Abstract
The microbial fluctuations along an increasing salinity gradient during two different salt production phases – initial salt harvesting (ISH) phase and peak salt harvesting (PSH) phase of Siridao solar salterns in Goa, India were examined through high-throughput sequencing of 16S rRNA genes on Illumina MiSeq platform. Elemental analysis of the brine samples showed high concentration of sodium (Na+) and chloride (Cl–) ions thereby indicating its thalassohaline nature. Comparison of relative abundance of sequences revealed that Archaea transited from sediment to brine while Bacteria transited from brine to sediment with increasing salinity. Frequency of Archaea was found to be significantly enriched even in low and moderate salinity sediments with their relative sequence abundance reaching as high as 85%. Euryarchaeota was found to be the dominant archaeal phylum containing 19 and 17 genera in sediments and brine, respectively. Phylotypes belonging to Halorubrum, Haloarcula, Halorhabdus, and Haloplanus were common in both sediments and brine. Occurence of Halobacterium and Natronomonas were exclusive to sediments while Halonotius was exclusive to brine. Among sediments, relative sequence frequency of Halorubrum, and Halorhabdus decreased while Haloarcula, Haloplanus, and Natronomonas increased with increasing salinity. Similarly, the relative abundance of Haloarcula and Halorubrum increased with increasing salinity in brine. Sediments and brine samples harbored about 20 and 17 bacterial phyla, respectively. Bacteroidetes, Proteobacteria, and Chloroflexi were the common bacterial phyla in both sediments and brine while Firmicutes were dominant albeit in sediments alone. Further, Gammaproteobacteria, Alphaproteobacteria, and Deltaproteobacteria were observed to be the abundant class within the Proteobacteria. Among the bacterial genera, phylotypes belonging to Rubricoccus and Halomonas were widely detected in both brine and sediment while Thioalkalispira, Desulfovermiculus, and Marinobacter were selectively present in sediments. This study suggests that Bacteria are more susceptible to salinity fluctuations than Archaea, with many bacterial genera being compartment and phase-specific. Our study further indicated that Archaea rather than Bacteria could withstand the wide salinity fluctuation and attain a stable community structure within a short time-frame.
Keywords: hypersaline environments, solar saltern, microbial diversity, metabarcoding, archaea, bacteria
Introduction
Hypersaline environments like salt lakes and solar salterns are characterized by the presence of high content of salt (>3.5% salinity) and therefore the diversity of organisms in these habitats is unique (Oren, 2002; Ventosa, 2006; Paul and Mormile, 2017). Halophilic organisms counter the high extracellular salinity by two different strategies. In the first strategy, Archaea and a few Bacteria (members exclusively belonging to order Halanaerobiales and genus Salinibacter) have adapted the “salt-in” strategy of accumulating K+ and Cl– ions while maintaining low Na+ concentration in response to the external osmotic pressure. In the second strategy, most Bacteria and Eukaryotes combat high salinity by accumulating or synthesizing organic solutes like glycine betaine, ectoine, and other derivatives of amino acids and sugars (Oren, 2002, 2013; Roberts, 2005). Apart from high salinity, these ecosystems further experience fluctuations in pH and temperature, facilitating, polyextremophilic organisms like halophilic alkaliphiles and halophilic alkalithermophiles to inhabit (Grant et al., 1999; Bowers and Wiegel, 2011; Mesbah and Wiegel, 2012).
Coastal solar salterns, employed primarily for the production of edible sodium chloride, contain a series of pans for concentrating seawater, thereby facilitating the sequential precipitation of calcite (CaCO3), gypsum (CaSO4) followed by halite (NaCl), leaving behind salts of magnesium and potassium (Javor, 2002). These ecosystems make an excellent model for studying the diversity patterns due to the maintenance of constant salinity over a period with minimal external disturbances. Further, the predictable chemistry and limited biodiversity of these systems make the coastal solar salterns an ideal platform for scaling the studies on the evaporative sequence of minerals, biological composition and activity over a broad range of salinities to much complex hypersaline ecosystems (Benlloch et al., 2002; Oren, 2009). In particular, studies on hypersaline environments have gathered interest in the past four decades (Rodriguez-Valera et al., 1981; Ventosa et al., 1982, 1998) due to their importance in understanding the metabolic, physiological and genetic adaptation of organisms in high salinity coupled with their potential biotechnological applications (Oren, 1999, 2010; Siglioccolo et al., 2011; Yin et al., 2015; Gunde-Cimerman et al., 2018).
Culture-independent studies carried out so far in assessing the prokaryotic diversity of coastal and inland solar salterns have identified that the relative sequence abundance of primary bacterial phyla distributed in low salinity samples (4–7%) were Proteobacteria, Cyanobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Tenericutes, a community composition similar to marine environments. In the intermediate salinity samples (13–18%), bacterial phylotypes affiliated to phyla Proteobacteria, Bacteroidetes, Actinobacteria, and Verrumicrobia were usually encountered while the archaeal members belonged to genera Halorubrum, Haloarcula, and Haloquadratum (phylum Euryarchaeota). High salinity samples (>25%) were dominated by halophilic archaea belonging to genera such as Haloquadratum and Halorubrum along with a minor contribution from bacterial phylotypes belonging to Bacteroidetes (genus Salinibacter) (Mouné et al., 2003; Pašić et al., 2005; Sørensen et al., 2005; Park et al., 2006; Wang et al., 2007; Baati et al., 2008, 2010; Tsiamis et al., 2008; Manikandan et al., 2009; Oren et al., 2009; Oh et al., 2010; López-López et al., 2010; Zafrilla et al., 2010; Tkavc et al., 2011; Trigui et al., 2011; Boujelben et al., 2012; Dillon et al., 2013; Fernandez et al., 2014a, b; Mani et al., 2015; Çinar and Mutlu, 2016; Di Meglio et al., 2016; Zhang et al., 2016; Kalwasińska et al., 2018; Leoni et al., 2020). In general, archaeal composition from the total prokaryotic community was observed to be between 27–46% at intermediate salinities (13–19%) and 80–90% at high salinities (>25%) (Ghai et al., 2011; Fernandez et al., 2014a, b). Though the general prokaryotic community distribution pattern has been established, a majority of these studies have employed techniques that suffer from limited phylogenetic resolution, like clone library analysis of 16S rRNA sequences or denaturing gradient gel electrophoresis (DGGE) and hence the data available on halophilic 16S rRNA gene diversity is still limited. Further, these studies have focused on the prokaryotic diversity of either brine or sediments and therefore a clear picture on the transition of the prokaryotic community between brine and sediments, through an increasing salinity from (∼5 to ∼37%) is still lacking. In the present work we have employed amplicon sequencing of 16S rRNA genes to study the dynamics of prokaryotic community composition of brine and sediment samples obtained from an artisanal solar saltern located in Goa, India at two different time periods of salt production.
Goa is a coastal state in India, having several solar salterns. These coastal salterns are located predominantly along the estuaries, due to easy access to seawater during tidal influxes. Goa receives an annual rainfall of 280–480 cm, maximally from the month of June to November thereby ceasing salt production. The salt pans are constructed in the months of December and January (preparatory phase) and salt production starts from February and continues till May. In the month of February, the salterns would begin the operation through the construction of compartments that are designated into reservoir pan (RP), evaporator pan (EP) and crystallizer pan (CP). Salt crystals produced in the month of February are usually not harvested and rather allowed to settle in the crystallizer pan forming a hard bed of salt crystals thereby facilitating the collection of clean salt without any impurities. This phase lasts for a period of 20 – 25 days and is termed as initial salt harvesting (ISH) phase. Due to the prevailing climatic conditions (low temperature and less wind) and low salinity of inlet water (2%), the time required for salt crystallization varies from 4 to 5 days. However, by March, with the onset of summer, compounded by strong winds and ample sunshine, the time required for salt crystallization in CP decreases to a day or two. This phase of salt production lasts until May and constitutes the peak salt harvesting (PSH) phase. This entire process is repeated on an annual basis and therefore the microbes in this ecosystem are subjected to varying salinity from 2% to 30%. This salinity fluctuation makes the salterns of Goa an ideal candidate for studying the microbial tolerance toward widely fluctuating salinities. The described salt production process is in contrast with the majority of solar salterns located elsewhere around the world, where the brine retention time normally varies from months to years compared to the salterns investigated in this study (Mani et al., 2012b).
In this study, we aimed at investigating the microbial community response toward sudden onset and sustained salinity over a time scale by profiling the prokaryotic composition of a coastal solar saltern between ISH and PSH. Given the nature of solar salterns of Goa, it was hypothesized that Archaea might be more susceptible to changes in the salinity fluctuations prevalent in solar salterns in comparison to Bacteria. Further, Bacteria were expected to dominate the salterns during the ISH and Archaea to dominate during the PSH phase owing to the sustained high salinity. Previous studies conducted in vertical depths of marine environments (Feng et al., 2009; Hamdan et al., 2013; Walsh et al., 2016) have shown that the prokaryotic community composition in water and sediment samples are distinct. Additionally, a study carried out by Walsh et al. (2016) suggested that water could act as a carrier in transporting the microbes over geographical distances due to the presence of rare OTUs in water, otherwise abundant in sediments. Therefore, we also sought to investigate the similarity in the community composition of brine and sediment samples since the compartments of salterns are interconnected and there is a constant influx of brine along a salinity gradient.
Materials and Methods
Sample Collection
Brine and surface sediments were collected from solar salterns (15°44′N, 73°86′E) located at Siridao in Goa, India (Supplementary Figure S1). Sampling was carried out in defined compartments namely RP, EP, and CP. Reservoir pans (RP) are primarily used for storing seawater during the tidal influxes. Once the salinity reaches around 5%, the brine is released into the evaporator pans (EP), where the brine is further concentrated until it reaches 23–25% salinity. When the salinity in the EP crosses 5% and 13%, calcium carbonate (CaCO3) and calcium sulfate (CaSO4) precipitates, respectively. This concentrated brine is finally fed to the crystallizer pans (CP) where the sodium chloride (NaCl) crystallizes around 28% salinity leaving behind magnesium (Mg2+) and potassium (K+) (Mani et al., 2012b). About 500 ml of Brine samples were collected in a sterile borosilicate glass bottles from the middle of the pans at a depth of 5–10 cm. Similarly, about 50 g of sediments were scooped using a sterile spatula at a depth of 0–5 cm from the surface and stored in polyethylene bags. Both brine and sediment samples were transported in ice and stored at −20°C. Replicate sampling (n = 5) was carried out at each sampling point and a composite sample representing each pan was prepared by mixing these replicates into a single sample. A similar approach was adopted for a simultaneous sampling of brine that was subjected to elemental analysis with exceptions of restricting the number of replicates to 3 and storing the samples at 4°C. Initial sampling was carried out in the month of February 2014, during the ISH phase and the second sampling in the month of May 2014 during the PSH phase.
Physico-Chemical Parameter Estimation and Elemental Analysis
The salinity of the brine samples was measured using a Baumé hydrometer while pH and temperature were documented onsite using a portable pH/temperature meter (Equiptronics, India). Prior to the salinity measurements, Baumé hydrometer was standardized against distilled water at 0% salinity, with reading corresponding to 0 Bé. Salinity and pH of the sediments were measured from the extracts obtained from 1:5 sediment: water mixture. The brine samples were subjected to elemental analysis after filtering through a 0.22 μm filter and diluting them appropriately. The concentration of monovalent cations, sodium (Na+) and potassium (K+) prevalent in the brine samples was measured through flame emission photometry (Systronics, India). Calcium (Ca2+) and magnesium (Mg2+) concentration were determined simultaneously by complexometric titration against ethylenediaminetetraacetic acid (EDTA) in the presence of Eriochrome Black T as an indicator (De Sousa, 1954). Chloride (Cl–) ion content was determined by argentometric titration against silver nitrate solution in the presence of potassium chromate as an indicator (Skoog et al., 1996).
Environmental DNA Extraction and Amplicon Sequencing of 16S rRNA Genes
The nucleic acid extraction protocol involved a combination of mechanical, enzymatic and chemical lysis of microbial cells (Zhou et al., 1996). About 5 g of sediment was washed thrice with 100 mM sodium phosphate buffer (pH 7.0)followed by bead beating in the presence of extraction buffer (100 mM tris hydrochloride (pH 8.0), 100 mM ethylenediaminetetraacetic acid (pH 8.0), 100 mM sodium phosphate (pH 8.0), 1.5 M sodium chloride, 2.5% cetyltrimethylammonium bromide and 1% polyvinylpyrrolidone). In the case of brine samples, about 200 mL was vacuum-filtered through a 0.22 μm polycarbonate filter and the obtained membrane was treated similarly to sediment samples. Following the addition of extraction buffer, samples were incubated at 65°C for 2 h after the addition of proteinase K (10 mg/ml) and 20% sodium dodecyl sulfate (SDS). After incubation, DNA was extracted with chloroform:isoamylalcohol (24:1 vol/vol) and precipitated with isopropanol. Obtained crude DNA extracts from brine and sediments were purified using PowerClean DNA Clean-Up Kit (MoBio Laboratories, United States).
