Abstract
Perforin-2 (P-2) is a recently described antimicrobial protein with unique properties to kill intracellular bacteria. We investigated P-2 expression pattern and cellular distribution in human skin and its importance in restoration of barrier function during wound healing process and infection with the common wound pathogen Staphylococcus aureus. We describe a novel approach for the measurement of P-2 mRNA within individual skin cells using an amplified fluorescence in situ hybridization (FISH) technique. The unique aspect of this approach is simultaneous detection of P-2 mRNA in combination with immune-phenotyping for cell surface proteins using fluorochrome-conjugated antibodies. We detected P-2 transcript in both hematopoietic (CD45+) and non-hematopoietic (CD45-) cutaneous cell populations, confirming the P-2 expression in both professional and non-professional phagocytes. Furthermore, we found induction of P-2 during wound healing. P-2 overexpression resulted in reduction of intracellular S. aureus, while infection of human wounds by this pathogen resulted in P-2 suppression revealing a novel mechanism by which S. aureus may escape cutaneous immunity to cause persistent wound infections.
Keywords: Perforin-2, skin, human wound, intracellular bacteria, Fish-Flow RNA Assay
Introduction
Wound healing is complex multistep process in which inflammatory response precedes proliferative phase, followed by epithelialization and remodeling [1, 2]. However, presence of bacterial infection often impedes wound closure and barrier restoration [3]. Staphylococcus aureus is the most common pathogen causing wound infections [3, 4] and is capable of evading host defense [5–9]. While S. aureus is the prevalent pathogen in multiple cutaneous conditions including non-healing wounds and atopic dermatitis [10–13], the mechanisms by which this pathogen persists in skin infection remain largely unknown.
Perforin-2 (P-2) is a membrane-attack-complex-perforin (MACPF) domain containing protein important for killing Gram-positive and Gram-negative intracellular bacteria [14–20]. The data suggest that upon bacterial infection P-2 vesicles translocate to bacterium containing endosomes or phagosomes or the plasma membrane. After membrane fusion the MACPF domain of the Perforin-2 is located next to the bacterial surface. P-2 polymerization and insertion into the bacterial wall results in pore formation and killing of the bacterium [15]. Importantly, we have previously shown that P-2 constitutive expression in the epidermis plays important role in protection from infection and invasion by S. aureus in a mouse model [16]. As a consequence of bacteremia, P-2−/− mice rapidly lost weight suggesting forthcoming mortality [16]. Here, we aimed to address specific questions regarding the role of the human P-2 in cutaneous wound healing.
We applied and optimized FISH-Flow method (by PrimeFlow, ThermoFisher/eBioscience) to the human full thickness skin samples to study the expression of P-2 on the single cell level. The unique aspect of this technique is that it amplifies the fluorescent signal rather than the target up to 8000-fold, resulting in the detection of as few as 1 copy of the target RNA, allowing detection of low expression genes such as P-2. In addition, we were able to differentiate between different major skin cell subsets important for wound healing (keratinocytes, fibroblasts, hematopoietic and immune cells) by staining for cell surface markers (CD45, TCR gamma delta, CD104, CD31, CCLR1 and CD325). While the current technique focuses on P-2 expression in healthy human skin cells, this method can be broadly applied to the variety of different genes and cutaneous conditions.
Here we report novel findings regarding differential cellular distribution of human P-2 and differential level of P-2 expression during acute wound healing in the absence and the presence of S. aureus infection. The major P-2 expressing cell populations in the intact skin include Gamma delta (GD) T cells, basal keratinocytes and endothelial cells. P-2 expression was induced by wounding in human skin and overexpression of P-2 resulted in decreased intracellular infection by S. aureus. The presence of infection resulted in P-2 suppression indicating a novel mechanism by which S. aureus escapes cutaneous immunity.
Materials and Methods
Cell lines, plasmids and transfection
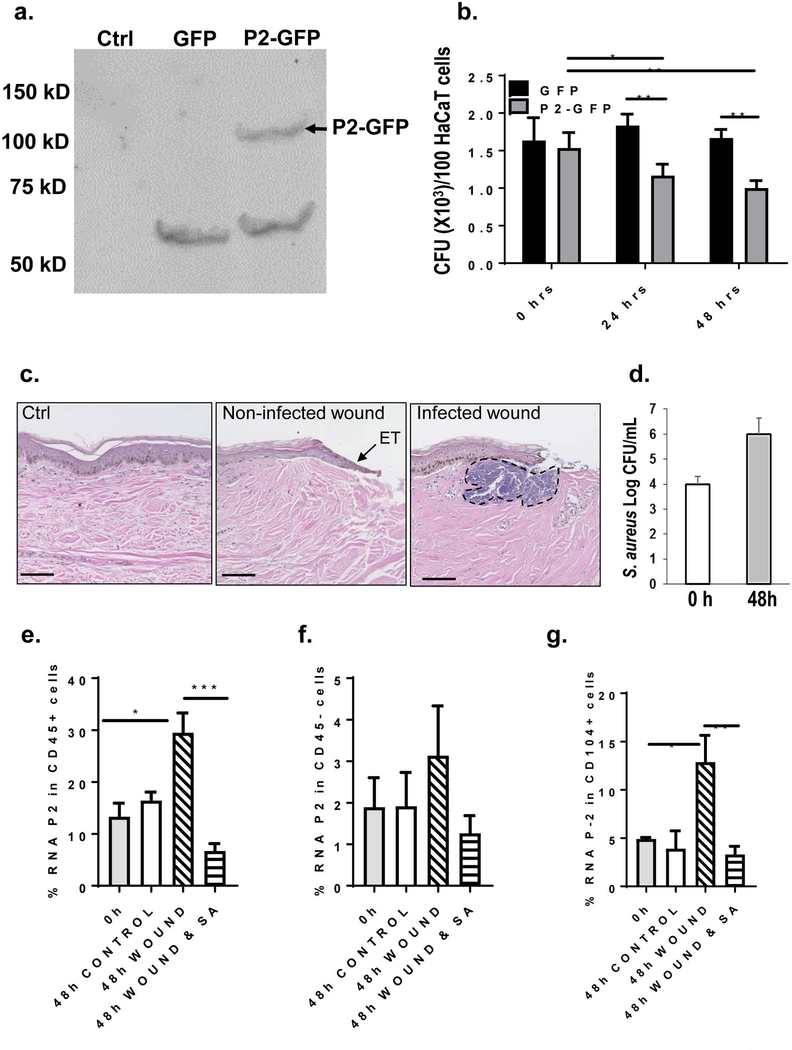
HEK-293 (CRL-1573) a cell line was obtained from American Type Culture Collection (ATCC), (Manassas, VA). Murine embryonic fibroblasts (MEFs) were isolated as previously described [16]. All cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 following ATCC recommendations for culture conditions. Coding sequence of human Mpeg1 (perforin-2) was assembled from several expressed sequence tag (EST) clones and tagged at the C-terminus with the green fluorescent protein (GFP) (Clontech, Thermo Fisher Scientific, Waltham, MA ) [16, 19]. Stable transfection with P-2-GFP expressing plasmid in human keratinocyte cell line, HaCaT cells, was established after selection with hygromycin (Thermo Fisher Scientific). Constitutive expression of P-2-GFP was confirmed by Western blotting. 20 μg of whole cells lysate was resolved on 4–20% SDS PAGE and membrane was probed with rabbit polyclonal anti-GFP antibody (Abcam, Cambridge, MA).
