Abstract
Long non-coding RNAs (lncRNAs), some of the most abundant epigenetic regulators, play an important role in esophageal squamous cell carcinoma (ESCC). In the current study, the functions and mechanisms of the lncRNA LINC00673 were investigated. The expression levels of LINC00673 and its potential target genes were assessed by quantitative real-time polymerase chain reaction (qPCR) in ESCC surgical specimens and ESCC cell lines. RNA fluorescence in situ hybridization (RNA FISH) was employed to detect the subcellular location and the levels of LINC00673 in ESCC samples from patients with different survival times. LINC00673 function in ESCC carcinogenesis was also evaluated in vivo and in vitro. Cell cycle synchronization was performed using serum withdrawal; the cell cycle was monitored by fluorescence analysis and cellular DNA was detected by flow cytometry. The molecular mechanisms underlying LINC00673 were explored via Western blotting, chromatin immunoprecipitation (ChIP), and ChIP-PCR. Up-regulated LINC00673 was associated with poor prognosis in ESCC patients and promoted the proliferation of ESCC cells both in vitro and in vivo. Compared to the control group, depletion of LINC00673 in ESCC cells arrested the cell cycle, at least, at the G1/S checkpoint. Knockdown of LINC00673 significantly enhanced posttranscriptional expression of CDKN2C, and histone 3 lysine 27 trimethylation (H3K27me3) was enriched at the promoter region of CDKN2C. After inhibiting EZH2, the CDKN2C expression levels were increased. The present findings are the first to reveal that LINC00673 represses CDKN2C expression and promotes ESCC cell proliferation by elevating EZH2-mediated H3K27me3 levels. These data suggest that LINC00673 regulates the cell cycle in ESCC and that it is a promising target for clinical therapy.
Keywords: ESCC, LINC00673, CDKN2C, H3K27me3, cell cycle
Introduction
Esophageal cancer (EC) is one of the most prevalent solid tumor cancers worldwide, ranking seventh, and sixth in morbidity and mortality, respectively (1). Notably, the incidence rate in East Asia, especially in Mongolia and China, ranks highest among regions (1). Furthermore, EC subtype incidence exhibits geographic variation; the most common histologic subtype in China is esophageal squamous cell carcinoma (ESCC), whereas the rates of esophageal adenocarcinoma (EAC) are rising rapidly in Europe and America (2, 3). Despite advances in surgical treatments over the past few decades, the poor prognosis of ESCC has not improved significantly (4, 5). Therefore, the mechanisms underlying human ESCC progression need to be further investigated.
Long non-coding RNAs (lncRNAs) are transcripts of more than 200 nucleotides that do not encode a protein due to a lack of an open reading frame. lncRNAs are critical regulators of multiple disease processes and may function as oncogenes or tumor suppressors by modulating gene expression at the epigenetic, transcriptional, and posttranscriptional levels (6–9). For instance, HOTAIR, and MALAT1 are associated with a variety of human cancers and have potential value in patients’ prognosis (10, 11). It is commonly recognized that lncRNAs, which function as signals of specific cellular states, could be used to identify the cytopathology, predict prognosis, or even guide therapeutic options for patients (10, 12). Nonetheless, compared with other solid tumors with a high incidence, a few studies of lncRNAs in ESCC have been reported; furthermore, the molecular mechanisms of lncRNAs are intricate, and they have not been fully elucidated.
Genomic copy number variations (CNVs) and abnormal expression of genes commonly occur in various tumors, including ESCC. Interestingly, the lncRNA LINC00673 is located at 17q24.3, a chromosomal region that has been found to be frequently amplified in ESCC (13, 14). In addition, LINC00673 has been reported to be associated with cell migration, invasion, and proliferation in squamous cell carcinomas, such as non-small cell lung carcinoma (NSCLC) (15–18), and tongue squamous cell carcinoma (TSCC) (19). Therefore, we investigated whether LINC00673 contributes to ESCC. In the present study, we explored the biological functions and molecular epigenetic mechanisms of LINC00673 in ESCC, and our findings indicate that LINC00673 might be a regulator of CDKN2C through histone methylation modification.
Materials and Methods
Patients and Tissue Samples
Paired cancerous tissues and adjacent non-cancerous samples were collected from 39 ESCC patients who underwent surgical resection at Jiangsu Cancer Hospital between 2014 and 2016. All ESCC samples were obtained during surgery and then immediately frozen in liquid nitrogen until RNA detection. In addition, 81 archival paraffin-embedded surgical specimen blocks were obtained from patients who underwent surgery for ESCC from 2004 to 2006 at the Department of Pathology, Fujian Province Tumor Hospital, China (Supplementary Table S1). Clinical information was collected, and the 8th version of the American Joint Committee on Cancer (AJCC) system was applied for tumor staging. Our study protocol was approved by the Ethical Committee of Southeast University.
Cell Culture
The human ESCC cell lines (KYSE30, KYSE510, and EC9706) were purchased from Guangdong Hybribio Biotech Ltd. (Guangdong, China), and an immortalized normal esophageal epithelial cell line (HET-1A) was purchased from Cinoasia Institute (Shanghai, China). KYSE30, KYSE510, EC9706, and HET-1A cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Scientific) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, United States) in a humidified incubator at 37°C with 5% CO2.
