Abstract
Friedreich's ataxia (FRDA) is the most common inherited form of ataxia in humans. It is caused by severe downregulation of frataxin (FXN) expression instigated by hyperexpansion of the GAA repeats located in intron 1 of the FXN gene. Despite numerous studies focused on identifying compounds capable of stimulating FXN expression, current knowledge regarding cis-regulatory elements involved in FXN gene expression is lacking. Using a combination of episomal and genome-integrated constructs, we defined a minimal endogenous promoter sequence required to efficiently drive FXN expression in human cells. We generated 19 constructs varying in length of the DNA sequences upstream and downstream of the ATG start codon. Using transient transfection, we evaluated the capability of these constructs to drive FXN expression. These analyses allowed us to identify a region of the gene indispensable for FXN expression. Subsequently, selected constructs containing the FXN expression control regions of varying lengths were site specifically integrated into the genome of HEK293T and human-induced pluripotent stem cells (iPSCs). FXN expression was detected in iPSCs and persisted after differentiation to neuronal and cardiac cells, indicating lineage independent function of defined regulatory DNA sequences. Finally, based on these results, we generated AAV encoding miniFXN genes and demonstrated in vivo FXN expression in mice. Results of these studies identified FXN sequences necessary to express FXN in human and mouse cells and provided rationale for potential use of endogenous FXN sequence in gene therapy strategies for FRDA.
Keywords: Friedreich's ataxia, transcription control, frataxin expression, gene therapy, endogenous promoter, induced pluripotent stem cells
Introduction
Friedreich's ataxia (FRDA, OMIM no. 229300; ORPHA:95) is an autosomal recessive inherited disease that causes impairment to several systems and organs, including but not limited to the peripheral and central nervous systems, heart, and pancreas.1–3 In the initial phases, FRDA manifests in areflexia, poor coordination, gait disturbance followed by speech impairment, skeletal abnormalities, frequently cardiomyopathy, and diabetes. The disease is progressive, leading to restricted mobility and significantly shortened life expectancy. FRDA incidence is ∼1:20,000–1:50,000 in the population of Indo European and North African descendent, with a carrier frequency of 1:60–1:100.4
The FXN gene, mutations of which are responsible for this disease, was identified in 1996, along with expansion of the trinucleotide repeat GAA representing the most common mutation type leading to FRDA.1 Encompassing typically hundreds of copies, these large expansions of the GAA repeats cause severe transcriptional downregulation of FXN expression.5,6 Importantly, the aberrantly expanded GAA trinucleotide repeats are located in the first intron of the gene, and thus, the coding sequence of frataxin, protein product of the FXN gene, is unaffected by the mutation. Because FRDA is a recessive disorder, both FXN alleles carry a mutation. In ∼96% of cases, it is a homozygous GAA expansion. In the remaining 4% of patients, a single allele containing the expanded GAAs is accompanied by a second allele bearing a point mutation.3,7,8
The FXN gene is located on chromosome 9q21.11 in relative proximity to the centromere region and spans ∼43 kbp. The pathogenic GAA repeats, likely evolved from an ancient Alu element, are located ∼1,500 bp from the ATG translation start codon.9,10 Although numerous studies point toward transcriptional initiation and/or elongation being affected by the expanded triplets, current knowledge regarding transcription control of the FXN locus in nonmutated alleles is limited.5,11–15 All studies reported thus far utilized episomal vectors containing the luciferase gene under the control of putative FXN promoter regions.10,16 The “strength” of the promoter and importance of certain regions were defined relative to the luciferase expression in constructs containing nonmodified fragments of the FXN gene. Therefore, the actual activity of these exogenous constructs in relation to the endogenous frataxin expression levels could not be determined. The initial reporter studies indicated that critical regions for FXN expression are located within a 1,255 bp region upstream of the start codon.10 Subsequent studies, also conducted using transfected episomal constructs, indicated that a fragment of intron 1, including a putative E-box motif located in the proximity of the GAA repeats, was necessary for expression.16 In addition, roles for p53, NRF2, SRF, and TFAP2 transcription factors on the expression of FXN gene were also demonstrated.17–21 Taken together, the results of these studies conducted with luciferase reporters indicated that the region required for efficient FXN transcription may encompass a relatively large fragment of the gene extending hundreds of base pairs upstream and downstream of the ATG start codon. However, the exact minimal promoter driving expression of FXN in its genomic context has not been elucidated.
Frataxin is a small, evolutionary conserved protein produced as a 210 amino acid precursor and cleaved into the mature 130 amino acid form upon import to the mitochondria.22 Ubiquitously expressed FXN participates in the biosynthesis of iron/sulfur clusters in the mitochondrial matrix, and its complete knockout is early embryonic lethal.22 Downregulation of FXN expression in FRDA cells results in progressive deterioration of intracellular homeostasis, metabolic changes, and oxidative damage consequently leading to cellular degeneration.3,23,24 Rescue of frataxin levels, via increase of transcription, protein stabilization, or exogenous gene or protein supplementation, represents a leading therapeutic strategy in FRDA.15,25 However, a growing number of reports indicate that an uncontrolled increase of frataxin levels significantly above physiological levels may lead to oxidative damage and cell death, which are changes similar to FXN downregulation.26–30 This suggests that a certain cellular tolerance exists for fluctuations in FXN levels, but moving out of a tolerable range (in any direction) may have detrimental consequences. Thus, defining the endogenous promoter may facilitate proper regulation of FXN expression, especially in light of current gene therapy efforts. In addition, excision of the intronic expanded GAAs has been demonstrated in proof-of-concept studies to be an efficient, permanent correction of the genetic defect underlying FRDA.31–33 Uncovering a potential regulatory element within intron 1 is of critical importance for judicial selection of cleavage sites for designer nucleases targeting the expanded GAAs.
Herein, we conducted a systematic study of the FXN promoter region via direct measurements of intracellular frataxin levels. We focused on defining a minimal endogenous FXN sequence capable of producing endogenous or near-endogenous levels of the protein. We conducted initial analyses using episomal vectors, followed by testing functionality of the FXN promoter in the genome of HEK293T cells, human-induced pluripotent stem cells (iPSCs), and neuronal and cardiac cells derived from iPSCs. Finally, we established a set of minimal frataxin constructs that contain an essential upstream ATG regulatory region with reduced intron 1 and remaining FXN sequences that can be easily packaged into AAV particles, suitable for evaluation as a gene therapy approach. We validated new AAV-packaged miniFXN genes by intracerebroventricular (ICV) delivery to mice and determined frataxin expression in tissues.
Materials and Methods
Design and cloning of miniFXN containing plasmids
All miniFXN constructs were derived from the longest miniFXN 1 gene (Supplementary Fig. S1) and cloned into the pCR4 vector (Invitrogen 45-0030). Progressive truncation of intron 1 was achieved by NsiI/KpnI (miniFXN 2), NsiI/PmlI (miniFXN 3), NsiI/AgeI (miniFXN 4), and NsiI/BssHII (miniFXN 5) digestion of the miniFXN 1 gene. A miniFXN 6 gene (lacking intron 1) was constructed by replacing the ClaI/BspEI fragment of miniFXN 1 (encompassing FXN exon 1–exon 5) with a fragment of the FXN cDNA lacking intronic sequence.
