In this study, Nitschke et al. investigated the mechanisms that regulate Ataxin-1 (ATXN1) expression, and identify miR760 as a negative regulator that binds to a conserved site in ATXN1's UTR to induce RNA degradation and translational inhibition. Their findings provide new insights into the regulation of ATXN1 levels, present additional evidence for miRNA-mediated gene regulation via 5' UTR binding, and raise the possibility that noncoding mutations in the ATXN1 locus may act as risk factors for yet to be discovered progressive ataxias.
Keywords: 5′ UTR, neurodegeneration, SCA1, miRNA
Abstract
Identifying modifiers of dosage-sensitive genes involved in neurodegenerative disorders is imperative to discover novel genetic risk factors and potential therapeutic entry points. In this study, we focus on Ataxin-1 (ATXN1), a dosage-sensitive gene involved in the neurodegenerative disease spinocerebellar ataxia type 1 (SCA1). While the precise maintenance of ATXN1 levels is essential to prevent disease, the mechanisms that regulate ATXN1 expression remain largely unknown. We demonstrate that ATXN1’s unusually long 5′ untranslated region (5′ UTR) negatively regulates its expression via posttranscriptional mechanisms. Based on recent reports that microRNAs (miRNAs) can interact with both 3′ and 5′ UTRs to regulate their target genes, we identify miR760 as a negative regulator that binds to a conserved site in ATXN1’s 5′ UTR to induce RNA degradation and translational inhibition. We found that delivery of Adeno-associated virus (AAV)-expressing miR760 in the cerebellum reduces ATXN1 levels in vivo and mitigates motor coordination deficits in a mouse model of SCA1. These findings provide new insights into the regulation of ATXN1 levels, present additional evidence for miRNA-mediated gene regulation via 5′ UTR binding, and raise the possibility that noncoding mutations in the ATXN1 locus may act as risk factors for yet to be discovered progressive ataxias.
Alterations in protein folding and levels underlie the pathogenesis of several neurodegenerative diseases including Alzheimer's and Parkinson's disease and polyglutamine expansion disorders. Despite the heterogeneity of these diseases, in each case the misfolded proteins accumulate over time, eventually causing toxicity in the affected neurons (Katsnelson et al. 2016). Interestingly, neurons are also sensitive to elevated levels of wild-type (WT) protein. Duplications of the amyloid precursor protein (APP), as observed in individuals with Trisomy 21 or APP locus duplications, cause autosomal dominant early-onset Alzheimer's disease (Rumble et al. 1989; Rovelet-Lecrux et al. 2006; Sleegers et al. 2006), and both duplications and triplications of α-synuclein are associated with familial Parkinson's disease (Singleton et al. 2003; Chartier-Harlin et al. 2004). While it may be expected that such severe changes cause disease, even subtle increases in protein levels can be pathogenic. As such, slight increases in APP and α-synuclein caused by mutations in regulatory promoter and enhancer elements are associated with Alzheimer's and Parkinson's disease, respectively (Theuns et al. 2006; Soldner et al. 2016).
An exemplar dosage-sensitive gene is Ataxin-1 (ATXN1), the causative gene involved in the neurodegenerative disease Spinocerebellar ataxia type 1 (SCA1) (Banfi et al. 1994; Zoghbi and Orr 1995). In SCA1, a CAG repeat expansion encoding the polyglutamine (polyQ) tract in ATXN1 stabilizes the protein, resulting in a toxic gain-of-function mechanism in the cerebellum (Orr et al. 1993; Cummings et al. 1999; Fryer et al. 2011; Rousseaux et al. 2018a). Various strategies to lower ATXN1 levels were successful in reducing ATXN1's accumulation and mitigating cerebellar SCA1 pathogenesis (Park et al. 2013; Keiser et al. 2014; Friedrich et al. 2018). Furthermore, it was shown that slight increases in the levels of WT ATXN1, independent of the polyQ expansion, could also cause ataxia and motor coordination deficits in mice, two hallmarks of SCA1 (Gennarino et al. 2015). Conversely, decreasing ATXN1 levels in the hippocampus increases the levels of BACE1, also implicating ATXN1 levels as a risk factor for Alzheimer's disease (Suh et al. 2019). These studies highlight the importance of maintaining ATXN1 protein levels within an optimal range as perturbances in its levels may, over time, increase susceptibility to disease.
Given the importance of ATXN1 levels in disease, the regulation of ATXN1 expression has been of particular interest in hopes of identifying therapeutic strategies as well as disease risk factors. Previous studies found ATXN1’s 3′ untranslated region (3′ UTR) to be critical for ATXN1’s posttranscriptional regulation. MicroRNA 101 (miR101), as well as the RNA-binding protein PUMILIO1 (PUM1), interact with ATXN1’s 3′ UTR to negatively regulate ATXN1 expression (Lee et al. 2008; Gennarino et al. 2015). Subsequently, loss-of-function mutations in PUM1 were shown to cause ataxia in patients with previously unknown disease etiology, presumably, in part, by increasing WT ATXN1 (Gennarino et al. 2018).