RNA Fluorescence in situ Hybridization and Scoring
Tissues were dehydrated with gradient alcohol after fixation with paraformaldehyde and then embedded in paraffin. Subsequently, the specimen was cut into slices and baked at 62°C for 2 h. The slices were successively incubated in xylene I for 15 min, xylene II for 15 min, anhydrous ethanol I for 5 min, anhydrous ethanol II for 5 min, 85% alcohol for 5 min, and 75% alcohol for 5 min and then washed with diethyl pyrocarbonate (DEPC)-treated water. The slices were boiled in an antigen repair solution for 10–15 min and then cooled to room temperature. Next, the slices were incubated with proteinase K (20 μg/ml) at 37°C and pre-hybridized with a hybridization buffer at 37°C for 1 h and overnight with a hybridization buffer containing the fluorescence in situ hybridization (FISH) probe in the dark in a humid chamber. The samples were washed with 2 × SSC for 10 min at 37°C, 1 × SSC for 2 × 5 min at 37°C, and 0.5 × SSC for 10 min at room temperature. The tissues were then incubated with anti-DIG-488 at 37°C for 50 min, and the nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Finally, the slices were sealed in fluorescence decay-resistant medium and images were obtained under a Nikon fluorescence microscope.
Tissue staining was observed by two researchers blinded to the specimen identity. When there was a significant disparity in scoring, a third observer was included to reach an agreement. According to the staining density and scope in ESCC tissues, the staining signals were categorized into three levels: 0 = negative; 1 = weak staining; and 2 = intensive staining.
shRNA and Plasmid Transfections
The sequences of short hairpin RNAs (shRNAs) targeting LINC00673 were cloned into the GV248 vector, which was purchased from GeneChem (Shanghai, China). Lentiviral plasmids were transfected into KYSE30 and KYSE510 cells according to the manufacturer’s instructions. Transfected cells were subsequently selected with puromycin (1 μg/ml) for 2 weeks. Small interfering RNAs (siRNAs) against EZH2, LINC00673, and the corresponding negative controls were synthesized by GenePharma (Shanghai, China). SiRNA transfection of the ESCC cell lines using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States) was performed according to the manufacturer’s protocol. The shRNA and siRNA sequences for the specific targets in this study are shown in Supplementary Table S2.
RNA Extraction and Quantitative Real-Time PCR
Total RNAs from specimens and cells were isolated with a TRIzol reagent (Invitrogen, United States) according to the manufacturer’s instructions. Reverse transcription of 1 μg RNA to cDNA was performed using a PrimeScriptTM RT Reagent Kit (Takara, Japan) under standard conditions. quantitative real-time polymerase chain reaction (qPCR) was performed to determine the expression levels of specific genes using SYBR Premix Ex Taq Kit (Takara, Japan), and β-actin was used as an internal control to normalize the data. All experiments were performed with a StepOne Plus system (Applied Biosystems, Foster City, CA, United States), and the primers used are listed in Supplementary Table S3. All data were calculated using the 2–ΔΔCt method, and each sample was detected in triplicate.
Cell Proliferation Assay
Esophageal squamous cell carcinoma cell proliferation was detected by Cell Counting Kit-8 (CCK8, Dojindo, Japan) according to the manufacturer’s instructions. Approximately 2.5 × 103 cells per well were seeded into 96-well plates, and all cells were adhered after 6 h. After the cells were incubated with 10 μl CCK8 solution at 37°C in the dark for 2 h, cell proliferation was evaluated by absorbance at 450 nm using an iMark Microplate Reader (Bio-Rad, United States).
Colony Forming Assay
Esophageal squamous cell carcinoma cells were seeded into fresh 6-cm dishes (103 cells/well) to monitor clonal capacity. 10 days later, the cells were fixed with 75% ethanol and incubated with a Crystal violet solution (Beyotime, Shanghai, China) for 20 min at room temperature and the number of colonies per dish was recorded.
Flow Cytometric Analysis
Esophageal squamous cell carcinoma cells were synchronized by serum starvation, trypsinized, washed twice with phosphate-buffered saline (PBS), and gently resuspended in cold 75% ethanol. Cell cycle distribution was examined by flow cytometry using a FACScan flow cytometer (Becton Dickinson & Co., San Jose, CA, United States).
Tumor Xenograft Mouse Model
A sample of approximately 5 × 106 ESCC cells in 0.2 ml PBS was injected subcutaneously into 4-week-old female athymic BALB/c nude mice (Model Animal Research Center of Nanjing University, China). Tumor formation was monitored every 3 days starting on the fourth day after injection, and the tumor volume was calculated using the following formula: V = 0.5 × L × W2. The mice were euthanized 4 weeks after injections, and the average tumor weights in each group were measured. All procedures were conducted in accordance with the institutional standard guidelines of the Medical School of Southeast University. The animal experiments in this study were approved by the Experimental Animal Ethics Committee of Southeast University.
Western Blotting
Protein lysates of ESCC cells were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to 0.22-mm NC membranes (Sigma), and incubated with specific antibodies: anti-CDK6, anti-CDK4, anti-CDK2, anti-cyclinD1, anti-cyclinD3, anti-p27, anti-p21, anti-CDKN2C (Abcam, Shanghai, China), anti-EZH2, anti-EED, anti-SUZ12, anti-EZH1, and anti-β-actin (Cell Signaling Technology). The dilution ratio of the primary antibodies was 1:1,000, although anti-β-actin was diluted to 1:8,000 for Western blotting. Protein bands were visualized with Super Signal Chemiluminescence Substrate (Thermo Scientific) and β-actin was used as a control.
Chromatin Immunoprecipitation and ChIP-PCR
The chromatin immunoprecipitation (ChIP) assay was performed using an EZ-Magna ChIPTM G Immunoprecipitation Kit (Millipore) following the manufacturer’s instructions. ChIP-grade anti-H3K27me3 (CST, United States; dilution ratio, 1:50) and normal mouse IgG (as a negative control; 1 μg per IP) were used for immunoprecipitation. The primers used for amplifying the precipitated DNA fragments are listed in Supplementary Table S3.
Statistical Analysis
The data were analyzed using the SPSS (Statistical Package for the Social Sciences) statistical program version 17 (SPSS, Inc., Chicago, IL, United States). An independent Student’s t test (two-tailed) was performed to determine statistical significance. The correlations between the target genes were examined with the chi-square test. The data are presented as the mean ± SD. P < 0.05 was considered statistically significant (∗P < 0.05, ∗∗P < 0.01).
