Abstract
Programmed cell death (PCD) pathways are found in many phyla, ranging from developmentally programmed apoptosis in animals to cell-autonomous programmed necrosis pathways that limit the spread of biotrophic pathogens in multicellular assemblies. Prominent examples for the latter include animal necroptosis and pyroptosis, plant hypersensitive response (HR), and fungal heterokaryon incompatibility (HI) pathways. PCD pathways in the different kingdoms show fundamental differences in execution mechanism, morphology of the dying cells, and in the biological sequelae. Nevertheless, recent studies have revealed remarkable evolutionary parallels, including a striking sequence relationship between the “HeLo” domains found in the pore-forming components of necroptosis and some types of plant HR and fungal HI pathways. Other PCD execution components show cross-kingdom conservation as well, or are derived from prokaryotic ancestors. The currently available data suggest a model, wherein the primordial eukaryotic PCD pathway used proteins similar to present-day plant R-proteins and caused necrotic cell death by direct action of Toll and IL-1 receptor (TIR) and HeLo-like domains.
Programmed cell death (PCD), either initiated cell autonomously in response to pathogens or stimulated from the outside through signaling molecules, is of crucial importance for the success of multicellular organisms. Most of current cell death research is focused on animals and plants, with mammals taking center stage, whereas substantial work is also devoted to model metazoans such as Drosophila melanogaster and Caenorhabditis elegans, or to Arabidopsis thaliana as the main model for plant cell death pathways. However, cell death pathways have also been described in other organisms such as filamentous fungi, and their molecular mechanisms are beginning to be unraveled. At first glance, PCD pathways in different kingdoms appear to work by fundamentally different rules, although a number of recent studies have revealed numerous mechanistic parallels and instances of clear evolutionary interkingdom relationships of cell death mediators. These findings suggest that at least a core pathway for PCD has existed in the common ancestor of metazoans, fungi, and plants. The following paragraphs will provide a synopsis of major cell death pathways in different kingdoms and will highlight the evolutionary processes leading to the diversification of PCD pathways observed today.
PROGRAMMED CELL DEATH SYSTEMS IN DIFFERENT KINGDOMS
PCD modalities can be classified as “necrotic” (i.e., accompanied by membrane rupture and release of intracellular material), and “non-necrotic” without such leakage (Ashkenazi and Salvesen 2014). Because metazoan apoptosis is the major—if not the only—example for the latter type, non-necrotic cell death is usually referred to as “apoptotic.” As compared with apoptosis, necrotic cell death is less intricate and far more widespread. Because cell death caused by major mechanical, chemical, or biological insults is typically associated with cell rupture, necrosis was initially considered to be a hallmark of non-PCD and it took a long time until “programmed necrosis” was accepted as a reality (Edinger and Thompson 2004). In animals, several different types of programmed necrosis exist, among which necroptosis and pyroptosis are the best understood and probably most important examples (Cookson and Brennan 2001; Bergsbaken et al. 2009; Vandenabeele et al. 2010). Cells dying by programmed necrosis release intracellular contents, several components of which are interpreted as “damage-associated molecular patterns” (DAMPs) by the immune system, resulting in inflammation (Schaefer 2014; Roh and Sohn 2018). Apoptosis, in contrast, is a more complicated process because it has to reliably kill the cell while at the same time preventing the leakage of intracellular material and DAMPs. This is no easy task, considering that cellular compartments such as the lysosome and the mitochondrion contain enzymes and oxidants with the potential to damage cell membranes. During apoptosis, the cell is broken into a number of smaller, membrane-enclosed vesicles called “apoptotic bodies,” which are subsequently removed by phagocytic processes (Elmore 2007; Nagata 2018). Apoptosis is therefore ideally suited for developmentally scheduled cell death, a physiological process not supposed to alert the immune system (Fuchs and Steller 2011).
Nonmetazoan forms of PCD are not easily classified as “apoptosis” or “programmed necrosis,” because the dying cells look morphologically different and the signaling cascades and death effectors appear—at least at first glance—unrelated to their metazoan counterparts. In the absence of circulating phagocytic cells, a permanent containment characteristic of metazoan apoptosis is unlikely; whether this containment is important in the absence of an inflammatory system is not clear. There is at least one class of PCD pathway in plants called the “hypersensitive response” (HR), which has been shown to be associated with DAMP release, therein resembling metazoan programmed necrosis pathways (Morel and Dangl 1997; Balint-Kurti 2019). The HR is a part of the so-called “effector triggered immunity” (ETI) system, which gets activated on the detection of pathogen-derived proteins within the host cell. The HR cell death is called “hypersensitive” because it exceeds the damage directly inflicted by the pathogen; its function is to limit the spread of biotrophic pathogens. The production and release of DAMPs—among them the small molecule salicylic acid (SA)—serves the purpose to alert other parts of the plants of the ongoing infection (Balint-Kurti 2019).
Outside of animals and plants, PCD pathways exist (Ameisen 2002), but only a few of them have been characterized in molecular detail. Filamentous fungi belonging to the Ascomycetes possess a number of functionally analogous, but molecularly diverse cell death pathways required for a process called “heterokaryon incompatibility” (HI) (Saupe 2000; Daskalov et al. 2017). Filamentous ascomycetes grow an extensive network of hyphae, which can both branch off and merge back—provided that the merging hyphae are genetically identical. Multiple HI systems prevent the successful fusion of hyphae emanating from genetically different individuals, thereby safeguarding against the spread of pathogens (Daskalov et al. 2017). For a successful hyphal fusion, each of the available HI systems has to be “disarmed” separately—usually by the two fusion partners being homozygous at a polymorphic sensor locus. Triggering only one of the HI systems is sufficient to cause localized cell death near the point of fusion. The formation of intrahyphal septa prevents the spreading of the cell death over the entire hyphal system. For several fungal species, in particular for the HI model organism Podospora anserina, multiple HI systems and their sensor proteins have been described (Saupe 2000; Daskalov et al. 2017; Gonçalves et al. 2017).
MOLECULAR FEATURES OF PROGRAMMED DEATH PATHWAYS
This section provides a synopsis of major PCD pathways, which are mechanistically understood to some degree. In particular, the molecular architecture of the key components is summarized, as this information is necessary to appreciate the evolutionary ancestry of PCD pathways.