Single cell suspension from human skin and ex vivo wounds
Discarded human skin tissue was obtained from voluntary reduction surgeries (n = 6) at the University of Miami (UM) Hospital and were found to be exempt from human subject research under CFR46.101.2 by the institutional review board at the UM Miller School Of Medicine.
Full thickness skin samples were minced with surgical scissors and incubated for 30 minutes in dispase II solution (Roche, Basel, Switzerland)(2.4 μg/mL) at 37°C. After this, tissue samples were incubated at 37°C for 3 hours with 2 mg/ml Collagenase D (Roche) at 37°C under constant agitation. Obtained cell suspensions were washed with DMEM (Gibco-Thermo Fisher Scientific) supplemented with 10% heat-inactivated FBS, 2 mM L -glutamine, 0.15% sodium hydrogencarbonate, 1 mM sodium pyruvate, nonessential amino acids, 100 μg/ml gentamycin. To separate epidermis and dermis skin samples were cut in 1–2 mm strips and incubated overnight with Dispase II (2.4 U/ml) at 4°C s. Epidermal cells were obtained by physical dissociation of epidermal sheets and vigorous pipetting. Dermis was additionally incubated for 1 h with 2 mg/ml collagenase D (Roche) at 37°C.
FISH-Flow P-2 RNA Assay and Flow cytometric analysis
The FISH-Flow assay workflow contains several steps: surface antibody staining; fixation and permeabilization followed by target probe hybridization, with P-2 mRNA-specific probe sets labeled with Alexa Fluor 647 (Type 1 Probe Sets); signal amplification using branch (b) DNA constructs and detection by flow cytometry (Supp. Figure 2).
Single cell suspensions obtained from full thickness, epidermal and dermal samples were first labeled with live/dead detection kit and then with following fluorescently labeled antibodies CD45-Alexa Fluor 700, TCR GD-PE-Cy7, CD31-PacBlue, CD104-FITC, CD325-PerCPCy5.5 and CCRL1-PE (Biolegend, San Diego, CA). Perforin-2 (P2) mRNA was detected using an amplified signal FISH technique (PrimeFlow; Affymetrix/eBioscience-Thermo Fisher Scientific). For mRNA detection, target probe hybridization was performed using type 1 (AlexaFluor647) probes for Perforin-2. Skin cells were incubated for 2 h with the target probes in a precisely calibrated incubator at 40°C. All samples were then incubated with the PreAmplification (PreAmp) reagent for 2-h and the Amplification (Amp) reagent for an additional 2-h at 40°C. After signal amplification, skin cells were incubated with label P-2 probes or control genes GAPDH, β-actin at 40°C for 1 h. Cells were washed and suspended in staining buffer prior to acquisition. Approximately 20,000 cell events were acquired from each sample on flow cytometer equipped with 405 nm, 488 nm, 642 nm, and 785 nm (SSC) lasers (Fortessa, BD, San Jose, CA). Spectral compensation was completed using single color control samples.
Immunofluorescence and immunohistochemistry
5–7 μm thick formalin-fixed, paraffin-embedded tissue sections were deparaffinized with xylene (EMD, Gibbstown, NJ), rehydrated, and H&E stained or processed for immunostaining as previously described [5, 21, 22]. Anti CD104 mouse monoclonal antibody 1:50 (BioLegend,) was used overnight at 4°C. Signal was visualized using Alexa-Fluor 488 secondary antibody (Invitrogen, Carlsbad, CA) and mounted with media containing 4’,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA) to visualize nuclei. Nikon Eclipse E800 microscope/NIS Elements BR 3.2 software was used for collection of digital images. 5–7 μm thick frozen tissue sections were fixed in cold acetone (−20°C) for 10 min washed in TBS (Tris-buffered saline) and incubated with 3% H2O2 in methanol for 10 min. Staining was performed as previously described [22, 23] using anti CD31 antibody (Serotec cat # MCA1746GA, 1:25) and anti CD45 (Dako cat # M0701, 1:100). Nikon Eclipse E800 microscope/NIS Elements BR 3.2 software was used for collection of digital images.
Bacterial strains and growth conditions
S. aureus isolate from diabetic foot ulcer (DFU) was utilized for the wound infection as described [5]. Todd-Hewitt broth supplemented with 0.2% yeast extract (THY) served as growth medium; tryptic soy agar with 5% sheep blood was used for colony forming units (CFU) quantification.
Intracellular S. aureus killing assay
HaCaT cell line constitutively expressing either P2-GFP or GFP, were seeded in DMEM without the antibiotics and challenged with S. aureus chronic wound isolate with multiplicity of infection (MOI) 50:1. After 1 hour challenge, cells were washed and remaining extracellular bacteria were lysed with lysostaphin and incubation continued for 24 or 48 h. Cells were than lysed in 0.1% Triton X-100 and lysate plated in technical triplicates on blood agar plates. CFU count was determined after overnight colony growth and expressed per 100 HaCaT cells.
Human ex vivo wound infection model
Healthy human skin samples were used to generate acute wounds. Wounded skin specimens were maintained at the air-liquid interface as previously described [5, 22, 24]. S. aureus isolate from DFU [5] was grown overnight in THY medium at 37 °C, then harvested by centrifugation, washed and re-suspended in DMEM. 104 CFU of S. aureus was used for the wound inoculation. Wounded, unwounded, and infected and non-infected skin samples were incubated at 37°C, 5% CO2 for two days. Tissue samples were either used for Prime-Flow analyses, CFU determination, fixed in 4% paraformaldehyde (Sigma-Aldrich) for H&E and histo-morphometric analyses [25].
