-lacZ | EC | (21,28) |
| 10XSTAT-DGFP | ISC, EB | (26, 39) |
| myolA-Gal4-UAS-GFP | ECs cells | (27) |
| myolA-lacZ | ECs cells | (27) |
| 24B-Gal4 (howGal4) | Visceral muscle-specific | (5,46) |
| ds-LacZ | EC and some ee cells | (44) |
| E(spl)mβ-CD2 | EB cells | (34) |
| mira-promoter-GFP | ISC | (25) |
| vkg-GFP | Basement membrane | (17) |
| esg-lacz | ISC, EB | (12,19) |
| cad-Gal4-UAS-GFP | Posterior midgut cells | (19) |
| wg-lacz | Circular muscle cells of midgut | (38) |
| collagenIV-GFP | Basement membrane structure | (17) |
| gstDl-lacZ | ISC and ee cells | (43) |
| D-p38b-lacZ | ISC and EB | (41) |
| SCNY-GFP | ISC and EC | (24) |
2. Materials
Prepare all solutions using ultrapure water and analytical grade reagents. Prepare and store all reagents at room temperature (unless otherwise indicated).
2.1. Drosophila Culture
Control and transgenic flies.
Fly culture incubator (18, 25, and 29°C).
Standard Drosophila vials.
Standard plastic bottles.
Foam plugs.
Drosophila food: Cornmeal, agar, sucrose, yeast, and acid medium.
Yeast paste.
Autoclave to prepare the food.
Morgue for dead flies.
Fly trap.
2.2. Lineage Tracing and Gene Manipulation
Stocks for generating tubulin-lacZ, PMML, and MARCM are easily available from the Bloomington Stock Center (http://www.flybase.org).
37°C water bath tank for heat shock.
18 and 25°C incubator to maintain fly crosses.
Useful stock and transgenes required in these experiments are given in Subheading 3.1.
2.3. BrdU Labeling
10 mM BrdU (Sigma) in dH2O. Store at −20°C (see Note 1).
Mouse anti-BrdU monoclonal antibody (Invitrogen).
2 N HCl (8.6 mL concentrated 12 N HCL stock per 50 mL H2O).
DNase (300 μL DNase +700 1×PBS).
2.4. Immunofluorescence Staining of Gut
2.4.1. Gut Dissection
Drosophila adult flies (3–5 days old).
Standard CO2 source for anesthetizing the flies.
Fine paintbrushes.
Two fine-tipped dissecting forceps (Roboz) for dissection.
Glass microslides (25×75 mm) for dissection.
Glass microcoverslips (22×60 mm).
Plastic dropper.
Kim wipes.
Dissecting microscope with attached light source.
Dissecting solution (Drosophila ringer’s solution): 130 mM NaCl, 4.7 mM KCl, 1.9 mM CaCl2, and 10 mM HEPES (4-(2-hydroxyethyl)-l-piperazineethanesulfonic acid), pH 6.9. Dissolve 7.5 g NaCl, 0.35 g KCl, 0.21 g CaCl2, and 2.38 g HEPES in approximately 1 L distilled water and stir to dissolve. Adjust to pH 7.2 with 1 N HCl and make the final volume of 1 L with distilled water. Store the dissecting solution in glass bottle at 4°C (see Note 2).
2.4.2. Gut Fixation
Phosphate-buffered saline (PBS): 130 mM Nad, 7 mM Na2HPO4, 3 mM NaH2PO4, and adjust to pH 7.4 with HCl. Store in glass bottles at room temperature. For longer stability of solution, store at 4°C.
Triton X-100 (Sigma) (see Note 3).
Gloves (Kimberly-Clark).
PBX solution: Dissolve the 0.1% Triton X-100 in 1× PBS plus 0.5% BSA. Store in at room temperature. For longer stability of solution, store at 4°C.
Formaldehyde (formalin) (37%) solution (Sigma) (see Note 4).
Fixation solution: (4% formaldehyde). In a 50-mL tube, add 5.4 mL 37% formaldehyde solution and then add 44.6 mL 1× PBX solution. Mix well at vortex. This solution should be prepared fresh every time but may be stored at 4°C for 1 week or longer at −20°C.
Parafilm to seal the Eppendorf tubes.
2.4.3. Blocking the Gut and Immunostaining
Normal goat serum (NGS; Vector laboratories). Store at 4°C.
Bovine serum albumin (BSA; Sigma). Use 0.5%.
Blocking solution (2% NGS): To make 2% NGS in 50-mL tube, add 1 mL NGS to 49.0 mL of 1× PBX. Mix well in vortex and store at 4°C (see Note 5).
Minivortex (VWR Scientific Products).
Tube shaker (Labquake).
