Abstract
Stem cell fate decisions are informed by physical and chemical cues presented within and by the extracellular matrix. Despite the generally attributed importance of extracellular cues in governing self-renewal, differentiation, and collective behavior, knowledge gaps persist with regard to the individual, synergistic, and competing effects that specific physiochemical signals have on cell function. To better understand basic stem cell biology, as well as to expand opportunities in regenerative medicine and tissue engineering, a growing suite of customizable biomaterials has been developed. These next-generation cell culture materials offer user-defined biochemical and biomechanical properties, increasingly in a manner that can be controlled in time and 3D space. This review highlights recent innovations in this regard, focusing on advances to culture and maintain stemness, direct fate, and to detect stem cell function using biomaterial-based strategies.
The stem cell niche consists of a highly dynamic extracellular microenvironment in which biochemical and mechanostructural cues displayed at specific times and locations guide anisotropic function (Gattazzo et al. 2014; Vining and Mooney 2017). Bidirectional interactions and signaling between stem cells and their surroundings govern cell proliferation, migration, and differentiation throughout development, homeostasis, and disease (Humphrey et al. 2014; Lane et al. 2014; Rompolas et al. 2016). Despite overwhelming evidence indicating the indispensable nature of these variable environmental cues, conventional biological studies continue to be conducted on uniform 2D substrates of supraphysiological stiffnesses (e.g., tissue-culture plastic, glass). Consequently, cells experience incomplete and often nonnatural local cues, resulting in potentially abnormal functional responses. Recognizing these deficiencies, a growing and exciting effort has focused on the development of advanced biomaterial culture platforms that better mimic the in vivo environment, with many offering spatial and temporal control over their biochemical and biomechanical properties. This review will highlight several of these efforts, providing an overview of the systems used to regulate, control, and better understand stem cell fate and function.
BIOMATERIAL ADVANCES TO CULTURE STEM CELLS AND MAINTAIN STEMNESS
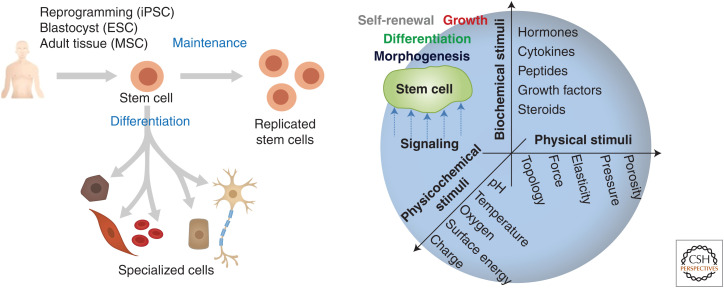
The stem cell niche is defined as the local microenvironment that regulates cell fate and function (Ehninger and Trumpp 2011). The niche is predominantly composed of the extracellular matrix (ECM) and various regulatory species including cytokines, hormones, and growth factors. Biochemical and physical cues embedded within or tethered to the ECM directly interact with niche-residing cells, modulating their behavior (Fig. 1). These behaviors vary depending on the state of the tissue and can include but are not limited to stem cell quiescence, self-renewal, and differentiation (Gattazzo et al. 2014).
Figure 1.
Stem cell fate and function are regulated through an integrated interpretation of biochemical and biophysical cues presented by and within the extracellular matrix (ECM). These environmental signals govern stem cell maintenance and lineage-specific differentiation.
Maintaining stem cells in their undifferentiated state while facilitating proliferation and self-renewal is often one of the most important goals for culture. In vivo, the ECM plays an essential role in the maintenance of stemness, defined by the cell's potential for self-renewal and differentiation (Dumont et al. 2015; Thomas et al. 2015; Ishii et al. 2018). In skeletal muscle, for example, ECM proteins like collagen, fibronectin, and laminin are required for the maintenance of satellite cells (Bentzinger et al. 2013). As these ECM proteins are numerous and likely act synergistically, one strategy to recapitulate the in vivo microenvironment in vitro has been to use decellularized ECM (dECM). In this approach, living cells and nuclear material are removed from tissues without affecting structural integrity and composition of the native cell-assembled ECM (Gilbert et al. 2006). The resultant dECM is then used as a scaffold for new cells, an effective strategy established many years ago (Pruniéras et al. 1983a,b). Pattabhi et al. (2014) showed that dECM deposited by naive human mesenchymal stem cells (hMSCs from bone marrow) enhanced the proliferation and preservation of stemness. Neural stem cells (NSCs) have shown similar trends; decellularized mouse brain sections supported long-term 3D culture and maintained stemness of NSCs (De Waele et al. 2015). In combination with 3D bioprinting techniques (Pati et al. 2014), dECM can be cast in customizable shapes that better match native tissue and organ structure.
Although dECM systems display biochemical, structural, and mechanical properties similar to that of the native ECM, challenges exist in independently modulating or tuning these effects. Toward this, synthetic chemical approaches have been developed to create culture systems whose physiochemical properties can be precisely controlled. These strategies have enabled investigation of how mechanical and structural aspects of cell scaffolds drive stem cell proliferation and undifferentiated maintenance. The Blau group first reported that muscle stem cell function is enhanced through culture on bioengineered substrates with tissue-like stiffness. Culturing cells on poly(ethylene glycol) (PEG)-based hydrogels with stiffnesses between 2 and 42 kPa, hydrogels whose stiffness matched that of native muscle (12 kPa) enhanced stem cell self-renewal following transplantation into mice (Gilbert et al. 2010). Similar studies have been reported with neural crest–derived ectodermal MSCs (eMSCs), in which polydimethylsiloxane (PDMS) substrates prepared with different stiffnesses by varied prepolymer ratios gave rise to varied Rho-ROCK signaling of cultured cells (Srinivasan et al. 2018). eMSCs cultured on soft substrates showed increased CD44 expression, modulating eMSC self-renewal and multipotency caused by the down-regulation of platelet-derived growth factor receptor β (PDGFRβ) signaling.
Although the effects of network mechanics on stem cell fate have largely focused on 2D culture, 3D platforms to expand and maintain stem cells are also of interest. In one such system, Lei and Schaffer cultured cell human pluripotent stem cells (hPSCs) within thermoreversible hydrogels based on poly(N-isoproylacrylamide)-co-poly(ethylene glycol) (PNIPAAm-PEG) (Lei and Schaffer 2013). The thermoreversibility of this system, transitioning from a soft solid to a liquid through temperature cycling between 4°C and 37°C, enables encapsulation, expansion, and subsequent collection of hPSCs at any time. With a ∼20-fold expansion of hPSCs per passage during 4–5 d cultivation, they showed a 1072-fold increase in cell number of >60 passages for 280 d. More recently, Heilshorn and colleagues developed a 3D hydrogel system based on elastic-like proteins (ELPs) to maintain neural progenitor cell (NPC) stemness (Madl et al. 2017). Screening hydrogels with a variety of physiologically relevant stiffnesses (∼0.5–50 kPa), they found that NSCs maintained stemness provided that cell-induced matrix remodeling was possible.
Maintenance of stemness has been correlated with low actomyosin contractility, typically associated with softer substrates (Winer et al. 2008; Chowdhury et al. 2010; Mih et al. 2012). Structural cues have been shown to have an important role in actomyosin contractility, generally through studies involving geometric confinement of cell adhesion. In this regard, the Chen and Watt groups have performed pioneering works highlighting the impact of geometric confinement on stem cell function (McBeath et al. 2004; Ruiz and Chen 2008; Connelly et al. 2010). Zhang and Kilian have exploited microcontact printing of self-assembled monolayers to isolated hMSCs on small adhesive islands for maintenance of stemness (Zhang and Kilian 2013). When tightly confined, hMSCs displayed enhanced expression of stem cell markers, STRO-1 and Endoglin, suggesting a route to use geometrically defined platforms to maintain multipotency in vitro. Somewhat related to 2D geometric confinement, cell substrate topography can also influence cell function (e.g., contact guidance, topotaxis) (Teixeira et al. 2003; Park et al. 2018). Dalby et al. (2007) showed that hMSC maintenance and directed differentiation could be influenced by surface topography; topographically defined surfaces containing disorderly arranged 120-nm-deep nanopits promoted osteogenesis, although those with regularly arranged pits promoted maintained expression of stem cell markers (e.g., STRO-1, ALCAM) (McMurray et al. 2011). Recognizing the effects that surface topography have on stem cell fate, de Boer and Watt and colleagues screened 2176 distinct surface topographies to see which best promoted maintenance of induced PSCs (iPSCs) (Reimer et al. 2016). Optimal topographies were those with small feature size, high wave number, and high feature density; these were capable of maintaining short-term iPSC pluripotency even when very stiff polystyrene substrates were used.
As discussed, maintenance and expansion of stem cells can be manipulated by tuning the biochemical, mechanical, and structural cues incorporated into culture platforms. Although modulation of individual effects has already been shown to have a profound influence on cell function, synergistic strategies that optimize several factors simultaneously will be critical in identifying culture conditions for a given application.
BIOMATERIAL-BASED STRATEGIES TO DIRECT CELL FATE
Physical and chemical cues presented by the ECM guide cell function. Here, strategies to probe and direct stem cell fate through tunable biomaterials are examined. Culture platforms whose properties can be dynamically modulated with external factors (e.g., light [Ruskowitz and DeForest 2018], temperature [Yamato et al. 2007], pH [Kocak et al. 2017], DNA [Murakami and Maeda 2005], enzymes [Ulijn 2006], and other fields [Kharkar et al. 2013; Manouras and Vamvakaki 2016; Uto et al. 2017b; Badeau and DeForest 2019]) are highlighted.