Results
Up-Regulated LINC00673 Is Associated With Poor Prognosis in ESCC Patients
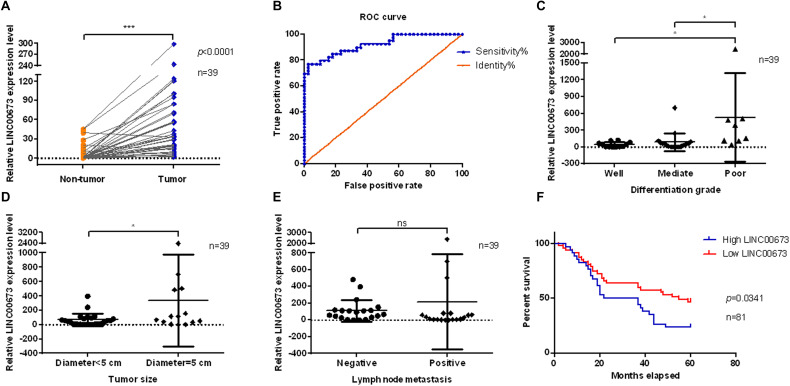
LINC00673 was evaluated in pairs of tumor tissues and corresponding adjacent non-tumor tissues from 39 ESCC patients using qPCR. LINC00673 showed a pattern of significantly higher expressions in ESCC tissues than in non-tumor tissues (Figure 1A). To confirm this result, we detected LINC00673 expression levels in tissue microarrays with 81 pairs of samples, consisting of ESCC tissues and corresponding adjacent non-tumor tissues, by RNA FISH (Supplementary Figure S1A and Supplementary Table S1). The results of the staining were consistent with the qPCR data (Supplementary Figure S1B). Furthermore, the value of LINC00673 in the diagnosis of ESCC was measured by a receiver operating characteristic (ROC) curve. The results showed a high true positive rate of the LINC00673 expression model for 39 ESCC patients (Figure 1B). In addition, the correlation between the expression levels of LINC00673 and the clinicopathological factors in the 39 ESCC patients was examined with the chi-square test. Up-regulated LINC00673 exhibited a close relationship with poor differentiation and large tumor size, but not with lymph node metastasis (Figures 1C–E). According to the RNA FISH scoring, the LINC00673 expression patterns correlated with differentiation grade, tumor size, and T stage, but not with age, sex, or other clinicopathological features (Table 1). Compared to patients with low LINC00673 levels (score ≤ 1), those with high LINC00673 levels (score > 1) displayed poor outcomes (Figure 1F). This finding suggests that an increased level of LINC00673 expression has a negative impact on the survival of ESCC patients. Taken together, our results demonstrate that LINC00673 is up-regulated in ESCC and can, to some extent, predict poor prognosis.
FIGURE 1.
Up-regulated LINC00673 is associated with poor prognosis in esophageal squamous cell carcinoma (ESCC) patients. (A) LINC00673 expression levels were evaluated using quantitative PCR (qPCR) in 39 pairs of ESCC tissues and corresponding non-tumor tissues. ***P < 0.001 (unpaired Student’s t test). (B) Receiver operating characteristic (ROC) curve of LINC00673 in 39 ESCC patients. (C–E) Relationships between LINC00673 expression and the differentiation grade, tumor size, and lymph node metastasis, respectively. (F) Kaplan–Meier curves show the survival of ESCC patients, as grouped by the LINC00673 expression levels. *P < 0.05.
TABLE 1.
Correlations between the expression patterns of LINC00673 detected by RNA fluorescence in situ hybridization (FISH) and related clinicopathological parameters.
| Clinicopathological features | LINC00673 (n = 81) |
|||
| Low | High | χ 2 | P value | |
| Sex | ||||
| Male | 33 | 27 | 0.8693 | 0.3511 |
| Female | 14 | 7 | ||
| Age (years) | ||||
| ≤60 | 29 | 23 | 0.3034 | 0.5818 |
| >60 | 18 | 11 | ||
| Tumor sub-sites | ||||
| Cervical portion | 9 | 4 | 0.7984 | 0.3716 |
| Thoracic portion | 38 | 30 | ||
| Differentiation grade | ||||
| I + II | 40 | 20 | 7.096 | 0.0077** |
| III | 7 | 14 | ||
| Tumor size (maximum diameter) | ||||
| ≤5 cm | 7 | 12 | 4.573 | 0.0325* |
| >5 cm | 40 | 22 | ||
| T stage | ||||
| I + II | 19 | 5 | 6.259 | 0.0124* |
| III + IV | 28 | 19 | ||
| N stage | ||||
| 0 + I | 31 | 22 | 0.01366 | 0.9069 |
| II + III | 16 | 12 | ||
| TNM stage | ||||
| I + II | 21 | 16 | 0.04496 | 0.8321 |
| III + IV | 26 | 18 | ||
*Significant differences are shown when the P value is less than 0.05. **Significant differences are shown when the P value is less than 0.01.