Aluminum foil.
Microcentrifuge tube rack (Fisher Scientific).
Primary antibodies: The primary antibodies available to study ISCs in adult Drosophila are listed in Table 1. We use anti-beta-galactosidase for lacZ reporter lines and anti-GFP for GFP-fusion protein lines (see Table 1) (see Note 6).
Secondary antibodies: Secondary antibodies can be purchased from many different companies. We use secondary antibodies of goat anti-mouse, goat anti-rat, goat anti-rabbit, and goat anti-guinea pig IgG conjugated to Alexa Fluor 488 or Alexa Fluor 594 or Texas Red from Invitrogen and Cell Signaling. Store all secondary antibodies in a dark place at 4°C. We used a 1:200–500 dilution of all secondary antibodies in 1× PBX with 0.5% BSA.
4,6-Diamidino-2-phenyldole dihydrochloride (DAPI; Invitrogen) to stain DNA. Dissolve in 1× PBS for counter-staining. Store in the dark at 4°C.
2.4.4. Mounting the Gut, Imaging, and Data Analysis
Microscope cover glass.
Microscope slides.
Quick-dry nail polish.
Dissecting microscope.
Fluorescence stereomicroscope with GFP filter.
Glycerol ultrapure (Sigma).
Mounting medium: 50% glycerol in 1× PBS (pH 7.4) containing 1% antifade (1,4-diazabicyclo [2,2,2] octane [DABCO]; Sigma) compounds. Store at room temperature (see Note 7).
Waterproof permanent marker to label the slides.
Microslide plastic folder (VWR Scientific Products).
Confocal microscope, we use Zeiss LSM510 for imaging.
Computer and appropriate software for image processing.
3. Methods
Carry out all procedures at room temperature unless otherwise specified.
3.1. Lineage Tracing and Gene Manipulation
Some of the important stocks will be needed to perform the experiments in this section: W1118; hs-FLP; FRT82B, tubP-GAL80; FRT19A, tubP-GAL80; FRTG13, tubP-GAL80; FRT82B, arm-lacZ; Arm-lacZ, FRT80B; FRT42D, arm-lacZ; Arm-lacZ, FRT40A; Arm-LacZ, FRT19A; FRT52B(y)-FRT-Gal4; FRT52B(w)-actin5C-FRT; X-15-29 (FRT-lacZ); X-15-33 (tub-FRI); UAS-lacZ; UAS-EGFP, and UAS-mCD8GFP.
3.1.1. Generation of lacZ-Marked Clones in Adult Gut
The lineage marking system to generate clones of lacZ-expressing cells has been developed by Harrison and Perrimon (57). In this labeling system, after mitotic recombination, α-tubulin promoter will be fused to lacZ to allow the transcription of the marker. Without heat shock, XI5 flies carry two inactive tubulin promoter-lacZ (X-15–33 and X-15–29). During mitotic division, randomly cells will go for flipase-mediated recombination at the two-flipase recombination target (FRT) sites, which results in generation of an active lacZ transgene (Fig. 3a). When this random recombination occurred in stem cell, the stem cells and its daughter cells will be marked with lacZ., and you can see the maintenance of the clones over long time. However, recombination in non-stem cell results in formation of a transient clone with short life span. The lacZ-marked lineage system is very efficient because marker genes are activated immediately but no background (see Note 8).
Fig. 3.
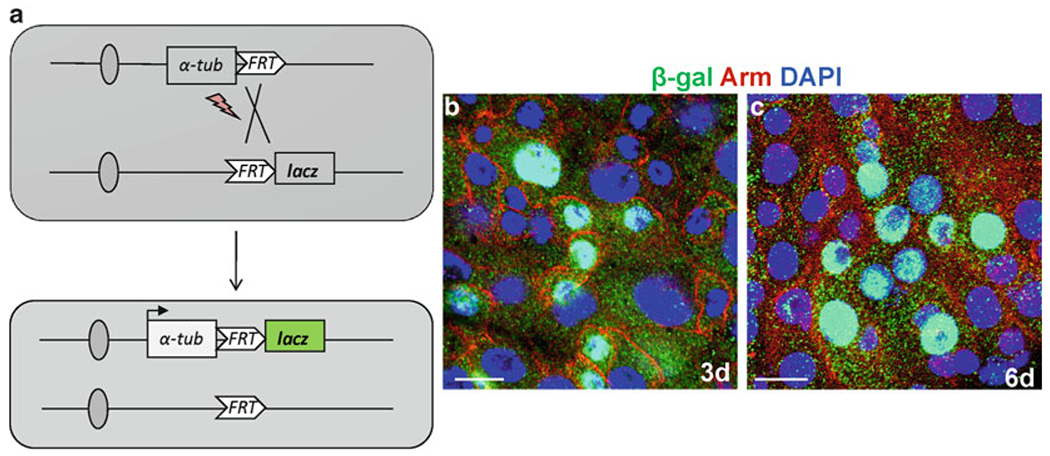
Tubulin-lacZ positive-labeling to generate ISCs clones in the midgut. (a) Diagram to show the lineage-tracing scheme to mark the randomly dividing cells by heat shock FLP-catalyzed site-specific recombination. (b, c) The representative examples of the induced clones, 3 day (b) and 6 day (c) in the midgut anti-β-Gal (green), anti-arm (red), and Dapi (blue). Scale bars: 10 μm.
