Here, Grunblatt et al. sought to understand the roles for MYCN amplification in SCLC progression and chemoresistance, and developed a genetically engineered mouse model of MYCN-overexpressing SCLC. Their findings show that MYCN overexpression drives SCLC chemoresistance and provide a therapeutic strategy to restore chemosensitivity.
Keywords: SCLC, MYCN, MYCL, chemoresistance, USP7
Abstract
Small cell lung cancer (SCLC) is an aggressive neuroendocrine cancer characterized by initial chemosensitivity followed by emergence of chemoresistant disease. To study roles for MYCN amplification in SCLC progression and chemoresistance, we developed a genetically engineered mouse model of MYCN-overexpressing SCLC. In treatment-naïve mice, MYCN overexpression promoted cell cycle progression, suppressed infiltration of cytotoxic T cells, and accelerated SCLC. MYCN overexpression also suppressed response to cisplatin–etoposide chemotherapy, with similar findings made upon MYCL overexpression. We extended these data to genetically perturb chemosensitive patient-derived xenograft (PDX) models of SCLC. In chemosensitive PDX models, overexpression of either MYCN or MYCL also conferred a switch to chemoresistance. To identify therapeutic strategies for MYCN-overexpressing SCLC, we performed a genome-scale CRISPR–Cas9 sgRNA screen. We identified the deubiquitinase USP7 as a MYCN-associated synthetic vulnerability. Pharmacological inhibition of USP7 resensitized chemoresistant MYCN-overexpressing PDX models to chemotherapy in vivo. Our findings show that MYCN overexpression drives SCLC chemoresistance and provide a therapeutic strategy to restore chemosensitivity.
Small cell lung cancer (SCLC) is a recalcitrant neuroendocrine carcinoma accounting for ∼15% of all lung cancers (Rudin et al. 2019; Poirier et al. 2020). Initially, approximately two out of three of patients respond well to platinum-based chemotherapy; however, responses are transient, and patients ultimately succumb to chemoresistant disease (Rossi et al. 2012). Recent FDA approval adding immune checkpoint inhibition to a chemotherapy doublet (Horn et al. 2018; Paz-Ares et al. 2019) has not changed the dismal prognosis for SCLC patients. Thus, understanding and targeting drivers of SCLC chemoresistance is of vital importance. A barrier to understanding chemoresistant SCLC has been the dearth of genomically characterized samples from chemotherapy-treated patients, as such tumors are not surgically removed in typical clinical care. A small-scale study of 30 chemotherapy-treated, relapsed SCLC patients implicated WNT pathway alterations in chemoresistance (Wagner et al. 2018). However, the largest genomic studies in SCLC have focused on untreated patients, where inactivating mutations or deletions in RB1 and TP53 are present in nearly 100% of cases and inactivating mutations in PTEN, NOTCH pathway components, and in chromatin regulators such as CREBBP, EP300, and KMT2D are also observed (Peifer et al. 2012; Rudin et al. 2012; George et al. 2015; Augert et al. 2017). SCLC also exhibits amplification of the MYC family of basic helix–loop–helix transcription factors, including MYCN, MYCL, and MYC (Peifer et al. 2012; Rudin et al. 2012; George et al. 2015). While there is a paucity of genomically characterized chemotherapy-treated tumor samples available, observations of an approximately threefold increased rate of high-level MYC, MYCN, or MYCL amplification in cell lines derived from chemotherapy-treated versus chemonaïve patients (Johnson et al. 1996) indirectly implicate MYC family amplification in chemoresistance. Also, a MYC transcriptional signature was associated with chemoresistance in human SCLC PDX models derived from chemonaïve versus treated patients (Drapkin et al. 2018). Despite these hints that MYC family activation could contribute to SCLC chemoresistance, this notion has yet to be demonstrated using rigorous in vivo models. MYC and MYCL have been shown to promote SCLC in mice (Huijbers et al. 2014; Kim et al. 2016; Mollaoglu et al. 2017), while mouse models of SCLC overexpressing MYCN are lacking. To investigate the contribution of MYCN to SCLC progression and therapy response, we overexpressed MYCN in a novel autochthonous mouse model. We also overexpressed MYCN in chemosensitive PDX models of SCLC. We studied roles for MYCN in SCLC progression and chemoresistance and employed a genetic screen to identify a druggable vulnerability for MYCN-overexpressing SCLC.
Results
MYCN overexpression promotes tumor progression in an autochthonous mouse model of SCLC
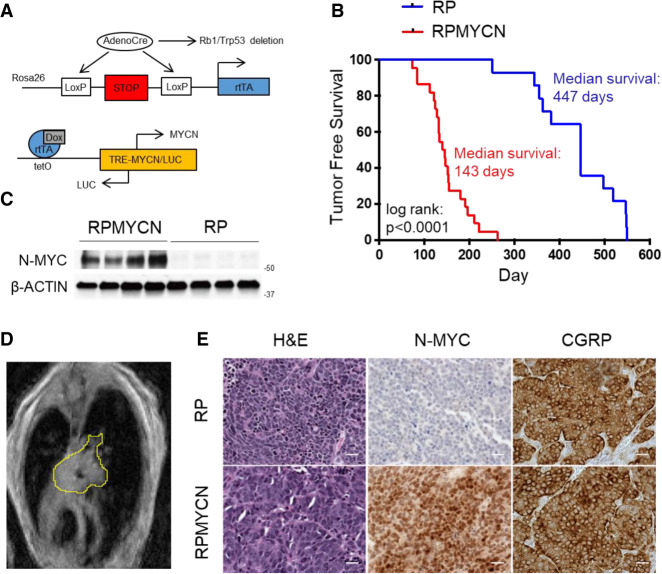
To overexpress MYCN in a controllable manner, we bred mice harboring Rosa26-M2rtTA (Hochedlinger et al. 2005) and Tre-MYCN/LUC alleles (Fig. 1A; Swartling et al. 2010) into an Rb1/Trp53 deleted model of SCLC (Meuwissen et al. 2003). The Tre-MYCN model has been used previously to model medulloblastoma and retinoblastoma (Swartling et al. 2010; Wu et al. 2017) and has the advantage that MYCN expression can be toggled based on the presence of doxycycline (DOX) in the feed. We infected Rb1lox/lox;Trp53lox/lox;M2rtTAlox/lox (here, RP) and Rb1lox/lox;Trp53lox/lox;M2rtTAlox/lox;Tre-MYCN/LUC (here, RPMYCN) mice intratracheally with adenovirus expressing Cre recombinase driven by a neuroendocrine promoter CGRP (Ad-CGRP-Cre) (Sutherland et al. 2011). One week after infection, we placed mice on a doxycycline diet to activate MYCN expression in infected lung neuroendocrine cells. RPMYCN mice maintained on DOX developed lung tumors significantly faster than RP mice, with a median tumor free survival of 143 d as compared with 447 d for RP mice (Fig. 1B). Western blotting confirmed overexpression of N-MYC in the RPMYCN model (Fig 1C). Magnetic resonance imaging (MRI) showed that RPMYCN tumors, like RP tumors, tended to be centrally located (Fig. 1D), a characteristic of human SCLC. Hematoxylin-eosin (H&E) stains of RPMYCN tumors examined by a clinical pathologist (A. Gazdar) showed histology of “classic” SCLC in eight out of nine tumors and “variant” SCLC in one out of nine tumors (Fig. 1E; Supplemental Fig. S1A). Notably, these findings with N-MYC overexpression differ from overexpression of a stabilizing T58A c-MYC allele, where the dominant tumor type was of variant histology (Mollaoglu et al. 2017). Immunostaining showed positive expression of CGRP, a marker of neuroendocrine cells, in both RP and RPMYCN models (Fig 1E). Immunostaining also confirmed increased expression of N-MYC in RPMYCN tumor samples (Fig 1E). SCLC can be classified into subtypes based on expression and activity of key transcription factors: ASCL1, NEUROD1, YAP1, and POU2F3 (Rudin et al. 2019). Immunohistochemistry showed broad ASCL1 expression in all RPMYCN samples along with scattered NEUROD1 and YAP1 staining in some tumors (Supplemental Fig. S1B). RNA-seq analysis showed that seven out of seven RPMYCN tumors exhibited high Ascl1 expression with two out of seven also expressing Neurod1; Yap1 levels were low in all samples while Pou2f3 expression levels were consistently below the minimum detection threshold (Supplemental Fig. S1C). Thus, MYCN overexpression in CGRP-positive cells promotes predominantly “classic” SCLC tumorigenesis with high expression of ASCL1.
Figure 1.
MYCN overexpression promotes SCLC in mouse models (A) Schematic of alleles used to generate the doxycycline-inducible MYCN overexpression mouse model. (B) Kaplan-Meier curve comparing survival of control RP mice with RPMYCN Ad-CGRP-Cre-infected mice. (n = 14 mice for RP; n = 22 mice for RPMYCN). Significance was determined using the log-rank (Mantel-Cox) test. (C) Immunoblot comparing levels of N-MYC expression in RP versus RPMYCN tumors. β-ACTIN was used as a loading control. (D) Representative magnetic resonance image (MRI) of a RPMYCN tumor, outlined in yellow. (E) Representative H&E and immunohistochemistry for N-MYC and neuroendocrine marker CGRP in RP versus RPMYCN tumors. Scale bar, 20 µm.
MYCN overexpression increases proliferation and protein synthesis in SCLC
To determine whether sustained MYCN expression is important for SCLC initiated with high levels of MYCN, we removed doxycycline from the diet of RPMYCN mice after tumors were detected using MRI. By day 14 OFF DOX, eight out of 12 tumors regressed by at least 25% from baseline and five out of 12 regressed by at least 40% (Fig. 2A,B). Western blotting of a parallel cohort of tumors showed a substantial reduction in levels of N-MYC 7 d following DOX removal (Fig. 2C). TUNEL analyses revealed significantly increased apoptosis upon DOX removal at this time point, while immunostaining for phospho-Ser 10 histone H3 (pH3), marking mitotic cells, showed decreased tumor cell proliferation (Fig. 2D,E). We also monitored a cohort of six RPMYCN mice for several weeks following withdrawal of DOX. In all mice, tumors eventually returned following initial regression (four out of six) or cytostatic (two out of six) early response to DOX removal (Supplemental Fig. S2A). We performed qPCR to assess MYC family member expression in these OFF DOX return tumors and found that one out of six tumors continued to express the MYCN transgene despite the absence of doxycycline (a breakdown of the RTTA system), while one out of six tumors exhibited high-level amplification of the murine Mycn locus (Supplemental Fig. S2B,C). The remaining four out of six return tumors did not express high levels of any MYC family members (Supplemental Fig. S2B,C). Thus, while RPMYCN tumors are initially dependent on sustained MYCN expression, they evolve the ability to regrow, in many cases without reactivating MYCN or overexpressing MYCL or MYC.
