Abstract
We have developed a platform for quantitative genetic interaction mapping using viral infectivity as a functional read-out, and constructed a viral host-dependency epistasis map (vE-MAP) of 356 human genes linked to HIV function, comprising >63,000 pair-wise genetic perturbations. The vE-MAP provides an expansive view of the genetic dependencies underlying HIV infection, and can be used to identify drug targets and study viral mutations. We found that the RNA deadenylase complex, CNOT, is a central player in the vE-MAP and show that knockout of CNOT1, 10 and 11 suppressed HIV infection in primary T-cells by upregulating innate immunity pathways. This phenotype was rescued by deletion of IRF7, a transcription factor regulating interferon stimulated genes, revealing a previously unrecognized host signaling pathway involved in HIV infection. The vE-MAP represents a generic platform that can be used to study the global effects of how different pathogens hijack and rewire the host during infection.
eTOC Blurb
Most experimental genetic screens studying pathogen infection study one genetic element at a time, however Gordon et al. adapted epistasis mapping to study combinations of perturbations impacting HIV infection, and use their approach to identify and characterize a role for CCR4NOT in mediating HIV infection in primary CD4+ T cells.
Graphical Abstract

Introduction
Genetic interaction mapping is a powerful approach for studying gene function and delineating functional modules and pathways. Perturbing gene combinations reveals epistatic and synergistic relationships that inform hypothesis generation regarding gene function. Furthermore, when carried out at a large scale, genetic interaction mapping can identify gene clusters that correspond to protein complexes (Beltrao et al., 2010). The bulk of genetic interaction data has been generated in basic model organisms such as yeast (Collins et al., 2007; Costanzo et al., 2016; Roguev et al., 2008; Ryan et al., 2012; Schuldiner et al., 2005), however this approach has recently been adapted for large-scale mammalian studies by us and others (Bassik et al., 2009; Du et al., 2017; Horlbeck et al., 2018; Laufer et al., 2013; Roguev et al., 2013; Shen et al., 2017). Genetic interaction mapping lends itself well to the study of pathways underlying cellular proliferation, and the majority of genetic interaction studies to date have used this as their readout. By contrast, the application of large-scale genetic interaction mapping to study pathogen infection is largely unexplored.
Human immunodeficiency virus (HIV) is a major public health concern, with an estimated 36.7 million people living with chronic infection and over 20.9 million people receiving continuous anti-retroviral treatment (UNAIDS, 2018). Due to the social and economic costs imposed by the HIV epidemic, a comprehensive description of the molecular mechanisms of HIV infection is of critical importance for the development of improved treatment and curative strategies. The vast majority of genetic studies of host-dependency factors mediating HIV infection have utilized individual gene perturbations through drug screening, siRNA or Cas9-based approaches. These studies suffer from two primary limitations: 1) they provide little information as to the pathway or complex to which a gene impacting viral infection might belong, and 2) by studying genes in isolation they fail to identify synergistic combinations of genes which may be targeted in combinatorial therapies. We hypothesized that the application of genetic interaction mapping to the study of HIV host factors would uncover complexes and pathways previously not functionally linked to HIV infection.
With others, we have pioneered the epistatic miniarray profile (E-MAP) approach to study genetic interactions in lower eukaryotes and mammalian systems (Collins et al., 2007; Du et al., 2017; Roguev et al., 2008; 2013; Schuldiner et al., 2005; 2006). Through disruption of a large matrix of gene pairs using combinatorial gene expression perturbations or mutations, and employing a high-throughput functional readout, the E-MAP approach permits the quantification of genetic interactions and enables identification of functional gene clusters. Here we describe our adaptation of the E-MAP approach for the study of genetic interactions impacting viral infection. The resulting viral host-dependency E-MAP (vE-MAP) pipeline is flexible and scalable, and may be utilized to study combinations of gene perturbations, drug treatments, and viral mutants.
Our vE-MAP was generated primarily by pairwise depletion of 356 human genes linked to HIV, using infection of cultured cells as a quantitative readout. By analyzing genetic interactions based on HIV infection data, we resolve functional modules of host genes consistent with published protein-protein interaction data, and demonstrate the utility of this approach to study the biological impact of small molecules and viral mutations. Using this map, we also identify a role for CCR4-NOT (CNOT), the major RNA deadenylase complex in eukaryotic cells (Miller and Reese, 2012), in mediating HIV infection. Validating this role for CNOT in primary CD4+ T-cells, we demonstrate that knockout of multiple CNOT components inhibits HIV infection by upregulating the innate immune response, a phenotype that is rescued by knockout of IRF7, a core activator of interferon stimulated genes. This work establishes a foundation for the study of genetic interactions impacting infection across many pathogens in cultured and primary cells.
Results
An arrayed pipeline for studying genetic interactions impacting HIV infection
The E-MAP approach entails screening a large matrix of pairwise mutation combinations, necessitating a large syngenic source of cells. We carried out our vE-MAP analysis in HeLa cells, which can be grown in large reproducible batches over months of screening, and validated selected gene candidates in primary CD4+ T-cells, the physiological target of HIV infection.
The analysis of genetic interactions is most straightforward when each perturbation is introduced homogenously across a population of cells, a requirement satisfied by using our previously published combinatorial RNA interference E-MAP strategy (Roguev et al., 2013). Similar to our previous work in yeast, this pipeline utilizes the S-score model for genetic interaction quantification: a modified t-test incorporating deviation from expected phenotype and reproducibility (Collins et al., 2006). In our HIV genetic interaction pipeline, a positive S-score indicates more HIV expression than expected, and a negative S-score indicates less HIV expression than expected. Hierarchical clustering of genetic interaction profiles highlights functional modules mediating the phenotype under study, and can also highlight relationships between these modules (Figure 1A). Knockdowns were performed in 384-well arrayed format using endonuclease-prepared small interfering RNA (esiRNA). This format is flexible and enables the combinatorial study of genetic perturbations, small molecules and viral mutants. We utilized esiRNA because it is a cost-effective, scalable approach to RNAi that produces fewer off-target effects than chemically synthesized individual siRNAs (Buchholz et al., 2006). Our esiRNA preparations generated robust knockdown when transfected via high-throughput liquid handling, and their transfection did not cause an innate immune response (Figure S1A and S1B).
Figure 1: An arrayed pipeline for studying genetic interactions impacting HIV infection.
A) The effect of pairwise knockdowns of human genes on HIV expression is measured using a high-throughput luciferase reporter system. Genetic interaction scores are calculated on the basis of luciferase signal normalized to cell count in each well. In the vast majority of wells, the normalized luciferase signal conforms to the expected combinatorial phenotype, indicating a lack of genetic interaction (neutral). In some wells, the luciferase signal is higher or lower than expected, indicating positive or negative genetic interactions, respectively. Genetic interaction profiles subjected to hierarchical clustering reveal functional gene modules. (B) Overview of the HIV vE-MAP pipeline: Cultured HeLa cells in each well are transfected with esiRNAs targeting two different genes and later infected with a HIV luciferase reporter virus. All combinations are analyzed in quadruplicate. Cell counts and HIV expression are measured by high-throughput microscopy and luminometry. (C and D) Typical library well behavior calculated across all plates ran in the HIV E-MAP, depicting single knockdown phenotypes for cell count (C) and cell count-corrected HIV luciferase expression (D). The 356 HIV-associated human gene knockdowns which passed our viability filter were utilized for vE-MAP analysis. See also Data S1 for pairwise knockdown vE-MAP data, and Figure S1 for esiRNA testing and library composition data.
The scale of an E-MAP increases quadratically with the number of genes tested, therefore it is crucial to select an enriched set of genes likely to mediate the function under study. To fulfill this requirement, we focused on our previously published global study of HIV-human protein-protein interactions (PPIs) in which we identified hundreds of known and putative HIV host-dependency factors (Jäger et al., 2012). We also included other HIV-associated human genes, since the strong protein interactions identified in biochemical screens may not capture all relevant host-pathogen interactions, and some host genes crucial for infection may not directly interact with viral proteins. Of the 366 HIV-related human genes we selected, 356 gene knockdowns met our viability threshold, and in total we quantified genetic interactions between 63,012 pairwise knockdown combinations (Figure S1C, S1D and Table S1). Controls were also included in each plate, including negative controls (water or GFP esiRNA), a transfection control esiRNA which causes cell death (Eg5/KIF11 esiRNA), and positive controls inhibiting HIV infection (the HIV reverse transcription inhibitor AZT) and HIV reporter expression (luciferase esiRNA).
Combinatorial treatments were generated by dispensing a second, query esiRNA on top of each plate. Each knockdown combination was tested in quadruplicate, leading to a total of >1,400 plates. After transfection of the HeLa cells, plates were incubated 72 hours for gene depletion, infected with VSV-G pseudotyped HIV luciferase reporter virus, and incubated for another 48 hours prior to analysis. Cells were counted by high-throughput microscopy and HIV luciferase expression was quantified using high-throughput luminometry. The influence of cell count on luciferase readouts was subtracted, and pairwise genetic interactions were scored using a modified S-score (Figure 1B).
As genetic interactions are rare, the phenotype of each single-gene perturbation was inferred by internally normalizing each plate, and calculating the median behavior of each library well across all plates. We evaluated the typical cell counts for single-gene perturbations and observed that our negative controls (water or GFP-targeted esiRNA) did not display any defect in cell growth, while the Eg5/KIF11 knockdown wells had few surviving cells, as expected for an essential gene (Figure 1C). We also evaluated the typical behavior of our controls for HIV infection and expression: as expected, the reverse transcriptase inhibitor AZT strongly inhibited HIV production, and esiRNA targeting firefly luciferase reduced HIV reporter expression (Figure 1D).
The reproducibility of our library plate controls over all >1,400 plates was consistent, as the interquartile ranges were relatively tight throughout the screen (Figure S2A and S2B). We also compared the phenotypes of reciprocal knockdown of the same gene combinations, and observed a positive relationship between these values (Figure S2C). Finally, confidence scores were calculated as a function of S-score (Figure S2D) (Collins et al., 2006). Overall, these results demonstrate the reliability of our combinatorial knockdown and viral phenotyping pipeline.
The vE-MAP highlights known protein complexes
An E-MAP can identify functional relationships between genes in two ways: 1) by quantifying individual genetic interactions between gene pairs, and 2) by quantifying the similarities in genetic interaction profiles between two or more genes (Figure 1A). Similar genetic interaction profiles suggest shared function, and clustering of E-MAP data according to interaction profiles (often using hierarchical clustering) can resolve biochemical complexes (Beltrao et al., 2010; Braberg et al., 2013; Collins et al., 2007; Costanzo et al., 2016; Du et al., 2017; Roguev et al., 2013; Schuldiner et al., 2005). Indeed, hierarchical clustering of the vE-MAP genetic interaction data (Figure 2A, Data S1) revealed numerous gene clusters that matched biochemical complexes known to mediate HIV infection, including ESCRT-0/1 (McDonald and Martin-Serrano, 2009), the COP9 signalosome, NEDD8 with functionally associated proteins CAND1 and RBX1 (Lydeard et al., 2013; Stanley et al., 2012; Zheng et al., 2002), and the PAF complex (Krogan et al., 2002; Sobhian et al., 2010; Squazzo et al., 2002) (Figure 2B, panels 2–5). In addition, we observed co-clustering of subunits from several other known protein complexes and pathways, including Cullin-2 with Elongin B and C (Lonergan et al., 1998; Lydeard et al., 2013), subunits 1 and 2 of the anaphase promoting complex (Zhou et al., 2016), and tRNA synthetases co-clustering with TNPO3 (whose yeast homolog, MTR10, has been implicated in nuclear transport of tRNAs) (Shaheen and Hopper, 2005) (Figure 2B, panels 1, 6 and 7). Notably, we were able to identify functional relationships using genetic interaction analysis even in the absence of strong single-knockdown phenotypes, as demonstrated with the COP9 signalosome (Figure 2B, panel 3), highlighting a strength of the vE-MAP approach relative to conventional single-perturbation genetic screens.
