The two most common causes of premature graft loss in renal transplantation, death with function and alloimmune-mediated injury, are inter-related; immunosuppressive medications used to prevent alloimmune injury also increase the risk of the most common causes of death with function (i.e., cardiac, infection, cancer). In the context of a heterogeneous population, the challenge is to administer the right therapy to the right recipient, personalizing the degree of immunosuppression in proportion to the individual’s alloimmune risk to minimize drug toxicity while maintaining therapeutic efficacy. Unfortunately, studies document that center practice accounts for the majority of the variation in immunosuppression selection rather than individualized alloimmune risk assessment as recommended in the Kidney Disease Improving Global Outcomes guidelines.1,2 Moreover, the standard of care has escalated to administering the highest level of immunosuppression tolerated to all recipients, resulting in clinicians being reactive, not proactive, in adjusting immunosuppression: increasing for rejection and decreasing in response to infection or off-target toxicities (Figure 1A). To permit individualized treatment and immune monitoring strategies, an essential requirement is the availability of reliable prognostic or predictive alloimmune risk biomarkers. However, attempts to identify “low-risk” recipients for immunosuppression minimization using traditional “immune” assessment criteria have been unsuccessful, underscoring the need for novel approaches to understand and quantify alloimmune risk.3
Figure 1.
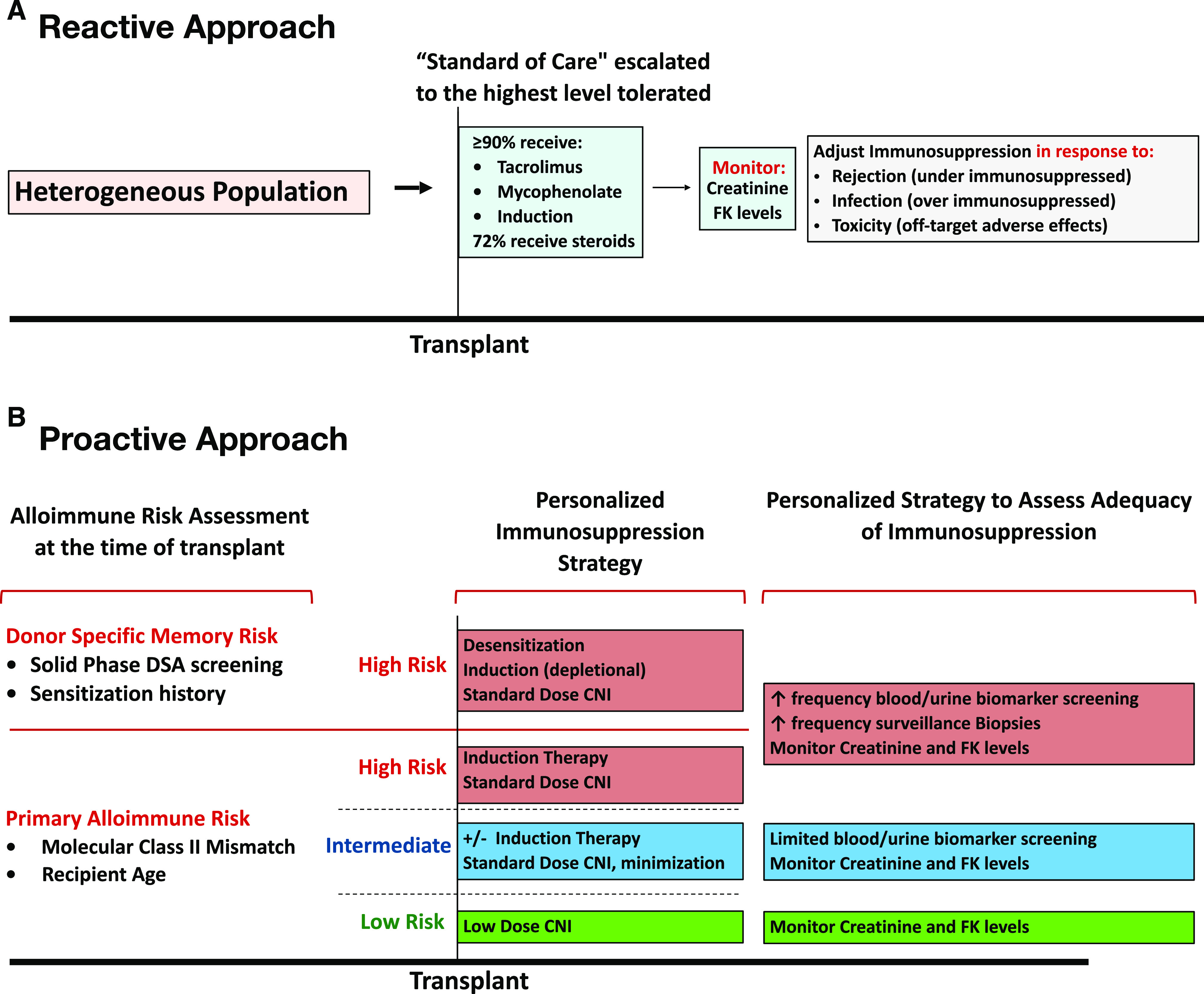
Precise alloimmune risk assessment enables a proactive immunosuppression and monitoring strategy. (A) Current immunosuppression and monitoring strategies are largely empirical and reactive. (B) In the future, HLA molecular mismatch may be part of a proactive approach where treatment and monitoring regimens are personalized to the underlying alloimmune risk. However, randomized, prospective trials are required to evaluate the risks and benefits of this approach. CNI, calcineurin inhibitors; DSA, donor-specific antibody; FK, tacrolimuis.