Apoptosis
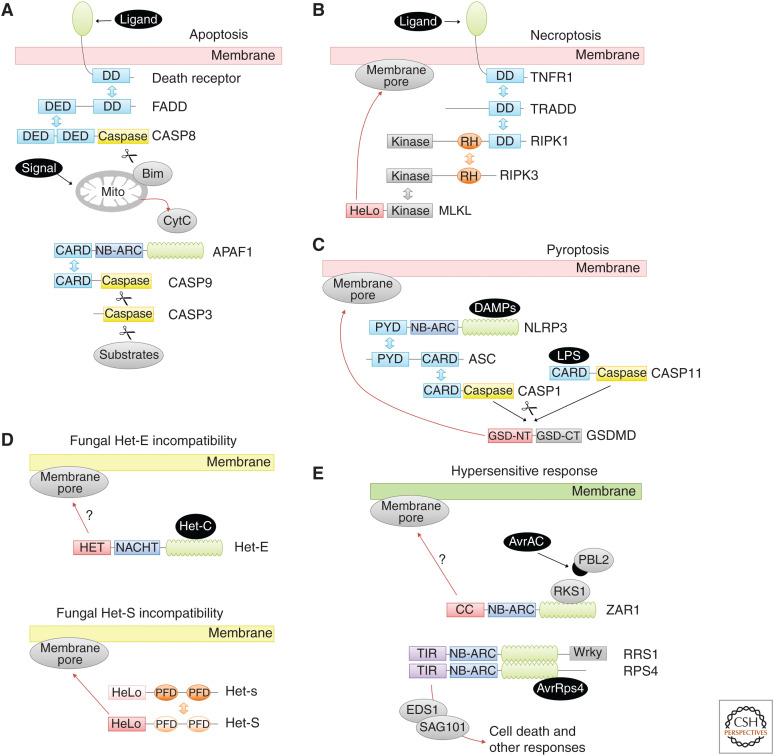
A simplified version of the apoptosis pathway is shown in Figure 1A, and detailed reviews can be found in Elmore (2007) and Nagata (2018). The crucial step in apoptotic cell death induction is the activation of caspase-3, a cysteine protease specifically cleaving a number of different substrates, which in combination bring about the apoptotic phenotype. Apoptosis has, quite appropriately, been called “death by a thousand cuts” (Martin and Green 1995). The activation of caspase-3 happens by proteolytic processing of an inactive precursor; the activating enzyme caspase-9 belongs to the same protease class as caspase-3 and several other proteases involved in cell death signaling (see below). Caspase-9, in turn, is activated by formation of a multiprotein complex called the “apoptosome.” The central component of the apoptosome is APAF1, an ATPase of the STAND class (Danot et al. 2009), which is able to sense the presence of cytochrome c released from mitochondria. On binding to cytochrome c, APAF1 undergoes a major conformational change, leading to the exposure of its amino-terminal oligomerization domain. This so-called CARD (caspase activation and recruitment domain) will then hetero-oligomerize with another CARD domain found at the amino terminus of the inactive precursor of caspase-9. In this oligomeric state, two proximal caspase-9 molecules can cleave and thereby activate each other. There are several pathways leading to apoptosome activation, either cell autonomously or responding to external stimuli via death receptors (Elmore 2007). These upstream pathways use other caspases (caspase-8, caspase-10) and other hetero-oligomerization domains, such as the “death domain” (DD) and the “death effector domain” (DED), which connect the apoptotic signaling components and can lead to the activation of caspases via induced oligomerization and cleavage.
Figure 1.
Signaling domains and their functions in programmed cell death (PCD) pathways. This figure shows simplified versions of major PCD pathways, focusing on the interaction properties of the domains. The arrangement of the proteins in homo-oligomeric complexes is not shown. Coloring: six-helix death-fold domains (death domain [DD], death effector domain [DED], caspase activation and recruitment domain [CARD], pyrin domain [PYD]) are shown in cyan, RIP homotypic interaction motifs (RHIMs) in orange, and Toll and IL-1 receptor (TIR) domains in purple. The central STAND ATPase domains (both NB-ARC and NACHT type) are blue and repetitive sensor domains (leucine-rich repeat [LRR], WD40) are green. Caspase domains are yellow and supposedly pore-forming domains (gasdermin amino-terminal domain, HET domain, and HeLo/coiled-coil [CC]) are shown in red. All other domain types are shown in gray. Homotypic oligomerization is indicated by double arrows colored by domain type. Proteolytic cleavage is indicated by a scissors symbol, whereas translocation events are shown as red arrows. Cell death stimuli are shown on a black background. (A) Apoptosis. Both the extrinsic pathway, triggered by ligand binding to a death receptor, and the intrinsic pathway initiated by a mitochondrial signal are shown. (B) Necroptosis. Only the main components of the canonical pathway, triggered by TNF-receptor type 1 (TNFR1) ligation, is shown here. (C) Pyroptosis. Both major pathways are shown: caspase-11 (human: caspase-4/5) triggering by intracellular lipopolysaccharide (LPS), and caspase-1 activation by signalosome signaling. (D) Fungal heterokaryon incompatibility (HI). Two HI systems are shown: Het-E (from one fusion partner) being triggered by Het-C (contributed by the other partner), and Het-S (from one fusion partner) being recruited to an amyloid formed by Het-s (from the other fusion partner). (E) Plant hypersensitive response. One example for each class of R-protein is shown: The CC-NB-LRR (CNL)-based ZAR1 resistosome is triggered by the ZAR1/RKS1 complex recognizing PBL2, which has been previously modified by the pathogen effector AvrAC. The dimer of the two TIR-NB-LRR (TNL)-based STAND proteins RPS4 and RRS1 is triggered by binding to the pathogen effector AvrRps4 and signals cell death via the EDS1/SAG101 complex.
Necroptosis
A simplified version of the core necroptosis pathway is shown in Figure 1B, and more detailed descriptions can be found in Newton and Manning (2016), Weinlich et al. (2017), and Petrie et al. (2019). The key step committing a cell to necroptosis is the oligomerization and activation of the protein kinase RIPK3. RIPK3 possesses a central RHIM (RIP homotypic interaction motif), which is important for recruitment of RIPK3 to other RHIM-containing proteins, in particular to the related protein kinase RIPK1. Recently, the RIPK1–RIPK3 “necrosome” complex was shown to form a RHIM-based amyloid fibril with alternating strands of RIPK1 and RIPK3 (Mompeán et al. 2018). Once part of the oligomeric necrosome, RIPK3 recruits and phosphorylates a protein called MLKL (mixed lineage kinase domain-like). MLKL is an inactive pseudo-kinase with an additional amino-terminal four-helix bundle (4HB) domain required for cell death execution. On phosphorylation of MLKL, the protein oligomerizes, associates with the cell membrane, causes ion influx, and eventually cells rupture. How exactly these events are timed and interconnected remains a matter of debate. It is nowadays assumed that membrane-associated MLKL oligomers form membrane pores, either on their own or with the help of other cellular factors (Petrie et al. 2019). Many aspects of necroptotic cell death can be mimicked by ectopic expression of the isolated MLKL amino-terminal domain, thereby obviating the need for upstream signaling and RIPK3 activity.
Pyroptosis
The two main pyroptosis pathways are shown in Figure 1C, more detailed descriptions can be found in Kovacs and Miao (2017), Man et al. (2017), and Shi et al. (2017). The key event in executing pyroptotic cell death is the proteolytic cleavage of gasdermin D (GSDMD) at a central position, separating the cell-killing amino-terminal domain from the inhibitory carboxy-terminal domain. Several (nonapoptotic) caspases are able to cleave GSDMD, depending on the initial trigger and the upstream signaling pathway. One subpathway responds to intracellular lipopolysaccharide (LPS) and cleaves GSDMD by caspase-11 in the mouse and caspase-4/5 in humans. Another pathway depends on inflammasome activation and uses the major proinflammatory caspase-1. Once activated, these caspases will not only cleave GSDMD but also other proteins with accessible cleavage sites, most importantly the proform of interleukin (IL)-1β, thereby forming the active mature form of this proinflammatory cytokine. The liberated amino-terminal domain of GSDMD is thought to undergo a major conformational change, leading to its oligomerization and formation of a large membrane pore, which allows the release of the processed IL-1β. Besides GSDMD, other members of the gasdermin family have cleavable amino-terminal domains that support pore formation (Feng et al. 2018). Recently, the structure of the gasdermin A3 pore has been solved by cryo-electron microscopy (EM) and was shown to form a 108-stranded β-barrel, consisting of 27 gasdermin units (Ruan et al. 2018). Other proteases have also been reported to cause gasdermin-dependent pyroptotic cell death (Xia et al. 2019). In cells infected by the bacterial pathogen Yersinia pestis, the apoptotic caspase-8 can be activated through a multiprotein complex called “RIPoptosome,” which in turn leads to GSDMD and GSDME processing by the activated caspase, resulting in pyroptosis (Orning et al. 2018; Sarhan et al. 2018). The neutrophil-specific elastase ELANE, a serine-protease unrelated to caspases, has been shown to cleave GSDMD at an alternative site, which also results in the generation of a cytotoxic amino-terminal fragment (Kambara et al. 2018).