Figure 2.
MYCN overexpression increases proliferation and protein synthesis in SCLC (A) Representative MRI of lungs from two RPMYCN mice (M1 and M2) at days 0 and 14 of doxycycline withdrawal. (B) Waterfall plot showing the percentage of change in tumor volume from baseline at day 14 after doxycycline removal for all OFF DOX tumors. (C) Immunoblot showing decrease in N-MYC expression in RPMYCN tumors collected after 7 d off of DOX. β-ACTIN was used as a loading control. (D) Representative immunohistochemistry for pH3 and TUNEL staining comparing RPMYCN ON DOX versus OFF DOX tumors. Scale bar, 20 µm. (E) Quantification of the percentage of pH3- and TUNEL-positive cells respectively across all ON DOX and OFF DOX samples. Data are means ± SEM. (n = 5 in all groups). Significance was determined using two-tailed unpaired Student's t-test. (*) P < 0.05; (***) P < 0.001. (F) Graph showing proliferation, determined using Cell Counting Kit-8 kit, of a representative RPMYCN-derived cell line (C1) cultured either with or without DOX over 4 d. Data are means ± SEM (n = 3 biological replicates each consisting of 3 technical replicates per condition). Significance was determined using two-tailed unpaired Student's t-test. (***) P < 0.001. (G) Quantification of propidium iodide cell cycle assay comparing live cells versus cells in the sub-G1 population for C1 cell line cultured either in the presence or absence of DOX. Data are means ± SEM (n = 3 biological replicates each consisting of three technical replicates per condition). (H) Quantification of propidium iodide cell cycle assay, focusing on live cells only and comparing G1, S, and G2/M populations for C1 cell line cultured either in the presence or absence of DOX. Data are means ± SEM (n = 3 biological replicates each consisting of three technical replicates per condition). (I) Representative immunoblot comparing levels of N-MYC, cyclins, and cell cycle regulators in the presence or absence of DOX for six RPMYCN-derived cell lines (C1–6). (J) Quantification of qRT-PCR analysis performed on C1 cell line cultured either with or without doxycycline. Top shows relative expression of pre-rRNA as determined by expression levels of the 47S rRNA internal transcribed spacer (ITS) region relative to β2m. The bottom shows relative expression of MYCN relative to Gapdh. Data are means ± SEM (n = 3 biological replicates each consisting of 3 technical replicates per condition). (K) Analysis of nascent protein synthesis in C1 cell line using a puromycin incorporation assay. β-ACTIN was used as a loading control.
To further assess the acute response to MYCN suppression in RPMYCN cells we established six SCLC cell lines from different RPMYCN tumors in the presence of DOX in the media. Six out of six RPMYCN cell lines responded to acute DOX removal with reduced N-MYC protein and decreased proportion of viable cells over 4 d OFF DOX, as shown using Cell Counting Kit-8 assays (Fig. 2F; Supplemental Fig. S3A). Flow cytometry analyses of DNA content at 72 h OFF DOX revealed that decreased viability was partly due to increased cell death, with sub-G1 populations substantially increased in five out of six cell lines following DOX removal (Fig. 2G; Supplemental Fig. S3B). Focusing only on live cells, MYCN suppression was associated with increases in the G1 population coupled with decreases in S and G2/M populations in five out of six cell lines (Fig. 2H; Supplemental Fig. S3B). Molecular correlates of these responses observed following DOX removal in at least five out of six of the independent lines tested included decreased expression of cyclins A and E and increased expression of the CDK inhibitor p27 (Fig. 2I). p130, encoded by RBL2, becomes functionally inactivated through cyclin/CDK-dependent phosphorylation (Hansen et al. 2001), is occasionally the target of inactivating mutations in human SCLC (George et al. 2015), and is a potent tumor suppressor in mouse models (Schaffer et al. 2010). Upon MYCN suppression with DOX removal, we observed decreased p130 phosphorylation, indicative of p130 activation, in six out of six RPMYCN cell lines (Fig. 2I). Decreased p130 phosphorylation is consistent with the decreased cyclin E/A expression and increased p27 expression that occurred with suppression of MYCN (Fig. 2I). Thus, MYCN overexpression results in functional inactivation of p130 and increased proliferation in SCLC.
MYC family members, including MYCN, promote ribosomal biogenesis and protein synthesis via direct binding to ribosomal DNA, transcribed by RNA Pol I, and via increased expression of RNA Pol II transcribed mRNAs for genes that promote translation (Boon et al. 2001; Grandori et al. 2005; Dang 2012; Beltran 2014; Kim et al. 2016). To examine the impact of MYCN overexpression on ribosomal biogenesis and protein synthesis in SCLC, we performed quantitative real time PCR analysis using primers targeting the internal transcribed spacer (ITS) sequence of the 47S rRNA subunit (Kim et al. 2016). Levels of pre-rRNA were drastically decreased with DOX removal in the RPMYCN cells (Fig. 2J; Supplemental Fig. S3C). To measure protein synthesis, we treated both ON DOX and OFF DOX cells with puromycin, which becomes incorporated into nascent proteins. Removal of DOX resulted in lower levels of puromycin incorporation in all six lines tested (Fig. 2K; Supplemental Fig. S3D). Taken together, these results show that MYCN overexpression in SCLC promotes proliferation and increases protein synthesis.
MYCN modulates tumor immune microenvironment in SCLC
To identify transcriptional changes resulting from MYCN overexpression, we performed RNA-seq analyses. We compared (1) mouse RPMYCN versus RP SCLCs, (2) RPMYCN SCLCs ON DOX versus 7 d OFF DOX in vivo, and (3) RPMYCN cell lines ON DOX versus 4 d OFF DOX in cell culture. Using EdgeR analyses (Robinson et al. 2010), we identified 1030 genes to be commonly differentially expressed with MYCN overexpression across all three comparisons (FDR < 0.05) (Fig. 3A). Analyzing the commonly differentially expressed genes using Enrichr (Kuleshov et al. 2016) querying the KEGG pathways database (Kanehisa and Goto 2000), we found that genes associated with ribosome biogenesis, RNA transport, and RNA polymerase were enriched with MYCN overexpression (Fig. 3B). Moreover, querying ENCODE and ChEA transcription factor occupancy databases (The ENCODE Project Consortium 2004; Lachmann et al. 2010), MYC, MAX, and E2F transcription factor binding was strongly enriched in the common genes that change with MYCN overexpression (Fig 3C). Additionally, CUT&RUN analyses (Skene and Henikoff 2017) of N-MYC occupancy in an RPMYCN-derived cell line (C3) in ON DOX versus OFF DOX conditions revealed decreased N-MYC genomic occupancy proximal to transcription start sites (TSS) following withdrawal of DOX for 4 d (Fig. 3D). Focusing on genes commonly changed with MYCN expression across all three of the RNA-seq comparisons, we found that 351 out of 546 (64%) of the genes with expression changes in the same direction as MYCN were N-MYC-bound, compared with 138 out of 459 (30%) of the genes that changed in the opposite direction as MYCN (Supplemental Fig. S4A,B). These data are consistent with MYCN primarily acting to increase expression of direct target genes. We next performed gene set enrichment analysis (GSEA) for each of the three comparisons individually. We queried the “Hallmark” gene set list from the molecular signatures database (MSigDB) (Subramanian et al. 2005; Liberzon et al. 2015) and found that MYC_TARGETS_V1, MYC_TARGETS_V2, and UNFOLDED_PROTEIN_RESPONSE gene sets were significantly up-regulated in MYCN-overexpressing samples across all three comparisons (Fig. 3E; Supplemental Fig. S5A–C). Examining the gene sets that were down-regulated with MYCN overexpression, several immune signaling pathways were represented, such as the INTERFERON_α_RESPONSE, INTERFERON_γ_RESPONSE, and INFLAMMATORY_RESPONSE (Fig. 3F; Supplemental Fig. S5A–C). Based on these results, we hypothesized that MYCN overexpression may alter the tumor immune microenvironment in SCLC. As the impact of overexpressing any MYC family member on specific immune cell types has yet to be examined in an immune-intact SCLC model system, we sought to identify the specific immune cell types that were changed with MYCN expression. We employed a previously described flow cytometry antibody panel and sequential gating strategy (Kargl et al. 2017) to identify specific immune cell populations from single-cell suspensions of RP, RPMYCN ON DOX, and RPMYCN OFF DOX mouse SCLC samples (Fig. 3G). MYCN status was not associated with changes in percentage of B cells, NK cells, or macrophages (Fig. 3H). In contrast, RPMYCN ON DOX tumors exhibited significantly lower percentage of CD3+ T cells, including both CD4+ helper and CD8+ cytotoxic T cells, and monocytes as compared with both RP and RPMYCN OFF DOX tumors (Fig. 3H). Furthermore, RPMYCN ON DOX tumors showed significantly higher percentage of neutrophils when compared with either RP or RPMYCN OFF DOX tumors (Fig. 3H). Neutrophils are often anti-correlated with T cells in tumors, and tumors with microenvironments high in neutrophils and low in T cells are correlated with poorer prognoses and outcomes (Gentles et al. 2015; Lizotte et al. 2016; Kargl et al. 2017). These data show that MYCN overexpression reduces the infiltration of T cells into SCLC tumors.
Figure 3.
