Abstract
We examined the effects of two direct-fed microbials (DFM) containing multiple microbial species and their fermentation products on energy status, nutrient digestibility, and ruminal fermentation, bacterial community, and metabolome of beef steers. Nine ruminally cannulated Holstein steers (mean ± SD body weight: 243 ± 12.4 kg) were assigned to three treatments arranged in a triplicated 3 × 3 Latin square design with three 21-d periods. Dietary treatments were 1) control (CON; basal diet), 2) Commence (PROB; basal diet plus 19 g/d of Commence), and 3) RX3 (SYNB; basal diet plus 28 g/d of RX3). Commence and RX3 are both multispecies DFM products. From day 16 to 20 of each period, feed and fecal samples were collected daily to determine the apparent total tract digestibilities of nutrients using indigestible neutral detergent fiber method. On day 21 of each period, blood samples were collected for analysis of plasma glucose and nonesterified fatty acid. Ruminal contents were collected at approximately 1, 3, 6, 9, 12, and 18 h after feeding on day 21 for analysis of volatile fatty acids (VFA), lactate, ammonia-N concentrations, bacterial community, and metabolome profile. Total tract digestibilities of nutrients did not differ (P > 0.05) among treatments. Compared with CON, steers fed either supplemental PROB or SYNB had greater (P = 0.04) plasma glucose concentrations. Compared with CON, total ruminal VFA, propionate, isovalerate, and valerate concentrations increased (P ≤ 0.05) or tended to increase (P ≤ 0.10) with either supplemental PROB or SYNB, but were not different (P > 0.05) between PROB and SYNB. Compared with CON, PROB reduced (P ≤ 0.05) the relative abundance of Prevotella 1 and Prevotellaceae UCG-001 but increased (P ≤ 0.05) the relative abundance of Rikenellaceae RC9, Succinivibrionaceae UCG-001, Succiniclasticum, and Ruminococcaceae UCG-002. Supplemental SYNB decreased (P ≤ 0.05) the relative abundance of Prevotella 1 and Prevotellaceae UCG-001 but increased (P ≤ 0.05) the relative abundance of Prevotella 7, Succinivibrio, Succiniclasticum, and Ruminococcaceae UCG-014. Compared with CON, metabolome analysis revealed that some amino acids were increased (P ≤ 0.05) in steers fed PROB. This study demonstrated that, compared with CON, supplementation of either PROB or SYNB altered the ruminal bacterial community and metabolome differently; however, their effects on the ruminal VFA profile and energy status of the steers were not different from each other.
Keywords: beef steer, direct-fed microbial, metabolome, rumen fermentation
Introduction
The rumen plays a central role in the overall metabolism, production, and health of ruminants (Morgavi et al., 2013). The rumen harbors microbial extracellular enzymes that hydrolyze dietary plant fiber that cannot be digested by the animal’s endogenous digestive enzymes to provide metabolic energy to the animals (Godoy-Vitorino et al., 2012). Thus, in the last decade, efforts to improve ruminant productivity have primarily focused on manipulating the ruminal microbial community and fermentation (DiLorenzo, 2011). One of such efforts includes the use of direct-fed microbials (DFM). Saccharomyces cerevisiae, the most extensively used DFM, is fed to modulate the composition and activities of the rumen microbial ecosystem, including favoring the activities of lactate-utilizing bacteria and fiber-degrading bacteria (Martin and Nisbet, 1992; Callaway and Martin, 1997). Some lactic acid utilizing bacteria, such as Megasphaera elsdenii and Propionibacterium freudenreichii, have also been evaluated as DFM with an attempt to enhance ruminal lactic acid metabolism toward production of propionate, a major precursor for glucose synthesis in ruminants (Yang et al., 2004; McAllister et al., 2011). Other studies have evaluated the use of lactic acid bacteria, such as Enterococcus lactis, Enterococcus faecium, and Lactobacillus casei, with the notion that increased production of lactate will lead to its increased fermentation (by lactate-utilizing bacteria) to propionate (Nocek et al., 2003). In recent years, most commercial DFM products are formulated to contain several species of the aforementioned microorganisms and their fermentation products in order to ensure efficacies and multifactorial response (McAllister et al., 2011).
Previous studies have applied culture-independent molecular techniques to reveal several mechanisms of action of DFM, including rumen bacterial community shift and selected growth of target bacteria (Fomenky et al., 2018; Ogunade et al., 2019a). It is well known that the effects of DFM products are inconsistent and heterogeneous due to various factors, including diet composition, differences in dose, strains, and physiological status of the animal; however, little emphasis has been placed on comparing multispecies DFM products under the same experimental condition (similar diet composition and physiological status of animals). Since DFM products are often fed to optimize rumen fermentation, we hypothesized that, compared with the control, dietary supplementation of two different multispecies DFM products would alter the rumen bacterial community and metabolome (comprehensive measurement of metabolites) differently, but their effects on the ruminal volatile fatty acids (VFA) profile would be similar to each other. The objective of this study was to compare the effects of two DFM products containing multiple microbial species, such as S. cerevisiae, E. lactis, Bacillus subtilis, E. faecium, and L. casei, and their fermentation products on energy status, nutrient digestibility, and ruminal fermentation, bacterial community, and metabolome of beef steers.
Materials and Methods
All experimental animals were managed according to the guidelines approved by the Institutional Animal Care and Use Committee of the University of Kentucky (IACUC number: 00674A2004).
Animal experiments and sample collection
Nine rumen-cannulated Holstein steers (mean ± SD body weight (BW): 243 ± 12.4 kg) were assigned to three treatments arranged in a 3 × 3 Latin square design with three 21-d periods and 10-d wash-out between periods. The steers were housed in individual pens and were fed (3% of BW on a dry matter (DM) basis) a total mixed ration (TMR) containing 79.7% corn silage and 20.3% concentrate mix on a DM basis (Table 1) once daily at 0900 hours. BWs were taken every 2 wk to adjust the amount of feed offered. Dietary treatments were 1) control (CON; basal diet without additive), 2) Commence (PROB; basal diet plus 19 g/d of Commence), and 3) RX3 (SYNB; basal diet plus 28 g/d of RX3). Commence is a blend of active S. cerevisiae, E. lactis, B. subtilis, E. faecium, and L. casei, and their fermentation products. RX3 is a blend of active S. cerevisiae and the fermentation products of S. cerevisiae, E. lactis, Bacillus licheniformis, and B. subtilis. Both additives were fed according to the manufacturer’s recommendation (PMI, Arden Hills, MN, USA). Approximately, 400 g of a premix (dried distillers grain with solubles), which was formulated to supply the appropriate supplemental level of the additives, was top-dressed daily on the TMR for the PROB and SYNB treatments, while a similar quantity of the premix with no additive was top-dressed for the CON treatment.
Table 1.
Ingredient and chemical composition of the basal diet1
| Ingredient (%DM) | % of dietary DM |
|---|---|
| Corn silage | 79.7 |
| Dehydrated distillers grain | 9.06 |
| Soybean meal | 9.28 |
| Limestone | 0.42 |
| Deccox2 | 0.03 |
| Vitamin and mineral premix3 | 1.51 |
| Nutrient analysis4 | |
| DM, % | 44.5 |
| CP, % | 14.7 |
| aNDF (amylase treated NDF), % | 38.6 |
| ADF, % | 21.5 |
| Ether extract, % | 3.50 |
| Ca, % | 0.87 |
| P, % | 0.63 |
| Total digestible nutrients , % | 72.6 |
| Net energy of maintenance, Mcal/kg | 1.72 |
| Net energy of gain, Mcal/kg | 1.10 |
1Chemical composition of complete diets calculated from analysis and concentration of individual ingredients.
2Contains 6% decoquinate for the prevention of coccidiosis (Zoetis Inc.).
3Guaranteed analysis: 15% Ca; 7.5% P; 20% salt; 1% Mg; 1% K; 3,600 mg/kg Mn; 12 mg/kg Co; 1,200 mg/kg Cu; 3,600 mg/kg Zn; 27 mg/kg Se; 60 mg/kg I; 660,000 IU/kg vitamin A; 660 IU/kg vitamin E; and 66,000 IU/kg vitamin D.
4The chemical composition of the top-dressed premix was not included in the nutrient analysis.