Produce the flies with the genotype yw,hs-FLP/+; X-15-33/X-15-29 by standard crosses.
To induce somatic recombination to generate the clones, take the 3–5-day-old adult flies and heat shock in circulating water bath at 37°C for 60 min (see Note 9).
After heat shock, return flies to 25°C.
Transfer the flies daily to fresh food vials with yeast granules.
Dissect the guts at different time intervals to check the changes in clone size over time.
Fix, stain, and examine the gut under confocal microscope as per protocol given in Subheading 3.3.
3.1.2. Generation of PMML Clones in Adult Gut
This system utilized the heat shock-inducible FLP to reconstitute a functional actin5C-gal4 gene from two complementary inactive alleles, actin5C FRT52B and FRT52B gal4. The actin5C-gal4 gene drives GFP expression to mark cells and at the same time activate or knock down the gene function by having UAS constructs in the marked cells (Fig. 4a) (58).
To use the PMML system to generate GFP-marked clones that also overexpress their respective genes, cross the hs-Flp UAS-srcEGFP; FRT52B(w) UAS-EGFP/Cyo virgin females with males of genotypes FRT52B(y)/Cyo, UAS-geneAFRT52B(y)/Cyo.
Collect the 3–5-day-old non-Cyo females and heat shock them in a water bath at 37°C for 60 min, one time.
After heat shock, return flies to room temperature or at 25°C.
Transfer the flies daily to fresh food vials with yeast granules.
Dissect the guts at different time intervals to check the changes in clone size over time.
Fix, stain and examine the gut under confocal microscope as per protocol given in Subheading 3.3.
3.1.3. Generation of MARCM Clones In Adult Gut
MARCM technique has been used to create individually labeled homozygous cells in an otherwise heterozygous background. MARCM relies on recombination during mitosis mediated by FLP-FRT recombination. MARCM contain six transgenes: two transgenes for two homologous FRT sites, one for the FLP recombinase, one for a UAS marker, one for a GAL4 driver, and one for the tubP-GAL80 transgene. The tubP-GAL80 has been placed distal to the FRT site in trans to the mutant gene of interest. In heterozygous cells, GAL80 blocks GAL4-induced UAS marker expression; however, in homozygous mutant cells, tubP-GAL80 transgene is inactive, and therefore, the marker will be expressed (59–62) (Fig. 5a). Here, we describe the brief protocol, for details see (59–62).
Fig. 5.
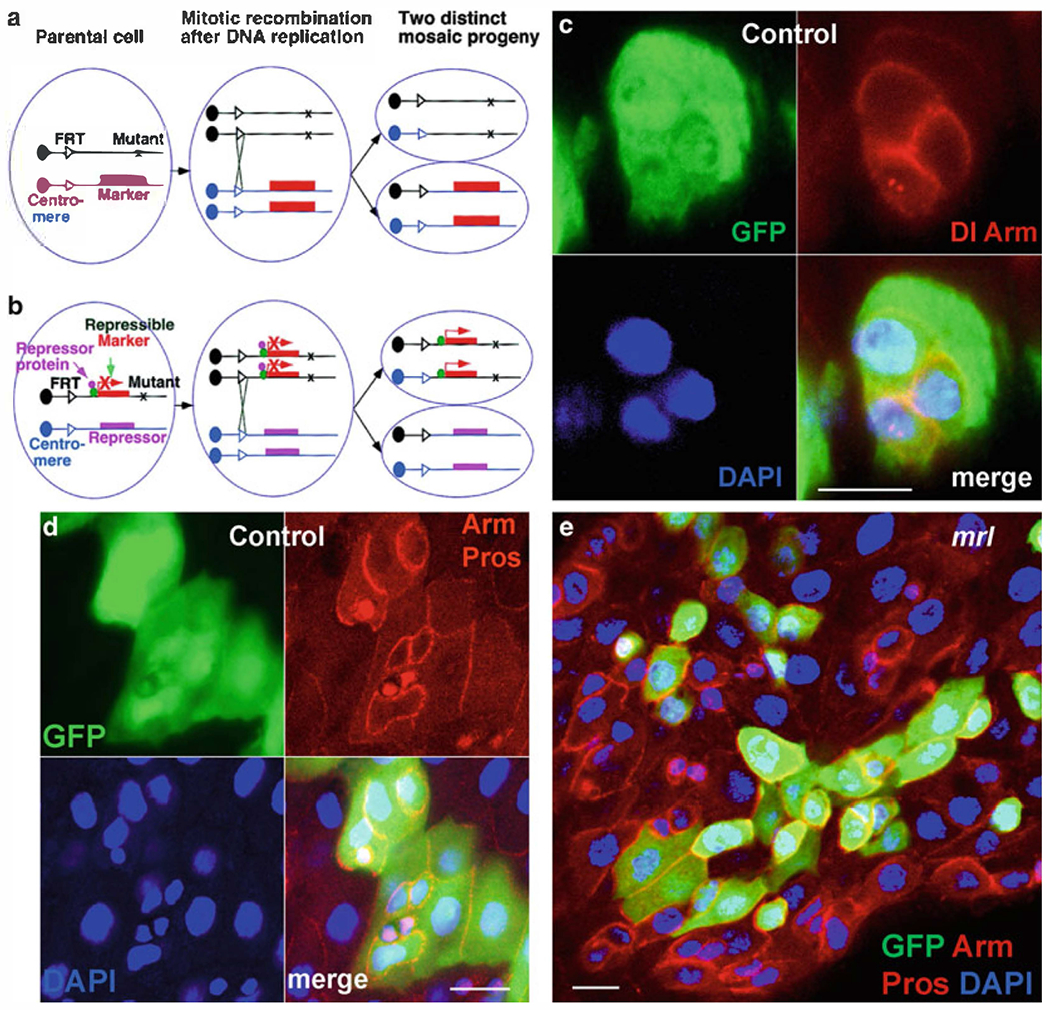