Amplification of the V4-V5 region of the 16S rRNA genes was performed using universal prokaryotic primers 515F (5′-GTG YCA GCM GCC GCG GTA-3′) and 909R (5′-CCC CGY CAA TTC MTT TRA GT-3′) (Wang and Qian, 2009; Tamaki et al., 2011). Each reaction mixture contained 10 X PCR buffer, 10 mM of each dNTPs, 2 mM MgCl2, 1U Taq Polymerase (ThermoFischer Scientific, United States), 10 mM primers (8-base barcoded), 8 μg BSA and 10 ng template DNA. PCR reactions were performed in duplicates with the following conditions: initial denaturation at 94°C for 10 min followed by 35 cycles of denaturation at 94°C for 60 s, annealing at 58°C for 60 s, elongation at 72°C for 90 s and a final extension at 72°C for 10 min. The PCR products were purified with MinElute kit (Qiagen, United States) and quantified using Qant-iT PicoGreen dsDNA Assay Kit (ThermoFischer Scientific, United States). The amplicons were normalized and multiplexed followed by library preparation and sequencing with 300-bp paired-end sequencing protocol on the Illumina MiSeq platform (Illumina, United States) at GenoScreen, France.
Bioinformatics and Statistical Analysis of 16S rRNA Amplicon Sequencing Data
Reads generated from the Illumina sequencing platform were processed sequentially using the PANAM pipeline1 (Hugoni et al., 2013; Taib et al., 2013). Quality processing steps involved removing reads containing bases with ambiguous nucleotides (N) and PHRED quality score <20. Further, reads with the length shorter than 200 bp and a mismatch in the forward primer sequence were also removed. This was followed by merging the paired-end reads using PANDAseq with a threshold value of 95% (Masella et al., 2012). Following the cleaning procedure, reads were checked for chimera using UCHIME and the clustering of the quality checked reads was carried out using UCLUSTwith a threshold value of 97% (Edgar, 2010). The final step involved taxonomic assignment by the phylogenetic affiliation procedure of PANAM. Briefly, the seed OTUs were compared against reference sequences obtained from the SSURefSILVA database using USEARCH followed by appending each sequence to a phyletic group along with five best hits and sorting the query sequences according to their assignment. Further, HMMalign was used to align the homologous reads with reference sequences from the corresponding profile and a phylogenetic tree was constructed for each profile with FASTTREE2 using Jukes-Cantor + Cat model and with a bootstrap threshold of 100 replicates. Following the phylogenetic tree construction, files containing the details of the taxonomy of bacterial and archaeal sequences (inferred by the lowest common ancestor) and their nearest neighbor were obtained. Alpha and Beta diversity analysis was performed after removing the singletons from the analyzed reads and randomly normalizing to 4100 sequences. The obtained rarefied reads were used to plot rarefaction curves depicting the sampling effort. Alpha diversity measures like species diversity (Shannon index), richness (Chao1), abundance-based coverage estimator (ACE), dominance, and evenness indices were calculated. Diversity indices like Shannon, Chao1, ACE and plotting of rarefaction curves were automated with PANAM. The remaining alpha diversity measures viz. Dominance and Buzas-Gibson’s evenness indices were estimated using PAST (PAlaeontologicalSTatistics) (Hammer et al., 2005). The overall similarity among the microbial community composition between ISH and PSH was analyzed based on Bray-Curtis dissimilarities and visualized using Non-Metric Dimensional Scaling (NMDS) (Minchin, 1987) in vegan package (Oksanen et al., 2013) implemented in R (R Development Core Team, 2014). Statistically significant variation in microbiome taxonomic composition was assed using analysis of similarity (ANOSIM) function in vegan. Unique and common operational taxonomic units (OTUs) shared between different sampling sites were identified through plotting Venn diagrams using jvenn (Bardou et al., 2014).
Results
Physico-Chemical Characteristics of the Saltern
The salinity of brine samples increased from 2.1% to 25.2% and 4% to 28% during ISH and PSH respectively (Table 1). Similarly, the salinity of sediment samples increased from 1.3% to 22.3% and 2.3% to 24.8% during ISH and PSH, respectively. The pH of sampling sites varied from 6.5 to 7.8 indicating that the samples are near neutral. The mean temperature of sampling sites during the ISH and PSH phase was 33°C and 38°C, respectively. Though some differences between the temperatures could be observed, the temperature profile of the salterns remained relatively stable throughout the operational stages, a typical feature of the tropical climatic zone (Couto-Rodríguez and Montalvo-Rodríguez, 2019). Elemental analysis of the brine samples indicated that the salterns are dominated with sodium and chloride ions indicating the thalassohaline nature. Most cations (sodium, magnesium and potassium) except for calcium and anion (chloride) increased with increasing salinity (Table 1). A decrease in the concentration of calcium in CP when compared with EP during both phases of salt production could be attributed to the precipitation of gypsum (calcium sulfate) in the intermediate salinity which was apparent during the sampling.
TABLE 1.
Physico-chemical parameters of brine and sedimentsa,b obtained from various compartments of solar saltern.
| Sampling sitesc | pH | Temp. (°C) | Salinity (%) | Na+ (g l–1) | K+ (g l–1) | Mg2+ (g l–1) | Ca2+ (g l–1) | Cl– (g l–1) |
| ISH | ||||||||
| RP | 7.1 | 33.2 | 2.1 | 9.9 | 0.4 | 0.2 | 0.5 | 18.4 |
| EP | 6.5 | 32.9 | 13.6 | 35.19 | 1.1 | 0.4 | 1.3 | 65.51 |
| CP | 7.4 | 33.4 | 25.2 | 68.2 | 4.2 | 8.6 | 0.3 | 119.28 |
| PSH | ||||||||
| RP | 7.2 | 38.2 | 4 | 10.8 | 0.4 | 0.2 | 0.7 | 19 |
| EP | 7.6 | 38.4 | 17.4 | 45.02 | 2.2 | 0.3 | 1.1 | 85.44 |
| CP | 7.8 | 38.1 | 28 | 72.45 | 4.5 | 8.6 | 0.3 | 141.8 |
aSalinity of sediment samples in (%): ISH – RP – 1.3%; EP – 9.1%; CP – 22.3%; PSH – RP – 2.3%; EP – 13.5%; CP – 24.8%. bElemental analysis was not carried out for the sediment samples. cISH and PSH phase sampling was carried out in the months of February and May 2014, respectively.
Diversity Patterns of the Solar Saltern
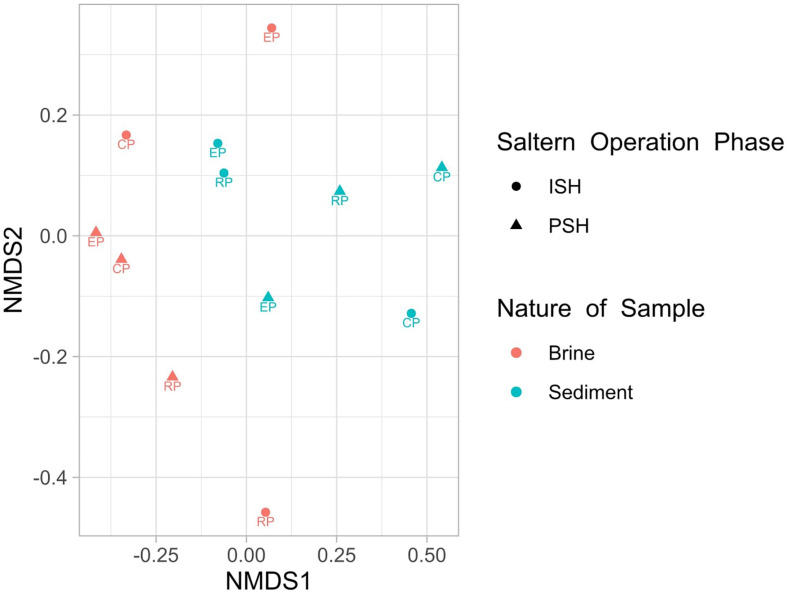
Quality filtering and processing the reads pertaining to the V4-V5 region of the 16S rRNA gene generated 403010 and 2930653 sequences corresponding to Bacteria and Archaea, respectively. Further, taxonomic affiliation with 97% similarity threshold yielded 2638 and 6681 OTUs related to bacterial and archaeal phylotypes respectively (Table 2). On comparing the archaeal and bacterial OTUs within the same sampling sites, it was observed that Archaea had at least two-fold higher OTUs than Bacteria. Similarly, when the OTUs that are commonly distributed across multiple sampling sites were analyzed, it was again observed that bacterial OTUs were outnumbered by archaeal OTUs. This indicated that the similar archaeal phylotypes were dwelling along an increasing salinity gradient in multiple sampling sites (Supplementary Figure S2). On a further note, only 17 OTUs were present consistently across all sampling sites and out of which, 16 belonged to Archaea (7 – Halorcula; 6 – Halorubrum; 1 – Halobacterium; 1 – Halobaculum; 1 – uncultured) while 1 belonged to Bacteria (Idiomarina). Rarefaction curves indicated that sediment samples harbored more OTUs than brine samples during both phases of salt production. The pattern of rarefaction curves further depicted that in most cases, the sampling depth was not sufficient to capture the entire diversity present in the solar saltern under investigation (Supplementary Figure S3). Chao1 and ACE values varied between 1095 to 2826 and 1175 to 3036, respectively. Based on the diversity indices and rarefaction curve patterns, the least species richness was observed for CP brine from ISH while EP sediment collected during the ISH phase displayed the highest species richness. Like rarefaction curves, Chao1 and ACE indices too indicated that sediments harbored a richer microbial community when compared to brine. Shannon diversity index values varied from 4.07 to 5.76 and the highest diversity was noticed among the sediment samples belonging to the ISH phase (Table 3). Dominance index varied from 0.016 to 0.074 in the brine samples and from 0.008 to 0.021 in the sediment samples during both phases of salt production. Buzas and Gibson’s evenness index varied from 0.103 to 0.239 for the brine samples and from 0.199 to 0.328 for the sediment samples during both phases of salt production. Dominance and Buzas and Gibson’s evenness indices implied that comparatively, the OTUs are distributed evenly and with equal dominance among sediments than brine. Similarities on the microbial community distribution between ISH and PSH phases were compared by ANOSIM and NMDS based on Bray-Curtis. NMDS plot indicated that there was no clear separation among the samples obtained indicating the similarity among the prokaryotic community composition between ISH and PSH which was further confirmed by ANOSIM (Figure 1). The statistical analysis revealed that there was no significant difference in the microbial community composition between brine (R = 0.03, P = 0.5) and sediments (R = 0.02, P = 0.6) obtained from both phases of salt production. Similarly, no significant difference was observed between brine and sediment samples obtained in ISH (R = 0.037, P = 0.6) and PSH (R = 0.307, P = 0.1).
TABLE 2.
Bacterial and archaeal features obtained after processing the amplicon sequencing data through PANAM pipeline.
| Saltern operation phase | Nature of the sample | Sampling sites | Total no of reads obtained | Total no of Bacterial reads | Total no of Archaeal reads | Total no of Bacterial OTUs | Total no of Archaeal OTUs |
| ISH | Brine | RP | 402348 | 262738 | 139610 | 360 | 451 |
| EP | 853366 | 419973 | 433393 | 291 | 519 | ||
| CP | 502356 | 46682 | 455674 | 46 | 523 | ||
| Sediment | RP | 5967 | 1021 | 4946 | 255 | 706 | |
| EP | 8997 | 1242 | 7755 | 170 | 915 | ||
| CP | 4282 | 1716 | 2566 | 180 | 223 | ||
| PSH | Brine | RP | 589275 | 430437 | 158818 | 262 | 436 |
| EP | 916698 | 172556 | 744142 | 34 | 602 | ||
| CP | 862086 | 160566 | 701520 | 161 | 750 | ||
| Sediment | RP | 14691 | 3746 | 10945 | 293 | 592 | |
| EP | 10320 | 3358 | 6962 | 371 | 588 | ||
| CP | 6458 | 3285 | 3173 | 215 | 376 |
TABLE 3.
