Abstract
The marine environment represents a well-off and diverse group of microbes, which offers an enormous natural bioactive compounds of commercial importance. These natural products have expanded rigorous awareness due to their widespread stability and functionality under harsh environmental conditions. The genus Planococcus is a halophilic bacterium known for the production of diverse secondary metabolites such as 2-acetamido-2-deoxy-α-d-glucopyranosyl-(1, 2)-β-d-fructofuranose exhibiting stabilizing effect and methyl glucosyl-3,4-dehydro-apo-8-lycopenoate displaying antioxidant activity. The genus Planococcus is reported generally for hydrocarbon degradation in comparison with biosurfactant/bioemulsifier secretion. Although Planococcus was proposed in 1894, it seized long stretch (till 1970) to get accommodated under the genus Planococcus authentically. Large-scale biosurfactant production from Planococcus was reported in 2014 with partial characterization. For the first time in 2019, we documented genomic and functional analysis of Planococcus sp. along with the physico-chemical properties of its biosurfactant. In 2020, again we screened biosurfactant for pharmacological applications. The present review discusses the comprehensive genomic insights and physical properties of Planococcus-derived biosurfactant. Moreover, we also highlight the prospects and challenges in biosurfactant production from Planococcus sp. Among ∼102 reports on biosurfactant produced by marine bacteria, 43 were of glycolipid and 59 were non-glycolipid type. Under other biosurfactant type, they were identified as lipopeptide (20) like surfactin (5), glycolipoprotein/lipoprotein (12), and other non-glycolipid (22). Planococcus sp. generally produces glycolipid-type biosurfactant (4) and exopolysaccharides (2). The single report documented in the literature is on biosurfactant production (glycolipid +non glycolipid) by diverse marine microbes (39) suggesting their novelty and diversity for biosurfactant secretion.
Keywords: Planococcus, biosurfactant, genomic insight, glycolipid, marine, hydrocarbon degradation
Richness and Uniqueness of Marine Ecosystem, and Challenges Faced to Explore Its Microbial Wealth
Marine ecosystems represent the largest one among different aquatic ecosystems of our planet where high salinity makes it different in comparison with another aquatic reservoir (Wang and D’Odorico, 2013). Salty water resulted through several mechanisms like rain, rock and sediment erosion processes, exchange of gases with the atmosphere, volcanic actions, and breakdown of metabolites has a prodigious impact on marine flora and fauna (Geist and Hawkins, 2016). Even though the marine biosphere appears to be a nutrient-rich ecosystem however, the actual nutrients available to lead a prosperous life for microbes are limited (Satpute et al., 2010; Merchant and Helmann, 2012). Under such nutrient-deficient and challenging circumstances, marine microbes withstand and compete through the production of a unique magical molecules (Gudiña et al., 2015). Therefore, the marine ecosystem grips an immense assure to invent an innovative bioactive compounds like enzymes, vitamins, antibiotics, drugs, biosurfactant (BS), and bioemulsifier (BE) (Satpute et al., 2010; Gudiña et al., 2016; Waghmode et al., 2019). Marine microbes produce not only diverse types of BSs but also other bioactive molecules under extreme surroundings of salinity, fluctuations in temperatures and pH, availability of limited nutrients, and increased UV exposure (de Carvalho, 2018; Naughton et al., 2019). Studies of marine microbes require access to the sea using well-equipped research ships which is costly and is a major hindrance (Baharum et al., 2010). Therefore, most of the marine microorganisms documented are from mangroves and coastal regions. Due to financial constraints and the inability to grow in common media, ∼0.01 to 0.1% marine microbes are culturable (Baharum et al., 2010; Antunes et al., 2019). Currently, with the advancement of metagenomics, the approaches have been used to identify new genes involved in the secretion of BS (Araújo et al., 2020). In this review, readers are served with an interesting but brief information on overall diversity of BS produced by marine microbes with a particular emphasis on Planococcus sp. Genomic insights of Planococcus sp. and pathways involved in BS synthesis are considerably evident from the literature. Sparse literature available on the physico-chemical characterization of Planococcus BS is included. We also highlight noteworthy applications of Planococcus-derived BS in hydrocarbon-oil biodegradation, and biomedical fields.
