Abstract
Stem cells maintain tissue homeostasis by driving cellular turnover and regeneration upon damage. They reside within specialized niches that provide the signals required for stem cell maintenance. Stem cells have been identified in many tissues and cancer types, but their behavior within the niche and their reaction to microenvironmental signals were inferred from limited static observations. Recent advances in live imaging techniques, such as live cell imaging and intravital microscopy, have allowed the visualization of stem cell behavior and dynamics over time in their (near) native environment. Through these recent technological advances, it is now evident that stem cells are much more dynamic than previously anticipated, resulting in a model in which stemness is a state that can be gained or lost over time. In this review, we will highlight how live imaging and intravital microscopy have unraveled previously unanticipated stem cell dynamics and plasticity during development, homeostasis, regeneration, and tumor formation.
Decades ago, clonogenic assays suggested that adult tissues are composed of heterogeneous cells that are hierarchically organized. Long-lived and self-renewing adult stem cells (SCs) are at the top of the hierarchy, whereas short-lived progenitors and differentiated cells with limited clonogenic capacity are at the bottom. Over the years, various techniques confirmed the existence of SC populations that maintain the dynamic equilibrium of tissue homeostasis in adult organs. In this review, we will highlight recent advances in the SC field, focusing on the new insights provided by live cell imaging and intravital microscopy (IVM). First, we will briefly describe the history of techniques used to study SCs and the advantages that live cell imaging brings to the field. Next, we will highlight how live imaging has revealed previously unanticipated dynamics and plasticity of SCs during development, homeostasis, and regeneration. Last, we will touch on the role of SCs in tumor initiation and progression.
REVEALING THE PRESENCE OF SCs—A PLETHORA OF TECHNIQUES
For a long time, clonogenic assays were used as the gold standard to identify adult SC populations in vitro (Withers and Elkind 1970; Barrandon and Green 1987). Similarly, transplantation studies enabled the first identification of adult SCs in vivo (Deome et al. 1959; Spangrude et al. 1988). These methods proved the capacity of specific subpopulations of cells, that is, the SCs, to give rise to progeny (form cell colonies in vitro) or repopulate organs (in vivo). Although powerful, these methods were based on repopulation of ablated or damaged niches, or introduction of cells in ectopic environments. These nonphysiologic contexts might affect and alter the behavior of the transplanted cells, triggering regeneration-like responses, rather than recapitulating homeostatic cell turnover.
The advent of lineage tracing contributed to resolve tissue hierarchy in an unperturbed tissue or tumor. Lineage tracing is the identification of all the progeny of a single cell by a heritable mark such as (multi)color fluorescent reporters, Brainbow (Livet 2007), or Confetti (Snippert et al. 2010b; Kretzschmar and Watt 2012). When crossed with a line expressing an inducible Cre-recombinase under a promoter of choice (e.g., tissue or cell-type-specific promoters), expression of the reporter construct can be precisely induced through the activation of the Cre-recombinase by Tamoxifen injection. In contrast to transplantation-based or in vitro approaches, static lineage tracing provides quantitative information on the number, localization, and differentiation status of the progeny of a mother cell in its intact and native in vivo environment at a specific chosen time. Nevertheless, most lineage-tracing approaches rely on static images that fail to describe the full dynamics of a complex tissue.
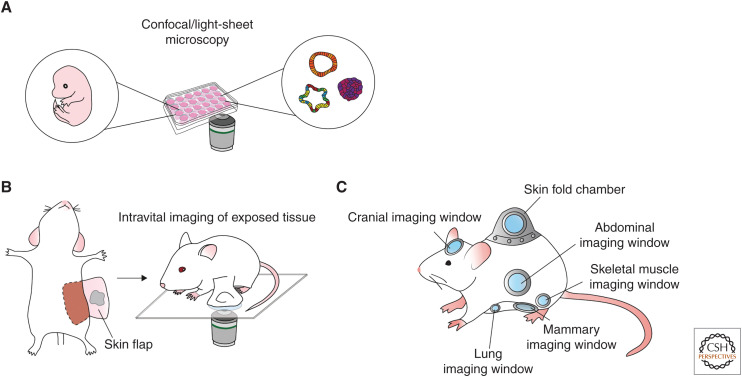
The implementation of fluorescent probes together with high-resolution microscopy technologies (such as confocal, multiphoton, and light-sheet microscopy) opened a whole world of opportunities to study real time (stem) cell dynamics in living organisms (Fig. 1A). Live cell imaging and IVM are unique compared with any other technique because they allow tracking of the same cells over time, thereby collecting coupled spatial and temporal information. For this reason, these techniques have made a strong contribution to the SC field, revealing SC dynamics in development, homeostasis, as well as in regeneration and cancer. Over the years, live cell imaging has been widely applied to in vitro 2D culture systems to study SC maintenance and differentiation. However, cell lines cultured in 2D fail to recapitulate the complexity of cell–cell interactions within living tissues. During the last decade, in vitro 3D cultured organoids allowed for the application of live cell imaging in more organotypic settings (Sato et al. 2009). Organoids represent miniature reconstructions of epithelial tissues that can be propagated indefinitely in vitro, whereby the epithelial hierarchy of the tissue of origin is maintained (i.e., adult SCs generate all the tissue-specific differentiated cells) (Kretzschmar and Clevers 2016). Although very useful to study cell–cell interactions over time, these 3D in vitro studies are largely lacking the context and natural niches in which the SCs reside.
Figure 1.
Overview of techniques for live cell imaging and intravital microscopy. (A) Confocal and light-sheet technologies to visualize stem cell dynamics in 3D organoids and embryo explants enabling high-throughput and high-resolution time-lapse imaging. (B,C) Intravital microscopy combined with multiphoton excitation allows in vivo stem cell imaging at great imaging depth at high resolution over time, either by surgically exposing the tissue for multiple hours (B) or by implantation of optical imaging windows for multiple days to weeks (C).
The development of IVM marks a huge step forward in the need to visualize cells in their native environment. Most IVM studies make use of microscopes equipped with a multiphoton laser that generates long wavelengths (i.e., within the infrared spectrum) and ultra-short, high-energy laser pulses, which confine excitation (by simultaneous absorption of two photons) to the focal plane, creating optical sectioning while minimizing light scattering and allowing high-imaging quality up to 1 mm imaging depth (Helmchen and Denk 2005; Dunn and Young 2006). In addition, the usage of low-energy wavelengths and excitation only at the focal plane reduces photobleaching and phototoxicity. Although the imaging quality of multiphoton excitation is not hampered as much as single-photon excitation at great imaging depth, the anatomical location of many different organs is too deep in the animal to obtain high-resolution images. To image these organs, tissues of interest can be surgically exposed; however, this does not allow imaging over multiple days (Fig. 1B). To overcome this, different optical imaging windows have been developed; small devices equipped with a cover glass that can be surgically implanted onto the organ of interest to reach deeper tissues and to image for longer times. This approach allows visual access to the mammary gland, intestine, liver, pancreas, kidney, spleen, lungs, and brain, enabling visualization of the dynamic behavior of individual cells in their natural environment (Fig. 1C; Huang et al. 1999; Trachtenberg et al. 2002; Kedrin et al. 2008; Ritsma et al. 2013; Alieva et al. 2014; Entenberg et al. 2018).
FILMING ORGAN DEVELOPMENT AND THE ORIGIN OF SCs
Tissue-specific SCs and their niches are formed during organogenesis in the developing embryo. Starting from the multipotent blastula, cells proliferate and migrate while undergoing a sequence of developmental decisions that force them in more and more differentiated lineages. This ultimately results in the formation of adult SCs that sustain the tissue for the lifetime of the organism. Although static analysis can reveal different (stem) cell populations at different embryonic stages, live imaging enables linking these populations over time. An early example of this is the imaging of hematopoietic stem cell (HSC) generation from the aortic hemogenic endothelial cells. Although it had been postulated before, live imaging in zebrafish embryos showed that HSCs emerge from the aortic floor into the subaortic space through a process called endothelial hematopoietic transition. This live imaging allowed for direct visualization of the emergence of HSCs in the embryo that had not been possible through static analyses (Boisset et al. 2010; Kissa and Herbomel 2010). The recently developed adaptive light-sheet technology now enables to even follow every single cell, and its fate in a developing postimplantation mouse embryo in real time (McDole et al. 2018). These emerging techniques have the power to reveal the role of SC dynamics and plasticity during organogenesis and adult SC formation.