Figure 2: The vE-MAP highlights known protein complexes.
(A and B) Clustering of HIV vE-MAP data highlights known human protein complexes. Genetic interaction scores (S-score) are visualized by a yellow (positive) to blue (negative) scale, and single knockdown phenotypes by a red (positive) to purple (negative) scale. (C and D) Genetic interaction clustering highlights structural submodules of the eIF3 complex (Wagner et al., 2016). (E) Enrichment of protein-protein interactions (Bioplex, Huttlin et al., 2017) as a function of correlation between genetic interaction profiles. This analysis indicates that genes with higher correlation in genetic interaction profiles are enriched for protein-protein interactions (PPIs).
Since correlation of genetic interaction profiles is evidence of shared function, clustering can also resolve structural submodules of protein complexes (Fraser et al., 2013). In our case, we observed clustering of structural submodules of the eIF3 ribosome initiation complex, with components eIF3H, F, and M comprising the eIF3 ‘left leg’; eIF3D and E comprising the ‘right arm’; eIF3B, G, and I forming the ‘yeast-like core’; and eif3A and C interacting as adjacent ‘head’ and ‘left arm’ submodules (Figure 2C and 2D) (Georges et al., 2015; Wagner et al., 2016). The ability of genetic interaction maps to resolve structural data has been observed in previous studies based on cellular proliferation (Braberg et al., 2013; Collins et al., 2007), and now we have demonstrated this is also the case in the context of a pathogen infection.
High correlation between genetic interaction profiles is predictive of protein interactions (Beltrao et al., 2010; Roguev et al., 2008), therefore genetic interaction datasets can be validated on a systems-level by comparison with protein-interaction datasets. We identified the overlap between the 356 genes analyzed in our HIV vE-MAP and the BioPlex human protein interaction network (Huttlin et al., 2017), and quantified enrichment of protein interacting pairs as a function of genetic interaction profile correlation. We observed that greater correlation of vE-MAP genetic interactions significantly increased the likelihood of physical interaction (Figure 2E), providing systems-level validation for our genetic interaction dataset.
The vE-MAP approach is amenable to small molecule and mutant virus queries
A key advantage of arrayed genetic interaction mapping is that it permits rapid and straightforward application of diverse additional treatments across our library plates. We therefore explored the use of our vE-MAP pipeline to identify genetic interactions with small molecule and HIV mutant queries (Figure 3A).
Figure 3: The vE-MAP approach is amenable to small molecule and mutant virus queries.
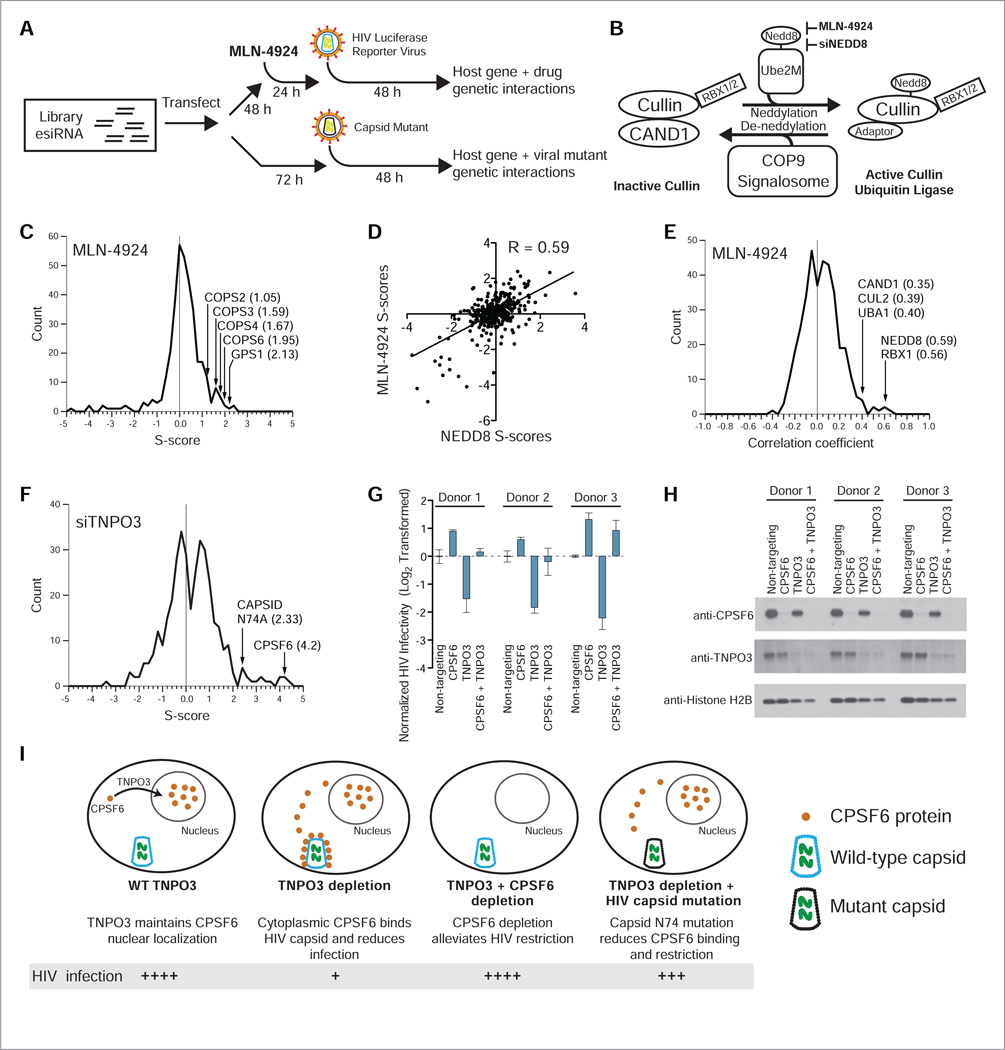
(A) Small molecules may be introduced during the 72-hour knockdown period prior to addition of reporter virus. Alternatively, a mutant virus may be introduced instead of the wild-type reporter virus. (B) Graphical representation of the NEDD8 conjugation pathway regulating activity of Cullin ubiquitin ligases. (C) The neddylation inhibitor MLN-4924 displays positive genetic interactions with all five subunits of the COP9 signalosome run in the vE-MAP. (D) Comparison of S-score profiles of MLN-4924 and NEDD8 knockdown (Pearson correlation = 0.59). (E) The MLN-4924 genetic interaction profile displays high correlation with NEDD8 and the NEDD8-associated genes RBX1/ROC1, CAND1, CUL2 and UBA1. (F) TNPO3 displays positive genetic interactions with CPSF6 and the HIV capsid point mutant N74A. (G and H) Double CPSF6 and TNPO3 knockout in primary CD4+ T-cells harvested from three healthy donors. Error bars: standard deviation (n = 3). Western blotting shows reproducible combinatorial knockout in all donors tested. (I) Model for circumvention of TNPO3-dependency by depletion of CPSF6 or mutation of HIV capsid.
We chose to study the small molecule MLN-4924 (Soucy et al., 2009) because it interferes with Cullin ubiquitin ligases. Several Cullin ubiquitin ligases have been reported to physically interact with HIV proteins, the most well-studied being Cullin-1, 4 and 5 (Hrecka et al., 2007; Jäger et al., 2012; Margottin et al., 1998; Yu et al., 2003). Cullin protein complexes are activated by covalent conjugation of the ubiquitin-like protein NEDD8, which is mediated by an E1-E2-E3 cascade inhibited by the small molecule MLN-4924 (Figure 3B) via inhibition of the NEDD8 E1 enzyme NAE (Soucy et al., 2009). We introduced MLN-4924 as a query treatment in our vE-MAP pipeline, and quantified its genetic interactions and genetic interaction profile correlations with the gene knockdowns in the vE-MAP. Consistent with its function as a neddylation inhibitor, MLN-4924 treatment displayed positive genetic interactions with subunits of the COP9 signalosome, a complex which mediates deneddylation (Lyapina et al., 2001; Schwechheimer et al., 2001; Wei and Deng, 2003) (Figure 3C and Table S2). Further supporting its on-target function, the genetic interactions observed with MLN-4924 treatment correlated strongly with those observed upon NEDD8 knockdown (Figure 3D). We also observed correlation between MLN-4924 treatment and knockdown of Cullin-2 (CUL2), as well as the Cullin-associated proteins CAND1 and RBX1 (Chuang et al., 2004; Enchev et al., 2015; Kamura et al., 1999; Zheng et al., 2002), indicating shared function between MLN-4924 treatment and depletion of these genes (Figure 3E). Finally, we observed correlation between MLN-4924 treatment and knockdown of the ubiquitin E1 enzyme UBA1, which can also act as a NEDD8 E1 enzyme under stress conditions (Leidecker et al., 2012) (Figure 3E). These data demonstrate that the vE-MAP platform can be used as a drug discovery and characterization platform for HIV and other pathogens.
We next queried our library with the HIV N74A capsid mutant, which shares functional dependencies with the host factors CPSF6 and TNPO3. When TNPO3 is depleted, CPSF6 translocates from the nucleus to the cytoplasm where it binds HIV capsid and restricts infection, and concomitant depletion of CPSF6 restores infectivity (De Iaco et al., 2013). Consistent with this, we observed a positive genetic interaction between TNPO3 and CPSF6 knockdown in the HIV vE-MAP (S = 4.2, one of the highest observed) (Figure 3F). Previous studies have shown that mutation of capsid residue N74, which reduces binding to CPSF6, circumvents TNPO3-dependence for HIV infection (De Iaco and Luban, 2011; Lee et al., 2010; Price et al., 2012; Zhou et al., 2012). Using the capsid N74A mutant in our system, we observed a positive genetic interaction with TNPO3 knockdown, supporting previous reports that this point mutant circumvents TNPO3 restriction (Figure 3F). This data demonstrates that genetic interactions involving viral mutants can be studied using our vE-MAP pipeline.
We also sought to validate the TNPO3-CPSF6 genetic interaction in primary CD4+ T-cells (Hendel et al., 2015; Hultquist et al., 2016). We purified CD4+ T-cells from three healthy donors and performed combinatorial knockout of TNPO3 and CPSF6 followed by infection with HIV GFP reporter virus, observing donor dependence in genetic interaction behavior with a clear positive genetic interaction observed in Donor 3. (Figure 3G–H). These results indicate that our combinatorial results obtained in HeLa cells can translate to primary human samples, however it is important to assess variability in multiple donors. A model of the functional dependencies between TNPO3, CPSF6 and HIV capsid and their roles in HIV infectivity are presented in Figure 3I.