The quantity of feed offered to each steer was recorded daily. Diet refused (as fed) was also measured daily. Diet DM refused and offered were obtained by drying daily samples of diets refused and offered in a forced-air oven at 56 °C for 48 h. Daily dry matter intake was determined by subtracting the daily DM refused from the daily DM offered. Samples of TMR collected weekly were dried for 48 h at 60 °C in a forced-air oven, ground to pass through a 1-mm screen (Wiley Mill; Arthur H. Thomas Co.), and sent to a commercial laboratory (Dairy One Forage Laboratory, Ithaca, NY) for analysis of the chemical composition.
Blood sample collection and analysis
Blood samples from each steer were collected from the coccygeal vessels before the morning feeding on the last day (day 21) of each period into a 10-mL vacutainer tube containing sodium heparin (Vacutainer, Becton Dickinson, Franklin Lakes, NJ). Immediately after collection, the blood samples were placed on ice, and thereafter centrifuged at 1,500 × g for 15 min at 4 °C to harvest the plasma. The plasma samples were then frozen at −20 °C until analysis for glucose and nonesterified fatty acids (NEFA). Plasma concentrations of glucose and NEFA were measured in duplicate. The plasma samples were analyzed in duplicate. Glucose concentrations (using 50 µL each of plasma samples) were measured using a quantitative colorimetric kit (G7521-1L; Pointe Scientific Inc., Canton, MI). Concentrations of NEFA (using 5 µL each of plasma samples) were measured using an enzyme-based assay (NEFA-C kit; Wako Diagnostics Inc., Richmond, VA) as modified by Johnson and Peters (1993). The intra- and inter-assay coefficients of variation for glucose were 2.65% and 4.92%, respectively, while those for NEFA were 3.61% and 5.84%, respectively.
Apparent total tract digestibility measurements
From day 16 to 20 of each period, TMR, refusal, and fecal samples were collected daily to determine apparent total tract digestibility of nutrients using indigestible neutral detergent fiber (iNDF) as the digestibility marker (Cole et al., 2011; Krizsan and Huhtanen, 2013). Samples of TMR and refusal were collected once daily and stored at −20 °C. Approximately, 75 g of fecal sample was collected from each steer four times daily at 0800, 1200, 1600, and 2000 hours from the ground, inside the pen, within few minutes after the animal defecated. Immediately after collection, fecal samples were stored at −20 °C. At the end of each period, TMR, refusal, and fecal samples were thawed and dried at 60 °C for 48 h in a forced-air oven, ground to pass through a 2-mm screen (Wiley Mill; Arthur H. Thomas Co.), pooled within steer for each day, and sent to a commercial laboratory (Dairy One Forage Laboratory, Ithaca, NY) for analysis of DM, crude protien (CP), acid detergent fiber (ADF), and neutral detergent fiber (NDF). For iNDF determination, 0.5 g of the TMR, refusal, and fecal samples was weighed into ANKOM bags (F57; Ankom Technology Corp.), incubated in rumen fluid, collected from three beef steers fed corn silage-based diet ad libitum, for 240 h using an ANKOM Daisy II incubator. The rumen fluid was changed approximately every 48 h. The residues were subsequently analyzed for NDF using an ANKOM fiber analyzer (ANKOM 200, Macedon, NY). Total iNDF consumed (g/d) was corrected for refusals. Total feces output (kg) was calculated as total iNDF consumed (g/d) divided by fecal iNDF concentration (g/kg). Digestibility was calculated as: (intake of nutrient − fecal output of nutrient)/intake of nutrient.
Rumen fluid collection and analyses
On day 21 of each period, representative samples (150 mL) of the ruminal contents were collected via the cannula at five different sites within the rumen at approximately 1, 3, 6, 9, 12, and 18 h after feeding. A subsample of the rumen content was manually homogenized as described by Ogunade et al. (2019a) and stored immediately at −80 °C until DNA extraction. Another subsample of the rumen content was strained through four layers of cheesecloth to separate solid and liquid fractions, and the liquid fraction was immediately measured for pH using a portable pH meter and, thereafter, stored at −20 °C until VFA, lactate, and ammonia-N analysis.
VFA, lactate, and ammonia-N analysis
Ruminal fluid samples collected at different time points were used for analyzing VFA, lactate, and ammonia nitrogen concentrations. A 1-mL aliquot of the rumen fluid sample was mixed with 0.1 mL of 500 g/L metaphosphoric acid and 0.1 mL of 85 mM 2-ethyl butyrate in a 1.5-mL centrifuge tube. The mixture was centrifuged at 39,000 × g at 23 °C for 15 min. The samples were analyzed for VFA concentrations (Xu et al., 2010) using a gas chromatograph with a flame ionization detector (Agilent Technologies, Inc., Santa Clara, CA). Ammonia nitrogen concentrations were analyzed in duplicate using a glutamate dehydrogenase procedure (Kun and Kearney, 1974) adapted to a Konelab 20XTi clinical analyzer (Thermo Fisher Scientific Inc., Beverly, MA; Trotta et al., 2018). The intra- and inter-assay coefficients of variation for ammonia nitrogen were 4.26% and 6.14%, respectively. Lactate was analyzed using a procedure described by Gutmann and Wahlefeld (1974), which was adapted to a microplate reader (Trotta et al., 2018).
Microbial DNA extraction and polymerase chain reaction amplification
Before DNA isolation, equal amounts of the rumen contents collected at different time points were pooled for each steer. Each pooled sample (0.25 g) underwent microbial DNA extraction using a Qiagen DNeasy Powersoil DNA Isolation kit following the manufacturer’s instructions (Qiagen, Frederick, MD). The lysing step was performed using the Disruptor Genie cell disruptor (Scientific Industries) during which the samples underwent a 15-min bead-beating lysis. Finally, the genomic DNA was eluted in 50 μL of 10 mM Tris-HCl. Subsequently, quantification was performed using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA) with the double-stranded DNA high-sensitivity assay.
Illumina iTag polymerase chain reaction (PCR) was the V4 region of the 16S rRNA gene, which was PCR-amplified using the Earth Microbiome Project’s 16S rRNA amplification protocol (Walters et al., 2016). The volume of each reaction was 25 μL and contained (final concentrations) 1X PCR buffer, 0.8 mM deoxynucleotide mix (Thermo Fisher, Wilmington, DE), 0.625 U Ex Taq DNA Polymerase (Takara Bio, Mountain View, CA), 0.2 μM 515F barcoded forward primer, 0.2 μM 806R reverse primer, and 10 ng of template DNA per reaction. PCR was carried out on a T100 Thermal Cycler (Bio-Rad, Hercules, CA) using the following cycling conditions: 98 °C for 3 min; then 35 cycles of 98 °C for 1 min, 55 °C for 40 s, and 72 °C for 1 min; final extension was at 72 °C for 10 min; then held at 4 °C. PCR products were visualized on a 2% agarose E-Gel with ethidium bromide (Thermo Fisher, Wilmington, DE) for bands at ~400 bp.
Library purification, verification, and sequencing
The PCR products were pooled in an approximate equimolar manner. The pooled PCR products were then run on a 2% agarose gel with GelStar Nucleic Acid Gel Stain (Lonza, Rockland, ME) for visualization. Bands of expected product length were cut from the gel using sterile scalpels and were subsequently purified using the QIAquick Gel Purification Kit (Qiagen, Frederick, MD). The pure library was then quantified using the Qubit 2.0 Fluorometer double-stranded DNA high-sensitivity assay (Life Technologies, Carlsbad, CA). Before submission for sequencing, libraries were quality checked using a 2100 Bioanalyzer high sensitivity DNA analysis kit (Agilent Technologies, Santa Clara, CA). The sequencing library was stored at −20 °C until it was shipped on dry ice to Laragen Inc. (Culver City, CA) for sequencing on an Illumina MiSeq v2 500-cycle kit cassette with 16S rRNA library sequencing primers set for 250 bp, paired-end reads.
Quality filtering and de-noising
Paired-end sequences were imported into the DADA2 pipeline for quality filtration, denoising, and chimera removal; 250 bp reads were filtered at a maximum expected error of 0.5. Trimmed sequences were then de-noised, merged, and underwent chimera removal within DADA2 software (https://github.com/benjjneb/dada2) (Callahan et al., 2016). Taxonomy was then assigned against the SILVA database (Caporaso et al., 2010). Amplicon sequence variants (ASV) were tabulated for use in downstream taxonomic summary and analysis. A reformatted ASV-table was converted to BIOM format for use with QIIME-1.9.1.