Observed microbial richness and diversity estimates among the sampling sites of salterns of Siridao based on 97% OTU clusters.
| Saltern operation phase | Nature of the sample | Sampling sites | OTUs observed | Shannon | Chao1 | ACEa | Dominance | Buzas-Gibson’s Evenness |
| ISH | Brine | RP | 810 | 5.26 | 1721 | 1818 | 0.017 | 0.239 |
| EP | 809 | 5.23 | 2191 | 2291 | 0.016 | 0.231 | ||
| CP | 568 | 4.07 | 1095 | 1175 | 0.074 | 0.103 | ||
| Sediment | RP | 960 | 5.75 | 2221 | 2294 | 0.008 | 0.328 | |
| EP | 1083 | 5.76 | 2826 | 3036 | 0.010 | 0.294 | ||
| CP | 983 | 5.70 | 2204 | 2231 | 0.011 | 0.306 | ||
| PSH | Brine | RP | 697 | 4.61 | 1795 | 1867 | 0.044 | 0.145 |
| EP | 635 | 4.36 | 1555 | 1578 | 0.052 | 0.123 | ||
| CP | 910 | 5.05 | 2267 | 2531 | 0.026 | 0.171 | ||
| Sediment | RP | 884 | 5.48 | 2026 | 2060 | 0.013 | 0.272 | |
| EP | 958 | 5.54 | 2466 | 2552 | 0.012 | 0.266 | ||
| CP | 590 | 4.76 | 1503 | 1633 | 0.021 | 0.199 |
aACE, Abundance-based coverage estimator. The diversity indices were calculated after normalizing the sequences obtained across all the sampling sites at 4100.
FIGURE 1.
Non-metric Multidimensional Scaling (NMDS) plot showing similarity in the microbial composition (Bray–Curtis distance) of solar salterns during ISH and PSH phase of salt production. RP stands for reservoir pan, EP for evaporator pan and CP for crystallizer pan.
Dynamics of Archaea and Bacteria Along the Salinity Gradient
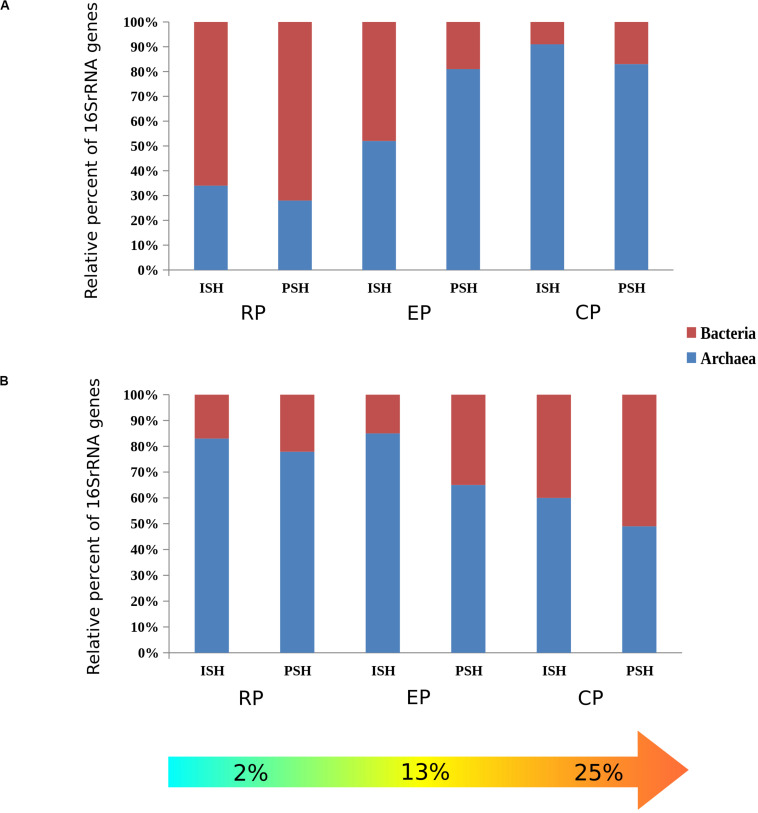
The relative abundance of archaeal and bacterial phylotypes distributed across the salinity gradient during both phases of salt production was calculated based on the taxonomic affiliation of the sequences. Among brine samples, the occurence of archaeal sequences increased along the salinity gradient from 34% to 91% (ISH) and 27% to 84% (PSH) while bacterial sequences decreased from 66% to 9% (ISH) and 73% to 17% (PSH) (Figure 2A). However, in the sediment samples, an opposite trend was observed. Composition of bacterial sequences increased along the salinity gradient from 17% to 40% (ISH) and 21% to 51% (PSH) while archaeal sequences decreased from 83% to 60% (ISH) and 74% to 49% (PSH) (Figure 2B). This data further indicated that there was no difference in the relative frequency of archaeal and bacterial phylotypes between two salt-producing seasons. As a whole, during both the phases of salt production, along the increasing salinity gradient, archaeal sequences outnumbered bacteria proving that Archaea is the dominant prokaryotic member (Table 2).
FIGURE 2.
Horizontal stacked bar charts describing the relative abundance of Archaea and Bacteria between sampling sites with different salinities and between different salt producing season (A) Brine and (B) Sediment. ISH stands for initial salt harvesting phase, PSH for peak salt harvesting phase, RP for reservoir pan, EP for evaporator pan and CP for crystallizer pan.
Community Composition of Archaea in Solar Salterns
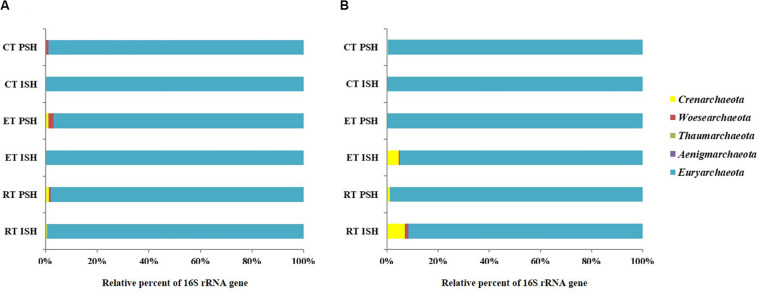
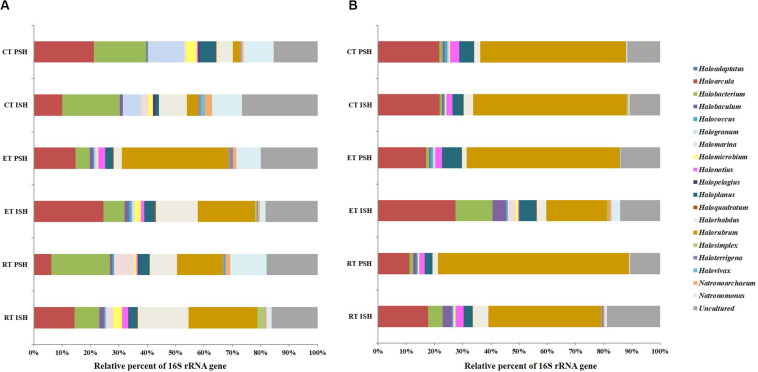
Archaeal sequences obtained from sediments were phylogenetically affiliated to five phyla namely Crenarchaeota, Woesearchaeota, Aenigmarchaeota, Euryarchaeota, and Thaumarchaeota, with Euryarchaeota being dominant and containing greater than 98% of the total sequences (Figure 3A). Sequences affiliated with Crenarchaeota, Aenigmarchaeota, and Thaumarchaeota were observed only in RP and EP. However, sequences belonging to Woesearchaeota were found across the salinity gradient from RP to CP albeit only during PSH. Halobacteria (including the predominant Halobacteriaceae, Haloferaceae, Natrialbaceae, and MSP41) was the dominant class of Euryarchaeota in sediment samples of Siridao salterns, contributing to 98.5% of the available sequences. The sequences were assigned to 19 halophilic archaeal genera. Further, major sequences at genus level (>1% among obtained sequences) across all salinities and during both phases of salt production in the sediments were identified as Haloarcula, Halorubrum, Halorhabdus, Halobacterium, Natronomonas and Haloplanus (Figure 4A). Halorubrum (24% in RP to 4% in CP and 15% in RP to 2% in CP during ISH and PSH respectively) and Halorhabdus (17% in RP to 9% in CP and 9% in RP to 5% in CP during ISH and PSH respectively) were found to decrease with increasing salinity during both phases of salt production. Sequences belonging to other genera like Haloarcula (14% in RP to 9% in CP and 8% in RP to 20% in CP during ISH and PSH respectively), Halobacterium (8% in RP to 20% in CP and 20% in RP to 18% in CP during ISH and PSH respectively), Haloplanus (2% in RP to 1% in CP and 3% in RP to 5% in CP during ISH and PSH respectively), Natronomonas (1% in RP to 10% in CP and 12% in RP to 10% in CP during ISH and PSH respectively) and Halomicrobium (3% in RP to 1% in CP and 1% in RP to 3% in CP during ISH and PSH respectively) were found to increase or decrease along the salinity gradient either during ISH or PSH (Supplementary Figure S4). Sequences affiliated with few genera were observed only at a particular salinity at specific sampling sites. For instance, Halogranum was observed exclusively in CP (6% and 12% during ISH and PSH respectively). Similarly, phylotypes associated with Halomarina was exclusively found to occur in the RP samples (2% and 6% during ISH and PSH respectively). Sequences affiliated with other genera like Halobaculum, Halonotius, and Natronoarchaeum were found to be sparsely occuring (between 1–2%) at intermittent sampling sites. About 15–26% of sequences across all compartments were assigned to uncultured haloarchaea (Figure 4A).
FIGURE 3.
Relative sequence abundance at phyla level of the archaeal communities in (A) sediments and (B) brine of salterns of Siridao. ISH stands for initial salt harvesting phase, PSH for peak salt harvesting phase, RP for reservoir pan, EP for evaporator pan and CP for crystallizer pan.
FIGURE 4.
Relative sequence abundance at genera level of the archaeal communities in (A) sediments and (B) brine of salterns of Siridao. ISH stands for initial salt harvesting phase, PSH for peak salt harvesting phase, RP for reservoir pan, EP for evaporator pan and CP for crystallizer pan.
In brine, the obtained archaeal sequences were phylogenetically affiliated to four phyla namely Crenarchaeota, Woesearchaeota, Aenigmarchaeota, and Euryarchaeota, with Euryarchaeota being dominant, containing greater than 91% of the total sequences (Figure 3B). Crenarchaeota were found to occur only in RP (7%) and EP (4.7%) samples obtained during the ISH phase. Similar to sediments, Halobacteria (including predominant Halobacteriaceae, Haloferaceae, Natrialbaceae, and MSP41) was the dominant class of Euryarchaeota in brine samples of Siridao salterns, contributing to 99% available sequences. Further, the sequences were assigned to 17 halophilic archaeal genera. Major sequences at genus level (>1% of all obtained sequences) across all salinities and during both phases of salt production in the brine samples were identified as Haloarcula, Halorubrum, Halonotius, Haloplanus and Halorhabdus (Figure 4B). Halorubrum (39% in RP to 54% in CP and 67% in RP to 51% in CP during ISH and PSH respectively) and Haloarcula (17% in RP to 21% in CP and 11% in RP to 21% in CP during ISH and PSH respectively) were found to increase along the salinity gradient. Phylotypes affiliated with Halobacterium (5% in RP to 1% in CP) and Halorhabdus (5% in RP to 3% in CP) were found to decrease across the salinity gradient albeit in ISH phase while Halonotius (2% in RP to 3% in CP) and Haloplanus (3% in RP to 5% in CP) were found to increase along the salinity gradient in the PSH phase (Supplementary Figure S4). Similar to the sediment samples, sequences affiliated with Halobaculum were found to occur (between 1–4%) sparsely at intermittent sampling sites while about 10–18% of sequences were assigned to uncultured haloarchaea.
The cosmopolitan halophilic archaeal phylotypes, across the increasing salinity gradient and during both phases of salt production, were found to be Haloarcula, Halorubrum, and Halobacterium, often constituting more than 75% of the total sequences obtained. As a whole, the community composition of halophilic archaea was found to be relatively stable with a few archaeal phylotypes (Haloarcula, Halorubrum, Halobacterium, Halorhabdus, Halogranum, Halomarina, Halobaculum, Haloplanus, and Natronomonas) equally colonizing the salterns across different salinities.