Statistical Analyses
Comparisons of flow cytometry cell frequencies was done by using the non-parametric two-tailed Mann-Whitney U test using the Prism software (GraphPad software). Error bars in all figures are reported as a SD of the mean. CFU count was determined after overnight colony growth and expressed per 100 cells. Statistical significance was measured by two-way Anova test, * p<0.05, ** p<0.01 and *** p<0.001
Results
Detection of human P-2 mRNA by FISH-Flow Assay
We have used a novel application of an amplified, fluorescent in situ hybridization (FISH) technique in combination with flow cytometry to analyze a P-2 mRNA expression in different epidermal skin cell subsets. First, we have validated FISH-Flow RNA Assay with human cell line, HEK 293 cells and MEF. Both cell lines were transfected with human P-2-GFP fusion protein or GFP only (Figure 1a-c). We have confirmed expression of control “house-keeping” genes: GAPDH and β actin (Figure 1b). Furthermore, we have tested the assay by comparing expression of human P-2 RNA (hP-2) in MEF-GFP and MEF-hP-2-GFP expressing cells. Human P-2-GFP fusion construct has been generated previously [16] and hP-2-GFP transfected MEF cells (MEF-hP2-GFP) have been extensively used in in vitro infection experiments [16, 17, 19]. As expected, we found that FISH-Flow Assay very efficiently and specifically detects hP-2 mRNA only in MEF-hP-2-GFPcell line while MEF-GFP cells were all negative for hP-2 RNA (Figure 1c). In addition, FISH-Flow Assay revealed the unique kinetics of mRNA and protein in the same cell. We found that hP-2 RNA is not expressed in all GFP positive cells, suggesting that post transcriptional modifications of mRNA may be the reasons for low P-2 mRNA levels. (Figure 1c). We conclude that FISH-Flow RNA Assay enables quantification of P-2 gene expression at the single-cell level. In addition, this method has the ability to elucidate the kinetics of mRNA and protein expression simultaneously.
Figure 1. Optimization of detection of human P-2 (hP-2) mRNA by FISH-Flow RNA Assay.
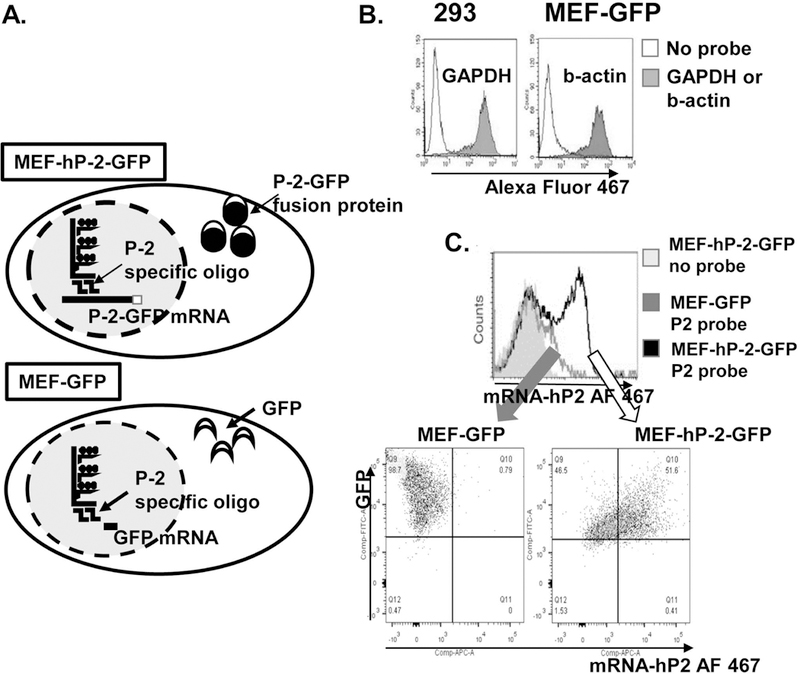
a. Human P-2 gene can be easily detected in a mouse embryonal fibroblast (MEF) cell line overexpressing human P-2-GFP fusion protein used as a positive control for the FISH-Flow Assay optimization. b. Flow cytometry data showing glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RNA expression in 293 cells and mouse b-actin in MEF cell line expressing GFP used as negative controls for the assay optimization. c. Representative overlay histogram showing hP-2 mRNA and gfp protein expression in MEF-GFP and MEF-hP2GFP cells by flow cytometry. The signal-to-noise ratio was calculated as the ratio between the mean fluorescence intensity (MFI) of the hP-2 gene and the MFI of the no probe control.
Both, non-hematopoietic and hematopoietic skin cells express P-2
We analyzed the frequency and P-2 expression in different cell subtypes within the CD45+ and CD45− cells obtained from human skin (Supp. Figure 1 and Figure 2). We confirmed the presence of epidermal and dermal CD45+ by immunohistochemistry and flow cytometry (Supp. Figure 1). Single cell suspensions were obtained from human skin (n=6) and labelled with antibody against CD45 molecule. Flow cytometric analysis of healthy human skin revealed that majority of the epidermal cells are large granular cells, mostly CD45 negative cells, keratinocytes. Dermal cell population contained higher proportion of CD45 positive cells (29%), as expected. Analysis of full thickness human skin showed the expected mixture of both, large and smaller cells and we found 83% +/−SEM 5.6 of CD45 negative cells and 16.8% +/− 5.8 of CD45 positive cells (Supp. Figure 1), confirming that utilization of the full thickness human skin samples for determination of cellular distribution of Perforin-2.
Figure 2. P-2 RNA is expressed in both CD45 negative and CD45 positive cells in healthy human skin.