Rumen metabolomics analysis
In-depth untargeted metabolome profile of the pooled rumen fluid samples was done using a chemical isotope labeling (CIL)/liquid chromatography-mass spectrometry (LC-MS)-based technique to target the amine and phenol-containing submetabolome. A subsample (1 mL) of the rumen fluid was centrifuged at 15,000 × g for 10 min and the supernatant was analyzed. The workflow of the differential 12C- and 13C-isotope dansylation LC-MS method for analyzing the amine/phenol-containing submetabolome has been reported in our previous study (Adeyemi et al., 2019). Sample amount normalization was done using liquid chromatography-ultraviolet (LC-UV) quantification of the dansyl-labeled metabolites (Wu and Li, 2012), and relative quantification of the metabolites based on peak ratio values was performed on an Agilent 1100 LC system (Palo Alto, CA) connected to a Bruker Impact HD quadrupole time-of-flight (QTOF) MS (Billerica, MA). Detailed information on dansylation protocol, LC-UV and LC-MS setup, and concentration measurement have been previously reported (Mung and Li, 2017). A total number of 35 LC-MS data files were generated (4 blank group samples, 4 quality control samples, 9 CON samples, 9 PROB samples, and 9 SYNB samples). The quality control sample was prepared by mixing an equal amount of a 12C-labeled and a 13C-labeled pooled sample and was injected every 10 sample runs to monitor instrument performance.
Data and statistical analysis
The experimental design was a triplicated 3 × 3 Latin square with nine experimental units per treatment. Variables that were measured repeatedly over time, such as VFA concentrations, lactate, ammonia-N, and pH, were analyzed using the GLIMMIX procedure (SAS version 9.4, SAS Institute Inc., Cary, NC). The model for analyzing the data included the effects of treatment, period, steer (random), sampling time, and the interaction between treatment and time. Normality was tested by examining the distribution of residuals. Denominator degrees of freedom were estimated using the Kenward–Roger option in the MODEL statement. Time was used in the repeated-measures statement. Autoregressive order 1 was selected based on Akaike information criterion values as the repeated measure covariance structure. The model for analyzing outcomes that were not measured repeatedly over time included the effects of treatment, period, and steer (random). For the bacterial diversity analysis, multiple rarefactions were conducted on sequences across all samples from a minimum depth of 100 to a maximum depth of 6,000 sequences, with a step size of 600, and 10 iterations at each step. Alpha diversities were then collated, plotted, and compared using a nonparametric Monte Carlo permutations (n = 999). Within-sample (α) diversity comparisons were done pairwise among the treatment groups based on the Chao1 estimates. Partial least squares discriminant analysis (PLS-DA) was performed using the cumulative sum scaling normalized operational taxonomic unit table with the mixOmics R package (Rohart et al., 2017). For all data, except the metabolomics data, post hoc mean comparisons were performed using the Tukey–Kramer procedure for pairwise multiple comparisons. Significance was declared at P ≤ 0.05, and trends toward significance were declared at 0.05 < P ≤ 0.10.
For the metabolomics data, the 35 LC-MS data files (in profile mode) were converted to text file (in centroid mode) using Bruker DataAnalysis software 4.4. Raw data processing and quality check were performed using IsoMS Pro 1.0 according to the procedures described by Mung and Li (2017). Peak pairs whose mean (sample)/mean (blank) was ≤ 4.0 were filtered out. Three-tier identification approach was used to perform metabolite identification (Li et al., 2013). In tier 1, peak pairs were searched against a CIL Library based on accurate mass and retention time (RT) (Huan and Li, 2015). In tier 2, linked identity (LI) library was used for high-confidence identification, based on accurate mass and predicted RT matches (Li et al., 2013). In tier 3, the remaining peak pairs were searched, based on accurate mass match, against the MyCompoundID library (MCID; www. MyCompoundID.org) (Li et al., 2013). Multivariate (PLS-DA scores plot) and univariate statistical analyses (volcano plot) were generated using IsoMS Pro 1.0 (Mung and Li, 2017). The volcano plot was constructed by plotting the fold change (FC; PROB/CON, SYNB/CON, SYNB/PROB) of each metabolite against false discovery rate adjusted P-value. Metabolites with FC ≥ 1.2 or ≤ 0.83 having P-value ≤ 0.05 were considered to be differentially increased or decreased, respectively.
Results
Rumen fermentation parameters
Compared with CON, supplementation of either PROB or SYNB increased (P ≤ 0.05) the total VFA, propionate, and valerate concentrations and tended to increase (P = 0.10) isovalerate concentration (Table 2). There were no differences (P > 0.05) between PROB and SYNB, except that supplemental PROB reduced (P = 0.05) ammonia-N concentration compared with CON and SYNB. There were no treatment or treatment × time interaction effects (P > 0.05) on ruminal pH, acetate, isobutyrate, butyrate, and lactate concentrations (Table 2).
Table 2.
Effects of dietary supplementation of DFM containing multiple microbial strains and their fermentation products on the rumen fermentation of beef steers
| Item | CON | PROB | SYNB | SEM | P-value |
|---|---|---|---|---|---|
| Acetate, mM | 25.8 | 27.8 | 30.5 | 2.02 | 0.32 |
| Propionate, mM | 8.32b | 11.1a | 12.3a | 0.95 | 0.03 |
| Valerate, mM | 0.99b | 1.76a | 1.55a | 0.18 | 0.01 |
| Isovalerate, mM | 0.43y | 0.55x | 0.58x | 0.05 | 0.10 |
| Butyrate, mM | 6.89 | 7.99 | 7.90 | 0.77 | 0.83 |
| Isobutyrate, mM | 0.32 | 0.35 | 0.39 | 0.04 | 0.58 |
| Total VFA, mM | 52.5b | 59.9a | 61.6a | 2.71 | 0.01 |
| Lactate, mM | 0.63 | 0.69 | 0.53 | 0.20 | 0.63 |
| Ammonia-N, mM | 3.72a | 2.13b | 3.21a | 0.42 | 0.05 |
| pH | 6.67 | 6.54 | 6.71 | 0.12 | 0.52 |
a,bWithin a row, treatment means with different superscripts differ, P ≤ 0.05.
x,yWithin a row, treatment means with different superscripts tend to differ, 0.05 < P ≤ 0.10.
Energy status and apparent nutrient digestibility
Compared with CON, steers fed diets supplemented with either PROB or SYNB had greater (P = 0.04) plasma glucose concentrations; however, there were no effects on plasma NEFA concentrations (P = 0.56; Table 3). Average daily intakes and apparent total tract digestibilities of DM, CP, NDF, and ADF did not differ (P > 0.05) among treatments (Table 4).
Table 3.
Effects of dietary supplementation of DFM containing multiple microbial species and their fermentation products on plasma glucose and NEFA concentration in beef steers
| Item | CON | PROB | SYNB | SEM | P-value |
|---|---|---|---|---|---|
| Plasma glucose, mg/Dl | 66.9b | 70.3a | 69.7a | 1.24 | 0.04 |
| NEFA, mEq/L | 0.13 | 0.12 | 0.13 | 0.005 | 0.56 |
a,bWithin a row, treatment means with different superscripts differ, P ≤ 0.05.
Table 4.
Effects of dietary supplementation of DFM containing multiple microbial species and their fermentation products on intake and apparent total tract digestibility of nutrients in beef steers (day 16 to 20)
| Item | CON | PROB | SYNB | SEM | P-value |
|---|---|---|---|---|---|
| Intake, kg/d | |||||
| DM | 6.83 | 7.02 | 6.91 | 0.32 | 0.81 |
| CP | 1.01 | 1.07 | 1.03 | 0.05 | 0.62 |
| ADF | 1.45 | 1.50 | 1.48 | 0.10 | 0.69 |
| NDF | 2.62 | 2.69 | 2.67 | 0.08 | 0.57 |
| Apparent digestibility, % | |||||
| DM | 65.6 | 65.6 | 63.9 | 1.12 | 0.26 |
| CP | 56.8 | 57.2 | 54.2 | 1.78 | 0.18 |
| ADF | 51.6 | 50.6 | 50.8 | 1.38 | 0.72 |
| NDF | 50.0 | 49.1 | 48.8 | 1.42 | 0.71 |
Relative abundance of bacteria
Following sequencing and quality control, a total of 950,000 filtered paired-end reads were generated. A range of 23,841 to 77,960 sequences per sample was retained after quality filtration and read merging. Prevotella 1 dominated (67 ± 22.4%) the ruminal bacterial community at the genus level, followed by Prevotella 7 (4.7 ± 16.93 %), Rikenellaceae RC9 gut group (3.3 ± 2.83%), and then Succinivibrio (1.8 ± 8.13%), Succinivibrionaceae UCG-001 (1.8 ± 7.74%), and Succiniclasticum (1.8 ± 2.40%; Supplementary Table S1).