MYCN modulates the SCLC tumor immune microenvironment (A) Venn diagram showing genes that are differentially regulated in MYCN-overexpressing samples for the following comparisons: RPMYCN (n = 7) versus RP (n = 7) tumors, RPMYCN ON DOX (n = 7) versus OFF DOX (n = 4) tumors, and RPMYCN ON DOX (n = 5) versus OFF DOX (n = 5) cell lines. Gene lists were determined using EdgeR analysis with an FDR cutoff of 0.05. (B) KEGG pathway analysis of the 1030 genes commonly differentially expressed across all three comparisons. Significance was determined using an adjusted P-value of P < 0.05. (C) CHEA and ENCODE binding analysis of the 1030 genes commonly regulated by MYCN across all three comparisons. Significance was determined using an adjusted P-value of P < 0.05. (D) Heat maps depicting N-MYC binding from CUT&RUN data in an RPMYCN-derived cell line (C3) in the presence versus absence of DOX. Heat map shows data 5 kb upstream of to 5 kb downstream from TSS. (E,F) Gene set enrichment analysis of RNA sequencing data for RPMYCN versus RP tumors, RPMYCN ON DOX versus OFF DOX tumors, and RPMYCN ON DOX versus OFF DOX cell line comparisons (n = 7 in all RPMYCN versus RP groups;. n = 7 in RPMYCN ON DOX tumors group; n = 4 in RPMYCN OFF DOX tumors group; n = 5 in all RPMYCN ON DOX versus OFF DOX cell line groups). Graphs show pathways from the Hallmark database that are either positively (E) or negatively (F) enriched in the respective MYCN-overexpressing conditions for each comparison. The size of the circles corresponds to −log(FDR) while the colors of the circles correspond to the normalized enrichment score (NES) for each pathway. (G) Representative dot plots showing the gating strategy used for FACS analysis of the tumor immune microenvironment. Starting from the top left, initial gates select for live cells (FVD) and then for leukocytes by CD45. Gating for specific cell surface marker combinations was then used to identify immune cell populations such as B cell (CD3− CD19+), CD4+ T cells (CD3+ CD4+), CD8+ T cells (CD3+ CD8+), and neutrophils (CD11b+ Ly6G+). (H) Quantification of the percentage of each immune cell population within the total population of leukocytes per tumor. Data are means ± SEM (RP: n = 5; RPMYCN ON DOX: n = 6; RPMYCN OFF DOX: n = 7). Significance was determined using two-tailed unpaired Student's t-test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
MYCN and MYCL promote cisplatin–etoposide resistance in an SCLC mouse model
Based on more frequent amplifications of MYC family members in cell lines derived from chemotherapy-treated versus chemonaive patients (Johnson et al. 1996), we hypothesized that MYCN overexpression may confer chemoresistance. To test this hypothesis, we monitored RP and RPMYCN mice for lung tumor burden using MRI and then treated tumor-bearing mice with either saline or cisplatin–etoposide (cis–eto) for 21 d (three weekly cycles of treatment). Sequential MRI scans at days 14 and 21 were used to calculate changes in tumor volume relative to baseline. While RP mice exhibited a cytostatic response to chemotherapy RPMYCN tumors continued to grow through chemotherapy at a rate comparable with saline-treated tumors (Fig. 4A–C). As MYC family members are amplified in SCLC in a mutually exclusive manner, we sought to extend our MYCN results to MYCL, which is the most frequently amplified MYC family member in SCLC (George et al. 2015). We made use of a previously described mouse model of SCLC (herein RPMYCL) in which intratracheal administration of adenovirus expressing Cre recombinase leads to the activation of a MYCL transgene in addition to loss of Rb and Trp53 (Huijbers et al. 2014). As with the RPMYCN model, RPMYCL tumors continued to grow through cis–eto treatment at a rate comparable with saline-treated tumors (Fig. 4A–C). As MYCL is expressed in the RP model and can undergo spontaneous amplification we sought to assess expression levels of MYC family members among a subset of the cis–eto-treated mice in the RP versus RPMYCL cohorts. RNA-seq analysis showed MYCL expression to be higher in RPMYCL compared with RP tumors (Supplemental Fig. S6A). The five RP cis–eto tumors expressed very low levels of MYC and MYCN, a finding that was consistent with immunohistochemistry analyses (Supplemental Fig. S6A,B). Thus, supraphysiological MYCL expression achieved with the MYCL transgene expression but not basal MYCL expression was associated with resistance to chemotherapy. To assess the functional effects of MYCN or MYCL overexpression on short term response to chemotherapy, we treated RP, RPMYCL, and RPMYCN mice with a single cycle of either cis–eto or saline and collected lung tumors 72 h later. RP tumors exhibited increased apoptosis (as determined by TUNEL staining) and decreased proliferation (as determined by pH3 staining) relative to saline controls in response to cis–eto. However, increased cell death and reduced proliferation were no longer observed in cis–eto-treated RPMYCN or RPMYCL tumors (Fig. 4D–F). Thus, in the GEM model of SCLC, MYCN or MYCL overexpression suppresses responses to chemotherapy.
Figure 4.

MYCN and MYCL drive chemoresistance in an SCLC mouse model (A) Representative MRI of RP, RPMYCL, and RPMYCN lungs at days 0, 14, and 21 of cis–eto treatment. Tumors are circled in yellow. (B) Quantification of the percentage of change in tumor volume between days 0 and 14 of treatment. Data are means ± SEM (RP SALINE: n = 8; RP CIS–ETO: n = 6; RPMYCN SALINE: n = 9; RPMYCN CIS–ETO: n = 7; RPMYCL SALINE: n = 8; RPMYCL CIS–ETO: n = 11). Significance was determined using two-tailed unpaired Student's t-test. (**) P < 0.01. (C) Quantification of the percentage of change in tumor volume between days 0 and 21 of treatment. Data are means ± SEM (RP SALINE: n = 8; RP CIS–ETO: n = 6; RPMYCN SALINE: n = 5; RPMYCN CIS–ETO: n = 7; RPMYCL SALINE: n = 8; RPMYCL CIS–ETO: n = 11). Significance was determined using two-tailed unpaired Student's t-test. (***) P<0.001. (D) Representative immunohistochemistry images for pH3 and TUNEL staining in a parallel cohort comparing RP, RPMYCL, and RPMYCN tumors at a 3-d time point of treatment with either saline or cis–eto. Scale bar, 20 µm. (E) Quantification of the percentage of TUNEL-positive cells. Data are means ± SEM (n = 5 in all groups). Significance was determined using two-tailed unpaired Student's t-test. (**) P < 0.01. (F) Quantification of the percentage of pH3-positive cells. Data are means ± SEM. (n = 5 in all groups). Significance was determined using two-tailed unpaired Student's t-test. (***) P < 0.001.
MYCN and MYCL overexpression drive cisplatin–etoposide resistance in PDX models of SCLC
The RP SCLC autochthonous model typically exhibits cytostatic responses to cis–eto treatment (Fig. 4A–C), with only rare tumor regressions. To better model cytoreductive responses to cis–eto seen in many SCLC patients, we overexpressed MYCN in highly chemosensitive patient-derived xenograft (PDX) models, FHSC14 and FHSC23, that we previously generated from chemonaive patients (Augert et al. 2019). Both PDX models exhibit flank tumors regressions with 3 wk of cis–eto treatment. FHSC14 and FHSC23 PDX tumors growing in NOD-SCID-γ (NSG) mice were dissociated and infected ex vivo with lentivirus expressing a MYCN cDNA or empty control. Within 16 h of PDX tumor extraction and addition of lentivirus, the infected cells were reinjected into the flanks of NSG mice. Once flank tumors reached 150 mm3, we treated mice with three weekly cycles of cis–eto or saline and measured tumor volume over the course of 21 d, and then extracted remaining tumor for molecular analyses. FHSC14 and FHSC23 empty vector-infected tumors strongly regressed upon cis–eto treatment (Fig.5A; Supplemental Fig. S7A). In contrast, in both FHSC14 and FHSC23 models, tumors from MYCN-infected cells robustly grew throughout the cis–eto treatment period (Fig. 5A; Supplemental Fig. S7A). For both models, we validated overexpression of N-MYC using immunoblotting (Fig. 5B; Supplemental Fig. S7B). Thus, overexpression of a single gene, MYCN, can switch two highly chemosensitive models of SCLC to become resistant. Having shown that MYCN overexpression can drive the development of chemoresistance in PDX models, we sought to extend our findings to MYCL. As in the MYCN-overexpressing model, while control empty vector tumors completely regressed in response to chemotherapy, the MYCL-overexpressing tumors also continued to grow over the course of the study (Fig. 5C,D; Supplemental Fig. S7C,D). To assess the acute effects of MYCN/L overexpression on chemotherapy response, we modified the experiment to treat with cis–eto or saline and collect tumors at 72 h after treatment. Control empty vector tumors exhibited a decrease in proliferation as assessed by pH3 staining and increased apoptosis as assessed by TUNEL, upon cis–eto treatment (Fig. 5E–H; Supplemental Fig. S7E–H). However, changes in proliferation or apoptosis relative to saline-treated controls were not observed in similarly treated MYCN- or MYCL-overexpressing tumors (Fig. 5E–H; Supplemental Fig. S7E–H). These data highlight the importance of MYC family members in driving chemoresistance in SCLC.
Figure 5.
MYCN and MYCL overexpression abrogates chemotherapy response in PDX model of SCLC (A) Graph showing flank tumor volumes of empty control versus MYCN-overexpressing FHSC14 PDX tumors over 21 d of treatment with either saline or three cycles of cis–eto. Data are means ± SEM (n = 5 in all groups). Significance was determined using a mixed model two-way ANOVA followed by a post hoc Tukey's multiple comparisons test. For each group, significance is presented relative to the respective saline condition. (**) P < 0.01. (B) Immunoblot showing successful MYCN overexpression in the FHSC14 PDX model. Lysate from a human SCLC cell line harboring a MYCN amplification (H69) was used as a positive control while lysate from the human SCLC cell line H209 was used as a negative control. GAPDH was used as a loading control. (C) Graph showing flank tumor volumes of empty control versus MYCL-overexpressing FHSC14 PDX tumors over 21 d of treatment with either saline or three cycles of cis–eto. Data are means ± SEM (n = 5 in all groups). Significance was determined using a mixed model two-way ANOVA followed by a post hoc Tukey's multiple comparisons test. For each group, significance is presented relative to the respective saline condition. (**) P < 0.01. (D) Immunoblot showing successful MYCL overexpression in the FHSC14 PDX model. Lysate from a human SCLC cell line harboring a MYCL amplification (H510) was used as a positive control while lysate from the human SCLC cell line H209 was used as a negative control. GAPDH was used as a loading control. (E) Representative immunohistochemistry images for TUNEL staining in a parallel cohort comparing FHSC14 empty, MYCN-overexpressing, and MYCL-overexpressing tumors at a 3-d time point after treatment with either saline or cis–eto. Scale bar, 20 µm. (F) Quantification of the percentage of TUNEL-positive cells. Data are means ± SEM (n = 5 in all groups). Significance was determined using two-tailed unpaired Student's t-test. (**) P < 0.01; (***) P < 0.001. (G) Representative immunohistochemistry images for pH3 staining in a parallel cohort comparing FHSC14 empty, MYCN-overexpressing, and MYCL-overexpressing tumors at a 3-d time point after treatment with either saline or cis–eto. Scale bar, 20 µm. (H) Quantification of the percentage of pH3-positive cells. Data are means ± SEM. (n = 5 in all groups). Significance was determined using two-tailed unpaired Student's t-test. (***) P < 0.001.