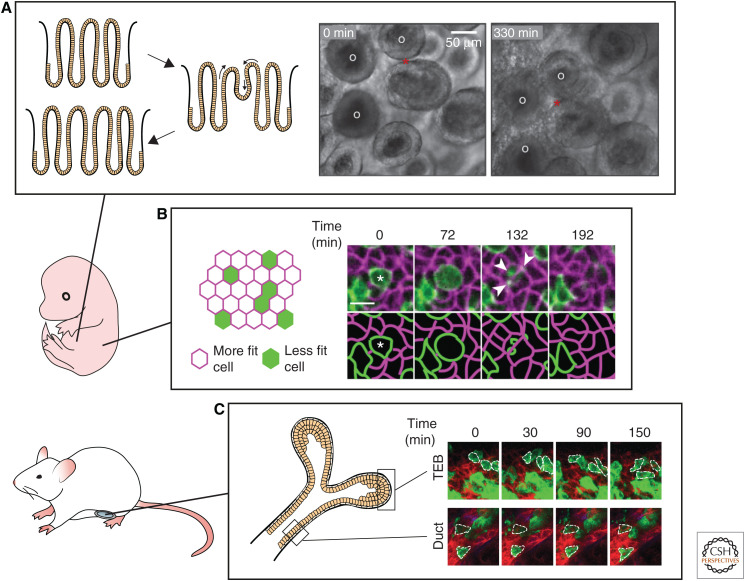
An excellent example showing that cell dynamics play a crucial role in the formation of SC niches can be found in the fetal intestine, which consists of a continuous sheet of nonproliferative Lgr5− villi and proliferative Lgr5+ intervillus regions. Adult crypts harboring the intestinal Lgr5+ SCs arise from these intervillus regions (Noah et al. 2011). However, how these crypts form and multiply while the developing intestine elongates remained unknown. Only recently, live imaging of intestinal explants from E16.5 mouse embryos showed that fetal villi undergo gross remodeling and fission thereby increasing intestinal length (Fig. 2A; Guiu et al. 2019). These fission events bring the nonproliferative villus cells into the proliferative intervillus region where they are exposed to SCs-inducing factors and become a new intestinal SC pool. As a result of this cellular plasticity, villus and intervillus cells have similar capacity to contribute to the growth of the intestinal epithelium during development, as shown by lineage-tracing experiments (Guiu et al. 2019). Thus, live imaging showed that adult intestinal SCs arise from equipotent precursors that get into the right niche through villus fission (Guiu et al. 2019).
Figure 2.
Intravital microscopy of organ development and the origin of stem cells. (A) Live imaging of intestinal explants (E16.5) shows the dynamics of crypt formation through extensive remodeling and fission events. Fission event is indicated with the red star, white circles indicate villi. Scale bar, 50 µm. (Photos in panel A are reprinted from Guiu et al. 2019, with permission, from Springer Nature © 2019.) (B) Live embryo imaging reveals that embryonic skin development is regulated by a process of cell competition. During early skin development, less fit cells (depicted in green) are actively eliminated and engulfed by neighboring more fit cells (depicted in magenta). Scale bar, 10 µm. (Photos in panel B are reprinted from Ellis et al. 2019, with permission, from Springer Nature © 2019.) (C) Intravital microscopy of the developing mammary gland uncovers the terminal end bud (TEB) cells as the driving forces of ductal extension and bifurcation, in contrast to the ductal cells that stay static and nonproliferative. (Photos in panel C are reprinted from Scheele et al. 2017, with permission, from the authors.)
Because adult SCs are dedicated to sustain tissue renewal for the entire lifetime of an organism, quality control mechanisms take place in the developing embryo to select and optimize tissue and organ development, including cell competition. This protective mechanism of cell competition, in which more fit “winner” cells actively sense and eliminate less fit “loser” cells was originally discovered in Drosophila (for review, see Amoyel and Bach 2014). Live embryo imaging, in combination with lineage tracing and single-cell transcriptomics, revealed two distinct modes of cell competition during skin development in mice (Fig. 2B; Ellis et al. 2019). In the early single-layered epithelium, loser cells are actively killed and subsequently engulfed by neighboring winner progenitors. Later, when the tissue begins to stratify, loser cells are expelled from the proliferative basal layer from which later the epidermal SCs will arise through differentiation. Loser cells more often divide perpendicular to the basal layer than their winner counterparts, a mechanism that also has been shown to play a role in SC competition in the aging skin (Liu et al. 2019). Together, these mechanisms serve as quality control for the barrier function of the skin (Ellis et al. 2019). Future live imaging studies will have to show how cell competition plays a role in the development of other mammalian organs.
Another developmental process during which cell dynamics is key, is branching morphogenesis. Branching morphogenesis leads to epithelial expansion generating extensively branched but compact organs, like the kidney, pancreas, lungs, and glands, including the salivary, mammary, and prostate glands (for review, see Varner and Nelson 2014). An organ where branching morphogenesis can be followed by IVM is the mammary gland as it occurs after birth. During embryonic development, a rudimentary tree is formed by multipotent progenitors that give rise to both basal and luminal cells (Lilja et al. 2018; Wuidart et al. 2018). After birth, multipotent progenitors are rapidly replaced by lineage-restricted progenitors but these cells are quiescent until puberty. During puberty, the lineage-restricted developmental progenitors that drive branching morphogenesis are localized in the terminal end buds (Davis et al. 2016; Scheele et al. 2017). IVM revealed that cells in the terminal end buds are the driving forces of ductal extension, whereas cells appeared to be static along the ducts (Fig. 2C; Scheele et al. 2017). Cells in the terminal end buds were shown to continuously proliferate and intermix, resulting in an equipotent pool of developmental progenitors in the terminal end buds equally contributing to ductal growth. Because of the proliferation in the end bud, cells at the edge are left behind to elongate the duct while the end bud is pushed further (Scheele et al. 2017). From these dynamic insights, in combination with static analysis of the branching networks in the mammary gland, kidney, and human prostate, a simple parameter-free model for branching morphogenesis could be deducted. Branched organs self-organize into a network of ducts following three rules: (1) equipotent ductal tips proliferate and stochastically branch, (2) they randomly explore their environment, and (3) they become proliferatively inactive when they come into close proximity with neighboring ducts (Hannezo et al. 2017).
IMAGING SC PLASTICITY DURING TISSUE HOMEOSTASIS
Over the past years, lineage tracing and clonal analysis have enabled quantitative modeling of SC behavior in adult organs, resulting in precise models of SC behavior of the different SC compartments. However, these analyses relied on static views of very dynamic processes, urging for techniques that allow for dynamic measures of SC fate (Krieger and Simons 2015). In this section, we will highlight how in vivo imaging exemplified the role of SC plasticity in space and time under homeostatic conditions.
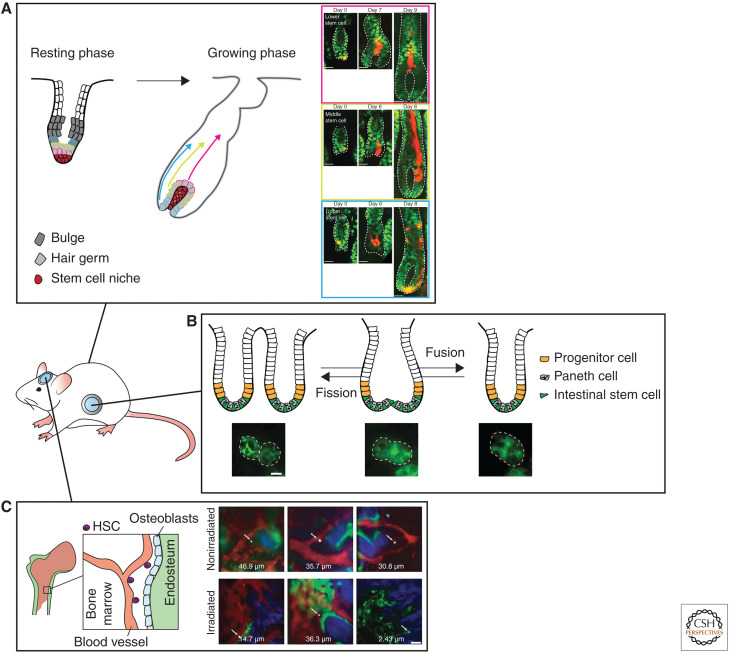
In the skin, lineage-tracing studies have led to a detailed model of tissue turnover, with distinct unipotent SC populations driving the homeostatic turnover in their specific compartment (Jaks et al. 2008; Jensen et al. 2009; Snippert et al. 2010a; Mascré et al. 2012; Lim et al. 2013; Page et al. 2013; Füllgrabe et al. 2015). However, this dogma of static SC compartments has been challenged by several studies. IVM of individual SCs over multiple generations revealed that each hair follicle SC has an equal potential to differentiate or divide (Rompolas et al. 2016). More recently, it was shown in mouse ear and paw skin that SC fate decisions (division or differentiation) are a direct consequence of neighboring cell behavior (Mesa et al. 2018). Differentiation of an SC and the subsequent transit into the upward layers of the skin, thereby leaving the niche partially vacant, is the trigger for neighboring SCs to divide symmetrically (Mesa et al. 2018). These two in vivo imaging studies clearly show that SC fate (symmetric or asymmetric division) is not a hard-wired intrinsic feature, but a flexible trait defined by the environment of an SC. Similarly, in the mouse hair follicle, in vivo imaging revealed that hair follicle SC fate is dictated by its location in the hair follicle niche (Rompolas et al. 2013). During the hair growth cycle (anagen), in vivo imaging with high spatial and temporal resolution uncovered what dynamic cellular processes take place, including long-range migration and major reorganization of epithelial SC progeny, that could not have been observed in static analyses (Rompolas et al. 2012). By tracking the same SC and their progeny over multiple days, it was shown that hair follicle SC location changes during the hair follicle growth cycle (Xin et al. 2018). As each hair follicle SC moves along its niche, it produces distinct differentiated cell types based on its location, indicating that hair follicle SCs have a flexible fate and differentiation potential determined by the direct environment (Fig. 3A; Xin et al. 2018). Upon hair follicle regression, the same SCs can act as phagocytes to clear the dying neighboring cells and this mechanism was shown to be essential to maintain tissue homeostasis and prevent overgrowth after hair follicle regression (Mesa et al. 2015). In conclusion, these studies show a previously unanticipated plasticity of SC commitment in the homeostatic hair follicle and interfollicular epidermis of the skin, enabling them to respond to changing conditions in the tissue.