Subunits of the CCR4-NOT complex mediate HIV infection
We next focused on subunits 1–3 of the CCR4-NOT (CNOT) complex, because although they had weak single-knockdown phenotypes that would have been disregarded in a single perturbation screen, they displayed numerous negative genetic interactions with many known HIV host-dependency factors, including factors related to proviral transcription (Jaehning, 2010; Mancebo et al., 1997; Ott et al., 2011; Sobhian et al., 2010), RNA granule biology (Lai et al., 2008; Yedavalli et al., 2004), and components of the ESCRT complex (McDonald and Martin-Serrano, 2009) (Figure 4A). In fact, the CNOT gene knockdowns had some of the richest genetic interaction profiles of our vE-MAP (Figure 4B). These observations indicated that depletion of CNOT subunits (most notably CNOT1) potentiated the reduction of HIV infection when combined with depletion of other HIV host-dependency factors in HeLa cells. We therefore explored the role of the CNOT complex in mediating HIV infection in human primary CD4+ T-cells.
Figure 4: Components of the CNOT complex are required for HIV infection.
(A) All three CNOT subunits in the vE-MAP library co-cluster and interact negatively with HIV host-dependency factors including RNA transcriptional machinery, the RNA granule gene DDX3X, and the ESCRT components HGS and TSG101. (B) CNOT subunits 1, 2 and 3 display a large number of negative genetic interactions in the HIV vE-MAP. (C) Combinatorial knockout of CNOT1 with CCNT1 in primary T-cells demonstrates an additive decrease in infection in four independent donors (72 hours post infection). Error bars: standard deviation (n = 3). (D) Representative western blot validates combinatorial knockout of the CNOT1 large isoform and CCNT1. See also Figure S3 for full western blot data and validation of a second negative genetic interaction between CNOT2 and DDX3X. (E) Map illustrating CNOT1 large and small isoforms predicted by CNOT1 western blots and the human genome sequence. Exon 8 encodes four in-frame start codons downstream of the canonical translational start site; see also Figure S4. The CNOT10/11 binding domain is highlighted at the N-terminus. (F and G) Polyclonal knockout of the CNOT1 large isoform is sufficient to reduce HIV infection in primary CD4+ T-cells. Error bars: standard deviation (n = 3). (H) Knockout of the CNOT1 large isoform reduces enrichment of CNOT10 and CNOT11 following immunoprecipitation of total CNOT1 (IP antibody epitope shown in panel E). Error bars: standard error (n = 4). See also Table S3. (I and J) Polyclonal knockout of CNOT10 and 11 causes a ~10-fold decrease in HIV infection. Error bars: standard deviation (n = 3). Asterisk: likely nonspecific band. See also Figure S5 for HIV infection data for other CNOT subunit knockouts.
We first validated two CNOT negative genetic interactions: CNOT1 + CCNT1 (Figure 4C, 4D and S3A) and CNOT2 + DDX3X (Figure S3B and S3C) in primary CD4+ T-cells. CCNT1 (Cyclin-T1) is a member of the pTEFb complex which binds HIV Tat and mediates HIV proviral transcription (Mancebo et al., 1997; Ott et al., 2011; Zhu et al., 1997), while DDX3X is an RNA helicase which binds HIV Rev (Naji et al., 2012; Yedavalli et al., 2004). We used Cas9-RNPs to knockout each gene alone or in combination in primary CD4+ T-cells harvested from four donors. In agreement with the vE-MAP results, we found that each combination led to an exacerbated decrease in HIV infectivity compared to the corresponding single-gene knockouts. These observations reinforce the notion that depletion of CNOT subunits potentiates a reduction in HIV infection in both cultured HeLa cells and primary human T-cells.
Our initial polyclonal knockouts of CNOT1 utilized guide RNAs targeting the first common exon (exon 2) of the open reading frame (Figure 4E; CNOT1 guide RNA-1). Interestingly, this knockout specifically reduced the amount of the large CNOT1 isoform but did not impact the smaller isoform visible by western blot (Figure 4D). This was inconsistent with isoform data in Uniprot, which indicated that exon 2 is shared between all reported CNOT1 splice isoforms. Instead, our data suggested that the smaller CNOT1 isoform may originate from an alternative downstream start site (Figure 4E). In support of this hypothesis, application of a guide RNA targeting exon 46 (CNOT1 guide RNA-2) decreased both of the CNOT1 isoforms (Figure 4F). Interestingly, the impact on HIV infection was identical regardless of which of these guide RNAs was utilized, indicating that knockout of the CNOT1 large isoform alone is sufficient to inhibit HIV infection (Figure 4G). Subsequent screening of guide RNAs targeting multiple CNOT1 exons supported an alternative start site in a region after exon 7, and sequence analysis indicated that the first in-frame start codons after exon 2 are located in exon 8 (Figure 4E and S4).
CNOT1 is by far the largest subunit of the CNOT complex and is the scaffold on which the rest of the CNOT subunits assemble. The CNOT1 N-terminus is known to mediate binding of CNOT10 and CNOT11, subunits of the CNOT complex in higher eukaryotes with unknown function (Bawankar et al., 2013; Mauxion et al., 2013; Xu et al., 2014). To verify that the large CNOT1 isoform specifically interacts with CNOT10 and 11 in CD4+ T-cells, we performed differential immunoprecipitations (IPs) of endogenous CNOT1 from primary CD4+ T-cells. We affinity-purified both CNOT1 isoforms by antibody IP targeting a common epitope, and compared the preys detected by mass spectrometry in wild-type cells versus cells in which the CNOT1 large isoform was knocked out (using CNOT1 guide RNA-1 or 3). Knockout of the CNOT1 large isoform reduced co-immunoprecipitation of CNOT10 and CNOT11 without impacting enrichment of other CNOT subunits (Figure 4H and Table S3). These data confirm that CNOT10 and 11 specifically associate with the CNOT1 large isoform.
Since knockout of the CNOT1 large isoform is sufficient to reduce HIV infection, we next knocked out CNOT10 and CNOT11 from primary CD4+ T-cells and challenged with HIV GFP reporter virus to determine whether they are also required for HIV infection. Knocking out CNOT10 or CNOT11 led to a ~10-fold decrease in HIV infection (Figure 4I and 4J), while knocking out CNOT2, 6L and 7 led to much smaller decreases in infectivity (Figure S5). We conclude that CNOT10 and 11 are the major mediators of CNOT1’s effect on HIV infection.
CNOT1, 10 and 11 are required for suppression of interferon-stimulated genes
To characterize the impact of CNOT1 and CNOT10 knockout on gene expression in primary CD4+ T-cells, we generated polyclonal knockouts in cells harvested from four healthy donors and profiled global transcriptomic changes by RNAseq. Disruption of CNOT1 and CNOT10 led to highly similar profiles of dysregulated genes (Spearman correlation = 0.9429) (Figure 5A and Table S4). Pathway analysis of genes upregulated by CNOT10 knockout (FDR < 0.01, Log2FC ≥ 1.0) indicated a strong enrichment for interferon-stimulated genes (ISGs), suggesting that expression of CNOT1 and CNOT10 may suppress innate immunity (Figure 5B). To determine whether this function is specific to CNOT1 and CNOT10, we deleted CNOT1, 2, 10 or 11 in primary CD4+ T-cells, and immunoblotted for the ISG IFIT1 (Figure 5C). Knockout of CNOT1, 10 or 11 dramatically upregulated expression of IFIT1, supporting a role for the CNOT10/11 submodule in regulating type 1 interferon response in primary CD4+ T-cells (Figure 5C). In a separate set of experiments, we also found that CNOT1 knockdown upregulates ISG15 and IFIT1 in A549 lung epithelial cells, and reduces spreading infection of influenza-A, suggesting that depletion of CNOT1 may cause a pan-viral restriction of infection (Figure S6).
Figure 5: CNOT1, 10 and 11 are required to suppress interferon-stimulated genes in T-cells.
(A) Polyclonal knockouts of the CNOT1 large isoform and of CNOT10 were generated in CD4+ T-cells, and the impact on RNA transcripts determined via RNAseq (NT = non-targeting, Spearman correlation = 0.9429). See also Table S4. (B) Pathway analysis was used to profile genes upregulated by CNOT10 polyclonal knockout (FDR < 0.01, Log2FC ≥ 1). (C) Knockout of the CNOT1 large isoform, CNOT10 and CNOT11, but not CNOT2, causes a dramatic upregulation of the interferon stimulated gene IFIT1 in primary CD4+ T-cells (interferon alpha-2a treatment added to wild-type cells as a positive control). (D) The top 30 genes (Library) upregulated by CNOT10 knockout were targeted for polyclonal knockout in primary CD4+ T-cells (utilizing three individual guide RNAs per gene) in combination with CNOT10 knockout or interferon alpha-2a treatment. The reduction in HIV infection caused by CNOT10 knockout was reversed by knocking out the IRF7 gene with either one of two guide RNAs (IRF7 guide 1 and IRF7 guide 3). None of the library gene knockouts reverted the effect of IFN alpha-2a. See also Figure S7 and Table S5. (E) Schematic of the CNOT complex. The N-terminal region of CNOT1, CNOT10 and CNOT11 are required for suppression of interferon stimulated genes.
We hypothesized that the reduction of HIV infection accompanying knockout of CNOT1, 10 or 11 resulted from upregulation of ISGs. To identify which ISGs mediate this function, we targeted each of the top 30 genes upregulated by CNOT10 knockout (FDR < 0.01) with 3 guide RNAs each. We knocked out these genes in stimulated primary CD4+ T-cells harvested from two donors, in combination with either CNOT10 knockout or interferon alpha-2a treatment. The CNOT10 knockout phenotype was only rescued by combinatorial knockout with IRF7 guides 1 or 3, while interferon treatment was not rescued by IRF7 knockout (Figure 5D, Figure S7, Table S5). This observation indicates that the impact of CNOT10 knockout on HIV is mediated through an IRF7-dependent pathway (Figure 5E).
Discussion
A role for the CCR4-NOT complex in regulating innate immunity and HIV infection
This study reports a role for CNOT1, and its poorly characterized binding partners CNOT10 and 11, in suppressing innate immunity in primary CD4+ T-cells to mediate HIV infection. The CNOT complex has many reported cellular functions, including regulation of RNA transcription, translation, and RNA degradation (Miller and Reese, 2012), and has previously been associated with regulation of innate immunity. A recent genome-wide screen identified CNOT2 and CNOT3 as required for suppression of ISG expression (Lumb et al., 2017), and hCaf1/CNOT7 has been reported to suppress expression of ISGs through interaction with STAT1 and restriction of its localization to the cytoplasm (Chapat et al., 2013). In agreement with this previous work, we observe a minor decrease in HIV infection accompanying CNOT2 knockout; however, CNOT7 knockout did not cause any HIV infection phenotype in primary CD4+ T-cells in our system, suggesting that CNOT7 does not regulate ISG expression in this cell type. In the context of HIV infection in primary T-cells, knockout of the CNOT subunits 10 and 11 had by far the strongest HIV phenotype, mediated by upregulation of innate immunity. Our data also supports the existence of an N-terminally truncated CNOT1 isoform, and we show that the CNOT10/11 module binds specifically to the N-terminus of full-length CNOT1. This finding supports the existence of two CNOT complexes, with one containing a CNOT10/11 module.
Given the negative interactions between CNOT1 and the HIV host-dependency factor CCNT1/Cyclin-T1, which is known to mediate HIV transcription (Mancebo et al., 1997; Ott et al., 2011; Zhu et al., 1997), it is tempting to speculate that CNOT too may directly support HIV transcription. Indeed, the CNOT complex has been associated with transcriptional regulation (Miller and Reese, 2012). While we cannot eliminate this possibility, the positive genetic interaction between CNOT10 and the transcription factor IRF7 in T-cells is consistent with an indirect effect of CNOT knockout via upregulation of innate immune genes by IRF7. More work is required to understand how these innate immune genes may mediate the impact of CNOT1 depletion on HIV.