Heterokaryon Incompatibility
A simplified depiction of two fungal HI systems is shown in Figure 1D, and more comprehensive descriptions can be found in Saupe (2000), Daskalov et al. (2017), and Gonçalves et al. (2017). A number of different HI pathways have been described in model fungi. Some of these pathways are “allelic,” meaning that they are triggered if the fused hyphae are heterozygous for a particular polymorphic sensor gene. Other systems are triggered by the interaction of two different gene products, each of them contributed by one of the fused cells. The best understood pathway is probably the allelic Het-S system in P. anserina (Seuring et al. 2012; Riek and Saupe 2016). Het-s and Het-S are two alleles of a gene encoding a potentially toxic two-domain protein. The carboxy-terminal domain can initiate formation of an amyloid structure, which is not toxic by itself, but can cluster multiple copies of the amino-terminal domain. On clustering, the amino-terminal domain can oligomerize, insert into the plasma membrane, and cause the loss of membrane integrity. The protein version encoded by the Het-s allele is able to initiate an amyloid structure by its carboxy-terminal “prion-forming domains” (PFDs); however, the amino-terminal “HeLo” domain of the Het-s protein is not able to permeabilize the membrane. In contrast, the protein encoded by the Het-S allele has a functional amino-terminal domain, but is not able to initiate amyloid formation owing to a mutation in the PFD region. When both Het-S and Het-s encoded proteins encounter each other during fusion of incompatible cells, the Het-s protein will initiate amyloid formation, whereas the Het-S protein can extend these amyloids, thereby triggering the membrane pore formed by the Het-S amino-terminal domain. A fundamentally different incompatibility mechanism, which is also relevant for the discussion of cell death evolution, is found in the non-allelic Het-E/Het-C system and its relatives. Here, the central component is the Het-E protein, a STAND-type ATPase with a similar architecture as the apoptosome component APAF1 and the NLR components of the inflammasome. The carboxy-terminal WD40-repeat region of Het-E can sense the presence of particular alleles of the (unrelated) Het-C gene product. On binding to Het-C, the STAND ATPase undergoes a conformational change leading to the exposure of the Het-E amino-terminal region, usually referred to as the “HET domain.” Unlike the situation in apoptosis and inflammasome activation, these HET domains are not thought to be recruitment domains, but rather to directly disintegrate the membrane, leading to a necrotic type of cell death (Paoletti and Clavé 2007). Many other fungal HI systems exist, but only a few of them have been studied for their cell-killing mechanism (Fig. 2).
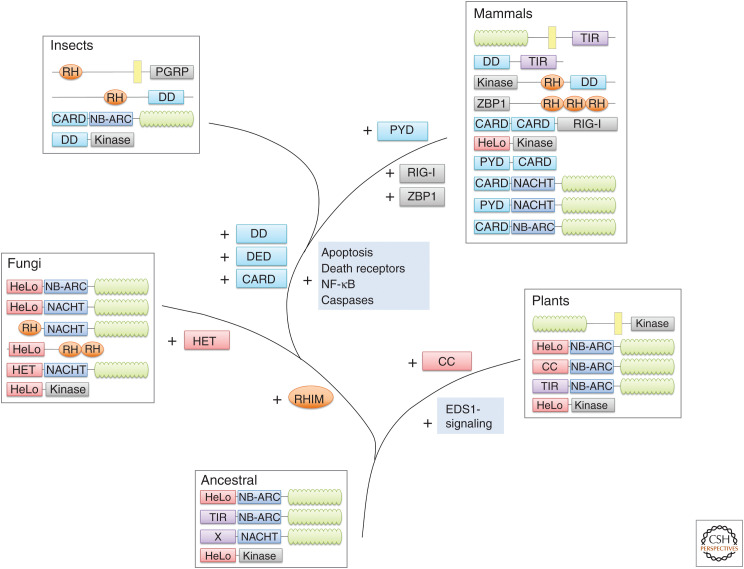
Figure 2.
Evolutionary origins of cell death protein architectures. This figure shows generic protein architectures found in metazoans, fungi, and plants—and what is predicted to have existed in an ancestral (early eukaryote) organism. Domain coloring identical to Figure 1. The ancestral system is predicted to mainly have used STAND ATPases, most likely of the NB-ARC type, with amino-terminal Toll and IL-1 receptor (TIR) and HeLo-like domains. However, NACHT-ATPases and a mixed-lineage kinase domain-like (MLKL) HeLo-domain kinase might also have been present. In the plant lineage, components of EDS1 and probably many other relevant proteins have been acquired. The coiled-coil [CC]-domain found in present plant CC-NB-LRR (CNL) proteins was either acquired or, more likely, evolved from the HeLo domain. RIP homotypic interaction motifs (RHIMs) are first seen in the opisthokont lineage, whereas the six-helix death-fold domains are specific for the metazoan lineage. A more detailed description of early cell death evolution is given in the main text.
Hypersensitive Response
Two examples of plant defense signaling leading to cell death are shown in Figure 1E. More detailed descriptions can be found in Coll et al. (2011), Jones et al. (2016), and Balint-Kurti (2019). Although plant defense mechanisms against pathogens have been studied intensively, and protective cell death by the HR is one important branch of this defense, the exact cell death mechanism and the death-specific signaling components are much less clear. The central orchestrators of intracellular defense pathways are the “R-proteins” (resistance proteins). Most R-proteins belong to the class of STAND-type ATPases with a leucine-rich repeat (LRR) domain at the carboxyl terminus and an effector/signaling domain at the amino terminus. Like all other STAND-type ATPases, R-proteins are thought to undergo a structural rearrangement on binding to its cognate ligand, which may be bacterial effectors secreted into the host cell or any other molecule indicative of biotic stress. Depending on the nature of the amino-terminal domain, different downstream signaling pathways can be engaged, eventually leading to the induction of defense genes, the generation and release of the long-range signaling molecule SA, and/or to the induction of hypersensitive cell death. The factors required for cell death execution are not known and depend on the R-protein, which might belong to one of several subtypes. The TNL proteins carry an amino-terminal Toll and IL-1 receptor (TIR) domain and signal cell death via the EDS1-SAG101-NRG1 pathway (Lapin et al. 2019) although the details of that mechanism are not known. A second class of R-proteins are the “CC-NB-LRR” (CNL) proteins, which carry an amino-terminal domain that was initially considered to form a coiled-coil (CC) structure, although this is not necessarily true. At least some members of the CNL-type R-proteins can induce cell death dependent on the amino-terminal domain (Collier et al. 2011), although the exact mechanism remains unresolved.
RECURRING MOTIFS AND MECHANISMS IN CELL DEATH SIGNALING
When comparing the mechanisms of the cell death pathways described above, in particular the domain architecture of the key proteins involved, a number of recurring features suggesting a common evolutionary history becomes apparent.