Biomaterials with Tunable Static Mechanics
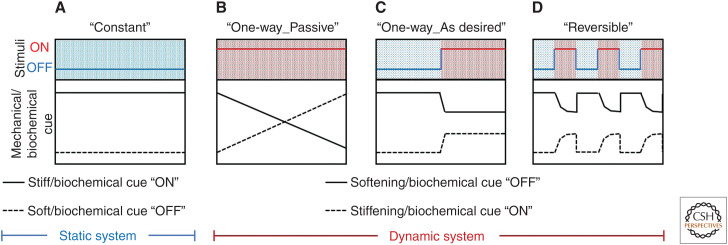
By varying material composition, cross-linking, and processing, static biomaterials can be readily formulated with stiffnesses that span all physiological tissues (Fig. 2A). These materials have helped reveal the important effects that network mechanics have on cell function. From this, stiff materials tend to promote attachment and spreading of adherent cells, whereas softer substrates yield soft tissue differentiation and tissue-like cell–cell associations depending on the cell type (Discher et al. 2005; Wells 2008). Although substrates of defined moduli can be used to maintain stemness (discussed above), stiffness can also be used as a variable to promote lineage-defined differentiation. In their landmark 2006 study, Engler and colleagues reported that hMSC expressed early neuro-, myo-, osteogenic markers on substrates whose moduli matched that of brain (0.1–1 kPa), muscle (8–17 kPa), and precalcified bone (25–40 kPa) (Engler et al. 2006). Stiffness-influenced stem cell function and directed differentiation is now widely reported.
Figure 2.
“Static” and “dynamic” biomaterials for directing stem cell fate. (A) Static biomaterials can provide a constant mechanical/biochemical cue that may be initially specified. (B–D) Dynamic biomaterials can be mechanically and biochemically modulated in the presence of live cells both (B) passively in response to the surrounding environment or (C) on demand through a triggered input. (D) Some dynamic biomaterials can reversibly present mechanical and/or biochemical cues in response to environmental stimuli.
Advanced processing techniques have also been used to create biomaterials with gradient stiffnesses (Hadden et al. 2017). These systems have helped establish the concept of durotaxis, in which cell migration is guided by gradients in substrate rigidity (Lo et al. 2000; Kidoaki and Matsuda 2008; Sunyer et al. 2016). Additionally, these platforms have enabled rapid screening of a broad range of physiological stiffness on cell function yielding the important observation that hMSC differentiation occurs in a dose-dependent manner (Hadden et al. 2017).
Softening Biomaterials
Dynamic changes in tissue stiffness that accompany development, homeostasis, and disease can direct stem cell function (Gattazzo et al. 2014). Culture platforms whose stiffness can be altered on demand can provide unique insights into these biological phenomena (Fig. 2B). Biomaterials that undergo hydrolytic, cell-mediated, or externally mediated softening have been widely reported.
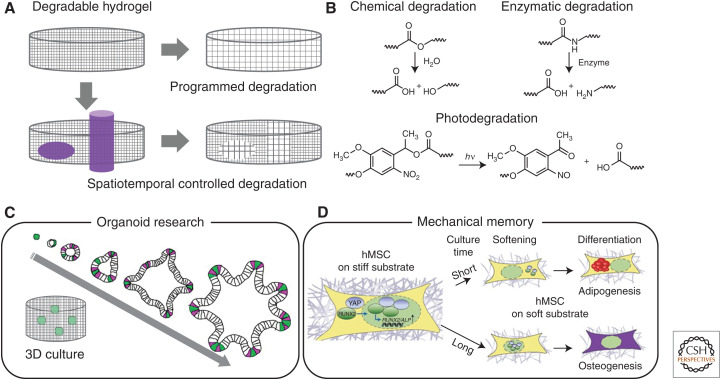
Biomaterials have been synthesized that show hydrolytic degradation (Fig. 3A,B); these materials undergo gradual softening under aqueous cell culture. Gjorevski and colleagues used one such material to control intestinal stem cell (ISC) behavior and intestinal organoid generation (Fig. 3C; Gjorevski et al. 2016). ISCs maintained in these initially stiff materials showed Yes-associated protein (YAP) activation associated with self-renewal and expansion, but could dissipate accumulated compressive forces and undergo organogenesis on partial material degradation.
Figure 3.
Softening biomaterials with programmed and on-demand degradation. (A) Biomaterials sensitive to water undergo near-uniform and slow gradation, whereas those sensitive cell-secreted enzymes respond on a cell-mediated timeframe. Photoresponsive biomaterials enable spatiotemporally dictated material softening. (B) Degradable moieties commonly used to control biomaterial mechanicals. (C) Softening biomaterials can govern intestinal organoid generation. (D) Mechanical memory of stem cell can be discovered by using photo-induced softening biomaterials. (Panel C was created from data in Gjorevski et al. (2016). Panel D was created using a figure in Eyckmans and Chen 2014, with permission, from Springer Nature combined with data in Yang et al. 2014.)
Biomaterials have also been designed that are degraded by cell-secreted enzymes, allowing for the gradual matrix remodeling on a cell-dictated timescale (Fig. 3B; Kharkar et al. 2013). In what has proven to be a particularly powerful strategy reported by Lutolf and colleagues, hydrogels cross-linked with matrix metalloproteinase (MMP)-sensitive peptides degrade at tunable rates governed by the sequence of the used peptide (Lutolf et al. 2003). The Burdick group established that the enzymatic degradative susceptibility of the biomaterial can influence hMSC differentiation (Khetan et al. 2013). High-throughput studies involving such enzymatically degradable materials have further revealed that proteolytic degradability favors cell self-renewal ability rather than proliferation as well as the role of dynamic matrix mechanics on somatic-cell reprogramming to iPSCs (Ranga et al. 2014). Similar materials have also been used to generate human intestinal organoids from PSCs (Cruz-Acuña et al. 2017, 2018).
Materials that respond to exogeneous stimuli have proven useful for on-demand softening (Fig. 2C). The Anseth group introduced the concept of photodegradable culture materials, in which directed light exposure could be used to specify when, where, and to what extent a material softens (Fig. 3A,B; Kloxin et al. 2009). User-triggered material softening in these systems has been shown to govern cell morphology; hMSCs transition from a rounded cell morphology in a stiff matrix to a more spread shape with numerous cell projections on material softening. In addition, these materials have permitted investigation of whether stem cell fate shows “mechanical memory” influenced by culture history (Fig. 3D; Yang et al. 2014); hMSCs cultured on initially stiff (∼10 kPa) matrices that were photo-softened (∼2 kPa) showed variable activation of YAP depending on the time previously spent on the stiff substrate.
Stiffening Biomaterials
Although significant efforts have been dedicated to materials that soften over time, biomaterials that stiffen are also of interest. Driving this innovation are observations that many diseases are associated with tissue stiffening, including liver cirrhosis and cardiac fibrosis (Georges et al. 2007). Recognizing that many of these changes in vivo are associated with initiation of collagen cross-linking (Levental et al. 2009), biomaterial scientists have developed platforms that also show increased synthetic cross-linking over time (Fig. 2B,C). This can be achieved by taking advantage of slow chemical reactions that proceed over the course of many days, as has been performed with stiffening hydrogels to enhance cardiomyocyte differentiation in vitro (Young and Engler 2011) or using cross-linking chemistries that can be exogenously triggered upon addition of external stimuli (e.g., temperature, pH, DNA, light).
One useful strategy for making stiffening biomaterials for stem cell culture has involved the usage of temperature-responsive polymers, primarily poly(N-isopropylacrylamide) (PNIPAAm). PNIPAAm-based materials have been shown to undergo 2D and 3D microenvironmental stiffening altered by the heating (Akimoto et al. 2016, 2018). Tuning PNIPAAm properties and material composition gives tunability over what the “soft” and “stiff” stiffnesses will be, as well as the point at which these temperature changes occur. Significant effort has gone into tuning polymer properties such that these changes can be triggered without much deviation from body temperature (Ebara et al. 2012; Uto et al. 2014, 2016, 2017a, 2018). Interestingly, the PNIPAAm-based stiffening can also involve a volumetric material change, which has been applied to create mechanically actuatable cell culture devices (Hashmi et al. 2014).
Materials that stiffen in response to pH changes have also been developed. In one example, Yoshikawa and colleagues reported thin micellar hydrogels based on ABA-type triblock copolymers composed of pH-sensitive poly(2-(diisopropylamino)ethyl methacrylate) as A blocks. These materials showed a 30-fold increase of Young's modulus during a modest pH change from 7.0 to 8.0 (Yoshikawa et al. 2011). Cells cultured on these materials throughout the stiffening processes showed a flatter morphology accompanied by increased stress fiber formation. As with temperature-sensitive materials, special care must be taken to ensure that the pH changes used that are required to elicit a material response do not themselves affect stem cell fate.
Sidestepping concerns that administered stimuli could inadvertently and directly affect stem cell function, efforts have been made to use fully cytocompatible inputs. Systems that can be cross-linked on addition of DNA is one way to achieve this. DNA strands can be covalently attached to polymers, permitting zipping together on addition of a complementary strand based on Watson–Crick–Franklin pairing (Jiang et al. 2010). Other strategies involving ion- (Gillette et al. 2010) and biological ligand-responsive chemistries (Miyata et al. 1999; Murphy et al. 2007) have been reported to create dynamic stiffening materials.