Figure 3.
In vivo imaging of stem cell plasticity during tissue homeostasis. (A) Intravital microscopy of the hair follicle stem cells reveals a previously unanticipated plasticity of the stem cells dictated by their positioning in the stem cell niche. Clones of progeny arising from a single stem cell are depicted in red. Scale bar, 20 µm. (Photos in panel A are reprinted from Xin et al. 2018, with permission, from Nature Publishing Group © 2018.) (B) Intravital microscopy of whole crypt dynamics reveals the presence of crypt fusion, as a counteracting mechanism for crypt fission. Scale bar, 20 µm. (Photos in panel B are reprinted from Bruens et al. 2017 courtesy of the Creative Commons CC BY-NC-ND 4.0 License.) (C) Intravital microscopy of the hematopoietic stem cells (indicated by white arrows) shows that the localization of the hematopoietic stem cells and its niche is dynamically regulated depending on the environmental cues (upper panel: normal conditions, lower panel: irradiation). Bone collagen is depicted in blue (second harmonics generation), osteoblasts in green, and vascularization in red. Scale bar, 50 µm. (Photos in panel C are reprinted from Lo Celso et al. 2009, with permission, from the authors.)
Such plasticity of cellular identity has also been observed in the testis during spermatogenesis. Sperm SCs mainly undergo incomplete divisions generating syncytia of undifferentiated spermatogonia. Classically, spermatogonia are subclassified into differentiated and undifferentiated subpopulations, based on relative expression levels of GFRα1 (SC marker) and receptor tyrosine kinase KIT (differentiation marker). Based on fixed samples, it was thought that SC activity was limited to the undifferentiated spermatogonia (mainly expressing GFRα1), whereas more differentiated spermatogonia (mainly expressing Kit) irreversibly lost SC potential (Shinohara et al. 2000). However, in vivo time-lapse imaging unraveled that these syncytia are not irreversibly committed, and can undergo fragmentation and subsequently cells can regain stemness (measured by reexpression of the GFRα1 SC marker) (Hara et al. 2014). Although cellular plasticity is rare in homeostatic spermatogenesis, it increases when the tissue is damaged upon administration of busulfan, a drug toxic to spermatogonia including SCs, leading to regeneration (Nakagawa et al. 2010; Hara et al. 2014). In conclusion, these in vivo imaging studies point toward a revised model of sperm SC dynamics, in which stemness is more flexible than previously anticipated.
In the intestine, an epithelial system with high-cellular turnover driven by SCs at the bottom of the crypt, live imaging has also greatly contributed to our understanding of the SC niche dynamics and plasticity. In contrast to the skin, only a single SC compartment is fueling tissue renewal. Lineage-tracing studies have been crucial to determine the SC identity and dynamics in the intestine. After the identification of the specific SC marker Lgr5 in the intestine, lineage-tracing studies revealed that these Lgr5+ cells at the base of the intestinal crypts generate all the differentiated cell types of the intestine along the crypt–villus axis (Barker et al. 2007). Further modeling of the clonal dynamics showed that the intestinal SCs in each crypt are equipotent, and by undergoing symmetric cell divisions they neutrally compete for niche space (Lopez-Garcia et al. 2010; Snippert et al. 2010b). Through this neutral competition, SC clones can be lost over time eventually leading to monoclonal crypts. Although the static lineage-tracing measurements revealed the long-term potential of the intestinal SCs, the short-term crypt dynamics were not resolved. To this end, multiday IVM was used to investigate the dynamic competition between different SCs within their niche. Strikingly, in vivo imaging revealed that positioning in the niche space is an important determinant of SCs to win the competition, and that this potential is reversible (Ritsma et al. 2014). Monitoring fluorescently labeled SCs (expressing the recombined Confetti construct) in the same crypts over multiple days revealed that “central” Lgr5+ SCs (i.e., residing at bottom of the crypt base) are likely to stay within the niche and give rise to progeny that will colonize the entire crypt and villus. In contrast, Lgr5+ SCs residing in the upper part of the crypt base (called “border cells”) are more susceptible to be displaced into the above transient-amplifying compartment (Ritsma et al. 2014). Moreover, IVM showed that SCs can change position within the niche over time, so that cells at a favorable position in the niche center can lose their potential when entering the border of the crypt, and vice versa (Ritsma et al. 2014). In addition, manipulation of the SC niche by, for example, reducing Wnt secretion (e.g., with administration of the porcupine inhibitor LGK974) down-regulates the expression of Lgr5 in the crypt base. Thereby, the number of SCs decreases leading to less competitors resulting in faster competition (Huels et al. 2018).
Live cell imaging of intestinal organoids was used to further elucidate cell dynamics and fate decisions in the intestine. Time-lapse phase contrast imaging combined with fluorescent lineage tracing confirmed that small intestinal organoids contain crypt-like buds harboring functional Lgr5+ SCs resembling the in vivo intestine (Sato et al. 2009). Lgr5+ SCs give rise to a wide range of specialized cell types that intermix into one intestinal epithelial lining, thus creating a mosaic pattern of cell types. Time-lapse imaging of organoids revealed that postmitotic positioning predicts long-term placement along the crypt axis and subsequent differentiation potential of the daughter cells (Carroll et al. 2017). This positioning occurs during cytokinesis on the apical surface of the epithelium (toward the lumen) in an actin-dependent manner (McKinley et al. 2018). During this process, neighboring cells can intrude within the cytokinetic furrow as a consequence of the elongated cell shape. Thus, neighboring cells can position themselves in between the daughter cells. Interference with cell shape abrogated cell intermixing, indicating that cell shape differences and apicobasal positioning are essential determinants of correct spatial organization of the intestinal epithelium (McKinley et al. 2018).
Another crucial factor determining tissue patterning is symmetry breaking, the process in which a pool of identical cells change their differentiation potential with respect to neighboring cells creating asymmetric tissue structures, such as the intestinal SC niche and crypt–villus axis. Large-scale quantitative light-sheet imaging of intestinal organoid development, combined with single-cell genomics, identified heterogeneity in YAP1 expression between identical cells as the driver of symmetry breaking in intestinal spheres (Serra et al. 2019). Cell-to-cell variability in YAP1 activation promotes lateral inhibition through Notch and DLL1, which in turn leads to the emergence of the first Paneth cell and subsequent organoid asymmetry and crypt-like bud formation (Serra et al. 2019). Together, these intravital and live imaging studies start to reveal the intricate and dynamic interplay between SCs and its niche, and show that previously unnoticed features such as cell shape, cell-to-cell variability, and relative cell position are crucial determinants of SC potential.
Cell dynamics in the intestine are further complicated by dynamics involving whole crypts. In the epithelium, crypts can be found in a bifurcating shape, which has been interpreted as crypts undergoing a fission event, during which one crypt splits into two. However, a recent IVM study revealed the presence of a counteracting mechanism called crypt fusion during which two independent crypts fuse into one (Fig. 3B; Bruens et al. 2017). Although producing opposite outcomes, crypt fission and fusion appear as morphologically identical in static images, highlighting the importance of in vivo imaging in studying dynamic processes. High-resolution live imaging in intestinal organoids was used to follow the cellular dynamics of crypt fission. This imaging showed that upon the initiation of fission, the more rigid Paneth cells form two clusters at either side of the crypt flanking a more flexible Lgr5+ SC cluster. Subsequently, an invagination is formed at this Lgr5+ site, which splits the crypt into two in a zipper-like fashion (Langlands et al. 2016). Together, these observations point toward a model in which the distribution and proportion of stiff cells determines the likelihood of in vivo tissue deformation events such as crypt fission (Almet et al. 2018). In conclusion, these studies emphasize the dynamic nature of SC potential and fate, and illustrate the power of live cell and IVM approaches to unravel new mechanisms of tissue organization and SC dynamics, otherwise indistinguishable when inspecting fixed tissue sections.
FOLLOWING SCs DURING REGENERATIVE PROCESSES
In many tissues, regenerative potential relies on the presence of SCs that respond to cues from the damaged niche by producing new cells repairing the damage. This response is highly dynamic as it relies on cell–cell interactions, microenvironmental changes, cell migration, and proliferation. Therefore, tissue regeneration is another example in which live cell imaging approaches can add important insights that cannot be captured using static models.
The bone marrow harboring the HSCs represents a highly dynamic SC niche that rapidly responds to various stress-inducing signals. Although the location of the HSC niche had been previously identified by immunohistochemistry, it still remained unknown how this niche was established. Live imaging experiments of the mouse calvarium revealed that HSCs injected in nonirradiated mice reside close to the vasculature but further away from the endosteum (Fig. 3C; Lo Celso et al. 2009). However, when the HSCs were injected in an irradiated setting or in a niche with impaired endogenous HSCs (carrying a c-Kit mutation) they localized more closely to endosteum. This position within the niche correlated with the differentiation state; long-term and proliferative HSCs localized closest to the endosteum and osteoblasts, whereas more mature subsets resided progressively further away (Lo Celso et al. 2009). Upon stress, however (such as acute infection), quiescent HSCs become motile and interact with wider osteoblastic regions, indicating that the location and behavior of the HSCs dynamically respond to the environmental cues (Rashidi et al. 2014).