STAND ATPases in Cell Death and Immunity
One of the most obvious recurring features in cell death signaling is the use of STAND-type ATPases as a signaling hub (Leipe et al. 2004; Danot et al. 2009). The name STAND, originally an acronym for “signal transducing ATPases with numerous domains,” encompasses two major subfamilies of large ATPases of similar core architecture, which are both important for cell death signaling. One class has a central “NB-ARC” ATPase domain (named after the proteins APAF1/R-Proteins/Ced-4), the other class has a central “NACHT” ATPase domain (named after NAIP/CIIA/Het-E/TEP1). Both classes contain carboxy-terminal sensor regions, typically consisting of repeat-forming domains, such as LRRs, WD40 repeats, or tetratricopeptide repeats (TPRs). At the amino terminus, different effector domains can be found. Both NB-ARC and NACHT ATPases work similarly. In the resting state, the proteins are monomeric and show a “closed” conformation, which shields the amino-terminal effector domain. On binding of the carboxy-terminal sensor domain to its cognate stimulus, the central ATPase domain undergoes a conformational change, causing an oligomerization of the ATPase and leading to the exposure of the amino-terminal effector domains. This process is best understood for the NB-ARC protein APAF1, the central component of the apoptosome (Cheng et al. 2016; Dorstyn et al. 2018). Here, the active conformation has the shape of a heptameric “wheel” formed by seven ATPase domains; the amino-terminal CARD domains of the seven APAF1 units are found clustered above the plane of the ATPase wheel. Ced-4, the APAF1 homolog from the nematode C. elegans, forms an octameric ring with a similar domain arrangement (Qi et al. 2010; Huang et al. 2013).
The vast majority of eukaryotic NB-ARC and NACHT proteins are known or suspected to be involved in innate immunity and cell death signaling. As mentioned before, the (sole) human NB-ARC protein APAF1 and its nematode homolog Ced-4 use their amino-terminal CARD domain for apoptosome formation. Mammals possess several NACHT proteins with amino-terminal CARD or pyrin domains (PYDs), which are the key components of “inflammasomes,” protein complexes similar to the apoptosome but activating caspase-1 rather than caspase-9 and thereby leading to pyroptosis (Broz and Dixit 2016).
The relationship between the mammalian STAND ATPases and the plant R-proteins is readily visible and has stimulated a number of analyses comparing animal and plant cell-autonomous immunity (Maekawa et al. 2011; Jones et al. 2016; Urbach and Ausubel 2017). Recently, it was shown by cryo-EM that plant R-proteins can also form wheel-like oligomers. In the example of ZAR1, an R-protein of the CNL class, the wheel has a pentameric structure called the “ZAR1 resistosome” to emphasize the analogy to the metazoan apoptosome and inflammasome complexes (Wang et al. 2019). In the activated ZAR1 resistosome, the five individual “CC-type” effector domains undergo a conformational rearrangement and form a pore-like α-barrel with an additional funnel-like structure formed by the first α-helices of each CC domain. This funnel appears to be crucial for cell death induction by activated ZAR1 (Wang et al. 2019). Although there is no formal proof that the CC domain of ZAR1 forms a membrane pore sufficient for ion influx or even cell rupture, it appears that the plant R-proteins use a more direct way to cell death than their metazoan counterparts, which rely on further downstream signaling.
Several of the fungal HI systems also make use of STAND ATPases. The Het-E protein mentioned above, but also Het-D and Het-R all contain a central NACHT ATPase domain, followed by a carboxy-terminal sensor domain consisting of WD40-repeats. The amino-terminal effector domains (HET-domains) are only found in filamentous fungi and are thought to directly form membrane pores on oligomerization (Paoletti and Clavé 2007). This mode of action would be analogous to what has been suggested for ZAR1 and possibly other CC-type R-proteins. When comparing the domain architectures of APAF1 (CARD, NB-ARC, WD40) with that of ZAR1 (CC, NB-ARC, LRR) or HET-E (HET, NACHT, WD40), it becomes obvious that these proteins have the same overall architecture but did not evolve by divergent evolution from a common ancestor. It can be assumed that early eukaryotes already contained STAND-ATPases because most extant bacteria encode several such ATPases. However, the classical bacterial STAND-ATPases do not belong to the NB-ARC or NACHT subtype and have no connection to cell death signaling (Leipe et al. 2004). A careful phylogenetic analysis of the STAND family concluded that plant and animal STAND proteins evolved independently from bacterial precursors, in at least two separate events (Urbach and Ausubel 2017). However, STAND ATPases and other innate immunity proteins are clearly subject to evolution by “domain swapping,” and when analyzing eukaryotic proteomes available nowadays, virtually all combinations of effector domains, ATPase subtype, and carboxy-terminal sensor domains can be observed. Therefore, alternative evolutionary hypotheses should not be discounted, for example, a pervasive “mixing and matching” of domains in early eukaryotic evolution, with a subsequent “fixing” of select domain architectures that proved most beneficial for the particular eukaryotic lineage.
Evolution of Effector and Oligomerization Domains
The formation of transient multiprotein complexes, leading to the recruitment of adaptor and effector proteins to activated receptors, is a recurring feature of apoptosis, necroptosis, and pyroptosis signaling. In metazoan systems, the prevalent oligomerization domain types are the DD, the DED, the CARD, and the PYD. All four domain types are distantly related to each other (Hofmann 1999; Park et al. 2007a; Kersse et al. 2011) and share a common structural fold consisting of six α-helices, often referred to as the “death fold” or “six-helix bundle fold.” The principal function of all four domain classes is to recruit another domain of the same class. Despite the relationship between DD, DED, CARD, and PYD, interactions across the class boundaries are rare. It is interesting to note that RHIM motifs and TIR domains fulfil analogous roles of “recruitment by oligomerization,” but are not related to the death-fold superfamily and do not share the six-helix bundle structure (Nanson et al. 2019).
Despite exhaustive bioinformatical searches (Hofmann 1999; unpubl. data), no members of the death-fold superfamily could be detected outside of metazoans, with the exception of viruses and other pathogens, which have probably acquired these domains from their metazoan hosts (Thome et al. 1997; Lamkanfi and Dixit 2010). It thus appears that the six-helix death fold arose during metazoan evolution. It is also remarkable that all characterized members of the death-fold superfamily reside in proteins involved in cell death and innate immunity signaling; other pathways requiring oligomerization appear to use other domains for this purpose. A possible reason for this pathway specificity might lie in the availability of multiple interaction surfaces of the fold, which can be used simultaneously and thus support the formation of higher-order oligomeric structures called “filaments” (Park et al. 2007b; Kersse et al. 2011; Hauenstein et al. 2015). Such filaments can be nucleated by a di- or trimerization and then grow by using the remaining interaction surfaces—analogous to an amyloid but without the β-stack structure typical of real amyloids. These cooperatively formed higher-order structures are instrumental for inflammasome formation (Hauenstein et al. 2015) and support the “all-or-nothing” characteristics required for life/death decisions.