While ATXN1’s regulation via its 3′ UTR has been extensively studied, not much is known about its regulation via the 5′ untranslated region (5′ UTR). 5′ UTRs are mainly known for their importance in translational regulation by dampening translation via upstream open reading frames and secondary structures (Mignone et al. 2002). Indeed, a recent study showed that ATXN1's 5′ UTR is involved in its translational control (Manek et al. 2019). Studies have also shown that 5′ UTRs can provide a platform for miRNAs to bind and regulate their target gene expression via a variety of other posttranscriptional mechanisms (Ørom et al. 2008; Moretti et al. 2010; Dewing et al. 2012; Zhou and Rigoutsos 2014). Importantly, genes that require fine regulation, such as growth factors, transcription factors, and proto-oncogenes have been shown to contain longer 5′ UTRs, providing more opportunity for regulation (Kozak 1987; Mignone et al. 2002).
As ATXN1’s 5′ UTR is exceptionally long with its exons spanning a genomic region of >433 kb (Banfi et al. 1994), we sought to investigate its role in regulating ATXN1 expression. We show that the 5′ UTR negatively regulates ATXN1 expression via posttranscriptional mechanisms and identify miR760 as a miRNA that interacts with a conserved binding site in ATXN1’s 5′ UTR to negatively regulate its expression through Argonaute 2 (AGO2)-mediated RNA degradation and translational inhibition. We further demonstrate that increasing miR760 expression in the cerebellum of a SCA1 mouse model reduces ATXN1 protein levels and ameliorates motor learning and coordination deficits. These findings highlight the importance of noncoding regions in regulating ATXN1 expression and raise the possibility that mutations in regulatory elements of ATXN1 may act as risk factors for non-CAG mediated ataxias.
Results
ATXN1 contains a long 5′ UTR that negatively regulates its expression
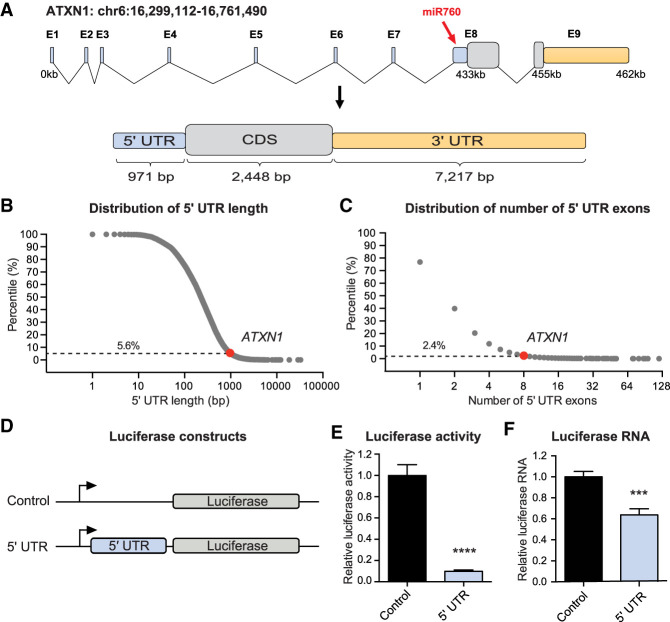
Human ATXN1 spans a genomic region of 462 kb on the short arm of chromosome 6 (Banfi et al. 1994). The vast majority of this region is occupied by ATXN1’s 5′ UTR, which consists of eight exons spanning a genomic region of 433 kb (Fig. 1A). Even after splicing, the 5′ UTR remains remarkably long, consisting of 971 bp (Fig. 1A). Human coding genes, on average, have a 5′ UTR length of 351 bp and contain two 5′ UTR exons (Fig. 1B,C). With 971 bp, ATXN1’s 5′ UTR ranks in the top 5.6th percentile (Fig. 1B), and its number of 5′ UTR exons (8 exons) ranks in the top 2.4th percentile (Fig. 1C) in comparison with all 5′ UTRs of human coding genes.
Figure 1.
ATXN1’s 5′ UTR negatively regulates ATXN1 expression. (A) Schematic overview of ATXN1’s genomic region and its spliced transcript (red arrow points to the miR760's binding site in ATXN1). (B,C) Scatter plots showing the distribution of 5′ UTR length (B) and exon number (C) of all known coding genes in the human genome. Dashed horizontal lines indicate the top percentile among which ATXN1 is ranked. (D) Schematic overview of Renilla luciferase constructs: control (pRL-TK) and 5′ UTR (pRL-TK_5′ UTR). (E,F) Relative Renilla luciferase activity (E) and Renilla luciferase RNA levels (F) following transient transfection of control and 5′ UTR constructs into HEK293T cells. Renilla luciferase activity and RNA were normalized to firefly luciferase activity and RNA, respectively. For each assay, a minimum of eight replicates were performed. Simple comparisons used Student's t-test. (***) P < 0.001; (****) P < 0.0001. All data are represented as means ± SEM.