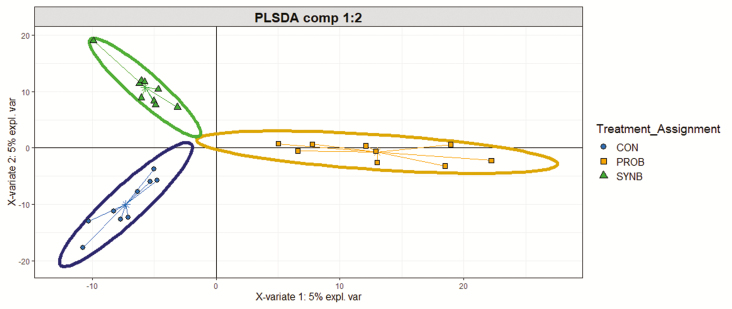
Treatments did not affect Chao1 estimate, a measure of within-sample (α) diversity (Supplementary Figure S1). PLS-DA scores plot revealed differential bacterial community composition among the three treatment groups (Figure 1). Treatment effects on the relative abundance of bacteria at the genus level that were affected by dietary treatments are shown in Table 5. Compared with CON, supplemental PROB reduced (P ≤ 0.05) the relative abundance of Prevotella 1 and Prevotellaceae UCG-001. Conversely, the relative abundance of Rikenellaceae RC9 gut group, Succinivibrionaceae UCG-001, Succiniclasticum, Ruminococcaceae UCG-014, and Ruminococcaceae UCG-002 were increased (P ≤ 0.05). Supplemental SYNB decreased (P ≤ 0.05) the relative abundance of Prevotella 1 and Prevotellaceae UCG-001 but increased (P ≤ 0.05) the relative abundance of Prevotella 7, Succinivibrio, Succiniclasticum, and Ruminococcaceae UCG-014. Compared with SYNB, supplemental PROB reduced (P ≤ 0.05) the relative abundance of Prevotella 7 and Succinivibrio but increased (P ≤ 0.05) the relative abundance of Ruminococcaceae UCG-002, Rikenellaceae RC9 gut group, and Succinivibrionaceae UCG-001.
Figure 1.
PLS-DA scores plot the ruminal bacterial community of beef steers fed diets supplemented with DFM containing multiple microbial species and their fermentation products.
Table 5.
Effects of dietary supplementation of DFM containing multiple microbial species and their fermentation products in the diet of beef steers on the relative abundance of ruminal bacteria at the genus level
| Genus (% of total sequences)1 | CON | PROB | SYNB | SEM | P-value |
|---|---|---|---|---|---|
| Prevotella 1 | 72.6a | 65.6b | 64.1b | 2.67 | 0.01 |
| Prevotella 7 | 0.30b | 0.96b | 12.8a | 3.51 | 0.01 |
| Rikenellaceae RC9 gut group | 1.96b | 5.62a | 2.44b | 0.96 | 0.01 |
| Succinivibrio | 0.07b | 0.34b | 4.96a | 1.17 | 0.02 |
| Succinivibrionaceae UCG-001 | 0.88b | 4.44a | 0.00b | 1.39 | 0.01 |
| Succiniclasticum | 0.19b | 1.34a | 1.69a | 0.42 | 0.01 |
| Ruminococcaceae UCG-014 | 0.57b | 1.57a | 1.66a | 0.41 | 0.04 |
| Ruminococcaceae UCG-002 | 0.83b | 1.43a | 0.79b | 0.23 | 0.05 |
| Prevotellaceae UCG-001 | 1.14a | 0.60b | 0.44b | 0.17 | 0.01 |
1Only bacterial genera (≥ 0.1% of total sequences) that were different in any of the treatment groups (P ≤ 0.05) are shown.
a,bWithin a row, treatment means with different superscripts differ, P ≤ 0.05.
CIL/LC-MS-based metabolomics analysis
An average of 3,392 ± 80 peak pairs per sample was detected. Peak pairs that were not present in at least 80.0% of samples in any group were removed. After filtering, 3,219 ± 59 peak pairs per sample were retained (Supplementary Table S2). Among them, 165 peak pairs were positively identified in tier 1, 201 peak pairs were putatively identified with high confidence in tier 2, and 2,096 peak pairs were matched against the MCID in tier 3.
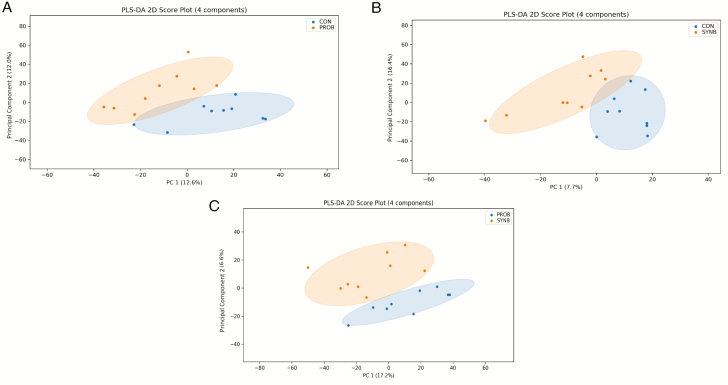
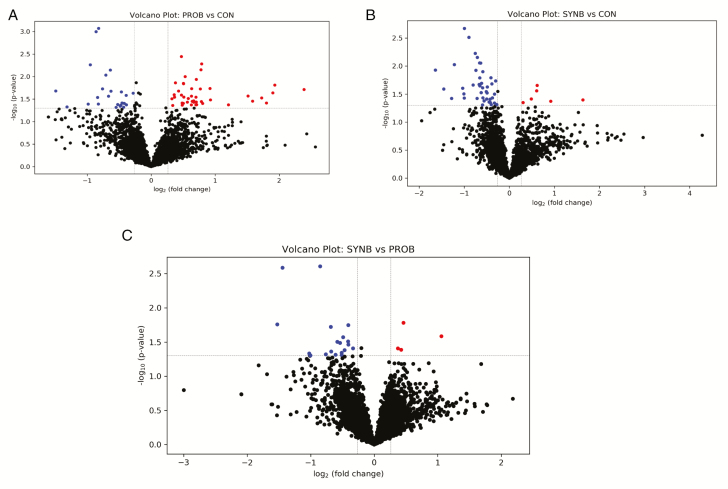
The PLS-DA scores plots showed separations between CON vs. each of PROB and SYNB and between PROB and SYNB (Figure 2a–c), indicating that PROB and SYNB supplementation differentially altered the ruminal metabolome of the beef steers. The result of the volcano plot analysis showed that, relative to CON, 44 peak pairs with FC ≥ 1.2, P-value ≤ 0.05 (in red), and 24 peak pairs with FC ≤ 0.83, P-value ≤ 0.05 (in blue, Figure 3a) were differentially altered by PROB supplementation. Among them, 9 peak pairs were positively identified in tier 1, 2 peak pairs were high-confidence-putatively identified in tier 2, and 38 peak pairs were putatively matched in tier 3 using MCID library (Supplementary Table S3). The metabolites that were positively and putatively identified with high confidence are shown in Table 6. Compared with CON, eight metabolites (taurine, glutamyl-proline, valine, phenylalanine, norleucine, histinyl-proline, tyrosine, and p-cresol) were differentially increased (FC ≥ 1.2, P ≤ 0.05) in steers fed PROB diet, whereas three metabolites (cystathionine, isomers of (R)-1-aminopropan-2-ol, and 3-(2,3-dihydroxyphenyl)propanoate) were differentially reduced (FC ≤ 0.83, P ≤ 0.05).
Figure 2.
PLS-DA scores plots (A. CON vs. PROB, B. CON vs. SYNB, and C. SYNB vs. PROB) of the ruminal metabolome of the beef steers fed diets supplemented with DFM containing multiple microbial species and their fermentation products.
Figure 3.