Evolution of TIR Domains
TIR domains have a particularly interesting evolutionary history. In mammals, TIR domains are found at the cytoplasmic end of Toll-like receptors (TLRs) and receptors for IL-1 cytokines—all of them receptors that alert a cell to a danger situation from the outside. Their function is the recruitment of other TIR domains found in “adaptor proteins” such as MYD88, TIRAP, TRAM, and TRIF. Typical downstream events of mammalian TIR signaling include the up-regulation of defense genes via transcription factors of the NF-κB (nuclear factor κ light chain enhancer) or IRF (interferon regulatory factor) families, but also necroptosis induced via the RHIM motif of TRIF. Despite a completely unrelated structure, the role of TIR domains is remarkably similar to members of the six-helix death-fold family. This analogy is further emphasized by the ability of TIR domains to form filaments akin to those of the death-fold domains (Ve et al. 2017; Nanson et al. 2019). Because there clearly is no evolutionary relationship between these domain classes, oligomerization through TIR and death-fold domains is another example of convergent evolution.
In contrast to animals, plants do not use TIR domains in their surface receptors, but rather as the amino-terminal effector domains of one class of intracellular R-proteins (TIR-NB-LRR or TNL type). Surprisingly, plants lack TIR-containing adaptor proteins and the TNL proteins signal via a complex of EDS1 (enhanced disease susceptibility 1) and SAG101 (senescence-associated gene 101) through an as-yet uncharacterized mechanism (Lapin et al. 2019). However, plant TIR domains share with their metazoan counterparts the capacity to mediate dimerization between different R-proteins (Bernoux et al. 2011; Williams et al. 2014; Zhang et al. 2017). An interesting twist to the enigma of TIR signaling was introduced recently by detecting an NAD-cleaving activity for the TIR domain of the human SARM1 (sterile α and TIR motif) protein (Essuman et al. 2017), as well as in several bacterial TIR proteins (Essuman et al. 2018). SARM1 has a role in mediating neurodegeneration and the NAD depletion by its TIR domain was suggested to be a main factor in this process (Essuman et al. 2017). It is unlikely that other human TIR domains share this enzymatic activity, because they either lack the active site glutamate residue or have a structure that places this residue outside the catalytic cleft. However, it remains possible that in the presence of suitable binding partners or posttranslational modifications, the structure may be converted into an active form. Plant TIR domains, in contrast, tend to show conservation in the active site region, and the few available structures appear to support catalysis. It is therefore an intriguing possibility that either NAD depletion, or the generation of (cyclic) ADP-ribose as the NAD degradation product, play a role in cell death signaling by R-proteins of the TIR-NB-LRR architecture. Given that TIR proteins are abundant in extant bacteria and appear to be generally catalytically active (Essuman et al. 2018), it is highly likely that both animal and plant TIR domains evolved from a prokaryotic ancestor and later gained the capacity for oligomerization, although (mostly) losing their catalytic activity in the process.
Evolutionary History of Necroptosis Execution
The execution phase of necroptosis is characterized by MLKL phosphorylation and oligomerization, initiated by formation of a mixed amyloid structure formed by the RHIM motifs of RIPK1 and RIPK3 (Mompeán et al. 2018; Petrie et al. 2019), or possibly by similar structures formed by RIPK3 with other RHIM proteins such as TRIF or ZBP1/DAI. Despite its fundamentally different structure, the role of RHIMs in necroptosis signaling is analogous to those of six-helix death-fold domains or TIR domains. All of these domains form homotypic oligomers and have the tendency to create higher-order superstructures. In the case of RHIM, this superstructure is a “real” amyloid (Li et al. 2012; Mompeán et al. 2018). In evolution, RHIM-based oligomerization is far more widespread than the few examples known in mammals. A bioinformatical analysis showed a large array of RHIM-containing proteins in nonmammalian metazoans. In many of these cases, the RHIM appears to substitute for TIR or six-helix death-fold domains, which are found in the mammalian version of these proteins (Kajava et al. 2014). This finding suggests that RHIM motifs work similar to TIRs and death-fold domains and actually can replace them.
Interestingly, the same bioinformatical analyses found an evolutionary relationship between RHIM motifs and the PFDs used in the Het-S system of HI (Kajava et al. 2014). Like RHIMs, the fungal PFDs form an amyloid of similar structure (Wasmer et al. 2008; Riek and Saupe 2016), supporting a common evolutionary origin of these two oligomerization systems. This evolutionary parallel is further underscored by the finding that the “HeLo domain,” the cell-killing moiety of the Het-S system, is related to the functionally analogous amino-terminal domain of MLKL (Daskalov et al. 2016). It is thus very likely that the metazoan necroptosis system evolved from a simpler precursor similar to Het-S, with amyloid-forming domain and killing domain within the same polypeptide. During vertebrate evolution, the two functionalities were probably split into separate proteins. It is difficult to decide from the available data whether the common precursor was using a one-component amyloid (as in Het-S) or a two-component version (as in RIPK1/RIPK3). Because even the one-component amyloid uses two alternating strands—PFD1 and PFD2 of Het-S—it appears more likely that the extant Het-S system is a degenerate version of a former two-component systems, now perfectly adapted to the task of detecting heterozygosity.
Evolutionary History of Pyroptosis and Caspases
The decisive step in pyroptosis execution is the cleavage of GSDMD by caspase-1 or other caspases with similar cleavage specificities. Caspase-1 itself is activated by inflammasomes, which can be induced by many proinflammatory stimuli. The fact that caspase-1 also activates IL-1β makes pyroptosis the major modality for releasing processed IL-1β and thus causing the inflammatory phenotype (Man et al. 2017; Green 2019). The other GSDMD-cleaving caspases 4, 5, and 11 are directly activated by intracellular LPS. The details of this activation are not fully understood. All of the GSDMD-cleaving caspases are thought to oligomerize via their amino-terminal CARD domains, but the involvement of filaments of higher-order structures has not been reported.
Neither gasdermins nor “proper” caspases are found outside of metazoans; the IL-1 cytokine family is even restricted to vertebrates. Thus, pyroptosis, like apoptosis, appears to be a relatively recent addition to the arsenal of PCD pathways. On the other hand, the metazoan caspases are related to two other classes of cysteine proteases, the metacaspases and paracaspases, which have a much wider evolutionary distribution and are even found in bacteria (Koonin and Aravind 2002). Initially, the finding of metacaspases in yeasts, together with the finding that overexpression of some mammalian apoptosis proteins killed yeast cells, gave rise to speculations that caspase-dependent cell death or even apoptosis might be conserved in lower eukaryotes (Váchová and Palková 2007). By now, it is known that metacaspases, despite their evolutionary relationship to caspases, have a totally different cleavage specificity (cleavage after arginine rather than aspartate) and cannot replace the function of proper metazoan caspases (Tsiatsiani et al. 2011). Nevertheless, metacaspases might have a more general role in DAMP processing during necrotic cell death. A recent study showed that plant metacaspases process a cytoplasmic immunomodulatory plant protein, the active part of which PEP1 (plant elicitor peptide 1) is secreted from damaged cells and acts as a defense signal (Hander et al. 2019). This situation is comparable to the IL-1β release during pyroptosis but without any sequence or structural similarity between PEP1 and the metazoan interleukin.