MARCM labeling system to generate ISCs clones in the midgut. (a) In conventional mosaic analysis, homozygous mutant cells are identified as unstained cells if the marker gene is placed distal to the FRT site on the homologous chromosome arm in trans to the mutant gene. (b) In the MARCM system, a transgene encoding the repressor of marker gene expression is placed distal to the FRT site on the homologous chromosome arm from the mutant gene. Only in homozygous mutant cells can the marker gene be expressed because of the loss of the repressor transgene. (c, d) Gut with GFP-marked wild-type MARCM clones. In (c) GFP (green), anti-delta (red, punctate expression) and anti-Arm (red-cell nest), and Dapi (blue). In (d) GFP (green), anti-Arm (red-cell nest), anti-Pros (red-nuclei of ee cells), and Dapi (blue). (e) Gut with GFP-marked Stat92E06346 mutant clones. GFP (green), anti-Arm (red-cell nest), anti-Pros (red-nuclei of ee cells), and Dapi (blue). Panel A in this figure is adapted with permission from Lee and Luo (59). Scale bars: 10 μm.
Cross the flies of genotype yw, hs-FLP UAS-GFP tub-Gal4+; FRT82B tub-Gal80, either with FRT82B flies or FRT82B-mutant flies.
Raised the Fl progenies at 18°C.
Take the 3–5-day-old flies from 18°C and heat shock at 37°C in a circulating water bath for 60 min, twice a day for 2 days, with an interval of 8 h in each heat shock (see Note 10).
After every heat shock, invert the vials and keep the flies at room temperature.
Dissect the guts at different time intervals to check the changes in clone size over time (see Note 11).
Fix, stain, and examine the gut under confocal microscope as per protocol given in Subheading 3.3.
3.2. Brdu Labeling
5-Bromo-2-deoxyuridine (BrdU) serves as marker of proliferation and is a uridine derivative and a structural analog of thymidine. It can be incorporated into DNA during the S-phase of the cell cycle. We used BrdU labeling to see the stem cell proliferation in adult Drosophila midgut.
Culture the adult 3–5-day-old flies with fly food containing 200 μL of 6 mg/mL BrdU in 20% sucrose for 2 days.
Transfer the flies every 2 days to new fly food vials.
For maximum labeling, feed the flies with Brdu 4–6 days.
After feeding the flies with BrdU, dissect the gut as per protocol given in Subheading 3.3.
Fix the guts as per protocol given in Subheading 3.3.
Treat the guts with DNase I for 1 h at 37°C to denature the DNA.
Remove the DNase I from the gut.
Rinse the guts three times with 1#x00D7;PBX.
Block and perform the staining as per protocol given in Subheading 3.3.
3.3. Immunofluorescence Staining of Midgut
The presented staining protocol is similar to one described for Drosophila testis with some modifications (53).