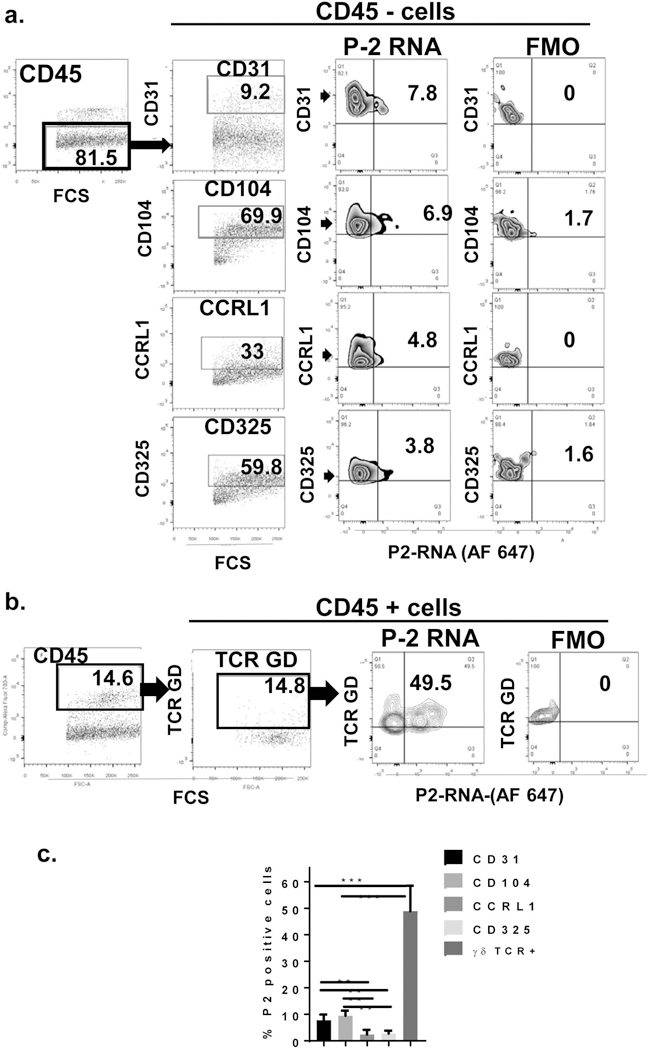
Single cell suspension from full thickness normal skin was obtained and labelled with antibodies against following surface markers: CD45, TCR GD, CD31, CD104, CCRL1 and CD325 and live/dead staining a) Live, CD45 negative and b) positive and cells were analyzed for the expression of specific markers, and P-2 RNA expression is determined within the each cell subset. Representative dot plots showing frequency of specific markers (cell subsets) and P-2 RNA. Fluorescence-Minus-One (FMO) is a sample that contains all of the fluorochromes without P-2 probe labelled with Alexa Fluor 647 and was used as a negative control c) Summary graph with frequency of P-2 positive cells within different cell subsets in the skin (n=6). All error bars are SD of the mean. **p<0.01, ***p<0.001.
We included several surface markers: TCR G=GD receptor, specifically expressed on subset of T cells, known as GD T cells [26–29]; CD31, platelet endothelial cell adhesion molecule (PECAM-1) expressed primarily on endothelial cells [30]; CD104, Integrin beta 4, interacts with α6 integrin and serves as receptor for laminin and component of hemidesmosome and is found in the basal layer [31–35]; C-C Motif Receptor-Like 1(CCRL1) and CD325 also known as Cadherin-2 (CDH2) or neural cadherin (NCAD) are novel markers of papillary and reticular fibroblasts, respectively [36, 37]. We found that both, hematopoietic (CD45 positive) as well as non-hematopoietic (CD45 negative) cells in the skin express P-2 (Figure 2a). We also found that all cell subsets analyzed (TCR GD+, CD104+, CCRL1+ and CD325+ cells) express P-2 (Figure 2a and 2b). Dermal fibroblasts come from 2 distinct lineages of cells, one populating the papillary dermis and the other lineage populating the reticular dermis. We have used distinct cell markers to distinguish between these two populations: CCRL1 (papillary) and CD325 (reticular) [36, 37]. We found low frequency of both fibroblasts populations expressing P-2. GD T cells (GD TCR+), basal keratinocytes (CD104+ cells) and endothelial cells (CD31+ cells) represent the major P-2 expressing cells population in the human skin (Figure 2c). This data identified cell type specificity in distribution and localization of P-2 mRNA in human skin.
Differential expression of P-2 within basal keratinocytes
Basal keratinocytes are known to express CD104, integrin beta 4 [32, 33, 35]. We confirmed the CD104 expression in the basal epidermal layer by immunostaining (Figure 3a.). Since we also confirmed by flow cytometry the differential level of expression of CD104 (Figure 3b) we correlated CD104 levels with expression of P-2 in the basal epidermal keratinocytes. We observed distinct levels of P-2 expression regarding the level of CD104. Frequency of P-2 RNA-expressing cells was increased within the cells that express very high level of CD104 marker while the CD104 negative, differentiated cells had the lowest expression of P-2 RNA (Figure 3b, p=0.0001). The primary cultured keratinocytes show similar findings (Figure 3c). Taken together, these data support the conclusion that P-2 is primarily expressed in basal keratinocytes expressing high level of CD104.
Figure 3. Differential expression of P-2 RNA in keratinocytes as assayed by FISH-Flow.
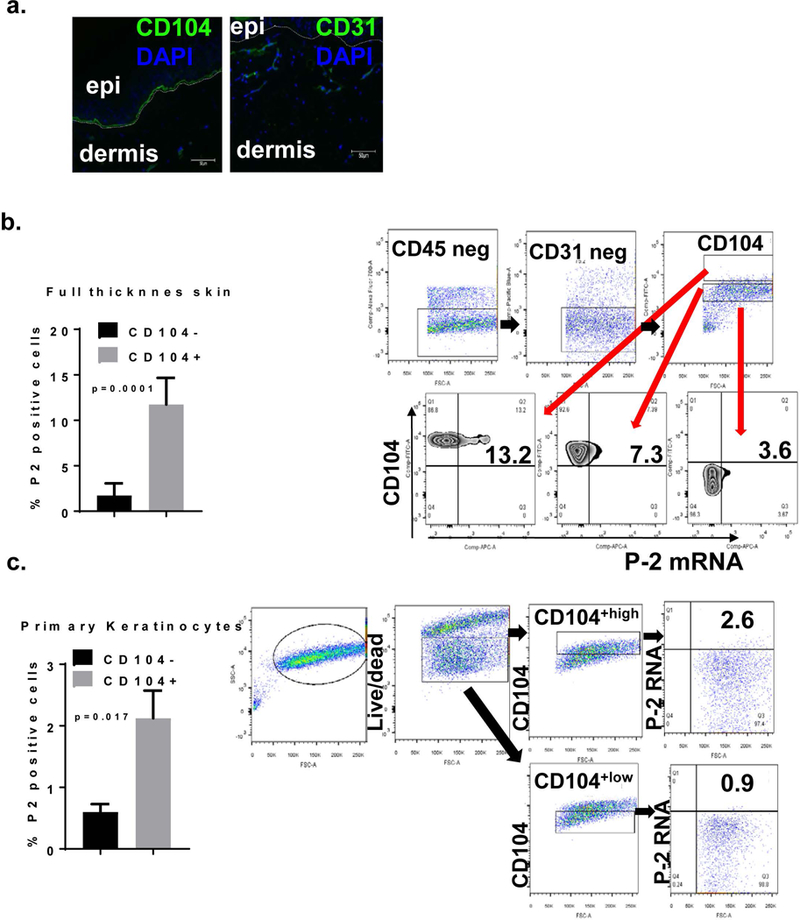
a. Immunofluorescence for CD104 and CD31 on normal skin. b. Full thickness normal skin samples were processed, single cell suspension obtained and labelled with CD45, CD31, CD104 and live/dead markers followed by in situ hybridization for. Cells were first analyzed for the lack of expression of CD45 and CD31 and then for CD104 expression. Electronic gates were set on the cells that express high, intermediate/low or negative levels CD104 and P-2 RNA levels were analyzed within these cell subsets. c. Primary human cultured keratinocytes were analyzed for P-2 RNA expression within CD104 +high and CD104+low subsets. All error bars are SD of the mean.