Frequently Reported Marine Microbial Communities for Biosurfactant Production
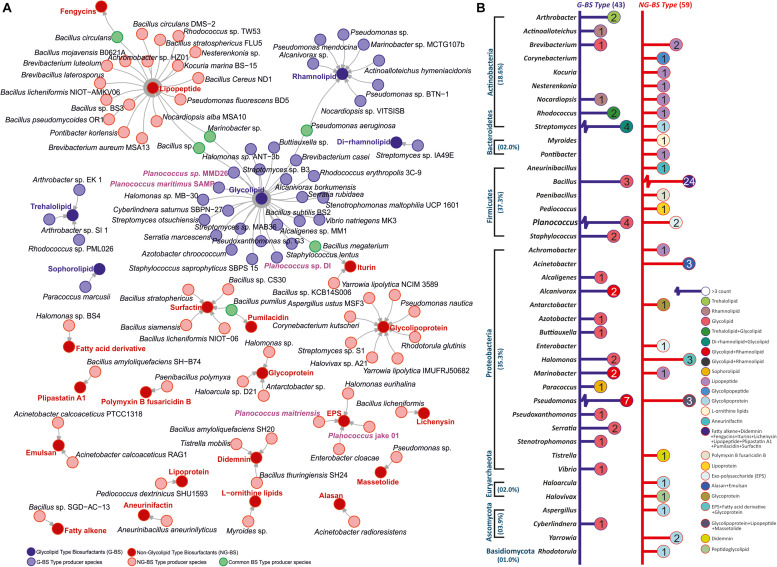
Marine environments have constantly fascinated interest as productive repository port of surface-active biomolecules. This section provides an outline of the respective marine microbial BS producers. Organisms from several phyla like Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, Euryarchaeota, Ascomycota, and Basidiomycota have been reported for the prduction of diverse types of BS (Figure 1A). Total ∼102 reports available on BS producing marine bacteria, representing 43 on glycolipid and 59 on non-glycolipid type. Among the six reports available on Planococcus sp., four are on glycolipid-type BS and two are on exopolysaccharides (EPS). The majority of the reports on BS are focused on Bacillus (25%) and Pseudomonas (11%) followed by Planococcus (6%) and then Halomonas (5%). Serratia, Arthrobacter, Staphylococcus, Brevibacterium, Acinetobacter, and Streptomyces were reported around 2–3% for novel BS production (Mapelli et al., 2017). Some of the recently reported stains include Marinobacter (Tripathi et al., 2019), Cyberlindnera (Balan et al., 2019), Acromobacter (Deng et al., 2016), Buttiauxella (Marzban et al., 2016), and Paracoccus (Antoniou et al., 2015). Nevertheless, BS production by marine microbes remains largely unexplored due to the difficulties to cultivate them in the laboratories (Gudiña et al., 2016; Tripathi et al., 2018; McCarthy et al., 2019). The global demand for BS is the foremost driving force to explore untapped marine microbial diversity (Floris et al., 2018; Twigg et al., 2019). There are many other marine microbes where the entire characterization of BS has not been revealed (Figure 1B). The type, quality, and yield of BS are dependent on microorganisms, their growth conditions, carbon, and nitrogen source, stability to withstand fluctuating pH, temperature, salinity, and agitation (Tripathi et al., 2018; Somoza-Coutiño et al., 2020).
FIGURE 1.
(A) Summary of different types of biosurfactant (BS)/bioemulsifier (BE) producing marine micobial species. (B) Phylum and genus taxon-based segregation of BS/BE producer literature reports (access till April 2020) from marine origin.
Planococcus Species – Untapped Marine Resource for the Production of Biosurfactant and Other Metabolites
Planococcus is a Gram-positive, psychrotolerant, halophilic bacterium, which was initially proposed by Migula (1994) in 1894. Despite having such old history, Planococcus was not reported in the public domain validly for several years. Kocur et al. (1970) proposed the revised description of the genus Planococcus. Initially, there were numerous acceptance and rejections whether to consider these flagellated cocci either under the genus Planococcus or Planosarcina. From 1949 to 1969, a lot of recommendations were given by several researchers to anticipate flagellated cocci under the roof of genus Planococcus considering few features like flagellated structure and % GC content (40–50%) (Kocur et al., 1970). Authors used seven strains having flagellated cocci isolated from a marine source and carried out morphological, cultural, biochemical, and physiological characteristics along with sensitivity to antibiotics and placed them under the genus of Planococcus (Van Hao and Komagata, 1985). The small cocci structures which were previously accommodated as a Micrococci group are presently popular as Planococcus (Cole, 1990; Engelhardt et al., 2001). This is how the successful journey of classification for the genus Planococcus began by considering the size (∼1.4 μm) and cocci (short) shape. Planococcus grows at 25°C within 3–5 days, results in the formation of orange, round, umbonate colonies (Mykytczuk et al., 2013). In 2001, the novel hydrocarbon-degrading P. alkanoclasticus sp. nov. strain MAE2 was isolated and characterized by the sediment of the intertidal beach (Engelhardt et al., 2001). This work was dedicated toward the complete characterization of novel strain by growing in a Bushnell-Haas medium enriched with crude oil (0.5 g). This novel strain was compared with three other species of Planococcus, namely Planococcus kocurii (NCIMB 629T), Planococcus citreus (NCIMB 1493T), and Planococcus okeanokoites (NCIMB 561T) through fatty acid composition and genotypic study (mol % G + C determination, sequencing of 16S rRNA gene) (Engelhardt et al., 2001).