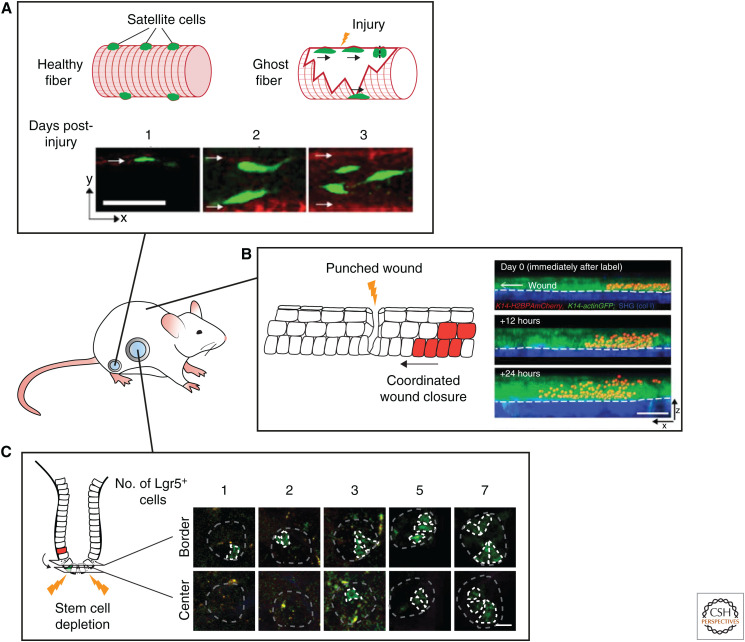
In the skeletal muscle, the SCs—called satellite cells—are reactivated upon damage and proliferate and differentiate to replace damaged muscle fibers. Although this is a relatively simple repair mechanism, only recently the dynamics of the response within the SC niche and the necessary cues have been studied by IVM. Under homeostasis, the satellite cells were found to be nondividing and immobile. However, upon injury remnants of the extracellular matrix of damaged fibers, called ghost fibers, guide satellite cells to proliferate and migrate along them. This results in the spreading of progenitors, which then form nascent myofibers replacing the damaged fibers. These findings show that the regeneration response in the skeletal muscle is not solely an SC intrinsic reaction but that the wound itself, in this case the ghost fibers, orchestrate the repair (Fig. 4A; Webster et al. 2016).
Figure 4.
Following stem cells during regenerative processes by intravital microscopy. (A) Intravital microscopy reveals the dynamics of satellite cell-mediated repair of injured muscle fibers. Remnants of damaged fibers form ghost fibers, which are used by satellite cells as guides for proliferation and migration. Scale bar, 50 µm. (Photos in panel A are reprinted from Webster et al. 2016, with permission, from Elsevier © 2016.) (B) Imaging of the cell dynamics during wound healing in the skin reveals the migratory and proliferative dynamics of the cells repairing the wound (migratory and proliferative cells are shown in red). Scale bar, 50 µm. (Photos in panel B are reprinted from Park et al. 2017, with permission, from Springer Nature © 2017.) (C) Multiday intravital microscopy shows the highly plastic nature of the intestinal cells. After ablation of all the stem cells, transit-amplifying cells fall back into the stem cell niche, adopt a stem cell fate, and repopulate the stem cell zone. Repopulating transit-amplifying cells are depicted in green. Scale bar, 20 µm. (Photos in panel C are adapted from Ritsma et al. 2014, with permission, from the authors.)
In contrast to skeletal muscle, the skin is easily accessible for IVM. Laser ablation of SCs during the hair growth cycle (anagen) combined with multiday IVM showed that the position of a hair follicle SC is predictive for its differentiation potential into an uncommitted, committed, or differentiated cell type (Rompolas et al. 2013). When hair follicle SCs are ablated, more differentiated epithelial cells can repopulate the SC compartment and sustain hair growth (Rompolas et al. 2012, 2013). In addition, in vivo time-lapse imaging has been used to study the cellular dynamics during the repair of a punch wound. During wound repair, directed division, differentiation, and migration are balanced to effectively repair the wound while maintaining tissue homeostasis (Park et al. 2017). Static analysis has shown that two zones appear after wounding: a migratory front that surrounds the wound edge surrounded by a ring of rapidly proliferation cells (Zhao et al. 2003; Usui et al. 2005; Safferling et al. 2013). However, IVM revealed that these zones spatially overlap, with some cells performing both behaviors simultaneously (Fig. 4B; Park et al. 2017). These zones do not only serve as a source for new cells for wound closure, they also restrict the area of unwounded epithelium that is used for re-epithelialization (Park et al. 2017). Together, this shows how homeostatic mechanisms can be repurposed and integrated with wound-specific behaviors to restore homeostasis after damage.
As in many other tissues, stemness in the intestine has been shown to be highly plastic. When Lgr5+ SCs are ablated, cells from higher up in the crypt can fall back into the SC niche where they dedifferentiate and reexpress the SC marker Lgr5 (Bankaitis et al. 2018). Multiday IVM revealed that on ablation of all SCs, repopulation is a sporadic event. Individual cells transfer from the transit-amplifying zone into the SC niche border where they clonally expand and refill the SC niche (Fig. 4C; Ritsma et al. 2014). When only one Lgr5+ SC is ablated, preexisting SCs rearrange to restore the alternation in pattern between Paneth and SCs within 2 hours and without cell division. Simultaneously, the damaged cell is forced out of the crypt by peristalsis-like motion of the crypt lumen (Choi et al. 2018). Thus, the intestinal crypt is highly dynamic and plastic, and the location within it defines stemness.
IMAGING OF SCs DURING TUMOR INITIATION AND PROGRESSION
Mechanisms used to maintain tissue homeostasis and regeneration, such as cell competition and cellular plasticity, are hijacked by cancer cells in the attempt to survive and infiltrate healthy tissues (Condeelis et al. 2005; Ellenbroek and van Rheenen 2014; Suijkerbuijk and van Rheenen 2017). IVM experiments have unraveled differences and similarities between healthy tissues and cancers, and provided unexpected findings to increase understanding of cancer initiation, tumor maintenance, and spreading, and possibly contribute to fine tune treatment strategies.
Tumor Initiation
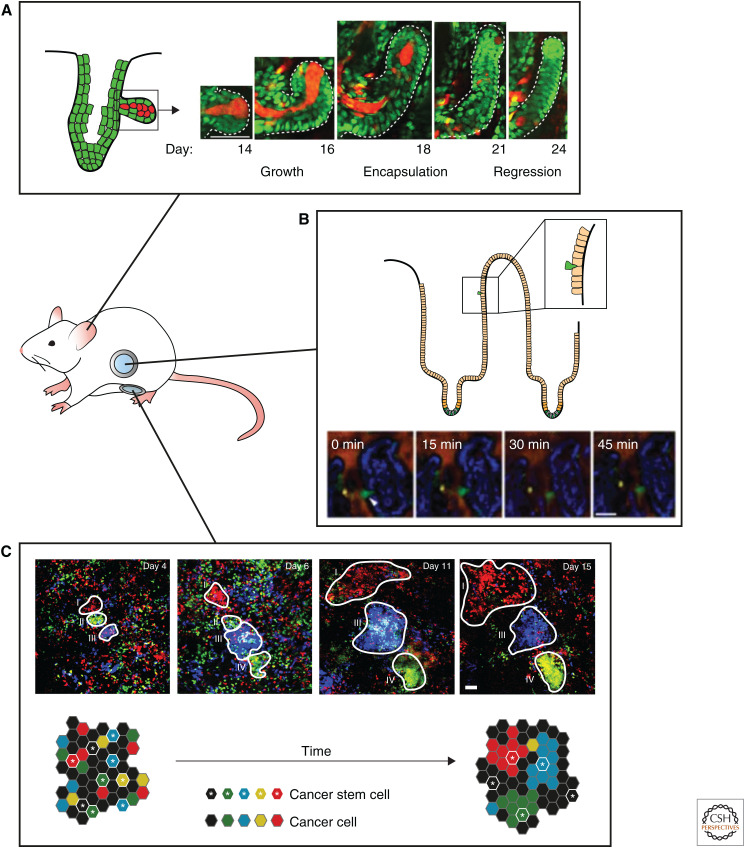
The human body has developed robust mechanisms to resist tumor growth and most of the time healthy tissues succeed in outcompeting mutant cells and reestablishing homeostasis. Both in intestine and in skin, it has been shown that tissue architecture and cellular turnover present protective mechanisms to oncogene-induced abnormalities. In the hair follicle, normal tissue dynamics lead to upward movement of the healthy hair follicle progenitor cells, resulting in relocation and subsequent differentiation of the mutant progenitor cells (Xin et al. 2018). Similarly, clonal analysis and in vivo imaging of cell fate choices in epidermal SCs harboring oncogenic Pik3ca mutations showed that oncogene-induced differentiation restricts clonal expansion of mutant cells, and eventually leads to the loss of oncogenic epithelial cells (Ying et al. 2018). In addition, the skin has been shown to exploit regeneration processes that actively exclude potentially harmful cancer cells. Cre-induced activation of oncogenic β-catenin in the hair follicle SCs of a genetic mouse model produces benign deformations (i.e., new axes of hair growth), which expand in a way reminiscent of early-stage embryonic hair follicle formation. IVM showed that wild-type (WT) cells are recruited to these new hair follicle branches and that Wnt signaling is activated in WT cells on the expression of Wnt ligands by neighboring mutant β-catenin cells, influencing the behavior of the WT cells (Deschene et al. 2014). Nevertheless, healthy tissue is able to counteract this ectopic growth by encapsulating the mutant cells (β-catenin or Hras) and actively eliminate them from the tissue (Fig. 5A; Brown et al. 2017). Together, these in vivo studies present a strong case for tissue autonomous mechanisms, such as tissue architecture and cell dynamics, that actively protect against the fixation of oncogenic mutations.