Interestingly, clinical data supports a role for not only IRF7 but also the CNOT complex in vaccine-mediated immunological response against HIV. Transcriptional profiling following administration of the RV144 HIV vaccine shows elevation of interferon-stimulated genes possessing putative IRF7 binding sites, suggesting that IRF7 contributes to protection from HIV acquisition (Fourati et al., 2019). Additionally, an intronic SNP in CNOT1, rs7188697 A/G, has been associated with response to an autologous dendritic cell-based therapeutic HIV vaccine, with the A/A genotype associated with a significant drop in HIV viral load following cell-based vaccine treatment and cessation of anti-retroviral therapy (Lu et al., 2004; Moura et al., 2014). Further investigation of IRF7-dependent HIV restriction factors, as well as deeper understanding of human variation at the CNOT1 locus, may enrich our knowledge of host defenses against HIV infection, and possibly illuminate molecular strategies to improve HIV vaccine efficacy in poor responders.
The application of genetic interaction mapping to study the host-pathogen interface
By quantifying individual genetic interactions and identifying highly correlated functional modules, vE-MAP analysis provides contextual information regarding the function of host factors and the effect of small molecules and viral mutations. The vE-MAP approach also represents a powerful tool to discover novel viral host-dependency factors by uncovering synergistic genetic interactions that escape detection in single-perturbation genetic screens. Overall, our study establishes a foundation for the interrogation of genetic dependencies underlying pathogen infection using large-scale, pairwise genetic interaction mapping.
Large-scale E-MAPs reveal clusters of genes with similar interaction patterns that can be indicative of functional modules. While previous E-MAPs have identified protein complexes involved in cellular proliferation (Braberg et al., 2013; Collins et al., 2007; Roguev et al., 2008; Schuldiner et al., 2005), our study demonstrates the potential of an E-MAP to identify host modules mediating pathogen infection. A striking example is our resolution of structural submodules of the eIF3 ribosome initiation complex, which we had previously found to be implicated in HIV replication (Jäger et al., 2012); the rich genetic interactions and clustering of eIF3 knockdowns in our vE-MAP further support its role in HIV infection. In addition, the vE-MAP was able to score genetic interactions for genes with modest single-perturbation phenotypes that might be disregarded in a single-perturbation genetic study. This strength of the vE-MAP is of particular importance for the investigation of host-pathogen interactions, because of functional redundancy within host pathways and because pathogens adopt dynamic routes to infection.
Beyond uncovering host regulators of infection, the vE-MAP approach may be used to investigate the impact of small molecules on viral infection. First, we can use epistatic genetic interactions to link compounds with the genes and molecular pathways they impact, thus helping clarify the function of poorly characterized compounds and poorly characterized genes. Second, we can use synergistic interactions to identify perturbations that potentiate the phenotype of small molecules, thus discovering new gene targets and small molecules that can inform combinatorial drug strategies. In the context of HIV cure research, genetic interaction analysis could be used to find the molecular targets of poorly characterized latency reversal agents, and to identify synergistic gene targets which potentiate reactivation of latent HIV.
The vE-MAP approach also holds exciting potential for the study of viral mutants. We were able to quantify known functional interactions between host proteins and the N74A HIV capsid mutant using our high-throughput pipeline, suggesting that our approach could be expanded to study large numbers of viral mutants. Viral proteins often interact with multiple host complexes, and the pairing of vE-MAP analysis with saturating mutagenesis could resolve host functional modules binding to distinct surfaces of a viral protein.
In addition to the arrayed genetic interaction strategy employed in this study where perturbations are physically combined in individual microwells, there is also a pooled approach where genetic perturbations are introduced by pairwise cloning of genetic elements (such as shRNAs or sgRNAs) in ultracomplex libraries which are then transduced into large cultures, selected with high-throughput phenotypic assays, and analyzed by next generation sequencing (Bassik et al., 2009; Du et al., 2017; Horlbeck et al., 2018; Shen et al., 2017). The pooled genetic interaction approach is highly scalable, but its implementation poses several challenges for viral studies, including the handling of large infected cultures and optimization of high-throughput selection methodology for viral phenotypes. Pooled approaches are also limited to cell intrinsic perturbations, precluding analysis of small molecules at scale. We find arrayed genetic interaction screening to be highly flexible, permitting the straightforward interrogation of gene perturbations, small molecule queries and viral mutants. Arrayed screening also permits the simultaneous interrogation of multiple phenotypes relevant to both host biology and the pathogen lifecycle using multiplexed optical readouts (e.g. fluorescence and luminescence assays) and biochemical readouts (e.g. viral titre). In subsequent pathogen E-MAP studies, utilization of the arrayed format will also permit spreading infection assays within individual microwells, enabling phenotypic interrogation of the entire pathogen lifecycle.
Our arrayed genetic interaction pipeline can be deployed to study any number of pathogens, and is most useful where an enriched set of host-dependency genes has been experimentally associated with the pathogen lifecycle. Enriched lists of host-dependency factors may be derived from genome-wide single perturbation screens, GWAS studies mapping human susceptibility to infection, protein interaction datasets, as well as other datasets. Notably, numerous host-pathogen protein-protein interaction datasets have been generated, constituting a rich resource to utilize for the construction of vE-MAP libraries studying other viral and bacterial pathogens (Batra et al., 2018; Davis et al., 2015; Diep et al., 2019; Eckhardt et al., 2018; Li et al., 2019; Mirrashidi et al., 2015; Penn et al., 2018; Ramage et al., 2015; Shah et al., 2018).
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nevan Krogan (nevan.krogan@ucsf.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Parental stocks of HeLa (female) and HEK293T/17 (female) human cell lines were obtained from the UCSF Cell Culture Facility (CCF), and the A549 (male) cell line was procured from the American Type Culture Collection (ATCC). No authentication was performed following cell line procurement from these repositories. HeLa cells were cultured in Eagle’s Medium Essential Medium with Earle’s Balanced Salt Solution, supplemented with 10% fetal calf serum, L-glutamine (2 mM), sodium pyruvate (110 μg/mL) and penicillin-streptomycin (110 μg/mL). HeLa cells stably expressing mCherry were cultured as above with the addition of 300 μg/mL Hygromycin-B. HEK293T/17 and A549 cells were cultured in Dulbecco’s Modified Eagle Medium supplemented with 10% fetal calf serum, L-glutamine (2 mM), sodium pyruvate (110 μg/mL) and penicillin-streptomycin (110 μg/mL). Cells were maintained in a jacketed incubator at 37°C and 5% CO2.
Primary Human CD4+ T-Cells
Blood from healthy anonymous donors was harvested from Trima Leukoreduction chambers (Vitalant, San Francisco CA). Sex or gender information was not retained for these anonymous donor samples, and human subjects approval was not required for use of anonymous blood samples. Primary CD4+ T-cells were purified by positive selection using a Fabian automated purification system (IBA Lifesciences). Stimulated primary CD4+ T-cells were cultured in RPMI-1640 supplemented with 10% fetal calf serum, L-glutamine (2 mM), sodium pyruvate (110 μg/mL), penicillin-streptomycin (110 μg/mL), 25mM HEPES, and 40 U/mL IL-2 IS (Miltenyi), and were maintained at a confluence of 1–3 million/mL. Cells were maintained in a jacketed incubator at 37°C and 5% CO2.
METHOD DETAILS
All experiments were performed without blinding.
Generation of mCherry HeLa Cells
The fluorescent protein mCherry was cloned into the retroviral vector pQCXIH and transduced into HeLa cells as follows. Transfection complexes were formed by mixing 200 μL Optimem (Thermo) with 21 μL TransIT-293 reagent (Mirus), incubated for 5 minutes, then mixed with 7 μg pQCXIH-mCherry plasmid. Transfection complexes were added to 10 cm dishes containing Phoenix helper cells (Kinsella and Nolan, 1996), and these were incubated overnight at 37°C. Media was replaced 24 hours post transfection and plates were moved to a 32°C incubator. Viral supernatant was harvested 48 hours post transfection, filtered through a 0.45 μm filter, mixed with Hexadimethrine bromide / Polybrene (Sigma-Aldrich; 5 μg/mL final concentration), and deposited on top of HeLa cells in a 6-well plate, which was sealed with parafilm and centrifuged at 1200g for 60 minutes in a tilt bucket rotor. Transduced cells were selected in 300 μg/mL Hygromycin-B, and clonal cells were selected for homogenous expression of cytoplasmic mCherry amenable to high-throughput microscopy and automated masking of individual cells.
esiRNA Synthesis
Endonuclease-prepared small interfering RNA (esiRNA) was prepared in 96-well format based on our previously published protocol (Roguev et al., 2013). PCR amplicons were chosen to minimize generation of off-target esiRNAs using either published pre-designed amplicons or the web design tool DEQOR (Henschel et al., 2004). The 5’ anchor sequence ‘GGGCGGGT-’ was appended to these gene-specific primers (Table S6). HeLa cell RNA was harvested using an RNeasy kit (Qiagen) and genomic DNA elimination and reverse transcription was accomplished using the QuantiTect reverse transcription kit (Qiagen). A first round of PCR (95°C 2 min; 95°C 30s, 65°C 30s 72°C 60s 40 cycles; 72°C 5 min) was utilized to amplify cDNA amplicons in 96-well format and add the anchor sequence using Taq polymerase (Sigma). Products from the first PCR were diluted 40x, and one microliter was added to a 50 μL second round PCR (95°C 2 min; 95°C 30s, 42°C 45s 72°C 60s 5 cycles; 95°C 30s, 60°C 45s, 72°C 60s 35 cycles; 72°C 5 min) with the T7 primer 5’-TAATACGACTCACTATAGGGAGACCACGGGCGGGT-3’ (800 nM final primer concentration) to amplify the amplicons and append a T7 promotor to each end. Agarose gel electrophoresis was used to confirm expected sizes for each amplicon, and amplicons with unexpected size or no amplification were rejected and resynthesized individually with redesigned gene-specific primers.
Unpurified DNA from the second-round PCR reaction was added directly to an in-vitro T7 RNA transcription reaction, composition below. RNAse-free 5X T7 Polymerase Buffer (400 mM Hepes pH 7.6, 120 mM MgCl2, 10 mM spermidine, 200 mM DTT) was made in advance, aliquoted and stored at −20°C.
| Reagent | Volume (μL) |
|---|---|
| H2O | 4.1 |
| 25 NTP mix (NEB - 25 mM each) | 14.4 |
| 5x T7 Buffer (Promega) | 10.0 |
| T7 Polymerase (Promega - 80 U/μL) | 0.5 |
| Yeast Inorganic Pyrophosphatase (YIPP) (NEB - 100 U/mL) | 0.5 |
| RNAsin (Promega) | 0.5 |
| Unpurified PCR #2 Product | 20.0 |
| Total Volume | 50.0 |
The T7 reaction was incubated at 37oC for 5.5 hours, then strands were annealed (90°C 1 min, 72°C 3 min, 50°C 3 min, 25°C 3 min, 4°C hold). Each reaction was treated with 2.5 units of RNAse-free DNAse in manufacturer-supplied buffer (Promega) at 37°C for 30 minutes, then purified in 96-well format using the RNeasy 96-well purification kit (Qiagen). Yield of each well was quantified using an 8-channel Nanodrop, and expected band sizes were confirmed by agarose electrophoresis.