Owing to its ability to modulate material properties with spatiotemporal control, light has also been used to create cell-culture platforms that undergo triggered stiffening (Fig. 2C). Using appropriately selected and cytocompatible wavelengths, photocontrol of material properties can be gained through dimerization, isomerization, or covalent chemical reactions. The light-induced dimerization characteristics of coumarin are widely used for biomaterial stiffening (Matsuda and Mizutani 2000; Matsuda et al. 2000). In one example, Tamate and colleagues reported that photostiffening of 3D hydrogels based on coumarin ABA-type triblock copolymers affected cell morphology and proliferation (Tamate et al. 2016). Similar strategies involving anthracene photodimerization have been used to study mechanobiology (Günay et al. 2019). Biomaterials secondarily cross-linked by photopolymerization after initial formation have proven a useful tool to study stem cell differentiation. The Burdick laboratory has established hyaluronic acid–based materials that can be photostiffened within minutes by several-fold and used to examine osteogenic/adipogenic differentiation of hMSCs (Guvendiren and Burdick 2012). These platforms have been used to show that the timepoint in which the culture material is stiffened can result in different differentiation pathways, as well as to investigate the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation (Caliari et al. 2016). Although low-wavelength ultraviolet (UV) light (<254 nm) results in DNA damage, recent studies have confirmed that near-UV light (>365 nm) does not affect stem cell function (Ruskowitz and DeForest 2019). As such, efforts to create stiffening materials that respond to visible (Hao et al. 2014; Truong et al. 2017) and near-infrared (NIR) light (Stowers et al. 2015) have been undertaken. In addition to decreased phototoxicity, NIR- and IR-responsive materials are more amenable to in vivo regulation.
Biomaterials with Reversible Stiffening
Although many of the material platforms highlighted above yield a one-way (irreversible) change to culture mechanics, strategies based on reversible reactions have also been reported that provide bidirectional stiffness control (Fig. 2D; Rosales and Anseth 2016). Such materials may be better suited to study the effects of reversible tissue dynamics (e.g., disease followed by healing, stiffness changes associated with pulsatile blood flow). Systems that show reversible cross-linking can offer cycled stiffness. Materials cross-linked with DNA can be altered based on addition of cross-linking and/or displacing strands, a strategy that has been used to investigate the temporal windows by which adult NSCs commit to different fates in response to ECM stiffness (Rammensee et al. 2017). Exploiting reversible changes in protonation has been used to cycle substrate mechanics with varied pH; hMSCs mechanically stressed by cycling material stiffness between 40 kPa and 2 kPa every 2 days showed enhanced proliferation, although higher frequencies in elasticity changes (Frank et al. 2016). Reversible stimuli-triggered multimerization of proteins can also be used to cycle biomaterial mechanics (Lyu et al. 2017; Wu et al. 2018), which has been used to study mechanosignaling-induced transcriptional reprogramming of hMSCs (Hörner et al. 2019).
Rather than making and breaking bonds to cycle material properties, other strategies have exploited isomerization reactions to alter cross-linking density and material stiffness. Azobenzene, a small molecule linker that transitions from a trans to cis isomer and back again in response to UV or blue light irradiation, has also been used to create materials with reversible stiffening (Rosales et al. 2015, 2018; Lee et al. 2018a). Cross-linkers based on stimuli-responsive protein–protein fusion proteins have been used to create materials whose stiffness can be cycled in response to arbitrary stimuli including calcium or blue light (Liu et al. 2018).
Other external forces have also been used to reversibly stiffen materials. Elastic substrates exposed to cyclic stretching have been used to direct fibroblast-to-myofibroblast transdifferentiation (Molkentin et al. 2017). Magnet-responsive materials have also shown promise in this regard; magnetic field–induced cycling of biomaterial stiffness has been used to modulate hMSC osteogenesis (Abdeen et al. 2016).
Biomaterials with Tunable Stress–Relaxation
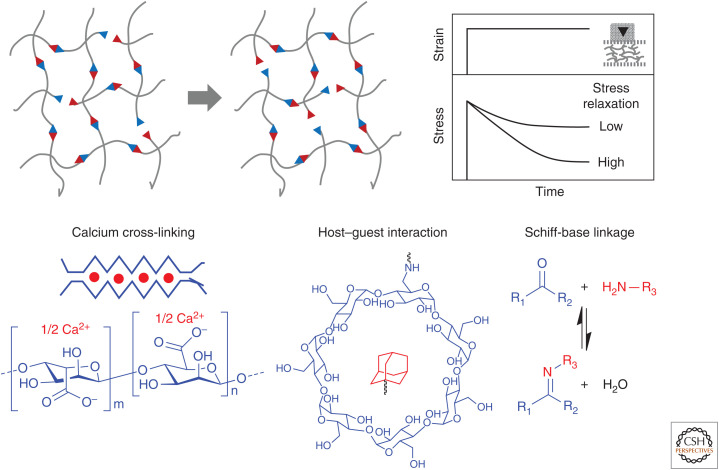
In addition to stiffness, biomaterial viscoelasticity and stress–relaxation have proven important parameters in the study and culture of stem cells (Fig. 4). Formation of biomaterials using equilibrium reactions of different strengths (e.g., guest–host chemistries [Rodell et al. 2013], hydrophobic interactions [Liu et al. 2011], hydrogen bonding [Tan et al. 2012], and dynamic covalent linkage [Yang et al. 2012]) enables creation of biomaterials with tunable viscoelasticity. By comparing stem cell function in materials that are fully elastic with those that show different degrees of stress relaxation, Chaudhuri and Mooney have shown the substantial impact that stress relaxation has on cell fate (Chaudhuri et al. 2015a,b). hMSCs cultured in alginate gels showed enhanced spreading, proliferation, and osteogenic differentiation when cultured in materials showing quicker relaxation. Similar effects have been observed in synthetic polymer culture materials, covalently cross-linked through dynamic covalent hydrazone bonds (McKinnon et al. 2014). Because the native ECM shows some degree of stress relaxation (Chaudhuri et al. 2015b), culture materials that isolate these effects are of prime interest.
Figure 4.
Adaptable biomaterials with stress–relaxation. Schematic illustration of hydrogel networks built from reversible linkages, as well as their stress–relaxability characterized by mechanical analysis. Reversible linkages can be formed either by noncovalent interactions (e.g., calcium cross-linking, host–guest interaction) or reversible covalent bonds (e.g., Schiff-base linkages).
Biochemically Defined Static Biomaterials
In addition to biophysical cues, biochemical cues presented by the ECM play a governing role in specifying stem cell fate. Although biologically derived materials such as Matrigel present many of the cues required to sustain and promote specific function, their batch-to-batch variability and ill-defined nature overall renders it challenging to conclusively attribute the influence of a single biochemical factor with function (Klotz et al. 2019). Chemically defined biomaterials are beneficial in this regard, particularly when their components are fully synthetic. Biomaterials based on synthetic polymers, for example, PEG and poly(lactic acid), offer tremendous advantages, providing a “blank slate” by which the influence of well-defined combinations of biochemical factors on cell functions can be readily investigated (Arakawa and DeForest 2017). Similar to conventional tissue culture, bioactive proteins, peptides, and small molecules can be readily added to the culture media to provide uniform and replenishable exposure to soluble cues. Unique to biomaterial-based strategies is the ability to tether species to surfaces and throughout materials, mimicking important aspects by which bioactive species are often presented by the ECM in vivo. When factors are immobilized onto and within biomaterials, the chemistries used and the physical residues for tethering must be carefully selected so as to maintain their bioactivity (Spicer et al. 2018). Static chemically patterned biomaterials can be generated through a variety of additive manufacturing techniques (Tibbitt and Anseth 2009; Tse and Engler 2010; Habib et al. 2013).
Dynamic Immobilization of Biochemical Factors
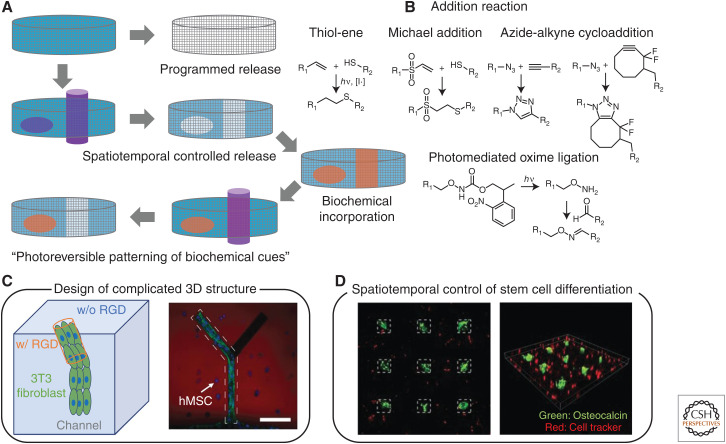
Although biomaterials are most often biochemically decorated at the time of synthesis, many strategies exist to control functionalization over time. This is generally achieved by formulating materials to contain reactive handles that can be later exploited for the immobilization of proteins, peptides, and small molecules. A large and growing number of chemistries has been used in this regard, although “bioorthogonal click” reactions have garnished significant attention as functionalization can be performed in the presence of living cells without directly affecting their function (Fig. 5A,B; Sletten and Bertozzi 2009). For example, materials decorated with azide groups can be postsynthetically functionalized with cycloalkyne-containing species, just as those with pendant tetrazines can be modified with strained alkenes (Blackman et al. 2008; Zhang et al. 2016). Spontaneous and triggered reaction strategies afford temporally controlled material functionalization. Functionalization of biomaterials using reactions that can be further controlled in space (e.g., light, temperature, ultrasound) affords full spatiotemporal regulation over biochemical factor immobilization. Light-based strategies are beneficial in this regard, being able to confine cell attachment, migration, outgrowth (Fig. 5C), differentiation, and other biochemical functions to specific regions within biomaterials (Luo and Shoichet 2004; Hahn et al. 2006; Hoffmann and West 2010; DeForest and Anseth 2011; Wylie et al. 2011; Mosiewicz et al. 2013). Photochemical-based strategies have been used to generate 2D and 3D patterns of immobilized cues in both discrete and graded patterns, whereby nonuniform sample irradiation enables heterogeneous material functionalization in manners that can mimic native tissue.