Planococcus sp. strain S5 degrades aromatic hydrocarbons such as salicylate or benzoate through the production of two noticeable enzymes. The authors noted that when strain S5 was grown on salicylate, it produces two types of dioxygenase enzymes (catechol 1,2-dioxygenase and catechol 2,3-dioxygenase) that catalyze the cleavage of oxidative ring catechol to cis and cis-muconic acid (Łabużek et al., 2003). They characterizd those enzymes by growing Planococcus in the presence of high concentration of phenol. Muthukamalam et al. (2017) suggested that enhanced quantity of enzymes produced in presence of hydrocarbon and certainly play a crucial role in degradation process. Abilities of BS production along with these enzyme can compliment microorganisms toward hydrocarbon biodegradation potential.
Kumar et al. (2007) isolated P. maitriensis Anita I (designated as NCBI Gen-Bank Accession number EF467308) from the coastline sea water of Bhavnagar district, Gujarat, India. This group reported on EPS production from P. maitriensis and its emulsifying efficacy. The strain produced EPS consisting of protein (24.44%), carbohydrate (12.06%), uronic acid (11%) along with sulfate (3.03%). This product was purified through centrifugation, precipitation (isopropanol), dialysis, and lyophilisation techniques. The EPS emulsifies xylene powerfully when compared with standard gums. The cell-free culture supernatant also showed a reduction of surface tension (ST) (72 to 46.07 mN/m) and interfacial tension (IFT) against hexane (45.57 ± 0.07 to 20.96 ± 1.56 mN/m) and xylene (33.84 ± 0.19 to 22.95 ± 0.11 mN/m). Thus, for the first time, physical properties of BS from Planococcus appeared in the literature.
Having this information at the background, Ebrahimipour et al. (2014) isolated Planococcus sp. from an oil-contaminated area (Iran) and further reported large-scale production of extracellular BS and utilization of crude oil / hexadecane. Analysis of some of the physical properties like emulsification activity, its stability at different temperatures, pH, and salt tolerance broadens the application of BS. Emulsifying activity of BS demonstrated 77% of bioemulsification activity against crude oil.
Major focus of Kavitha et al. (2015) appeared on investigating the influence of the phase-separated disintegration process in pre-treatment by using P. jake. This experiment was performed in two phases. For the first time authors had demonstrated the effects of CaCl2 (0.06 g/g SS) on deflocculation and further pretreatment with BS producing bacteria in phase separation for biodegradation process. Authors had assessed BS production abilities of culture through various screening assays.
In 2016, Ganapathy et al. (2016) isolated P. maritimus MKU009 from Pichavaram (South East Coast), India, and identified the methyl glucosyl-3,4-dehydro-apo-8-lycopenoate as a novel carotenoid. Four genes (crtN, crtB, crtP, crtI) were reported for carotenoid production located in the first contig of the genome assembly. Such genome-level information is certainly useful to reveal the functional genomics of carotenoid synthesis and metabolic engineering (Han et al., 2019). Thus, metabolic engineering approaches would be supportive to explore additional genetic level information of bioactive metabolites including BS. Yang et al. (2018) had isolated a Planococcus culture from oil-contaminated soil, Qinghai-Tibetan Plateau. Genomic research on Planococcus strain Y42 illustrated the theoretical mechanism of oil degradation which would be helpful for biodegradation of various hydrocarbons.
Recently, we reported P. maritimus strain SAMP for BS production. Based on the physical properties like critical micelle concentration (CMC), ST, IFT, CA (contact angle), wetting properties, and emulsification index proved the BS as a powerful substance. The CMC value of terpene containing BS was found to be 1.3 mg/mL and reduced ST of PBS from 72 to 30 mN/m. The analytical characterization revealed the existence of sugar and lipid moieties in BS. Possibly this was the first report detailing the physico-chemical properties of BS derived form Planococcus sp. (Waghmode et al., 2019).
Response surface methodology (RSM), a statistical Technique is being routinely used to design experiments, build models, evaluate the effect of an interactive variables to plot their optimal conditions and enhance the yield of microbial products. In recent times, Hema et al. (2019) used RSM to enhance the yield of BS (glycolipid) from the wild strain, Planococcus MMD26.
Bioprospecting of Planococcus and Its Biosurfactants, Exopolysaccharide, Bioactive Compounds for Industrial Applications
Biosurfactants have an extensive scope in diverse industries such as pharmaceutics, cosmetics, food, agriculture, textile, petroleum, and wastewater. BSs are well identified as multi-purposeful agents, as antimicrobial, emulsifying, stabilizing, wetting, and moisturizing. Currently, Planococcus-derived BSs attained a prospective significance for environmental applications in remediation of organic/inorganic contaminants, pharmaceutical products, and microbial-enhanced oil recovery (MEOR).