Plasmids miniFXN 7 and miniFXN 10 were obtained by EcoRI/PacI digestion of miniFXN 3 to generate constructs with shorter 5′ untranslated region (5′UTR) sequences. Plasmids miniFXN 8 and miniFXN 11 were obtained by EcoRI/PacI digestion of the miniFXN 4, construct miniFXN 12 was derived from miniFXN 5 by EcoRI/PacI digestion, and plasmid miniFXN 9 was obtained from miniFXN 6 by EcoRI/PacI digestion. Construct miniFXN 13 (lacking 5′UTR) was obtained by cloning of the polymerase chain reaction (PCR) product generated with primers Exon1_end_F (5′ AGCAGCATGTGGACTCTC) and Flagxho_R (5′ GCTCTCGAGTCACTTATCGTCGTCATCCTTGTAATC), complementary to the 5′end of exon 1 and Flag tag sequence. Construct miniFXN 8 was used as a PCR template. Plasmids miniFXN 14 and miniFXN 15 are derived from minFXN 7 and miniFXN 8 to introduce MluI (at the 5′ end) and XhoI (at the 3′ end) for subsequent cloning into pAAV-minCMV-mCherry (Addgene no. 27970) for AAV generation. MiniFXN 16 was derived from miniFXN 15 by AgeI/BssHII digestion. Further shortening of the 5′UTR sequence in miniFXN 17, 18, and 19 was achieved by fragment replacement mutagenesis. In all constructs, sequences of frataxin and all regulatory elements were verified by Sanger sequencing at Genewiz or the UAB Heflin Center for Genomic Sciences.
HEK293T transfection and Western blot
HEK293T cells were cultured in DMEM high-glucose medium (Life Technologies 11965) supplemented with 10% FBS (HyClone SH30396.03) at 37°C, 5% CO2. A day before transfection, 1 × 10e6 cells were seeded per well of six-well plate. Up to 2.5 μg of plasmid DNA adjusted for the same copy number of plasmids depending on total length of the constructs was combined with 5 μL of Lipofectamine 2000 (Invitrogen 2066194) in 200 μL of Opti-MEM (Life Technologies 31985-070) and added to the cells. After 48 h, cells were harvested and proteins isolated and electrophoresed on sodium dodecyl sulphate-polyacrylamide gel electrophoresis, as described in Li et al.5,31 Frataxin was detected using an FXN-specific antibody (GeneTex, GTX 54036; 1:2,000) and a secondary anti-rabbit peroxidase-linked antibody (GE Healthcare; LNA934; 1:10,000).
Integration of miniFXN constructs into AAVS1 safe harbor locus
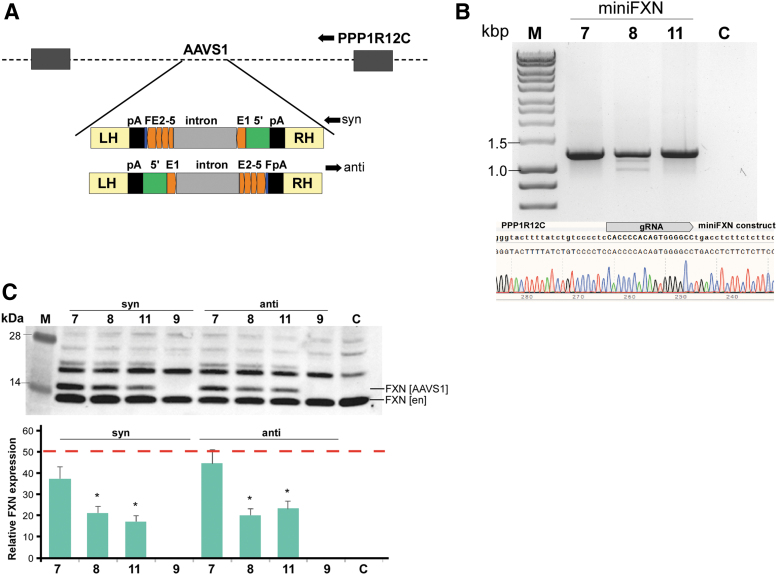
Selected miniFXN genes 7, 8, 9, and 11 were cloned into the EcoRV site of pZDonor-AAVS1 puromycin vector (Sigma, PZD0020) in two opposite orientations relative to the left and right homology arms (i.e., PPP12R1C transcription direction; Fig. 3A). The miniFXN containing inserts were excised by ZraI/PshAI digestion. HEK293T cells were transfected using Lipofectamine 2000 with 2 μg of pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene no. 42230) encoding SpCas9 and expressing gRNA and 2 μg of pZDonor-AAVS1 puromycin containing miniFXN constructs. Two days after transfection, cells were split into fresh wells of a 6-well plate and subjected to selection by 2 μg/mL puromycin for 1 week. After subsequent passage, cells were analyzed for proper integration using PCR (primers AAVS1_Forward/Puro_Reverse; 5′ CGGAACTCTGCCCTCTAACG and 5′ TGAGGAAGAGTTCTTGCAGCT) and Western blot to determine frataxin expression.
Figure 3.
MiniFXN genes are expressed efficiently after integration into the genome. (A) Schematic of CompoZr pZdonor-AAVS1 vector harboring miniFXN gene in two opposite orientations (termed syn- and anti-) relative to the transcription of the endogenous PPP1R12C gene. LH and RH are left and right homology arms allowing for recombination with the genomic AAVS1 site. The miniFXN genes are flanked by SV40 polyadenylation signals (pA, black bars); (B) junction PCR and DNA sequencing confirming proper integration of the miniFXN genes into the AAVS1 site. Location of the gRNA used for integration of the miniFXN constructs is indicated. MiniFXN genes 7, 8, 9, and 11 (as shown in Figure 2) were separately integrated into the AAVS1 site. M—DNA molecular-weight marker (HyperLadder™ 1kb). (C) FXN expression from miniFXN genes 7, 8, 9, and 11 integrated into the AAVS1 site in syn- and antiorientations relative to endogenous PPP1R12C transcription. C—untransfected control HEK293T cells. M—molecular-weight protein marker. Quantitation of FXN expression driven from miniFXN genes integrated into the AAVS1 locus (FXN [AAVS1]) determined relative to the amount of endogenous FXN expression (FXN [en]); red dashed line indicates expression of FXN from the AAVS1 locus at a level of 50% of endogenous FXN. Quantitative data shown as an average of three independent Western blot analyses were compared by one-way ANOVA, and levels of miniFXN genes 8 and 11 were statistically different than miniFXN gene 7; *p < 0.05. ANOVA, analysis of variance; PCR, polymerase chain reaction.
The same vectors were used to integrate miniFXN 7 into the AAVS1 locus in the FRDA iPSC line 4676. The miniFXN 7 was integrated in the -cis orientation relative to PPP12R1C transcription. Transfection of iPSCs (3 × 10e5 cells) was performed using 4 μL of the Lipofectamine Stem Transfection Reagent (Thermo Fisher Scientific STEM00008), 4 μg of pX330-U6-Chimeric_BB-CBh-hSpCas9, and 4 μg of pZDonor-AAVS1 puromycin vector according to the manufacturer's recommendations.
The transfection of vectors for integration delivers a limited number of DNA molecules to each cell. Considering the relatively low efficiency of Cas9 cleavage, single-integration events per cell are significantly more likely than double, homozygous integration events. Prior studies also indicated that homozygous AAVS1 integration is rarely observed.34
Culturing iPSCs, neuronal and cardiac differentiation
The iPSCs were cultured using mTeSR-1 (StemCell Technologies) as we described.32 Cells were differentiated into cardiomyocytes using the Gibco® PSC Cardiomyocyte Differentiation Kit (Gibco, A2921201) according to the manufacturer's recommendations. Neuronal cells were obtained via Ngn2 overexpression-mediated differentiation according to Ref.35 Plasmids FUW-TetO-Ng2-P2A-T2A-puromycin and FUW-rtTA were obtained from Dr. Thomas Sudhof's laboratory.