Figure 5.
Dynamic biomaterials with tunable biochemical cues. (A) Hydrogels can present dynamic biological cues by removal and addition of bioactive molecules. Photochemistry allows spatiotemporal removal and sequential presentation of biochemical cues for 2D and 3D environment of hydrogels. (B) Reactions commonly used to conjugate biomolecules to biomaterials include thiol-ene, Michael-type, azide-alkyne, and oxime ligations. Dynamic regulation of biochemical cues within 3D hydrogels enable (C) creation of complex multicellular 3D structures and (D) spatiotemporal regulation of stem cell differentiation. Scale bar, 100 μm. (Panel C created from data in DeForest and Anseth 2011. Panel D created from data in DeForest and Tirrell 2015.)
Controlled Release of Biochemical Factors
Just as biochemical cues can be tethered to biomaterials to regulate stem cell fate, their controlled soluble presentation can also influence function. Many biomaterials use restricted diffusion and affinity interactions to tune the release rate of bioactive species (Lin and Metters 2006; Peppas et al. 2006; Lin and Anseth 2009). Although such strategies are relatively powerful and simple to implement, release profiles must be defined a priori and cannot be adjusted postformulation. Triggered release can be obtained by tethering species to biomaterials through degradable bonds, yielding soluble presentation on bond severance. Hydrolytically sensitive bonds yield gradual release in aqueous environments, although those sensitive to pH, reductants, or enzymes have been used to release many bioactive species in response to a variety of biologically relevant external stimuli (Chen et al. 2013; Purcell et al. 2014; Ham et al. 2016; Guo et al. 2017). Recent strategies have established logic-based release of bioactive cues only in response to complex combinations of environmental inputs following Boolean YES/OR/AND operations (Badeau et al. 2018; Gawade et al. 2019; Ruskowitz et al. 2019). Alternatively, full spatiotemporal release of bioactive factors can be afforded using photodegradable moieties (Kloxin et al. 2009; Azagarsamy and Anseth 2013). In one implementation, use of wavelength-selective chemistries enabled sequential delivery of BMP-2 and BMP-7 to hMSCs to drive osteogenic differentiation (Azagarsamy and Anseth 2013).
Reversible Immobilization of Biochemical Cues
Toward recapitulating full dynamic biochemical signaling in vitro, some progress has been made toward creation of biomaterials that can be reversibly functionalized with bioactive factors. Metal affinity interactions (e.g., Ni-NTA with 6xHis-tagged species), DNA dimerization, and enzyme-based strategies have been developed that provide triggered control over species immobilization and release (Liu et al. 2010; Kolodziej et al. 2011; Ham et al. 2016). Full spatiotemporal control can be obtained when light-based reactions are used to trigger species binding/unbinding. Use of bond-forming/breaking reactions initiated with different wavelengths of light has permitted dynamic control of cell adhesion through reversibly immobilized peptides (DeForest and Anseth 2012). Photoexchange reactions have been used to control hMSC spreading and transforming growth factor-β signaling of mouse embryonic fibroblasts (Gandavarapu et al. 2014; Grim et al. 2018). Sequential photochemistries have been used to dynamically control attachment, proliferation, growth, differentiation, and intracellular signaling in 3D and with subcellular resolution using patterned protein growth factors (Fig. 5C,D; DeForest and Tirrell 2015; Shadish et al. 2019). Reversibly patterned vitronectin presentation has permitted spatiotemporally mediated control over osteogenic differentiation dynamics, whereas user-defined release of epidermal growth factor (EGF) has been exploited to pattern EGF receptor activation and downstream signaling.
DETECTING STEM CELL BEHAVIOR USING BIOMATERIAL APPROACHES
For the past many years, biochemical methods such as fluorescent immunostaining, gene and protein expression by polymerase chain reaction (PCR), and enzyme-linked immunosorbent assay (ELISA), respectively, have served as the biological workhorses for analyzing stem cell fate and function. Despite the wealth of information that these techniques provide, many such traditional methods generally require destructive protocols (e.g., fixation, lysis) and provide only a static snapshot of what is occurring throughout dynamic biological processes. Although biomaterial strategies have discovered profound use in the culture and direction of stem cell fates, their tunability also enables newfound routes to probe and detect microenvironmental effects on cell function in a manner that is dynamic and nondestructive, providing real-time information of cell function that is complementary to that given using FRET-based biosensors, fluorescence-activated cell sorting (FACS), and in vivo lineage tracing.
Cell sorting is an essential tool in the field of stem cell biology for defining and collecting target cells with high purity. Because antibody labels may not be available for all cell types and may potentially interfere with expansion and differentiation, label-free strategies for cell sorting have gained popularity. Here, intrinsic cell markers including size, electrical polarizability, and hydrodynamic properties are used to identify or separate specific cells (Gossett et al. 2010). Although hardware engineering has yielded analytical advances in this regard, several biomaterial-based approaches have also been reported for achieving label-free cell sorting. Photodegradable hydrogels have been used to capture cells through 2D surface interactions, and then release through material degradation (Shin et al. 2014). To extend the overall throughput of these collection strategies, similar approaches involving cell-adhesive degradable microbeads have been used (Siltanen et al. 2013). Photodegradable gels have also been used for optically specified collection encapsulated cells from within 3D materials of well-defined properties (DeForest and Anseth 2011; Tamura et al. 2014).
In addition to collecting cells, biomaterial-based strategies have been exploited to understand how cells interact with their surroundings. Given their importance in stem cell maintenance and decisions of fate, several efforts have sought to study and quantify traction forces exerted by cells on their local environment (Polacheck and Chen 2016). Early strategies used a wrinkling silicone membrane to measure contractile cellular forces (Harris et al. 1980). More recently, traction force microscopy (TFM) has been developed to analyze cellular contraction forces, providing spatial information in a quantitative manner (Lee et al. 1994). In traditional TFM, cells are seeded on elastic materials containing immobilized fluorescent beads. In conjunction with knowledge of the material stiffness, measurements of individual bead displacements provide information about cell-generated forces. To study both the magnitude and direction of forces by cells plated in 2D, Salaita's group combined molecular tension probes and fluorescence polarization microscopy in a technique known as molecular force microscopy (MFM) (Brockman et al. 2018). Subcellular force determination has also been achieved by seeding cells onto micropost arrays, in which post displacement gives information about the magnitude and direction of cell traction forces (Tan et al. 2003). By varying post length and density, as well as underlying material composition, cell function can be assayed under varied mechanical inputs, including those that direct differentiation (Fu et al. 2010). Nanopillar electrode arrays have also been generated to assay cell action potentials using high-throughput electroporation, which has been applied to study hPSC-derived cardiomyocyte electrophysiology and stem cell differentiation (Kim et al. 2015; Lin et al. 2017; Lee et al. 2018b).
Biomaterial strategies can also provide information about how cells are behaving within 3D materials. TFM has been extended for application with 3D cell culture, where both cells and fluorescent beads are embedded within 3D gels and tracked/analyzed with finite-element techniques (Legant et al. 2010). These techniques have been applied to examine the effect of matrix degradation and cellular traction force on hMSC differentiation (Khetan et al. 2013). By tracking the Brownian motion of embedded particles, “microrheology” techniques provide information about spatial change of material mechanics remodeling of cell-laden hydrogel and how this changes through stem cell proliferation and migration (Schultz et al. 2015). Fluorogenic biomaterials have also been developed that will fluoresce on cell-mediated enzymatic degradation, enabling visualization of 3D cell migratory paths (Lee et al. 2007; Leight et al. 2013). Collectively, such biomaterial strategies offer new and powerful routes to understand basic stem cell function.
CONCLUDING REMARKS
As highlighted in this review, a growing collection of customizable biomaterials exists that provide user-defined control over the biochemical and biomechanical parameters comprising the stem cell niche. Such materials offer tremendous potential with respect to advancing our understanding of cell physiology, maintaining stemness, and directing desired fates including differentiation. Whether to reliably culture and expand stem cells before transplantation, promote their enhanced engraftment in direct cell therapy, or to aid in the engineering of complex functional tissue for transplantation, these systems are poised to translate stem cell therapies into the clinic as well as further revolutionize our understanding of stem cell biology.
ACKNOWLEDGMENTS
This work was supported by a CAREER Award (DMR 1652141 to C.A.D.) and a grant (DMR 1807398 to C.A.D.) from the National Science Foundation. The work was also supported in part by AMED-PRIME, AMED (JP18gm5810017 to K.U.), and the International Research Fellow of Japan Society for the Promotion of Science (Postdoctoral Fellowships for Research in Japan to C.A.K.).