Knockdown of LINC00673 Inhibits ESCC Cell Proliferation in vitro and in vivo
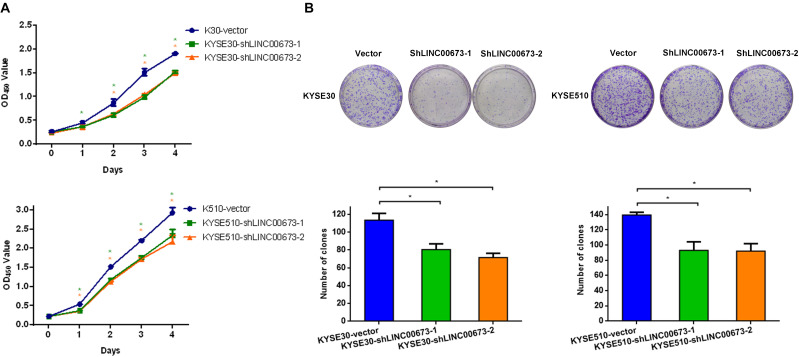
The strong association of LINC00673 expression with the tumor size, differentiation grade, and poor prognosis in ESCC prompted us to investigate whether LINC00673 affects ESCC cell proliferation. Thus, we constructed KYSE30-shLINC00673 cell lines, KYSE510-shLINC00673 cell lines, and corresponding controls and then measured the efficiency of shRNA-LINC00673 (Supplementary Figures S2A,B). Compared to the corresponding controls, the depletion of LINC00673 suppressed the ESCC cell proliferation capacity (Figure 2A). Similarly, the down-regulation of LINC00673 reduced the clone-forming capacity (Figure 2B).
FIGURE 2.
LINC00673 knockdown suppresses the proliferation of esophageal squamous cell carcinoma (ESCC) cells in vitro. (A) Cell proliferation was assessed by the CCK8 method in KYSE30 and KYSE510 cells. Bars show the mean ± SD of OD450 from triplicate samples. (B) Cell proliferation was assessed using a colony forming assay. Bar plots show the average number of colonies (±SD) from triplicate samples. *P < 0.05.
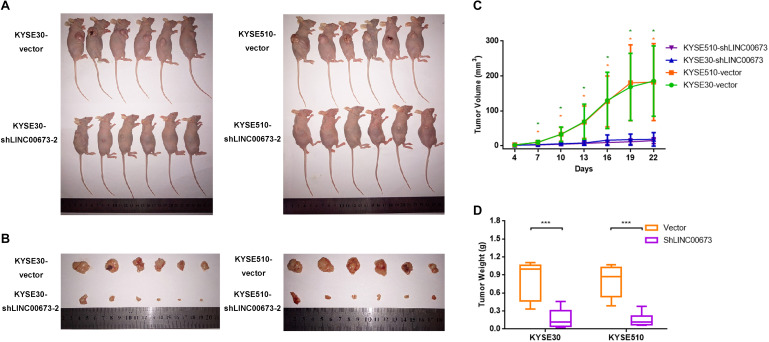
To further investigate the role of LINC00673 in the tumorigenesis of ESCC in vivo, we performed an in vivo tumor formation assay by injecting KYSE30-shLINC00673 cell lines, KYSE510-shLINC00673 cell lines, and their corresponding control cells into BALB/c nude mice (n = 6 per group). Tumor growth was measured every 3 days starting on the fourth day after injection, and the mice were euthanized 4 weeks after injection. The results suggested that compared to control cells, smaller tumors developed in mice from cells with stable knockdown of LINC00673 (Figures 3A–C). The tumor weight was also reduced in the LINC00673 knockdown groups compared to the control groups (Figure 3D). Overall, these data suggest that LINC00673 knockdown suppresses ESCC cell proliferation both in vitro and in vivo.
FIGURE 3.
LINC00673 knockdown inhibits esophageal squamous cell carcinoma (ESCC) cell proliferation in vivo. (A,B) Stable KYSE30-shLINC00673 and KYSE510-shLINC00673 cells were injected subcutaneously into nude mice. After 4 weeks, the tumors were resected. (C) Tumor growth was measured by Vernier calipers every 3 days starting on the fourth day after injection. Line chart show the tumor growth curve. (D) Box plot showing the tumor weights of the control and LINC00673 knockdown groups. *P < 0.05, ***P < 0.001.
Knockdown of LINC00673 Arrests the Cell Cycle at the G1/S Checkpoint in ESCC Cells
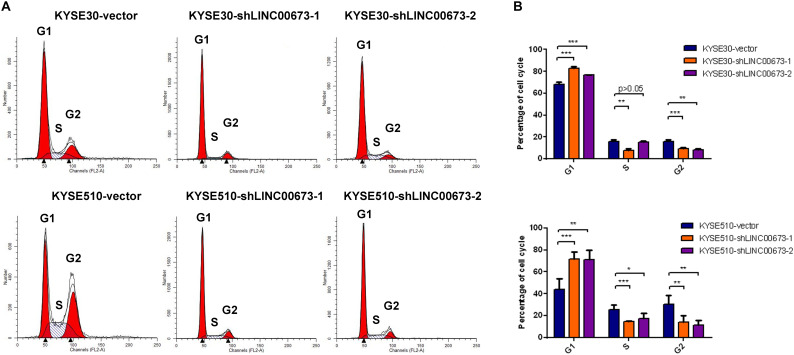
To determine the contribution of the cell cycle to the suppression of ESCC cell proliferation both in vitro and in vivo, we examined the cell cycle for various expression levels of LINC00673 by flow cytometry. Compared to the control groups, KYSE30 and KYSE510 cells with down-regulated LINC00673 showed higher proportions of cells in the G1 phase (Figures 4A,B), indicating that LINC00673 depletion suppresses cell proliferation by blocking the cell cycle at the G1/S checkpoint. In addition, the relatively low proportions of KYSE30-shLINC00673 and KYSE510-shLINC00673 cells in the G2 phase were shown by the results. It needs further investigation to confirm whether LINC00673 depletion will promote ESCC cells to enter the M phase. In the current study, the data showed that LINC00673 down-regulation inhibited cell proliferation in ESCC; therefore, we preferentially focused on the investigation of G1/S checkpoint arrest rather than the G2/M transition.
FIGURE 4.
LINC00673 knockdown arrests the cell cycle at the G1/S checkpoint in esophageal squamous cell carcinoma (ESCC) cells. (A,B) Cell cycle examined by a flow cytometry assay. Bar plots show the proportion of cells in the G1 phase, S phase, and G2 phase. *P < 0.05, **P < 0.01, and ***P < 0.001.