Figure 5.
Imaging of stem cells during tumor initiation and progression. (A) Repeated intravital imaging of a hair follicle harboring an oncogenic β-catenin mutation shows ectopic hair follicle outgrowth. Counteracting mechanisms result in encapsulation of the mutant cells, thereby actively eliminating them from the tissue. Mutant cells are depicted in red, wild-type cells are depicted in green. (Photos in panel A are reprinted from Brown et al. 2017, with permission, from Springer Nature © 2017.) (B) Time-lapse intravital microscopy in the intestine shows that transformed cells (RasV12 mutant cells depicted in green) are actively expelled from the healthy intestinal epithelium. (Photos in panel B are reprinted from Kon et al. 2017, with permission, from Springer Nature © 2017.) (C) Multiday intravital microscopy of labeled tumor cells in a growing mammary carcinoma shows different growth patterns. Only a small population of the tumor cells (cancer stem cells) drives the outgrowth of clones. Analysis of the clonal growth patterns reveals that cancer stem cell properties are plastic, and can be lost and gained over time. Scale bars, 50 µm. (Photos in panel C are reprinted from Zomer et al. 2013, with permission, from John Wiley & Sons © 2013.)
The removal of aberrant cells from the epithelium has also been shown to occur in the intestine. As in the skin, differentiated cells are thought to be incapable of inducing tumor formation because normal tissue dynamics push them upward to the tip of the villus where they undergo apoptosis giving them no time to form aberrant clones (Barker et al. 2009). In addition, neutral competition in the intestinal SC niche may be a protective mechanism against the acquisition of mutations, and subsequent tumor initiation (van Rheenen and Bruens 2017). Several studies have investigated the effect of specific mutations on the SC competition in the niche (Vermeulen et al. 2013; Snippert et al. 2014). Indeed, introduction of sporadic mutations, such as Apc loss, Kras activation, or p53 mutations in the SC compartment, leads to a bias in the competition between WT and mutant SCs. As a result, the mutant clones have a slightly increased chance to expand and colonize a crypt (Vermeulen et al. 2013; Snippert et al. 2014). Nevertheless, because the number of WT SCs exceeds the number of mutant SCs, many mutated SCs are still replaced by the progeny of WT SCs, confirming the hypothesis that intestinal tissue architecture protects against mutation accumulation. This notion was further confirmed by a recent IVM study using a porcupine inhibitor to reduce the Wnt secretion, resulting in a reduction of the number of Lgr5+ SCs. Lineage tracing in an Apc-deficient background showed that the fixation speed of Apc-deficient SCs was increased, leading to accelerated adenoma formation (Huels et al. 2018). Together, these studies show that clonal drift and SC competition are protective against mutation fixation in the SC compartment.
In addition to SC competition in the crypt, it has been shown that cell competition—discussed in the section “Filming Organ Development and the Origin of SCs”—plays a role in eliminating transformed cells from the villus epithelium. Time-lapse IVM in the intestine showed that normal epithelial cells can recognize RasV12 transformed cells resulting in active elimination of the transformed cells (Fig. 5B; Kon et al. 2017). On contact with neighboring normal cells, the actin-binding protein epithelial protein lost in neoplasm (EPLIN) accumulates in transformed cells during a process called EDAC (epithelial defense against cancer) (Ohoka et al. 2015; Kon et al. 2017). EDAC induces Warburg-effect-like metabolic changes in the transformed cells via up-regulation of pyruvate dehydrogenase kinase 4 (PDK4), leading to enhanced aerobic glycolysis and down-regulation of mitochondrial function. This PDK-mediated mitochondrial dysfunction results in apical extrusion of RasV12-transformed cells from the intestinal epithelium (Kon et al. 2017).
Even though many transformed cells are eliminated and do not have the chance to initiate tumorigenesis, some transformed cells escape these control mechanisms and give rise to cancer. When cancer progresses, the healthy tissue inevitably succumbs and loses cell–cell competition within the niche. IVM of leukemic cell colonization in the bone marrow revealed that unlike healthy HSCs, these cells are highly motile, migrate irrespective of any bone marrow subcompartment, and do not remain cohesive after cell division (Hawkins et al. 2016). Infiltrating leukemic cells force remodeling of the bone marrow and actively induce loss of osteoblastic SCs (Hawkins et al. 2016; Duarte et al. 2018). Thus, because of the lack of a supporting niche, the fitness of resident healthy HSCs may be dramatically reduced, possibly leading to a progressive loss of normal hematopoiesis (Duarte et al. 2018). Together, these studies highlight the importance of the interaction between the SCs and their niche during cancer initiation and progression.
Tumor Maintenance by Cancer Stem Cells
Although SCs are believed to be the tumor-initiating cells in most tissues, it still remains unclear whether SCs are driving tumor progression and metastasis. At premalignant stages, lineage-tracing studies have shown that tissue hierarchy is largely maintained; cells with stem-like properties drive clonal expansion leading to multiclonal tumors, supporting the cancer SC (CSC) hypothesis (Driessens et al. 2012; Schepers et al. 2012). According to the CSC theory, the majority of tumor cells within a tumor mass have limited proliferative potential and die after a few rounds of cell divisions, although a small proportion of tumor cells, the CSCs, can self-renew and sustain tumor (and metastasis) growth (Batlle and Clevers 2017). In line with this hypothesis, a recent lineage-tracing study in human primary colon cancer xenografts showed evidence for a small subpopulation of CSCs, driving long-term tumor growth and progression (Lenos et al. 2018). These CSC clones were predominantly found at the tumor edge in close proximity to osteopontin-producing cancer-associated fibroblasts (Lenos et al. 2018). CSC behavior, however, changed over time depending on the location of the CSC within the tumor, indicating that functional CSC properties can be defined by microenvironmental factors (Lenos et al. 2018).
What remains unclear from these static lineage-tracing studies is whether CSCs show a similar degree of plasticity compared with their healthy SC counterparts and how CSC plasticity contributes to tumor growth, progression, and resistance. To understand the role of plasticity in tumor maintenance, several live imaging studies have been conducted to follow CSC dynamics in their intact environment. For example, multicolor in vivo Confetti tracing of unperturbed mammary tumors (MMTV-PyMT tumor model) revealed that most cancer cells provide lineages that disappear over time and that only a minor population of cancer cells (i.e., CSCs) is able to generate large, long-lived single-colored Confetti clones during both adenoma and carcinoma stages (Fig. 5C; Zomer et al. 2013). In addition, analysis of the clonal dynamics over time showed that cancer cells can form clones with delayed onset of growth or clones that suddenly undergo regression. This indicated that stem-like characteristics are highly dynamic and can be either acquired (upon plasticity) or lost (with differentiation) over time (Zomer et al. 2013). Another study, in which human colorectal cancer organoids were followed by multiday confocal imaging, showed that non-CSCs (identified by the lack of expression of the fluorescent ASCL2-specific SC reporter STAR) were able to form organoids, and underwent cellular plasticity, gaining SC identity (visualized by reexpression of the STAR reporter) to further fuel organoid growth (Oost et al. 2018).
CSC plasticity not only plays a role in tumor growth, but also in the metastatic cascade. It has been extensively reported that cells that undergo epithelial-to-mesenchymal transition (EMT) may gain self-renewal potential (i.e., stemness), which enables them to efficiently fuel metastatic growth at distant sites (Kalluri and Weinberg 2009). This view was recently challenged by static lineage-tracing experiments showing that metastases are not seeded by cells that have expressed specific mesenchymal markers (e.g., Fsp1) (Fischer et al. 2015; Zheng et al. 2015). However, it remains unknown whether all mesenchymal cells express the classical mesenchymal markers (such as N-cadherin, vimentin, fibronectin, and Fsp1), especially in light of recent findings revealing the existence of multiple EMT stages in skin and in mammary tumors, ranging from epithelial to completely mesenchymal states (Pastushenko et al. 2018). Cells can be in different hybrid EMT states leading to differences in cellular plasticity, invasiveness, and metastatic potential (Pastushenko et al. 2018). IVM of EMT and mesenchymal-to-epithelial-transition (MET) in mammary tumors using a fluorescent E-cadherin expression reporter showed that only a small population of cells undergoes EMT in an unperturbed system (Beerling et al. 2016). This small population of mesenchymal cells is motile and able to disseminate to a distant site, both in early- and late-stage tumors (Beerling et al. 2016; Harper et al. 2016). However, at the distant site, mesenchymal cells switch back to an epithelial state already after a few cell divisions, rendering potential differences in self-renewal capacity between seeding epithelial and mesenchymal cells irrelevant (Beerling et al. 2016).
CSC identity and plasticity are also important in the context of treatment efficiency and treatment resistance. A recent study in basal cell carcinoma showed that these tumor cells can escape targeted therapy (smoothened inhibitor Vismodegib) by adopting a different cellular identity (Biehs et al. 2018). Normally, basal cell carcinoma cells have an identity that closely resembles hair follicle bulge cells. However, on treatment with Vismodegib, a subset of basal cell carcinoma cells initiates a transcriptional program that resembles that of interfollicular and isthmus SCs (Biehs et al. 2018). This involves transcriptional changes leading to a rapid Wnt activation, which renders these cells insensitive to the Smoothened inhibitor (Biehs et al. 2018). Thus, cell identity switches may be a common way for tumor cells to escape drug-induced cell death.