P-2 kills intracellular S. aureus
In order to determine how P-2 in the cells affects their ability to kill intracellular bacteria we generated an epithelial cell line, constitutively expressing P-2-GFP fusion protein (Figure 4a). We used cells that express P-2 to determine the rate of intracellular S. aureus bacterial killing (Figure 4b). Human keratinocyte cell line, HaCaT cells, were incubated with S. aureus for 1 h to allow attachment and infection, followed by elimination of all extracellular bacteria by lystostaphin treatment. We observed significant difference in the number of intracellular bacteria between the cells that overexpress P-2-GFP and control cells that express only GFP (Figure 4b). Although the overexpression of P-2-GFP protein levels are not necessarily comparable to % of cells positive for P-2 mRNA these experiments are designed for functional validation. Indeed, overexpression of P-2 resulted in reduction of intracellular S. aureus at time point 24 h and 48 h compared to initial bacterial load (p<0.01 and p<0.001), although we did not observe complete elimination of bacteria. These results suggest that keratinocytes can efficiently kill intracellular S. aureus by action of P-2.
Figure 4. P-2 has antimicrobial activity against intracellular S. aureus in vitro, while S. aureus suppresses P-2 in human ex vivo wounds.
a) Immunoblot showing transient overexpression of GFP or human P2-GFP fusion protein in human keratinocytes cell line (HaCaT cells), Ctrl=non-transfected HaCaT cells. 20 μg of whole cells lysate was resolved on 4–20% SDS PAGE and membrane was probed with anti-GFP antibody. Arrow indicates the P2-GFP band with the expected molecular weight of 100 kD. b) HaCaT cells transiently transfected with either P2-GFP (in gray) or GFP (in black) were seeded 24 h post transfection in DMEM without the antibiotics and challenged with S. aureus with multiplicity of infection (MOI) 50:1. After 1 h challenge, cells were washed and extracellular bacteria were lysed with lysostaphin after which the HaCaT cells were either lysed with 0.1% Triton X-100 (time point zero) or further incubated in the presence of lysostaphin for 24 or 48 h. HaCaT cells were than hypotonicaly lysed and lysates plated in technical triplicates on blood agar plates for CFU enumeration. Statistical significance was measured by two-way Anova test, * p<0.05, ** p<0.01. c) Representative histology showing unwounded human skin (Ctrl), non-infected and S. aureus infected wound 48 h post-wounding and infection. Arrow indicates epithelial tongue (ET) in non-infected wounds, black dashed line indicates bacterial aggregates in the infected wound. d) CFU quantification of S. aureus at the time of infection (0 h) and from the infected wounds 48 h post-infection (48 h); error bars indicate SD, n=4. Unwounded and wounded, infected and non-infected tissue was collected at baseline and 48 h after wounding for FISH-Flow. Single cells suspension was obtained using Collagenase D and labelled with antibody against CD45 and live/dead staining. Using FISH-Flow RNA assay, P-2 RNA levels were analyzed within e) hematopoietic (CD45+) and f) non-hematopoietic (CD45-) cells and g CD104+ cells. Bar graph showing SEM of P-2 mRNA positive cells within CD45+ (C), CD45− skin cells (D) and CD45-CD104+ cell population (E) (n=4). Comparisons of flow cytometry frequencies were done by using non-parametric two tail Mann-Whitney U test. * p<0.05, ***p<0.001.
Wounding upregulates expression of P-2 while S. aureus suppresses it
In order to further determine effect of wounding and wound infection on P-2 we utilized ex vivo human wound model. Ex vivo human skin model is widely used and validated model to study epithelialization in human skin [3, 5, 24]. We utilized this model to determine P-2 expression during wound healing in the presence or absence of S. aureus infection. All S. aureus infected wounds have shown growth of ~2 Log CFU (Figure 4c and d) resulting in inhibition of epithelial tongue formation (Figure 4c), confirming our previous data showing inhibition of re-epithelialization by S. aureus [5]. We found an increase in P2 expression in hematopoietic cells 48 h after wounding compared to control skin and non-wounded tissue (Figure 4e) On the other hand, S. aureus infection inhibited P-2 expression during the acute wound healing in CD45+ resident skin cells and has shown trend of inhibition in non-hematopoietic cells (Figure 4e and 4f). Furthermore we identified CD104+ population as a major target of S. aureus mediated P-2 suppression in a CD45− cells (Figure 4g).
Discussion
Normal innate skin mechanisms consist of multiple cell types and antimicrobial peptides (AMPs) that function as sentinels of immune system and prevent overgrowth of microorganisms. Recently, our group had discovered novel AMP, Perforin-2 (P-2) [16, 19]. P-2 is highly conserved innate protein with unique properties to kill intracellular (both Gram-positive and –negative) bacteria. P-2, encoded by mpeg1 gene, is transmembrane protein inserted in membrane vesicles, that forms pores on targets (intracellular bacteria) entrapped by membrane vesicles [14, 15, 18, 20, 38]. In professional phagocytic cells P-2 is expressed constitutively. In other cells P-2 expression can be induced by interferons (IFN), LPS and TNF-α, or by bacterial infection [16, 39].