Genome wide CRISPR–Cas9 screen reveals USP7 as a synthetic vulnerability for MYCN
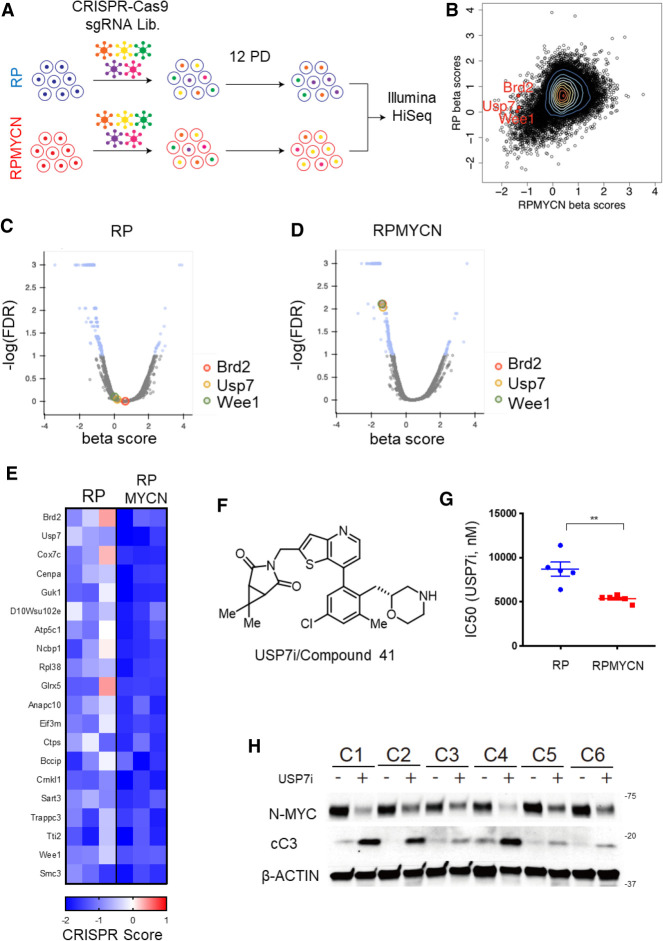
Having shown that MYCN drives tumor progression and chemoresistance in SCLC, we sought to find means to therapeutically target MYCN-overexpressing SCLC. To uncover unique vulnerabilities in MYCN-driven SCLC, we performed genome-scale CRISPR–Cas9 small guide RNA (sgRNA) inactivation screens on 3 RP and 3 RPMYCN SCLC cell lines derived from GEMM tumors. We infected each cell line with lentivirus containing the Murine GeCKO Lentiviral sgRNA Library v2 vector system, which expresses a pool of 130,209 sgRNAs targeting 20,611 genes (Sanjana et al. 2014). Cells were infected at a multiplicity of infection (MOI) of <1 and then subjected to puromycin selection. Following selection, a portion of cells for each line was collected as an initial time point (P0) and the remainder grown out for 12 population doublings prior to collection (P12). Using deep sequencing, we compared the abundance of sgRNAs in the P0 and P12 populations for each RP or RPMYCN cell line (Fig. 6A). For each gene, we generated a “CRISPR score,” defined as log2 (sgRNA abundance at P12/sgRNA abundance at P0) averaged across all sgRNAs targeting a given gene (Wang et al. 2015). We also employed MAGeCK-MLE analysis to determine which sgRNAs were preferentially depleted in the three RPMYCN as compared with the three RP cell lines (Li et al. 2014). We were particularly interested in druggable genes that are essential in the MYCN-overexpressing cells and found that the sgRNAs targeting WEE1, BRD2, and USP7 were preferentially depleted after 12 population doublings in the RPMYCN compared with the RP cell lines (Fig. 6B–E). USP7 was of particular interest as this gene encodes a deubiquitinase that directly deubiquitinates N-MYC, resulting in increased protein stability (Tavana et al. 2016). Given this, we hypothesized that USP7 could be a synthetic lethal target for MYCN-driven SCLC. To test this, we treated RP and RPMYCN cell lines with a novel inhibitor of USP7, (3((7-(5-chloro-3-methyl-2-(((R)-morpholin-2-yl)methyl)phenyl)thieno[3,2-b]pyridin-2-yl)methyl)‐6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2,4-dione), referred to here as USP7i, discovered by RAPT Therapeutics, Inc. (Fig. 6F). The discovery, synthesis, and complete characterization of this orally bioavailable, potent USP7 inhibitor is described elsewhere as compound 41 (Leger et al. 2020). Consistent with the genetic screen results, RPMYCN cell lines exhibited increased sensitivity to USP7 inhibition as compared with RP cell lines (Fig. 6G; Supplemental Fig. S8A,B). Analysis by Western blotting showed that treatment of RPMYCN cell lines with USP7i led to decreased levels of N-MYC protein abundance as well as increased levels of cleaved caspase 3, indicative of apoptosis (Fig. 6H). Thus, USP7 is a druggable synthetic vulnerability in MYCN-driven SCLC.
Figure 6.
CRISPR inactivation screens reveal USP7 as a MYCN synthetic vulnerability. (A) Schematic outlining the strategy used in a genome scale CRISPR–Cas9 sgRNA screen comparing cell lines derived from RP and RPMYCN tumors (n = 3 cell lines in both groups). (B) Contour plot comparing MAGeCK analysis generated β scores for RP-derived lines (Y-axis) versus RPMYCN-derived lines (X-axis). Each data point (shown as a black circle) represents a single gene in the sgRNA library. A positive β score indicates that guide RNAs targeting a given gene are present in a higher proportion after 12 population doublings while a negative β score indicates that guide RNAs targeting a given gene are present in a lower proportion after 12 population doublings. A selection of known druggable targets that exhibit a significantly lower β score in RPMYCN lines as compared with RP lines are highlighted in red. Genes with fewer than five associated sgRNAs were omitted. (C) Volcano plot showing genes that are significantly enriched or depleted in RP lines following 12 population doublings as determined by MAGeCK-MLE analysis. (D) Volcano plot showing genes that are significantly enriched or depleted in RPMYCN lines following 12 population doublings as determined by MAGeCK-MLE analysis. (E) Heat map showing CRISPR scores of 20 selected genes for each RP- and RPMYCN-derived cell line. Genes were selected based on statistical significance from MAGECK analyses and ordered based on greatest difference in RPMYCN and RP CRISPR scores. (F) Chemical structure of the novel USP7 inhibitor, USP7i, also known as compound 41, developed by RAPT Therapeutics. (G) Comparison of IC50 for USP7i between RP- and RPMYCN-derived cell lines. Data are means ± SEM from n = 5 cell lines (RP) and n = 6 cell lines (RPMYCN). Data from each individual cell line are from three biological replicates each consisting of three technical replicates. Significance was determined using two-tailed unpaired Student's t-test. (**) P < 0.01. (H) Immunoblot comparing levels of N-MYC and cleaved CASPASE 3 (cC3) in six RPMYCN-derived cell lines (C1–6) either with or without USP7i treatment. β-ACTIN was used as a loading control.
USP7 inhibition resensitizes MYCN-overexpressing chemoresistant tumors to cisplatin/etoposide
Having found that pharmacologic inhibition of USP7 can selectively target MYCN-driven SCLC, we next assessed whether inhibition of USP7 could restore sensitivity to chemotherapy in MYCN-overexpressing, chemoresistant PDX models. To improve on the model system described in Figure 5 that we use to overexpress single genes in chemosensitive PDX models, we switched to a two-step system in which we sorted productively infected cells following expansion in vivo, ensuring essentially complete lentiviral transduction, before propagating for therapeutic experiments. We infected the FHSC14 model with lentivirus containing either a MYCN cDNA-IRES-ZsGreen vector, a MYCL cDNA-IRES-ZsGreen vector, or an empty control IRES-ZsGreen vector, and then injected the cells into the flanks of NSG mice. Once flank tumors developed, we dissociated tumor cells and used fluorescence activated cell sorting (FACS) to obtain a pure population of ZsGreen-positive MYCN-overexpressing, MYCL-overexpressing, or empty vector control FHSC14 cells (see Materials and Methods). We then injected these populations into the flanks of NSG mice to allow for tumor development and we validated pure populations of infected cells by sorting for ZsGreen-positivity (Supplemental Fig. S9A). For therapeutic studies using this system, once tumors reached 150 mm3, we treated with either saline, cis–eto, 100 mg/kg USP7i, or a combination of 100 mg/kg USP7i and cis–eto (USP7i-cis–eto) and measured tumor volume over 14 d. While cis–eto caused near complete tumor regression in empty vector controls, treatment with USP7i alone had no effect on tumor volume (Fig. 7A,B). In the MYCL-overexpressing tumors, neither cis–eto nor USP7i alone or in combination significantly slowed tumor growth (Fig. 7B). However, in the MYCN-overexpressing tumors, while cis–eto alone did not blunt tumor growth given MYCN-driven chemoresistance, we found that combination USP7i-cis–eto treatment resulted in complete tumor regression in a manner similar to the empty vector control tumors treated with cis–eto (Fig. 7A). The combination of high dose USP7i and cis–eto resulted in weight loss as shown in Supplemental Fig. S10A. A validation cohort of FHSC14 MYCN and FHSC14 empty control mice used a reduced 50 mg/kg dose of USP7i and extension to 21 d, with cis–eto and USP7i treatments administered during weeks one and three, a dosing regimen that was better tolerated (Supplemental Fig. S10B). Again, MYCN-overexpressing tumors were resistant to cis–eto but strongly regressed upon combined USP7i-cis–eto treatment (Fig. 7C). These data conclusively show that inhibition of USP7 can resensitize MYCN-overexpressing SCLC to cytotoxic chemotherapy. Observations that USP7i did not resensitize MYCL-overexpressing SCLC to chemotherapy (Fig. 7B) support the notion that reduction of N-MYC following USP7i in the MYCN-overexpressing model was critical for the observed responses. To assess the short-term effects of the various treatments, we repeated the experiment but treated for just one cycle of cis–eto and/or USP7i, collecting tumors after 7 d. Treatment with USP7i alone or with cis–eto dramatically reduced levels of N-MYC in MYCN-overexpressing tumors (Fig. 7D). Immunostaining revealed that MYCN-overexpressing tumors were resistant to the increased apoptosis and reduced proliferation upon cis–eto treatment that was observed in empty vector controls. However, MYCN-overexpressing tumors in the combined USP7i-cis–eto treatment group exhibited reduced proliferation and increased apoptosis (Fig. 7E–H). These phenotypes are consistent with the strong tumor regression seen with combined USP7 inhibition and chemotherapy in the MYCN-overexpressing model.