Volcano plots (A. CON vs. PROB, B. CON vs. SYNB, and C. SYNB vs. PROB.) showing the differential ruminal metabolites in beef steers fed diets supplemented with DFM containing multiple microbial species and their fermentation products. FC ≥ 1.2, P-value ≤ 0.05 (in red): significantly increased; FC ≤ 0.83, P-value ≤ 0.05 (in blue): significantly reduced relative to CON.
Table 6.
Identified metabolites that were affected by supplemental PROB
| Item1 | Normalized RT2 | FC3 | P-value | Identification level4 |
|---|---|---|---|---|
| Taurine | 157.6 | 1.50 | 0.04 | Tier 1 |
| Glutamyl-proline | 389.6 | 1.42 | 0.03 | Tier 1 |
| Valine | 662.4 | 1.26 | 0.04 | Tier 1 |
| Phenylalanine | 783.2 | 1.73 | 0.01 | Tier 1 |
| Cystathionine | 830.8 | 0.70 | 0.04 | Tier 1 |
| Norleucine | 837.8 | 1.63 | 0.01 | Tier 1 |
| Histidinyl-proline | 1,098.2 | 1.25 | 0.03 | Tier 1 |
| Tyrosine | 1,363.3 | 1.62 | 0.04 | Tier 1 |
| Para-cresol | 1,482.6 | 1.42 | 0.01 | Tier 1 |
| Isomer of (R)-1-aminopropan-2-ol | 413 | 0.51 | 0.04 | Tier 2 |
| 3-(2,3-Dihydroxyphenyl)propanoate | 756.9 | 0.72 | 0.02 | Tier 2 |
1Only metabolites with both FC ≥ 1.2 or ≤ 0.83, relative to Control, and P ≤ 0.05 are shown.
2Normalized RT shows the corrected RT of the peak pair with Universal RT Calibrant data.
3FC: fold change relative to Control.
4Tier 1—Positive Identification (CIL Library);Tier 2—High Confidence Putative Identification (LI Library).
For comparison of CON vs. SYNB, the volcano plot analysis showed that 44 peak pairs with FC ≥ 1.2, P-value ≤ 0.05 (in red), and 24 peak pairs with FC ≤ 0.83, P-value ≤ 0.05 (in blue, Figure 3b). None of these peak pairs could be positively or high-confidence-putatively identified (Supplementary Table S4). For comparison of SYNB vs. PROB, the volcano plot analysis showed that four peak pairs with FC ≥ 1.2, P-value ≤ 0.05 (in red), and 19 peak pairs with FC ≤ 0.83, P-value ≤ 0.05 were altered (in blue, Figure 3c). Among those that were positively and putatively identified with high confidence, relative concentrations of phenylalanine, histinyl-proline, glutamyl-proline, valine, and para-cresol were differentially increased (FC ≥ 1.2, P-value ≤ 0.05) by PROB supplementation (data not shown).
Discussion
Alterations in ruminal fermentation and bacterial population structure caused by dietary supplementation of DFM products containing multiple microbial species such as S. cerevisiae and lactic acid bacteria have been widely reported in ruminants (McAllister et al., 2011, Fomenky et al., 2018; Ogunade et al., 2019b). However, results of DFM supplementation in animals cannot be compared due to the diversity of DFM products, differences in their composition, processing, doses, and animal factors. Moreover, the continuing development of different DFM products emphasize the need for more research studies to understand their underlying mechanisms.
Dietary supplementation of either PROB or SYNB increased the total ruminal VFA concentration possibly due to increased ruminal microbial growth or activities, rather than increased substrate availability because DM intake was not different among the treatments. VFA are the major products of microbial fermentation in the rumen (Dijkstra, 1994). They are essential for the overall metabolism of the ruminant animals because they contribute up to 75% of the total metabolic energy needs of ruminants (Bergman, 1990). Thus, the increased production of VFA in the rumen often results in improved metabolic status and productivity of the animals (Kolver and de Veth, 2002; Oba and Allen, 2003).
Lactate was not influenced in this study even though PROB contains a blend of lactic acid-producing bacteria (E. lactis, E. faecium, and L. casei) possibly because the diet fed did not support lactate accumulation as shown by the low lactate concentrations and high ruminal pH values. Increased concentrations of ruminal propionate, valerate, and isovalerate, which are glucogenic precursors for net synthesis of glucose, often translate to the improved energy status of ruminants (Aschenbach et al., 2010). This explains greater plasma glucose concentrations in beef steers fed either supplemental PROB or SYNB. Plasma glucose concentration is often utilized as an indicator of energy metabolism in ruminants (Aschenbach et al., 2010). Glucose is the primary energy source for ruminants and its increased concentration is often an evidence of improved energy and nutritional status of the animals (Grummer, 1993). NEFA is formed from the degradation of fatty acids in the adipose tissues and liver cells for energy production when glucose supply is inadequate (Aschenbach et al., 2010). Regardless of dietary treatment, plasma NEFA concentration was low, indicating that the diet fed was adequate to meet the energy demand of the steers. Although comparisons of results of research studies that evaluated the use of DFM products should be done with caution due to factors mentioned earlier, similar results have been reported by studies that tested multispecies DFM products in dairy cows. For example, Nocek et al. (2003) reported that dietary supplementation of a DFM product containing two strains of Enterococcus (5 × 109 cfu/d) and S. cerevisiae (2 × 109 cfu/d) to lactating Holstein cows during the transition period resulted into higher blood levels of glucose and lower serum levels of beta-hydroxybutyric acid. Similarly, Nocek and Kautz (2006) observed no changes in plasma NEFA concentration, but there were higher blood glucose and lower beta-hydroxybutyric acid levels in transition dairy cows. These results were also supported by the results of our previous studies that demonstrated improved energy status and performance of newly weaned beef steers fed supplemental PROB (Adeyemi et al., 2019, 2020). Adeyemi et al. (2020) demonstrated that supplementation with PROB altered the plasma metabolome toward increased concentrations of monosaccharides, such as glucose, galactose, fructose, and glyceraldehyde, and decreased concentration of acetoacetate, indicating an improved energy status of the animals. Adeyemi et al. (2019) reported improved growth and feed efficiency of beef steers fed supplemental PROB possibly as a result of improved energy status of the animals.
Apparent total tract digestibilities of DM, CP, NDF, and ADF were not different among treatments. Effects of DFM on diet digestibility have been inconsistent; however, a large number of research studies have either reported no or negative effects of DFM products on nutrient digestibility. For example, in a recent study that evaluated a similar multispecies DFM product in dairy cows (Oh et al., 2019), dietary supplementation of a blend of S. cerevisiae, Lactococcus lactis, B. subtilis, E. faecium, and L. casei had no effects on total tract digestibility of nutrients. Beauchemin et al. (2003) observed no effects of dietary supplementation of a DFM product containing 6 × 109 cfu/g of E. faecium on total tract digestibility. In fact, dietary supplementation of 6 × 109 cfu/g of E. faecium decreased in situ digestibility of corn, barley, and alfalfa hay in steers (Beauchemin et al., 2003). In another study (Raeth-Knight et al., 2007), no effects on apparent digestibilities of DM, CP, NDF, or starch digestibility were observed in dairy cows fed a diet supplemented with multispecies DFM product containing 1 × 109 cfu/g of Lactobacillus acidophilus, 2 × 109 cfu/g of P. freudenreichii, and 5 × 108 cfu/g of L. acidophilus. Lack of effects on apparent total tract digestibility of nutrients in this study showed that the improved energy status of the steers fed supplemental PROB or SYNB in this study was not a reflection of feed digestibility.
Analysis of the ruminal bacterial composition revealed a Prevotella-dominated community. Species of Prevotella grow rapidly when fermentable carbohydrates are available (Bekele et al., 2010; Pitta et al., 2010). Corn silage is a source of fermentable carbohydrates that can be utilized by Prevotella as energy sources to produce succinate and acetate as the major fermentation end products (Flint et al., 2008; Dodd et al., 2011). As observed in this study, Lettat et al. (2013) reported the dominance of metabolically active Prevotella species in the rumen of dairy cows fed 100% corn silage-based diet. Moreover, Prevotella is considered the most dominant bacterial group in the rumen (Nagaraja and Titgemeyer, 2007; Stevenson and Weimer, 2007; Kim et al., 2011).