Community Composition of Bacteria in Solar Salterns
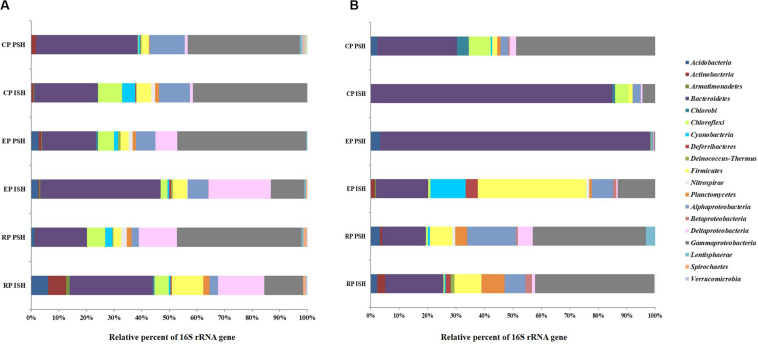
Bacterial sequences obtained from sediment samples were taxonomically affiliated to 20 phyla (Figure 5A). The major sequences at phylum level (>1% of all obtained sequences), across all salinities and during both phases of salt production in the sediments were identified as Bacteroidetes, Firmicutes and Proteobacteria. Sequences belonging to certain phyla like Chloroflexi (5% in RP to 8% in CP and 6% in RP to 0.2% in CP during ISH and PSH, respectively) were found to increase along the salinity gradient during ISH and vice versa during PSH. Relative sequence frequency of Firmicutes (11% in RP to 2% in CP) was found to be high along the increasing salinity gradient albeit exclusively during ISH phase. Sequences belonging to Acidobacteria (6% in RP and 2% in EP during ISH and PSH respectively) and Actinobacteria (6% in RP during ISH) were found to be abundant in RP and EP. Sequences affiliated with phylum Proteobacteria, class Gammaproteobacteria (13% in RP to 41% in CP and 45% in RP to 40% in CP during ISH and PSH respectively) and Alphaproteobacteria (2% in RP to 11% in CP and 2% in RP to 12% in CP during ISH and PSH respectively) increased while Deltaproteobacteria (16% in RP to 1% in CP and 13% in RP to 1% in CP during ISH and PSH respectively) decreased along the increasing salinity gradient during both phases of salt production (Supplementary Figure S4). Unlike Archaea, there were no dominant bacterial phylotypes across salinity gradient and throughout the salt production process (Table 4). Phylotypes belonging to Bacillus (except CP PSH) and Halomonas (except RP PSH) were found frequently across all sampling locations while Rubricoccus was observed consistently across the salinity gradient during ISH phase. In contrast, phylotypes affiliated to Thioalkalispira and Desulfovermiculus were observed in RP and EP while Marinobacter and Marinicella were observed in EP and RP sediment samples, across both phases of salt production. Apart from the culturable representatives, we could also observe sequences with similarity to the uncultured Bacteroidetes clone, ML602J-37 (1.6% in RP to 14.4% in CP and 1.5% in RP to 19% in CP during ISH and PSH respectively) along the salinity gradient. Similarly, sequences affiliated to uncultured Chitinophagaceae were observed to increase along salinity gradient during ISH (18.6% in RP to 28.9% in EP) and PSH (10.8% in RP to 15% in CP) phases. Further, we could affiliate about 14.3% sequences obtained from CP PSH to Uncultured Ectothiorhodospiraceae
FIGURE 5.
Relative sequence abundance at phyla level of the bacterial communities in (A) sediments and (B) brine of salterns of Siridao. ISH stands for initial salt harvesting phase, PSH for peak salt harvesting phase, RP for reservoir pan, EP for evaporator pan and CP for crystallizer pan.
TABLE 4.
Bacterial diversity at the genus level among the various sampling sites of salterns of Siridao.
| Sampling Sites | Sediment |
Brine |
||
| Genus | Relative abundance in percent | Genus | Relative abundance in percent | |
| ISH – RP | Uncultured Chitinophagaceae | 18.6% | Halomonas | 25.8% |
| Delsulfovermiculus | 12.9% | Planctomyces | 4.8% | |
| Rubricoccus | 5.6% | Uncultured Clostridiaceae | 4.3% | |
| Uncultured Xanthomonadales | 5.1% | Fulvivirga | 3% | |
| Bacillus | 3.9% | Gracilimonas | 3% | |
| Acidothermus | 2.7% | ML602J-37 | 3% | |
| Dialister | 2.3% | Salegentibacter | 2.5% | |
| Uncultured Acidobacteriaceae | 2.1% | Marinicella | 2.2% | |
| Uncultured Anaerolineaceae | 2.1% | Alcanivorax | 1.8% | |
| Thioalkalispira | 1.8% | Uncultured Deferribacterales | 1.8% | |
| Halomonas | 1.6% | Uncultured Xanthomonadaceae | 1.5% | |
| ML602J-37 | 1.6% | Idiomarina | 1.6% | |
| Truepera | 1.2% | |||
| Methylophaga | 1.1% | |||
| ISH – EP | Uncultured Chitinophagaceae | 28.9% | Bacillus | 21.6% |
| Desulfovermiculus | 18.8% | Halomonas | 7.9% | |
| Rubricoccus | 5.6% | Psychroflexus | 7.7% | |
| ML602J-37 | 4.3% | Cladithrix | 4.1% | |
| Rhodovulum | 3.2% | ML602J-37 | 3.8% | |
| Bacillus | 2.3% | Uncultured Saprospiraceae | 3.2% | |
| Thiohalorhabdus | 1.9% | Lysinibacillus | 1.8% | |
| Halomonas | 1% | Uncultured Paenibacillaceae | 1.8% | |
| Tropicimonas | 1.6% | |||
| Marinobacter | 1.6% | |||
| Phaeobacter | 1.2% | |||
| Rathybacter | 1.1% | |||
| Ruegaria | 1% | |||
| ISH – CP | ML602J-37 | 16.7% | Rubricoccus | 79% |
| Halomonas | 12.3% | Uncultured Chitinophagaceae | 3% | |
| Marinobacter | 11.2% | ML602J-37 | 2.7% | |
| Uncultured Anaerolineaceae | 5.6% | Halomonas | 2.2% | |
| Bacillus | 3.4% | |||
| Uncultured Saprospiraceae | 3.2% | |||
| Sediminimonas | 3.1% | |||
| Uncultured Caldilineaceae | 2.6% | |||
| Uncultured Rhodobacteraceae | 2% | |||
| Marinicella | 1.7% | |||
| Rubricoccus | 1.2% | |||
| PSH – RP | Thioalkalispira | 15.3% | Acidoferrobacter | 13.6% |
| Uncultured Chitinophagaceae | 10.8% | Uncultured Clostridiaceae | 6.1% | |
| Uncultured Anaerolineaceae | 5.8% | Magnetospira | 5% | |
| Delsulfovermiculus | 3.3% | Idiomarina | 4.5% | |
| Desulfosalsimonas | 2.3% | Halomonas | 4% | |
| Uncultured Gemmatimonadaceae | 2% | Fulvivirga | 3.9% | |
| Uncultured Marinilabiaceae | 2% | Gramella | 3.9% | |
| Robiginitalea | 2% | Uncultured Victivallaceae | 3.2% | |
| Bacillus | 1.5% | Planctomyces | 2.3% | |
| Uncultured Desulfobacteraceae | 1.5% | Acanthopleuribacter | 1.9% | |
| ML602J-37 | 1.5% | Thiohalophilus | 1.6% | |
| PSH – EP | Halovibrio | 11.8% | Rubricoccus | 94.3% |
| Thioalkalispira | 10.1% | Uncultured Acidobacteriaceae | 3.2% | |
| Marinobacter | 7.1% | |||
| ML602J-37 | 5.6% | |||
| Rubricoccus | 5.3% | |||
| Robiginitalea | 2.8% | |||
| Uncultured Anaerolineaceae | 2.7% | |||
| Uncultured Chitinophagaceae | 2.5% | |||
| Halomonas | 2.2% | |||
| Uncultured Caldilineaceae | 1.8% | |||
| Marinicella | 1.4% | |||
| Uncultured Desulfobacteraceae | 1.3% | |||
| Bacillus | 1.2% | |||
| Uncultured Gemmatimonadaceae | 1.2% | |||
| Desulfosalsimonas | 1.1% | |||
| Uncultured Acidobacteriaceae | 1% | |||
| PSH – CP | ML602J-37 | 19.6% | Halovibrio | 31.7% |
| Uncultured Chitinophagaceae | 15% | Rubricoccus | 17.3% | |
| Uncultured Ectothiorhodospiraceae | 14.3% | Uncultured Chitinophagaceae | 5.5% | |
| Marinobacter | 10.5% | Uncultured Caldilineaceae | 3.1% | |
| Ruegeria | 4.8% | Uncultured Acidobacteriaceae | 2% | |
| Albidovulum | 1.8% | Uncultured Anaerolineaceae | 1.8% | |
| Marinicella | 1.4% | ML602J-37 | 1.7% | |
| Halomonas | 1.3% | |||
| Rathybacter | 1.1% | |||
16S rRNA gene sequences greater than 1% relative abundance alone were considered.
Bacterial sequences obtained from brine samples were taxonomically affiliated to 17 phyla (Figure 5B). The major sequences at phylum level (>1% of all obtained sequences) across all salinities and during both phases of salt production in the brine were identified as Bacteroidetes and Proteobacteria. Sequences belonging to Firmicutes (9% in RP to 1% in CP and 7% in RP to 1% in CP during ISH and PSH respectively) and Proteobacteria (52.5% in RP to 7.9% in CP and 62.5% in RP to 49.9% in CP during ISH and PSH respectively) were found to decrease while Bacteroidetes (20% in RP to 85% in CP and 15% in RP to 25% in CP during ISH and PSH respectively) was found to increase along a salinity gradient during both phases of salt production. Sequences affiliated to Planctomycetes (4% in RP to 1% in CP) and Acidobacteria (3% in RP to 2% in CP) were found to decrease along the salinity gradient during the PSH phase. Actinobacteria (2% in RP-ISH), Cyanobacteria (12% in EP-ISH) and Deferribacteres (4% in EP-ISH) were found to be abundant at intermittent sampling sites. Within the phylum Proteobacteria, Gammaproteobacteria (41% in RP to 4% in CP and 39% in RP to 44% in CP during ISH and PSH respectively) were found to decrease along the salinity gradient in the ISH phase and vice versa during PSH phase. Alphaproteobacteria (7% in RP to 2% in CP and 17% in RP to 2% in CP during ISH and PSH respectively) was found to decrease along the salinity gradient during both phases of salt production while the same was observed for Deltaproteobacteria (5% in RP to 2% in CP), however only during PSH (Supplementary Figure S4). Like sediment samples, there were no common phylotypes (>1% of all obtained sequences) observed across all the sampling sites. Sequences belonging to Halomonas (25.8% in RP to 2.2% in CP) decreased along the salinity gradient in the ISH phase while Rubricoccus was particularly dominant in the EP (94.3%) and CP (79% in ISH and 17.3% in PSH) during both phases of salt production (Table 4). Sequences affiliated to Bacillus and Halovibrio were observed majorly at defined salinities and during a particular phase of salt production. For example, Bacillus was dominant (21.6%) in EP PSH brine while Halovibrio was dominant (31.7%) in CP PSH brine. However, sequences belonging to Idiomarina, Planctomyces, and Fulvivirga were confined exclusively to RP brine during both phases of salt production. Unlike sediment samples, sequences of ML602J-37 (3% in RP and 2% in CP) in brine were found to decrease along the salinity gradient in the ISH phase (Table 4). Similarly, sequences belonging to uncultured Chitinophagaceae was observed in CP (3% and 5.5% in ISH and PSH) samples while Uncultured Clostridiaceae was observed in RP (4.3% and 6.3% in ISH and PSH) samples.
Based on the taxonomic classification of reads obtained, Bacteroidetes and Proteobacteria were the dominant bacterial phyla distributed across the salinity gradient and omnipresent during both phases of salt production. Overall, sediments exhibited a higher diversity compared to brine indicated through the presence of sequences belonging to higher number of phyla and a correspondingly greater number of bacterial genera. Though Halomonas, Rubricoccus, and Bacillus were the dominant bacterial phylotypes in the solar saltern under study, they could not be observed at all sampling sites. Similarly, phylotypes belonging to Marinobacter and Marinicella were exclusive to sediment samples while Halovibrio was exclusive to brine.
Discussion
Scant information is available regarding the microbial diversity of highly transient hypersaline environments like the salterns of Siridao, Goa. In this study, the prokaryotic community composition of a solar saltern has been profiled over a salinity gradient using a metabarcoding approach employing high-throughput sequencing technology.
Coastal Salterns of Siridao Are One of the Most Diverse Solar Salterns Around the World
In this study, we obtained about 10-fold higher reads in brine samples compared to sediments. Much lower read numbers from sediment samples could be attributed to the difficulty in the extraction of nucleic acids, despite trying several commercial kits and in-house DNA extraction protocols, the quality of DNA obtained was relatively inferior compared to brine samples. These sediments were predominantly composed of clay and therefore were recalcitrant to DNA extraction procedures. This difficulty was further compounded by the high salinity of the sediment samples (Cai et al., 2006). Despite the low read numbers, the number of OTUs obtained in the sediments were comparable to similar studies carried out at salt marshes in Odiel estuary, Spain (Vera-Gargallo et al., 2019) and a solar saltern located at West Bengal, India (Sen and Mukhopadhyay, 2019) focusing on the microbial diversity of hypersaline soils.