To systematically understand cell identity switches as an escape mechanism in different tumor types and identify new potential drug combinations, more high-throughput methods are required. Now, with proper culture conditions, organoids can be efficiently generated from patient biopsies. The recent establishment of human tumor organoid libraries represents a breakthrough in cancer biology and expands the possibilities to look at tumor genetic heterogeneity (van de Wetering et al. 2015; Fujii et al. 2016; Seino et al. 2018). Moreover, these patient-derived biobanks offer large platforms to test the response to old and new (combinations of) drugs. For example, a panel of human colorectal cancer organoids in combination with confocal live cell imaging was used to link appearance of chromosomal instability to specific colorectal cancer mutations, by looking at segregation errors over time in human colorectal cancer organoids carrying increasing mutational load (Drost et al. 2015; Bolhaqueiro et al. 2018). Similar approaches were used to monitor drug responses on a panel of KRAS-mutant versus KRAS-WT patient-derived colorectal cancer organoids and revealed that KRAS-mutant organoids are significantly less sensitive to EGFR inhibitors (Verissimo et al. 2016). Together with the development of new high-throughput screening platforms and microscopy, organoids systems will provide a powerful tool to understand cancer progression and mechanisms of cellular drug resistance (Rios and Clevers 2018). Moreover, human tumor-derived organoids can be engineered with fluorescent markers, and subsequently be transplanted orthotopically into animals, providing a unique opportunity to track human CSC dynamics at a cellular resolution by IVM. The relevance of this unique combination of tools was recently shown by a study using engineered human colorectal cancer organoids to elucidate the contribution of defined mutations to metastasis formation (Fumagalli et al. 2017). The authors found that metastatic ability is directly caused by mutations that allow growth independent of certain niche signals, thereby identifying the cellular mechanisms of key drivers of progression in colorectal cancer (Fumagalli et al. 2017).
CONCLUDING REMARKS
The combination of in vitro live cell imaging, lineage tracing, and IVM has been essential to define the SC dynamics of healthy and tumorigenic tissues in the murine setting. Overall (in vivo), live cell imaging studies challenge the concept of fixed SCs populations with a determined and hardwired self-renewal capacity. It now appears that instead of a static one-way route, cellular hierarchy is much more plastic than previously thought, and stemness is a state that can be gained or lost, especially during regeneration responses and tumor growth.
Because most IVM data is based on imaging in animal models, the next challenge will be to understand whether these dynamics reflect the human situation. In vitro, the advent of organoid technology has allowed mapping cell dynamics of primary human cells in a 3D cellular organization, closely resembling the tissue of origin. However, these cultures are mostly limited to the epithelial component, and do not allow to study the interactions between cells and their microenvironment. Recent efforts have succeeded in integrating some components of the microenvironment such as mesenchymal stroma (Ootani et al. 2009; Spence et al. 2011; McCracken et al. 2014; Múnera et al. 2017) and immune cells (Dijkstra et al. 2018; Neal et al. 2018) together with patient-derived organoids. To go one step further, orthotopic transplantation of human (tumor) organoids can be combined with in vivo live cell imaging techniques. For instance, healthy human colon organoids—derived from either iPS cells or adult tissues—can engraft and reconstruct the human colon epithelium in a mouse, allowing to study human tissue dynamics in an orthotopic environment using IVM (Múnera et al. 2017; Sugimoto et al. 2018). In addition, orthotopic transplantation of human tumor-derived organoids will open new avenues to study the dynamic processes occurring during tumor progression including the metastatic cascade. In conclusion, the combination of state-of-the-art live imaging techniques will have the unique potential to ultimately resolve the intricate dynamics of SCs and their niches.
ACKNOWLEDGMENTS
We regret that owing to space limitations we could only describe a selection of papers and therefore have not been able to reference all literature relevant to the topic. We thank members of the laboratory of J.v.R. for helpful discussions and their critical reading of the manuscript. J.v.R. is supported by CancerGenomics.nl, a European Research Council, Grant CANCER-RECURRENCE 648804, and the Doctor Josef Steiner Foundation.
Footnotes
Editors: Cristina Lo Celso, Kristy Red-Horse, and Fiona M. Watt
Additional Perspectives on Stem Cells: From Biological Principles to Regenerative Medicine available at www.cshperspectives.org
REFERENCES
- Alieva M, Ritsma L, Giedt RJ, Weissleder R, van Rheenen J. 2014. Imaging windows for long-term intravital imaging: general overview and technical insights. Intravital 3: e29917. doi: 10.4161/intv.29917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almet AA, Hughes BD, Landman KA, Näthke IS, Osborne JM. 2018. A multicellular model of intestinal crypt buckling and fission. Bull Math Biol 80: 335–359. doi: 10.1007/s11538-017-0377-z [DOI] [PubMed] [Google Scholar]
- Amoyel M, Bach EA. 2014. Cell competition: how to eliminate your neighbours. Development 141: 988–1000. doi: 10.1242/dev.079129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis ED, Ha A, Kuo CJ, Magness ST. 2018. Reserve stem cells in intestinal homeostasis and injury. Gastroenterology 155: 1348–1361. doi: 10.1053/j.gastro.2018.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007. doi: 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608–611. doi: 10.1038/nature07602 [DOI] [PubMed] [Google Scholar]
- Barrandon Y, Green H. 1987. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci 84: 2302–2306. doi: 10.1073/pnas.84.8.2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batlle E, Clevers H. 2017. Cancer stem cells revisited. Nat Med 23: 1124–1134. doi: 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, Schäfer R, van Diest P, Voest E, van Oudenaarden A, et al. 2016. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep 14: 2281–2288. doi: 10.1016/j.celrep.2016.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehs B, Dijkgraaf GJP, Piskol R, Alicke B, Boumahdi S, Peale F, Gould SE, de Sauvage FJ. 2018. A cell identity switch allows residual BCC to survive Hedgehog pathway inhibition. Nature 562: 429–433. doi: 10.1038/s41586-018-0596-y [DOI] [PubMed] [Google Scholar]
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. 2010. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464: 116–120. doi: 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Bolhaqueiro ACF, van Jaarsveld RH, Ponsioen B, Overmeer RM, Snippert HJ, Kops G. 2018. Live imaging of cell division in 3D stem-cell organoid cultures. Methods Cell Biol 145: 91–106. doi: 10.1016/bs.mcb.2018.03.016 [DOI] [PubMed] [Google Scholar]
- Brown S, Pineda CM, Xin T, Boucher J, Suozzi KC, Park S, Matte-Martone C, Gonzalez DG, Rytlewski J, Beronja S, et al. 2017. Correction of aberrant growth preserves tissue homeostasis. Nature 548: 334–337. doi: 10.1038/nature23304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruens L, Ellenbroek SIJ, van Rheenen J, Snippert HJ. 2017. In vivo imaging reveals existence of crypt fission and fusion in adult mouse intestine. Gastroenterology 153: 674–677.e3. doi: 10.1053/j.gastro.2017.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll TD, Langlands AJ, Osborne JM, Newton IP, Appleton PL, Näthke I. 2017. Interkinetic nuclear migration and basal tethering facilitates post-mitotic daughter separation in intestinal organoids. J Cell Sci 130: 3862–3877. doi: 10.1242/jcs.211656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Rakhilin N, Gadamsetty P, Joe DJ, Tabrizian T, Lipkin SM, Huffman DM, Shen X, Nishimura N. 2018. Intestinal crypts recover rapidly from focal damage with coordinated motion of stem cells that is impaired by aging. Sci Rep 8: 10989. doi: 10.1038/s41598-018-29230-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condeelis J, Singer RH, Segall JE. 2005. The great escape: when cancer cells hijack the genes for chemotaxis and motility. Annu Rev Cell Dev Biol 21: 695–718. doi: 10.1146/annurev.cellbio.21.122303.120306 [DOI] [PubMed] [Google Scholar]
- Davis FM, Lloyd-Lewis B, Harris OB, Kozar S, Winton DJ, Muresan L, Watson CJ. 2016. Single-cell lineage tracing in the mammary gland reveals stochastic clonal dispersion of stem/progenitor cell progeny. Nat Commun 7: 13053. doi: 10.1038/ncomms13053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deome KB, Faulkin LJ Jr, Bern HA, Blair PB. 1959. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res 19: 515–520. [PubMed] [Google Scholar]
- Deschene ER, Myung P, Rompolas P, Zito G, Sun TY, Taketo MM, Saotome I, Greco V. 2014. β-Catenin activation regulates tissue growth non-cell autonomously in the hair stem cell niche. Science 343: 1353–1356. doi: 10.1126/science.1248373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, et al. 2018. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174: 1586–1598.e12. doi: 10.1016/j.cell.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. 2012. Defining the mode of tumour growth by clonal analysis. Nature 488: 527–530. doi: 10.1038/nature11344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, et al. 2015. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521: 43–47. doi: 10.1038/nature14415 [DOI] [PubMed] [Google Scholar]