Figure 7.
USP7 inhibition resensitizes MYCN-overexpressing tumors to chemotherapy (A) Graph showing flank tumor volumes of empty control versus MYCN-overexpressing FHSC14 PDX tumors over 14 d of treatment with either saline, cis–eto, 100 mg/kg USP7i, or 100 mg/kg USP7i + cis–eto. Data are means ± SEM (n = 5 in all groups except for EMPTY CIS–ETO and MYCN USP7i, where n = 4). Significance was determined using a mixed model two-way ANOVA followed by a post hoc Tukey's multiple comparisons test. For each group, significance is presented relative to the respective saline condition. (**) P < 0.01. (B) Graph showing flank tumor volumes of empty control versus MYCL-overexpressing FHSC14 PDX tumors over 14 d of treatment with either saline, cis–eto, 100 mg/kg USP7i, or 100 mg/kg USP7i + cis–eto. Data are means ± SEM (n = 5 in all groups except for EMPTY CIS–ETO, MYCL SALINE, and MYCL USP7i + CIS–ETO, where n = 4; EMPTY samples are the same as in A). Significance was determined using a mixed model two-way ANOVA followed by a post hoc Tukey's multiple comparisons test. For each group, significance is presented relative to the respective saline condition. (*) P < 0.05; (**) P < 0.01. (C) Graph showing flank tumor volumes of empty control versus MYCN-overexpressing FHSC14 PDX tumors over 21 d of treatment with either saline, cis–eto, 50 mg/kg USP7i, or 50 mg/kg USP7i + two cycles of cis–eto (weeks 1 and 3, respectively). Data are means ± SEM (n = 5 in all groups). Significance was determined using a mixed model two-way ANOVA followed by a post hoc Tukey's multiple comparisons test. For each group, significance is presented relative to the respective saline condition. (**) P < 0.01. (D) Immunoblot comparing levels of N-MYC expression across treatment groups after 7 d of treatment in a parallel cohort. GAPDH was used as a loading control. (E) Representative immunohistochemistry images for pH3 staining comparing treatment groups. Scale bar, 20 µm. (F) Quantification of the percentage of pH3-positive cells. Data are means ± SEM (n = 5 in all groups). Significance was determined using two-tailed unpaired Student's t-test. (***) P < 0.001. (G) Representative immunohistochemistry images for TUNEL staining comparing treatment groups. Scale bar, 20 µm. (H) Quantification of the percentage of TUNEL-positive cells. Data are means ± SEM. (n = 5 in all groups). Significance was determined using two-tailed unpaired Student's t-test. (***) P < 0.001.
Discussion
Here, we report the first autochthonous mouse model of MYCN-driven SCLC. MYCN overexpression dramatically accelerated SCLC, with RPMYCN mice developing lung tumors on average almost a year earlier than the RP model. This model provides a valuable tool for further studies of MYCN in SCLC tumor biology and treatment response.
In addition to supporting MYCN control of the cell cycle and ribosomal biogenesis, consistent with previous data from other systems (Boon et al. 2001; Dang 2012; Beltran 2014), our transcriptional analyses also revealed multiple immune signaling pathways changed in SCLC with MYCN overexpression. Cell sorting analyses revealed a decrease in infiltrating T-lymphocytes along with an increase in infiltrating neutrophils in MYCN-overexpressing SCLC. Other recent studies have also found correlations between high levels of MYCN expression and decreased cytotoxic immune cell signatures in neuroblastoma (Zhang et al. 2017; Wei et al. 2018). In neuroblastoma, MYCN overexpression dampened expression of NK cell activating receptors (Brandetti et al. 2017). However, this is unlikely to be the case in SCLC, as we found no difference in levels of infiltrating NK cells between MYCN-high and MYCN-low tumors (Fig. 3H). It is known that other MYC family members can directly regulate expression levels of PD-L1 to inhibit T-cell responses in other cancers (Casey et al. 2016); however, our RNA-seq analyses did not reveal changes in PD-L1 with MYCN overexpression in SCLC. Our results constitute the first evidence that MYCN functionally modulates the tumor microenvironment in SCLC and the novel MYCN-overexpressing model provides a valuable tool for future studies to interrogate how MYCN overexpression suppresses T-cell infiltration. SCLC is known to have a less active tumor microenvironment relative to other lung cancers. While ∼50% of the tumor microenvironment in nonsmall cell lung cancer consists of CD45+ cells, immune cell populations make up only ∼15% of the SCLC tumor microenvironment (Busch et al. 2016). Notably, the immune component of the SCLC tumor microenvironment is normally characterized by high levels of T lymphocytes and low levels of innate immune cells such as neutrophils (Busch et al. 2016). As such, the suppression of T-cell infiltration with MYCN overexpression may have important implications for response to immune checkpoint inhibitors that rely on modulation of T-cell-mediated killing. A subset of SCLC patients respond to immune checkpoint modulation (Horn et al. 2018; Paz-Ares et al. 2019). Further studies are required to determine whether in the small subset of SCLC that harbors MYCN amplification, MYCN may reflect a therapeutic target to achieve enhanced responses toward immune checkpoint inhibition.
Our study compared Rb/p53 with Rb/p53/MYCN GEM as well as isogenic PDX models in which we overexpressed either MYCN or MYCL. In both GEM and isogenic PDX systems, MYCN and MYCL conferred clear chemoresistance. We note that prior MYC- and MYCL-overexpressing mouse model studies have included chemotherapy treatments (Mollaoglu et al. 2017; Böttger et al. 2019). However, in both studies (Mollaoglu et al. 2017; Böttger et al. 2019), lack of Rb/p53 comparator impaired assessment of the contribution of MYC or of MYCL overexpression to chemotherapy response and resistance. Our inclusion of a control Rb/p53 arm was critical for our assessments that MYCN overexpression and MYCL overexpression drives chemoresistance. A limitation of the GEM models relates to the level of chemosensitivity observed, as tumor regressions in response to cis–eto are rare in this model system. The absence of highly chemosensitive models of SCLC that regress with chemotherapy, in which specific genes could be manipulated and functionally assessed, has hindered study of genetic drivers of SCLC chemoresistance. Our observation that two highly chemosensitive PDX models of SCLC completely switch to becoming chemoresistant with MYCN or MYCL overexpression definitively show that these oncogenes can indeed drive chemoresistance in SCLC. Reducing cell culture time was a critical aspect of our novel approach to genetically perturb PDX models of SCLC, as it has been shown that PDX models of SCLC passaged in vivo more fully recapitulate the expression of the original tumor versus derived cell lines (Daniel et al. 2009). While the experiments in this study employed a 16-h culture period for infections, our current protocols now reduce the time in ex vivo culture to >4 h. With the two-step system for model propagation (Fig. 7; Supplemental Fig. S9), we can generate single-gene-perturbed PDX models that can be cryopreserved, shared, and repropagated at any time for functional or therapeutic studies. The Rudin laboratory has also recently demonstrated the utility of genetically engineering PDX models to harbor single gene perturbations (Hulton et al. 2020). Given that the response of GEM models to chemotherapy does not include strong regressions as seen in human SCLC patients, employment of chemosensitive PDX models that mimic the response of the originating patient are critical. We anticipate that this system will also enable functional screens to systematically identify drivers of SCLC chemoresistance.
We performed a comprehensive genome wide CRISPR–Cas9 screen on both RP- and RPMYCN-derived cell lines in order to uncover druggable vulnerabilities for MYCN-overexpressing SCLC. Our results show that multiple druggable targets such as WEE1, BRD2, and USP7 are synthetically lethal with MYCN overexpression. WEE1 has been studied as a potential therapeutic target in SCLC (Sen et al. 2017; Lallo et al. 2018). Likewise bromodomain and extraterminal (BET) domain inhibitors, such as JQ1, that target proteins including BRD2 and BRD4, have shown efficacy in targeting MYCL- and MYCN-overexpressing cell lines (Lenhart et al. 2015; Kato et al. 2016; Wang et al. 2017). While the use of USP7 inhibitors to target MYCN-driven tumors has been studied in neuroblastoma (Tavana et al. 2016), this approach has not previously been assessed in PDX or GEM models of SCLC or in the context of chemoresistance. By using a novel inhibitor of USP7, we demonstrated that MYCN-overexpressing SCLC is highly sensitive to loss of USP7. Furthermore, pharmacological inhibition of USP7 can completely reverse MYCN-driven chemoresistance in PDX models, providing a genotype-specific strategy to target a subset of chemoresistant SCLC.
A key finding was that not only MYCN overexpression, but also MYCL overexpression drives chemoresistance in SCLC. Even though most SCLC tumors express MYCL, a transcriptional target of the ASCL1 transcription factor (Borromeo et al. 2016), physiological MYCL expression was not associated with chemoresistance in our mouse models. Supra-physiological levels of MYCL may be needed to achieve chemoresistance, such as those achieved with high-level genomic amplification. In untreated SCLC, amplifications in MYCL, MYCN and MYC occur at approximate frequencies of 9%, 4%, and 6%, respectively (George et al. 2015). The frequency of MYCN, MYCL and MYC amplifications in chemoresistant extensive-stage SCLC patients is not well defined, as the largest human SCLC genomic data sets are of untreated patients at limited stage (Rudin et al. 2012; George et al. 2015). In a series of PDX models, a MYC transcriptional signature was associated with chemoresistance even without clear enrichment for MYC family genomic amplifications, suggesting that genomic amplifications may not be the only means to achieve high MYC family activation (Drapkin et al. 2018). Recent studies of SCLC PDX models have revealed a high degree of intratumoral heterogeneity in SCLC, indicating that chemoresistance could be associated with multiple mechanisms in different tumor cell subsets even within a single patient (Simpson et al. 2020; Stewart et al. 2020). Large-scale genomic analyses of extensive stage chemoresistant SCLC, ideally with inclusion of paired pretreatment samples would help define the frequency of key genetic alterations that drive chemoresistance and constitute therapeutic targets to resensitize tumors toward chemotherapy.