Various microbial communities can efficiently degrade water-insoluble substrates like oils, hydrocarbons through different mechanisms. Firstly, organisms adhere to the oil droplets, aggregating around, and then solubilize them by breaking into smaller chains (Bach and Gutnick, 2004). After creating these miniature droplets, microbial cells interact more efficiently and subsequently degrade hydrocarbons (Karlapudi et al., 2018; Paniagua-Michel and Fathepure, 2018) through BS/BE (Banat et al., 2014). It is an imperative to note that being a marine strain, researchers have mainly targetted the genus Planococcus and/or its BS for bioremediation purposes. Engelhardt et al. (2001) highlighted the abundance of Planococcus sp. MAE2 in a variety of marine/oceanic surroundings, including few Antarctic niches. Authors reported the potential of MAE2 strain in the biodegradation of branched n-alkanes present in crude oil (C11 to C33), but not aromatic hydrocarbons. This study was not associated with any kind of BS production. Thus, the existence of such hydrocarbon-degrading Planococcus sp. is extremely valuable to control contamination by hydrocarbons in marine reservoirs (Park et al., 2018; Somee et al., 2018). Similarly, the strain S5 has been described for degradation of aromatic hydrocarbons like salicylate or benzoate. The S5 strain possesses plasmid (pLS5) having the capacity to degrade salicylate through a metabolic pathway (Łabużek et al., 2003), and the fact was proved through curing experiment. Supportive data for degradation of oil also appeared from the genomic analysis (Yang et al., 2018). Authors have successfully used the Planococcus Y42-derived BS for bioremediation and oil recovery purpose. This work illustrated the exploitation of Planococcus strains for hydrocarbon degradation through understanding the draft genome information (Engelhardt et al., 2001; Jung et al., 2018; Waghmode et al., 2019). The genome of Planococcus sp. PAMC21323 contains the detoxification enzymes of nitroalkanes, aromatic hydrocarbons, and heavy metal ions. Planococcus sp. PAMC21323 has three extradioldioxygenases (Plano 0315, 2898, and 2901) that catalyze the cleavage of aromatic ring structure. Therefore, the genome sequence of this psychrotrophic strain showed the potential application for bioremediation in harsh environments (Santos et al., 2016; Jung et al., 2018).
In addition to bioremediation of hydrocarbons, Planococcus sp. are also known to participate in MEOR processes. Ebrahimipour et al. (2014) endorsed Planococcus sp. for large-scale production of BS and proposed as an extraordinary aspirant for MEOR. Production of BS is associated with the growth of the organism and it has an advantage for a one-stage continuous culture system during scale-up processes.
Planococcus sp. has been reported for production of biopolymer, EPS. In 2007, Kumar et al., proposed that the EPS produced by P. maitriensis Anita I can spread oil and is equivalent in properties with well-known mild nonionic synthetic detergents like Triton X-100 and Tween 80. Therefore, authors recommended its applications in bioremediation, MEOR and cosmetics.
Researcher have utilized the functionality of Planococcus for biogas production. Kavitha et al. (2015) isolated a new strain P. jake 01 and demonstrated its application for biogas production. The, pre-treatment procedures in biogas production through bacteria are significant to diminish the suspended solids and chemical oxygen demand of deflocculated pre-treated sludge. These processes are highly efficient than flocculated sludge (treatment with bacteria only).
Biosurfactants are the most significant and precious microbial products. In the medical field, the BSs act as antibacterial, antiviral, antifungal, anticancer, antioxidants, anti-adhesive, immunomodulator, stimulate dermal fibroblasts, vaccines, and gene therapy (Williams, 2009; Manikandan and Senthilkumar, 2017). In 2016, Ganapathy et al. (2016) reported the production of methyl glucosyl-3,4-dehydro-apo-8-lycopenoate as a novel carotenoid with antioxidant property from P. maritimus MKU009 strain. The foremost study reported by us is on cytotoxic activity of terpene containing Planococcus-derived BS against HeLa, MCF-7, and HCT cells lines at concentrations 41.41 ± 4.214 μg/mL, 42.793 ± 6.072 μg/mL, and 31.233 ± 5.083 μg/mL, respectively. The BS was also exploited against Plasmodium falciparum to determine the killing efficacies of the whole organism. Thus, we have reported the EC50 (34 μM ± 0.26) value against P. falciparum. BS also exhibited anti-tubercular activity IC50 value at 64.11 ± 1.64 μg/mL and MIC at 160.8 ± 1.64 μg/mL concentration (Waghmode et al., 2020). Exploring such applications of Planococcus-derived BS is noteworthy and can encourage the researcher to explore BS further for pharmacological applications.