Footnotes
Editors: Cristina Lo Celso, Kristy Red-Horse, and Fiona M. Watt
Additional Perspectives on Stem Cells: From Biological Principles to Regenerative Medicine available at www.cshperspectives.org
REFERENCES
- Abdeen AA, Lee J, Bharadwaj NA, Ewoldt RH, Kilian KA. 2016. Temporal modulation of stem cell activity using magnetoactive hydrogels. Adv Healthc Mater 5: 2536–2544. doi: 10.1002/adhm.201600349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimoto AM, Hasuike E, Tada H, Nagase K, Okano T, Kanazawa H, Yoshida R. 2016. Design of tetra-arm PEG-crosslinked thermoresponsive hydrogel for 3D cell culture. Anal Sci 32: 1203–1205. doi: 10.2116/analsci.32.1203 [DOI] [PubMed] [Google Scholar]
- Akimoto AM, Niitsu EH, Nagase K, Okano T, Kanazawa H, Yoshida R. 2018. Mesenchylmal stem cell culture on poly(N-isopropylacrylamide) hydrogel with repeated thermo-stimulation. Int J Mol Sci 19: 1253. doi: 10.3390/ijms19041253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa CK, DeForest CA. 2017. Polymer design and development In Biology and engineering of stem cell niches (ed. Vishwakarma A, Karp JM), pp. 295–314. Academic, Boston. [Google Scholar]
- Azagarsamy MA, Anseth KS. 2013. Wavelength-controlled photocleavage for the orthogonal and sequential release of multiple proteins. Angew Chem Int Ed Engl 52: 13803–13807. doi: 10.1002/anie.201308174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badeau BA, DeForest CA. 2019. Programming stimuli-responsive behavior into biomaterials. Annu Rev Biomed Eng 21: 241–265. doi: 10.1146/annurev-bioeng-060418-052324 [DOI] [PubMed] [Google Scholar]
- Badeau BA, Comerford MP, Arakawa CK, Shadish JA, DeForest CA. 2018. Engineered modular biomaterial logic gates for environmentally triggered therapeutic delivery. Nat Chem 10: 251–258. doi: 10.1038/nchem.2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, von Maltzahn J, Soleimani VD, Yin H, Rudnicki MA. 2013. Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell 12: 75–87. doi: 10.1016/j.stem.2012.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman ML, Royzen M, Fox JM. 2008. Tetrazine ligation: Fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J Am Chem Soc 130: 13518–13519. doi: 10.1021/ja8053805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman JM, Blanchard AT, Pui-Yan Ma V, Derricotte WD, Zhang Y, Fay ME, Lam WA, Evangelista FA, Mattheyses AL, Salaita K. 2018. Mapping the 3D orientation of piconewton integrin traction forces. Nat Methods 15: 115–118. doi: 10.1038/nmeth.4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliari SR, Perepelyuk M, Cosgrove BD, Tsai SJ, Lee GY, Mauck RL, Wells RG, Burdick JA. 2016. Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci Rep 6: 21387. doi: 10.1038/srep21387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Gu L, Darnell M, Klumpers D, Bencherif SA, Weaver JC, Huebsch N, Mooney DJ. 2015a. Substrate stress relaxation regulates cell spreading. Nat Commun 6: 6365. doi: 10.1038/ncomms7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O, Gu L, Klumpers D, Darnell M, Bencherif SA, Weaver JC, Huebsch N, Lee HP, Lippens E, Duda GN, et al. 2015b. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater 15: 326–334. doi: 10.1038/nmat4489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zheng M, Meng F, Cheng R, Deng C, Feijen J, Zhong Z. 2013. In situ forming reduction-sensitive degradable nanogels for facile loading and triggered intracellular release of proteins. Biomacromolecules 14: 1214–1222. doi: 10.1021/bm400206m [DOI] [PubMed] [Google Scholar]
- Chowdhury F, Li Y, Poh YC, Yokohama-Tamaki T, Wang N, Tanaka TS. 2010. Soft substrates promote homogeneous self-renewal of embryonic stem cells via downregulating cell-matrix tractions. PLoS ONE 5: e15655. doi: 10.1371/journal.pone.0015655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JT, Gautrot JE, Trappmann B, Tan DW-M, Donati G, Huck WTS, Watt FM. 2010. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol 12: 711–718. doi: 10.1038/ncb2074 [DOI] [PubMed] [Google Scholar]
- Cruz-Acuña R, Quirós M, Farkas AE, Dedhia PH, Huang S, Siuda D, García-Hernández V, Miller AJ, Spence JR, Nusrat A, et al. 2017. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat Cell Biol 19: 1326–1335. doi: 10.1038/ncb3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz-Acuña R, Quirós M, Huang S, Siuda D, Spence JR, Nusrat A, García AJ. 2018. PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery. Nat Protoc 13: 2102–2119. doi: 10.1038/s41596-018-0036-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, Wilkinson CDW, Oreffo ROC. 2007. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat Mater 6: 997–1003. doi: 10.1038/nmat2013 [DOI] [PubMed] [Google Scholar]
- DeForest CA, Anseth KS. 2011. Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nat Chem 3: 925–931. doi: 10.1038/nchem.1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest CA, Anseth KS. 2012. Photoreversible patterning of biomolecules within click-based hydrogels. Angew Chem Int Ed Engl 51: 1816–1819. doi: 10.1002/anie.201106463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeForest CA, Tirrell DA. 2015. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat Mater 14: 523–531. doi: 10.1038/nmat4219 [DOI] [PubMed] [Google Scholar]
- De Waele J, Reekmans K, Daans J, Goossens H, Berneman Z, Ponsaerts P. 2015. 3D culture of murine neural stem cells on decellularized mouse brain sections. Biomaterials 41: 122–131. doi: 10.1016/j.biomaterials.2014.11.025 [DOI] [PubMed] [Google Scholar]
- Discher DE, Janmey P, Wang Y. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science 310: 1139–1143. doi: 10.1126/science.1116995 [DOI] [PubMed] [Google Scholar]
- Dumont NA, Wang YX, Rudnicki MA. 2015. Intrinsic and extrinsic mechanisms regulating satellite cell function. Development 142: 1572–1581. doi: 10.1242/dev.114223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebara M, Uto K, Idota N, Hoffman JM, Aoyagi T. 2012. Shape-memory surface with dynamically tunable nano-geometry activated by body heat. Adv Mater 24: 273–278. doi: 10.1002/adma.201102181 [DOI] [PubMed] [Google Scholar]
- Ehninger A, Trumpp A. 2011. The bone marrow stem cell niche grows up: mesenchymal stem cells and macrophages move in. J Exp Med 208: 421–428. doi: 10.1084/jem.20110132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. 2006. Matrix elasticity directs stem cell lineage specification. Cell 126: 677–689. doi: 10.1016/j.cell.2006.06.044 [DOI] [PubMed] [Google Scholar]
- Eyckmans J, Chen CS. 2014. Stem cell differentiation: sticky mechanical memory. Nat Mater 13: 542–543. doi: 10.1038/nmat3989 [DOI] [PubMed] [Google Scholar]
- Frank V, Kaufmann S, Wright R, Horn P, Yoshikawa HY, Wuchter P, Madsen J, Lewis AL, Armes SP, Ho AD, et al. 2016. Frequent mechanical stress suppresses proliferation of mesenchymal stem cells from human bone marrow without loss of multipotency. Sci Rep 6: 24264. doi: 10.1038/srep24264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. 2010. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Meth 7: 733–736. doi: 10.1038/nmeth.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandavarapu NR, Azagarsamy MA, Anseth KS. 2014. Photo-click living strategy for controlled, reversible exchange of biochemical ligands. Adv Mater 26: 2521–2526. doi: 10.1002/adma.201304847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattazzo F, Urciuolo A, Bonaldo P. 2014. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim Biophys Acta 1840: 2506–2519. doi: 10.1016/j.bbagen.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawade PM, Shadish JA, Badeau BA, DeForest CA. 2019. Logic-based delivery of site-specifically modified proteins from environmentally responsive hydrogel biomaterials. Adv Mater 31: e1902462. doi: 10.1002/adma.201902462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges PC, Hui JJ, Gombos Z, McCormick ME, Wang AY, Uemura M, Mick R, Janmey PA, Furth EE, Wells RG. 2007. Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am J Physiol Gastrointest Liver Physiol 293: G1147–G1154. doi: 10.1152/ajpgi.00032.2007 [DOI] [PubMed] [Google Scholar]
- Gilbert TW, Sellaro TL, Badylak SF. 2006. Decellularization of tissues and organs. Biomaterials 27: 3675–3683. [DOI] [PubMed] [Google Scholar]
- Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM. 2010. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science 329: 1078–1081. doi: 10.1126/science.1191035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette BM, Jensen JA, Wang M, Tchao J, Sia SK. 2010. Dynamic hydrogels: switching of 3D microenvironments using two-component naturally derived extracellular matrices. Adv Mater 22: 686–691. doi: 10.1002/adma.200902265 [DOI] [PubMed] [Google Scholar]
- Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, Clevers H, Lutolf MP. 2016. Designer matrices for intestinal stem cell and organoid culture. Nature 539: 560–564. doi: 10.1038/nature20168 [DOI] [PubMed] [Google Scholar]