Since the first twin studies in the 1950s, it has been appreciated that donor-recipient HLA mismatch correlates with alloimmune risk and drives the need for immunosuppression. Advancements in HLA typing methods have revealed the breadth of HLA diversity and have been vital to understanding HLA antibody specificity when interpreting crossmatch results. Nevertheless, when HLA mismatch is used as a correlate of primary alloimmune risk, most centers still rely on the HLA-A/B/DR antigen mismatch result that has been available since the 1980s. It is now known that each HLA-A/B/DR/DQ antigen mismatch is actually the result of a range from 1 to >100 surface-exposed polymorphic amino acid mismatches.4 For example, a single HLA-DR antigen mismatch for one recipient may only result in one or two nonself amino acids exposed to the immune system from that molecule, whereas another recipient with the same single HLA-DR antigen mismatch may be exposed to 20- to 50-fold more nonself amino acids. Because this is true for each single HLA molecule mismatched, far more precision in HLA mismatch assessment is readily available when analyzed at the molecular level, leading to a rebirth in quantifying the degree of donor-recipient relatedness at individual HLA loci.
Although multiple methods of analyzing the HLA donor-recipient molecular mismatch have now been reported, the most widely published has been the HLA Matchmaker method.5 Here, small clusters of surface-exposed amino acids named “eplets” are identified and compared across each donor-recipient HLA mismatch. The degree of eplet mismatch has been reported to correlate with de novo donor-specific antibody (dnDSA) development, antibody-mediated rejection, transplant glomerulopathy, T cell–mediated rejection, and allograft failure.6–9
In JASN, Senev et al.10 studied a large cohort of renal transplant recipients and found a significant correlation between eplet mismatch and dnDSA, T cell–mediated rejection, antibody-mediated rejection, and graft failure. HLA-DQ dnDSAs were the most common, and the HLA-DQ eplet mismatch was highly correlated with dnDSA development (C statistic 0.89). Despite only ten HLA-DR dnDSA events, HLA-DR eplet mismatch was also significantly associated with HLA-DR dnDSA development and T cell–mediated rejection (Supplemental Tables 2 and 7 in ref. 10), highlighting the need to consider the degree of both HLA-DR and -DQ eplet mismatch. Although the absolute number of dnDSA events was low (4.6%), eplet mismatch outperformed antigen mismatch as a correlate of dnDSA development, emphasizing the benefits of HLA molecular mismatch assessment. Like Senev et al.,10 our group also reported that eplet mismatch had a continuous linear relationship with dnDSA development when analyzed in an HLA locus–specific fashion (i.e., each eplet mismatch at a given HLA locus increases the risk for dnDSA toward that locus).11 Nevertheless, in order to develop a tool for enrichment or stratification in clinical trial design, we developed statistical thresholds in order to generate alloimmune risk groups (low, intermediate, high).11 These same single-molecule alloimmune risk groups were also significantly correlated with dnDSA development in the Senev et al.10 cohort reported here. In fact, only three of 369 recipients (0.8%) developed donor-specific antibody if their single-molecule mismatch scores fell below previously identified “low-risk” thresholds for both HLA-DR and -DQ, whereas the prevalence was more than eightfold greater (6.8%) in patients in the “high-risk” single-molecule HLA-DR/DQ alloimmune risk category (Supplemental Figure 7 in ref. 10). Unfortunately, medication adherence and/or drug-level data were not known for the rare patients who developed dnDSA despite a low-risk mismatch to understand the drivers of donor-specific antibody formation in these low-risk patients.