3.3.1. Gut Dissection
Anesthetize the flies on the CO2 source surface.
Use a clean glass microslide and put under the dissecting microscope.
Use a plastic dropper to place a few drops of Ringer’s solution on the slide.
Take the flies from fine forceps and place flies on a slide containing Ringer’s solution.
With the help of two fine forceps, dissect the gut in fresh Ringer’s solution.
Use one pair of forceps, turn the body upside down, hold the top of the abdomen, and pull out the external genitalia with the other pair of forceps.
Take off the gut, which is usually cream color, and remove the other fly parts such as ovary (in case of female flies), malpighian tubules, and anterior gut.
Transfer the dissected gut into a tube with Ringer’s solution. Optional: guts after dissection can be transferred directly to the fixation solution.
10–20 flies gut should be ideal for one tube.
Wait the gut tissues to settle down at the base and mark the tube for specific genotype.
3.3.2. Fixation of Gut
Take out the dissecting solution from the tube and keep the gut in the tube.
Prepare 4% formaldehyde (fixing solution) in 1× PBX. Optional: 4% formaldehyde can also be prepared in 1×PBS.
Add 200–500 μL 4% formaldehyde in the tube with gut.
Fix the gut by incubating the tube on a shaker at room temperature for 20–40 min (see Note 12).
After specified time above, stop the shaker and place the tube in the tube rack to allow the gut to settle down in the tube.
Remove the fixative solution and rinse the guts three times, for 2 min each in 1× PBX.
3.3.3. Blocking and Staining of Gut
To block the gut tissue, prepare the blocking solution in 1× PBX.
Add 200–500 μL blocking solution to guts and incubate the guts overnight at 4°C or 30 min at room temperature (see Note 13).
Take out the blocking solution.
Dilute the primary antibody in 1× PBX containing 0.5% BSA.
Mix the tube containing antibodies on minivortex (see Note 14).
Add 50–100 μL of diluted primary antibody to the guts and wrap the tube with Parafilm (see Note 15).
Incubate the gut tissues with the primary antibody, preferably overnight at 4°C or at room temperature (see Note 16).
After specified incubation time above, take out the primary antibodies and save at 4°C for reuse (see Note 17).
Rinse the gut three times with 1× PBX.
Then wash the gut on shaker at room temperature for 15 min (three times) in 1×PBX.
Prepare secondary antibody to desired concentration in 1×PBX (see Note 18).
Add 200–400 μL of diluted secondary antibody to the gut tissues.
Wrap the tube with aluminum foil to avoid the exposure of light.
Incubate the gut with secondary antibodies on shaker at room temperature for 2 h or overnight at 4°C (see Note 19).
Remove the secondary antibody from the tube.
Rinse the gut three times with 1× PBX.
Then wash the gut on shaker at room temperature for 15 min (three times) in 1× PBX.
After final wash, rinse the gut three times in 1× PBS.
To counter stain the gut with DNA, prepare the DAPI (from 1 mg/mL stock) in 11× PBS (see Note 20).
Add the DAPI to gut tissues and incubate for 5 min at room temperature.
Remove the DAPI solution and store in a dark place at 4°C for to reuse.
Rinse the gut with 1× PBS three times for 2 min each.
Wait for tissues to settle down and prepare for mounting.
3.3.4. Mounting, Microscopy, and Data Analysis
Prepare mounting medium in 1× PBS.
Add 20–100 μL of mounting medium to the gut tissues.
Place the gut tissues with mounting medium in a dark overnight at 4°C to allow tissues to equilibrate.
With the help of pipette, transfer the gut to a glass microslide frosted at one end.
Arrange the gut under the dissecting microscope using low levels of light.
Carefully place a cover slip to the slide containing the gut (see Note 21).
Remove excess mounting medium using Kimwipes.
Seal the edges of the microcoverslip with nail polish (see Note 22).
Use a permanent marker to label the slide for the specific genotype.
Put the prepared slides in a slide box, wrap the box with aluminum foil, and store the slides in the dark at 4°C until observation (see Note 23).
First, confirm the staining by examining the slides by fluorescence microscopy (see Note 24).
For best quality imaging, use confocal microscopy. To capture images, we used a Zeiss LSM 510 confocal laser-scanning microscope attached to computer.
LSM 5 Image Browser used to download the images (see Note 25).
Images processed using Adobe Photoshop.
Some of the examples using the above techniques and staining protocols are presented in Figs. 2–4. However, for best results, optimize the condition as per requirements of the experiments. The above protocols can be used to identify other stem cells in adult Drosophila.