AAV packaging and HEK293T transduction
A triple-plasmid transfection protocol was used to produce recombinant AAV vectors. Briefly, the transfected plasmids include a capsid-specific pXR helper plasmid (i.e., pXR9), the adenoviral helper plasmid pXX680, and either AAV-miniFXN 8 or AAV-miniFXN 7 plasmid (encoding a frataxin minigene driven by different endogenous promoters), flanked by inverted terminal repeats (TRs) derived from the AAV2 genome. Vectors were purified from media supernatant by PEG precipitation followed by iodixanol density gradient ultracentrifugation. Recombinant vectors were subsequently subjected to buffer exchange, desalting using ZebaSpin® columns (40,000 MWCO, Thermo Scientific, catalog no. 87770), and concentrated using Pierce Protein PES centrifugation columns (100,000 MWCO, Thermo Scientific, catalog no. 88524). Following purification, vector genome titers were determined via quantitative PCR using a Roche LightCycler 480 (Roche Applied Sciences). Primers were designed to specifically recognize the AAV2 inverted terminal repeats (forward, 5′-AACATGCTACGCAGAGAGGGAGTGG-3′; reverse, 5′-CATGAGACAAGGAACCCCTAGTGATGGAG-3′). Transduction of HEK293T cells was performed using 100,000 viral genomes (vg) per cell in a six-well plate. Seventy-two h after transduction, cells were harvested for Western blot analyses of frataxin expression.
ICV injection of miniFXN-AAV into mice
All animal procedures were approved by the University of Alabama at Birmingham Animal Care and Use Committee (protocol 21279) and in accordance with the NIH Guide for Care and Use of Laboratory Animals. Bilateral ICV injections were performed as previously described36 in C57BL/6 mouse pups on postnatal day 0 or 1. Briefly, newborn C57BL/6 mice were cryoanesthetized and subsequently placed on a cold plate. A Hamilton Neuros Model 1701 RN 10 μL syringe with 33-gauge needle was used to pierce the skull ∼0.8–1 mm lateral from the sagittal suture, halfway between the lambda and bregma to a depth of ∼3 mm. Three microliters of PBS or miniFXN AAV9 (at 2.6e9 vg/uL–2e10 vg/μL) was injected into each cerebral ventricle. Neonatal mice were kept with their mothers until sacrificed at day 14 postinjection. The brain, spinal cord, heart, and liver were isolated, snap-frozen in liquid nitrogen, and stored at −80°C for protein expression analyses.
Results
Location of putative regulatory elements in the FXN promoter region
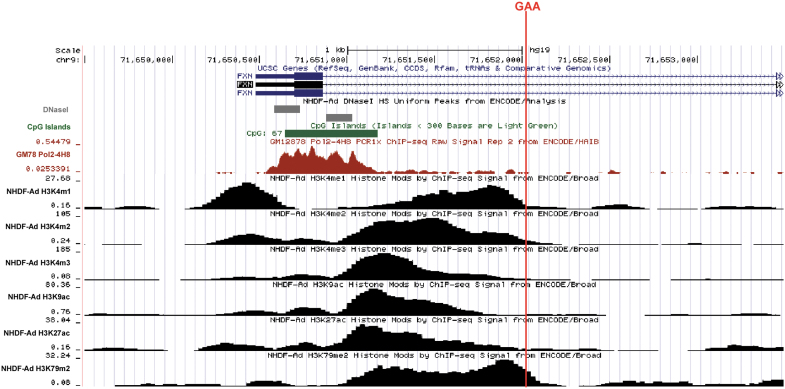
FXN belongs to the group of genes with CpG promoters lacking typical TATA box and containing a CpG island in the vicinity of the transcription start site (TSS, Fig. 1).37–39 Although the major TSS has been annotated 220 bp (−220) upstream of the ATG, alternative TSSs were reported.37–39 Exon 1 contains 165 bp and encodes 55 amino acids (aa) of the N-terminal fragment of precursor frataxin. Intron 1 contains 10,419 bp, excluding the polymorphic GAAs, and the repeat tract begins 1,339 bp downstream of the exon1/intron 1 junction (Fig. 1). Analyses of the TSS region using data sets available from the Encode project demonstrate accumulation of RNA polymerase II (RNAPII) and presence of a DNAse I hypersensitive region in the vicinity of the putative TSSs. In addition, acetylation of histone at lysine 27 (H3K27ac) and methylation of histone H3 at lysine 4 (H3K4me1,2,3) chromatin marks typically present in regulatory/enhancer regions are enriched downstream of the TSS, encompass exon 1, and extend into the proximal region of intron 1 (Fig. 1). These data gathered from various cell types (all non-FRDA samples), together with prior studies, demonstrate that both exon 1 and sequences in intron 1 (adjacent to exon 1) may play a role in the initiation of the FXN transcription.16
Figure 1.
Regulatory elements and chromatin landscape in the vicinity of the FXN TSS. University of California, Santa Cruz Genome Browser (GRCh37/hg19) presentation of chromatin modifications and other regulatory elements present in the FXN gene of an unaffected individual. Location of the GAA repeats is indicated by a red line. When possible, data obtained by the Encode consortium using NHDF are presented. Location of DNaseI hypersensitive sites and RNA polymerase II are shown. The presence of transcription-specific histone H3 covalent modifications (H3: K4me1, K4me2, K4me3, K9ac, K27ac, and K79me2) is presented. FXN, frataxin; NHDF, normal dermal human fibroblasts; TSS, transcription start site.
Minimal intron 1 sequence is required for FXN expression
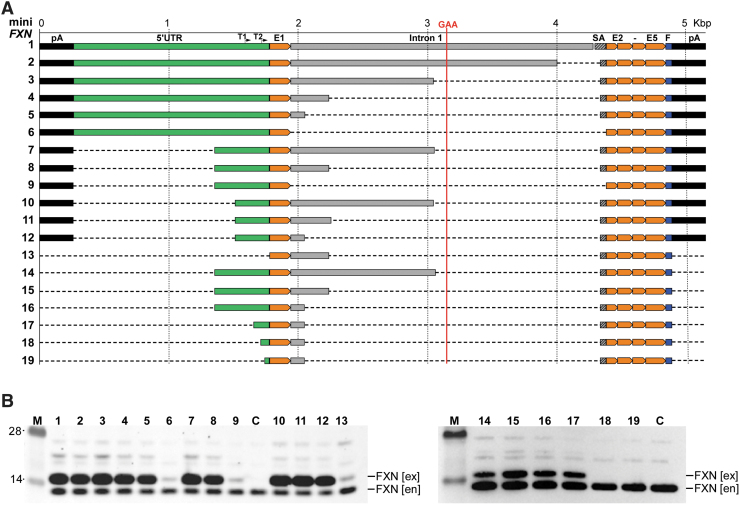
To conduct a systematic analysis of the endogenous sequences required for efficient expression of frataxin, we generated 19 vectors containing different lengths of the putative promoter region for the FXN gene (Fig. 2A, 1–19). Importantly, no reporter gene was fused to these constructs; instead, the entire FXN gene sequence was transcribed, and levels of frataxin were used as a quantitative readout of the promoter strength. Episomal vectors were constructed on the pCR4 backbone and included an additional termination/polyadenylation signal (pA) upstream of the 5′ end of the mini-FXN genes to insulate from any unlikely transcription initiated from the vector sequences. The longest miniFXN gene (miniFXN 1) construct was prepared by assembling different PCR products of the normal human FXN gene promoter region. It contained a fragment from 1,513 bp upstream to 2,500 bp downstream of FXN start codon, linked to 95 bp of intron 1 sequences upstream of exon 2 (for efficient splicing). The remaining exons 2–5 were cloned as a continuous cDNA sequence without respective intronic sequences followed by an in-frame C-terminal Flag epitope tag, allowing for distinction between endogenous and mini-FXN gene encoded proteins (Fig. 2). The total length of the miniFXN 1 construct, including the Flag tag and excluding the PA signal, is 4,634 bp. This version of the minFXN gene expressed frataxin at high levels (Fig. 2B, lanes 1 and C). Subsequently, we conducted a series of deletions that included both regions upstream of the ATG start codon, intron 1 at the exon 1/intron1 junction and intron 1 at the intron1/exon 2 junction (miniFXN 1–13). The lengths of the minFXN genes varied from 1,032 to 4,634 bp. All constructs are depicted in Fig. 2A, and the sequence of miniFXN 1 is included in Supplementary Fig. S1.