Materials and methods
Genetically engineered mouse models
Rb1lox/lox mice were from Tyler Jacks (MIT) while Trp53lox/lox mice were generated by Dr. Anton Berns (Netherlands Cancer Institute). The Rblox/lox;Trp53lox/lox;MyclOE mice (RPMYCL) were obtained from Dr. Anton Berns (Huijbers et al. 2014). The TRE-MYCN/LUC model was provided by Dr. William Weiss (Swartling et al. 2010) and the Rosa26 lox-stop-lox M2rtTA allele, originally generated by Rudolph Jaenisch's laboratory (Hochedlinger et al. 2005) was obtained from Jackson labs (strain 006965). We bred the TRE-MYCN/LUC mice to RP mice in order to generate Rb1lox/lox;Trp53lox/lox;M2rtTAlox/lox;TRE-MYCN/LUClox/lox mice (RPMYCN). Tumor formation was initiated by intratracheal infection of adult mice with Ad-CGRP-Cre (RP and RPMYCN) or Ad-CMV-Cre (RPMYCL) at a titer of 1.25 × 109 pfu per mouse (DuPage et al. 2009). Ad-CGRP-Cre and Ad-CMV-Cre were obtained from the University of Iowa Gene Vector Core. To induce transgene activation, all RPMYCN mice were given feed containing doxycycline (DOX feed, Teklad) beginning at 1 wk following intratracheal infection and continuing throughout the course of the study. After being placed on DOX feed, mice described in Figure 1 were monitored weekly and were euthanized upon showing signs of severe labored breathing. Overall time of survival was plotted using a Kaplan-Meier curve. For the RPMYCN ON DOX versus OFF DOX study described in Figure 2, A–D, RPMYCN mice were switched from DOX feed to standard mouse chow following tumor development (as assessed by MRI) in order to generate the OFF DOX cohort. In all studies, lung tumor fragments were collected for molecular analyses (snap-frozen), histology, and immunohistochemistry (fixed in neutral buffered formalin). All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the Fred Hutchinson Cancer Research Center.
Cell line generation and cell culture
Lung tumor tissue from RP and RPMYCN mouse models was collected shortly following euthanasia using a sterile syringe and needle. Tissue was dissociated by pipetting and then cultured in DMEM complete medium (Dulbecco's modified Eagle medium containing 15% fetal bovine serum, 1% penicillin–streptomycin, 1% sodium pyruvate, β-mercaptoethanol, and recombinant insulin) containing 1 µg/mL doxycycline after which growth of stable lines was observed. Human SCLC cell lines H510, H69, and H209 were obtained from ATCC (Phelps et al. 1996).
Western blot analysis
Protein was extracted from lung tumor tissues and cell lines by dissociation in RIPA buffer (Cell Signaling, 9806). Dissociation was performed by either serial passaging through small-gauge needles or by mechanical disruption. Immunoblotting by Western blot was performed according to standard procedures. The following antibodies were used: anti-N-MYC(Cell Signaling 51705), anti-β ACTIN (Cell Signaling 4970), anti-CYCLIN E (Santa Cruz Biotechnology sc-247), anti-CYCLIN A (Santa Cruz Biotechnology sc-271645), anti-p27 (Aviva OAAI00157), anti-p130 (Santa Cruz Biotechnology sc-374521), anti-L-MYC (in-house), anticleaved CASPASE3 (Cell Signaling 9661), and anti-GAPDH (Santa Cruz Biotechnology sc-32233).
Histology and immunohistochemistry
Mouse lung tumor and PDX tumor fragments were fixed in neutral-buffered formalin for 48 h prior to processing to paraffin blocks. Paraffin blocks were sectioned at a thickness of 4 µm. Hematoxylin and eosin (H&E) staining was done according to standard procedures. For immunohistochemistry analysis, paraffin sections were dewaxed in xylene and then rehydrated by passage through a graded series of ethanol into Tris-buffered saline-Tween 20 (TBS-T). Antigen unmasking was performed by heating in a tiered steamer in Trilogy buffer (Cell Marque 920-P) according to the manufacturer's protocol. Endogenous peroxidases were blocked with 3.5% H2O2 and sections were incubated overnight at 4°C in primary antibody following blocking with 5% goat serum (Jackson ImmunoResearch 005-000-121). The following primary antibodies were used: anti-CGRP (Cell Signaling 14959), anti-N-MYC (Cell Signaling 51705), antiphospho Ser10 histone H3 (EMD Millipore 06-570), anti-ASCL1 (BD, 556604), anti-NEUROD1 (Abcam ab109224), anti-YAP1 (Cell Signaling, 14074), and anti-MYC (Santa Cruz Biotechnology sc-764). Sections were then incubated with biotin-conjugated secondary antibodies (Vector Laboratories BA-1000) after which a biotin–peroxide complex was formed using the VectaStain ABC kit (Vector Laboratories PK-4000) Detection was carried out using 3,3′-diaminobenzidine substrate (Vector Laboratories SK-4100). TUNEL assays were performed using the POD in situ cell death detection kit (Roche 11684817910) according to the manufacturer's protocol. Slides were imaged using a Nikon E800 microscope and quantified at 100× magnification with three fields per section.
Cell proliferation analysis
Cells were seeded at 5000 cells per well in 96-well plates in DMEM complete medium containing either 1 µg/mL doxycycline (ON DOX condition) or no doxycycline (OFF DOX condition). Relative cell viability was assessed at days 1, 2, 3, and 4 using the Cell Counting Kit-8 kit (Dojindo CK04) according to the manufacturer's protocol.
Cell cycle analysis
Cells were plated in six-well plates in DMEM complete medium containing either 1 µg/mL doxycycline (ON DOX condition) or no doxycycline (OFF DOX condition). After 72 h of incubation, cells were collected and fixed using ethanol and stained with propidium iodide (Sigma P4170). Stained cells were analyzed using a FACS Canto II system. Gating and analysis were done using FlowJo software.
qPCR and RT-qPCR
RNA was extracted from cell lines and tumor tissue by mechanical disruption in TRIzol reagent (Invitrogen 15596026) and isolated according to the manufacturer's protocol. cDNA was generated using the iScript reverse transcription kit (Bio-Rad 1708840) according to the manufacturer's protocol. DNA was extracted from tumor tissue and tails using a salt precipitation protocol (Chen et al. 2015). qPCR reactions were set up using the SYBR Green-based All-in-One qPCR reagent (GeneCopoeia AOPR-4000) and run on a 7900 real-time PCR system (Applied Biosystems). Primers were used against MYCN (human and murine), Mycl, Myc, 47S ITS, Gapdh, Slc25a4, and β2m (Supplemental Table S1).
Puromycin incorporation assay
Cells were plated in six-well plates in DMEM complete medium containing either 1 µg/mL doxycycline (ON DOX condition) or no doxycycline (OFF DOX condition). Cells were incubated with 1 µM puromycin (Sigma P8833) for 30 min after which protein was extracted. Immunoblotting was then done using an anti-puromycin antibody (EMD Millipore MABE343).
RNA-seq analysis
RNA was extracted from mouse lung tissue and cell lines by mechanical disruption in TRIzol reagent and isolated according to the manufacturer's protocol. Indexed RNA-seq libraries were generated using the Ultra RNA library preparation kit for Illumina (New England BioLabs E753L) according to the manufacturer's protocol. Single-end sequencing was run on an Illumina HiSeq 2500 and reads were aligned to the mm9 genome build using TopHat (Trapnell et al. 2009). Counts were generated from TopHat alignments using the HTSeq software package and fragment per kilobase per million (FPKM) expression values were generated using CuffDiff (Trapnell et al. 2013; Anders et al. 2015). Genes with low counts (less than five mapped reads) across conditions were discarded after which, differentially expressed genes were identified using the EdgeR software package (Robinson et al. 2010) with an FDR cutoff of 0.05 set to determine significance. Lists of differentially expressed genes were analyzed using the Enrichr (Kuleshov et al. 2016) application. Pathway and transcriptional analyses were done by querying the Kyoto Encyclopedia of Genes and Genomes (KEGG) (Kanehisa and Goto 2000), ChIP Enrichment Analysis (Lachmann et al. 2010), and Encyclopedia of DNA Elements (The ENCODE Project Consortium 2004) respectively. Gene set enrichment analysis (GSEA) was run using the FPKM values of each sample and analysis was done by querying the Hallmark gene sets in the molecular signatures database.
CUT&RUN analysis
An RPMYCN-derived cell line (C3) was grown in the presence or absence of DOX for 4 d after which cells were collected and bound to concanavalin A beads (Polysciences, Inc. 86057-3) and permeabilized by suspension in a digitonin-containing buffer. Cells were incubated overnight with an antibody against N-MYC (Santa Cruz Biotechnologies sc-53993) and libraries were prepared for CUT&RUN analysis according to a previously described protocol (Skene and Henikoff 2017). Spike-in normalization was done using a yeast DNA spike-in. Sequences were aligned to the mouse mm9 genome assembly using Bowtie2 (Langmead and Salzberg 2012) and peaks were called using MACS2 (Zhang et al. 2008). Following peak calling, processing was carried out using bedtools and the deepTools2 software package (Ramírez et al. 2016). Peaks were identified as associated with a specific gene if they were within 5 kb of the TSS.
FACS analysis
Single-cell suspensions were generated from saline-perfused mouse lungs using mechanical disruption, followed by a 15-min digestion at 37°C in RPMI1640 containing 80 U/mL DNase (Worthington Biochemical LS002007) and 300 U/mL collagenase type 1 (Worthington Biochemical LS004196). Digested lungs were sheared through a 10-mL pipette, strained through 70-mm nylon mesh, washed with RPMI1640 medium, lysed (RBCs), strained through 40-µm mesh, and ultimately resuspended in Dulbecco's PBS + 2% FBS. Single-cell suspensions were incubated with mouse TruStain FcX (BioLegend 101319) for at least 15 min on ice prior to a 30-min immunostaining with fluorochrome-conjugated antibody cocktails (antibodies used are outlined in Supplemental Table S2). After the extracellular staining, cell pellets were washed using 2% FBS + PBS, and Fixable viability dye eFluor 780 (eBioscience 501129035) was used to exclude dead cells according to the manufacturer's protocol. Stained cells were then washed and fixed with IC fixation buffer (eBioscience 501129058). Samples were analyzed using a Symphony II analyzer. Compensation and gating analysis were performed using FlowJo software.