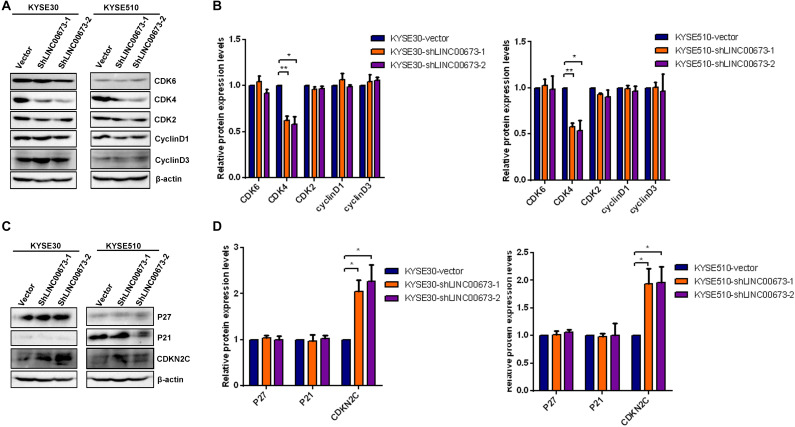
To determine how LINC00673 regulates cell cycle progression, the expression levels of the G1/S checkpoint regulators CDK2, CDK4, CDK6, cyclin D1, and cyclin D3 were examined by Western blotting. Among these proteins, the expression of CDK4 was significantly down-regulated in the LINC00673 knockdown KYSE30 and KYSE510 cells compared to the control groups (Figures 5A,B). As the CDK inhibitors P27Kip1, P21Cip1, and CDKN2C are important regulators of the cell cycle (Supplementary Figure S3), we hypothesized that LINC00673 promotes ESCC cell cycle progression by suppressing p21Cip1, p27Kip1, or CDKN2C expression. As expected, only the CDKN2C protein expression increased when LINC00673 was knocked down in KYSE30 and KYSE510 cells (Figures 5C,D).
FIGURE 5.
LINC00673 knockdown improved CDKN2C levels in esophageal squamous cell carcinoma (ESCC) cells. (A,C) The levels of the G1/S checkpoint regulator expression were detected by Western blotting analysis in stable KYSE30-shLINC00673, KYSE510-shLINC00673, and corresponding control cells. (B,D) Bar plots showing the relative expression levels of the G1/S checkpoint associated regulators. The image was processed by ImageJ software. *P < 0.05, **P < 0.01.
Consistent with the depleted level of CDK4, CDKN2C, a cyclin-dependent kinase inhibitor, showed an increasing expression trend compared to the control groups. These findings indicate that LINC00673 knockdown enhances CDKN2C expression, further regulating cell cycle progression in ESCC cells.
H3K27me3 Is Enriched at the Promoter Region of CDKN2C to Silence CDKN2C Expression
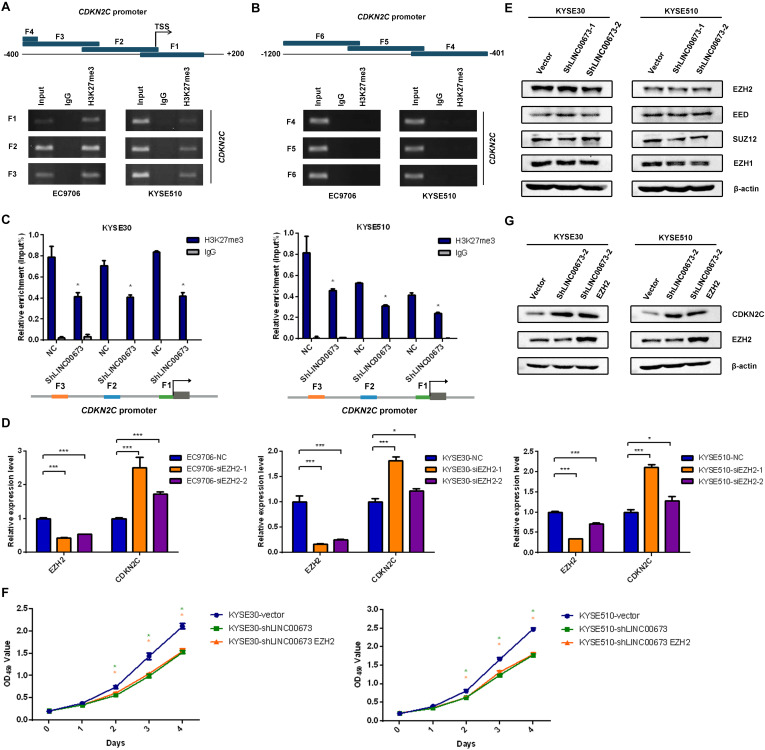
To clarify whether LINC00673 regulates the cell cycle by inhibiting CDKN2C, we conducted a ChIP assay using antibodies against H3K27me3 in EC9706 and KYSE510 cells, as transcriptional repression is characterized by H3K27me3, which serves as a typical suppressor marker associated with inactive gene promoters (20–24). Primers for the CDKN2C promoter region were designed to probe DNA fragments via PCR (the primers, covering -1,200 to + 150 bp, are shown in Supplementary Table S3). In this assay, the sequences between the CDKN2C transcription initiation site (TSS) and the upstream 400 bp were amplified as DNA fragments interacting with H3K27me3 (Figures 6A,B), which indicated that H3K27me3 was indeed enriched at the promoter region of CDKN2C. Furthermore, we performed ChIP-qPCR of H3K27me3 in KYSE30-shLINC00673 and KYSE510-shLINC00673 cells and found that the enrichment of H3K27me3 at the CDKN2C promoter region decreased in cells with LINC00673 down-regulation compared to the control groups (Figure 6C). This result suggests that LINC00673 regulates CDKN2C expression through H3K27me3.