Supplementation of PROB increased the relative abundance of Succinivibrio, Rikenellaceae RC9 gut group, and Succiniclasticum, whereas supplemental SYNB increased the relative abundance of Succinivibrionaceae UCG-001 and Succiniclasticum. Succinivibrio and Succinivibrionaceae UCG-001 contribute to the production of succinate, a precursor for propionate and valerate synthesis by other microbes in the rumen (Holman and Gzyl, 2019). Rikenellaceae RC9 gut group can produce succinate and propionate as fermentation end products (Graf, 2014). Succiniclasticum can ferment succinate solely to propionate (van Gylswyk, 1995, Stewart et al., 1997). Greater relative abundances of these ruminal bacteria possibly explain the increased propionate concentrations in steers fed supplemental PROB and SYNB. The effects of DFM on ruminal fermentation have been shown to be influenced by several factors including diet. In animals fed a high grain diet, certain DFMs containing S. cerevisiae and/or lactic acid bacteria have been demonstrated to alter ruminal fermentation by stimulating the population of lactate-consuming bacteria, which can ferment lactate to propionate (Nagaraja and Titgemeyer, 2007; McAllister et al., 2011), thereby increasing ruminal pH. In high roughage diet, S. cerevisiae has been demonstrated to create an ecological condition that favors the growth of cellulolytic bacteria by scavenging oxygen and supplying growth factors essential for their growth (Robinson and Erasmus, 2009). The results of this study showed that the corn silage-based diet fed favored the dominance of Prevotella, which can produce acetate and succinate from fermentation of saccharides (Ueki et al., 2007). Thus, it is reasonable to infer that supplemental PROB and SYNB stimulated the relative abundance of ruminal bacteria that can produce succinate and those that can ferment succinate to propionate, valerate, and isovalerate. In a previous study, the relative abundance of Succinivibrio and Succinivibrionaceae were found to be positively correlated with propionate and valerate (Xue et al., 2018). This further explains why supplemental PROB and SYNB modulated the rumen fermentation toward increased concentration of ruminal glucogenic precursors for net synthesis of glucose. The fact that either PROB or SYNB increased the relative abundance of cellulolytic bacteria such as Ruminococcaceae UCG-014 and Ruminococcaceae UCG-002 is consistent with previous studies that demonstrated that some DFM containing strains of S. cerevisiae improved ruminal cellulolytic activities by favoring the growth of cellulolytic bacteria (Newbold et al., 1996; Chaucheyras-Durand and Fonty, 2002). It is important to note that although both additives altered the general structure of the bacterial community differently, similar ruminal environment and fermentation in terms of fermentation acids and pH were achieved.
Increased levels of amino acids such as taurine, valine, norleucine, phenylalanine, glutamyl-proline, histidinyl-proline, and tyrosine in steers fed supplemental PROB relative to CON are probably an indication of reduced deamination of amino acids in the rumen, which supports the lower levels of ammonia-N observed in steers fed supplemental PROB, compared with CON. The fact that the same results were not observed with supplemental SYNB is probably a result of the differential effects of the two additives on the ruminal bacterial community. As earlier stated, supplemental PROB reduced the relative abundance of Prevotella, relative to CON. In contrast, supplemental SYNB reduced the relative abundance of Prevotella 1 and increased that of Prevotella 7. In addition to being carbohydrate-fermenters and succinate-producers, species of Prevotella generally can degrade protein and peptides in the rumen (Stewart, 1997) because they possess a significant dipeptidyl peptidase activity (Wallace et al., 1997). Species of Prevotella have been reported to act synergistically with hyper-ammonia-producing bacteria to convert peptides and amino acids to ammonia (Madeira et al., 1997). Indeed, ruminal ammonia levels were increased in ruminants fed supplemental Prevotella bryantii (Chiquette et al., 2008; Fraga et al., 2018). Excessive deamination of amino acids to ammonia-N in the rumen often causes inefficient N retention by ruminants (Calsamiglia et al., 2010; Yang et al., 2010); thus, increased levels of amino acids could, in turn, contribute to an efficient N utilization due to their increased supply for intestinal absorption.
The biological significance of reduced ruminal concentration of cystathionine, a sulfur-containing amino acid formed as an intermediate in the conversion of methionine to ammonia, ketobutyrate, and cysteine (Beatty and Zhou, 2005), by supplemental PROB is not known because the concentrations of methionine and cysteine were not affected. Limited or no information is available in the literature on the significance of the other metabolites (para-cresol, isomer of (R)-1-aminopropan-2-ol and 3-(2,3-dihydroxyphenyl)propanoate) altered by PROB supplementation in this study. More in-depth studies are needed to understand better how these metabolites are associated with rumen fermentation and future advances in metabolomics technology should also focus on identifying some of the metabolites that were putatively matched in MCID library.
Conclusions
This study demonstrated that supplementation of PROB or SYNB differentially altered the ruminal bacterial community toward increased relative abundance of bacteria (Succinivibrio, Rikenellaceae RC9 gut group, and Succinivibrionaceae UCG-001) that can produce succinate and that of Succiniclasticum that can ferment succinate to propionate. This resulted in a significant improvement in the plasma glucose levels of beef steers fed supplemental PROB or SYNB; however, there were no effects on apparent digestibilities of nutrients. In contrast to SYNB, analysis of the amine/phenol-metabolome of the rumen fluid revealed that supplemental PROB reduced ruminal amino acid deamination, which was supported by reduced ammonia-N concentration. The differential bacterial community and metabolome shift caused by the additives resulted in similar concentrations of ruminal fermentation acids. These results also revealed that the positive effect of supplemental PROB and SYNB on the energy status of the steers, under the condition of this study, was achieved via mechanisms such as altered ruminal fermentation pattern and growth of target ruminal bacteria, other than feed digestibility.
Supplementary Material
Acknowledgments
The study was funded by Land O’ Lakes Inc. Additional funding support was provided by the United States Department of Agriculture’s National Institute of Food and Agriculture Evans-Allen project 1008985.
Glossary
Abbreviations
- ADF
acid detergent fiber
- ASV
amplicon sequence variants
- BW
body weight
- CIL
chemical isotope labeling
- CON
basal diet with no additive
- CP
crude protein
- DFM
direct fed microbial
- DM
dry matter
- DNA
deoxyribonucleic acid
- FC
fold change
- iNDF
indigestible neutral detergent fiber
- LC-MS
liquid chromatography-mass spectrometry
- LC-UV
liquid chromatography-ultraviolet
- LI
Linked identity
- MCID
my compound identification library
- NDF
neutral detergent fiber
- NEFA
nonesterified fatty acids
- PCR
polymerase chain reaction
- PLS-DA
partial least squares discriminant analysis
- PROB
a blend of Saccharomyces cerevisiae, Enterococcus lactis, Bacillus subtilis, Enterococcus faecium, and Lactobacillus casei, and their fermentation products fed at 19 g/steer/d
- rRNA
ribosomal ribonucleic acid
- RT
retention time
- SD
standard deviation
- SYNB
a blend of live Saccharomyces cerevisiae and the fermentation products of S. cerevisiae, Enterococcus lactis, Bacillus licheniformis, and Bacillus subtilis fed at 28 g/steer/d
- TMR
total mixed ration
- VFA
volatile fatty acid
Conflict of interest statement
The authors declare no conflict of interest.