The diversity indices obtained from the salterns of Goa were comparable against the diversity of similar thalassohaline ecosystems like solar salterns located in USA (Youssef et al., 2012), India (Sen and Mukhopadhyay, 2019), Turkey (Çinar and Mutlu, 2016), and Spain (Gómez-Villegas et al., 2018), soda ash concentration ponds in Ethiopia (Simachew et al., 2016) and graduation towers from Poland (Kalwasińska et al., 2018). When the diversity between the two salt production phases was compared, samples from the ISH phase indicated an overall higher diversity owing to the moderately halophilic and halotolerant regime prevailing before the start of the salt production process. However, a sustained high salinity during the PSH restricted the growth of non-halophilic microorganisms, otherwise prevalent in ISH, resulting in the lower diversity values. In general, sediments exhibited a greater diversity in terms of OTU to sequence ratio compared to brine. This is in support of the view that sediments support a broad range of microorganisms because of the availability of a wide range of organic nutrients and hypersaline soils are no exception to this pattern (Sander and Kalff, 1993; Kimbrel et al., 2018). Though the statistical analysis indicated that the taxonomic composition between the brine and sediments during different salt production phases are similar, we acknowledge the low statistical power involved in the analysis due to the composite sampling approach resulting in single sample per sampling location.
High diversity prevailing in these ecosystems could be owed to the stable tropical climate and high annual precipitation resulting in the introduction of diverse microbes (Peter et al., 2014; Couto-Rodríguez and Montalvo-Rodríguez, 2019). Salterns of Siridao receive annual rainfall in the range of 280–480 cm primarily from the beginning of June till November, flooding the salterns and this event provides an opportunity for the introduction of non-halophilic microbes through runoffs from adjacent environments and urban dwellings. Further, this is in accordance with a study carried out by Youssef et al. (2012), indicating that hypersaline environments that are in constant salinity fluctuations tend to possess a diverse non-halophilic, halotolerant and extremely halophilic members.
Archaea Dominates the Low and Moderate Salinity Sediments
It is commonly observed that there is an increase in archaeal phylotypes in brine along the increasing salinity gradient with a simultaneous decrease in Bacteria (Benlloch et al., 2002; Casamayor et al., 2002; Baati et al., 2008; Ghai et al., 2011). Though our results corroborated with the established trend in brine, we could observe Archaea being dominant over Bacteria in sediments too, during both phases of salt production. However, microbial diversity studies in hypersaline soils and sediments suggest that there is a dominance of Bacteria, especially among the samples obtained from low and moderate salinities (Sørensen et al., 2005; Pandit et al., 2015). Further, the studies carried out by Robertson et al. (2009) on the hypersaline microbial mats at Guerrero Negro solar salterns, Wang et al. (2007) on the saline sediments obtained from the solar salterns of Taiwan, and Vera-Gargallo et al. (2019) on the hypersaline sediments at the Odiel salt marshes of Spain have estimated that Bacteria constituted about 90, 53, and 78% among the total prokaryotes respectively. Contrary to the existing literature, bacterial phylotypes were outnumbered by archaea in sediments of solar salterns of Siridao. With the onset of salinity, Archaea could possibly compete with Bacteria and colonize the saltern compartments at a faster rate. Once the prokaryotic community structure is established with Archaea being the dominant prokaryote, the structure remains less perturbed during the continued operation of the saltern (Barin et al., 2015).
One of the most remarkable findings of our study is the prevalence of archaeal sequences among the RP sediment samples. Though the occurrence of halophilic archaea in the low salinity environments has been documented before, the large number of sequences observed in the salterns of Siridao is unprecedented. Recently, Kimbrel et al. (2018) have shown that at 7.5% salinity, there were about 20% and 18% of archaeal sequences in brine and sediments obtained from a solar saltern respectively. Similarly, Elshahed et al. (2004) have reported the isolation of halophilic archaeal clones belonging to Halogeometricum, Natronomonas, Halococcus, and Haloferax from low salinity (0.7 – 1%) brine and sediment samples. Walsh et al. (2005) have also reported the isolation of archaeal clones belonging to Euryarchaeota from low salinity sediments. Purdy et al., 2004 have reported the isolation of Haloferax and Halogeometricum from low salinity estuary sediments while Braganca and Furtado (2009) have reported the isolation of Haloarcula and Halobacterium from various low salinity eco-niches in close vicinity to the study site. One possible reason for the survival of halophilic archaea in the low salinity samples of Siridao salterns could be attributed to the predominant red clay in saltern sediments (Fukushima et al., 2007). These clay particles could act as micro-niches for halophilic archaea because they contain micropores which eventually are filled up by brine along with the microbes. Further, provided that the clay particles are negatively charged and could hold up cations like Na+ and K+, establishing a conducive environment for the survival of halophilic archaea even during unfavorable conditions (Rožić et al., 2000). Despite their gene abundance at low salinities, the viability of halophilic archaea remains questionable and should be assessed by other molecular techniques.
Another interesting finding of the study is the high sequence percentage of Archaea prevalent at the intermediate salinities. Studies carried out by Ghai et al. (2011) on a 19% salinity brine sample and Fernandez et al. (2014a) on a 13% salinity brine sample have shown that about 46% and 27% of the total 16S rRNA gene sequences could be attributed to Archaea respectively. This indicates that the Archaea plays a significant part in the community composition of prokaryotes at intermediate salinities. In our study, we could also detect a significant proportion of Archaea at intermediate salinities albeit at a higher rate than previously reported ranging between 55% and 85% in brine and sediments, respectively. As the saltern compartments are interconnected and the prevalent Archaea in RP could provide a microbial seed bank, microorganisms could be transported from RP to CP via EP through a constant influx of brine (Lennon and Jones, 2011; Kimbrel et al., 2018). This could provide Archaea a head start over Bacteria to rapidly colonize and dominate in EP and CP.
Halophilic Archaea Tolerate Wide Salinity Fluctuations and Show Minimal Dependency on Magnesium
Halorubrum, the most diverse cultivable halophilic archaeal genus containing about 39 culturable representatives (Parte, 2018) and reported to be a constituent member of solar salterns and hypersaline environments around the world (Ochsenreiter et al., 2002; Burns et al., 2004; Baati et al., 2008; Çinar and Mutlu, 2016; Kambourova et al., 2016) was very abundant genus in the salterns of Siridao too. Given its low dependency on magnesium (0.005–0.6 M) and a wide range of NaCl tolerance (1.0–5.2 M) (Oren, 2018), Halorubrum appears to be the most suitable to colonize the highly dynamic Siridao salterns. Haloarcula, though not frequently reported to be encountered in metagenomic studies (Ochsenreiter et al., 2002; Dillon et al., 2013; Plominsky et al., 2014; Çinar and Mutlu, 2016) was seen in the salterns of Siridao. Their incidence could be attributed to the ability to metabolize a wide range of substrates and possessing very similar characteristics to Halorubrum in terms of magnesium (0.005–0.6 M) and NaCl (1.7–5.2 M) dependency (Ventosa et al., 2019). One of the interesting findings of the study is the occurence of Halobacterium in Siridao salterns, especially in sediments. Despite, Halobacterium being one of the earliest described halophilic archaeal genus and isolated from various hypersaline environments like salterns, salt lakes, salted fish and salted hides, it has been scarcely found in metagenomic studies (Maturrano et al., 2006; Zafrilla et al., 2010; Baati et al., 2011; Plominsky et al., 2014). Since Halobacterium possess higher NaCl (2.5–5.2 M) and magnesium (0.05–0.6 M) requirements coupled with versatile anaerobic respiration, CP sediments appear to be a suitable habitat for their prevalence (Oren and Ventosa, 2017). Consistent with previous studies, Natronomonas, an extremely halophilic archaeon (requiring at least 2 M NaCl) was found in CP sediment. Though Natronomonas has been frequently correlated with halophilic alkaine environments, a few species have been recovered from neutrophilic environments as well. Therefore, sequences affiliated to Natronomonas recovered from Siridao saltern could possibly be neutrophilic, low-magnesium dependent phylotypes (Minegishi and Kamekura, 2015). Other genera found in this study, Halorhabdus (Mouné et al., 2003; Baati et al., 2011; Makhdoumi-Kakhki et al., 2012; Baricz et al., 2014) and Haloplanus (Baricz et al., 2014; Vera-Gargallo and Ventosa, 2018; Couto-Rodríguez and Montalvo-Rodríguez, 2019) are the common dwellers of solar salterns around the world while Halogranum (Youssef et al., 2012) and Halomicrobium (Youssef et al., 2012; Castelán-Sánchez et al., 2019) have been rarely documented. Interestingly, almost all of the halophilic archaea reported in Siridao salterns have been recorded to possess wide NaCl tolerance (0–5.2 M) with minimal or no dependency on magnesium.
The prevalence of Haloquadratum, a cosmopolitan halophilic archaeal genus in the crystallizer pans around the world (Maturrano et al., 2006; Baati et al., 2011; Ghai et al., 2011; Dillon et al., 2013) was found to be negligible (<0.3% of available reads) in Siridao salterns. Few studies have also reported the low or negligible occurrence of Haloquadratum in salterns that are very similar in operation with Siridao salterns and other hypersaline environments (Pašić et al., 2005; Wang et al., 2007; Manikandan et al., 2009; Kalwasińska et al., 2018). This may be explained by the lower level of magnesium prevalent in the saltern samples. Solar salterns of Goa are characterized by low retention time resulting in the brine replenishment daily and therefore the concentration of magnesium remains tightly regulated. This is in contrast to the well-studied Bras del Port saltern in Spain and Cabo Rojo salterns in Puerto Rico, where the brine is replenished every year and once in two months respectively (Pašić et al., 2014; Couto-Rodríguez and Montalvo-Rodríguez, 2019). Further, a study carried out by Podell et al. (2014) has shown that Haloquadratum displayed a positive correlation with elevated magnesium concentration and negative correlation with Halorubrum and Haloarcula. Therefore, our study indicates that low retention time, low magnesium concentration, and competition from other fast-growing halophilic archaea for space prevent Haloquadratum from becoming a dominant halophilic archaeal genus. The low retention time of brine could also be attributed to the relatively low incidence of Halonotius, which requires at least 2.7 M NaCl for growth (Oren, 2016) and uncultured Nanohaloarchaeota, frequently encountered in hypersaline environments (Narasingarao et al., 2012; Finstad et al., 2017; Mora-Ruiz et al., 2018). Another possibility of the absence of Nanohaloarchaeota could be owed upon the absence of a specific host because these organisms have been found to dwell as symbionts with other halophilic archaea (Hamm et al., 2019). A previous study carried out in Ribandar salterns employing culture-dependent techniques, located at a distance of 10 km from the site under study, reported the isolation of members belonging to Halococcus during ISH phase and Halorubrum, Halococcus, Haloferax, and Haloarcula during PSH phase (Mani et al., 2012a). However, in Siridao salterns, Halococcus constituted a minor fraction (<1%) and Haloferax was not encountered in any of the samples studied. This shows the variation in the halophilic archaeal community structure between the closely located solar salterns despite similar operational procedures and geophysical conditions.
Major Shifts in Bacterial Diversity Between Salt Production Phases and Compartments
Like several other salterns and hypersaline environments, Siridao salterns support diverse communities of phylotypes belonging to Bacteroidetes and Gammaproteobacteria (Sørensen et al., 2005; Baati et al., 2011; Zhang et al., 2016). The occurence of Alphaproteobacteria, Betaproteobacteria and Firmicutes decreased with increasing salinity which was consistent with the results obtained by Oueriaghli et al. (2018). A study carried out in the same salterns under investigation, employing DGGE had recovered sequences affiliated with Gammaproteobacteria, supporting their importance at Siridao salterns (Mani et al., 2015).
Interestingly, Rubricoccus, one of the dominant bacterial genera in the Siridao salterns, has a growth range between 1–5% NaCl (w/v) and was originally isolated from seawater (Park et al., 2011). However, this particular bacterium has seldom been found in hypersaline environments and not been reported in any metagenomic studies to the best of our knowledge. The prevalence of this genus in EP and CP could indicate novel phylotypes related to Rubricoccus dwelling in salterns of Siridao that are yet to be explored. Another abundant genus like Halomonas is a versatile moderately halophilic bacterium with a very wide salinity range [0–5.2 M NaCl (w/v)] (Vreeland, 2015) and are commonly encountered in salterns (Tsiamis et al., 2008; Boujelben et al., 2014; Oueriaghli et al., 2014). Sediments showed the occurrence of Marinobacter, the denitrifying bacterium Halovibrio, sulfate-reducing anaerobic bacterium Desulfovermiculus and sulfur-oxidizing microaerophilic bacterium Thioalkalispira. As Siridao salterns are in the vicinity of urban dwellings and industries, hydrocarbons and organic acids may eventually get concentrated in the salterns. This would provide an opportunity for the moderately halophilic and facultative anaerobic bacteria capable of degrading hydrocarbon like Marinobacter (Handley and Lloyd, 2013; Bowman and McMeekin, 2015) and Halovibrio (Sorokin et al., 2006) to flourish in EP and CP sediments. Though the presence of halophilic Desulfovermiculus in hypersaline environments has been recorded before (Roychoudhury et al., 2013; McGonigle et al., 2019), the incidence of Thioalkalispira, originally isolated from alkaline soda lake sediments and with a pH range of 8.4 -10 is truly surprising (Sorokin, 2008; Sorokin et al., 2013). Together with Desulfovermiculus and Thioalkalispira, the presence of sulfur-oxidizing Acidoferrobacter in RP indicates the active sulfur metabolism occurring in the RP and EP sediments. Bacillus has been frequently reported from the solar salterns of India (Kumar et al., 2015; Suganthi et al., 2018; Sen and Mukhopadhyay, 2019). Given the dynamic nature of the solar salterns of India being operated and halotolerance of Bacillus in surviving salinities up to 25% NaCl (w/v) (Garabito et al., 1998), this genus could be dominating the salterns during the non-operational phases. In the case of Siridao salterns, their diminishment could be due to the sustained salinity during PSH, paving the way for moderately and extremely halophilic bacteria to colonize the salterns.