- Duarte D, Hawkins ED, Akinduro O, Ang H, De Filippo K, Kong IY, Haltalli M, Ruivo N, Straszkowski L, Vervoort SJ, et al. 2018. Inhibition of endosteal vascular niche remodeling rescues hematopoietic stem cell loss in AML. Cell Stem Cell 22: 64–77.e6. doi: 10.1016/j.stem.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Young PA. 2006. Principles of multiphoton microscopy. Nephron Exp Nephrol 103: e33–e40. doi: 10.1159/000090614 [DOI] [PubMed] [Google Scholar]
- Ellenbroek SI, van Rheenen J. 2014. Imaging hallmarks of cancer in living mice. Nat Rev Cancer 14: 406–418. doi: 10.1038/nrc3742 [DOI] [PubMed] [Google Scholar]
- Ellis SJ, Gomez NC, Levorse J, Mertz AF, Ge Y, Fuchs E. 2019. Distinct modes of cell competition shape mammalian tissue morphogenesis. Nature 569: 497–502. doi: 10.1038/s41586-019-1199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entenberg D, Voiculescu S, Guo P, Borriello L, Wang Y, Karagiannis GS, Jones J, Baccay F, Oktay M, Condeelis J. 2018. A permanent window for the murine lung enables high-resolution imaging of cancer metastasis. Nat Methods 15: 73–80. doi: 10.1038/nmeth.4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al. 2015. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527: 472–476. doi: 10.1038/nature15748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, et al. 2016. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell 18: 827–838. doi: 10.1016/j.stem.2016.04.003 [DOI] [PubMed] [Google Scholar]
- Füllgrabe A, Joost S, Are A, Jacob T, Sivan U, Haegebarth A, Linnarsson S, Simons BD, Clevers H, Toftgård R, et al. 2015. Dynamics of Lgr6+ progenitor cells in the hair follicle, sebaceous gland, and interfollicular epidermis. Stem Cell Reports 5: 843–855. doi: 10.1016/j.stemcr.2015.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli A, Drost J, Suijkerbuijk SJ, van Boxtel R, de Ligt J, Offerhaus GJ, Begthel H, Beerling E, Tan EH, Sansom OJ, et al. 2017. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci 114: E2357–E2364. doi: 10.1073/pnas.1701219114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiu J, Hannezo E, Yui S, Demharter S, Ulyanchenko S, Maimets M, Jorgensen A, Perlman S, Lundvall L, Mamsen LS, et al. 2019. Tracing the origin of adult intestinal stem cells. Nature 570: 107–111. doi: 10.1038/s41586-019-1212-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannezo E, Scheele C, Moad M, Drogo N, Heer R, Sampogna RV, van Rheenen J, Simons BD. 2017. A unifying theory of branching morphogenesis. Cell 171: 242–255.e27. doi: 10.1016/j.cell.2017.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Nakagawa T, Enomoto H, Suzuki M, Yamamoto M, Simons BD, Yoshida S. 2014. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 14: 658–672. doi: 10.1016/j.stem.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper KL, Sosa MS, Entenberg D, Hosseini H, Cheung JF, Nobre R, Avivar-Valderas A, Nagi C, Girnius N, Davis RJ, et al. 2016. Mechanism of early dissemination and metastasis in Her2+ mammary cancer. Nature 540: 588–592. doi: 10.1038/nature20609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins ED, Duarte D, Akinduro O, Khorshed RA, Passaro D, Nowicka M, Straszkowski L, Scott MK, Rothery S, Ruivo N, et al. 2016. T-cell acute leukaemia exhibits dynamic interactions with bone marrow microenvironments. Nature 538: 518–522. doi: 10.1038/nature19801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F, Denk W. 2005. Deep tissue two-photon microscopy. Nat Methods 2: 932–940. doi: 10.1038/nmeth818 [DOI] [PubMed] [Google Scholar]
- Huang Q, Shan S, Braun RD, Lanzen J, Anyrhambatla G, Kong G, Borelli M, Corry P, Dewhirst MW, Li CY. 1999. Noninvasive visualization of tumors in rodent dorsal skin window chambers. Nat Biotechnol 17: 1033–1035. doi: 10.1038/13736 [DOI] [PubMed] [Google Scholar]
- Huels DJ, Bruens L, Hodder MC, Cammareri P, Campbell AD, Ridgway RA, Gay DM, Solar-Abboud M, Faller WJ, Nixon C, et al. 2018. Wnt ligands influence tumour initiation by controlling the number of intestinal stem cells. Nat Commun 9: 1132. doi: 10.1038/s41467-018-03426-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, Toftgård R. 2008. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet 40: 1291–1299. doi: 10.1038/ng.239 [DOI] [PubMed] [Google Scholar]
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, Watt FM. 2009. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4: 427–439. doi: 10.1016/j.stem.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. 2009. The basics of epithelial–mesenchymal transition. J Clin Invest 119: 1420–1428. doi: 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedrin D, Gligorijevic B, Wyckoff J, Verkhusha VV, Condeelis J, Segall JE, van Rheenen J. 2008. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods 5: 1019–1021. doi: 10.1038/nmeth.1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464: 112–115. doi: 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Kon S, Ishibashi K, Katoh H, Kitamoto S, Shirai T, Tanaka S, Kajita M, Ishikawa S, Yamauchi H, Yako Y, et al. 2017. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat Cell Biol 19: 530–541. doi: 10.1038/ncb3509 [DOI] [PubMed] [Google Scholar]
- Kretzschmar K, Clevers H. 2016. Organoids: modeling development and the stem cell niche in a dish. Dev Cell 38: 590–600. doi: 10.1016/j.devcel.2016.08.014 [DOI] [PubMed] [Google Scholar]
- Kretzschmar K, Watt FM. 2012. Lineage tracing. Cell 148: 33–45. doi: 10.1016/j.cell.2012.01.002 [DOI] [PubMed] [Google Scholar]
- Krieger T, Simons BD. 2015. Dynamic stem cell heterogeneity. Development 142: 1396–1406. doi: 10.1242/dev.101063 [DOI] [PubMed] [Google Scholar]
- Langlands AJ, Almet AA, Appleton PL, Newton IP, Osborne JM, Näthke IS. 2016. Paneth cell-rich regions separated by a cluster of Lgr5+ cells initiate crypt fission in the intestinal stem cell niche. PLoS Biol 14: e1002491. doi: 10.1371/journal.pbio.1002491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenos KJ, Miedema DM, Lodestijn SC, Nijman LE, van den Bosch T, Romero Ros X, Lourenço FC, Lecca MC, van der Heijden M, van Neerven SM, et al. 2018. Stem cell functionality is microenvironmentally defined during tumour expansion and therapy response in colon cancer. Nat Cell Biol 20: 1193–1202. doi: 10.1038/s41556-018-0179-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilja AM, Rodilla V, Huyghe M, Hannezo E, Landragin C, Renaud O, Leroy O, Rulands S, Simons BD, Fre S. 2018. Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat Cell Biol 20: 677–687. doi: 10.1038/s41556-018-0108-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim X, Tan SH, Koh WL, Chau RM, Yan KS, Kuo CJ, van Amerongen R, Klein AM, Nusse R. 2013. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science 342: 1226–1230. doi: 10.1126/science.1239730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Matsumura H, Kato T, Ichinose S, Takada A, Namiki T, Asakawa K, Morinaga H, Mohri Y, De Arcangelis A, et al. 2019. Stem cell competition orchestrates skin homeostasis and ageing. Nature 568: 344–350. doi: 10.1038/s41586-019-1085-7 [DOI] [PubMed] [Google Scholar]
- Livet J. 2007. The brain in color: transgenic “Brainbow” mice for visualizing neuronal circuits. Med Sci (Paris) 23: 1173–1176. doi: 10.1051/medsci/200723121173 [DOI] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Côté D, Rowe DW, Lin CP, Scadden DT. 2009. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature 457: 92–96. doi: 10.1038/nature07434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia C, Klein AM, Simons BD, Winton DJ. 2010. Intestinal stem cell replacement follows a pattern of neutral drift. Science 330: 822–825. doi: 10.1126/science.1196236 [DOI] [PubMed] [Google Scholar]
- Mascré G, Dekoninck S, Drogat B, Youssef KK, Brohee S, Sotiropoulou PA, Simons BD, Blanpain C. 2012. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature 489: 257–262. doi: 10.1038/nature11393 [DOI] [PubMed] [Google Scholar]
- McCracken KW, Catá EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence JR, Zavros Y, et al. 2014. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 516: 400–404. doi: 10.1038/nature13863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDole K, Guignard L, Amat F, Berger A, Malandain G, Royer LA, Turaga SC, Branson K, Keller PJ. 2018. In toto imaging and reconstruction of post-implantation mouse development at the single-cell level. Cell 175: 859–876.e33. doi: 10.1016/j.cell.2018.09.031 [DOI] [PubMed] [Google Scholar]
- McKinley KL, Stuurman N, Royer LA, Schartner C, Castillo-Azofeifa D, Delling M, Klein OD, Vale RD. 2018. Cellular aspect ratio and cell division mechanics underlie the patterning of cell progeny in diverse mammalian epithelia. eLife 7: e36739. doi: 10.7554/eLife.36739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa KR, Rompolas P, Zito G, Myung P, Sun TY, Brown S, Gonzalez DG, Blagoev KB, Haberman AM, Greco V. 2015. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature 522: 94–97. doi: 10.1038/nature14306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa KR, Kawaguchi K, Cockburn K, Gonzalez D, Boucher J, Xin T, Klein AM, Greco V. 2018. Homeostatic epidermal stem cell self-renewal is driven by local differentiation. Cell Stem Cell 23: 677–686.e4. doi: 10.1016/j.stem.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, Zarzoso-Lacoste A, et al. 2017. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell 21: 51–64.e6. doi: 10.1016/j.stem.2017.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. 2010. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328: 62–67. doi: 10.1126/science.1182868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, Liu IH, Chiou SH, Salahudeen AA, Smith AR, et al. 2018. Organoid modeling of the tumor immune microenvironment. Cell 175: 1972–1988.e16. doi: 10.1016/j.cell.2018.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah TK, Donahue B, Shroyer NF. 2011. Intestinal development and differentiation. Exp Cell Res 317: 2702–2710. doi: 10.1016/j.yexcr.2011.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohoka A, Kajita M, Ikenouchi J, Yako Y, Kitamoto S, Kon S, Ikegawa M, Shimada T, Ishikawa S, Fujita Y. 2015. EPLIN is a crucial regulator for extrusion of RasV12-transformed cells. J Cell Sci 128: 781–789. doi: 10.1242/jcs.163113 [DOI] [PubMed] [Google Scholar]