Functional Genomics Insights of Planococcus Species and Biosynthetic Pathway for the Production of Biosurfactant
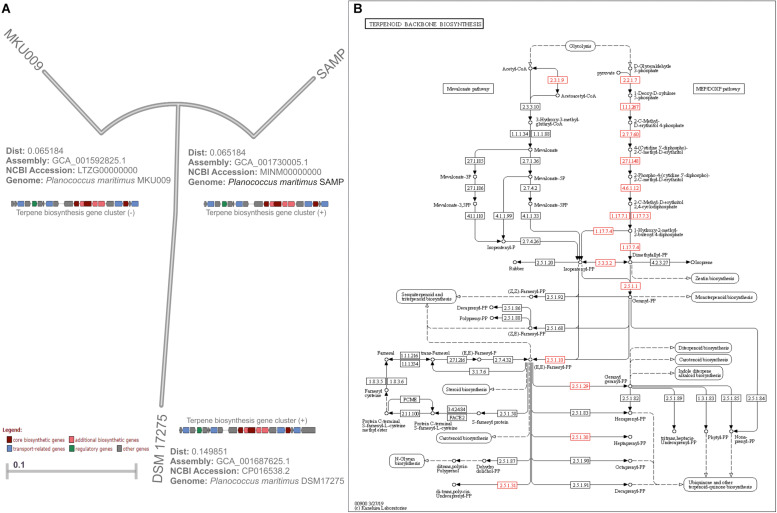
The genomic study provides profound insight into secondary metabolites produced by various bacterial communities, and functional genomics offers the biochemical characteristics of the bioactive compounds (Van Lanen and Shen, 2006; Shaligram et al., 2016; Waghmode et al., 2017). The whole-genome sequencing study of P. maritimus strains revealed an uniform genome size of about 3,220,000 bp for SAMP, 3,280,721 bp for DSM 17275, and 3,251,644 bp for MKU009 (Figure 2A). The similar GC content is reported for all the strains that are about 47.27%. The unique vital genes associated with the SAMP strain was glucose-1-phosphate thymidylyl transferase; for strain DSM 17275, it was cell wall binding repeat two family proteins, and for strain MKU009, it was DUF559 domain-containing protein. The entire gene cluster was screened to identify the secondary metabolite genes. However, the entire gene sets involved in the synthesis of terpene was identified in lipid synthesis pathway. The comparative genome analysis has helped us to identify the genomics potential of all the three strains. The presence of terpene synthesis cluster was observed in genomes of all three strains. From genome analysis, terpene biosynthetic pathway was traced out and it could be the backbone molecule for production of BS (Figure 2B). The location of the gene cluster in all three genomes is different; the entire gene cluster was on the positive strand for two strains DSM17275 and SAMP while for strain MKU009 on the negative strand (Waghmode et al., 2019).
FIGURE 2.
(A) Bonafide genome tree of Planococcus maritimus genomes available from NCBI database (access till April 2020). (B) KEGG database-based metabolic pathway of terpene biosynthesis derived from Planococcus maritimus strain SAMP. Note: Part of the gene cluster image adopted from Waghmode et al. (2019).
Conclusion
Marine microorganisms emerged as a rich source for various natural bioactive compounds. Among the marine microbial world, Planococcus-derived BS offers great interest to the scientific community due to its outstanding functional properties including ST, IFT, wetting, emulsification, stability to a wide range of pH, temperature, salt tolerance, etc. The pharmacological activities presented by BS suggest its potential usage as multipurpose ingredients in various microbial-based commercial products. Furthermore, sampling and consequent enrichment culturing techniques of BS producing Planococcus are essential to explore the potential of BS in many other fields. The accessibility of efficient fast sequencing methods and bioinformatic tools could be helpful in the analysis of biosynthetic pathways. Rapid tracing of the genes involved in BS production and inducers triggering the expression of those particular genes are mandatory to make feasible commercial-scale production. Such metabolic set-up will enable the exposition of the unknown BS biosynthesis pathways of marine microbes. It also facilitates the expansion of synthetic biology-derived concepts to develop competent recombinant strains. These interdisciplinary research tools will contribute toward the identification, production, purification, and application of BS not only from Planococcus sp. but also from other microbes for industrial applications.
Future Prospects
Little is known about the potential of BS production and hydrocarbon degradation by Planococcus sp. of marine origin. Few reports discussed the partial physico-chemical characterization of Planococcus BS. More efforts are mandatory to evaluate BS production on a larger scale and to find their role in various technologies. More information is essential for (1) predicting the structural details of Planococcus-derived BS, (2) identifying their properties, (3) scale-up and (4) cost-effective production studies. To reduce the cost of BS production, commercially feasible biological and engineering solutions can be employed. The use of low-cost substrates appears to be a promising approach to enhance the productivity of BS commercially. More efforts are indispensable to explore marine BS for additional pharmacological applications. Molecular docking studies also provide new avenues to discover the possible target sites for effective interactions of BS. A detailed investigation in this particular area would offer lead molecules for bio-therapeutics. However, applications of marine microbial originated BS in various industrial sectors are awaited and thus provide enormous opportunities to utilize them in food, cosmetics, agriculture, etc.
Author Contributions
SW, MS, DS, and SS contributed to the literature search, the design of the figure, and the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- BS
biosurfactant
- BE
bioemulsifier
- CMC
critical micelle concentration
- CA
contact angle
- EPS
exopolysaccharide
- IFT
interfacial tension
- MEOR
microbial-enhanced oil recovery
- RSM
response surface methodology
- ST
surface tension.