At majority of the sampling sites, uncultured phylotypes belonging to phylum Bacteroidetes were found to be abundant, sometimes outnumbering their culturable counterparts. Interestingly, uncultured Bacteroidetes clone ML602J-37, initially recovered from alkaline Mono lake in USA (Humayoun et al., 2003), was abundant in EP and CP sediments. Though it has been recovered from other neutral environments like soil samples from Antarctica (Shravage et al., 2007), stromatolites from Argentina (Toneatti et al., 2017), and Keke Salt Lake from China (Han et al., 2017), the presence of ML602J-37 in solar saltern samples requires a detailed investigation of their role in the metabolic activity of sediments. Similarly, abundance of uncultured Chitinophagaceae in sediments indicates novel phylotypes thriving in salterns of Siridao that are yet to be explored.
Salinibacter (Antón et al., 2008; Baati et al., 2008; Di Meglio et al., 2016) and Spiribacter (Fernandez et al., 2014a, b; León et al., 2014, 2017), most widely represented bacterial genera at moderate and high salinities across global hypersaline environments failed to be identified in the Siridao salterns. This could be attributed to the relatively low salinities and transient nature of the Siridao salterns since the occurence of Salinibacter has been shown to strongly correlate with increasing and sustained salinity (Pandit et al., 2015). On the other hand, though Spiribacter could tolerate wide salinity fluctuations (0-2 M NaCl), its growth peaks at very specific salinity (0.8 M NaCl) making it difficult to flourish in highly fluctuating salinities (León et al., 2018). Few studies carried out in the salterns of Chile, Argentina, Poland, and Spain have also reported the absence or low incidence of Salinibacter (Kalwasińska et al., 2018; Mora-Ruiz et al., 2018; Oueriaghli et al., 2018). Further, considering the absence of dominant archaeal phylotype of Haloquadratum and bacterial phylotypes such as Salinibacter and Spiribacter, we are in complete agreement with the views drawn by Gomariz et al. (2015) that solar salterns, despite being called ‘steady-state’ systems, undergo various microbial fluctuations at similar salinities and therefore general conclusions should be drawn with caution.
Conclusion
Artisanal solar salterns of Goa are of rich historical and cultural importance. Their transient nature makes these salterns unique ecosystems for studying the effect of salinity fluctuations on the prokaryotic community structure. On analyzing the microbial community structure between two salt production seasons, indicated the prevalence of a similar prokaryotic composition. Further, this study has clearly shown that Archaea was stable and dominant despite encountering wide changes in salinity between two different salt production phases, while significant fluctuations were observed in bacterial community structure. Among the archaeal phylotypes, Halorubrum and Haloarcula were dominant while Rubricoccus and Halomonas were the dominant bacterial phylotypes. Thus, the prokaryotic community structure of salterns of Siridao is mainly driven by halophilic archaea and bacteria that can withstand wide fluctuations in salinity and have a low dependency on ionic constituents. A thorough assessment of these ecosystems through metagenomic approaches can help us in understanding the adaptations and functioning of the microbiome in transient conditions.
Data Availability Statement
The raw sequencing data generated in this study have been deposited in the SRA under BioProject PRJNA614987.
Author Contributions
JB, GB, and DD conceived the study. KM and JB conducted sampling. KM and MH performed DNA extraction, PCR and library preparation for Illumina sequencing. KM, NT, and GB carried out the data analysis and statistical calculations. KM, MH, and NT drafted the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. KM thanks Centre Franco-Indien pour la Promotion de la Recherche Avancée (CEFIPRA) for providing financial assistance through the Raman-Charpak Fellowship (2013) and Council of Scientific and Industrial Research India for awarding the Senior Research Fellowship (SRF) (09/919 (0017)/2012-EMR-I.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01891/full#supplementary-material
References
- Antón J., Peña A., Santos F., Martínez-García M., Schmitt-Kopplin P., Rosselló-Mora R. (2008). Distribution, abundance and diversity of the extremely halophilic bacterium Salinibacter ruber. Saline Syst. 4:15. 10.1186/1746-1448-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baati H., Guermazi S., Amdouni R., Gharsallah N., Sghir A., Ammar E. (2008). Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles 12 505–518. 10.1007/s00792-008-0154-x [DOI] [PubMed] [Google Scholar]
- Baati H., Guermazi S., Gharsallah N., Sghir A., Ammar E. (2010). Novel prokaryotic diversity in sediments of Tunisian multipond solar saltern. Res. Microbiol. 161 573–582. 10.1016/j.resmic.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Baati H., Jarboui R., Gharsallah N., Sghir A., Ammar E. (2011). Molecular community analysis of magnesium-rich bittern brine recovered from a Tunisian solar saltern. Can. J. Microbiol. 57 975–981. 10.1139/w11-088 [DOI] [PubMed] [Google Scholar]
- Bardou P., Mariette J., Escudié F., Djemiel C., Klopp C. (2014). jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15:293. 10.1186/1471-2105-15-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baricz A., Coman C., Andrei A. Ş, Muntean V., Keresztes Z. G., Pãuşan M., et al. (2014). Spatial and temporal distribution of archaeal diversity in meromictic, hypersaline Ocnei Lake (Transylvanian Basin, Romania). Extremophiles 18 399–413. 10.1007/s00792-013-0625-6 [DOI] [PubMed] [Google Scholar]
- Barin M., Aliasgharzad N., Olsson P. A., Rasouli-Sadaghiani M. (2015). Salinity-induced differences in soil microbial communities around the hypersaline Lake Urmia. Soil Res. 53 494–504. 10.1071/SR14090 [DOI] [Google Scholar]
- Benlloch S., López-López A., Casamayor E. O., Øvreås L., Goddard V., Daae F. L., et al. (2002). Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4 349–360. 10.1046/j.1462-2920.2002.00306.x [DOI] [PubMed] [Google Scholar]
- Boujelben I., Gomariz M., Martínez-García M., Santos F., Peña A., López C., et al. (2012). Spatial and seasonal prokaryotic community dynamics in ponds of increasing salinity of Sfax solar saltern in Tunisia. Antonie Van Leeuwenhoek 101 845–857. 10.1007/s10482-012-9701-7 [DOI] [PubMed] [Google Scholar]
- Boujelben I., Martínez-García M., van Pelt J., Maalej S. (2014). Diversity of cultivable halophilic archaea and bacteria from superficial hypersaline sediments of Tunisian solar salterns. Antonie Van Leeuwenhoek 106 675–692. 10.1007/s10482-014-0238-9 [DOI] [PubMed] [Google Scholar]
- Bowers K. J., Wiegel J. (2011). Temperature and pH optima of extremely halophilic archaea: a mini-review. Extremophiles 15 119–128. 10.1007/s00792-010-0347-y [DOI] [PubMed] [Google Scholar]
- Bowman J. P., McMeekin T. A. (2015). “Marinobacter,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. Timmis K. N. (Hoboken, NJ: John Wiley & Sons, Inc; ), 1–6. [Google Scholar]
- Braganca J. M., Furtado I. (2009). Isolation and characterization of haloarchaea from low-salinity coastal sediments and brines of Goa. Curr. Sci. 96 1182–1184. [Google Scholar]
- Burns D. G., Camakaris H. M., Janssen P. H., Dyall-Smith M. L. (2004). Combined use of cultivation-dependent and cultivation-independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl. Environ. Microbiol. 70 5258–5265. 10.1128/AEM.70.9.5258-5265.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai P., Huang Q., Zhang X., Chen H. (2006). Adsorption of DNA on clay minerals and various colloidal particles from an Alfisol. Soil Biol. Biochem. 38 471–476. 10.1016/j.soilbio.2005.05.019 [DOI] [Google Scholar]
- Casamayor E. O., Massana R., Benlloch S., Øvreås L., Díez B., Goddard V. J., et al. (2002). Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4 338–348. 10.1046/j.1462-2920.2002.00297.x [DOI] [PubMed] [Google Scholar]
- Castelán-Sánchez H. G., Elorrieta P., Romoacca P., Liñan-Torres A., Sierra J. L., Vera I., et al. (2019). Intermediate-salinity systems at high altitudes in the peruvian andes unveil a high diversity and abundance of bacteria and viruses. Genes 10:891. 10.3390/genes10110891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çinar S., Mutlu M. B. (2016). Comparative analysis of prokaryotic diversity in solar salterns in eastern Anatolia (Turkey). Extremophiles 20 589–601. 10.1007/s00792-016-0845-7 [DOI] [PubMed] [Google Scholar]
- Couto-Rodríguez R. L., Montalvo-Rodríguez R. (2019). Temporal analysis of the microbial community from the crystallizer ponds in cabo rojo, puerto rico, using metagenomics. Genes 10:422. 10.3390/genes10060422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa A. (1954). La détermination rapide du calcium et du magnésium dans l’eau de mer. Anal. Chim. Acta 11 221–224. 10.1016/S0003-2670(00)87721-1 [DOI] [Google Scholar]
- Di Meglio L., Santos F., Gomariz M., Almansa C., López C., Antón J., et al. (2016). Seasonal dynamics of extremely halophilic microbial communities in three Argentinian salterns. FEMS Microbiol. Ecol. 92:fiw184. 10.1093/femsec/fiw184 [DOI] [PubMed] [Google Scholar]
- Dillon J. G., Carlin M., Gutierrez A., Nguyen V., McLain N. (2013). Patterns of microbial diversity along a salinity gradient in the Guerrero Negro solar saltern, Baja CA Sur, Mexico. Front. Microbio. 4:399. 10.3389/fmicb.2013.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Elshahed M. S., Najar F. Z., Roe B. A., Oren A., Dewers T. A., Krumholz L. R. (2004). Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide-and sulfur-rich spring. Appl. Environ. Microbiol. 70 2230–2239. 10.1128/AEM.70.4.2230-2239.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. W., Li X. R., Wang J. H., Hu Z. Y., Meng H., Xiang L. Y., et al. (2009). Bacterial diversity of water and sediment in the Changjiang estuary and coastal area of the East China Sea. FEMS Microbiol. Ecol. 70 236–248. 10.1111/j.1574-6941.2009.00772.x [DOI] [PubMed] [Google Scholar]
- Fernandez A. B., Ghai R., Martin-Cuadrado A. B., Sanchez-Porro C., Rodriguez-Valera F., Ventosa A. (2014a). Prokaryotic taxonomic and metabolic diversity of an intermediate salinity hypersaline habitat assessed by metagenomics. FEMS Microbiol. Ecol. 88 623–635. 10.1111/1574-6941.12329 [DOI] [PubMed] [Google Scholar]
- Fernandez A. B., Vera-Gargallo B., Sánchez-Porro C., Ghai R., Papke R. T., Rodriguez-Valera F., et al. (2014b). Comparison of prokaryotic community structure from Mediterranean and Atlantic saltern concentrator ponds by a metagenomic approach. Front. Microbio. 5:196. 10.3389/fmicb.2014.00196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finstad K. M., Probst A. J., Thomas B. C., Andersen G. L., Demergasso C., Echeverría A., et al. (2017). Microbial community structure and the persistence of cyanobacterial populations in salt crusts of the hyperarid Atacama desert from genome-resolved Metagenomics. Front. Microbio. 8:1435. 10.3389/fmicb.2017.01435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T. T., Usami R., Kamekura M. (2007). A traditional Japanese-style salt field is a niche for haloarchaeal strains that can survive in 0.5% salt solution. Aquat. Biosyst. 3:2. 10.1186/1746-1448-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garabito M. J., Márquez M. C., Ventosa A. (1998). Halotolerant Bacillus diversity in hypersaline environments. Can. J. Microbiol. 44 95–102. 10.1139/w97-125 [DOI] [Google Scholar]