Another question of evolutionary relevance concerns the origins of the gasdermin family. Of the six gasdermins known in the human genome, four are relatively closely related to each other (GSDMA, GSDMB, GSDMC, GSDMD). This subfamily is fast evolving, with some additional members in rodents and other mammals. Common to all these proteins is the architecture with an amino-terminal toxic domain, a carboxy-terminal inhibitory domain, and a protease cleavage site in the middle. Two more gasdermins (DFNA5/GSDME and Pejvakin/PJVK) form a second subfamily, which is somewhat more distantly related, in particular in the quite divergent inhibitory region. GSDME, which unlike gasdermin A-D has homologs in fish, behaves like a classical gasdermin; it is cleaved by caspases and its amino-terminal domain is able to cause pyroptosis (Wang et al. 2017; Jiang et al. 2019). In contrast, the sixth gasdermin, Pejvakin/PJVK, does not appear to be toxic (Feng et al. 2018) but has homologs in invertebrates. The exact role of PJVK is not known; the human gene is implicated in nonsyndromic hearing loss (Harris et al. 2017) and has been proposed to regulate pexophagy, the autophagic removal of peroxisomes (Defourny et al. 2019). Based on these considerations, it appears likely that the metazoan gasdermin family evolved from a Pejvakin-like precursor and possibly acquired the cell-killing activity of the amino-terminal later on. However, it remains possible that the proto-gasdermin gene encoded a cell-killing protein and that Pejvakin lost this activity. As a third possibility, Pejvakin might still be able to kill cells, but requires a specific activation mode awaiting to be discovered. A much older evolutionary history—or possibly a horizontal acquisition—is suggested by the recently published structure of the mouse gasdermin A3 pore (Ruan et al. 2018), which is probably a good model for other gasdermin pores as well. Both the pore structure and the conformational change undergone by the GSDMA3 amino-terminal domain on pore formation strongly resemble the pores of bacterial cytolysins such as Pneumolysin and Perfringolysin O, but also the pores of mammalian perforin and the membrane attack complex (MAC) of the complement system (Ruan et al. 2018). Despite the structural similarities of these pores, there is no overt sequence similarity, which makes it difficult to judge whether these proteins are truly related or just further examples of convergent evolution.
HOW IT ALL BEGAN
An inventory of present-day cell death components, their domain architecture, and their interrelationship should, in principle, allow the reconstruction of key evolutionary events shaping cell death signaling. However, the fast evolution of some components, the pervasive domain shuffling, and the incomplete knowledge of death signaling outside the classical model organisms make this task difficult if not impossible. The sequence of evolutionary events proposed here is in accordance with the available data, but there is no certainty that it reflects the history correctly.
Because STAND-type ATPases are abundant in both pro- and eukaryotes, and fulfill similar purposes in multiple kingdoms, it is safe to assume that STAND-based signaling was available in early eukaryotes including the last common ancestor to animals, fungi, and plants. At the carboxyl terminus of extant STAND ATPases, many different sensor domains are found, typically repeat domains. In plants, most of the sensing appears to be performed by LRR domains, which are also common in mammalian STAND ATPases. However, when including invertebrates and fungi (which are more closely related to animals than plants), a much greater sensor diversity is observed, including WD40, TPR, and ankyrin repeats. One published study concluded that the NB-ARC subtype (as used in plant R-proteins) and the NACHT subtype (as used in animal inflammasomes and fungal Het-E) evolved independently from a bacterial ancestor (Urbach and Ausubel 2017). This study assumed bacteria-specific subfamilies of the STAND superfamily as the precursor, from which eukaryotic NACHT and NB-ARC subtypes evolved. Whereas this is undoubtedly possible, there are plenty of bacterial NB-ARC and NACHT members in present-day genomes. This observation means that either the two independent events leading to NACHT and NB-ARC happened before the split of eukaryotes, or that the NACHT and NB-ARC proteins seen in extant bacteria are late horizontal acquisitions from a eukaryotic source. Most likely, early eukaryotes coded for NACHT and NB-ARC-type ATPases with a wide range of possible sensor motifs.
A most relevant question of cell death evolution is what kind of effector domains were used by these early signaling hubs. One prime candidate are the TIR domains, they clearly predate the advent of eukaryotes, and they were most likely enzymatically active like the extant bacterial versions (Essuman et al. 2018). Actually, the NAD-degrading function of TIR domains in the context of a STAND ATPase might have formed the basis for a very early eukaryotic cell death system, triggered by some pathogen- or damage-derived molecule and leading to cell death by NAD depletion. Another early cell death system could have used a HeLo-like domain as the effector. Although HeLo domains have not been detected in bacteria, several copies of this domain exist in plants, fungi, animals, and other eukaryotes. Because the HeLo domain appears to cause membrane pores autonomously, at least in fungi, this domain is an equally good candidate for an early PCD system. Interestingly, the two most likely architectures resemble the two classes of plant R-proteins: TIR-NB-LRR and CC-NB-LRR. However, it is well possible that the ancient versions of this system used another subtype of STAND ATPase, or another type of sensor repeat.
Cell death systems relying on caspase activity, such as apoptosis and pyroptosis, most likely did not develop before the advent of metazoans, although it cannot be excluded that there have been pyroptosis-like systems using another type of protease. However, there are no indications for such a system in the presently available data. Apoptosis signaling, at least in its present mammalian form, relies not only on caspases but also on numerous six-helix death-fold domains, all of which appear only in metazoans, suggesting that apoptosis is probably the youngest of the PCD systems discussed here. In contrast, necroptosis appears to be ancient, at least the downstream events. Necroptotic cell death is caused by the HeLo domain of MLKL, which is clearly ancient, whereas RHIM motifs are only found in animals and the related PFD of Het-s has not been found outside of fungi. Nevertheless, we recently identified an MLKL pseudokinase in plants, whose amino-terminal domain forms a 4HB similar to that of mammalian MLKL and is also able to cause cell death (Mahdi et al. 2019). Although plants appear to lack RIPK3 homologs and other RHIM-based signaling proteins, this finding suggests that at least the execution step of necroptosis predates the split of animals and plants.
When considering the later evolutionary steps of cell death pathways, there are several examples of intermediate steps being added, probably to allow more regulatory layers, or to further amplify the reaction—making PCD induction an “all-or-nothing” decision. One example is the (proposed) conversion of TIR domains from its original catalytic form, thought to exert a direct killing effect, into a signaling domain that recruits further TIR-containing proteins in large numbers. A similar change might have occurred to the HeLo domain, thought to be a direct cell-killing domain in fungi, into a possible signaling domain in plants. On a smaller scale, the addition of regulatory layers is also visible in apoptosis evolution among metazoans. Whereas in nematodes, the Ced-4 apoptosome directly recruits the effector caspase Ced-3, the mammalian APAF1 apoptosome recruits and activates an intermediate caspase (caspase-9), each molecule of which can then activate multiple copies of the mammalian effector caspase-3. It might even be possible that the RHIM motifs and six-helix death-fold domains, in their original form, did have a direct cell-killing effect and only later evolved into signaling and recruitment domains. However, this remains speculative because there are no data to support that idea. Taken together, it becomes clear that PCD pathways across kingdoms use a similar signaling logic and evolved from a common ancestral pathway, probably much simpler than the intricate multilayer systems observed in multicellular organisms.
ACKNOWLEDGMENTS
We thank Shuhua Chen for valuable discussions. This work was funded by a grant from the Deutsche Forschungsgemeinschaft (SFB 670).