- Oost KC, van Voorthuijsen L, Fumagalli A, Lindeboom RGH, Sprangers J, Omerzu M, Rodriguez-Colman MJ, Heinz MC, Verlaan-Klink I, Maurice MM, et al. 2018. Specific labeling of stem cell activity in human colorectal organoids using an ASCL2-responsive minigene. Cell Rep 22: 1600–1614. doi: 10.1016/j.celrep.2018.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, Sugihara H, Fujimoto K, Weissman IL, Capecchi MR, et al. 2009. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med 15: 701–706. doi: 10.1038/nm.1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page ME, Lombard P, Ng F, Göttgens B, Jensen KB. 2013. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell 13: 471–482. doi: 10.1016/j.stem.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Gonzalez DG, Guirao B, Boucher JD, Cockburn K, Marsh ED, Mesa KR, Brown S, Rompolas P, Haberman AM, et al. 2017. Tissue-scale coordination of cellular behaviour promotes epidermal wound repair in live mice. Nat Cell Biol 19: 155–163. doi: 10.1038/ncb3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, Van Keymeulen A, Brown D, Moers V, Lemaire S, et al. 2018. Identification of the tumour transition states occurring during EMT. Nature 556: 463–468. doi: 10.1038/s41586-018-0040-3 [DOI] [PubMed] [Google Scholar]
- Rashidi NM, Scott MK, Scherf N, Krinner A, Kalchschmidt JS, Gounaris K, Selkirk ME, Roeder I, Lo Celso C. 2014. In vivo time-lapse imaging shows diverse niche engagement by quiescent and naturally activated hematopoietic stem cells. Blood 124: 79–83. doi: 10.1182/blood-2013-10-534859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios AC, Clevers H. 2018. Imaging organoids: a bright future ahead. Nat Methods 15: 24–26. doi: 10.1038/nmeth.4537 [DOI] [PubMed] [Google Scholar]
- Ritsma L, Steller EJ, Ellenbroek SI, Kranenburg O, Borel Rinkes IH, van Rheenen J. 2013. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat Protoc 8: 583–594. doi: 10.1038/nprot.2013.026 [DOI] [PubMed] [Google Scholar]
- Ritsma L, Ellenbroek SIJ, Zomer A, Snippert HJ, de Sauvage FJ, Simons BD, Clevers H, van Rheenen J. 2014. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature 507: 362–365. doi: 10.1038/nature12972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Deschene ER, Zito G, Gonzalez DG, Saotome I, Haberman AM, Greco V. 2012. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487: 496–499. doi: 10.1038/nature11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Greco V. 2013. Spatial organization within a niche as a determinant of stem-cell fate. Nature 502: 513–518. doi: 10.1038/nature12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM, Greco V. 2016. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352: 1471–1474. doi: 10.1126/science.aaf7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safferling K, Sütterlin T, Westphal K, Ernst C, Breuhahn K, James M, Jäger D, Halama N, Grabe N. 2013. Wound healing revised: a novel reepithelialization mechanism revealed by in vitro and in silico models. J Cell Biol 203: 691–709. doi: 10.1083/jcb.201212020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265. doi: 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]
- Scheele CL, Hannezo E, Muraro MJ, Zomer A, Langedijk NS, van Oudenaarden A, Simons BD, van Rheenen J. 2017. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature 542: 313–317. doi: 10.1038/nature21046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337: 730–735. doi: 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, Ohta Y, Matano M, Nanki K, Kawasaki K, et al. 2018. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22: 454–467.e6. doi: 10.1016/j.stem.2017.12.009 [DOI] [PubMed] [Google Scholar]
- Serra D, Mayr U, Boni A, Lukonin I, Rempfler M, Challet Meylan L, Stadler MB, Strnad P, Papasaikas P, Vischi D, et al. 2019. Self-organization and symmetry breaking in intestinal organoid development. Nature 569: 66–72. doi: 10.1038/s41586-019-1146-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara T, Orwig KE, Avarbock MR, Brinster RL. 2000. Spermatogonial stem cell enrichment by multiparameter selection of mouse testis cells. Proc Natl Acad Sci 97: 8346–8351. doi: 10.1073/pnas.97.15.8346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, et al. 2010a. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327: 1385–1389. doi: 10.1126/science.1184733 [DOI] [PubMed] [Google Scholar]
- Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, et al. 2010b. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143: 134–144. doi: 10.1016/j.cell.2010.09.016 [DOI] [PubMed] [Google Scholar]
- Snippert HJ, Schepers AG, van Es JH, Simons BD, Clevers H. 2014. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep 15: 62–69. doi: 10.1002/embr.201337799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL. 1988. Purification and characterization of mouse hematopoietic stem cells. Science 241: 58–62. doi: 10.1126/science.2898810 [DOI] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. 2011. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470: 105–109. doi: 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto S, Ohta Y, Fujii M, Matano M, Shimokawa M, Nanki K, Date S, Nishikori S, Nakazato Y, Nakamura T, et al. 2018. Reconstruction of the human colon epithelium in vivo. Cell Stem Cell 22: 171–176.e5. doi: 10.1016/j.stem.2017.11.012 [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk SJE, van Rheenen J. 2017. From good to bad: intravital imaging of the hijack of physiological processes by cancer cells. Dev Biol 428: 328–337. doi: 10.1016/j.ydbio.2017.04.015 [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. 2002. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature 420: 788–794. doi: 10.1038/nature01273 [DOI] [PubMed] [Google Scholar]
- Usui ML, Underwood RA, Mansbridge JN, Muffley LA, Carter WG, Olerud JE. 2005. Morphological evidence for the role of suprabasal keratinocytes in wound reepithelialization. Wound Repair Regen 13: 468–479. doi: 10.1111/j.1067-1927.2005.00067.x [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. 2015. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161: 933–945. doi: 10.1016/j.cell.2015.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rheenen J, Bruens L. 2017. Cellular protection mechanisms that minimise accumulation of mutations in intestinal tissue. Swiss Med Wkly 147: w14539. [DOI] [PubMed] [Google Scholar]
- Varner VD, Nelson CM. 2014. Cellular and physical mechanisms of branching morphogenesis. Development 141: 2750–2759. doi: 10.1242/dev.104794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verissimo CS, Overmeer RM, Ponsioen B, Drost J, Mertens S, Verlaan-Klink I, Gerwen BV, van der Ven M, Wetering MV, Egan DA, et al. 2016. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife 5: e18489. doi: 10.7554/eLife.18489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L, Morrissey E, van der Heijden M, Nicholson AM, Sottoriva A, Buczacki S, Kemp R, Tavare S, Winton DJ. 2013. Defining stem cell dynamics in models of intestinal tumor initiation. Science 342: 995–998. doi: 10.1126/science.1243148 [DOI] [PubMed] [Google Scholar]
- Webster MT, Manor U, Lippincott-Schwartz J, Fan CM. 2016. Intravital imaging reveals ghost fibers as architectural units guiding myogenic progenitors during regeneration. Cell Stem Cell 18: 243–252. doi: 10.1016/j.stem.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers HR, Elkind MM. 1970. Microcolony survival assay for cells of mouse intestinal mucosa exposed to radiation. Int J Radiat Biol Relat Stud Phys Chem Med 17: 261–267. doi: 10.1080/09553007014550291 [DOI] [PubMed] [Google Scholar]
- Wuidart A, Sifrim A, Fioramonti M, Matsumura S, Brisebarre A, Brown D, Centonze A, Dannau A, Dubois C, Van Keymeulen A, et al. 2018. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat Cell Biol 20: 666–676. doi: 10.1038/s41556-018-0095-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin T, Gonzalez D, Rompolas P, Greco V. 2018. Flexible fate determination ensures robust differentiation in the hair follicle. Nat Cell Biol 20: 1361–1369. doi: 10.1038/s41556-018-0232-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Z, Sandoval M, Beronja S. 2018. Oncogenic activation of PI3K induces progenitor cell differentiation to suppress epidermal growth. Nat Cell Biol 20: 1256–1266. doi: 10.1038/s41556-018-0218-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Song B, Pu J, Forrester JV, McCaig CD. 2003. Direct visualization of a stratified epithelium reveals that wounds heal by unified sliding of cell sheets. FASEB J 17: 397–406. doi: 10.1096/fj.02-0610com [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. 2015. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527: 525–530. doi: 10.1038/nature16064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer A, Ellenbroek SI, Ritsma L, Beerling E, Vrisekoop N, Van Rheenen J. 2013. Brief report: intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells 31: 602–606. doi: 10.1002/stem.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]