Footnotes
Funding. SS expresses special thanks to Rashtriya Uchchatar Shiksha Abhiyan (Reference No: RUSA-CBS-TH-3.2) for the financial support.
References
- Antoniou E., Fodelianakis S., Korkakaki E., Kalogerakis N. (2015). Biosurfactant production from marine hydrocarbon-degrading consortia and pure bacterial strains using crude oil as carbon source. Front. Microbiol. 6:274. 10.3389/fmicb.2015.00274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes J., Leão P., Vasconcelos V. (2019). Marine biofilms: diversity of communities and of chemical cues. Env. Microbiol. Rep. 11 287–305. 10.1111/1758-2229.12694 [DOI] [PubMed] [Google Scholar]
- Araújo S. C. D. S., Silva-Portela R. C. B., de Lima D. C., da Fonsêca M. M. B., Araújo W. J., da Silva U. B., et al. (2020). MBSP1: a biosurfactant protein derived from a metagenomic library with activity in oil degradation. Sci. Rep. 10 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach H., Gutnick D. L. (2004). Potential applications of bioemulsifiers in the oil industry. petroleum biotechnology developments and perspectives. Pet. Biotechnol. 151 233–281. 10.1016/s0167-2991(04)80150-2 [DOI] [Google Scholar]
- Baharum S. N., Beng E. K., Mokhtar M. A. A. (2010). Marine microorganisms: potential application and challenges. J. Biol. Sci. 10 555–564. 10.3923/jbs.2010.555.564 [DOI] [Google Scholar]
- Balan S. S., Kumar C. G., Jayalakshmi S. (2019). Physicochemical, structural and biological evaluation of Cybersan (trigalactomargarate), a new glycolipid biosurfactant produced by a marine yeast. Cyberlindnera saturnus strain SBPN-27. Process Biochem. 80 171–180. 10.1016/J.PROCBIO.2019.02.005 [DOI] [Google Scholar]
- Banat I. M., Satpute S. K., Cameotra S. S., Patil R., Nyayanit N. V. (2014). Cost effective technologies and renewable substrates for biosurfactants’ production. Front. Microbiol. 5:697. 10.3389/fmicb.2014.00697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R., Jr. (1990). Micrococcus and Staphylococcus. in Diagnostic Procedure in Veterinary Bacteriology and Mycology. Cambridge, MA: Academic Press, 201–209. [Google Scholar]
- de Carvalho C. C. (2018). Marine biofilms: a successful microbial strategy with economic implications. Front. Mar. Sci. 5:126 10.3389/fmars.2018.00126 [DOI] [Google Scholar]
- Deng M. C., Li J., Hong Y. H., Xu X. M., Chen W. X., Yuan J. P., et al. (2016). Characterization of a novel biosurfactant produced by marine hydrocarbon-degrading bacterium Achromobacter sp. HZ 01. J. Appl. Microbiol. 120 889–899. 10.1111/jam.13065 [DOI] [PubMed] [Google Scholar]
- Ebrahimipour G., Gilavand F., Karkhane M., Kavyanifard A., Teymouri M., Marzban A. (2014). Bioemulsification activity assessment of an indigenous strain of halotolerant Planococcus and partial characterization of produced biosurfactants. Int. J. Environ. Sci. Technol. 11 1379–1386. 10.1007/s13762-014-0548-5 [DOI] [Google Scholar]
- Engelhardt M. A., Daly K., Swannell R. P., Head I. M. (2001). Isolation and characterization of a novel hydrocarbon-degrading, Gram-positive bacterium, isolated from intertidal beach sediment, and description of Planococcus alkanoclasticus sp. nov. J. Appl. Microbiol. 90 237–247. 10.1046/j.1365-2672.2001.01241.x [DOI] [PubMed] [Google Scholar]
- Floris R., Rizzo C., Giudice A. L. (2018). Metabolomics-New Insights into Biology and Medicine. London: IntechOpen. [Google Scholar]
- Ganapathy A., Jayavel S., Natesan S. (2016). Draft genome sequence of carotenoid producing yellow pigmented Planococcus maritimus MKU009. J. Genomics 4 23–25. 10.7150/jgen.15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geist J., Hawkins S. J. (2016). Habitat recovery and restoration in aquatic ecosystems: current progress and future challenges. Aquat. Conserv. Mar. Freshwater Ecosyst. 26 942–962. 10.1002/aqc.2702 [DOI] [Google Scholar]
- Gudiña E. J., Fernandes E. C., Teixeira J. A., Rodrigues L. R. (2015). Antimicrobial and anti-adhesive activities of cell-bound biosurfactant from Lactobacillus agilis CCUG31450. RSC Adv. 5 909–960. [Google Scholar]