Acknowledgments
M.K.S. is supported by the Knight’s Templar Eye Foundation and start-up support from the University of Dayton, OH. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
4. Notes
BrdU is a mutagen and should be handled with care. BrdU solution should be made fresh every time.
1×PBS can be used in the place of Drosophila Ringer’s solution.
Use of Triton X-100 allows the cell membrane to be permeable to the antibody. Avoid using more than specified concentration as it can disrupt the epithelial membrane of the gut, which results in no or poor staining of membrane proteins.
Formaldehyde is a fixative and carcinogenic; therefore, skin contact should be avoided. Avoid using bare hands to retrieve the tissue pieces; wear gloves during handling.
In addition to NGS, calf serum or bovine serum albumin can also be used for blocking the tissues. Blocking solution can be contaminated because it contains serum. It is stable for a week at 4°C and for several months frozen. If the solution is becoming cloudy, that means it is contaminated and should not be used for blocking the tissues.
Store the primary antibodies at 4°C with 0.02% sodium azide. For specificity and quality, primary antibodies can be stored at −20°C with 50% glycerol and at −80°C for long-term storage. Avoid frequent freezing and thawing, which degrade proteins.
As an alternative, Vectashield H-1000 from Vector Laboratories can also be used as a mounting medium.
This method has limitation because it is not ideal for manipulating the gene’s activity in the clones.
A 37°C bacteria incubator can also be used in the place of circulating water bath. A single heat shock at 37°C for 1 h is enough to induce the clones. After heat shock, transfer the flies to a fresh, dry vial to avoid flies to stick with wet vial as water vapor is formed after heat shock.
With this system, clones are activated slowly. The advantage of this system that clones can mis-express a gene or homozygose a mutation (52).
If there are very few clones induced, make sure the heat shock regimes are timed properly. In addition, double-check your cross to verify the genotypes. In Drosophila midgut, transient clones are present until 7 days after heat shock; it is advised to examine the clones at different time intervals (3, 6, 12, 20 days) (11).
For best results, limit the fixation time to 25–40 min as prolonged fixation can cross-link antigens and mask epitopes.
Blocking reduces background staining by preventing nonspecific protein-antibody interactions. The blocking can be done for extended time (over weekend) at 4°C without any visible loss of staining quality.
Gut can be stained with several primary antibodies raised in different species in the same tube. Sometimes, mouse and rat antibodies cross-react; avoid longer incubation time of primary antibodies.
To prevent the leakage of the antibodies during shaking, wrap the tube with Parafilm.
Alternatively incubate the primary antibodies at room temperature for 2 h. Sometimes, it can be incubated for 2 days at 4°C. However, long incubation times can create background staining.
Primary antibodies can be reused many times with same quality of staining.
Store the secondary antibodies in the dark at 4°C. For optimal staining, order new secondary antibodies if they are more than a year old. However, most of the secondary antibodies are good for several years, if stored properly. It can be stored at −20°C.
For best results, incubate the tissues with secondary antibody for 2 h at room temperature. Incubating tissues for overnight at 4°C can produce high background for polyclonal antibodies.
DAPI is a potential carcinogen and is a popular nuclear counterstain for use in multicolor fluorescent techniques. It may be harmful by inhalation, ingestion, or skin absorption. Wear proper gloves and avoid breathing the dust and vapors. Dispose all tips and tubes in appropriate container.
Carefully place the edge of a coverslip next to the drop of mounting medium. With the help of forceps, lower the coverslip onto the gut tissues.
To prevent movement of the coverslip and evaporation of the media during imaging, first, put nail polish to the four corners of the cover slip and let it dry. Then, put nail polish to the edges. The edges of the coverslips can also be sealed by using halocarbon oil 27.
For best results, store the slides in the dark at 4°C and collect the images within 2–3 days.
If there is problem in staining such as no staining and weak or high background in staining, this might be due to missing some steps, primary and/or secondary antibody may be bad due to improper storage, improper dilution, and/or incubation time. To avoid the above problems, use some known antibody with known expression pattern to ensure that the primary and/or secondary antibody is working properly.
After capturing the images, one can process the images in several different ways depending upon the available software.