Figure 2.
Transient expression of miniFXN constructs in HEK293T cells. (A) Schematic representation of all 19 constructs generated and tested in this work. All constructs are depicted to scale (shown above top line). Location of GAA repeats is indicated as a vertical red line. Positions of T1 and T2 TSSs are indicated. The 5′UTR sequence upstream of the ATG start codon is shown in green, intron 1 (including splice acceptor sequence, SA) in gray. All FXN exons (E1, E2–E5) are depicted in orange. Flag tag (F) is indicated in blue and polyadenylation signals (pA) in black. (B) All 19 minFXN genes were transfected to HEK293T cells in equimolar amounts and cell lysates were prepared 48 h after transfection. Western blot with FXN-specific antibodies detects both endogenous (en) and exogenous (ex) frataxin, migrating slower due to the presence of C-terminal Flag tag. Numbers above the gel lanes correspond to the miniFXN constructs depicted in (A). C represents untransfected HEK293T cells used as a control. M indicates molecular-weight protein marker (SeeBlue™ Plus2 Pre-stained Protein Standard). Representative images of three independent experiments are shown. miniFXN, frataxin minigene; 5′UTR, 5′ untranslated region.
Prior studies conducted using luciferase-conjugated reporters indicated that intron 1 of FXN is necessary for expression of this gene. Two out of 13 episomal constructs (miniFXN 6 and 9, Fig. 2A) screened initially in the HEK293T cells did not contain any intronic sequences. In agreement with prior work, these constructs expressed frataxin at low but detectable levels, irrespective of the length of the sequences upstream of the ATG codon (Fig. 2B, lanes 6 and 9), indicating that in fact intron 1 is necessary for FXN expression. Overall, quantitative analyses of Western blot data revealed that miniFXN 6, 9, and 13 express lower levels of frataxin compared with the longest miniFXN 1 gene (analysis of variance [ANOVA], p < 0.01).
The entire intron 1 of the FXN gene exceeds 10 kbp, and currently cannot be included in AAV-based gene therapy vectors. To determine the minimal size of the intron necessary for efficient FXN expression, we conducted a series of sequential restriction endonuclease digestions reducing the size of intron 1 from the initial 2,336 bp in miniFXN 1 to 2,063, 1,108, 297, and finally 110 bp (Fig. 2A, mini FXN 2–5). Importantly, we systematically decreased the intron 1 sequence directionally from the 3′ to 5′ end (toward exon 1–intron 1 junction) leaving a short fragment (95 bp miniFXN 1 or 57 bp miniFXN 2–5) at the 5′ end exon 2 splice acceptor site to assure appropriate splicing of the modified, shortened intron. The results of the transfection experiment indicate that shortening the intron up to 110 bp has no significant effect of frataxin levels when expressed from the episomal vector (Fig. 2B).
Minimal length of the FXN 5′UTR required for efficient frataxin expression
Next, to define the DNA sequence upstream of the ATG codon required for efficient FXN expression, we shortened the 5′UTR (defined here as any sequence upstream of the ATG start codon) from the initial 1,513 bp (construct miniFXN 3) to 429 bp (miniFXN 7) and 246 bp (miniFXN 10). All three vectors contained a constant 1,108 bp of the intron 1 sequence (Fig. 2A). Again, transfection of these vectors into HEK293T cells demonstrated an equally efficient expression irrespective of the length of the 5′UTR (Fig. 2B, lanes 3, 7, 10). We verified these results by comparing miniFXN genes 4, 8, and 11 harboring the same three variants of the 5′UTR, but containing a shorter (297 bp) intron 1 fragment. No significant differences in FXN levels were observed upon transfection of these vectors (Fig. 2B); however, complete removal of the entire 5′UTR upstream of the ATG completely abolished frataxin expression (miniFXN gene 13). To further narrow down the 5′UTR required for FXN expression, we decreased the length of the sequence upstream of the ATG from 429 bp (miniFXN 16) to 115, 74, and 36 bp (miniFXN 17, 18, and 19; Fig. 2A). Western blot analyses of the transfected constructs demonstrated that while 115 bp fragment upstream of the ATG codon is sufficient to efficiently drive frataxin expression, deletion of the neighboring 41 bp reduced FXN expression to background levels. Similarly, frataxin was not detected upon transfection of the construct harboring only 36-bp-long 5′UTR (Fig. 2B, lane 19). In summary, considering episomal expression of frataxin, a construct containing only 912 bp of the endogenous FXN locus (miniFXN 17; excluding Flag tag and pA sequences) is capable of efficiently expressing the entire frataxin.
Genome-integrated miniFXN transgenes in HEK293T cells express frataxin
Transfection of plasmid DNA into the cells results in hundreds of copies of a vector available for transcription. In addition, plasmid DNA does not form a proper chromatin structure, and therefore, direct translation of the results obtained using exogenous vectors to the genome-integrated expression systems may not be straightforward. To test whether our miniFXN genes can in fact drive expression of frataxin from a single genomic location, we targeted these constructs into the AAVS1 safe harbor locus. Selected miniFXN genes were recloned into the CompoZr pZdonor-AAVS1 puromycin vector in two different orientations relative to the integration site (Fig. 3A). We selected three miniFXN genes, 7, 8, and 11, harboring various lengths of the 5′UTR and intron 1 (Fig. 2A). A construct lacking intron 1 (miniFXN 9) was used as a negative control. Integration in both orientations, relative to transcription of the endogenous PPP1R12C gene, which hosts the AAVS1 locus, was conducted to minimize potential read-through effects of endogenous transcription of PPP1R12C. In addition to two different orientations, the integrated transgenes contained two pA signals upstream and downstream of the minFXN genes to further minimize potential for transcription read through (Fig. 3A).