- Ghai R., Pašić L., Fernández A. B., Martin-Cuadrado A. B., Mizuno C. M., McMahon K. D. (2011). New abundant microbial groups in aquatic hypersaline environments. Sci. Rep. 1:135. 10.1038/srep00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomariz M., Martínez-García M., Santos F., Rodriguez F., Capella-Gutiérrez S., Gabaldon T., et al. (2015). From community approaches to single-cell genomics: the discovery of ubiquitous hyperhalophilic Bacteroidetes generalists. ISME J. 9 16–31. 10.1038/ismej.2014.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Villegas P., Vigara J., León R. (2018). Characterization of the microbial population inhabiting a solar saltern pond of the odiel marshlands (SW Spain). Mar. Drugs 16:332. 10.3390/md16090332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S., Grant W. D., Jones B. E., Kato C., Li L. (1999). Novel archaeal phylotypes from an East African alkaline saltern. Extremophiles 3 139–145. 10.1007/s007920050109 [DOI] [PubMed] [Google Scholar]
- Gunde-Cimerman N., Plemenitaš A., Oren A. (2018). Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 42 353–375. 10.1093/femsre/fuy009 [DOI] [PubMed] [Google Scholar]
- Hamdan L. J., Coffin R. B., Sikaroodi M., Greinert J., Treude T., Gillevet P. M. (2013). Ocean currents shape the microbiome of Arctic marine sediments. ISME J. 7 685–696. 10.1038/ismej.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm J. N., Erdmann S., Eloe-Fadrosh E. A., Angeloni A., Zhong L., Brownlee C., et al. (2019). Unexpected host dependency of Antarctic Nanohaloarchaeota. Proc. Natl. Acad. Sci. U.S.A. 116 14661–14670. 10.1073/pnas.1905179116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø, Harper D. A. T., Ryan P. D. (2005). PAST-PAlaeontological Statistics, ver. 1.35. Palaeontol. Electrónica 4 1–9. [Google Scholar]
- Han R., Zhang X., Liu J., Long Q., Chen L., Liu D., et al. (2017). Microbial community structure and diversity within hypersaline Keke Salt Lake environments. Can. J. Microbiol. 63 895–908. 10.1139/cjm-2016-0773 [DOI] [PubMed] [Google Scholar]
- Handley K. M., Lloyd J. R. (2013). Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by Marinobacter species. Front. Microbiol. 4:136. 10.3389/fmicb.2013.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugoni M., Taib N., Debroas D., Domaizon I., Dufournel I. J., Bronner G., et al. (2013). Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc. Natl. Acad. Sci. U.S.A. 110 6004–6009. 10.1073/pnas.1216863110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humayoun S. B., Bano N., Hollibaugh J. T. (2003). Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69 1030–1042. 10.1128/AEM.69.2.1030-1042.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javor B. J. (2002). Industrial microbiology of solar salt production. J. Ind. Microbiol. Biotechnol. 28 42–47. 10.1038/sj/jim/7000173 [DOI] [PubMed] [Google Scholar]
- Kalwasińska A., Deja-Sikora E., Burkowska-But A., Szabó A., Felföldi T., Kosobucki P., et al. (2018). Changes in bacterial and archaeal communities during the concentration of brine at the graduation towers in Ciechocinek spa (Poland). Extremophiles 22 233–246. 10.1007/s00792-017-0992-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambourova M., Tomova I., Boyadzhieva I., Radchenkova N., Vasileva-Tonkova E. (2016). Unusually high archaeal diversity in a crystallizer pond, Pomorie salterns, Bulgaria, revealed by phylogenetic analysis. Archaea 2016:7459679. 10.1155/2016/7459679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbrel J. A., Ballor N., Wu Y. W., David M. M., Hazen T. C., Simmons B. A., et al. (2018). Microbial community structure and functional potential along a hypersaline gradient. Front. Microbiol. 9:1492. 10.3389/fmicb.2018.01492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R. M., Kaur G., Kumar A., Bala M., Singh N. K., Kaur N., et al. (2015). Taxonomic description and genome sequence of Bacillus campisalis sp. nov., a member of the genus Bacillus isolated from a solar saltern. Int. J. Syst. Evol. Microbiol. 65 3235–3240. 10.1099/ijsem.0.000400 [DOI] [PubMed] [Google Scholar]
- Lennon J. T., Jones S. E. (2011). Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9 119–130. 10.1038/nrmicro2504 [DOI] [PubMed] [Google Scholar]
- León M. J., Aldeguer-Riquelme B., Antón J., Sánchez-Porro C., Ventosa A. (2017). Spiribacter aquaticus sp. nov., a novel member of the genus Spiribacter isolated from a saltern. Int. J. Syst. Evol. Microbiol. 67 2947–2952. 10.1099/ijsem.0.002053 [DOI] [PubMed] [Google Scholar]
- León M. J., Fernández A. B., Ghai R., Sánchez-Porro C., Rodriguez-Valera F., Ventosa A. (2014). From metagenomics to pure culture: isolation and characterization of the moderately halophilic bacterium Spiribacter salinus gen. nov., sp. nov. Appl. Environ. Microbiol. 80 3850–3857. 10.1128/aem.00430-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León M. J., Hoffmann T., Sánchez-Porro C., Heider J., Ventosa A., Bremer E. (2018). Compatible solute synthesis and import by the moderate halophile Spiribacter salinus: physiology and genomics. Front. Microbio. 9:108. 10.3389/fmicb.2018.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoni C., Volpicella M., Fosso B., Manzari C., Piancone E., Dileo M. C., et al. (2020). A differential metabarcoding approach to describe taxonomy profiles of bacteria and archaea in the saltern of margherita di savoia (Italy). Microorganisms 8:936. 10.3390/microorganisms8060936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-López A., Yarza P., Richter M., Suárez-Suárez A., Antón J., Niemann H., et al. (2010). Extremely halophilic microbial communities in anaerobic sediments from a solar saltern. Environ. Microbiol. Rep. 2 258–271. 10.1111/j.1758-2229.2009.00108.x [DOI] [PubMed] [Google Scholar]
- Makhdoumi-Kakhki A., Amoozegar M. A., Kazemi B., Pašić L., Ventosa A. (2012). Prokaryotic diversity in Aran-Bidgol salt lake, the largest hypersaline playa in Iran. Microbes Environ. 27 87–93. 10.1264/jsme2.me11267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani K., Chandrasekaran S., Salgaonkar B. B., Mutnuri S., Bragança J. M. (2015). Comparison of bacterial diversity from solar salterns and a simulated laboratory study. Ann. Microbio. 65 995–1005. 10.1007/s13213-014-0944-6 [DOI] [Google Scholar]
- Mani K., Salgaonkar B. B., Braganca J. M. (2012a). Culturable halophilic archaea at the initial and crystallization stages of salt production in a natural solar saltern of Goa, India. Aquat. Biosyst. 8:15. 10.1186/2046-9063-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani K., Salgaonkar B. B., Das D., Bragança J. M. (2012b). Community solar salt production in Goa, India. Aquat.Biosyst. 8:30. 10.1186/2046-9063-8-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikandan M., Kannan V., Pašić L. (2009). Diversity of microorganisms in solar salterns of Tamil Nadu, India. World J. Microbiol. Biotechnol. 25 1007–1017. 10.1007/s11274-009-9980-y [DOI] [Google Scholar]
- Masella A. P., Bartram A. K., Truszkowski J. M., Brown D. G., Neufeld J. D. (2012). PANDAseq: paired-end assembler for Illumina sequences. BMC Bioinformatics 13:31. 10.1186/1471-2105-13-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maturrano L., Santos F., Rosselló-Mora R., Antón J. (2006). Microbial diversity in Maras salterns, a hypersaline environment in the Peruvian Andes. Appl. Environ. Microbiol. 72 3887–3895. 10.1128/AEM.02214-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle J. M., Bernau J. A., Bowen B. B., Brazelton W. J. (2019). Robust archaeal and bacterial communities inhabit shallow subsurface sediments of the Bonneville Salt Flats. mSphere 4:e00378-19. 10.1128/mSphere.00378-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah N. M., Wiegel J. (2012). Life under multiple extreme conditions: diversity and physiology of the halophilic alkalithermophiles. Appl. Environ. Microbiol. 78 4074–4082. 10.1128/AEM.00050-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minchin P. R. (1987). An evaluation of the relative robustness of techniques for ecological ordination. Vegetatio 69 89–107. 10.1007/BF00038690 [DOI] [Google Scholar]
- Minegishi H., Kamekura M. (2015). “Natronomonas,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. Kamekura M. (Hoboken, NJ: John Wiley & Sons, Inc; ), 1–8. 10.1002/9781118960608.gbm00493.pub2 [DOI] [Google Scholar]
- Mora-Ruiz M. D. R., Cifuentes A., Font-Verdera F., Pérez-Fernández C., Farias M. E., González B., et al. (2018). Biogeographical patterns of bacterial and archaeal communities from distant hypersaline environments. Syst. Appl. Microbiol. 41 139–150. 10.1016/j.syapm.2017.10.006 [DOI] [PubMed] [Google Scholar]
- Mouné S., Caumette P., Matheron R., Willison J. C. (2003). Molecular sequence analysis of prokaryotic diversity in the anoxic sediments underlying cyanobacterial mats of two hypersaline ponds in Mediterranean salterns. FEMS Microbiol. Ecol. 44 117–130. 10.1016/S0168-6496(03)00017-5 [DOI] [PubMed] [Google Scholar]
- Narasingarao P., Podell S., Ugalde J. A., Brochier-Armanet C., Emerson J. B., Brocks J. J., et al. (2012). De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 6 81–93. 10.1038/ismej.2011.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenreiter T., Pfeifer F., Schleper C. (2002). Diversity of Archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles 6 267–274. 10.1007/s00792-001-0253-4 [DOI] [PubMed] [Google Scholar]
- Oh D., Porter K., Russ B., Burns D., Dyall-Smith M. (2010). Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds. Extremophiles 14 161–169. 10.1007/s00792-009-0295-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., et al. (2013). Vegan: Community Ecology Package. The Comprehensive R Archive Network (CRAN). Available online at: http://CRAN.R-project.org/package=vegan (accessed June 20, 2020). [Google Scholar]
- Oren A. (1999). Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63 334–348. 10.1128/MMBR.63.2.334-348.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. (2002). Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J. Ind. Microbiol. Biot. 28 56–63. 10.1038/sj/jim/7000176 [DOI] [PubMed] [Google Scholar]
- Oren A. (2009). Saltern evaporation ponds as model systems for the study of primary production processes under hypersaline conditions. Aquat. Microbiol. Ecol. 56 193–204. 10.3354/ame01297 [DOI] [Google Scholar]
- Oren A. (2010). Industrial and environmental applications of halophilic microorganisms. Environ. Technol. 31 825–834. 10.1080/09593330903370026 [DOI] [PubMed] [Google Scholar]
- Oren A. (2013). Salinibacter: an extremely halophilic bacterium with archaeal properties. FEMS Microbiol. Lett. 342 1–9. 10.1111/1574-6968.12094 [DOI] [PubMed] [Google Scholar]
- Oren A. (2016). “Halonotius,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. Whitman W. B. (Hoboken, NJ: John Wiley & Sons, Inc; ), 1–5. 10.1002/9781118960608.gbm01343 [DOI] [Google Scholar]
- Oren A. (2018). “Halorubrum,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds McGenity T. J., Grant W. D. (Hoboken, NJ: John Wiley & Sons, Inc; ), 1–48. [Google Scholar]
- Oren A., Sørensen K. B., Canfield D. E., Teske A. P., Ionescu D., Lipski A., et al. (2009). Microbial communities and processes within a hypersaline gypsum crust in a saltern evaporation pond (Eilat, Israel). Hydrobiologia 626 15–26. 10.1007/s10750-009-9734-8 [DOI] [Google Scholar]
- Oren A., Ventosa A. (2017). “Halobacterium,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. Grant W. D. (Hoboken, NJ: John Wiley & Sons, Inc; ), 1–12. [Google Scholar]
- Oueriaghli N., Castro D. J., Llamas I., Béjar V., Martínez-Checa F. (2018). Study of bacterial community composition and correlation of environmental variables in Rambla Salada, a hypersaline environment in South-Eastern Spain. Front. Microbiol. 9:1377. 10.3389/fmicb.2018.01377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oueriaghli N., González-Domenech C. M., Martínez-Checa F., Muyzer G., Ventosa A., Quesada E., et al. (2014). Diversity and distribution of Halomonas in Rambla Salada, a hypersaline environment in the southeast of Spain. FEMS Microbiol. Ecol. 82 460–474. 10.1111/1574-6941.12237 [DOI] [PubMed] [Google Scholar]
- Pandit A. S., Joshi M. N., Bhargava P., Shaikh I., Ayachit G. N., Raj S. R., et al. (2015). A snapshot of microbial communities from the Kutch: one of the largest salt deserts in the World. Extremophiles 19 973–987. 10.1007/s00792-015-0772-z [DOI] [PubMed] [Google Scholar]
- Park S., Yoshizawa S., Kogure K., Yokota A. (2011). Rubricoccus marinus gen. nov., sp. nov., of the family ‘Rhodothermaceae’, isolated from seawater. Int. J. Syst. Evol. Microbiol. 61 2069–2072. 10.1099/ijs.0.026294-0 [DOI] [PubMed] [Google Scholar]