References
- 1.Reya T, Morrison SJ, Clarke MF, Weissman IL (2001) Stem cells, cancer, and cancer stem cells. Nature 414:105–111 [DOI] [PubMed] [Google Scholar]
- 2.Blanpain C, Fuchs E (2009) Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol 10:207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simons BD, Clevers H (2011) Strategies for homeostatic stem cell self-renewal in adult tissues. Cell 145:851–862 [DOI] [PubMed] [Google Scholar]
- 4.Amcheslavsky A, Jiang J, Ito N, Ip YT (2011) Tuberous Sclerosis Complex and Myc coordinate intestinal stem cell growth and division in Drosophila. J Cell Biol 193:695–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu N, Wang SQ, Tan D, Gao Y, Lin G, Xi R (2011) EGFR, Wingless and JAK/STAT signaling cooperatively maintain Drosophila intestinal stem cells. Dev Biol 354:31–43 [DOI] [PubMed] [Google Scholar]
- 6.Singh SR, Zheng Z, Wang H, Oh SW, Chen X, Hou SX (2010) Competitiveness for the niche and mutual dependence of the germline and somatic stem cells in the Drosophila testis are regulated by the JAK/STAT signaling. J Cell Physiol 223:500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh SR, Burnicka-Turek O, Chauhan C, Hou SX (2011) Spermatogonial stem cells, infertility and testicular cancer. J Cell Mol Med 15:468–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barker N, van Es JH, Kuipers J, Kujala P, van den Bom M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449:1003–1007 [DOI] [PubMed] [Google Scholar]
- 9.Zhu L, Gibson P, Currie DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ (2009. ) Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature 457:603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sangiorgi E, Capeccltii MR (2008) Bmil is expressed in vivo in intestinal stem cells. Nat Genet 40:915–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohlstein B, Spradling A (2006) The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439:470–474 [DOI] [PubMed] [Google Scholar]
- 12.Micdtelli CA, Peirrimon N (2006) Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439:475–479 [DOI] [PubMed] [Google Scholar]
- 13.Ohlstein B, Spradling A (2007) Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315:988–992 [DOI] [PubMed] [Google Scholar]
- 14.Biteau B, Hochminth CE, Jasper H (2008) JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 3:442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi NH, Kim JG, Yang DJ, Kim YS, Yoo MA (2008) Age-related changes in Drosophila midgut are associated with PVE2, a PDGF/VEGF-like growth factor. Aging Cell 7:318–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi YJ, Hwang MS, Park JS, Bae SK, Kim YS, Yoo MA (2008) Age-related upregulation of Drosophila caudal gene via NF-kappaB in the adult posterior midgut. Biochim Biophys Acta 1780:1093–1100 [DOI] [PubMed] [Google Scholar]
- 17.Amchesiavsky A, Jiang J, Ip YT (2009) Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatterjee M, Ip YT (2009) Pathogenic stimulation of intestinal stem cell response in Drosophila. J Cell Physiol S220:664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apidianakis Y, Pitsouli C, Perrimon N, Rahmc L (2009) Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Nad Acad Sci USA 106:20883–20888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B (2009) Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe 5:200–211 [DOI] [PubMed] [Google Scholar]
- 21.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B (2009) Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev 23:2333–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchon N, Broderick NA, Kuraishi T, Lemaitre B (2010) Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol 8:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, Leibbrandt A, Simoes RD, Gruber S, Puc U, Ebersberger I, Zoranovic T, Neely GG, von Haeseler A, Ferrandon D, Penninger JM (2009) Genomewide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science 325:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buszczak M, Paterno S, Spradling AC (2009) Drosophila stem cells share a common requirement for the histone H2B ubiquitin protease scrawny. Science 323:248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bardin AJ, Perdigoto CN, Southall TD, Brand AH, Schweisguth F (2010) Transcriptional control of stem cell maintenance in the Drosophila intestine. Development 137:705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang H, Patel PH, Kohlmaier A, Greniey MO, McEwen DG, Edgar BA (2009) Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137:1343–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang H, Edgar BA (2009) EGFR signaling regulates the proliferation of Drosophila adult midgut progenitors, Development 136:483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang H, Greniey MO, Bravo MJ, Blumhagen RZ, Edgar BA (2011) EGFR/Ras/MAPK signaling media tes adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8:84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apidianakis Y, Rahme LG (2011) Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech 4:21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw RL, Kohlmaier A, Poleseilo C, Veelken C, Edgar BA, Tapon N (2010) The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137:4147–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee WC, Beebe K, Sudmeier L, Miccheili CA (2009) Adenomatous polyposis coli regulates Drosophila intestinal stem cell proliferation. Development 136:2255–2264 [DOI] [PubMed] [Google Scholar]