Double-stranded RNA amplicons were digested into esiRNA fragments using GST-RNAse III . Briefly, 60 μg dsRNA was mixed with sufficient RNAse III (titrated in advance) in RNAse III buffer and 20 mM MnCl2. This reaction was incubated at 37°C for 30 minutes, placed on ice, and inactivated by addition of 50 mM EDTA (final concentration). 10X RNAse III Buffer (500 mM Tris pH 7.5, 10 mM DTT, 500 mM NaCl) was made in advance, aliquoted and stored at −20°C prior to use.
Digested esiRNA was purified by mixing the RNAse III reaction with an equal volume of RNeasy buffer RLT, then another volume of ethanol to a final 33% ethanol concentration. This was passed through a single RNeasy column setup to eliminate any residual undigested dsRNA, and flow-through was collected. Additional ethanol was added to the flow through for a final 75% ethanol concentration, and this was passed through a second RNeasy column to bind esiRNAs, washed twice with 500 μL buffer RPE, and eluted in RNAse-free water. Complete digestion was confirmed for all esiRNAs by electrophoresis on 15% TBE-PAGE gels and ethidium bromide staining. Final esiRNA concentrations were quantified by Nanodrop (Thermo), and normalized using a BioMek FX 96-well pipetting robot (Beckman Coulter).
In small scale experiments our esiRNAs effectively reduced mRNA expression of target genes by 65–88% (Figure S1A). We also confirmed that esiRNA preparations did not cause upregulation of the interferon regulated genes OAS1 and MX1, in contrast to a blunt-ended 30bp fragment or interferon treatment itself (Figure S1B).
vE-MAP esiRNA Transfection and HIV Infection
Barcoded library plates (384-well assay plates; Greiner) were pre-arrayed with 2 ng esiRNA per well in a total of 10 μl nuclease-free water, using a Biomek FX liquid handler. These plates were sealed and stored short term at −20°C. Plates were thawed on the day of transfection in batches of 40, and query esiRNA was diluted in cold OptiMEM for a final concentration of 0.4 ng/μl and dispensed (5 μl/well) into library plates; plates were then mixed using a MixMate (Eppendorf) and centrifuged. RNAiMAX was diluted in warm OptiMEM to generate a master mix (90 ml OptiMEM and 337 μl RNAiMAX) so each well would contain a final concentration of 0.0375 μl RNAiMAX. This RNAiMAX/OptiMEM mixture was mixed gently and incubated at RT for 5 min, then dispensed onto each plate (10 μl per well) and incubated for 20 minutes. While this incubation was taking place, HeLa cells (expressing cytoplasmic mCherry) were trypsinised and resuspended in antibiotic-free phenol red-free MEM (Gibco) supplemented with 20% FBS (Gibco) to a final of 1.2 × 104 cells/ml. 25 μl of dilute cells were then dispensed per well of each plate, then placed in an incubator. Three days post-transfection, VSV-G pseudotyped HIV luciferase reporter virus was diluted in complete MEM media and added to each plate, and rocked two minutes to facilitate mixing.
Automated Microscopy
Five days post-transfection (and two days post-virus addition), plates were imaged using a CellInsight microscope (Thermo) connected to an Orbitor robotic plate handler and a Cytomat (Thermo) automated plate incubator. HeLa cells expressing cytoplasmic mCherry were imaged using a 2.5x objective, and cells were masked and counted using Thermo high content analysis software. Cell count by automated microscopy was chosen in lieu of metabolism-based viability reagents to save on consumables costs and because recent publications have demonstrated discrepancies between cell count and reported metabolism-based readouts (Chan et al., 2013).
HIV Expression Luminescence Assay
To maximize the sensitivity of our HIV expression detection, we employed a luciferase reporter virus, and adapted a home-made luciferase assay with wide dynamic range which is amenable to high-throughput quantification (Connor et al., 1995; Siebring-van Olst et al., 2013).
Post-imaging, all plates were processed for viral luciferase reporter readout in batches of 10 using a home-made glow luciferase assay modified from a published method (Siebring-van Olst et al., 2013). Every two weeks a 2X base buffer was prepared in ddH2O: 40 mM Tricine pH 7.8, 200 μM EDTA, 2.14 mM MgCO3 pH 7.8, 5.35 mM MgSO4, 2% Triton-X-100, with Antifoam A (Sigma; A6582). Directly before running plates, water and freshly thawed reagents were supplemented to this base buffer for a final 1X buffer with the following final concentrations: 34 mM DTT, 500 μM ATP, 400 μM D-luciferin. All plates were read on an EnSpire multimode plate reader (PerkinElmer).
Analysis of Genetic Interaction Data
Since genetic perturbations could affect the growth of the cells as well as virus infectivity, we quantified cell count in each well using automated microscopy, and utilized this data to normalize the viral luciferase signal. The impact of cell count on luciferase signal was quantified by cell titration in the presence of constant virus, and these data were fitted to a one-phase exponential decay function with the following parameters: y0 = 28913, A = −30223, tau = 680.18. This function was then used to internally normalise the luminescence data of every plate run in the HIV vE-MAP screen. Count-normalised luminescence data was then analysed using a previously published genetic interaction analysis tool kit (Collins et al., 2006; 2010; Roguev et al., 2013). Briefly, highly toxic library esiRNA or those exhibiting highly inconsistent phenotypic profiles were filtered out, individual plate batches were scaled using the median of the entire dataset, plate effects were corrected using a parabolic surface fit, and the resulting data was normalized using a Parzen window smoothing. The data was then then log-transformed and genetic interaction scores (S-scores) were computed. S-score profiles were clustered using Cluster 3.0, and the resulting heatmaps were visualized using TreeView (de Hoon et al., 2004; Eisen et al., 1998; Saldanha, 2004).
To calculate enrichment of protein-protein interactions from Bioplex (Huttlin et al., 2017) as a function of correlation between genetic interaction profiles, pairwise Pearson correlation coefficients were computed for all gene pairs in the vE-MAP. Enrichment analysis was then performed using a sliding cut-off of correlation coefficients in the range (−1,1) with a step of 0.05. Random data was generated by permuting the real data 1000 times and final values were calculated by taking the median of the results at each correlation coefficient threshold.
Genetic interaction confidence was assessed based on the observation that S-scores for non-interacting genes are described well by a t-distribution parametrized by the experimental noise. This distribution can be estimated from the experimental data and used to compute the probability of a pairwise genetic interaction score emerging from a non-interacting pair of genes (Collins et al., 2006).
HIV Virus Production
The VSV-G pseudotyped luciferase HIV virus used in our screen was produced by co-transfecting HEK 293T/17 cells with plasmids encoding VSV-G and HIV-1 NL4–3 delta Env luciferase reporter virus (Connor et al., 1995) using linear 25 kDa polyethylenimine (PEI; Polysciences Inc.) (Fukumoto et al., 2010). DNA (1.4 μg pMD2.G, 23.8 μg HIV DNA) was diluted in 625 μl lactate buffered saline (LBS; pH 4.0), and separately 25 μl acidified PEI (5 mg/mL stock) was diluted in 625 μl LBS. Dilute LBS was added to dilute DNA, vortexed aggressively, incubated for 15 minutes RT, then added dropwise onto cultures in a T175 flask and rocked to mix. Media was exchanged 24 hours post transfection, and 48 hours post transfection viral supernatant was centrifuged for 5 min, 300g, sterile-filtered through a 0.22 μm filter unit, aliquoted, frozen on dry ice, and stored at −80°C. VSV-G HIV virus yields were quantified using a p24 fluorescence-linked antigen quantification (FLAQ) assay (Gesner et al., 2014). The quantity of Capsid mutant virus added as query was normalized to wild-type by quantification of p24 in the supernatant.
Wild-type HIV NL4–3 and Nef:IRES:GFP virus used to infect primary CD4+ T-cells was produced using the commercial transfection reagent PolyJet (Signagen). Virus plasmid (12 μg) and 100 μl PolyJet were diluted separately in two tubes of 625 μl serum-free DMEM, then dilute PolyJet was added to dilute DNA and vortexed to mix. After 15 minutes RT the transfection complexes were added to T175 flasks containing 293T/17 cells, and were gently rocked to mix. Virus was harvested 48 hours post transfection, spun at 400g for 5 minutes and filtered through a 0.45 μm filter. For every flask of cells 25 mL viral supernatant was precipitated by addition of 2.4 mL 4M NaCl and 5.5 mL 50% PEG-6000 for two hours at 4 degrees. Precipitated virus was pelleted for 40 minutes at 3500 rpm, resuspended in complete RPMI media at 50x concentration, and frozen in aliquots.
Polyclonal Knockouts in Primary CD4+ T-cells
Polyclonal knockouts were performed in primary CD4+ T-cells as previously described (Hendel et al., 2015; Hultquist et al., 2016). Chemically synthesized guide RNA (crRNA and tracrRNA) was ordered from Dharmacon (Table S9). Blood from healthy donors was harvested from Trima Leukoreduction chambers (Vitalant, San Francisco CA). Primary CD4+ T-cells were purified by positive selection using a Fabian automated purification system (IBA Lifesciences) and stimulated by plate-bound CD3 and suspension CD28 (5 μg/mL) for 48–72 hours prior to nucleofection. For single guide-RNA knockouts, 1 μL crRNA and 1 μL tracrRNA (160 μM each) were mixed and incubated for 35 minutes at 37 degrees to make 80 μM guide RNA, then 2 μL Cas9 nuclease (40 μM) was added with mixing motion to make 4 μL of Cas9-RNP complex and incubated at 37 degrees for an additional 20 minutes. For double knockouts, Cas9-RNP was made for each guide, then final RNP complexes were mixed in combinations of 3 μL + 3 μL (single knockout treatments in double knockout experiments were comprised of non-targeting RNP mixed with a gene-targeting RNP). Cas9-RNP complexes were electroporated into 100,000–500,000 stimulated primary CD4+ T-cells using the Lonza Primary P3 nucleofection kit and a 96-well shuttle connected to a 4D Nucleofector system. After nucleofection, cells were re-stimulated using T-cell activation and expansion beads and 80 IU/mL IL-2 IS (Miltenyi), and cells were split every 2–3 days. Virus infection experiments were performed 5–7 days post electroporation, whereby cells were replica-plated in triplicate and infected with GFP reporter virus. Polyclonal knockout efficiency was assayed by western blotting and/or TIDE analysis of genomic DNA PCR amplicons with Sanger sequencing (Brinkman et al., 2014). TIDE primers were designed using COSMID (Cradick et al., 2014), and in cases of amplification failure primers were redesigned using the Benchling genome sequence viewer (Table S8).
HIV Infection Quantitation by Flow Cytometry
Primary CD4+ T-cells were infected with concentrated Nef:IRES:GFP reporter virus in 96-well U-bottom plates. Cells were fixed 72-hours post infection by addition of 2–4% formaldehyde in PBS, and percent infection was quantified based on GFP expression. Flow cytometry was run using an Attune flow cytometer equipped with a plate sampler (Thermo).
Quantitative PCR Using Primer-Probes
Quantitative real-time PCR to quantify human gene knockdown was performed on HeLa cells transfected in 384-well format to approximate the screening conditions used during EMAP acquisition. Wells were harvested 72-hours post transfection and RNA was purified using an RNeasy kit (Qiagen). Primer-probe assays (IDT) (Table S7) were used to detect gene expression levels with the Sensifast Probe one-step kit (Bioline). All experiments were run using an ABI 7900 qRT-PCR machine (Applied Biosystems). The amplification efficiency of primer-probe sets was assayed and incorporated in relative quantification calculations.