Given this unusual length and the known importance of 5′ UTRs in regulating gene expression (Mignone et al. 2002), we cloned the spliced 5′ UTR of ATXN1 upstream of a luciferase reporter construct and transfected the construct into HEK293T cells to investigate its impact on gene expression (Fig. 1D). Compared with the control, which expresses luciferase alone, the 5′ UTR drastically reduced luciferase activity, indicating a negative regulatory function (Fig. 1E). While a recent study found that ATXN1’s 5′ UTR regulates its translation via out-of-frame upstream AUGs (Manek et al. 2019), we were in parallel interested in studying the effect the 5′ UTR may have on mRNA levels. To investigate whether the 5′ UTR has an effect on mRNA stability, we measured luciferase RNA levels and found that compared with the control, ATXN1’s 5′ UTR significantly decreased luciferase RNA levels (Fig. 1F), suggesting that in addition to translation efficiency, the 5′ UTR may regulate the stability of ATXN1’s mRNA.
miR760 negatively regulates ATXN1's expression
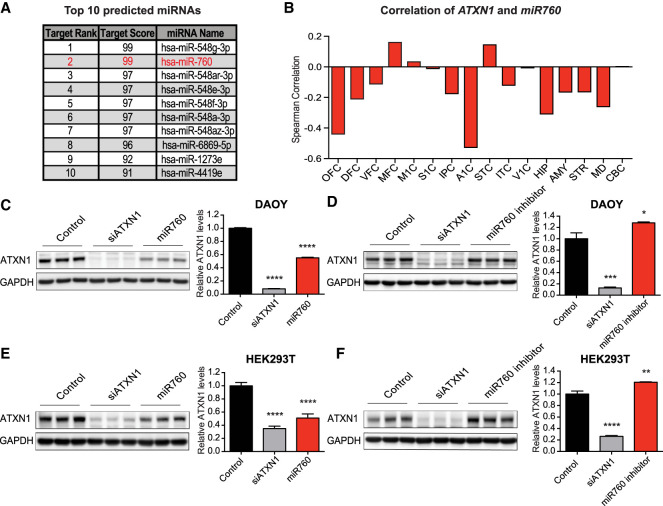
miRNAs are small noncoding RNAs that regulate gene expression typically via binding to the 3′ UTR of their target genes (Bartel 2004). However, recent studies provide evidence that miRNAs can also bind to the 5′ UTR to regulate target gene expression (Lytle et al. 2007; Ørom et al. 2008; Dewing et al. 2012; Zhou and Rigoutsos 2014). To identify miRNAs that may bind to ATXN1’s 5′ UTR and regulate ATNX1 expression, we used miRDB, an online miRNA target prediction database (Wong and Wang 2015; Liu and Wang 2019). The analysis identified miRNA 548g-3p (miR548g-3p) and miRNA 760 (miR760) as the strongest predicted candidates to interact with ATXN1’s 5′ UTR (Fig. 2A). We further used the COMETA approach (Gennarino et al. 2012) and found that while ATXN1 is highly coexpressed with the targets of miR760, it is not coexpressed with the targets of miR548g-3p or other putative miRNAs (Supplemental Fig. S1), suggesting a regulatory function of miR760 on ATXN1. Additionally, we found that miR760 is expressed in multiple human brain regions (Supplemental Fig. S2A,B) and that the levels of miR760 and ATXN1 were negatively correlated across the majority of these brain regions (Fig. 2B). In comparison, ATXN1 and other putative miRNAs did not show the same extent of negative correlation (Supplemental Fig. S2C–G). Together, these findings support the idea that ATXN1 is a potential direct target of miR760 in the human brain (Fig. 2B).