Integration of constructs was facilitated by Cas9 cleavage (Fig. 3A) and, after puromycin selection, integration was verified by PCR and sequencing (Fig. 3B). Frataxin levels produced from all constructs were measured by Western blot and quantified relatively to the expression of endogenous FXN. All three constructs containing intron 1 (7, 8, and 11) expressed frataxin efficiently in both orientations, while miniFXN 9 (lacking intron 1) did not (Fig. 3C). Quantitative analysis revealed that the longest construct (miniFXN 7) expressed FXN at the highest level (∼40% of the endogenous level) compared with the two shorter variants (both ∼20% irrespectively of the orientation; Fig. 3C, p < 0.05). Considering that endogenous frataxin is expressed from two FXN alleles and AAVS1 integration occurred preferentially into a single safe harbor site, the amount of FXN expressed from the miniFXN 7 gene is near endogenous levels typical for HEK293T cells. In summary, we demonstrated that miniFXN constructs containing as little as 246 bp of the 5′UTR and 297 bp of intron 1 can efficiently express FXN following integration into the genome.
MiniFXN gene is expressed in pluripotent and terminally differentiated cells
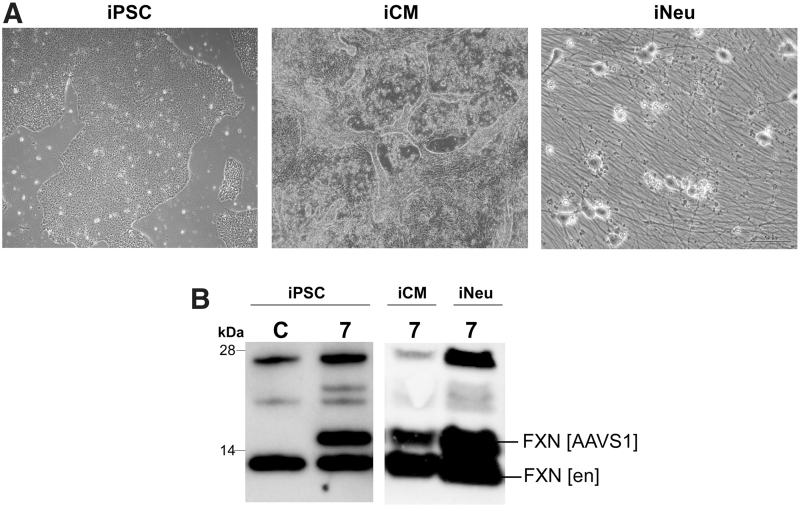
Expression of a transgene using a novel promoter sequence requires testing its capability to sustain activity in different cell types. As partial or complete silencing of a promoter can occur in different cell types, we tested whether a miniFXN gene is expressed in iPSCs as well as induced neurons (iNeu) and induced cardiomyocytes (iCM) differentiated from FRDA iPSCs (Fig. 4A). Based on results obtained in HEK293T cells, we selected the miniFXN 7 gene, demonstrating the highest frataxin expression after genome integration (Fig. 3C). After integration into the AAVS1 site, the miniFXN 7 gene expressed ∼100% of the endogenous FXN levels in iPSCs. A decrease of expression (∼35% of endogenous FXN levels) was maintained after differentiation to iCMs, while miniFXN 7 expressed FXN at near endogenous levels in iNeu cells (Fig. 4C), thus indicating that the regulatory elements driving miniFXN expression are likely to be universal and functioning in different cell types.
Figure 4.
MiniFXN gene expression in FRDA iPSCs, neurons, and cardiomyocytes differentiated from the iPSCs. (A) MiniFXN 7 (Figure 2A) was integrated into the AAVS1 locus in FRDA iPSCs (iPSC). Cells were selected with puromycin and differentiated into beating cardiomyocytes (iCM cells) and neurons (iNeu cells). Images of iPSCs and iCMs were taken at 40 × and iNeu at 400 × magnification. (B) FXN expression was analyzed by Western blot with anti-frataxin antibodies in iPSC, iCM, and iNeu (after 4 weeks of differentiation). FXN [en] designates endogenous frataxin, and FXN [AAVS1] indicates frataxin expressed from the AAVS1-integrated miniFXN 7. C represents nonintegrated iPSCs used as a control. FRDA, Friedreich's ataxia; iPSCs, induced pluripotent stem cells.
In summary, miniFXN genes containing frataxin-specific regulatory sequences are capable of efficient expression even when integrated into the genome. A correlation between the length of the regulatory (5′UTR and intron 1) sequences and frataxin levels was observed, however, all miniFXN constructs harboring a 5′UTR sequence longer than 115 bp and an intron 1 fragment longer than 110 bp expressed substantial amounts of FXN.
MiniFXN genes express frataxin after AAV-mediated transduction in mice
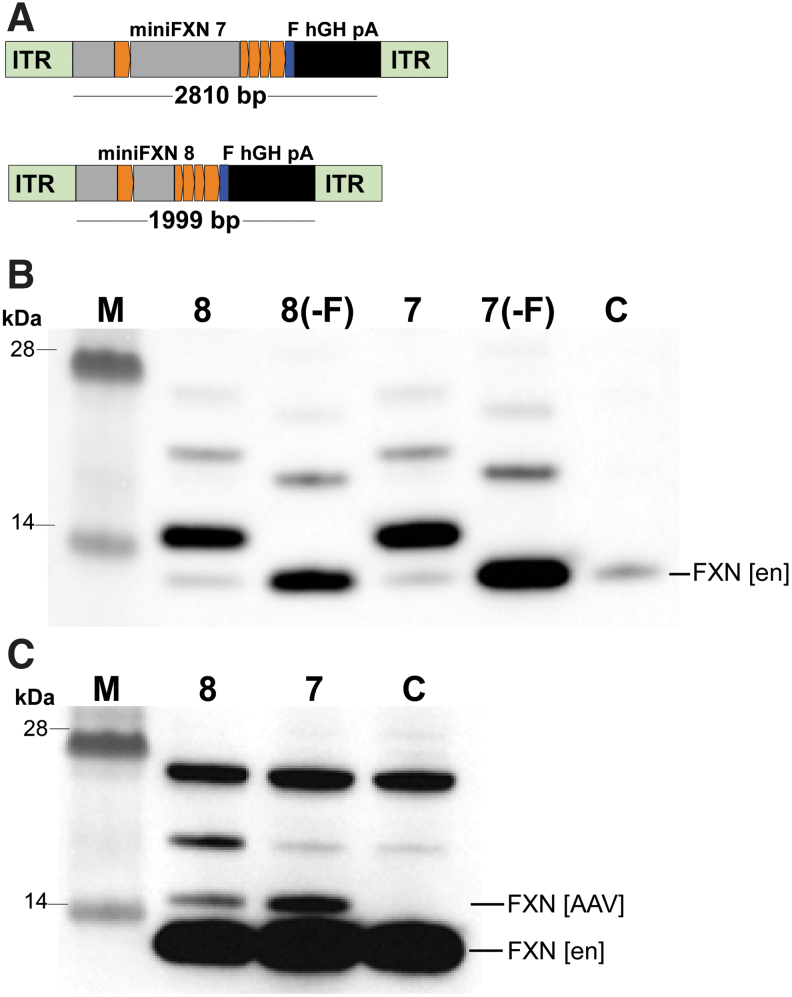
Next, to demonstrate that miniFXN genes can be considered alternatives to ubiquitous or tissue-specific strong promoters in gene therapy vectors for FRDA as a way to avoid potential toxic effects of frataxin overexpression, we constructed pAAV-cherry vectors (Addgene 27970) (Fig. 5A). The total lengths of the miniFXN genes cloned into the AAV vector were 2,810 bp and 1,999 bp for miniFXN 7 and 8, respectively. Direct transfection of the constructs into HEK293T cells verified efficient FXN expression (Fig. 5B). Based on previous preclinical gene therapy efforts in FRDA, AAV9 vectors packaging different transgene constructs were prepared.40,41 Transduction of HEK293T cells at ∼10e5 viral genomes (vg) per cell resulted in low but measurable expression of exogenous frataxin compared with endogenous FXN (Fig. 5C). Considering the low transduction efficiency of AAV in vitro and high endogenous FXN expression in HEK293T cells, the amount of frataxin produced from the miniFXN genes should significantly increase levels of frataxin in FRDA cells. Similar to results obtained using genome-integrated minFXN constructs, we observed that expression from the larger AAV-miniFXN 7 tends to be higher than from the shorter AAV-miniFXN 8 variant, however, quantitative analyses did not reach statistical significance (Fig. 5C).