Quantitative PCR Using Intercalating Dye
Quantitative PCR using intercalating dye was performed in conjunction with influenza functional assays. RNA was purified using PureLink RNA Mini Kit (Life Technologies) according to the manufacturer’s instructions. 1 μg of RNA was reverse transcribed with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) using Oligo(dT)20 primers (Life Technologies). Quantitative PCR analysis was performed using cDNA and iQ SYBR Green Supermix (BioRad) on the LightCycler® 480 instrument (Roche).
Mutagenesis of HIV Capsid
Two-step PCR was utilized to generate DNA fragments carrying capsid mutations flanked by arms with sequence homology to the unique BssHII and SpeI sites in the HIV NL4–3 luciferase reporter virus plasmid (primer sequences in Table S8). The NL4–3 backbone was digested using these restriction enzymes and ligation-free cloning was completed using InFusion (Clontech).
CNOT1 Antibody Immunoprecipitation
To perform large-scale knockout of the CNOT1 large isoform, ten million stimulated T-cells were pelleted and resuspended in 100 μL Lonza P3 primary cell buffer, then mixed with 15 μL Cas9RNP complex targeting the CNOT1 large isoform (using crRNAs CNOT1–1 or CNOT1–3) or a nontargeting control. Nucleofections were performed in cuvette format using the Lonza 4D nucleofector system, and cells were restimulated following nucleofection using T-cell stimulation and expansion beads (Miltenyi). Following one week of cell expansion, total endogenous CNOT1 was immunoprecipitated as follows. Primary CD4+ T-cells from each donor were counted and subjected to centrifugation three times at 400g for 10 minutes, initially to collect cells and after each of two washes with Dulbecco’s Phosphate-Buffered Saline without calcium and magnesium. Cell lysate was adjusted to 50 million cells per milliliter in IP Buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA) supplemented with 0.5% Nonidet P 40 Substitute (Fluka Analytical) and cOmplete mini EDTA-free protease and PhosSTOP phosphatase inhibitor cocktails (Roche) and incubated on a tube rotator for 30 minutes before centrifugation at 18,000g for 10 minutes. Equal volumes of cleared lysate from each donor, ~50 million cells, were first incubated with 5 μg rabbit anti-CNOT1 monoclonal antibody for 2 hours. For each sample, 40 μl Protein A Dynabead slurry (Invitrogen) was equilibrated in IP Buffer supplemented with 0.05% Nonidet P-40 Substitute (2 × 1 mL) and then added to lysates incubating with antibody for an additional 2 hours. After removing lysates, Dynabeads were washed three times with IP Buffer supplemented with 0.05% Nonidet P-40 Substitute (3 × 1 mL) and transferred to a new tube with a final wash in 1 mL IP Buffer. After each addition of buffer, Dynabeads were nutated for 5 minutes before proceeding. Proteins were eluted by gently agitating Dynabeads with 50 μL 0.1 M Glycine pH 2.5 on a vortex mixer at room temperature for 30 minutes. After removing from Dynabeads, eluates were neutralized with 2.5 μL 1 M Tris pH 9.0. Prior to submitting 20 μl for trypsin digest, 10% of each sample was assessed by silver stain. All centrifugation and incubation steps were performed at 4°C unless otherwise indicated. Additionally, during incubation steps, tubes were on a tube rotator or nutating mixer.
To prepare samples for LC-MS/MS analysis, immunoprecipitated proteins were denatured and reduced in 2 M urea, 10 mM NH4HCO3, 2 mM DTT for 30 min with constant shaking on a Thermomixer R incubator set at 60°C, alkylated in the dark with 2 mM iodoacetamide for 45 min at room temperature and digested overnight at 37°C with 80 ng trypsin (Promega). Following digestion, peptides were acidified with formic acid and desalted using C18 ZipTips (Millipore) according to the manufacturer’s specifications. Samples were resuspended in 4% formic acid, 4% acetonitrile solution, and separated by an 80 minute reversed-phase gradient over a nanoflow C18 column (Dr. Maisch). Peptides were directly injected into an Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo), with all MS1 spectra collected in the orbitrap, and MS2 spectra collected in the ion trap. Raw MS data were searched against the human proteome (Uniprot canonical protein sequences downloaded May 28, 2015) using MaxQuant (Cox and Mann, 2008; Cox et al., 2014). Differential analysis of non-targeting knockouts versus CNOT large isoform knockouts across four donors was performed using MSstats (Choi et al., 2014).
RNAseq on CNOT1 and CNOT10 T-cell Knockouts
Stimulated primary T-cells harvested from four healthy donors were electroporated with Cas9-RNPs complexed with CNOT1–1, CNOT10, or non-targeting guide RNAs as detailed above. Following electroporation to generate polyclonal knockouts, cells were restimulated and expanded for one week, after which DNAse-treated RNA was purified using an RNeasy Mini Kit and on-column DNAse digestion (Qiagen). RNA-Seq libraries were prepared using the Ovation RNA-seq Universal kit (NuGEN). Reverse transcribed DNA was fragmented with the Covaris S-series System, and during library construction uninformative transcripts were depleted using Insert Dependent Adaptor Cleavage (InDA-C) technology (InDA-C uses species-specific probes to eliminate undesirable transcripts like ribosomal RNA). The resulting RNA-seq libraries were analyzed by Bioanalyzer and quantified by qPCR (KAPA). High-throughput sequencing was done using a HiSeq 4000 (Illumina).
RNAseq analysis was performed as follows. Input sequences were provided in FASTQ format. Trimming of known adapters and low-quality regions of reads was performed using Fastq-mcf (Aronesty, 2011). Sequence quality control was assessed using the program FastQC and RSeQC (Andrews; Wang et al., 2012). Alignment of the provided samples to the reference genome was performed using STAR 2.5.2a (Dobin et al., 2013). Reads were aligned to the human hg38 reference assembly indicated in the header of the differential expression file. Reads were assigned to genes using “featureCounts”, part of the Subread suite (Liao et al., 2014). Gene-level counts were arrived at using Ensembl gene annotation, in GTF format. We calculated differential expression P-values using edgeR, an R package available through Bioconductor (Robinson et al., 2010). We first filtered out genes where there were not at least two samples with at least 5 (raw) reads. This is our minimum threshold for expression detectability. After excluding these genes, we re-normalized the remaining ones using calcNormFactors in edgeR (Robinson and Oshlack, 2010). Calculation of P-values is performed in edgeR for the differential expression between conditions (Robinson et al., 2010). The pairwise comparisons between the conditions were performed accounting for the donor source for each sample as a potential confounder of the relationship between gene expression and the condition. EdgeR uses a negative binomial distribution as a model for expected gene expression. Specifically, edgeR estimates the genewise dispersions by conditional maximum likelihood, conditioning on the total count for that gene. An empirical Bayes procedure is used to shrink the dispersions towards a consensus value, effectively borrowing information between genes. Differential expression is assessed for each gene using an exact test analogous to Fisher’s exact test, but adapted for overdispersed data (Robinson and Oshlack, 2010; Robinson and Smyth, 2007; 2008; Smyth and Verbyla, 1996). Finally we use the built-in R function “p.adjust” to calculate the FDR (false discovery rate) for each P-value, using the Benjamini-Hochberg method (Benjamini and Hochberg, 1995). Gene set enrichment analysis was performed utilizing Enrichr (Chen et al., 2013; Kuleshov et al., 2016).
Influenza-A Infection Assays
A549 cells were seeded into 6-well plates at 0.3*106 cells per well (in 1.5 mL media). The following day they were transfected with siRNA using Lipofectamine RNAiMAX reagent (Life Technologies, Cat. No. 13778–075). We mixed 250 μL OptiMEM + 5 μL 20 μM siRNA in one tube, mixed 250 μL OptiMEM + 5 μL RNAiMAX in a separate tube, and finally combined and mixed the siRNA and transfection reagent, incubated for 20 minutes at room temperature, then added to our cells. For single-cycle infection assays, cells were exposed to GFP-expressing influenza virus 48 hours after transfection in Opti-MEM media at MOI=0.1 for 1h at 37°C and 5% CO2, then virus was removed and cells incubated in complete media for a further 8 hours before the percentage of infected cells (% GFP) was determined by flow cytometry. For spreading infection assays, cells were exposed to GFP-expressing influenza virus in Opti-MEM media at MOI=0.01 with 0.5 μg/mL trypsin for 1 hour at 37°C and 5% CO2, before virus was removed and cells incubated in complete media. Viral supernatant was collected at 24 hours and 48 hours post infection, and applied to fresh target cells. Percentage of infected target cells (% GFP) was determined 8 hours following infection by flow cytometry. Influenza-A PR8 GFP reporter virus (PR8-GFP) was used for all influenza infection assays (Martínez-Sobrido et al., 2010).
QUANTIFICATION AND STATISTICAL ANALYSIS
All genetic interaction knockdown combinations in HeLa cells were performed in quadruplicate, with a few exceptions where plate failures necessitated utilizing triplicate data. Genetic interaction quantification was performed as detailed in ‘Method Details: Analysis of Genetic Interaction Data’ utilizing a previously published algorithm and analysis toolkit (Collins et al., 2006; 2010; Roguev et al., 2013). In primary T-cell polyclonal knockout experiments, cells from each donor were electroporated with Cas9-RNP complexes and replica-plated in triplicate to perform infectivity assays. Flow cytometry analysis of percent HIV-infected cells was performed using FlowJo, and mathematical calculations including normalization to non-targeting control, log2-transformation, mean, and standard deviation was performed in Microsoft Excel and Graphpad Prism. Quantification of polyclonal knockout efficiency was performed using Sanger sequencing and TIDE (Brinkman et al., 2014). CNOT1 endogenous immunoprecipitation experiments were performed in primary CD4+ T-cells harvested from four donors, and differential quantification and standard error were calculated based on mass spectrometer intensity values using MSstats (Choi et al., 2014). RNAseq on CNOT1 and CNOT10 knockouts were performed in primary T-cells harvested from four healthy donors, and differential quantification of gene transcripts was performed as described in ‘Method Details: RNAseq on CNOT1 and CNOT10 T-cell Knockouts’. Pathway analysis of RNAseq data was performed using Enrichr (Chen et al., 2013; Kuleshov et al., 2016). Quantitative PCR assays were performed on triplicate samples, threshold cycle (Ct) values were calculated using software on the platform utilized (ABI 7900 for HeLa experiments or Roche LightCycler 480 for A549 experiments), and differential quantification of expression levels and standard deviation calculated using Microsoft Excel. Influenza infection data was gathered across three experiments, percent infected was calculated by flow cytometry, and mean and standard deviation calculated using Microsoft Excel and Graphpad Prism. Details regarding number of donors, replicates, or other statistical analyses not indicated here is specified per experiment in figure legends and text.
DATA AND CODE AVAILABILITY
The accession number for the RNAseq data reported in this paper is GEO: GSE144142. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144142. Genetic interaction scores, differential proteomics analysis, processed RNAseq data, and genetic interaction data in primary T-cells are attached as supplemental data. The analysis pipelines supporting the current study are available from the lead contact on request.
Supplementary Material
Table S1: Single knockdown behavior for each esiRNA in the HIV vE-MAP (related to Figures 1C, 1D, and 2B)
Table S2: Genetic interaction scores (S-score) quantified with MLN-4924 query treatment (related to Figure 3C–3E)
Table S3: Differential CNOT1 immunoprecipitation proteomics data with CNOT1 large isoform knockout (related to Figure 4H).