Footnotes
Editors: Kim Newton, James M. Murphy, and Edward A. Miao
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Ameisen JC. 2002. On the origin, evolution, and nature of programmed cell death: a timeline of four billion years. Cell Death Differ 9: 367–393. doi: 10.1038/sj.cdd.4400950 [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Salvesen G. 2014. Regulated cell death: signaling and mechanisms. Annu Rev Cell Dev Biol 30: 337–356. doi: 10.1146/annurev-cellbio-100913-013226 [DOI] [PubMed] [Google Scholar]
- Balint-Kurti P. 2019. The plant hypersensitive response: concepts, control and consequences. Mol Plant Pathol 20: 1163–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, Cookson BT. 2009. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol 7: 99–109. doi: 10.1038/nrmicro2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN. 2011. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9: 200–211. doi: 10.1016/j.chom.2011.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P, Dixit VM. 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16: 407–420. doi: 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- Cheng TC, Hong C, Akey IV, Yuan S, Akey CW. 2016. A near atomic structure of the active human apoptosome. eLife 5: e17755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. 2011. Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256. doi: 10.1038/cdd.2011.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, Moffett P. 2011. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24: 918–931. doi: 10.1094/MPMI-03-11-0050 [DOI] [PubMed] [Google Scholar]
- Cookson BT, Brennan MA. 2001. Pro-inflammatory programmed cell death. Trends Microbiol 9: 113–114. doi: 10.1016/S0966-842X(00)01936-3 [DOI] [PubMed] [Google Scholar]
- Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. 2009. Wheel of life, wheel of death: a mechanistic insight into signaling by STAND proteins. Structure 17: 172–182. doi: 10.1016/j.str.2009.01.001 [DOI] [PubMed] [Google Scholar]
- Daskalov A, Habenstein B, Sabaté R, Berbon M, Martinez D, Chaignepain S, Coulary-Salin B, Hofmann K, Loquet A, Saupe SJ. 2016. Identification of a novel cell death-inducing domain reveals that fungal amyloid-controlled programmed cell death is related to necroptosis. Proc Natl Acad Sci 113: 2720–2725. doi: 10.1073/pnas.1522361113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalov A, Heller J, Herzog S, Fleißner A, Glass NL. 2017. Molecular mechanisms regulating cell fusion and heterokaryon formation in filamentous fungi. Microbiol Spectr 5. doi: 10.1128/microbiolspec.FUNK-0015-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defourny J, Aghaie A, Perfettini I, Avan P, Delmaghani S, Petit C. 2019. Pejvakin-mediated pexophagy protects auditory hair cells against noise-induced damage. Proc Natl Acad Sci 116: 8010–8017. doi: 10.1073/pnas.1821844116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Akey CW, Kumar S. 2018. New insights into apoptosome structure and function. Cell Death Differ 25: 1194–1208. doi: 10.1038/s41418-017-0025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. 2004. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol 16: 663–669. doi: 10.1016/j.ceb.2004.09.011 [DOI] [PubMed] [Google Scholar]
- Elmore S. 2007. Apoptosis: a review of programmed cell death. Toxicol Pathol 35: 495–516. doi: 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essuman K, Summers DW, Sasaki Y, Mao X, DiAntonio A, Milbrandt J. 2017. The SARM1 Toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 93: 1334–1343.e5. doi: 10.1016/j.neuron.2017.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essuman K, Summers DW, Sasaki Y, Mao X, Yim AKY, DiAntonio A, Milbrandt J. 2018. TIR domain proteins are an ancient family of NAD+-consuming enzymes. Curr Biol 28: 421–430.e4. doi: 10.1016/j.cub.2017.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Fox D, Man SM. 2018. Mechanisms of gasdermin family members in inflammasome signaling and cell death. J Mol Biol 430: 3068–3080. doi: 10.1016/j.jmb.2018.07.002 [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. 2011. Programmed cell death in animal development and disease. Cell 147: 742–758. doi: 10.1016/j.cell.2011.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves AP, Heller J, Daskalov A, Videira A, Glass NL. 2017. Regulated forms of cell death in fungi. Front Microbiol 8: 1837. doi: 10.3389/fmicb.2017.01837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DR. 2019. The coming decade of cell death research: five riddles. Cell 177: 1094–1107. doi: 10.1016/j.cell.2019.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hander T, Fernández-Fernández AD, Kumpf RP, Willems P, Schatowitz H, Rombaut D, Staes A, Nolf J, Pottie R, Yao P, et al. 2019. Damage on plants activates Ca2+-dependent metacaspases for release of immunomodulatory peptides. Science 363: eaar7486. doi: 10.1126/science.aar7486 [DOI] [PubMed] [Google Scholar]
- Harris SL, Kazmierczak M, Pangršič T, Shah P, Chuchvara N, Barrantes-Freer A, Moser T, Schwander M. 2017. Conditional deletion of pejvakin in adult outer hair cells causes progressive hearing loss in mice. Neuroscience 344: 380–393. doi: 10.1016/j.neuroscience.2016.12.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauenstein AV, Zhang L, Wu H. 2015. The hierarchical structural architecture of inflammasomes, supramolecular inflammatory machines. Curr Opin Struct Biol 31: 75–83. doi: 10.1016/j.sbi.2015.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K. 1999. The modular nature of apoptotic signaling proteins. Cell Mol Life Sci 55: 1113–1128. doi: 10.1007/s000180050361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Jiang T, Choi W, Qi S, Pang Y, Hu Q, Xu Y, Gong X, Jeffrey PD, Wang J, et al. 2013. Mechanistic insights into CED-4-mediated activation of CED-3. Genes Dev 27: 2039–2048. doi: 10.1101/gad.224428.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Gu H, Zhao Y, Sun L. 2019. Teleost gasdermin E is cleaved by caspase 1, 3, and 7 and induces pyroptosis. J Immunol 203: 1369–1382. doi: 10.4049/jimmunol.1900383 [DOI] [PubMed] [Google Scholar]
- Jones JD, Vance RE, Dangl JL. 2016. Intracellular innate immune surveillance devices in plants and animals. Science 354: aaf6395. doi: 10.1126/science.354.6316.1174-b [DOI] [PubMed] [Google Scholar]
- Kajava AV, Klopffleisch K, Chen S, Hofmann K. 2014. Evolutionary link between metazoan RHIM motif and prion-forming domain of fungal heterokaryon incompatibility factor HET-s/HET-s. Sci Rep 4: 7436. doi: 10.1038/srep07436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H, Liu F, Zhang X, Liu P, Bajrami B, Teng Y, Zhao L, Zhou S, Yu H, Zhou W, et al. 2018. Gasdermin D exerts anti-inflammatory effects by promoting neutrophil death. Cell Rep 22: 2924–2936. doi: 10.1016/j.celrep.2018.02.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. 2011. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci 36: 541–552. doi: 10.1016/j.tibs.2011.06.006 [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. 2002. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ 9: 394–404. doi: 10.1038/sj.cdd.4400991 [DOI] [PubMed] [Google Scholar]
- Kovacs SB, Miao EA. 2017. Gasdermins: effectors of pyroptosis. Trends Cell Biol 27: 673–684. doi: 10.1016/j.tcb.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. 2010. Manipulation of host cell death pathways during microbial infections. Cell Host Microbe 8: 44–54. doi: 10.1016/j.chom.2010.06.007 [DOI] [PubMed] [Google Scholar]