- Gossett DR, Weaver WM, Mach AJ, Hur SC, Tse HTK, Lee W, Amini H, Di Carlo D. 2010. Label-free cell separation and sorting in microfluidic systems. Anal Bioanal Chem 397: 3249–3267. doi: 10.1007/s00216-010-3721-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grim JC, Brown TE, Aguado BA, Chapnick DA, Viert AL, Liu X, Anseth KS. 2018. A reversible and repeatable thiol–ene bioconjugation for dynamic patterning of signaling proteins in hydrogels. ACS Cent Sci 4: 909–916. doi: 10.1021/acscentsci.8b00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günay KA, Ceccato TL, Silver JS, Bannister KL, Bednarski OJ, Leinwand LA, Anseth KS. 2019. PEG–anthracene hydrogels as an on-demand stiffening matrix to study mechanobiology. Angew Chem Int Ed Engl 58: 9912–9916. doi: 10.1002/anie.201901989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Kim H, Ovadia EM, Mourafetis CM, Yang M, Chen W, Kloxin AM. 2017. Bio-orthogonal conjugation and enzymatically triggered release of proteins within multi-layered hydrogels. Acta Biomater 56: 80–90. doi: 10.1016/j.actbio.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvendiren M, Burdick JA. 2012. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat Commun 3: 792. doi: 10.1038/ncomms1792 [DOI] [PubMed] [Google Scholar]
- Habib SJ, Chen BC, Tsai FC, Anastassiadis K, Meyer T, Betzig E, Nusse R. 2013. A localized Wnt signal orients asymmetric stem cell division in vitro. Science 339: 1445–1448. doi: 10.1126/science.1231077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadden WJ, Young JL, Holle AW, McFetridge ML, Kim DY, Wijesinghe P, Taylor-Weiner H, Wen JH, Lee AR, Bieback K, et al. 2017. Stem cell migration and mechanotransduction on linear stiffness gradient hydrogels. Proc Natl Acad Sci 114: 5647–5652. doi: 10.1073/pnas.1618239114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MS, Taite LJ, Moon JJ, Rowland MC, Ruffino KA, West JL. 2006. Photolithographic patterning of polyethylene glycol hydrogels. Biomaterials 27: 2519–2524. doi: 10.1016/j.biomaterials.2005.11.045 [DOI] [PubMed] [Google Scholar]
- Ham HO, Qu Z, Haller CA, Dorr BM, Dai E, Kim W, Liu DR, Chaikof EL. 2016. In situ regeneration of bioactive coatings enabled by an evolved Staphylococcus aureus sortase A. Nat Commun 7: 11140. doi: 10.1038/ncomms11140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Shih H, Muňoz Z, Kemp A, Lin CC. 2014. Visible light cured thiol-vinyl hydrogels with tunable degradation for 3D cell culture. Acta Biomater 10: 104–114. doi: 10.1016/j.actbio.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Wild P, Stopak D. 1980. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208: 177–179. doi: 10.1126/science.6987736 [DOI] [PubMed] [Google Scholar]
- Hashmi B, Zarzar LD, Mammoto T, Mammoto A, Jiang A, Aizenberg J, Ingber DE. 2014. Developmentally-inspired shrink-wrap polymers for mechanical induction of tissue differentiation. Adv Mater 26: 3253–3257. doi: 10.1002/adma.201304995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JC, West JL. 2010. Three-dimensional photolithographic patterning of multiple bioactive ligands in poly(ethylene glycol) hydrogels. Soft Matter 6: 5056–5063. doi: 10.1039/c0sm00140f [DOI] [Google Scholar]
- Hörner M, Raute K, Hummel B, Madl J, Creusen G, Thomas OS, Christen EH, Hotz N, Gübeli RJ, Engesser R, et al. 2019. Phytochrome-based extracellular matrix with reversibly tunable mechanical properties. Adv Mater 31: 1806727. doi: 10.1002/adma.201806727 [DOI] [PubMed] [Google Scholar]
- Humphrey JD, Dufresne ER, Schwartz MA. 2014. Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol 15: 802–812. doi: 10.1038/nrm3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Sakurai H, Suzuki N, Mabuchi Y, Sekiya I, Sekiguchi K, Akazawa C. 2018. Recapitulation of extracellular LAMININ environment maintains stemness of satellite cells in vitro. Stem Cell Rep 10: 568–582. doi: 10.1016/j.stemcr.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang FX, Yurke B, Schloss RS, Firestein BL, Langrana NA. 2010. Effect of dynamic stiffness of the substrates on neurite outgrowth by using a DNA-crosslinked hydrogel. Tissue Eng Part A 16: 1873–1889. doi: 10.1089/ten.tea.2009.0574 [DOI] [PubMed] [Google Scholar]
- Kharkar PM, Kiick KL, Kloxin AM. 2013. Designing degradable hydrogels for orthogonal control of cell microenvironments. Chem Soc Rev 42: 7335–7372. doi: 10.1039/C3CS60040H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetan S, Guvendiren M, Legant WR, Cohen DM, Chen CS, Burdick JA. 2013. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater 12: 458–465. doi: 10.1038/nmat3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidoaki S, Matsuda T. 2008. Microelastic gradient gelatinous gels to induce cellular mechanotaxis. J Biotechnol 133: 225–230. doi: 10.1016/j.jbiotec.2007.08.015 [DOI] [PubMed] [Google Scholar]
- Kim TH, Yea CH, Chueng STD, Yin PTT, Conley B, Dardir K, Pak Y, Jung GY, Choi JW, Lee KB. 2015. Large-scale nanoelectrode arrays to monitor the dopaminergic differentiation of human neural stem cells. Adv Mater 27: 6356–6362. doi: 10.1002/adma.201502489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz BJ, Oosterhoff LA, Utomo L, Lim KS, Vallmajo-Martin Q, Clevers H, Woodfield TBF, Rosenberg AJWP, Malda J, Ehrbar M, et al. 2019. A versatile biosynthetic hydrogel platform for engineering of tissue analogues. Adv Healthc Mater 8: 1900979. doi: 10.1002/adhm.201900979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloxin AM, Kasko AM, Salinas CN, Anseth KS. 2009. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science 324: 59–63. doi: 10.1126/science.1169494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocak G, Tuncer C, Bütün V. 2017. pH-responsive polymers. Polym Chem 8: 144–176. doi: 10.1039/C6PY01872F [DOI] [Google Scholar]
- Kolodziej CM, Chang CW, Maynard HD. 2011. Glutathione S-transferase as a general and reversible tag for surface immobilization of proteins. J Mater Chem 21: 1457–1461. doi: 10.1039/C0JM02370A [DOI] [Google Scholar]
- Lane SW, Williams DA, Watt FM. 2014. Modulating the stem cell niche for tissue regeneration. Nat Biotech 32: 795–803. doi: 10.1038/nbt.2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K. 1994. Traction forces generated by locomoting keratocytes. J Cell Biol 127: 1957–1964. doi: 10.1083/jcb.127.6.1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Moon JJ, Miller JS, West JL. 2007. Poly(ethylene glycol) hydrogels conjugated with a collagenase-sensitive fluorogenic substrate to visualize collagenase activity during three-dimensional cell migration. Biomaterials 28: 3163–3170. doi: 10.1016/j.biomaterials.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Lee IN, Dobre O, Richards D, Ballestrem C, Curran JM, Hunt JA, Richardson SM, Swift J, Wong LS. 2018a. Photoresponsive hydrogels with photoswitchable mechanical properties allow time-resolved analysis of cellular responses to matrix stiffening. ACS Appl Mater Interfaces 10: 7765–7776. doi: 10.1021/acsami.7b18302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Choi HK, Yang L, Chueng STD, Choi JW, Lee KB. 2018b. Nondestructive real-time monitoring of enhanced stem cell differentiation using a graphene-au hybrid nanoelectrode array. Adv Mater 30: 1802762. doi: 10.1002/adma.201802762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant WR, Miller JS, Blakely BL, Cohen DM, Genin GM, Chen CS. 2010. Measurement of mechanical tractions exerted by cells in three-dimensional matrices. Nat Methods 7: 969–971. doi: 10.1038/nmeth.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Schaffer DV. 2013. A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc Natl Acad Sci 110: E5039–E5048. doi: 10.1073/pnas.1309408110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leight JL, Alge DL, Maier AJ, Anseth KS. 2013. Direct measurement of matrix metalloproteinase activity in 3D cellular microenvironments using a fluorogenic peptide substrate. Biomaterials 34: 7344–7352. doi: 10.1016/j.biomaterials.2013.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SFT, Csiszar K, Giaccia A, Weninger W, et al. 2009. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139: 891–906. doi: 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Anseth KS. 2009. PEG hydrogels for the controlled release of biomolecules in regenerative medicine. Pharm Res 26: 631–643. doi: 10.1007/s11095-008-9801-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Metters AT. 2006. Hydrogels in controlled release formulations: network design and mathematical modeling. Adv Drug Deliv Rev 58: 1379–1408. doi: 10.1016/j.addr.2006.09.004 [DOI] [PubMed] [Google Scholar]