The objective of this study was to determine P-2 expression patterns in human skin, and its role in wound epithelialization and response to infection with S. aureus, the most common cutaneous and wound pathogen. We applied a novel approach to analyze P-2 expression in human skin on a single cell resolution. FISH-Flow™ RNA Assay reveals the dynamics of RNA and protein expression within individual cells, facilitating unprecedented analysis of their correlation. This novel assay uses FISH enabling simultaneous detection of RNA transcripts in a single cell using a standard flow cytometer. Target-specific probe set hybridizes to specific regions across the target P-2 RNA sequence. Subsequent signal amplification requires that each half of a given oligonucleotide pair (total of 20–40 oligonucleotide pair) binds to the target P-2 mRNA in adjacent positions. This is superior approach as it specifically measures P-2 transcript thus overcoming the limitation of qPCR analyses which could also detect P-2 DNA due to intron-less nature of the mpeg1 gene.
We have shown that flow cytometric analysis of a single cell suspension obtained from different human skin compartments (epidermis, dermis or full thickness skin) can be easily combined with in situ hybridization protocol. In agreement with our previous findings [16], constitutive expression of P-2 was confirmed in hematopoietic (CD45+) as well as in non-hematopoeitic (CD45+) skin cells. Specifically, we have shown that P-2 is expressed in human skin within different cell subsets: keratinocytes, GD T cells, fibroblasts and endothelial cells. To the best of our knowledge this has not been reported previously.
We have further analyzed P-2 expression in both hematopoietic and non-hematopoietic cells. Since human epidermal keratinocytes (HEK) as well as human dermal fibroblast (HDF) are well known protagonists of the skin’s innate immune system and can act potentially as a reservoir of intracellular bacteria, i.e. as “nonprofessional phagocytes” we have further analyzed the expression of P-2 in these cell subsets. In agreement with other reports, we have documented CD104 preferential expression on basal keratinocytes. CD104 positive cells have been described as a minor population of multipotent epithelial stem/progenitor cells with the capacity for self-renewal and whose descendants give rise to epithelial cell lineages in vitro [40]. These findings support our hypothesis that P-2 constitutive expression in epidermis may play important role in protection from infection and invasion by pathogenic microorganisms. However, it is somewhat surprising that it P-2 is mostly expressed in “deeper” epidermis, i.e. basal keratinocytes and understanding its role is the focus of further investigation.
GD T cells are often the major T cell population in epithelial tissues including the skin, gut, and lung where they have been implicated in maintaining tissue integrity, defending against pathogens, and regulating inflammation [41, 42]. We found that almost 50% of all human GDT cells constitutively express P-2, suggesting that direct engagement of GDT in pathogen clearance through activation and polymerization of P-2. GDT cells have recently been identified as a potent source of innate IL-17 and implicated in host protection in murine models of S. aureus infection [43–46].
S. aureus is the most common cutaneous pathogen capable of evading host defense [5–9] [3, 4]. S. aureus residing inside human keratinocytes was associated with atopic dermatitis [6], while S. aureus colonization in chronic diabetic foot ulcers contributes to impaired DNA repair mechanisms, which subsequently serves as a facilitator for the persistent, unresolved inflammatory state found in these wounds. Despite the fact that S. aureus is the most prevalent pathogen in multiple cutaneous diseases [10–13], the mechanisms by which this pathogen persists in skin remain largely unknown.
Constitutive presence of P-2 in both non-hematopoietic and hematopoietic skin cells is suggesting a role in infection surveillance, while overexpression of P-2 contributed to reduction of S. aureus counts invading epithelial cells. However we observed differential expression pattern of P-2 after wound infection with S. aureus. Also, we have shown previously that S. aureus infection delays wound closure in ex vivo model [5], which could implicate S. aureus direct and indirect effect of on P-2 expression. Further studies focusing on S. aureus mediated P-2 suppression in skin are necessary to decipher the mechanism by which this suppression occurs. The dominant position of P-2 in bactericidal activity suggests that pathogenic bacteria must have mechanisms to evade, suppress or block P-2 activation. Recently, it was shown that enteropathogenic E. coli and Y. pseudotuberculosis inhibit P-2-dependent killing by blocking its intracellular trafficking through bacterial effector protein Cif [17]. Also, it has been shown that individuals with P-2 haplo-insufficiency suffer to higher rates of chronic and recurring infectious disease than the general population [20].
In addition we have observed induction of P-2 expression within CD45+ cells 48 h after wounding compared to P-2 expression in CD45+ cells isolated from unwounded skin. Based on these findings, we postulate that P-2 is necessary for both: bacterial clearance as well as the wound healing process. A corollary to this premise is that aberrant expression of P-2 leads to modification of wound environment leading to defective bacterial clearance, sustained inflammation and inhibition of wound healing thus contributing towards the chronicity of infection. In summary, the novel approach to study P-2 expression on a single-cell resolution revealed that P-2 is expressed in the skin and has an antimicrobial role. Lastly, S. aureus is capable of suppressing a P-2 in human wounds suggesting a novel mechanism by which this pathogen may cause persistent cutaneous infections.
Supplementary Material
Supplementary Figure 1. Human skin single cell analysis of hematopoietic (CD45+) and non-hematopoietic (CD45−) cells by flow cytometry.
a) Immunohistochemistry for CD45 in normal skin sample. b). Epidermis, dermis and full thickness skin samples were processed as described in Material and Methods and single cell suspension was labelled with antibody against CD45 molecule. Cells were analyzed by flow cytometry according to the granularity (SSC) and size (FSC) and expression of CD45 surface molecule. Representative dot plots for cells obtained from epidermis, dermis and full thickness skin specimens showing percent of CD45+ cells within each compartment in normal skin. c). Bar graph showing SEM of CD45 negative and positive cells within total cells in full thickness skin specimen (n=6).
Supplementary Figure 2. Illustration of a probe set design containing both left and right oligonucleotide pairs, and signal amplifications reagents: pre-amplifier, amplifier, and fluorescently labeled probes.
Acknowledgements
This work was supported by NIH/NINR R01NR015649 (MTC and NS). We dedicate this work to late Dr Eckhard Podack without whom studies of Perforin-2 would not be possible. We are thankful to Dr Liang (Lisa) Liang for technical support and all the members of Tomic-Canic and Strbo laboratories.
Footnotes
Conflict of interest Statement
Authors declare no conflict of interest.