FIGURE 6.
H3K27me3 was enriched at the promoter region of CDKN2C. (A,B) Diagram of the promoter of the CDKN2C gene, with the transcription start site (TSS) indicated. The blue block represents the location of the F1–F6 fragments detected by chromatin immunoprecipitation (ChIP). ChIP assays were performed using an H3K27me3-specific antibody, followed by PCR amplification of six individual fragments representing CDKN2C promoter regions (F1–F6). Input and products immunoprecipitated by IgG were used as positive and negative controls, respectively. (C) ChIP assays were performed using an H3K27me3-specific antibody in KYSE30-shLINC00673, KYSE510-shLINC00673, and corresponding control cells, followed by quantitative PCR (qPCR) amplification of three individual fragments near the CDKN2C promoter (F1–F3). *P < 0.05 (unpaired Student’s t test). (D) CDKN2C expression levels were detected using qPCR after silencing EZH2 in EC9706, KYSE30, and KYSE510 cells. *P < 0.05, ***P < 0.001. (E) The major subunits of the PRC2 complex expression levels were detected by Western blotting analysis in stable KYSE30-shLINC00673, KYSE510-shLINC00673, and corresponding control cells. (F) Cell proliferation was assessed by the CCK8 method in KYSE30-shLINC00673 and KYSE510-shLINC00673 cells after transfection with EZH2. Bars show the mean ± SD of OD450 from triplicate samples. (G) EZH2 and CDKN2C expression levels were detected by Western blotting analysis after the transfection of EZH2 in KYSE30-shLINC00673 and KYSE510-shLINC00673 cells.
Several researchers have reported the interaction between LINC00673 and enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) in solid tumors (17, 25–27), and EZH2 is generally considered to execute methyltransferase effects against H3K27 (28). Thus, we hypothesized that LINC00673 regulates H3K27me3 levels by recruiting EZH2, affecting the downstream cell cycle regulator CDKN2C. When EZH2 expression was transiently knocked down in three ESCC cell lines—EC9706, KYSE30, and KYSE510—an increased expression of CDKN2C was detected, as expected (Figure 6D). In addition, Western blotting was conducted to confirm whether LINC00673 influences the major subunits of the PRC2 complex, and the results showed that the expression levels of the major6 subunits, such as EZH2, SUZ12, EED, and EZH1, were not affected by the LINC00673 expression pattern (Figure 6E and Supplementary Figures S4A,B).
To assess whether EZH2 overexpression can reverse the inhibited cell proliferation and up-regulated CDKN2C levels in LINC00673 knockdown cells, EZH2 pcDNA3.1 plasmids were transfected into KYSE30-shLINC00673 and KYSE510-shLINC00673 cells. Data from CCK8 and the Western blotting assays suggested that up-regulated EZH2 in LINC00673 knockdown cells did not reverse cell proliferation or decrease CDKN2C expression (Figures 6F,G and Supplementary Figures S4C,D), indicating that CDKN2C was regulated by histone modification due to a decrease in EZH2 recruitment rather than a decrease in EZH2 expression. Our hypothesis is that LINC00673 can elevate H3K27me3 levels at the CDKN2C promoter by recruiting EZH2. However, due to the knockdown of LINC00673, EZH2 cannot become massively enriched near the CDKN2C promoter. Therefore, cell proliferation in KYSE30-shLINC00673 and KYSE510-shLINC00673 cells was not reversed by the overexpression of EZH2. In general, these data show that LINC00673 recruits EZH2 to regulate H3K27me3 levels at the CDKN2C promoter region and further modulates the ESCC cell cycle and proliferation.
Discussion
Esophageal squamous cell carcinoma is characterized by striking geographic variation, intricate etiology, and poor clinical outcomes in China. Various pathogenic factors, such as dietary habits and chemical carcinogens, might cause genetic mutations and epigenetic variations, yet little is known about the epigenetic regulatory mechanisms underlying ESCC progression (29). To improve prognosis and better understand this complicated disease, this study was designed to elucidate the molecular regulatory mechanisms involved in ESCC development.
Chemical modifications of the fundamental macromolecule DNA and proteins participate in the control and adaptability of almost all biological processes. Methylation, known as one of the most prevalent modifications, is widespread throughout all forms of life (30), and many tumor suppressor genes are silenced by DNA methylation in cancers (31). For example, hypomethylation of repetitive elements in mammals can induce the activation and transposition of endogenous retroviral elements, thus promoting instability in the genome and tumorigenesis (30, 32). Histone methylation levels in facultative heterochromatin are also associated with genes that are differentially expressed during cell differentiation or development (30, 33), as characterized by gene-inactive H3K27me3 modification. Developmental genes contain so-called bivalent domains that possess both gene-active H3K4me3 and gene-repressive H3K27me3 modifications. This “poised” state has been shown to allow these genes to respond to differentiation cues quickly and accurately (30, 34). Furthermore, histone methylation, as represented by H3K27me3, H3K9me3, and H4K20me3, is related to the outcome of ESCC patients (23, 35). This evidence suggests that epigenetic modifications, such as methylation, may play an important role in tumor progression.