- Lin ZC, McGuire AF, Burridge PW, Matsa E, Lou HY, Wu JC, Cui B. 2017. Accurate nanoelectrode recording of human pluripotent stem cell–derived cardiomyocytes for assaying drugs and modeling disease. Microsyst Nanoeng 3: 16080. doi: 10.1038/micronano.2016.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YCC, Rieben N, Iversen L, Sørensen BS, Park J, Nygård J, Martinez KL. 2010. Specific and reversible immobilization of histidine-tagged proteins on functionalized silicon nanowires. Nanotechnology 21: 245105. doi: 10.1088/0957-4484/21/24/245105 [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu B, Riesberg JJ, Shen W. 2011. In situ forming physical hydrogels for three-dimensional tissue morphogenesis. Macromol Biosci 11: 1325–1330. doi: 10.1002/mabi.201100119 [DOI] [PubMed] [Google Scholar]
- Liu L, Shadish JA, Arakawa CK, Shi K, Davis J, DeForest CA. 2018. Cyclic stiffness modulation of cell-laden protein–polymer hydrogels in response to user-specified stimuli including light. Adv Biosyst 2: 1800240. doi: 10.1002/adbi.201800240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CM, Wang HB, Dembo M, Wang Y. 2000. Cell movement is guided by the rigidity of the substrate. Biophys J 79: 144–152. doi: 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Shoichet MS. 2004. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nat Mater 3: 249–253. doi: 10.1038/nmat1092 [DOI] [PubMed] [Google Scholar]
- Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. 2003. Synthetic matrix metalloproteinase-sensitive hydrogels for the conduction of tissue regeneration: engineering cell-invasion characteristics. Proc Natl Acad Sci 100: 5413–5418. doi: 10.1073/pnas.0737381100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu S, Fang J, Duan T, Fu L, Liu J, Li H. 2017. Optically controlled reversible protein hydrogels based on photoswitchable fluorescent protein Dronpa. Chem Commun (Camb) 53: 13375–13378. doi: 10.1039/C7CC06991J [DOI] [PubMed] [Google Scholar]
- Madl CM, LeSavage BL, Dewi RE, Dinh CB, Stowers RS, Khariton M, Lampe KJ, Nguyen D, Chaudhuri O, Enejder A, et al. 2017. Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat Mater 16: 1233–1242. doi: 10.1038/nmat5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manouras T, Vamvakaki M. 2016. Field responsive materials: photo-, electro-, magnetic- and ultrasound-sensitive polymers. Polym Chem 8: 74–96. doi: 10.1039/C6PY01455K [DOI] [Google Scholar]
- Matsuda T, Mizutani M. 2000. Molecular design of photocurable liquid biodegradable copolymers. II: Synthesis of coumarin-derivatized oligo(methacrylate)s and photocuring. Macromolecules 33: 791–794. doi: 10.1021/ma991405a [DOI] [Google Scholar]
- Matsuda T, Mizutani M, Arnold SC. 2000. Molecular design of photocurable liquid biodegradable copolymers. I: Synthesis and photocuring characteristics. Macromolecules 33: 795–800. doi: 10.1021/ma991404i [DOI] [Google Scholar]
- McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. 2004. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6: 483–495. doi: 10.1016/S1534-5807(04)00075-9 [DOI] [PubMed] [Google Scholar]
- McKinnon DD, Domaille DW, Cha JN, Anseth KS. 2014. Biophysically defined and cytocompatible covalently adaptable networks as viscoelastic 3d cell culture systems. Adv Mater 26: 865–872. doi: 10.1002/adma.201303680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, Murawski K, Kingham E, Oreffo ROC, Dalby MJ. 2011. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater 10: 637–644. doi: 10.1038/nmat3058 [DOI] [PubMed] [Google Scholar]
- Mih JD, Marinkovic A, Liu F, Sharif AS, Tschumperlin DJ. 2012. Matrix stiffness reverses the effect of actomyosin tension on cell proliferation. J Cell Sci 125: 5974–5983. doi: 10.1242/jcs.108886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Asami N, Uragami T. 1999. A reversibly antigen-responsive hydrogel. Nature 399: 766–769. doi: 10.1038/21619 [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Bugg D, Ghearing N, Dorn LE, Kim P, Sargent MA, Gunaje J, Otsu K, Davis J. 2017. Fibroblast-specific genetic manipulation of p38 mitogen-activated protein kinase in vivo reveals its central regulatory role in fibrosis. Circulation 136: 549–561. doi: 10.1161/CIRCULATIONAHA.116.026238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosiewicz KA, Kolb L, van der Vlies AJ, Martino MM, Lienemann PS, Hubbell JA, Ehrbar M, Lutolf MP. 2013. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat Mater 12: 1072–1078. doi: 10.1038/nmat3766 [DOI] [PubMed] [Google Scholar]
- Murakami Y, Maeda M. 2005. DNA-responsive hydrogels that can shrink or swell. Biomacromolecules 6: 2927–2929. doi: 10.1021/bm0504330 [DOI] [PubMed] [Google Scholar]
- Murphy WL, Dillmore WS, Modica J, Mrksich M. 2007. Dynamic hydrogels: translating a protein conformational change into macroscopic motion. Angew Chem Int Ed Engl 46: 3066–3069. doi: 10.1002/anie.200604808 [DOI] [PubMed] [Google Scholar]
- Park J, Kim DH, Levchenko A. 2018. Topotaxis: a new mechanism of directed cell migration in topographic ECM gradients. Biophys J 114: 1257–1263. doi: 10.1016/j.bpj.2017.11.3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati F, Jang J, Ha DH, Won Kim S, Rhie JW, Shim JH, Kim DH, Cho DW. 2014. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat Commun 5: 3935. doi: 10.1038/ncomms4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabhi SR, Martinez JS, Keller TCS. 2014. Decellularized ECM effects on human mesenchymal stem cell stemness and differentiation. Differentiation 88: 131–143. doi: 10.1016/j.diff.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppas NA, Hilt JZ, Khademhosseini A, Langer R. 2006. Hydrogels in biology and medicine: from molecular principles to bionanotechnology. Adv Mater 18: 1345–1360. doi: 10.1002/adma.200501612 [DOI] [Google Scholar]
- Polacheck WJ, Chen CS. 2016. Measuring cell-generated forces: a guide to the available tools. Nat Meth 13: 415–423. doi: 10.1038/nmeth.3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruniéras M, Régnier M, Woodley D. 1983a. Methods for cultivation of keratinocytes with an air–liquid interface. J Invest Dermatol 81: S28–S33. doi: 10.1111/1523-1747.ep12540324 [DOI] [PubMed] [Google Scholar]
- Pruniéras M, Regnier M, Fougère S, Woodley D. 1983b. Keratinocytes synthesize basal-lamina proteins in culture. J Invest Dermatol 81: S74–S81. doi: 10.1111/1523-1747.ep12540736 [DOI] [PubMed] [Google Scholar]
- Purcell BP, Lobb D, Charati MB, Dorsey SM, Wade RJ, Zellars KN, Doviak H, Pettaway S, Logdon CB, Shuman JA, et al. 2014. Injectable and bioresponsive hydrogels for on-demand matrix metalloproteinase inhibition. Nat Mater 13: 653–661. doi: 10.1038/nmat3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammensee S, Kang MS, Georgiou K, Kumar S, Schaffer DV. 2017. Dynamics of mechanosensitive neural stem cell differentiation. Stem Cells 35: 497–506. doi: 10.1002/stem.2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP. 2014. 3D niche microarrays for systems-level analyses of cell fate. Nat Commun 5: 4324. doi: 10.1038/ncomms5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer A, Vasilevich A, Hulshof F, Viswanathan P, van Blitterswijk CA, de Boer J, Watt FM. 2016. Scalable topographies to support proliferation and Oct4 expression by human induced pluripotent stem cells. Sci Rep 6: 18948. doi: 10.1038/srep18948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodell CB, Kaminski AL, Burdick JA. 2013. Rational design of network properties in guest–host assembled and shear-thinning hyaluronic acid hydrogels. Biomacromolecules 14: 4125–4134. doi: 10.1021/bm401280z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Kawaguchi K, Park S, Gonzalez D, Brown S, Boucher J, Klein AM, Greco V. 2016. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352: 1471–1474. doi: 10.1126/science.aaf7012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales AM, Anseth KS. 2016. The design of reversible hydrogels to capture extracellular matrix dynamics. Nat Rev Mater 1: 15012. doi: 10.1038/natrevmats.2015.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales AM, Mabry KM, Nehls EM, Anseth KS. 2015. Photoresponsive elastic properties of azobenzene-containing poly(ethylene-glycol)-based hydrogels. Biomacromolecules 16: 798–806. doi: 10.1021/bm501710e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales AM, Rodell CB, Chen MH, Morrow MG, Anseth KS, Burdick JA. 2018. Reversible control of network properties in azobenzene-containing hyaluronic acid-based hydrogels. Bioconjug Chem 29: 905–913. doi: 10.1021/acs.bioconjchem.7b00802 [DOI] [PubMed] [Google Scholar]
- Ruiz SA, Chen CS. 2008. Emergence of patterned stem cell differentiation within multicellular structures. Stem Cells 26: 2921–2927. doi: 10.1634/stemcells.2008-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskowitz ER, DeForest CA. 2018. Photoresponsive biomaterials for targeted drug delivery and 4D cell culture. Nat Rev Mater 3: 17087. doi: 10.1038/natrevmats.2017.87 [DOI] [Google Scholar]