Figure 5.
Expression of miniFXN constructs using AAV vectors in human cells. (A) Constructs miniFXN 7 and 8 were cloned into a pAAV vector (Addgene 27970) in two formats: with and without Flag tag (-F). Importantly, pAAV constructs harbor polyA (pA) signals. (B) All four constructs were transfected to HEK293T cells to verify FXN expression. Western blots were performed with a frataxin-specific antibody; FXN [en] indicates migration of the endogenous frataxin; C—control, untransfected cells, M—molecular-weight protein marker. No statistically significant difference in FXN expression between these constructs was observed. (C) AAV9 vectors were prepared using constructs miniFXN 7 and 8 containing the Flag tag and transduced to HEK293T cells at 10e5 viral genomes (vg)/cell. Expression of frataxin was analyzed by Western blot. FXN [AAV] indicates AAV-expressed FXN. C—control, untransduced cells. M—molecular-weight protein marker.
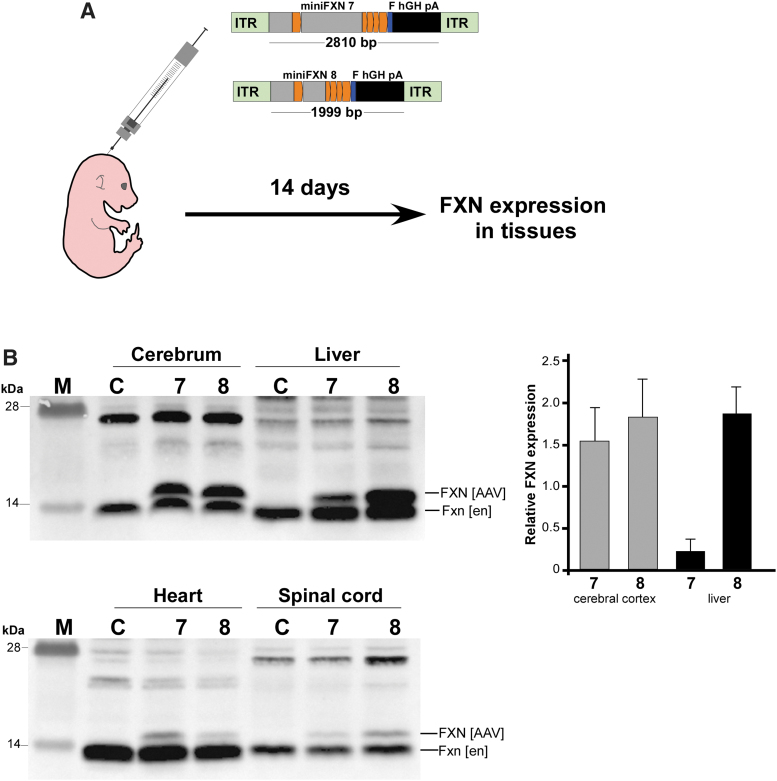
Finally, to determine whether miniFXN genes can express frataxin in vivo, we delivered ∼1e11 vg directly into the cerebrospinal fluid of neonatal animals via ICV injection (Fig. 6A). Animals were sacrificed 2 weeks after AAV9 delivery and whole-cell extracts from the cerebrum, spinal cord, heart, and liver were analyzed to evaluate the expression level of exogenous frataxin. Both miniFXN 7 and 8 AAV were highly expressed in the brain and liver, and also detectable in the spinal cord and heart (Fig. 6B). Normalization to endogenous mouse Fxn levels revealed an ∼1.5–2-fold overexpression of AAV FXN in the cerebral cortex, irrespective of the encoded minFXN gene. In the liver, miniFXN 7 was expressed at ∼20% of the endogenous, while miniFXN 8 was expressed at levels ∼two-fold greater than endogenous Fxn. AAV-delivered FXN was detectable in the heart and spinal cord, but at levels not exceeding 10% of endogenous Fxn (Fig. 6B). These relative levels of exogenous versus endogenous frataxin do not directly reflect the transcriptional activity of the miniFXN gene; however, these data clearly demonstrate that miniFXN constructs are capable of frataxin expression in vivo. Importantly, for potential future applications, the sequence lengths of both constructs are well within the packaging capacity of AAV vectors, allowing for future development of a miniFXN variant capable of delivering potentially therapeutically relevant levels of FXN.
Figure 6.
Expression of AAV-miniFXN gene products in mouse tissues upon ICV delivery. (A) Schematic of the experimental design. AAV9-miniFXN constructs 7 and 8 (at ∼1 × 10e11 vg/animal) were delivered to WT B6 neonatal (d0–1) mice via ICV injection. After 14 days, animals were sacrificed and tissues analyzed for FXN expression. (B) Expression of AAV-encoded frataxin (FXN [AAV]) in the brain, liver, heart, and spinal cord. Endogenous Fxn is indicated as Fxn [en]. M—molecular-weight protein marker. Quantitation of FXN expression relative to endogenous mouse Fxn in the cerebellar cortex and liver is shown on the right. Data based on analyses of at least four animals per group. ICV, intracerebroventricular.
Discussion
The goals of this work were two-fold: (1) to define a minimal regulatory region that will allow for efficient expression of FXN in human cells using endogenous sequences of this gene; and (2) to determine the extent of DNA sequence that can be removed from intron 1 of FXN without affecting the expression of this gene. The studies were conducted first using episomes, and then by analyses of the transcription control regions after site-specific integration of miniFXN constructs into the human genome. Furthermore, we determined whether frataxin expression driven by a minimal endogenous promoter is sustained in proliferating (human iPSCs, HEK293T) or terminally differentiated cells of distinct lineages (iCM, iNeu). Finally, we constructed AAV vectors encoding the FXN cDNA under the control of endogenous promoter sequences and demonstrated that they can express FXN in vitro (in cells) as well as in vivo (in mice). The outcome of this work is of high significance for both gene therapy and genome editing efforts in FRDA.