References
- [1].Eming SA, Martin P, Tomic-Canic M, Wound repair and regeneration: mechanisms, signaling, and translation, Sci Transl Med 6(265) (2014) 265sr6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pastar I, Stojadinovic O, Yin NC, Ramirez H, Nusbaum AG, Sawaya A, Patel SB, Khalid L, Isseroff RR, Tomic-Canic M, Epithelialization in Wound Healing: A Comprehensive Review, Adv Wound Care (New Rochelle) 3(7) (2014) 445–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Pastar I, Nusbaum AG, Gil J, Patel SB, Chen J, Valdes J, Stojadinovic O, Plano LR, Tomic-Canic M, Davis SC, Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection, PLoS One 8(2) (2013) e56846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Valencia IC, Kirsner RS, Kerdel FA, Microbiologic evaluation of skin wounds: alarming trend toward antibiotic resistance in an inpatient dermatology service during a 10-year period, J Am Acad Dermatol 50(6) (2004) 845–9. [DOI] [PubMed] [Google Scholar]
- [5].Ramirez HA, Pastar I, Jozic I, Stojadinovic O, Stone RC, Ojeh N, Gil J, Davis SC, Kirsner RS, Tomic-Canic M, Staphylococcus Aureus Triggers Induction of MIR-15B-5P to Diminish DNA Repair and De-Regulate inflammatory Response in Diabetic Foot Ulcers, J Invest Dermatol (2017). [DOI] [PMC free article] [PubMed]
- [6].Soong G, Paulino F, Wachtel S, Parker D, Wickersham M, Zhang D, Brown A, Lauren C, Dowd M, West E, Horst B, Planet P, Prince A, Methicillin-resistant Staphylococcus aureus adaptation to human keratinocytes, MBio 6(2) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Williams MR, Nakatsuji T, Gallo RL, Staphylococcus aureus: Master Manipulator of the Skin, Cell Host Microbe 22(5) (2017) 579–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Williams MR, Nakatsuji T, Sanford JA, Vrbanac AF, Gallo RL, Staphylococcus aureus Induces Increased Serine Protease Activity in Keratinocytes, J Invest Dermatol 137(2) (2017) 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Iwamoto K, Moriwaki M, Niitsu Y, Saino M, Takahagi S, Hisatsune J, Sugai M, Hide M, Staphylococcus aureus from atopic dermatitis skin alters cytokine production triggered by monocyte-derived Langerhans cell, J Dermatol Sci 88(3) (2017) 271–279. [DOI] [PubMed] [Google Scholar]
- [10].Kalan L, Loesche M, Hodkinson BP, Heilmann K, Ruthel G, Gardner SE, Grice EA, Redefining the Chronic-Wound Microbiome: Fungal Communities Are Prevalent, Dynamic, and Associated with Delayed Healing, MBio 7(5) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].van Asten SA, La Fontaine J, Peters EJ, Bhavan K, Kim PJ, Lavery LA, The microbiome of diabetic foot osteomyelitis, Eur J Clin Microbiol Infect Dis 35(2) (2016) 293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA, The Neuropathic Diabetic Foot Ulcer Microbiome Is Associated With Clinical Factors, Diabetes 62(3) (2013) 923–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fadini GP, Menegazzo L, Rigato M, Scattolini V, Poncina N, Bruttocao A, Ciciliot S, Mammano F, Ciubotaru CD, Brocco E, Marescotti MC, Cappellari R, Arrigoni G, Millioni R, Vigili de Kreutzenberg S, Albiero M, Avogaro A, NETosis delays diabetic wound healing in mice and humans, Diabetes (2016). [DOI] [PubMed]
- [14].McCormack R, de Armas L, Shiratsuchi M, Podack ER, Killing machines: three pore-forming proteins of the immune system, Immunol Res 57(1–3) (2013) 268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Podack ER, Munson GP, Killing of Microbes and Cancer by the Immune System with Three Mammalian Pore-Forming Killer Proteins, Front Immunol 7 (2016) 464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McCormack RM, de Armas LR, Shiratsuchi M, Fiorentino DG, Olsson ML, Lichtenheld MG, Morales A, Lyapichev K, Gonzalez LE, Strbo N, Sukumar N, Stojadinovic O, Plano GV, Munson GP, Tomic-Canic M, Kirsner RS, Russell DG, Podack ER, Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria, Elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McCormack RM, Lyapichev K, Olsson ML, Podack ER, Munson GP, Enteric pathogens deploy cell cycle inhibiting factors to block the bactericidal activity of Perforin-2, Elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].McCormack R, Podack ER, Perforin-2/Mpeg1 and other pore-forming proteins throughout evolution, J Leukoc Biol 98(5) (2015) 761–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McCormack R, de Armas LR, Shiratsuchi M, Ramos JE, Podack ER, Inhibition of intracellular bacterial replication in fibroblasts is dependent on the perforin-like protein (perforin-2) encoded by macrophage-expressed gene 1, J Innate Immun 5(2) (2013) 185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].McCormack RM, Szymanski EP, Hsu AP, Perez E, Olivier KN, Fisher E, Goodhew EB, Podack ER, Holland SM, MPEG1/perforin-2 mutations in human pulmonary nontuberculous mycobacterial infections, JCI Insight 2(8) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jozic I, Vukelic S, Stojadinovic O, Liang L, Ramirez HA, Pastar I, Tomic Canic M, Stress Signals, Mediated by Membranous Glucocorticoid Receptor, Activate PLC/PKC/GSK-3beta/beta-catenin Pathway to Inhibit Wound Closure, J Invest Dermatol 137(5) (2017) 1144–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pastar I, Stojadinovic O, Krzyzanowska A, Barrientos S, Stuelten C, Zimmerman K, Blumenberg M, Brem H, Tomic-Canic M, Attenuation of the transforming growth factor beta-signaling pathway in chronic venous ulcers, Mol Med 16(3–4) (2010) 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ramirez HA, Liang L, Pastar I, Rosa AM, Stojadinovic O, Zwick TG, Kirsner RS, Maione AG, Garlick JA, Tomic-Canic M, Comparative Genomic, MicroRNA, and Tissue Analyses Reveal Subtle Differences between Non-Diabetic and Diabetic Foot Skin, PLoS One 10(8) (2015) e0137133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stojadinovic O, Tomic-Canic M, Human ex vivo wound healing model, Methods Mol Biol 1037 (2013) 255–64. [DOI] [PubMed] [Google Scholar]