Table S4: Differential transcriptome analysis of CNOT1 and CNOT10 knockouts in primary CD4+ T-cells (related to Figure 5A)
Table S5: HIV infection data for polyclonal knockouts in primary CD4+ T-cells +/− CNOT10 knockout or interferon alpha-2a treatment (related to Figure 5D)
Data S1: HIV vE-MAP Symmetric Knockdown Matrix (related to Figures 2, 3, 4A–4B, S1, S2). The vE-MAP can be visualized using Java Treeview software.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-BTF3 | Abcam | ab203517, RRID:AB_2783867 |
| Rabbit polyclonal anti-CCNT1 | Bethyl | A303–499A-M, RRID:AB_2781257 |
| Rabbit monoclonal anti-CNOT1 | Cell Signaling | 44613, RRID:AB_2783868 |
| Rabbit polyclonal anti-CNOT2 | Sigma/Atlas | HPA067711, RRID:AB_2685893 |
| Rabbit polyclonal anti-CNOT10 | Bethyl | A304–899A, RRID:AB_2621094 |
| Rabbit polyclonal anti-CNOT10 (Figure S7) | Sigma/Atlas | HPA041450 RRID:AB_2677487 |
| Rabbit polyclonal anti-CNOT11 | Sigma/Atlas | HPA069823, RRID:AB_2621094 |
| Rabbit polyclonal anti-CPSF6 | Proteintech | 15489, RRID:AB_10694140 |
| Mouse monoclonal anti-DDX3X | Biolegend | 658602, RRID:AB_2562957 |
| Mouse monoclonal anti-gamma-adaptin | Peden Lab, University of Sheffield. Originally published by Ahle et al., 1988 EMBO J. | mab100/3 RRID: AB_772494 (commercial source for this antibody) |
| Mouse monoclonal anti-GAPDH | Sigma | G8795, RRID:AB_1078991 |
| Rabbit monoclonal anti-GAPDH (Figure S6) | Cell Signaling | 5174T, RRID:AB_10622025 |
| Rabbit monoclonal anti-Histone H2B | Cell Signaling | 12364S, RRID:AB_2714167 |
| Rabbit monoclonal anti-IFIT1 | Cell Signaling | 14769S, RRID:AB_2783869 |
| Rabbit monoclonal anti-IRF7 | Cell Signaling | 13014S, RRID:AB_2737060 |
| Mouse monoclonal anti-TNPO3 | Abcam | ab54353, RRID:AB_883189 |
| Bacterial and Virus Strains | ||
| HIV-1 pNL4–3 delta-Envelope Firefly Luciferase-in-Nef | Connor et al., 1995 | N/A |
| HIV-1 pNL4–3 delta-Envelope Firefly Luciferase-in-Nef Capsid N74A mutant | This manuscript | N/A |
| HIV-1 pNL4–3 | NIH AIDS Reagent Program | 114 |
| HIV-1 pNL4–3 Nef:IRES:GFP | Kirchhoff Lab | N/A |
| Influenza-A PR8-GFP | Martínez-Sobrido et al., 2010 | N/A |
| Biological Samples | ||
| Leukoreduction chambers from healthy human donors | Vitalant | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cas9-NLS purified protein | QB3 Macrolab | N/A |
| Deposited Data | ||
| RNAseq data for primary T-cell knockouts of CNOT1 and CNOT10 | This manuscript | GEO: GSE144142 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144142 |
| Experimental Models: Cell Lines | ||
| Homo sapiens: HeLa | UCSF CCF | N/A |
| Homo sapiens: 293T/17 | UCSF CCF | N/A |
| Homo sapiens: A549 | ATCC | ATCC-CCL-185 |
| Oligonucleotides | ||
| Primers for esiRNA synthesis (see Table S6) | Sigma Aldrich | N/A |
| Primers and probes for qPCR analysis (see Table S7) | IDT | N/A |
| Primers for cloning HIV mutant virus (see Table S8) | IDT | N/A |
| Primers for TIDE analysis of genomic editing (see Table S8) | IDT | N/A |
| Synthesized crRNA for Cas9-RNP knockouts (see Table S9) | Dharmacon | N/A |
| Synthesized siRNA for A549 knockdowns (see Table S10) | Dharmacon | L-015369–01-0005, D-001810–10 |
| Software and Algorithms | ||
| Cluster 3.0 | de Hoon et al., 2004; Eisen et al., 1998 | http://bonsai.hgc.jp/~mdehoon/software/cluster/ |
| edgeR | Robinson and Oshlack, 2010 | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Enrichr | Chen et al., 2013; Kuleshov et al., 2016 | https://amp.pharm.mssm.edu/Enrichr/ |
| EMAP toolbox for MATLAB | Collins et al., 2006; Collins et al., 2010 | https://sourceforge.net/projects/emap-toolbox/ |
| Fastq-mcf | Aronesty, 2011 | https://expressionanalysis.github.io/ea-utils/ |
| FastQC | Andrews | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| MaxQuant | Cox and Mann, 2008 | http://www.coxdocs.org/doku.php?id=:maxquant:start |
| MSstats | Choi et al., 2014 | http://msstats.org |
| RSeQC | Wang et al., 2012 | http://rseqc.sourceforge.net/#download-rseqc |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR/releases |
| Subread (featureCounts) | Liao et al., 2014 | http://subread.sourceforge.net/ |
| TreeView | Saldanha, 2004 | http://jtreeview.sourceforge.net/ |
Highlights.
We adapted the E-MAP approach to study genetic interactions underlying viral infection.
We studied host gene pairs, as well as gene-drug and gene-viral mutant combinations.
The HIV vE-MAP highlighted a role for the CNOT complex in mediating infection.
CNOT mediates HIV infection in primary CD4+ T cells by suppression of innate immunity.
Acknowledgements
This work was supported by an NIH Postdoctoral Individual National Service Award (5F32GM108303–02 to DEG), the HARC Center (P50 GM082250 to ADF, AM, NJK), the Host Pathogen Map Initiative (U19 AI135990 to AM and NJK), and the FluOMICS consortia (U19 AI135972 to IM and NJK). AW and GC were supported by a Science Foundation Ireland Principal Investigator Award (SFI 10/1N.1/B3.19). This project has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement 666010. AM holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund, is an investigator at the Chan Zuckerberg Biohub and has received funding from the Innovative Genomics Institute (IGI) and the Parker Institute for Cancer Immunotherapy (PICI). Library preparation and QCs for sequencing were conducted by Jim McGuire and Natasha Carli, PhD at the Gladstone Genomics Core. Flow cytometry was performed at the Gladstone Flow Cytometry Core and the UCSF Laboratory for Cell Analysis. Flow cytometry facilities were supported by NIH grants (P30 AI027763), (S10 RR028962) and the James B. Pendleton Charitable Trust. Illumina sequencing was performed at the UCSF Center for Advanced Technology. Many thanks to Michael McGregor and Paige Haas for their support in running flow cytometry plates, as well as Francoise Chanut for her support in editing this manuscript.
Declaration of Interests
The Krogan Laboratory has received research support from Vir Biotechnology and F. Hoffmann-La Roche. AM is a co-founder of Spotlight Therapeutics and Arsenal Therapeutics. AM serves on the scientific advisory board of PACT Pharma, is an advisor to Sonoma Biotherapeutics and was a former advisor to Juno Therapeutics. The Marson laboratory has received sponsored research support from Juno Therapeutics, Epinomics, Sanofi and a gift from Gilead.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahle S, Mann A, Eichelsbacher U, and Ungewickell E (1988). Structural relationships between clathrin assembly proteins from the Golgi and the plasma membrane. EMBO J. 7, 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S FastQC. Bioinformatics.babraham.ac.uk.
- Aronesty E (2011). EA-Utils: Command-line tools for processing biological sequencing data. Code.Google.com.
- Bassik MC, Lebbink RJ, Churchman LS, Ingolia NT, Patena W, LeProust EM, Schuldiner M, Weissman JS, and McManus MT (2009). Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat. Methods 6, 443–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra J, Hultquist JF, Liu D, Shtanko O, Dollen, Von J,Satkamp L, Jang GM, Luthra P, Schwarz TM, Small GI, et al. (2018). Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication. Cell 175, 1917–1930.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawankar P, Loh B, Wohlbold L, Schmidt S, and Izaurralde E (2013). NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biology 10, 228–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Cagney G, and Krogan NJ (2010). Quantitative genetic interactions reveal biological modularity. Cell 141, 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series BStatistical Methodology 57, 289–300. [Google Scholar]
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al. (2013). From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell 154, 775–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, and van Steensel B (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research 42, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz F, Kittler R, Slabicki M, and Theis M (2006). Enzymatically prepared RNAi libraries. Nat. Methods 3, 696–700. [DOI] [PubMed] [Google Scholar]
- Chan GKY, Kleinheinz TL, Peterson D, and Moffat JG (2013). A simple high-content cell cycle assay reveals frequent discrepancies between cell number and ATP and MTS proliferation assays. PLoS ONE 8, e63583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapat C, Kolytcheff C, Le Romancer M, Auboeuf D, La Grange, De P, Chettab K, Sentis S, and Corbo L (2013). hCAF1/CNOT7 regulates interferon signalling by targeting STAT1. Embo J. 32, 688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Chang C-Y, Clough T, Broudy D, Killeen T, MacLean B, and Vitek O (2014). MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526. [DOI] [PubMed] [Google Scholar]
- Chuang H-W, Zhang W, and Gray WM (2004). Arabidopsis ETA2, an apparent ortholog of the human cullin-interacting protein CAND1, is required for auxin responses mediated by the SCF(TIR1) ubiquitin ligase. Plant Cell 16, 1883–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. (2007). Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446, 806–810. [DOI] [PubMed] [Google Scholar]
- Collins SR, Roguev A, and Krogan NJ (2010). Quantitative genetic interaction mapping using the E-MAP approach. Meth. Enzymol. 470, 205–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, and Weissman JS (2006). A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 7, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RI, Chen BK, Choe S, and Landau NR (1995). Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206, 935–944. [DOI] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. (2016). A global genetic interaction network maps a wiring diagram of cellular function. Science 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, and Mann M (2014). Accurate proteomewide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics 13, 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Qiu P, Lee CM, Fine EJ, and Bao G (2014). COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol Ther Nucleic Acids 3, e214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis ZH, Verschueren E, Jang GM, Kleffman K, Johnson JR, Park J, Dollen, Von J, Maher MC, Johnson T, Newton W, et al. (2015). Global mapping of herpesvirus-host protein complexes reveals a transcription strategy for late genes. Molecular Cell 57, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, and Miyano S (2004). Open source clustering software. Bioinformatics 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- De Iaco A, and Luban J (2011). Inhibition of HIV-1 infection by TNPO3 depletion is determined by capsid and detectable after viral cDNA enters the nucleus. Retrovirology 8, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Santoni F, Vannier A, Guipponi M, Antonarakis S, and Luban J (2013). TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm. Retrovirology 10, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep J, Ooi YS, Wilkinson AW, Peters CE, Foy E, Johnson JR, Zengel J, Ding S, Weng K-F, Laufman O, et al. (2019). Enterovirus pathogenesis requires the host methyltransferase SETD3. Nat Microbiol 4, 2523–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Roguev A, Gordon DE, Chen M, Chen S-H, Shales M, Shen JP, Ideker T, Mali P, Qi LS, et al. (2017). Genetic interaction mapping in mammalian cells using CRISPR interference. Nat. Methods 14, 577–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt M, Zhang W, Gross AM, Dollen, Von J, Johnson JR, Franks-Skiba KE, Swaney DL, Johnson TL, Jang GM, Shah PS, et al. (2018). Multiple Routes to Oncogenesis Are Promoted by the Human Papillomavirus-Host Protein Network. Cancer Discov 8, 1474–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, and Botstein D (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.a. 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enchev RI, Schulman BA, and Peter M (2015). Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol 16, 30–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourati S, Ribeiro SP, Blasco Tavares Pereira Lopes F, Talla A, Lefebvre F, Cameron M, Kaewkungwal J, Pitisuttithum P, Nitayaphan S, Rerks-Ngarm S, et al. (2019). Integrated systems approach defines the antiviral pathways conferring protection by the RV144 HIV vaccine. Nat Commun 10, 863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JS, Gross JD, and Krogan NJ (2013). From systems to structure: bridging networks and mechanism. Molecular Cell 49, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georges, des A, <Dhote V, Kuhn L, Hellen CUT, Pestova TV, Frank J, and Hashem Y (2015). Structure of mammalian eIF3 in the context of the 43S preinitiation complex | Nature. Nature 525, 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesner M, Maiti M, Grant R, and Cavrois M (2014). Fluorescence-linked Antigen Quantification (FLAQ) Assay for Fast Quantification of HIV-1 p24Gag. Bio Protoc 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, et al. (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschel A, Buchholz F, and Habermann B (2004). DEQOR: a web-based tool for the design and quality control of siRNAs. Nucleic Acids Research 32, W113–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Xu A, Wang M, Bennett NK, Park CY, Bogdanoff D, Adamson B, Chow ED, Kampmann M, Peterson TR, et al. (2018). Mapping the Genetic Landscape of Human Cells. Cell 174, 953–967.