Literature Cited
- Adeyemi, J. A., Harmon D. L., Compart D. M. P., and Ogunade I. M.. . 2019. Effects of a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products in the diet of newly weaned beef steers: growth performance, whole-blood immune gene expression, serum biochemistry, and plasma metabolome1. J. Anim. Sci. 97:4657–4667. doi: 10.1093/jas/skz308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemi, J. A., Peters S. O., De Donato M., Cervantes A. P., and Ogunade I. M.. . 2020. Effects of a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products on plasma carbonyl-metabolome and fecal bacterial community of beef steers. J. Anim. Sci. Biotechnol. 11:14. doi: 10.1186/s40104-019-0419-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschenbach, J. R., Kristensen N. B., Donkin S. S., Hammon H. M., and Penner G. B.. . 2010. Gluconeogenesis in dairy cows: the secret of making sweet milk from sour dough. IUBMB Life. 62:869–877. doi: 10.1002/iub.400 [DOI] [PubMed] [Google Scholar]
- Beatty, R., and Zou G. C.. . 2005. Redox regulation and reaction mechanism of human cystathionine beta synthase: a PLP-dependent homo-sensor protein. Arch. Biochem. Biophys. 443:144–156. doi: 10.1016/j.abb.2004.08.037 [DOI] [PubMed] [Google Scholar]
- Beauchemin, K. A., Yang W. Z., Morgavi D. P., Ghorbani G. R., Kautz W., and Leedle J. A.. . 2003. Effects of bacterial direct-fed microbials and yeast on site and extent of digestion, blood chemistry, and subclinical ruminal acidosis in feedlot cattle. J. Anim. Sci. 81:1628–1640. doi: 10.2527/2003.8161628x [DOI] [PubMed] [Google Scholar]
- Bekele, A. Z., Koike S., and Kobayashi Y.. . 2010. Genetic diversity and diet specificity of ruminal Prevotella revealed by 16S rRNA gene-based analysis. FEMS Microbiol. Lett. 305:49–57. doi: 10.1111/j.1574-6968.2010.01911.x [DOI] [PubMed] [Google Scholar]
- Bergman, E. N. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 70:567–590. doi: 10.1152/physrev.1990.70.2.567 [DOI] [PubMed] [Google Scholar]
- Callahan, B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J., and Holmes S. P.. . 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, T. R., and Martin S. A.. 1997. Effects of cellobiose and monensin on in vitro fermentation of organic acids by mixed ruminal bacteria. J. Dairy Sci. 80:1126. doi: 10.3168/jds.S0022-0302(97)76039-9 [DOI] [PubMed] [Google Scholar]
- Calsamiglia, S., Ferret A., Reynolds C. K., Kristensen N. B., and van Vuuren A. M.. . 2010. Strategies for optimizing nitrogen use by ruminants. Animal 4:1184–1196. doi: 10.1017/S1751731110000911 [DOI] [PubMed] [Google Scholar]
- Caporaso, J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., . et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaucheyras-Durand, F., and Fonty G.. . 2002. Influence of a probiotic yeast (Saccharomyces cerevisiae CNCM I-1077) on microbial colonization and fermentation in the rumen of newborn lambs. Microb. Ecol. Health Dis. 14:30–36. doi: 10.1080/089106002760002739 [DOI] [Google Scholar]
- Chiquette, J., Allison M. J., and Rasmussen M. A.. . 2008. Prevotella bryantii 25A used as a probiotic in early-lactation dairy cows: effect on ruminal fermentation characteristics, milk production, and milk composition. J. Dairy Sci. 91:3536–3543. doi: 10.3168/jds.2007-0849 [DOI] [PubMed] [Google Scholar]
- Cole, N. A., McCuistion K., Greene L. W., and McCollum F. T.. . 2011. Effects of concentration and source of wet distillers grains on digestibility of steam-flaked corn-based diets fed to finishing steers. Prof. Anim. Sci. 27:302–311. doi: 10.15232/S1080-7446(15)30493-9 [DOI] [Google Scholar]
- Dijkstra, J. 1994. Production and absorption of volatile fatty acids in the rumen. Livest. Prod. Sci. 39:61–69. doi: 10.1016/0301-6226(94)90154-6 [DOI] [Google Scholar]
- DiLorenzo, N. 2011. Manipulation of the rumen microbial environment to improve performance of beef cattle. In: Proceedings of 22nd Annual Florida Ruminant Nutrition Symposium, Gainesville, FL. p. 118.
- Dodd, D., Mackie R. I., and Cann I. K.. . 2011. Xylan degradation, a metabolic property shared by rumen and human colonic Bacteroidetes. Mol. Microbiol. 79:292–304. doi: 10.1111/j.1365-2958.2010.07473.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint, H. J., Bayer E. A., Rincon M. T., Lamed R., and White B. A.. . 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121–131. doi: 10.1038/nrmicro1817 [DOI] [PubMed] [Google Scholar]
- Fomenky, B. E., Do D. N., Talbot G., Chiquette J., Bissonnette N., Chouinard Y. P., Lessard M., and Ibeagha-Awemu E. M.. . 2018. Direct-fed microbial supplementation influences the bacteria community composition of the gastrointestinal tract of pre- and post-weaned calves. Sci. Rep. 8:14147. doi: 10.1038/s41598-018-32375-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga, M., Fernández S., Perelmuter K., Pomiés N., Cajarville C., and Zunino P.. . 2018. The use of Prevotella bryantii 3C5 for modulation of the ruminal environment in an ovine model. Braz. J. Microbiol. 49 (Suppl 1):101–106. doi: 10.1016/j.bjm.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy-Vitorino, F., Goldfarb K. C., Karaoz U., Leal S., Garcia-Amado M. A., Hugenholtz P., Tringe S. G., Brodie E. L., and Dominguez-Bello M. G.. . 2012. Comparative analyses of foregut and hindgut bacterial communities in hoatzins and cows. ISME J. 6:531–541. doi: 10.1038/ismej.2011.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, J. 2014. The family rikenellaceae. In: Rosenberg, E., E. F. DeLong, S. Lory, E. Stackebrandt, F. Thompson, editors. The prokaryotes. Berlin (Heidelberg): Springer; pp. 857–859. [Google Scholar]
- Grummer, R. R. 1993. Etiology of lipid-related metabolic disorders in periparturient dairy cows. J. Dairy Sci. 76:3882–3896. doi: 10.3168/jds.S0022-0302(93)77729-2 [DOI] [PubMed] [Google Scholar]
- Gutmann, I., and Wahlefeld A. W.. . 1974. L(+) lactate determination with lactate dehydrogenase and NAD. In: Bergmeyer, H., editor. Methods of enzymatic analysis. Vol. 3. New York (NY): Academic Press; p. 1464–1468. [Google Scholar]
- van Gylswyk, N. O. 1995. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Bacteriol. 45:297–300. doi: 10.1099/00207713-45-2-297 [DOI] [PubMed] [Google Scholar]
- Holman, D. B., and Gzyl K. E.. . 2019. A meta-analysis of the bovine gastrointestinal tract microbiota. FEMS Microbiol. Ecol. 95:fiz072. doi: 10.1093/femsec/fiz072 [DOI] [PubMed] [Google Scholar]
- Huan, T., and Li L.. . 2015. Quantitative metabolome analysis based on chromatographic peak reconstruction in chemical isotope labeling liquid chromatography mass spectrometry. Anal. Chem. 87:7011–7016. doi: 10.1021/acs.analchem.5b01434 [DOI] [PubMed] [Google Scholar]
- Johnson, M. M., and Peters J. P.. . 1993. Technical Note: An improved method to quantify nonesterified fatty acids in bovine plasma. J. Anim. Sci. 71:753–756. doi: 10.2527/1993.713753x [DOI] [PubMed] [Google Scholar]
- Kim, M., Morrison M., and Yu Z.. . 2011. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 76:49–63. doi: 10.1111/j.1574-6941.2010.01029.x [DOI] [PubMed] [Google Scholar]
- Kolver, E. S., and de Veth M. J.. . 2002. Prediction of ruminal pH from pasture-based diets. J. Dairy Sci. 85:1255–1266. doi: 10.3168/jds.S0022-0302(02)74190-8 [DOI] [PubMed] [Google Scholar]
- Krizsan, S. J., and Huhtanen P.. . 2013. Effect of diet composition and incubation time on feed indigestible neutral detergent fiber concentration in dairy cows. J. Dairy Sci. 96:1715–1726. doi: 10.3168/jds.2012-5752 [DOI] [PubMed] [Google Scholar]
- Kun, E., and Kearney E. B.. . 1974. Ammonia. In: Bergmeyer H. U., Gawehn, K, editors. Methods of enzymatic analysis. London and New York: Academic Press; pp. 1802–1806. [Google Scholar]
- Lettat, A., Hassanat F., and Benchaar C.. . 2013. Corn silage in dairy cow diets to reduce ruminal methanogenesis: effects on the rumen metabolically active microbial communities. J. Dairy Sci. 96:5237–5248. doi: 10.3168/jds.2012-6481 [DOI] [PubMed] [Google Scholar]