- Gudiña E. J., Teixeira J. A., Rodrigues L. R. (2016). Biosurfactants produced by marine microorganisms with therapeutic applications. Mar. Drugs. 14:38. 10.3390/md14020038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Long L., Ding S. (2019). Expression and characterization of carotenoid cleavage oxygenases from Herbaspirillum seropedicae and rhodobacteraceae bacterium capable of biotransforming isoeugenol and 4-vinylguaiacol to vanillin. Front. Microbiol. 10:1869. 10.3389/fmicb.2019.01869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hema T., Kiran G. S., Sajayyan A., Ravendran A., Raj G. G., Selvin J. (2019). Response surface optimization of a glycolipid biosurfactant produced by a sponge associated marine bacterium Planococcus sp. MMD26. Biocatal. Agric. Biotechnol. 18:101071 10.1016/j.bcab.2019.101071 [DOI] [Google Scholar]
- Jung J. H., Joe M. H., Kim D. H., Park H., Choi J. I., Lim S. (2018). Complete genome sequence of Planococcus sp. PAMC21323 isolated from Antarctica and its metabolic potential to detoxify pollutants. Stand. Genomic Sci. 13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlapudi A. P., Venkateswarulu T. C., Tammineedi J., Kanumuri L., Ravuru B. K., Dirisala V., et al. (2018). Role of biosurfactants in bioremediation of oil pollution-a review. Pet 4 241–249. 10.1016/j.petlm.2018.03.007 [DOI] [Google Scholar]
- Kavitha S., Saranya T., Kaliappan S., Adishkumar S., Yeom I. T., Rajesh Banu J. (2015). Accelerating the sludge disintegration potential of a novel bacterial strain Planococcus jake 01 by CaCl2 induced deflocculation. Bioresour. Technol. 175 396–405. 10.1016/j.biortech.2014.10.122 [DOI] [PubMed] [Google Scholar]
- Kocur M., Zdena P., Hodgkiss W., Martinec T. (1970). The taxonomic status of the genus Planococcus Migula 1894. Int. J. Syst. Evol. Microbiol. 20 241–248. 10.1099/00207713-20-3-241 [DOI] [Google Scholar]
- Kumar A. S., Mody K., Jha B. (2007). Evaluation of biosurfactant/bioemulsifier production by a marine bacterium. Bull. Environ. Contam. Toxicol. 79 617–621. 10.1007/s00128-007-9283-7 [DOI] [PubMed] [Google Scholar]
- Łabużek S. Y. L. W. I. A., Hupert-Kocurek K., Skurnik M. I. K. A. E. L. (2003). Isolation and characterisation of new Planococcus sp. strain able for aromatic hydrocarbons degradation. Acta Microbiol. Pol. 52 395–404. [PubMed] [Google Scholar]
- Manikandan P., Senthilkumar P. K. (2017). On overview of saltpan halophilic bacterium. Antimicrob. Agents 3 1–5. [Google Scholar]
- Mapelli F., Scoma A., Michoud G., Aulenta F., Boon N., Borin S., et al. (2017). Biotechnologies for marine oil spill cleanup: indissoluble ties with microorganisms. Trends Biotechnol. 35 860–870. 10.1016/j.tibtech.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Marzban A., Ebrahimipour G., Danesh A. (2016). Bioactivity of a novel glycolipid produced by a halophilic Buttiauxella sp. and improving submerged fermentation using a response surface method. Molecules 21:1256. 10.3390/molecules21101256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy A. H., Peck L. S., Hughes K. A., Aldridge D. C. (2019). Antarctica: the final frontier for marine biological invasions. Global Change Biol. 25 2221–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant S. S., Helmann J. D. (2012). Elemental economy: microbial strategies for optimizing growth in the face of nutrient limitation. Adv. Microb. Physiol. 60 91–210. 10.1016/b978-0-12-398264-3.00002-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migula W. (1894). Ubereinneues system der bacterien. Arb. Bakteriol. Inst. Karlsruhe. 1 235–238. [Google Scholar]
- Muthukamalam S., Sivagangavathi S., Dhrishya D., Sudha Rani S. (2017). Characterization of dioxygenases and biosurfactants produced by crude oil degrading soil bacteria. Brazilian J. Microbiol. 48 637–647. 10.1016/j.bjm.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytczuk N. C., Foote S. J., Omelon C. R., Southam G., Greer C. W., Whyte L. G. (2013). Bacterial growth at −15°C; molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J. 7 1211–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton P. J., Marchant R., Naughton V., Banat I. M. (2019). Microbial biosurfactants: current trends and applications in agricultural and biomedical industries. J. App. Microbiol. 127 12–28. 10.1111/jam.14243 [DOI] [PubMed] [Google Scholar]
- Paniagua-Michel J., Fathepure B. Z. (2018). “Microbial consortia and biodegradation of petroleum hydrocarbons in marine environments,” in Microbial Action on Hydrocarbons, eds Kumar V., Kumar M., Prasad R. (Singapore: Springer; ). [Google Scholar]