The renaissance of HLA mismatch evaluation is rapidly evolving and is likely to become the standard of care. Still, further research is necessary to evaluate the full utility of HLA molecular mismatch in cohorts with greater ethnic diversity because the report by Senev et al.10 was 98% white. In addition, prior to widespread adoption, prospective studies are needed to evaluate the utility of HLA molecular mismatch as a prognostic/predictive biomarker in the clinical setting. As well, the optimal computational approach to evaluating HLA molecular mismatch is yet to be determined. Nevertheless, measuring alloimmune risk in a more precise way at the time of transplantation, after it is fully developed, will transform transplant medicine by informing allocation policy and clinical trial design as well as personalized therapy and immune monitoring strategies (Figure 1B).
Disclosures
All authors have nothing to disclose.
Funding
C. Wiebe and P.W. Nickerson are funded by an operating grant from the Paul I. Terasaki Research Fund.
Acknowledgments
The content of this article reflects the personal experience and views of the author(s) and should not be considered medical advice or recommendations. The content does not reflect the views or opinions of the American Society of Nephrology (ASN) or JASN. Responsibility for the information and views expressed herein lies entirely with the author(s).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related article, “Eplet Mismatch Load and De Novo Occurrence of Donor-Specific Anti-HLA Antibodies, Rejection, and Graft Failure after Kidney Transplantation: An Observational Cohort Study,” on pages 2193–2204.
References
- 1.Dharnidharka VR, Naik AS, Axelrod DA, Schnitzler MA, Zhang Z, Bae S, et al. : Center practice drives variation in choice of US kidney transplant induction therapy: A retrospective analysis of contemporary practice. Transpl Int 31: 198–211, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kidney Disease: Improving Global Outcomes (KDIGO) Transplant Work Group: 2009. transplant recipient guideline. Available at: https://kdigo.org/wp-content/uploads/2017/02/KDIGO-2009-Transplant-Recipient-Guideline-English.pdf. Accessed July 16, 2020
- 3.Hricik DE, Formica RN, Nickerson P, Rush D, Fairchild RL, Poggio ED, et al. ; Clinical Trials in Organ Transplantation-09 Consortium : Adverse outcomes of tacrolimus withdrawal in immune-quiescent kidney transplant recipients. J Am Soc Nephrol 26: 3114–3122, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kramer CSM, Koster J, Haasnoot GW, Roelen DL, Claas FHJ, Heidt S: HLA-EMMA: A user-friendly tool to analyse HLA class I and class II compatibility on the amino acid level. HLA 96: 43–51, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duquesnoy RJ, Askar M: HLAMatchmaker: A molecularly based algorithm for histocompatibility determination. V. Eplet matching for HLA-DR, HLA-DQ, and HLA-DP. Hum Immunol 68: 12–25, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiebe C, Pochinco D, Blydt-Hansen TD, Ho J, Birk PE, Karpinski M, et al. : Class II HLA epitope matching-A strategy to minimize de novo donor-specific antibody development and improve outcomes. Am J Transplant 13: 3114–3122, 2013. [DOI] [PubMed] [Google Scholar]
- 7.Wiebe C, Rush DN, Nevins TE, Birk PE, Blydt-Hansen T, Gibson IW, et al. : Class II eplet mismatch modulates tacrolimus trough levels required to prevent donor-specific antibody development. J Am Soc Nephrol 28: 3353–3362, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sapir-Pichhadze R, Tinckam K, Quach K, Logan AG, Laupacis A, John R, et al. : HLA-DR and -DQ eplet mismatches and transplant glomerulopathy: A nested case-control study. Am J Transplant 15: 137–148, 2015. [DOI] [PubMed] [Google Scholar]
- 9.Wiebe C, Rush DN, Gibson IW, Pochinco D, Birk PE, Goldberg A, et al. : Evidence for the alloimmune basis and prognostic significance of borderline T cell-mediated rejection [published online ahead of print March 17, 2020]. Am J Transplant 10.1111/ajt.15860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senev A, Coemans M, Lerut E, Van Sandt V, Kerkhofs J, Daniëls L, et al. : Eplet mismatch load and de novo occurrence of donor-specific anti-HLA antibodies, rejection, and graft failure after kidney transplantation: An observational cohort study. J Am Soc Nephrol 31: 2193–2204, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiebe C, Kosmoliaptsis V, Pochinco D, Gibson IW, Ho J, Birk PE, et al. : HLA-DR/DQ molecular mismatch: A prognostic biomarker for primary alloimmunity. Am J Transplant 19: 1708–1719, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]