Gene therapy approaches frequently rely on strong, ubiquitously expressing promoters, and thus typically lack gene-/tissue-specific regulation of expression characteristic for a delivered therapeutic gene.42 These promoters may also be subjected to progressive silencing in certain cell types.43,44 In addition, uncontrolled expression may be of particular importance in the case of FRDA and other diseases where overexpression of the gene may lead to toxic side effects, potentially reducing the therapeutic benefit of the therapy.26–28,30 Prior studies in Drosophila as well as more recent work in human cell models demonstrated that overexpression of frataxin affects cellular metabolism, increases oxidative stress and labile iron pool levels.26–28,30 Considering that similar phenotypic effects are observed upon frataxin depletion, it reaffirms the notion that fine-tuning of frataxin levels is important for cellular fitness. On the contrary, initial proof-of-concept gene therapy studies demonstrated that significant overexpression of human FXN delivered by AAV in conditional mouse models of FRDA rescues severe phenotypes associated with complete loss of frataxin.40,41 Additional studies in different model systems will be required to determine whether a toxic limit of FXN overexpression exists in various human cells, preferably disease relevant, and if long-term overexpression results in any adverse changes. Thus, developing therapeutic approaches that rely on expression of frataxin using endogenous FXN control regions may represent a safer, alternative strategy for gene therapy vector design, especially considering that therapeutic effects should be achieved by restoring the frataxin level to ∼50% of levels detected in unaffected individuals (i.e., FXN level in asymptomatic carriers).45 In fact, HSV-1-mediated delivery of the entire 135 kbp FXN locus into brains of FXN knockdown mice rescued behavioral phenotypes in these animals.46 The results presented herein demonstrate that a minimal FXN promoter sequence is capable of efficiently expressing frataxin.
The pathogenic GAAs are located in intron 1, within an ancient Alu element. Recent studies demonstrated that excision of the tract, together with flanking sequences, using editing nucleases, increases FXN expression to near unaffected control levels.31–33 This strongly suggests that no critical regulatory elements exist in the direct vicinity of the GAAs. However, intron 1 of the FXN gene encodes numerous interspersed repeat elements making genome editing of this region a challenging task. Thus, identifying and defining FXN gene expression control sequences allow for judicial selection of regions amenable to genome editing to minimize possible off-target, or undesirable, effects. Our studies demonstrated that at least a fragment of intron 1 is absolutely necessary for FXN expression. Surprisingly, the size of intron 1 can be reduced to only 110 bp without a pronounced effect on FXN expression. In addition, the minimal 5′UTR necessary for FXN expression contains 115 bp upstream of the ATG. Shortening this fragment by 41 bp completely abolishes expression of FXN. In silico analyses by TRANSFAC (Supplementary Fig. S2) demonstrated that this critical sequence may be a binding region for the TFAP2 transcription factor. Interestingly, prior studies on FXN regulation indicated that TFAP2 is important for transcription of this gene and siRNA-mediated TFAP2 knockdown reduces frataxin expression.17 Our data confirmed those results and implicate TFAP2 as a transcription factor critical for FXN transcription.
Defining endogenous expression control regions is a laborious process requiring numerous constructs designed typically by a systematic mutagenesis approach. In addition, conducting these studies in episomal vectors discounts the potential effect of genomic/epigenetic context and includes a variable factor of uncontrolled transfection efficiency, making any comparisons between experiments difficult. Also, including reporter fusions (e.g., luciferase) allows for precise measurements of specific but only relative changes in DNA sequence. Moreover, it eliminates the possibility of direct assessment of the role of putative regulatory elements on the expression of the endogenous gene of interest. To overcome some of the abovementioned issues, we integrated selected miniFXN genes into the genome to determine whether their activity would be sustained in a chromatin context and if their expression would be detectable with a minimal number of site specifically integrated copies. The iPSCs and iPS-derived cells present unique systems to determine whether miniFXN expression would persist in a pluripotent state and in different lineages of human cells. Expanding this approach to various cell types from all lineages will allow us to ascertain whether certain transgenes driven by an endogenous promoter will be active ubiquitously or in a cell-specific manner. Although miniFXN constructs described in this work express frataxin at levels comparable with the endogenous locus in different lineages, we cannot rule out a possibility that certain sequences excluded from the miniFXN-genes participate in fine-tuning of frataxin expression. This is especially relevant to potential long-range enhancers that may be located thousands of base pairs away from the core promoter region.
Finally, we performed in vivo analyses to determine whether regulatory sequences of the miniFXN genes can drive protein expression in different tissues and potentially be utilized in gene therapy applications. We demonstrated that both selected miniFXN constructs can be packaged into AAV capsids and delivered to human cells and mouse tissues. Human FXN was expressed in all four different mouse tissues tested following ICV injection, thus confirming that endogenous FXN sequences can be considered potential exogenous promoters driving FXN expression from AAV vectors. Recent progress and successes in gene therapy approaches have instigated research projects aimed at improving delivery methods, vectors, and targeting the virus-specific immune response.47 Additional studies will be necessary to identify and characterize novel promoters, especially focusing on endogenous sequences that can drive the expression where the target gene is controlled in a precise, tissue-specific manner. This is especially important in cases where toxicity of the overexpressed protein should be considered, as in FRDA. Further work is needed to precisely define regulatory elements, especially tissue-specific control regions. Assuring that these sequences can be introduced into AAV vectors and persistently expressed in different human tissues represents a great challenge that, when overcome, will bring us to a successful gene therapy strategy for FRDA.
Supplementary Material
Acknowledgments
We thank all FRDA patients for contributing research material that allowed us to conduct these studies. We would also like to thank Laura Wheeler for critical reading and editing of the article.
Authors' Contributions
J.L., J.S.N., and M.N. conceived the study, designed experiments, and wrote the article. J.L., Y.L., and T.J.G. carried out cell culture, cloning, transfection, editing, and virus generation experiments. J.W. conducted animal ICV injections and tissue procurement. A.A., J.S.N., and M.N. provided conceptualization and funding acquisition.
Author Disclosure
J.L., Y.L., J.W., T.J.G, J.S.N., and M.N. have no competing financial interests. A.A. is a cofounder and advisor to StrideBio, Inc.
Funding Information
This study was supported by the National Institutes of Health (R01NS081366) and Muscular Dystrophy Association (MDA418838) awarded to M.N. and NIH grants (UG3 AR075336; R01GM127708; R01NS099371) awarded to A.A.