PDX model generation
The generation and characterization of the FHSC14 PDX model has been described previously (Augert et al. 2019). The FHSC23 PDX model was generated from the circulating tumor cells of a chemonaive SCLC patient using previously described methodology (Hodgkinson et al. 2014). Both models are propagated in the flanks of NOD-SCID-γ (NSG) mice. To generate MYCN- and MYCL-overexpressing PDX models described in Figure 5 and Supplemental Figure S7, human MYCN or MYCL cDNA was cloned into the pLX304 lentiviral vector backbone (Addgene 25890) using Gibson assembly to generate pLX304 MYCN and pLX304 MYCL plasmids (Yang et al. 2011). A pLX304 plasmid containing a stuffer sequence (pLX304 EMPTY) was used as a control vector. Lentivirus was generated by cotransfection of 293TN cells with either pLX304 EMPTY, MYCN, or MYCL plasmid along with packaging plasmids psPAX2 (Addgene, 12260) and pMD2.G (Addgene, 12259) using a calcium phosphate transfection protocol. Viral supernatants were collected 64 h after transfection, filtered through a 0.45-µm filter, and concentrated using a Centricon-70 system (EMD Millipore UFC710008). Virus was resuspended in sterile PBS. For infection, PDX tumors were resected and digested for 20 min at 37°C in 1 mg/mL collagenase (Sigma C5138) in PBS. Digested tissue was sequentially strained through a 70-µm and 40-µm mesh to generate a single-cell suspension that was then plated in DMEM-F12 medium. Cells were then incubated with virus for a total of 4 h after which they were collected, spun down, mixed 1:1 with Matrigel (Corning CB-40234) and reinjected into NSG mice. To generate the MYCN- and MYCL-overexpressing PDX models described in Figure 7, human MYCN or MYCL cDNA was cloned into a pLVX-IRES-ZsGreen lentiviral vector backbone using Gibson assembly to generate pLVX MYCN, pLVX MYCL, or pLVX empty vectors (Pelish et al. 2015). Virus production and infection of FHSC14 cells was carried out as described above. Once tumors from infected FHSC14 cells grew out, the tumors were collected, digested as described above and sorted using the FITC channel of an Aria 2 cell sorter. Sorted cells were reinjected into NSG mice and allowed to grow out. Tumors from these mice were then collected, digested and put through another round of sorting to generate pure populations of FHSC14 MYCN, FHSC14 MYCL, and FHSC14 empty control cells (Supplemental Fig. S9A) that were then reinjected into NSG mice.
Drug treatment studies
For the experiments described in Figure 4, RP, RPMYCL, and RPMYCN mice with sufficient tumor burden (as determined by MRI) were randomly assigned to saline or cis–eto treatment groups. Mice in the saline group were treated weekly with saline while mice in the cis–eto group were treated with three weekly cycles of chemotherapy consisting of 5 mg/kg cisplatin (Sigma 479306) on day 1 and 10 mg/kg etoposide (Sigma E1383) on days 1,2, and 3. Thoracic MR images were collected on days 0, 14, and 21 and tumor volume was calculated using ImageJ. For the experiments described in Figures 5 and 7 and Supplemental Figure S5, NSG mice were implanted with 1.0 × 106 disaggregated cells from the modified FHSC14 or FHSC23 PDX models as described above. Flank tumors were measured using calipers and tumor volume was calculated using the formula for a prolate ellipsoid: V = D × (d2/2), where V is volume, D is the major axis of the tumor and d is the minor axis of the tumor (Hodgkinson et al. 2014). Once flank tumor volume reached 150–250 mm3, mice were randomized into saline, cis–eto, USP7i, or USP7i-cis–eto treatment groups. Mice in the saline and cis–eto groups were treated as described above. Mice in the USP7i group were treated with a daily dose of either 100 mg/kg (Fig. 7A,B) or 50 mg/kg (Fig. 7C) from days 0 to 7 and days 15 to 21 (Fig. 7C). Mice in the USP7i-cis–eto treatment group were given both USP7i and cis–eto treatments as described above. In this group, both USP7i and cis–eto treatment regimens were not given for days 8–14. For the experiments described in Figure 6 and Supplemental Figure S8, cells were seeded at 5000 cells per well in 96-well plates in DMEM complete medium containing 1 µg/mL doxycycline and treated with doses of USP7i for 24 h after which viability was assessed using Cell Titer Glo reagent (Promega G7573) according to the manufacturer's protocol. Dose response curves were generated in GraphPad Prism 8 using a four-parameter nonlinear regression curve fit.
Genome wide CRISPR–Cas9 screen
Cells from 3 RP and 3 RPMYCN cell lines were cultured in DMEM complete media containing doxycycline as described above. ∼400 × 106 cells from each line were infected at MOI < 1 with lentivirus containing the mouse GeCKO lentiviral sgRNA library v2 pool. Cells were then placed under puromycin selection for 72 h after which 65 × 106 cells from each line were collected as a P0 reference while the remaining cells were passaged for 12 population doublings to generate a P12 end point. DNA was extracted using a previously described salt precipitation protocol (Chen et al. 2015). sgRNA libraries were generated using a previously described PCR protocol and purified by running out on an agarose gel electrophoresis (Chen et al. 2015). The libraries were then quantified using a Kapa Biosystems library quantification kit (Thermo Fisher KK4824) and sequenced using an Illumina HiSeq 2500. Demultiplexed reads were trimmed and aligned to the mouse GeCKO sgRNA library using Bowtie (Langmead et al. 2009). Using the sgRNA read counts, we generated CRISPR scores (as in [Wang et al. 2015]). Nontargeting control guides were randomly binned in sets of six to generate a set of 166 NTC sgRNAs, prior to performing MAGeCK- MLE analysis to identify essential genes in RP and RPMYCN lines (Li et al. 2014).
Statistics
All plots and statistics were generated using GraphPad Prism 8. For Kaplan-Meier curves, significance was determined using a log-rank (Mantel-Cox) test. For immunohistochemistry, immunofluorescence, cell cycle, cell proliferation, FACS, and MRI analyses significance between groups was determined using a two-tailed unpaired Student's t-test. Mixed model two-way ANOVA was used to compare groups in the PDX model drug treatment studies. Significance was then determined using Tukey's multiple comparisons test.
Competing interest statement
Y.O. and P.L. are employees of RAPT Therapeutics, Inc., which developed the USP7 inhibitor employed in this manuscript.
Supplementary Material
Acknowledgments
We thank the staff of the Fred Hutchinson Genomics and Bioinformatics Shared Resource for help with the generation and analysis of sequencing data, as well as Matthew Fitzgibbon for help with CUT&RUN data analysis. We also acknowledge support from the staff of the Fred Hutchinson Experimental Histopathology and Comparative Medicine Shared Resources. We thank Adi Gazdar for analysis and grading of H&E slides. We are grateful to Robert Eisenman and Dirk Brockstedt for critical review of the manuscript. E.G. was supported by National Institutes of Health (NIH) training fellowship F30CA232475. NIH grants U01CA235625 and R01CA200547 to D.M. supported this work. This research was funded in part through the NIH/National Cancer Institute Cancer Center Support Grant P30 CA015704.
Author contributions: E.G. designed and executed experiments; collected, analyzed, and interpreted data; and contributed to writing the manuscript. N.W. and H.Z. designed and executed experiments and collected, analyzed, and interpreted data. X.L., J.P.N., R.T., A.H.I., E.C.E., and D.J. executed experiments and collected and analyzed data. Y.O. and P.L. provided the USP7 inhibitor and contributed to writing the manuscript. R.B. and J.B.H. performed bioinformatic analysis. K.D.E. and R.M. provided patient samples for PDX model generation. A.M.H. helped design the study and analyzed and interpreted data. D.M. designed the study, analyzed and interpreted data, and wrote the manuscript. All authors reviewed and provided input on the contents of the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.340133.120.