- [25].Pastar I, Khan AA, Stojadinovic O, Lebrun EA, Medina MC, Brem H, Kirsner RS, Jimenez JJ, Leslie C, Tomic-Canic M, Induction of specific microRNAs inhibits cutaneous wound healing, J Biol Chem 287(35) (2012) 29324–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hayday AC, [gamma][delta] cells: a right time and a right place for a conserved third way of protection, Annu Rev Immunol 18 (2000) 975–1026. [DOI] [PubMed] [Google Scholar]
- [27].Garman RD, Doherty PJ, Raulet DH, Diversity, rearrangement, and expression of murine T cell gamma genes, Cell 45(5) (1986) 733–42. [DOI] [PubMed] [Google Scholar]
- [28].Heilig JS, Tonegawa S, Diversity of murine gamma genes and expression in fetal and adult T lymphocytes, Nature 322(6082) (1986) 836–40. [DOI] [PubMed] [Google Scholar]
- [29].Jameson J, Havran WL, Skin gammadelta T-cell functions in homeostasis and wound healing, Immunol Rev 215 (2007) 114–22. [DOI] [PubMed] [Google Scholar]
- [30].Newman PJ, Berndt MC, Gorski J, White GC 2nd, Lyman S, Paddock C, Muller WA, PECAM-1 (CD31) cloning and relation to adhesion molecules of the immunoglobulin gene superfamily, Science 247(4947) (1990) 1219–22. [DOI] [PubMed] [Google Scholar]
- [31].Aho S, Uitto J, Direct interaction between the intracellular domains of bullous pemphigoid antigen 2 (BP180) and beta 4 integrin, hemidesmosomal components of basal keratinocytes, Biochem Biophys Res Commun 243(3) (1998) 694–9. [DOI] [PubMed] [Google Scholar]
- [32].Biffo S, Sanvito F, Costa S, Preve L, Pignatelli R, Spinardi L, Marchisio PC, Isolation of a novel beta4 integrin-binding protein (p27(BBP)) highly expressed in epithelial cells, J Biol Chem 272(48) (1997) 30314–21. [DOI] [PubMed] [Google Scholar]
- [33].Gulati N, Krueger JG, Suarez-Farinas M, Mitsui H, Creation of differentiation-specific genomic maps of human epidermis through laser capture microdissection, J Invest Dermatol 133(11) (2013) 2640–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Radoja N, Gazel A, Banno T, Yano S, Blumenberg M, Transcriptional profiling of epidermal differentiation, Physiol Genomics 27(1) (2006) 65–78. [DOI] [PubMed] [Google Scholar]
- [35].Sivamani RK, Schwartz MP, Anseth KS, Isseroff RR, Keratinocyte proximity and contact can play a significant role in determining mesenchymal stem cell fate in human tissue, FASEB J 25(1) (2011) 122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Janson DG, Saintigny G, van Adrichem A, Mahe C, El Ghalbzouri A, Different gene expression patterns in human papillary and reticular fibroblasts, J Invest Dermatol 132(11) (2012) 2565–72. [DOI] [PubMed] [Google Scholar]
- [37].Woodley DT, Distinct Fibroblasts in the Papillary and Reticular Dermis: Implications for Wound Healing, Dermatol Clin 35(1) (2017) 95–100. [DOI] [PubMed] [Google Scholar]
- [38].McCormack R, Bahnan W, Shrestha N, Boucher J, Barreto M, Barrera CM, Dauer EA, Freitag NE, Khan WN, Podack ER, Schesser K, Perforin-2 Protects Host Cells and Mice by Restricting the Vacuole to Cytosol Transitioning of a Bacterial Pathogen, Infect Immun 84(4) (2016) 1083–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xiong P, Shiratsuchi M, Matsushima T, Liao J, Tanaka E, Nakashima Y, Takayanagi R, Ogawa Y, Regulation of expression and trafficking of perforin-2 by LPS and TNF-alpha, Cell Immunol 320 (2017) 1–10. [DOI] [PubMed] [Google Scholar]
- [40].McQualter JL, Brouard N, Williams B, Baird BN, Sims-Lucas S, Yuen K, Nilsson SK, Simmons PJ, Bertoncello I, Endogenous fibroblastic progenitor cells in the adult mouse lung are highly enriched in the sca-1 positive cell fraction, Stem Cells 27(3) (2009) 623–33. [DOI] [PubMed] [Google Scholar]
- [41].Macleod AS, Havran WL, Functions of skin-resident gammadelta T cells, Cell Mol Life Sci 68(14) (2011) 2399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Strbo N, Yin N, Stojadinovic O, Innate and Adaptive Immune Responses in Wound Epithelialization, Adv Wound Care (New Rochelle) 3(7) (2014) 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Murphy AG, O’Keeffe KM, Lalor SJ, Maher BM, Mills KH, McLoughlin RM, Staphylococcus aureus infection of mice expands a population of memory gammadelta T cells that are protective against subsequent infection, J Immunol 192(8) (2014) 3697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cho JS, Pietras EM, Garcia NC, Ramos RI, Farzam DM, Monroe HR, Magorien JE, Blauvelt A, Kolls JK, Cheung AL, Cheng G, Modlin RL, Miller LS, IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice, J Clin Invest 120(5) (2010) 1762–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Maher BM, Mulcahy ME, Murphy AG, Wilk M, O’Keeffe KM, Geoghegan JA, Lavelle EC, McLoughlin RM, Nlrp-3-driven interleukin 17 production by gammadeltaT cells controls infection outcomes during Staphylococcus aureus surgical site infection, Infect Immun 81(12) (2013) 4478–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sheridan BS, Romagnoli PA, Pham QM, Fu HH, Alonzo F 3rd, Schubert WD, Freitag NE, Lefrancois L, gammadelta T cells exhibit multifunctional and protective memory in intestinal tissues, Immunity 39(1) (2013) 184–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Human skin single cell analysis of hematopoietic (CD45+) and non-hematopoietic (CD45−) cells by flow cytometry.
a) Immunohistochemistry for CD45 in normal skin sample. b). Epidermis, dermis and full thickness skin samples were processed as described in Material and Methods and single cell suspension was labelled with antibody against CD45 molecule. Cells were analyzed by flow cytometry according to the granularity (SSC) and size (FSC) and expression of CD45 surface molecule. Representative dot plots for cells obtained from epidermis, dermis and full thickness skin specimens showing percent of CD45+ cells within each compartment in normal skin. c). Bar graph showing SEM of CD45 negative and positive cells within total cells in full thickness skin specimen (n=6).
Supplementary Figure 2. Illustration of a probe set design containing both left and right oligonucleotide pairs, and signal amplifications reagents: pre-amplifier, amplifier, and fluorescently labeled probes.