Supplementary Material
References
- 1. Campuzano V, Montermini L, Molto MD, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 1996;271:1423–1427 [DOI] [PubMed] [Google Scholar]
- 2. Marmolino D. Friedreich's ataxia: past, present and future. Brain Res Rev 2011;67:311–330 [DOI] [PubMed] [Google Scholar]
- 3. Delatycki MB, Bidichandani SI. Friedreich ataxia- pathogenesis and implications for therapies. Neurobiol Dis 2019;132:104606. [DOI] [PubMed] [Google Scholar]
- 4. Vankan P. Prevalence gradients of Friedreich's ataxia and R1b haplotype in Europe co-localize, suggesting a common Palaeolithic origin in the Franco-Cantabrian ice age refuge. J Neurochem 2013;126(Suppl. 1):11–20 [DOI] [PubMed] [Google Scholar]
- 5. Li Y, Lu Y, Polak U, et al. Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus. Hum Mol Genet 2015;24:6932–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Napierala JS, Li Y, Lu Y, et al. Comprehensive analysis of gene expression patterns in Friedreich's ataxia fibroblasts by RNA sequencing reveals altered levels of protein synthesis factors and solute carriers. Dis Model Mech 2017;10:1353–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Galea CA, Huq A, Lockhart PJ, et al. Compound heterozygous FXN mutations and clinical outcome in Friedreich ataxia. Ann Neurol 2016;79:485–495 [DOI] [PubMed] [Google Scholar]
- 8. Clark E, Butler JS, Isaacs CJ, et al. Selected missense mutations impair frataxin processing in Friedreich ataxia. Ann Clin Transl Neurol 2017;4:575–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clark RM, Dalgliesh GL, Endres D, et al. Expansion of GAA triplet repeats in the human genome: unique origin of the FRDA mutation at the center of an Alu. Genomics 2004;83:373–383 [DOI] [PubMed] [Google Scholar]
- 10. Greene E, Entezam A, Kumari D, et al. Ancient repeated DNA elements and the regulation of the human frataxin promoter. Genomics 2005;85:221–230 [DOI] [PubMed] [Google Scholar]
- 11. Chutake YK, Costello WN, Lam CC, et al. FXN promoter silencing in the humanized mouse model of Friedreich ataxia. PLoS One 2015;10:e0138437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chutake YK, Lam C, Costello WN, et al. Epigenetic promoter silencing in Friedreich ataxia is dependent on repeat length. Ann Neurol 2014;76:522–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Butler JS, Napierala M. Friedreich's ataxia—a case of aberrant transcription termination? Transcription 2015;6:33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumari D, Biacsi RE, Usdin K. Repeat expansion affects both transcription initiation and elongation in Friedreich ataxia cells. J Biol Chem 2010;286:4209–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gottesfeld JM. Molecular mechanisms and therapeutics for the GAA.TTC expansion disease Friedreich ataxia. Neurotherapeutics 2019;16:1032–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Greene E, Mahishi L, Entezam A, et al. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res 2007;35:3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li K, Singh A, Crooks DR, et al. Expression of human frataxin is regulated by transcription factors SRF and TFAP2. PLoS One 2010;5:e12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shimizu R, Lan NN, Tai TT, et al. p53 directly regulates the transcription of the human frataxin gene and its lack of regulation in tumor cells decreases the utilization of mitochondrial iron. Gene 2014;551:79–85 [DOI] [PubMed] [Google Scholar]
- 19. Petrillo S, Piermarini E, Pastore A, et al. Nrf2-inducers counteract neurodegeneration in Frataxin-silenced motor neurons: disclosing new therapeutic targets for Friedreich's ataxia. Int J Mol Sci 2017;18. [DOI] [PMC free article] [PubMed]
- 20. Jasoliya M, Sacca F, Sahdeo S, et al. Dimethyl fumarate dosing in humans increases frataxin expression: a potential therapy for Friedreich's Ataxia. PLoS One 2019;14:e0217776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sahdeo S, Scott BD, McMackin MZ, et al. Dyclonine rescues frataxin deficiency in animal models and buccal cells of patients with Friedreich's ataxia. Hum Mol Genet 2014;23:6848–6862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pastore A, Puccio H. Frataxin: a protein in search for a function. J Neurochem 2013;126(Suppl. 1):43–52 [DOI] [PubMed] [Google Scholar]
- 23. Stepanova A, Magrane J. Mitochondrial dysfunction in neurons in Friedreich's ataxia. Mol Cell Neurosci 2019;102:103419. [DOI] [PubMed] [Google Scholar]
- 24. Lupoli F, Vannocci T, Longo G, et al. The role of oxidative stress in Friedreich's ataxia. FEBS Lett 2018;592:718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang S, Napierala M, Napierala JS. Therapeutic prospects for Friedreich's ataxia. Trends Pharmacol Sci 2019;40:229–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vannocci T, Notario Manzano R, Beccalli O, et al. Adding a temporal dimension to the study of Friedreich's ataxia: the effect of frataxin overexpression in a human cell model. Dis Models Mech 2018;11:dmm.032706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vannocci T, Dinarelli S, Girasole M, et al. A new tool to determine the cellular metabolic landscape: nanotechnology to the study of Friedreich's ataxia. Sci Rep 2019;9:19282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Navarro JA, Llorens JV, Soriano S, et al. Overexpression of human and fly frataxins in Drosophila provokes deleterious effects at biochemical, physiological and developmental levels. PLoS One 2011;6:e21017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Llorens JV, Navarro JA, Martinez-Sebastian MJ, et al. Causative role of oxidative stress in a Drosophila model of Friedreich ataxia. FASEB J 2007;21:333–344 [DOI] [PubMed] [Google Scholar]
- 30. Seguin A, Bayot A, Dancis A, et al. Overexpression of the yeast frataxin homolog (Yfh1): contrasting effects on iron-sulfur cluster assembly, heme synthesis and resistance to oxidative stress. Mitochondrion 2009;9:130–138 [DOI] [PubMed] [Google Scholar]
- 31. Li Y, Polak U, Bhalla AD, et al. Excision of expanded GAA repeats alleviates the molecular phenotype of Friedreich's ataxia. Mol Ther 2015;23:1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li J, Rozwadowska N, Clark A, et al. Excision of the expanded GAA repeats corrects cardiomyopathy phenotypes of iPSC-derived Friedreich's ataxia cardiomyocytes. Stem Cell Res 2019;40:101529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ouellet DL, Cherif K, Rousseau J, et al. Deletion of the GAA repeats from the human frataxin gene using the CRISPR-Cas9 system in YG8R-derived cells and mouse models of Friedreich ataxia. Gene Ther 2017;24:265–274 [DOI] [PubMed] [Google Scholar]
- 34. Oceguera-Yanez F, Kim SI, Matsumoto T, et al. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods 2016;101:43–55 [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y, Pak C, Han Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 2013;78:785–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim JY, Grunke SD, Levites Y, et al. Intracerebroventricular viral injection of the neonatal mouse brain for persistent and widespread neuronal transduction. J Vis Exp 2014:51863. [DOI] [PMC free article] [PubMed]
- 37. Puspasari N, Rowley SM, Gordon L, et al. Long range regulation of human FXN gene expression. PLoS One 2011;6:e22001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chutake YK, Costello WN, Lam C, et al. Altered nucleosome positioning at the transcription start site and deficient transcriptional initiation in Friedreich ataxia. J Biol Chem 2014;289:15194–15202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Biase I, Chutake YK, Rindler PM, et al. Epigenetic silencing in Friedreich ataxia is associated with depletion of CTCF (CCCTC-binding factor) and antisense transcription. PLoS One 2009;4:e7914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perdomini M, Belbellaa B, Monassier L, et al. Prevention and reversal of severe mitochondrial cardiomyopathy by gene therapy in a mouse model of Friedreich's ataxia. Nat Med 2014;20:542–547 [DOI] [PubMed] [Google Scholar]
- 41. Piguet F, de Montigny C, Vaucamps N, et al. Rapid and complete reversal of sensory ataxia by gene therapy in a novel model of Friedreich ataxia. Mol Ther 2018;26:1940–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ingusci S, Verlengia G, Soukupova M, et al. Gene therapy tools for brain diseases. Front Pharmacol 2019;10:724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bestor TH. Gene silencing as a threat to the success of gene therapy. J Clin Invest 2000;105:409–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alhaji SY, Ngai SC, Abdullah S. Silencing of transgene expression in mammalian cells by DNA methylation and histone modifications in gene therapy perspective. Biotechnol Genet Eng Rev 2019;35:1–25 [DOI] [PubMed] [Google Scholar]
- 45. Campuzano V, Montermini L, Lutz Y, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum Mol Genet 1997;6:1771–1780 [DOI] [PubMed] [Google Scholar]
- 46. Lim F, Palomo GM, Mauritz C, et al. Functional recovery in a Friedreich's ataxia mouse model by frataxin gene transfer using an HSV-1 amplicon vector. Mol Ther 2007;15:1072–1078 [DOI] [PubMed] [Google Scholar]
- 47. Li C, Samulski RJ. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet 2020 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.