References
- Anders S, Pyl, PT., Huber, W. 2015. HTSeq- a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166-169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augert A, Zhang Q, Bates B, Cui M, Wang X, Wildey G, Dowlati A, MacPherson D. 2017. Small cell lung cancer exhibits frequent inactivating mutations in the histone methyltransferase KMT2D/MLL2: CALGB 151111 (alliance). J Thorac Oncol 12: 704–713. 10.1016/j.jtho.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augert A, Eastwood E, Ibrahim AH, Wu N, Grunblatt E, Basom R, Liggitt D, Eaton KD, Martins R, Poirier JT, et al. 2019. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci Signal 12: eaau2922 10.1126/scisignal.aau2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H. 2014. The N-myc oncogene: maximizing its targets, regulation, and therapeutic potential. Mol Cancer Res 12: 815–822. 10.1158/1541-7786.MCR-13-0536 [DOI] [PubMed] [Google Scholar]
- Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voûte PA, Schwab M, et al. 2001. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J 20: 1383–1393. 10.1093/emboj/20.6.1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, Girard L, Minna JD, Gazdar AF, Cobb MH, et al. 2016. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep 16: 1259–1272. 10.1016/j.celrep.2016.06.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttger F, Semenova EA, Song JY, Ferone G, van der Vilet J, Cozijnsen M, Bhaskaran R, Bombardelli L, Piersma SR, Pham TV, et al. 2019. Tumor heterogeneity underlies differential cisplatin sensitivity in mouse models of small-cell lung cancer. Cell Rep 27: 3345–3358.e4. 10.1016/j.celrep.2019.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandetti E, Veneziani I, Melaiu O, Pezzolo A, Castellano A, Boldrini R, Ferretti E, Fruci D, Moretta L, Pistoia V, et al. 2017. MYCN is an immunosuppressive oncogene dampening the expression of ligands for NK-cell activating receptors in human high-risk neuroblastoma. Oncoimmunology 6: e1316439 10.1080/2162402X.2017.1316439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch SE, Hanke ML, Kargl J, Metz HE, MacPherson D, Houghton AM. 2016. Lung cancer subtypes generate unique immune responses. J Immunol 197: 4493–4503. 10.4049/jimmunol.1600576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, et al. 2016. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352: 227–231. 10.1126/science.aac9935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. 2015. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160: 1246–1260. 10.1016/j.cell.2015.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. 2012. MYC on the path to cancer. Cell 149: 22–35. 10.1016/j.cell.2012.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M, Peacock CD, et al. 2009. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res 69: 3364–3373. 10.1158/0008-5472.CAN-08-4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin BJ, George J, Christensen CL, Mino-Kenudson M, Dries R, Sundaresan T, Phat S, Myers DT, Zhong J, Igo P, et al. 2018. Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov 8: 600–615. 10.1158/2159-8290.CD-17-0935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T. 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4: 1064–1072. 10.1038/nprot.2009.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2004. The ENCODE (encyclopedia of DNA elements) project. Science 306: 636–640. 10.1126/science.1105136 [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. 2015. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 21: 938–945. 10.1038/nm.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, Leenders F, Lu X, Fernández-Cuesta L, Bosco G, et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature 524: 47–53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ. 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318. 10.1038/ncb1224 [DOI] [PubMed] [Google Scholar]
- Hansen K, Farkas T, Lukas J, Holm K, Rönnstrand L, Bartek J. 2001. Phosphorylation-dependent and independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J 20: 422–432. 10.1093/emboj/20.3.422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. 2005. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121: 465–477. 10.1016/j.cell.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, Polanski R, Burt DJ, Simpson KL, Morris K, et al. 2014. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 20: 897–903. 10.1038/nm.3600 [DOI] [PubMed] [Google Scholar]
- Horn L, Mansfield AS, Szczęsna A, Havel L, Krzakowski M, Hochmair MJ, Huemer F, Losonczy G, Johnson ML, Nishio M, et al. 2018. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med 379: 2220–2229. 10.1056/NEJMoa1809064 [DOI] [PubMed] [Google Scholar]
- Huijbers IJ, Bin Ali R, Pritchard C, Cozijnsen M, Kwon MC, Proost N, Song J-Y, Vries H, Badhai J, Sutherland K, et al. 2014. Rapid target gene validation in complex cancer mouse models using re-derived embryonic stem cells. EMBO Mol Med 6: 212–225. 10.1002/emmm.201303297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulton CH, Costa EA, Shah NS, Quintanal-Villalonga A, Heller G, de Stanchina E, Rudin CM, Poirier JT. 2020. Direct genome editing of patient-derived xenografts using CRISPR–Cas9 enables rapid in vivo functional genomics. Nat Cancer 1: 359–369. 10.1038/s43018-020-0040-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BE, Russell E, Simmons AM, Phelps R, Steinberg RM, Ihde DC, Gazdar AF. 1996. MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer. J Cell Biochem 63: 210–217. 10.1002/jcb.240630516 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargl J, Busch SE, Yang GH, Kim KH, Hanke ML, Metz HE, Hubbard JJ, Lee SM, Madtes DK, McIntosh MW, et al. 2017. Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nat Commun 8 10.1038/ncomms14381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato F, Fiorentino FP, Alibés A, Perucho M, Sánchez-Céspedes M, Kohno T, Yokota J. 2016. MYCL is a target of a BET bromodomain inhibitor, JQ1, on growth suppression efficacy in small cell lung cancer. Oncotarget 7: 77378–77388. 10.18632/oncotarget.12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Wu N, Kim YC, Cheng PF, Basom R, Kim D, Dunn CT, Lee AY, Kim K, Lee CS, et al. 2016. Genetic requirement for Mycl and efficacy of RNA pol I inhibition in mouse models of small cell lung cancer. Genes Dev 30: 1289–1299. 10.1101/gad.279307.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma'ayan A. 2010. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26: 2438–2444. 10.1093/bioinformatics/btq466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallo A, Frese KK, Morrow CJ, Sloane R, Gulati S, Schenk MW, Trapani F, Simms N, Galvin M, Brown S, et al. 2018. The combination of the PARP inhibitor Olaparib and the WEE1 inhibitor AZD1775 as a new therapeutic option for small cell lung cancer. Clin Cancer Res 24: 5153–5164. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg S. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger PR, Hu DX, Biannic B, Bui M, Han X, Karbarz E, Maung J, Okano A, Osipov M, Shibuya GM, et al. 2020. Discovery of potent, selective, and orally bioavailable inhibitors of USP7 with in vivo anti-tumor activity. J Med Chem 63: 5398–5420. 10.1021/acs.jmedchem.0c00245. [DOI] [PubMed] [Google Scholar]
- Lenhart R, Kirov S, Desilva H, Cao J, Lei M, Johnston K, Peterson R, Schweizer L, Purandare A, Ross-MacDonald P, et al. 2015. Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol Cancer Ther 14: 2167–2174. 10.1158/1535-7163.MCT-15-0037 [DOI] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15: 554 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 1: 417–425. 10.1016/j.cels.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizotte P, Ivanova EV, Awad MM, Jones RE, Keogh L, Liu H, Dries R, Almonte C, Herter-Sprie GS, Santos A, et al. 2016. Multiparametric profiling of non–small-cell lung cancers reveals distinct immunophenotypes. JCI Insight 1: e89014 10.1172/jci.insight.89014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. 2003. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 4: 181–189. 10.1016/S1535-6108(03)00220-4 [DOI] [PubMed] [Google Scholar]
- Mollaoglu G, Guthrie MR, Böhm S, Brägelmann J, Can I, Ballieu PM, Marx A, George J, Heinen C, Chalishazar MD, et al. 2017. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell 31: 270–285. 10.1016/j.ccell.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, Hochmair MJ, Özgüroğlu M, Ji JH, et al. 2019. Durvalumab plus platinum–etoposide versus platinum–etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomized, controlled, open-label, phase 3 trial. Lancet 394: 1929–1939. 10.1016/S0140-6736(19)32222-6 [DOI] [PubMed] [Google Scholar]
- Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. 2012. Integrative genomic analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44: 1104–1110. 10.1038/ng.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelish HE, Liau BB, Nitulescu II, Tangpeerachaikul A, Poss ZC, Da Silva DH, Caruso BT, Arefolov A, Fadeyi O, Christie AL, et al. 2015. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature 526: 273–276. 10.1038/nature14904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps RM, Johnson BE, Ihde CD, Gazdar AF, Carbone DP, McClintock PR, Linnoila RI, Matthews MJ, Bunn PA, Carney D, et al. 1996. NCI-Navy medical oncology branch cell line data base. J Cell Biochem Suppl 63: 32–91. 10.1002/jcb.240630505 [DOI] [PubMed] [Google Scholar]
- Poirier JT, George J, Owonikoko TK, Berns A, Brambilla E, Byers LA, Carbone D, Chen HJ, Christensen CL, Dive C, et al. 2020. New approaches to SCLC therapy: from the laboratory to the clinic. J Thorac Oncol 15: 520–540. 10.1016/j.jtho.2020.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Di Maio M, Chiodini P, Rudd RM, Okamoto H, Skarlos DV, Früh M, Qian W, Tamura T, Samantas E, et al. 2012. Carboplatin- or cisplatin-based chemotherapy in first-line treatment of small-cell lung cancer: the COCIS meta-analysis of individual patient data. J Clin Oncol 30: 1692–1698. 10.1200/JCO.2011.40.4905 [DOI] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. 2012. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44: 1111–1116. 10.1038/ng.2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, Heymach JV, Johnson JE, Lehman JM, MacPherson D, et al. 2019. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 19: 289–297. 10.1038/s41568-019-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer BE, Park KS, Yiu G, Conklin JF, Lin C, Burkhart DL, Karnezis AN, Sweet-Cordero A, Sage J. 2010. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 70: 3877–3883. 10.1158/0008-5472.CAN-09-4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen T, Tong P, Diao L, Li L, Fan Y, Hoff J, Heymach JV, Wang J, Byers LA. 2017. Targeting AXL and mTOR pathway overcomes primary and acquired resistance to WEE1 inhibition in small-cell lung cancer. Clin Cancer Res 23: 6239–6253. 10.1158/1078-0432.CCR-17-1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson KL, Stoney R, Frese KK, Simms N, Rowe W, Pearce SP, Humphrey S, Booth L, Morgan D, Dynowski M, et al. 2020. A biobank of small cell lung cancer CDX models elucidates inter- and intratumoral phenotypic heterogeneity. Nat Cancer 1: 437–451. 10.1038/s43018-020-0046-2 [DOI] [PubMed] [Google Scholar]
- Skene PJ, Henikoff S. 2017. An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. Elife 6: e21856 10.7554/eLife.21856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CS, Gay CM, Xi Y, Sivajothi S, Sivakamasundari V, Fujimoto J, Bolisetty M, Hartsfield PM, Balasubramaniyan V, Chalishazar MD, et al. 2020. Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer. Nat Cancer 1: 423–436. 10.1038/s43018-019-0020-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. 2011. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19: 754–764. 10.1016/j.ccr.2011.04.019 [DOI] [PubMed] [Google Scholar]
- Swartling FJ, Grimmer MR, Hackett CS, Northcott PA, Fan QW, Goldenberg DD, Lau J, Masic S, Nguyen K, Yakovenko S, et al. 2010. Pleiotropic role for MYCN in medulloblastoma. Genes Dev 24: 1059–1072. 10.1101/gad.1907510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavana O, Li D, Dai C, Lopez G, Banerjee D, Kon N, Chen C, Califano A, Yamashiro DJ, Sun H, et al. 2016. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat Med 22: 1180–1186. 10.1038/nm.4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AH, Devarakonda S, Skidmore ZL, Krysiak K, Ramu A, Trani L, Kunisaki J, Masood A, Waqar SN, Spies NC, et al. 2018. Recurrent WNT pathway alterations are frequent in relapsed small cell lung cancer. Nat Commun 9: 3787 10.1038/s41467-018-06162-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Hong B, Li X, Deng K, Li H, Yan Lui VW, Lin W. 2017. JQ1 synergizes with the Bcl-2 inhibitor ABT-263 against MYCN amplified small cell lung cancer. Oncotarget 8: 86312–86324. 10.18632/oncotarget.21146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Kuznetsov IB, Zhang S, Song YK, Asgharzadeh S, Sindiri S, Wen X, Patidar R, Najaraj S, Walton A, et al. 2018. Clinically relevant cytotoxic immune cell signatures and clonal expansion of T-cell receptors in high-risk MYCN-not-amplified human neuroblastoma. Clin Cancer Res 24: 5673–5684. 10.1158/1078-0432.CCR-18-0599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Jia D, Bates B, Basom R, Eberhart CG, MacPherson D. 2017. A mouse model of MYCN-driven retinoblastoma reveals MYCN-independent tumor reemergence. J Clin Invest 127: 888–898. 10.1172/JCI88508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, et al. 2011. A public genome-scale lentiviral expression library of human ORFs. Nat Methods 8: 659–661. 10.1038/nmeth.1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Wu X, Basu M, Dong C, Zheng P, Liu Y, Sandler AD. 2017. MYCN amplification is associated with repressed cellular immunity in neuroblastoma: an in silico immunological analysis of TARGET database. Front Immunol 8: 1473 10.3389/fimmu.2017.01473 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.