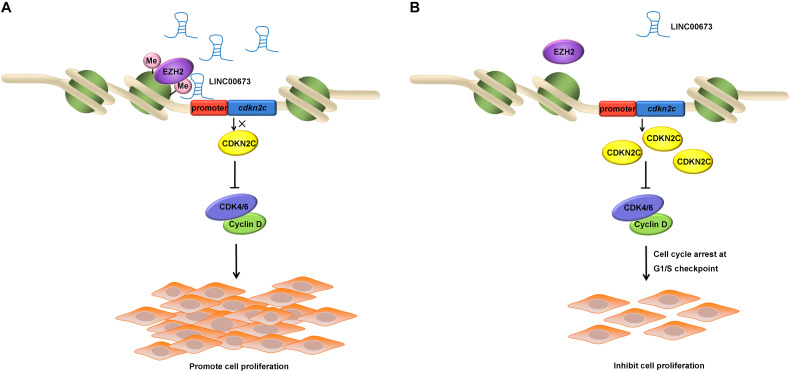
After confirming that depleted LINC00673 was able to up-regulate CDKN2C and promote the cell cycle, we conducted a ChIP assay using antibodies against H3K27me3 in EC9706 and KYSE510 cells and found that H3K27me3 was enriched at the promoter region of CDKN2C. Some key molecules that may regulate CDKN2C expression via methylation modification—DNMT1, DNMT3A, and EZH2 (36–38)—were knocked down in ESCC cells; among these molecules, only EZH2 knockdown could up-regulate CDKN2C. It has been reported that EZH2 recruits LINC00673 (17, 25–27), and it is reasonable to consider that EZH2 is recruited to the CDKN2C promoter by LINC00673, increasing the level of H3K27me3 to regulate CDKN2C expression and, thus, affecting the proliferation of ESCC cells (Figures 7A,B).
FIGURE 7.
Schematic of the regulatory relationships among LINC00673, EZH2, and CDKN2C in ESCC cells. (A,B) Illustrated the effect of up-regulated or down-regulated LINC00673 on ESCC cell proliferation, respectively.
An interesting phenomenon was noted in our study: compared with the control group, ESCC cells with LINC00673 down-regulation had a very dramatic inhibitory effect on tumor formation in vivo, whereas this repressive effect of decreased LINC00673 on tumor cell proliferation was not that obvious in vitro. These results suggest that the tumor microenvironment may be responsible for the difference between in vivo and in vitro results. In fact, information exchange in the tumor microenvironment can significantly affect tumor progression and malignant biological behaviors (39–41). Moreover, lncRNAs secreted by tumor-derived exosomes can alter the cellular physiology of cancer cells by elevating their expressions of membrane molecules and soluble factors (42, 43).
In conclusion, our study confirmed that LINC00673 plays a role in promoting tumor proliferation by regulating G1/S checkpoint regulator expression. Furthermore, our data demonstrate that LINC00673 acts as a scaffold protein that recruits EZH2 to repress CDKN2C levels in ESCC. Therefore, LINC00673 may be a promising molecular target for the treatment of ESCC.
Data Availability Statement
All datasets generated or analyzed in this study are included in the article/Supplementary Material.
Ethics Statement
Our study protocol was approved by the Ethical Committee of Southeast University. The patients provided their written informed consent to participate in this study.
Author Contributions
HF conceived and designed the experiments. MZ conducted the experiments and wrote the manuscript. YL, YM, SY, and YX analyzed the experiment data. RY, SL, and QZ provided the ESCC clinical specimens. YQ performed the animal experiment. TL and RS analyzed the RNA FISH data. All authors reviewed the manuscript before submission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors appreciate the help of Professor Zehui Hong, Zhujiang Zhao, and Xiufang Liu.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Nos. 81672414 and 81972664) and the Key Project of Cutting-edge Clinical Technology of Jiangsu Province (BE2017759). This work was also supported by the Postgraduate Research and Practice Innovation Program of Jiangsu Province (No. KYCX180058).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2020.01546/full#supplementary-material
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Bernard WS, Christopher PW. World Cancer Report. Lyon: International Agency for Research on Cancer. (2014). [Google Scholar]
- 3.Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. (2017) 390:2383–96. 10.1016/s0140-6736(17)31462-9 [DOI] [PubMed] [Google Scholar]
- 4.Abnet CC, Arnold M, Wei WQ. Epidemiology of esophageal squamous cell carcinoma. Gastroenterology. (2018) 154:360–73. 10.1053/j.gastro.2017.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. (2015) 149:1700–15. 10.1053/j.gastro.2015.08.054 [DOI] [PubMed] [Google Scholar]
- 6.Serviss JT, Johnsson P, Grander D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet. (2014) 5:234. 10.3389/fgene.2014.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. (2013) 45:1895–910. 10.1016/j.biocel.2013.05.030 [DOI] [PubMed] [Google Scholar]
- 8.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: background, mechanisms, and therapeutic approaches. Physiol Rev. (2016) 96:1297–325. 10.1152/physrev.00041.2015 [DOI] [PubMed] [Google Scholar]
- 9.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. (2015) 21:1253–61. 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 10.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. (2016) 29:452–63. 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutschner T, Hammerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. (2013) 91:791–801. 10.1007/s00109-013-1028-y [DOI] [PubMed] [Google Scholar]
- 12.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. (2018) 172:393–407. 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao JJ, Shi ZZ, Zhao ZX, Zhang Y, Gong T, Li CX, et al. Characterization of genetic rearrangements in esophageal squamous carcinoma cell lines by a combination of M-FISH and array-CGH: further confirmation of some split genomic regions in primary tumors. BMC Cancer. (2012) 12:367. 10.1186/1471-2407-12-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu X, Moon JW, Li S, Xu W, Wang X, Liu Y, et al. Amplification and overexpression of CTTN and CCND1 at chromosome 11q13 in esophagus squamous cell carcinoma (ESCC) of North Eastern Chinese Population. Int J Med Sci. (2016) 13:868–74. 10.7150/ijms.16845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu W, Xu Q, Kuang D, Wang Z, Lu Q, Lin Q, et al. Long noncoding RNA SLNCR1 regulates nonsmall cell lung cancer migration, invasion and stemness through interactions with secretory phospholipase A2. Mol Med Rep. (2019) 20:2591–6. 