- Li, L., Li R., Zhou J., Zuniga A., Stanislaus A. E., Wu Y., Huan T., Zheng J., Shi Y., Wishart D. S., . et al. 2013. MyCompoundID: using an evidence-based metabolome library for metabolite identification. Anal. Chem. 85:3401–3408. doi: 10.1021/ac400099b [DOI] [PubMed] [Google Scholar]
- Madeira, H. M., Peng L., and Morrison M.. . 1997. Biochemical and mutational analysis of a gingipain-like peptidase activity from Prevotella ruminicola B(1)4 and its role in ammonia production by ruminal bacteria. Appl. Environ. Microbiol. 63:670–675. PMID: 9023945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, S. A., and Nisbet D. J.. 1992. Effect of direct-fed microbials on rumen microbial fermentation. J. Dairy Sci. 75:1736. doi: 10.3168/jds.S0022-0302(92)77932-6 [DOI] [PubMed] [Google Scholar]
- McAllister, T., Beauchemin K. A., Alazzeh A., Baah J., Teather R., and Stanford K.. . 2011. Review: The use of direct fed microbials to mitigate pathogens and enhance production in cattle. Can. J. Anim. Sci. 91:193–211. doi: 10.4141/cjas10047 [DOI] [Google Scholar]
- Moffett, J. R., and Namboodiri M. A.. . 2003. Tryptophan and the immune response. Immunol. Cell Biol. 81:247–265. doi: 10.1046/j.1440-1711.2003.t01-1-01177.x [DOI] [PubMed] [Google Scholar]
- Morgavi, D. P., Kelly W. J., Janssen P. H., and Attwood G. T.. . 2013. Rumen microbial metagenomics and its application to ruminant production. Animal 7:184–201. doi: 10.1017/S1751731112000419 [DOI] [PubMed] [Google Scholar]
- Mung, D., and Li L.. . 2017. Development of chemical isotope labeling LC-MS for milk metabolomics: comprehensive and quantitative profiling of the amine/phenol submetabolome. Anal. Chem. 89:4435–4443. doi: 10.1021/acs.analchem.6b03737 [DOI] [PubMed] [Google Scholar]
- Nagaraja, T. G., and Titgemeyer E. C.. . 2007. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J. Dairy Sci. 90:17–38. doi: 10.3168/jds.2006-478 [DOI] [PubMed] [Google Scholar]
- Newbold, C. J., Wallace R. J., and McIntosh F. M.. . 1996. Mode of action of the yeast Saccharomyces cerevisiae as a feed additive for ruminants. Br. J. Nutr. 76:249–261. doi: 10.1079/bjn19960029 [DOI] [PubMed] [Google Scholar]
- Nocek, J. E., and Kautz W. P.. 2006. Direct-fed microbial supplementation on ruminal digestion, health, and performance of pre- and postpartum dairy cattle. J. Dairy Sci. 89:260–266. doi: 10.3168/jds.S0022-0302(06)72090-2 [DOI] [PubMed] [Google Scholar]
- Nocek, J. E., Kautz W. P., Leedle J. A. Z., and Block E.. 2003. Direct-fed microbial supplementation on the performance of dairy cattle during the transition period. J. Dairy Sci. 86:331–335. doi: 10.3168/jds.S0022-0302(03)73610-8 [DOI] [PubMed] [Google Scholar]
- Oba, M., and Allen M. S.. . 2003. Effects of diet fermentability on efficiency of microbial nitrogen production in lactating dairy cows. J. Dairy Sci. 86:195–207. doi: 10.3168/jds.S0022-0302(03)73600-5 [DOI] [PubMed] [Google Scholar]
- Ogunade, I. M., Lay J., Andries K., McManus C. J., and Bebe F.. . 2019a. Effects of live yeast on differential genetic and functional attributes of rumen microbiota in beef cattle. J. Anim. Sci. Biotechnol. 10:68. doi: 10.1186/s40104-019-0378-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogunade I. M., Schweickart H., McCoun M., Cannon K., and McManus C.. . 2019b. Integrating 16S rRNA sequencing and LC-MS-based metabolomics to evaluate the effects of live yeast on rumen function in beef cattle. Animals (Basel). 28:683 doi: 10.3390/ani9010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, J., Harper M., Melgar A., Compart D. M. P., and Hristov A. N.. . 2019. Effects of Saccharomyces cerevisiae-based direct-fed microbial and exogenous enzyme products on enteric methane emission and productivity in lactating dairy cows. J. Dairy Sci. 102:6065–6075. doi: 10.3168/jds.2018-15753 [DOI] [PubMed] [Google Scholar]
- Pitta, D. W., Pinchak E., Dowd S. E., Osterstock J., Gontcharova V., Youn E., Dorton K., Yoon I., Min B. R., Fulford J. D., . et al. 2010. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb. Ecol. 59:511–522. doi: 10.1007/s00248-009-9609-6 [DOI] [PubMed] [Google Scholar]
- Raeth-Knight, M. L., Linn J. G., and Jung H. G.. . 2007. Effect of direct-fed microbials on performance, diet digestibility, and rumen characteristics of Holstein dairy cows. J. Dairy Sci. 90:1802–1809. doi: 10.3168/jds.2006-643 [DOI] [PubMed] [Google Scholar]
- Robinson, P. H., and Erasmus L. J.. . 2009. Effects of analyzable diet components on responses of lactating dairy cows to Saccharomyces cerevisiae based yeast products: a systematic review of the literature. Anim. Feed Sci. Technol. 149:185–198. doi: 10.1016/j.anifeedsci.2008.10.003 [DOI] [Google Scholar]
- Rohart, F., Gautier B., Singh A., and Lê Cao K. A.. 2017. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13:e1005752. doi: 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, D. M., and Weimer P. J.. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 75:165–174. doi: 10.1007/s00253-006-0802-y [DOI] [PubMed] [Google Scholar]
- Stewart, C. S., Flint H. J., and Bryant M. P.. . 1997. The rumen bacteria . In: Hobson, P. S., and Stewart C. S., editors. The rumen microbial ecosystem. 2nd ed. London (UK):Blackie Academic and Professional; pp. 10–72 [Google Scholar]
- Trotta, R. J., Klotz J. L., and Harmon D. L.. . 2018. Effects of source and level of dietary energy supplementation on in vitro digestibility and methane production from tall fescue-based diets. Anim. Feed Sci. Technol. 242:41–47. doi: 10.1016/j.anifeedsci.2018.05.010 [DOI] [Google Scholar]
- Ueki, A., Akasaka H., Satoh A., Suzuki D., and Ueki K.. . 2007. Prevotella paludivivens sp. nov., a novel strictly anaerobic, Gram-negative, hemicellulose-decomposing bacterium isolated from plant residue and rice roots in irrigated rice-field soil. Int. J. Syst. Evol. Microbiol. 57(Pt 8):1803–1809. doi: 10.1099/ijs.0.64914-0 [DOI] [PubMed] [Google Scholar]
- Wallace, R. J., McKain N., Broderick G. A., Rode L. M., Walker N. D., Newbold C. J., and Kopecny J.. . 1997. Peptidases of the rumen bacterium, Prevotella ruminicola. Anaerobe 3:35–42. doi: 10.1006/anae.1996.0065 [DOI] [PubMed] [Google Scholar]
- Walters, W., Hyde E. R., Berg-Lyons D., Ackermann G., Humphrey G., and Parada A. E.. . 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems. 1:e00009–15. doi: 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y., and Li L.. . 2012. Determination of total concentration of chemically labeled metabolites as a means of metabolome sample normalization and sample loading optimization in mass spectrometry-based metabolomics. Anal. Chem. 84:10723–10731. doi: 10.1021/ac3025625 [DOI] [PubMed] [Google Scholar]
- Xu, M., Rinker M., McLeod K. R., and Harmon D. L.. . 2010. Yucca schidigera extract decreases in vitro methane production in a variety of forages and diets. Anim. Feed Sci. Technol. 159:18–26. doi: 10.1016/j.anifeedsci.2010.05.005 [DOI] [Google Scholar]
- Xue, M., Sun H., Wu X., and Liu J.. . 2018. Assessment of rumen microbiota from a large dairy cattle cohort reveals the pan and core bacteriomes contributing to varied phenotypes. Appl. Environ. Microbiol. 84. doi: 10.1128/AEM.00970-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. Z., Beauchemin K. A., Vedres D. D., Ghorbani G. R., Colombatto D., and Morgavi D. P.. . 2004. Effects of direct-fed microbial supplementation on ruminal acidosis, digestibility and bacterial protein synthesis in continuous culture. Anim. Feed Sci. Technol. 114:179–193. doi: 10.1016/j.anifeedsci.2003.12.010 [DOI] [Google Scholar]
- Yang, J. Y., Seo J., Kim H. J., Seo S., and Ha J. K.. . 2010. Nutrient synchrony: is it a suitable strategy to improve nitrogen utilization and animal performance? Asian-Australas. J. Anim. Sci. 23:972–979. doi: 10.5713/ajas.2010.r.04 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.