- Park J. S., Park Y. J., Kim B. S., Lee B. H., Park M. G., Yang J. (2018). “Evaluation using ethyl formate and nitrogen to control Planococcus citri (Risso) on import bananas,” in Proceedings of the Journal of the Korean Society of Applied Insectology Conference, 2018, Joint International Conference on Crop Protection, 509–509. [Google Scholar]
- Santos D. K. F., Rufino R. D., Luna J. M., Santos V. A., Sarubbo L. A. (2016). Biosurfactants: multifunctional biomolecules of the 21st century. Int. J. Mol. Sci. 17:401. 10.3390/ijms17030401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute S. K., Banat I. M., Dhakephalkar P. K., Banpurkar A. G., Chopade B. A. (2010). Biosurfactants, bioemulsifiers and exopolysaccharides from marine microorganisms. Biotechnol. Adv. 28 436–450. 10.1016/j.biotechadv.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Shaligram S., Kumbhare S. V., Dhotre D. P., Muddeshwar M. G., Kapley A., Joseph N., et al. (2016). Genomic and functional features of the biosurfactant producing Bacillus sp. AM13. Funct. Integr. Genom. 16 557–566. 10.1007/s10142-016-0506-z [DOI] [PubMed] [Google Scholar]
- Somee M. R., Shavandi M., Dastgheib S. M. M., Amoozegar M. A. (2018). Bioremediation of oil-based drill cuttings by a halophilic consortium isolated from oil-contaminated saline soil. 3 Biotech. 8:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoza-Coutiño G., Wong-Villarreal A., Blanco-González C., Pérez-Sariñana B., Mora-Herrera M., Mora-Herrera S. I., et al. (2020). A bacterial strain of Pseudomonas aeruginosa B0406 pathogen opportunistic, produce a biosurfactant with tolerance to changes of pH, salinity and temperature. Microb. Pathogenesis 139:103869. 10.1016/j.micpath.2019.103869 [DOI] [PubMed] [Google Scholar]
- Tripathi L., Irorere V. U., Marchant R., Banat I. M. (2018). Marine derived biosurfactants: avast potential future resource. Biotechnol. Lett. 40 1441–1457. 10.1007/s10529-018-2602-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L., Twigg M. S., Zompra A., Salek K., Irorere V. U., Gutierrez T., et al. (2019). Biosynthesis of rhamnolipid by a Marinobacter species expands the paradigm of biosurfactant synthesis to a new genus of the marine microflora. Microb. Cell Fact. 18:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg M., Tripathi L., Zompra K., Salek K., Irorere V., Gutierrez T., et al. (2019). Surfactants from the sea: rhamnolipid production by marine bacteria. Access Microbiol. 1:192. [Google Scholar]
- Van Hao M. A. I., Komagata K. (1985). A new species of Planococcus, P. kocurii isolated from fish, frozen foods, and fish curing brine. J. Gen. Appl. Microbiol. 31 441–455. 10.2323/jgam.31.441 [DOI] [Google Scholar]
- Van Lanen S. G., Shen B. (2006). Microbial genomics for the improvement of natural product discovery. Curr. Opin. Microbiol. 9 252–260. 10.1016/j.mib.2006.04.002 [DOI] [PubMed] [Google Scholar]
- Waghmode S., Dama L., Hingamire T., Bharti N., Doijad S., Suryavanshi M. (2017). Draft genome sequence of a biosurfactant producing, Bacillus aquimaris strain SAMM MCC 3014 isolated from Indian Arabian coastline sea water. J. Genom. 5 124–127. 10.7150/jgen.21724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmode S., Suryavanshi M., Dama L., Kansara S., Ghattargi V., Das P., et al. (2019). Genomic insights of halophilic Planococcus maritimus SAMP MCC 3013 and detail investigation of its biosurfactant production. Front. Microbiol. 10:235. 10.3389/fmicb.2019.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghmode S., Swami S., Sarkar D., Suryavanshi M., Roachlani S., Choudhari P., et al. (2020). Exploring the pharmacological potentials of biosurfactant derived from Planococcus maritimus SAMP MCC 3013. Curr. Microbiol. 77 452–459. 10.1007/s00284-019-01850-1 [DOI] [PubMed] [Google Scholar]
- Wang L., D’Odorico P. (2013). Decomposition and mineralization. Encycl. Ecol. 2 280–285. 10.1016/b978-0-12-409548-9.00688-6 [DOI] [Google Scholar]
- Williams K. J. (2009). Biosurfactants for cosmetic application: overcoming production challenges. Basic Biotechnol. e - J. 5 78–83. [Google Scholar]
- Yang R., Liu G., Chen T., Zhang W., Zhang G., Chang S. (2018). The complete genomic sequence of a novel cold-adapted bacterium, Planococcus maritimus Y42, isolated from crude oil-contaminated soil. Stand. Genomic Sci. 13:23. [DOI] [PMC free article] [PubMed] [Google Scholar]