- 32.Ren F, Wang B, Yue T, Yun EY, Ip YT, Jiang J (2010) Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci USA 107:21064–21069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathur D, Bost A, Driver I, Ohlstein B (2010) A transient niche regulates the specification of Drosophila intestinal stem cells. Science 327:210–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maeda K, Takemura M, Umemori M, Adachi-Yamada T (2008) E cadherin prolongs the moment for interaction between intestinal stem cell and its progenitor cell to ensure Notch signaling in adult Drosophila midgut. Genes Cells 13:1219–1227 [DOI] [PubMed] [Google Scholar]
- 35.Wang P, Hou SX (2010) Regulation of intestinal stem cells in mammals and Drosophila. J Cell Physiol 222:33–37 [DOI] [PubMed] [Google Scholar]
- 36.Cordero J, Vidal M, Sansom O (2009) APC as a master regulator of intestinal homeostasis and transformation: from flies to vertebrates. Cell Cycle 8:2926–2931 [PubMed] [Google Scholar]
- 37.Beebe K, Lee WC, Micchelli CA (2010) JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev Biol 338:28–37 [DOI] [PubMed] [Google Scholar]
- 38.Lin G, Xu N, Xi R (2008) Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455:1119–1123 [DOI] [PubMed] [Google Scholar]
- 39.Liu W, Singh SR, Hou SX (2010) JAK-STAT is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J Cell Biochem 109:992–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin G, Xu N, Xi R (2010) Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of Drosophila intestinal stem cells. J Mol Cell Biol 2:37–49 [DOI] [PubMed] [Google Scholar]
- 41.Park JS, Kim YS, Yoo MA (2009) The role of p38b MAPK in age-related modulation of intestinal stem cell proliferation and differentiation in Drosophila. Aging 1:637–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park JS, Kim YS, Kim JG, Lee SH, Park SY, Yamaguchi M, Yoo MA (2010) Regulation of the Drosophila p38b gene by transcription factor DREF in the adult midgut. Biochim Biophys Acta 1799:510–519 [DOI] [PubMed] [Google Scholar]
- 43.Hochmuth CE, Biteau B, Bohmann D, Jasper H (2011) Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila. Cell Stem Cell 8:188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karpowicz P, Perez J, Perrimon N (2010) The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 137:4135–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Staley BK, Irvine KD (2010) Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr Biol 20:1580–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biteau B, Jasper H (2011) EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development 138:1045–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takashima S, Mkrtchyan M, Younossi-Hartenstein A, Merriam JR, Hartenstein V (2008) The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature 454:651–655 [DOI] [PubMed] [Google Scholar]
- 48.Fox DT, Spradling AC (2009) The Drosophila hindgut lacks constitutively active adult stem cells but proliferates in response to tissue damage. Cell Stem Cell 5:290–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Losick VP, Morris LX, Fox DT, Spradling A (2011) Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell 21:159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh SR, Liu W, Hou SX (2007) The adult Drosophila malpighian tubules are maintained by multipotent stem cells. Cell Stem Cell 1:191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh SR, Zeng X, Zheng Z, Hou SX (2011) The adult Drosophila gastric and stomach organs are maintained by a multipotent stem cell pool at the foregut/midgut junction in the cardia (proventriculus). Cell Cycle 10:1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fox DT, Morris LX, Nystul T, Spradling AC (2009) Lineage analysis of stem cells. In: The Stem Cell Research Community (ed.) StemBook. doi: 10.3824/stembook.1.33.1 [DOI] [PubMed] [Google Scholar]
- 53.Singh SR, Hou SX (2008) Immunohistological techniques for studying the Drosophila male germline stem cell. Methods Mol Biol 450:45–59 [DOI] [PubMed] [Google Scholar]
- 54.Liu W, Hou SX (2008) Genetic tools used for cell lineage tracing and gene manipulation in Drosophila germline stem cells. Methods Mol Biol 450:61–70 [DOI] [PubMed] [Google Scholar]
- 55.Zeng X, Chauhan C, Hou SX (2010) Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in Drosophila. Genesis 48:607–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zeng X, Singh SR, Hou D, Hou SX (2010) Tumor suppressors Sav/Scrib and oncogene Ras regulate stem-cell transformation in adult Drosophila malpighian tubules. J Cell Physiol 224:766–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrison DA, Perrimon N (1993) A simple and efficient generation of marked clones in Drosophila. Curr Biol 3:424–433 [DOI] [PubMed] [Google Scholar]
- 58.Kirilly D, Spana EP, Perrimon N, Padgett RW, Xie T (2005) BMP signaling is required for controlling somatic stem cell self-renewal in the Drosophila ovary. Dev Cell 9:651–662 [DOI] [PubMed] [Google Scholar]
- 59.Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22:451–161 [DOI] [PubMed] [Google Scholar]
- 60.Lee T, Luo L (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24:251–254 [DOI] [PubMed] [Google Scholar]
- 61.Wu JS, Luo L (2006) A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc 1:2583–2589 [DOI] [PubMed] [Google Scholar]
- 62.Shrestha BR, Grueber WB (2011) Generation and staining of MARCM clones in Drosophila. Cold Spring Harb Protoc 2011(8):973–979, pii: pdb.prot5659 [DOI] [PubMed] [Google Scholar]