- Lapin D, Kovacova V, Sun X, Dongus JA, Bhandari DD, von Born P, Bautor J, Guarneri N, Rzemieniewski J, Stuttmann J, et al. 2019. A coevolved EDS1-SAG101-NRG1 module mediates cell death signaling by TIR-domain immune receptors. Plant Cell. doi: 10.1105/tpc.19.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. 2004. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 343: 1–28. doi: 10.1016/j.jmb.2004.08.023 [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, et al. 2012. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150: 339–350. doi: 10.1016/j.cell.2012.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Kufer TA, Schulze-Lefert P. 2011. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12: 817–826. doi: 10.1038/ni.2083 [DOI] [PubMed] [Google Scholar]
- Mahdi L, Huang M, Zhang X, Nakano RT, Kopp LB, Saur IML, Jacob F, Kovacova V, Lapin D, Parker JE, et al. 2019. Plant mixed lineage kinase domain-like proteins limit biotrophic pathogen growth. bioRxiv 681015. doi: 10.1101/681015 [DOI] [Google Scholar]
- Man SM, Karki R, Kanneganti TD. 2017. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev 277: 61–75. doi: 10.1111/imr.12534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Green DR. 1995. Protease activation during apoptosis: death by a thousand cuts? Cell 82: 349–352. doi: 10.1016/0092-8674(95)90422-0 [DOI] [PubMed] [Google Scholar]
- Mompeán M, Li W, Li J, Laage S, Siemer AB, Bozkurt G, Wu H, McDermott AE. 2018. The structure of the necrosome RIPK1-RIPK3 core, a human hetero-amyloid signaling complex. Cell 173: 1244–1253.e10. doi: 10.1016/j.cell.2018.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Dangl JL. 1997. The hypersensitive response and the induction of cell death in plants. Cell Death Differ 4: 671–683. doi: 10.1038/sj.cdd.4400309 [DOI] [PubMed] [Google Scholar]
- Nagata S. 2018. Apoptosis and clearance of apoptotic cells. Annu Rev Immunol 36: 489–517. doi: 10.1146/annurev-immunol-042617-053010 [DOI] [PubMed] [Google Scholar]
- Nanson JD, Kobe B, Ve T. 2019. Death, TIR, and RHIM: self-assembling domains involved in innate immunity and cell-death signaling. J Leukoc Biol 105: 363–375. doi: 10.1002/JLB.MR0318-123R [DOI] [PubMed] [Google Scholar]
- Newton K, Manning G. 2016. Necroptosis and inflammation. Annu Rev Biochem 85: 743–763. doi: 10.1146/annurev-biochem-060815-014830 [DOI] [PubMed] [Google Scholar]
- Orning P, Weng D, Starheim K, Ratner D, Best Z, Lee B, Brooks A, Xia S, Wu H, Kelliher MA, et al. 2018. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362: 1064–1069. doi: 10.1126/science.aau2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, Clavé C. 2007. The fungus-specific HET domain mediates programmed cell death in Podospora anserina. Eukaryot Cell 6: 2001–2008. doi: 10.1128/EC.00129-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Lo YC, Lin SC, Wang L, Yang JK, Wu H. 2007a. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol 25: 561–586. doi: 10.1146/annurev.immunol.25.022106.141656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. 2007b. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell 128: 533–546. doi: 10.1016/j.cell.2007.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie EJ, Czabotar PE, Murphy JM. 2019. The structural basis of necroptotic cell death signaling. Trends Biochem Sci 44: 53–63. doi: 10.1016/j.tibs.2018.11.002 [DOI] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. 2010. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell 141: 446–457. doi: 10.1016/j.cell.2010.03.017 [DOI] [PubMed] [Google Scholar]
- Riek R, Saupe SJ. 2016. The HET-S/s prion motif in the control of programmed cell death. Cold Spring Harb Perspect Biol 8: a023515. doi: 10.1101/cshperspect.a023515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh JS, Sohn DH. 2018. Damage-associated molecular patterns in inflammatory diseases. Immune Netw 18: e27. doi: 10.4110/in.2018.18.e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J, Xia S, Liu X, Lieberman J, Wu H. 2018. Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557: 62–67. doi: 10.1038/s41586-018-0058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarhan J, Liu BC, Muendlein HI, Li P, Nilson R, Tang AY, Rongvaux A, Bunnell SC, Shao F, Green DR, et al. 2018. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc Natl Acad Sci 115: E10888–E10897. doi: 10.1073/pnas.1809548115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe SJ. 2000. Molecular genetics of heterokaryon incompatibility in filamentous ascomycetes. Microbiol Mol Biol Rev 64: 489–502. doi: 10.1128/MMBR.64.3.489-502.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L. 2014. Complexity of danger: the diverse nature of damage-associated molecular patterns. J Biol Chem 289: 35237–35245. doi: 10.1074/jbc.R114.619304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuring C, Greenwald J, Wasmer C, Wepf R, Saupe SJ, Meier BH, Riek R. 2012. The mechanism of toxicity in HET-S/HET-s prion incompatibility. PLoS Biol 10: e1001451. doi: 10.1371/journal.pbio.1001451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Gao W, Shao F. 2017. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci 42: 245–254. doi: 10.1016/j.tibs.2016.10.004 [DOI] [PubMed] [Google Scholar]
- Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schröter M, et al. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386: 517–521. doi: 10.1038/386517a0 [DOI] [PubMed] [Google Scholar]
- Tsiatsiani L, Van Breusegem F, Gallois P, Zavialov A, Lam E, Bozhkov PV. 2011. Metacaspases. Cell Death Differ 18: 1279–1288. doi: 10.1038/cdd.2011.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach JM, Ausubel FM. 2017. The NBS-LRR architectures of plant R-proteins and metazoan NLRs evolved in independent events. Proc Natl Acad Sci 114: 1063–1068. doi: 10.1073/pnas.1619730114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váchová L, Palková Z. 2007. Caspases in yeast apoptosis-like death: facts and artefacts. FEMS Yeast Res 7: 12–21. doi: 10.1111/j.1567-1364.2006.00137.x [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. 2010. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 11: 700–714. doi: 10.1038/nrm2970 [DOI] [PubMed] [Google Scholar]
- Ve T, Vajjhala PR, Hedger A, Croll T, DiMaio F, Horsefield S, Yu X, Lavrencic P, Hassan Z, Morgan GP, et al. 2017. Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling. Nat Struct Mol Biol 24: 743–751. doi: 10.1038/nsmb.3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, Shao F. 2017. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 547: 99–103. doi: 10.1038/nature22393 [DOI] [PubMed] [Google Scholar]
- Wang J, Hu M, Wang J, Qi J, Han Z, Wang G, Qi Y, Wang HW, Zhou JM, Chai J. 2019. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science 364: eaav5870. [DOI] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. 2008. Amyloid fibrils of the HET-s (218-289) prion form a β solenoid with a triangular hydrophobic core. Science 319: 1523–1526. doi: 10.1126/science.1151839 [DOI] [PubMed] [Google Scholar]
- Weinlich R, Oberst A, Beere HM, Green DR. 2017. Necroptosis in development, inflammation and disease. Nat Rev Mol Cell Biol 18: 127–136. doi: 10.1038/nrm.2016.149 [DOI] [PubMed] [Google Scholar]
- Williams SJ, Sohn KH, Wan L, Bernoux M, Sarris PF, Segonzac C, Ve T, Ma Y, Saucet SB, Ericsson DJ, et al. 2014. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science 344: 299–303. doi: 10.1126/science.1247357 [DOI] [PubMed] [Google Scholar]
- *.Xia S, Hollingsworth LR IV, Wu H. 2019. Mechanism and regulation of gasdermin-mediated cell death. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a036400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bernoux M, Bentham AR, Newman TE, Ve T, Casey LW, Raaymakers TM, Hu J, Croll TI, Schreiber KJ, et al. 2017. Multiple functional self-association interfaces in plant TIR domains. Proc Natl Acad Sci 114: E2046–E2052. doi: 10.1073/pnas.1621248114 [DOI] [PMC free article] [PubMed] [Google Scholar]