- Park S. J., Kang C. H., Rhee S. K. (2006). Characterization of the microbial diversity in a Korean solar saltern by 16S rRNA gene analysis. J. Microbiol. Biotechnol. 16 1640–1645. [Google Scholar]
- Parte A. C. (2018). LPSN — list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int. J. Syst. Evol. Microbiol. 68 1825–1829. 10.1099/ijsem.0.002786 [DOI] [PubMed] [Google Scholar]
- Pašić L., Ambrožič Avguštin J., Starčič Erjavec M., Herzog-Velikonja B., Podlesek Z., Žgur-Bertok D. (2014). Two tales of prokaryotic genomic diversity: Escherichia coli and Halophiles. Food Technol. Biotechnol. 52 158–169. [Google Scholar]
- Pašić L., Bartual S. G., Ulrih N. P., Grabnar M., Velikonja B. H. (2005). Diversity of halophilic archaea in the crystallizers of an Adriatic solar saltern. FEMS Microbiol. Ecol. 54 491–498. 10.1016/j.femsec.2005.06.004 [DOI] [PubMed] [Google Scholar]
- Paul V. G., Mormile M. R. (2017). A case for the protection of saline and hypersaline environments: a microbiological perspective. FEMS Microbiol. Ecol. 93:fix091. 10.1093/femsec/fix091 [DOI] [PubMed] [Google Scholar]
- Peter H., Hörtnagl P., Reche I., Sommaruga R. (2014). Bacterial diversity and composition during rain events with and without Saharan dust influence reaching a high mountain lake in the Alps. Environ. Microbiol. Rep. 6 618–624. 10.1111/1758-2229.12175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plominsky A. M., Delherbe N., Ugalde J. A., Allen E. E., Blanchet M., Ikeda P., et al. (2014). Metagenome sequencing of the microbial community of a solar saltern crystallizer pond at Cáhuil Lagoon, Chile. Genome Announc. 2:e01172-14. 10.1128/genomeA.01172-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S., Emerson J. B., Jones C. M., Ugalde J. A., Welch S., Heidelberg K. B., et al. (2014). Seasonal fluctuations in ionic concentrations drive microbial succession in a hypersaline lake community. ISME J. 8 979–990. 10.1038/ismej.2013.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy K. J., Cresswell-Maynard T. D., Nedwell D. B., McGenity T. J., Grant W. D., Timmis K. N., et al. (2004). Isolation of haloarchaea that grow at low salinities. Environ. Microbiol. 6 591–595. 10.1111/j.1462-2920.2004.00592.x [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2014). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Roberts M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1:5. 10.1186/1746-1448-1-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson C. E., Spear J. R., Harris J. K., Pace N. R. (2009). Diversity and stratification of archaea in a hypersaline microbial mat. Appl. Environ. Microbiol. 75 1801–1810. 10.1128/AEM.01811-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Valera F., Ruiz-Berraquero F., Ramos-Cormenzana A. (1981). Characteristics of the heterotrophic bacterial populations in hypersaline environments of different salt concentrations. Microb. Ecol. 7 235–243. 10.1007/BF02010306 [DOI] [PubMed] [Google Scholar]
- Roychoudhury A. N., Cowan D., Porter D., Valverde A. (2013). Dissimilatory sulphate reduction in hypersaline coastal pans: an integrated microbiological and geochemical study. Geobiology 11 224–233. 10.1111/gbi.12027 [DOI] [PubMed] [Google Scholar]
- Rožić M., Cerjan-Stefanović Š., Kurajica S., Vančina V., Hodžić E. (2000). Ammoniacal nitrogen removal from brine by treatment with clays and zeolites. Brine Res. 34 3675–3681. 10.1016/S0043-1354(00)00113-5 [DOI] [Google Scholar]
- Sander B. C., Kalff J. (1993). Factors controlling bacterial production in marine and freshwater sediments. Microb. Ecol. 26 79–99. 10.1007/BF00177045 [DOI] [PubMed] [Google Scholar]
- Sen U., Mukhopadhyay S. K. (2019). Microbial community composition of saltern soils from Ramnagar, West Bengal, India. Ecol. Genet. Genomics 12:100040 10.1016/j.egg.2019.100040 [DOI] [Google Scholar]
- Shravage B. V., Dayananda K. M., Patole M. S., Shouche Y. S. (2007). Molecular microbial diversity of a soil sample and detection of ammonia oxidizers from Cape Evans, Mcmurdo Dry Valley, Antarctica. Microbiol. Res. 162 15–25. 10.1016/j.micres.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Siglioccolo A., Paiardini A., Piscitelli M., Pascarella S. (2011). Structural adaptation of extreme halophilic proteins through decrease of conserved hydrophobic contact surface. BMC Struct. Biol. 11:50. 10.1186/1472-6807-11-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simachew A., Lanzén A., Gessesse A., Øvreås L. (2016). Prokaryotic community diversity along an increasing salt gradient in a soda ash concentration pond. Microb. Ecol. 71 326–338. 10.1007/s00248-015-0675-7 [DOI] [PubMed] [Google Scholar]
- Skoog D. A., West D. M., Holler F. J. (1996). Fundamentals of Analytical Chemistry, 7th Edn Orlando, FL: Saunders College Publishing. [Google Scholar]
- Sørensen K. B., Canfield D. E., Teske A. P., Oren A. (2005). Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 11 7352–7365. 10.1128/AEM.71.11.7352-7365.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y. (2008). “Diversity of halophilic sulfur-oxidizing bacteria in hypersaline habitats,” in Microbial Sulfur Metabolism, eds Dahl C., Friedrich C. G. (Heidelberg: Springer; ), 225–237. 10.1007/978-3-540-72682-1_18 [DOI] [Google Scholar]
- Sorokin D. Y., Banciu H., Robertson L. A., Kuenen J. G., Muntyan M. S., Muyzer G. (2013). “Halophilic and haloalkaliphilic sulfur-oxidizing bacteria,” in The Prokaryotes: Prokaryotic Physiology and Biochemistry, eds Rosenberg E., DeLong F., DeLong E., Lory S., Stackebrandt E., Thompson F. (Heidelberg: Springer; ), 529–554. 10.1007/978-3-642-30141-4_77 [DOI] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Galinski E. A., Belloch C., Tindall B. J. (2006). Extremely halophilic denitrifying bacteria from hypersaline inland lakes, Halovibrio denitrificans sp. nov. and Halospina denitrificans gen. nov., sp. nov., and evidence that the genus name Halovibrio Fendrich 1989 with the type species Halovibrio variabilis should be associated with DSM 3050. Int. J. Syst. Evol. Microbiol. 56 379–388. 10.1099/ijs.0.63964-0 [DOI] [PubMed] [Google Scholar]
- Suganthi C., Mageswari A., Shankar M., Gothandam K. M., Karthikeyan S. (2018). Genome sequence of Bacillus vallismortis TD3, a salt-tolerant strain isolated from the sediments of a solar saltern in Tamil Nadu, India. Microbiol. Resour. Announc. 7 e817–e818. 10.1128/MRA.00817-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taib N., Mangot J. F., Domaizon I., Bronner G., Debroas D. (2013). Phylogenetic affiliation of SSU rRNA genes generated by massively parallel sequencing: new insights into the freshbrine protist diversity. PLoS One 8:58950. 10.1371/journal.pone.0058950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki H., Wright C. L., Li X., Lin Q., Hwang C., Wang S., et al. (2011). Analysis of 16S rRNA amplicon sequencing options on the Roche/454 next-generation titanium sequencing platform. PLoS One 6:e25263. 10.1371/journal.pone.0025263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkavc R., Gostinčar C., Turk M., Visscher P. T., Oren A., Gunde-Cimerman N. (2011). Bacterial communities in the ‘petola’microbial mat from the Sečovlje salterns (Slovenia). FEMS Microbiol. Ecol. 75 48–62. 10.1111/j.1574-6941.2010.00985.x [DOI] [PubMed] [Google Scholar]
- Toneatti D. M., Albarracín V. H., Flores M. R., Polerecky L., Farías M. E. (2017). Stratified bacterial diversity along physico-chemical gradients in high-altitude modern stromatolites. Front. Microbiol. 8:646. 10.3389/fmicb.2017.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigui H., Masmoudi S., Brochier-Armanet C., Barani A., Grégori G., Denis M., et al. (2011). Characterization of heterotrophic prokaryote subgroups in the Sfax coastal solar salterns by combining flow cytometry cell sorting and phylogenetic analysis. Extremophiles 15 347–358. 10.1007/s00792-011-0364-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiamis G., Katsaveli K., Ntougias S., Kyrpides N., Andersen G., Piceno Y., et al. (2008). Prokaryotic community profiles at different operational stages of a Greek solar saltern. Res. Microbiol. 159 609–627. 10.1016/j.resmic.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Ventosa A. (2006). “Unusual micro-organisms from unusual habitats: hypersaline environments,” in Prokaryotic Diversity: Mechanisms and Significance, eds Logan N., Lappin-Scott H., Oyston P. (Cambridge: Cambridge University Press; ), 223–254. 10.1017/cbo9780511754913.015 [DOI] [Google Scholar]
- Ventosa A., Haba R. R., Sánchez-Porro C. (2019). “Haloarcula,” in Bergey’s Manual of Systematics of Archaea and Bacteria, ed. Ventosa A. (Hoboken, NJ: John Wiley & Sons, Inc; ), 1–12. [Google Scholar]
- Ventosa A., Nieto J. J., Oren A. (1998). Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62 504–544. 10.1128/MMBR.62.2.504-544.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventosa A., Quesada E., Rodriguez-Valera F., Ruiz-Berraquero F., Ramos-Cormenzana A. (1982). Numerical taxonomy of moderately halophilic Gram-negative rods. Microbiology 128 1959–1968. 10.1099/00221287-128-9-1959 [DOI] [Google Scholar]
- Vera-Gargallo B., Chowdhury T. R., Brown J., Fansler S. J., Durán-Viseras A., Sánchez-Porro C., et al. (2019). Spatial distribution of prokaryotic communities in hypersaline soils. Sci. Rep. 9:1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Gargallo B., Ventosa A. (2018). Metagenomic insights into the phylogenetic and metabolic diversity of the prokaryotic community dwelling in hypersaline soils from the Odiel Saltmarshes (SW Spain). Genes 9:152. 10.3390/genes9030152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreeland R. H. (2015). “Halomonas,” in Bergey’s Manual of Systematics of Archaea and Bacteria, eds Whitman W. B., Rainey F., Kämpfer P., Trujillo M., Chun J., De Vos P., et al. (Chichester: John Wiley & Sons Ltd; ), 1–19. [Google Scholar]
- Walsh D. A., Papke R. T., Doolittle W. F. (2005). Archaeal diversity along a soil salinity gradient prone to disturbance. Environ. Microbiol. 7 1655–1666. 10.1111/j.1462-2920.2005.00864.x [DOI] [PubMed] [Google Scholar]
- Walsh E. A., Kirkpatrick J. B., Rutherford S. D., Smith D. C., Sogin M., D’Hondt S. (2016). Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 10 979–989. 10.1038/ismej.2015.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. Y., Ng C. C., Chen T. W., Wu S. J., Shyu Y. T. (2007). Microbial diversity analysis of former salterns in southern Taiwan by 16S rRNA-based methods. J. Basic Microbiol. 47 525–533. 10.1002/jobm.200700250 [DOI] [PubMed] [Google Scholar]
- Wang Y., Qian P. Y. (2009). Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS One 4:e7401. 10.1371/journal.pone.0007401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Chen J. C., Wu Q., Chen G. Q. (2015). Halophiles, coming stars for industrial biotechnology. Biotechnol. Adv. 33 1433–1442. 10.1016/j.biotechadv.2014.10.008 [DOI] [PubMed] [Google Scholar]
- Youssef N. H., Ashlock-Savage K. N., Elshahed M. S. (2012). Phylogenetic diversities and community structure of members of the extremely halophilic Archaea (order Halobacteriales) in multiple saline sediment habitats. Appl. Environ. Microbiol. 78 1332–1344. 10.1128/AEM.07420-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafrilla B., Martínez-Espinosa R. M., Alonso M. A., Bonete M. J. (2010). Biodiversity of Archaea and floral of two inland saltern ecosystems in the Alto Vinalopó Valley, Spain. Saline Syst. 6:10. 10.1186/1746-1448-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ma G., Deng Y., Dong J., Van Stappen G., Sui L. (2016). Bacterial diversity in Bohai Bay solar saltworks, China. Curr.Microbiol. 72 55–63. 10.1007/s00284-015-0916-5 [DOI] [PubMed] [Google Scholar]
- Zhou J., Bruns M. A., Tiedje J. M. (1996). DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62 316–322. 10.1128/aem.62.2.316-322.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data generated in this study have been deposited in the SRA under BioProject PRJNA614987.