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka K, Gierszewska M, Srivastava S, Kozaczkiewicz L, Swanson SK, Florens L, Washburn MP, and Skowronski J (2007). Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. U.S.a. 104, 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, Doudna J, Simon V, Krogan NJ, and Marson A (2016). A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep 17, 1438–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehning JA (2010). The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim. Biophys. Acta 1799, 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. (2012). Global landscape of HIV-human protein complexes. Nature 481, 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, and Conaway JW (1999). The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes & Development 13, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsella TM, and Nolan GP (1996). Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7, 1405–1413. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, and Greenblatt JF (2002). RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Molecular and Cellular Biology 22, 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Research 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M-C, Lee Y-HW, and Tarn W-Y (2008). The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol. Biol. Cell 19, 3847–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer C, Fischer B, Billmann M, Huber W, and Boutros M (2013). Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat. Methods 10, 427–431. [DOI] [PubMed] [Google Scholar]
- Lee K, Ambrose Z, Martin TD, Oztop I, Mulky A, Julias JG, Vandegraaff N, Baumann JG, Wang R, Yuen W, et al. (2010). Flexible use of nuclear import pathways by HIV-1. Cell Host & Microbe 7, 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidecker O, Matic I, Mahata B, Pion E, and Xirodimas DP (2012). The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle 11, 1142–1150. [DOI] [PubMed] [Google Scholar]
- Li M, Johnson JR, Truong B, Kim G, Weinbren N, Dittmar M, Shah PS, Dollen, Von J, Newton BW, Jang GM, et al. (2019). Identification of antiviral roles for the exon-junction complex and nonsense-mediated decay in flaviviral infection. Nat Microbiol 4, 985–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lonergan KM, Iliopoulos O, Ohh M, Kamura T, Conaway RC, Conaway JW, and Kaelin WG (1998). Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Molecular and Cellular Biology 18, 732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Arraes LC, Ferreira WT, and Andrieu J-M (2004). Therapeutic dendritic-cell vaccine for chronic HIV-1 infection. Nat. Med. 10, 1359–1365. [DOI] [PubMed] [Google Scholar]
- Lumb JH, Li Q, Popov LM, Ding S, Keith MT, Merrill BD, Greenberg HB, Li JB, and Carette JE (2017). DDX6 Represses Aberrant Activation of Interferon-Stimulated Genes. Cell Rep 20, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, and Deshaies RJ (2001). Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Schulman BA, and Harper JW (2013). Building and remodelling Cullin–RING E3 ubiquitin ligases. EMBO Rep. 14, 1050–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al. (1997). P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes & Development 11, 2633–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, and Benarous R (1998). A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Molecular Cell 1, 565–574. [DOI] [PubMed] [Google Scholar]
- Martínez-Sobrido L, Cadagan R, Steel J, Basler CF, Palese P, Moran TM, and García-Sastre A (2010). Hemagglutinin-pseudotyped green fluorescent protein-expressing influenza viruses for the detection of influenza virus neutralizing antibodies. J. Virol. 84, 2157–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauxion F, Prève B, and Séraphin B (2013). C2ORF29/CNOT11 and CNOT10 form a new module of the CCR4-NOT complex. RNA Biology 10, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B, and Martin-Serrano J (2009). No strings attached: the ESCRT machinery in viral budding and cytokinesis. Journal of Cell Science 122, 2167–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JE, and Reese JC (2012). Ccr4-Not complex: the control freak of eukaryotic cells. Crit. Rev. Biochem. Mol. Biol. 47, 315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Dollen, Von J, Rosenberg O, Gulbahce N, Jang G, Johnson T, et al. (2015). Global Mapping of the Inc-Human Interactome Reveals that Retromer Restricts Chlamydia Infection. Cell Host & Microbe 18, 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura R, Pontillo A, D’Adamo P, Pirastu N, Campos Coelho A, and Crovella S (2014). Exome analysis of HIV patients submitted to dendritic cells therapeutic vaccine reveals an association of CNOT1 gene with response to the treatment. Journal of the International AIDS Society 17, 18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naji S, Ambrus G, Cimermancic P, Reyes JR, Johnson JR, Filbrandt R, Huber MD, Vesely P, Krogan NJ, Yates JR, et al. (2012). Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell Proteomics 11, M111.015313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Geyer M, and Zhou Q (2011). The control of HIV transcription: keeping RNA polymerase II on track. Cell Host & Microbe 10, 426–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn BH, Netter Z, Johnson JR, Dollen, Von J, Jang GM, Johnson T, Ohol YM, Maher C, Bell SL, Geiger K, et al. (2018). An Mtb-Human Protein-Protein Interaction Map Identifies a Switch between Host Antiviral and Antibacterial Responses. Molecular Cell 71, 637–648.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AJ, Fletcher AJ, Schaller T, Elliott T, Lee K, Kewalramani VN, Chin JW, Towers GJ, and James LC (2012). CPSF6 defines a conserved capsid interface that modulates HIV-1 replication. PLoS Pathog 8, e1002896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage HR, Kumar GR, Verschueren E, Johnson JR, Dollen, Von J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, et al. (2015). A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Molecular Cell 57, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, and Oshlack A (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, and Smyth GK (2007). Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23, 2881–2887. [DOI] [PubMed] [Google Scholar]
- Robinson MD, and Smyth GK (2008). Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9, 321–332. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park H-O, Hayles J, et al. (2008). Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322, 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Talbot D, Negri GL, Shales M, Cagney G, Bandyopadhyay S, Panning B, and Krogan NJ (2013). Quantitative genetic-interaction mapping in mammalian cells. Nat. Methods 10, 432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, et al. (2012). Hierarchical Modularity and the Evolution of Genetic Interactomes across Species. Molecular Cell 46, 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ (2004). Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. (2005). Exploration of the Function and Organization of the Yeast Early Secretory Pathway through an Epistatic Miniarray Profile. Cell 123, 507–519. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, and Krogan NJ (2006). Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods 40, 344–352. [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, and Deng XW (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Shah PS, Link N, Jang GM, Sharp PP, Zhu T, Swaney DL, Johnson JR, Dollen, Von J, Ramage HR, Satkamp L, et al. (2018). Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 175, 1931–1945.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, and Hopper AK (2005). Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.a. 102, 11290–11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. (2017). Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat. Methods 14, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebring-van Olst E, Vermeulen C, de Menezes RX, Howell M, Smit EF, and van Beusechem VW (2013). Affordable Luciferase Reporter Assay for Cell-Based High-Throughput Screening. J Biomol Screen 18, 453–461. [DOI] [PubMed] [Google Scholar]
- Smyth GK, and Verbyla AP (1996). A conditional likelihood approach to residual maximum likelihood estimation in generalized linear models. Journal of the Royal Statistical Society Series B-Methodological 58, 565–572. [Google Scholar]
- Sobhian B, Laguette N, Yatim A, Nakamura M, Levy Y, Kiernan R, and Benkirane M (2010). HIV-1 Tat assembles a multifunctional transcription elongation complex and stably associates with the 7SK snRNP. Molecular Cell 38, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. (2009). An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736. [DOI] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, and Hartzog GA (2002). The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. Embo J. 21, 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley DJ, Bartholomeeusen K, Crosby DC, Kim DY, Kwon E, Yen L, Cartozo NC, Li M, Jäger S, Mason-Herr J, et al. (2012). Inhibition of a NEDD8 Cascade Restores Restriction of HIV by APOBEC3G. PLoS Pathog 8, e1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS (2018). 2017 Global HIV Statistics. [Google Scholar]
- Wagner S, Herrmannová A, Šikrová D, and Valášek LS (2016). Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: the yeast-like core and the octamer. Nucleic Acids Research 44, 10772–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. [DOI] [PubMed] [Google Scholar]
- Wei N, and Deng XW (2003). The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19, 261–286. [DOI] [PubMed] [Google Scholar]
- Xu K, Bai Y, Zhang A, Zhang Q, and Bartlam MG (2014). Insights into the structure and architecture of the CCR4-NOT complex. Front Genet 5, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VSRK, Neuveut C, Chi Y-H, Kleiman L, and Jeang K-T (2004). Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119, 381–392. [DOI] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, and Yu X-F (2003). Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060. [DOI] [PubMed] [Google Scholar]
- Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, and Zhang H (2002). CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Molecular Cell 10, 1519–1526. [DOI] [PubMed] [Google Scholar]
- Zhou L, Sokolskaja E, Jolly C, James W, Cowley SA, and Fassati A (2012). Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. Molecular Biology International 2012, e1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, He M, Shah AA, and Wan Y (2016). Insights into APC/C: from cellular function to diseases and therapeutics. Cell Div 11, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, and Price DH (1997). Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes & Development 11, 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Single knockdown behavior for each esiRNA in the HIV vE-MAP (related to Figures 1C, 1D, and 2B)
Table S2: Genetic interaction scores (S-score) quantified with MLN-4924 query treatment (related to Figure 3C–3E)
Table S3: Differential CNOT1 immunoprecipitation proteomics data with CNOT1 large isoform knockout (related to Figure 4H).
Table S4: Differential transcriptome analysis of CNOT1 and CNOT10 knockouts in primary CD4+ T-cells (related to Figure 5A)
Table S5: HIV infection data for polyclonal knockouts in primary CD4+ T-cells +/− CNOT10 knockout or interferon alpha-2a treatment (related to Figure 5D)
Data S1: HIV vE-MAP Symmetric Knockdown Matrix (related to Figures 2, 3, 4A–4B, S1, S2). The vE-MAP can be visualized using Java Treeview software.
Data Availability Statement
The accession number for the RNAseq data reported in this paper is GEO: GSE144142. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144142. Genetic interaction scores, differential proteomics analysis, processed RNAseq data, and genetic interaction data in primary T-cells are attached as supplemental data. The analysis pipelines supporting the current study are available from the lead contact on request.