10.3892/mmr.2019.10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu W, Zhang H, Niu Y, Wu Y, Sun W, Li H, et al. Long non-coding RNA linc00673 regulated non-small cell lung cancer proliferation, migration, invasion and epithelial mesenchymal transition by sponging miR-150-5p. Mol Cancer. (2017) 16:118. 10.1186/s12943-017-0685-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma C, Wu G, Zhu Q, Liu H, Yao Y, Yuan D, et al. Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget. (2017) 8:32696–705. 10.18632/oncotarget.16158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi X, Ma C, Zhu Q, Yuan D, Sun M, Gu X, et al. Upregulation of long intergenic noncoding RNA 00673 promotes tumor proliferation via LSD1 interaction and repression of NCALD in non-small-cell lung cancer. Oncotarget. (2016) 7:25558–75. 10.18632/oncotarget.8338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Liu Y, Gong Z, Zhang S, Guo C, Li X, et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget. (2017) 8:16621–32. 10.18632/oncotarget.14200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res. (2018) 6:10. 10.1186/s40364-018-0122-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol. (2015) 37:42–8. 10.1016/j.ceb.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 22.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. (2007) 129:823–37. 10.1016/j.cell.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 23.Lin S, Zhou M, Li Y, Chen Y, Xu W, Xia W, et al. H3K27 trimethylation and H3K9 dimethylation as poor prognostic markers for patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. (2019) 12:2657–64. [PMC free article] [PubMed] [Google Scholar]
- 24.Liu F, Gu L, Cao Y, Fan X, Zhang F, Sang M. Aberrant overexpression of EZH2 and H3K27me3 serves as poor prognostic biomarker for esophageal squamous cell carcinoma patients. Biomarkers. (2016) 21:80–90. 10.3109/1354750x.2015.1118537 [DOI] [PubMed] [Google Scholar]
- 25.Meng XF, Zhao LY, Chu XF. LncRNA LINC00673 inhibits p53 expression by interacting with EZH2 and DNMT1 in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. (2019) 23:2075–83. 10.18632/aging.101920 [DOI] [PubMed] [Google Scholar]
- 26.Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu YB, et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. (2017) 8:95542–53. 10.18632/oncotarget.20980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F, et al. Long noncoding RNA LINC00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol Ther. (2017) 25:1014–26. 10.1016/j.ymthe.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Yan KS, Lin CY, Liao TW, Peng CM, Lee SC, Liu YJ, et al. EZH2 in cancer progression and potential application in cancer therapy: a friend or foe? Int J Mol Sci. (2017) 18:1172. 10.3390/ijms18061172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kojima T, Doi T. Immunotherapy for esophageal squamous cell carcinoma. Curr Oncol Rep. (2017) 19:33. 10.1007/s11912-017-0590-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michalak EM, Burr ML, Bannister AJ. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat Rev Mol Cell Biol. (2019) 20:573–89. 10.1038/s41580-019-0143-1 [DOI] [PubMed] [Google Scholar]
- 31.Schubeler D. Function and information content of DNA methylation. Nature. (2015) 517:321–6. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- 32.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. (2012) 150:12–27. 10.1016/j.cell.2012.06.013 [DOI] [PubMed] [Google Scholar]
- 33.McCabe MT, Mohammad HP, Barbash O, Kruger RG. Targeting histone methylation in cancer. Cancer J. (2017) 23:292–301. 10.1097/ppo.0000000000000283 [DOI] [PubMed] [Google Scholar]
- 34.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. (2006) 125:315–26. 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 35.Zhou M, Li Y, Lin S, Chen Y, Qian Y, Zhao Z, et al. H3K9me3, H3K36me3, and H4K20me3 expression correlates with patient outcome in esophageal squamous cell carcinoma as epigenetic markers. Dig Dis Sci. (2019) 64:2147–57. 10.1007/s10620-019-05529-2 [DOI] [PubMed] [Google Scholar]
- 36.Cheng P, Wang YF, Li G, Yang SS, Liu C, Hu H, et al. Interplay between menin and Dnmt1 reversibly regulates pancreatic cancer cell growth downstream of the Hedgehog signaling pathway. Cancer Lett. (2016) 370:136–44. 10.1016/j.canlet.2015.09.019 [DOI] [PubMed] [Google Scholar]
- 37.Cui H, Zhao C, Gong P, Wang L, Wu H, Zhang K, et al. DNA methyltransferase 3A promotes cell proliferation by silencing CDK inhibitor p18INK4C in gastric carcinogenesis. Sci Rep. (2015) 5:13781. 10.1038/srep13781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meseure D, Vacher S, Alsibai KD, Nicolas A, Chemlali W, Caly M, et al. Expression of ANRIL-polycomb complexes-CDKN2A/B/ARF genes in breast tumors: identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol Cancer Res. (2016) 14:623–33. 10.1158/1541-7786.mcr-15-0418 [DOI] [PubMed] [Google Scholar]
- 39.Le MT, Hamar P, Guo C, Basar E, Perdigao-Henriques R, Balaj L, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. (2014) 124:5109–28. 10.1172/jci75695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu T, Dai Y. Tumor microenvironment and therapeutic response. Cancer Lett. (2017) 387:61–8. 10.1016/j.canlet.2016.01.043 [DOI] [PubMed] [Google Scholar]
- 41.Hui L, Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Lett. (2015) 368:7–13. 10.1016/j.canlet.2015.07.039 [DOI] [PubMed] [Google Scholar]
- 42.Todorova D, Simoncini S, Lacroix R, Sabatier F, Dignat-George F. Extracellular vesicles in angiogenesis. Circ Res. (2017) 120:1658–73. 10.1161/circresaha.117.309681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Z, Yang S, Zhou Q, Wang G, Song J, Li Z, et al. Emerging role of exosome-derived long non-coding RNAs in tumor microenvironment. Mol Cancer. (2018) 17:82. 10.1186/s12943-018-0831-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated or analyzed in this study are included in the article/Supplementary Material.