- Ruskowitz ER, DeForest CA. 2019. Proteome-wide analysis of cellular response to ultraviolet light for biomaterial synthesis and modification. ACS Biomater Sci Eng 5: 2111–2116. doi: 10.1021/acsbiomaterials.9b00177 [DOI] [PubMed] [Google Scholar]
- Ruskowitz ER, Comerford MP, Badeau BA, DeForest CA. 2019. Logical stimuli-triggered delivery of small molecules from hydrogel biomaterials. Biomater Sci 7: 542–546. doi: 10.1039/C8BM01304G [DOI] [PubMed] [Google Scholar]
- Schultz KM, Kyburz KA, Anseth KS. 2015. Measuring dynamic cell–material interactions and remodeling during 3D human mesenchymal stem cell migration in hydrogels. Proc Natl Acad Sci 112: E3757–E3764. doi: 10.1073/pnas.1511304112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadish JA, Benuska GM, DeForest CA. 2019. Bioactive site-specifically modified proteins for 4D patterning of gel biomaterials. Nat Mater 18: 1005–1014. doi: 10.1038/s41563-019-0367-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D-S, You J, Rahimian A, Vu T, Siltanen C, Ehsanipour A, Stybayeva G, Sutcliffe J, Revzin A. 2014. Photodegradable hydrogels for capture, detection, and release of live cells. Angew Chem Int Ed Engl 53: 8221–8224. doi: 10.1002/anie.201404323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siltanen C, Shin DS, Sutcliffe J, Revzin A. 2013. Micropatterned photodegradable hydrogels for the sorting of microbeads and cells. Angew Chem Int Ed Engl 52: 9224–9228. doi: 10.1002/anie.201303965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten EM, Bertozzi CR. 2009. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed Engl 48: 6974–6998. doi: 10.1002/anie.200900942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer CD, Pashuck ET, Stevens MM. 2018. Achieving controlled biomolecule–biomaterial conjugation. Chem Rev 118: 7702–7743. doi: 10.1021/acs.chemrev.8b00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Chang SY, Zhang S, Toh WS, Toh YC. 2018. Substrate stiffness modulates the multipotency of human neural crest derived ectomesenchymal stem cells via CD44 mediated PDGFR signaling. Biomaterials 167: 153–167. doi: 10.1016/j.biomaterials.2018.03.022 [DOI] [PubMed] [Google Scholar]
- Stowers RS, Allen SC, Suggs LJ. 2015. Dynamic phototuning of 3D hydrogel stiffness. Proc Natl Acad Sci 112: 1953–1958. doi: 10.1073/pnas.1421897112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyer R, Conte V, Escribano J, Elosegui-Artola A, Labernadie A, Valon L, Navajas D, García-Aznar JM, Muñoz JJ, Roca-Cusachs P, et al. 2016. Collective cell durotaxis emerges from long-range intercellular force transmission. Science 353: 1157–1161. doi: 10.1126/science.aaf7119 [DOI] [PubMed] [Google Scholar]
- Tamate R, Ueki T, Kitazawa Y, Kuzunuki M, Watanabe M, Akimoto AM, Yoshida R. 2016. Photo-dimerization induced dynamic viscoelastic changes in ABA triblock copolymer-based hydrogels for 3D cell culture. Chem Mater 28: 6401–6408. doi: 10.1021/acs.chemmater.6b02839 [DOI] [Google Scholar]
- Tamura M, Yanagawa F, Sugiura S, Takagi T, Sumaru K, Matsui H, Kanamori T. 2014. Optical cell separation from three-dimensional environment in photodegradable hydrogels for pure culture techniques. Sci Rep 4: 4793. doi: 10.1038/srep04793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. 2003. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc Natl Acad Sci 100: 1484–1489. doi: 10.1073/pnas.0235407100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Xiao C, Sun J, Xiong D, Hu X. 2012. Biological self-assembly of injectable hydrogel as cell scaffold via specific nucleobase pairing. Chem Commun (Camb) 48: 10289–10291. doi: 10.1039/c2cc35449g [DOI] [PubMed] [Google Scholar]
- Teixeira AI, Abrams GA, Bertics PJ, Murphy CJ, Nealey PF. 2003. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J Cell Sci 116: 1881–1892. doi: 10.1242/jcs.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K, Engler AJ, Meyer GA. 2015. Extracellular matrix regulation in the muscle satellite cell niche. Connect Tissue Res 56: 1–8. doi: 10.3109/03008207.2014.947369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbitt MW, Anseth KS. 2009. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng 103: 655–663. doi: 10.1002/bit.22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong VX, Li F, Forsythe JS. 2017. Versatile bioorthogonal hydrogel platform by catalyst-free visible light initiated photodimerization of anthracene. ACS Macro Lett 657–662. doi: 10.1021/acsmacrolett.7b00312 [DOI] [PubMed] [Google Scholar]
- Tse JR, Engler AJ. 2010. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol 47: 10.16.11–10.16.16. [DOI] [PubMed] [Google Scholar]
- Ulijn RV. 2006. Enzyme-responsive materials: a new class of smart biomaterials. J Mater Chem 16: 2217–2225. doi: 10.1039/b601776m [DOI] [Google Scholar]
- Uto K, Ebara M, Aoyagi T. 2014. Temperature-responsive poly(ε-caprolactone) cell culture platform with dynamically tunable nano-roughness and elasticity for control of myoblast morphology. Int J Mol Sci 15: 1511–1524. doi: 10.3390/ijms15011511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto K, Mano SS, Aoyagi T, Ebara M. 2016. Substrate fluidity regulates cell adhesion and morphology on poly(ε-caprolactone)-based materials. ACS Biomater Sci Eng 2: 446–453. doi: 10.1021/acsbiomaterials.6b00058 [DOI] [PubMed] [Google Scholar]
- Uto K, Aoyagi T, DeForest CA, Hoffman AS, Ebara M. 2017a. A combinational effect of “bulk” and “surface” shape-memory transitions on the regulation of cell alignment. Adv Healthc Mater 6: 1601439. doi: 10.1002/adhm.201601439 [DOI] [PubMed] [Google Scholar]
- Uto K, Tsui JH, DeForest CA, Kim DH. 2017b. Dynamically tunable cell culture platforms for tissue engineering and mechanobiology. Prog Polym Sci 65: 53–82. doi: 10.1016/j.progpolymsci.2016.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uto K, Aoyagi T, DeForest CA, Ebara M. 2018. Dynamic alterations of hepatocellular function by on-demand elasticity and roughness modulation. Biomater Sci 6: 1002–1006. doi: 10.1039/C8BM00047F [DOI] [PubMed] [Google Scholar]
- Vining KH, Mooney DJ. 2017. Mechanical forces direct stem cell behaviour in development and regeneration. Nat Rev Mol Cell Biol 18: 728–742. doi: 10.1038/nrm.2017.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RG. 2008. The role of matrix stiffness in regulating cell behavior. Hepatology 47: 1394–1400. doi: 10.1002/hep.22193 [DOI] [PubMed] [Google Scholar]
- Winer JP, Janmey PA, McCormick ME, Funaki M. 2008. Bone marrow-derived human mesenchymal stem cells become quiescent on soft substrates but remain responsive to chemical or mechanical stimuli. Tissue Eng Part A 15: 147–154. doi: 10.1089/ten.tea.2007.0388 [DOI] [PubMed] [Google Scholar]
- Wu X, Huang W, Wu WH, Xue B, Xiang D, Li Y, Qin M, Sun F, Wang W, Zhang WB, et al. 2018. Reversible hydrogels with tunable mechanical properties for optically controlling cell migration. Nano Res 11: 5556–5565. doi: 10.1007/s12274-017-1890-y [DOI] [Google Scholar]
- Wylie RG, Ahsan S, Aizawa Y, Maxwell KL, Morshead CM, Shoichet MS. 2011. Spatially controlled simultaneous patterning of multiple growth factors in three-dimensional hydrogels. Nat Mater 10: 799–806. doi: 10.1038/nmat3101 [DOI] [PubMed] [Google Scholar]
- Yamato M, Akiyama Y, Kobayashi J, Yang J, Kikuchi A, Okano T. 2007. Temperature-responsive cell culture surfaces for regenerative medicine with cell sheet engineering. Prog Polym Sci 32: 1123–1133. doi: 10.1016/j.progpolymsci.2007.06.002 [DOI] [Google Scholar]
- Yang B, Zhang Y, Zhang X, Tao L, Li S, Wei Y. 2012. Facilely prepared inexpensive and biocompatible self-healing hydrogel: a new injectable cell therapy carrier. Polym Chem 3: 3235–3238. doi: 10.1039/c2py20627g [DOI] [Google Scholar]
- Yang C, Tibbitt MW, Basta L, Anseth KS. 2014. Mechanical memory and dosing influence stem cell fate. Nat Mater 13: 645–652. doi: 10.1038/nmat3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa HY, Rossetti FF, Kaufmann S, Kaindl T, Madsen J, Engel U, Lewis AL, Armes SP, Tanaka M. 2011. Quantitative evaluation of mechanosensing of cells on dynamically tunable hydrogels. J Am Chem Soc 133: 1367–1374. doi: 10.1021/ja1060615 [DOI] [PubMed] [Google Scholar]
- Young JL, Engler AJ. 2011. Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials 32: 1002–1009. doi: 10.1016/j.biomaterials.2010.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Kilian KA. 2013. The effect of mesenchymal stem cell shape on the maintenance of multipotency. Biomaterials 34: 3962–3969. doi: 10.1016/j.biomaterials.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Zhang H, Trout WS, Liu S, Andrade GA, Hudson DA, Scinto SL, Dicker KT, Li Y, Lazouski N, Rosenthal J, et al. 2016. Rapid bioorthogonal chemistry turn-on through enzymatic or long wavelength photocatalytic activation of tetrazine ligation. J Am Chem Soc 138: 5978–5983. doi: 10.1021/jacs.6b02168 [DOI] [PMC free article] [PubMed] [Google Scholar]