Figure 2.
miR760 negatively regulates ATXN1 levels. (A) Top 10 miRNAs predicted by miRDB (http://mirdb.org) to bind to ATXN1’s 5′ UTR. miR760, which also emerged from the COMETA analysis marked in red. (B) Spearman's correlation of ATXN1 and miR760 expression in 16 human brain regions: orbital prefrontal cortex (OFC), dorsolateral prefrontal cortex (DFC), ventrolateral prefrontal cortex (VFC), medial prefrontal cortex (MFC), primary motor cortex (M1C), primary somatosensory cortex (S1C), posterior inferior parietal cortex (IPC), primary auditory cortex (A1C), superior temporal cortex (STC), inferior temporal cortex (ITC), primary visual cortex (V1C), hippocampus (HIP), amygdala (AMY), striatum (STR), mediodorsal nucleus of the thalamus (MD), and cerebellar cortex (CBC). The CBC bar represents a value of 0.002. (C–F) Representative Western blots (left) and quantifications (right) of ATXN1 levels in DAOY (C,D) and HEK293T (E,F) cells treated with control, siATXN1, or miR760 mimics (C,E) or control, siATXN1, or miR760 inhibitors (D,F). For each quantification, ATXN1 levels were normalized to GAPDH. For each assay, a minimum of three replicates were performed. Multigroup comparisons used one-way ANOVAs. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001. All data are represented as means ± SEM.
In an unbiased manner, we tested the effect of the top 10 predicted miRNAs from miRDB (Fig. 2A) on ATXN1 protein levels. We performed gain-of-function studies in cell culture by transfecting commercially available miRNA mimics, which imitate the mature miRNA of interest, into the DAOY human medulloblastoma cell line. miR760, most miR548 family members, and miR1273e were able to reduce ATXN1's protein levels (Fig. 2C; Supplemental Fig. S3). However, when using miRNA inhibitors, which sequester the endogenous miRNA from its target, only inhibitors against miR760 were able to increase ATXN1 levels (Fig. 2D; Supplemental Fig. S4). To validate our findings, we repeated the miR760 mimics and inhibitor experiments in HEK293T cells, a human embryonic kidney cell line. We confirmed that miR760 mimics decrease ATXN1 levels while miR760 inhibitors increase ATXN1 levels (Fig. 2E,F). As miR760 is endogenously expressed in both cell lines (Supplemental Fig. S5), these data indicate that endogenous miR760 negatively regulates ATXN1 expression.
miR760 regulates ATXN1 by binding to a specific site in its 5′ UTR
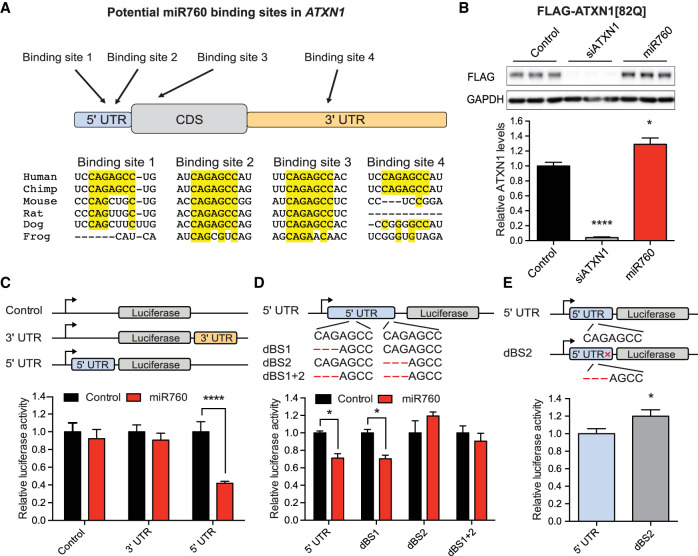
Next, we tested whether miR760 directly regulates ATXN1’s expression by binding to its 5′ UTR or whether its effect is mediated by binding to other regions in ATXN1. miRNAs bind to their target mRNA via complementary base pairing of the 7 nucleotide (nt) seed region located at position 2–8 in the 5′ end of the miRNA (Bartel 2009). ATXN1 contains four sites with complete complementarity to the seed region of miR760 (5′-GGCUCUG-3′). Two of the sites are located in the 5′ UTR, one in the coding region (CDS) and one in the 3′ UTR of ATXN1 (Fig. 3A). Importantly, of these four putative sites, only binding site 2 in the 5′ UTR and binding site 3 in the CDS are highly conserved across multiple species (Fig. 3A).
Figure 3.
miR760 regulates ATXN1 expression by binding to a conserved site in ATXN1’s 5′ UTR. (A) Schematic overview of ATXN1’s transcript with potential miR760-binding sites. Below is the species conservation of each potential binding site. (B) Representative Western blot (top) and quantification (bottom) of ATXN1 levels in DAOY cells stably expressing a transgenic Flag-ATXN1[82Q] construct missing the 5′ and 3′ UTR transfected with control or miR760 mimics. Flag-ATXN1 protein levels were normalized to GAPDH. (C, top) Schematic overview of the control (pRL-TK), ATXN1 5′ UTR (pRL-TK_5′ UTR) and ATXN1 3′ UTR (pRL-TK_3′ UTR) luciferase constructs. (Bottom) Relative Renilla luciferase activity following transfection of these constructs into HEK293T cells in the presence of control or miR760 mimics. (D, top) Schematic overview of ATXN1 5′ UTR constructs with 3-nt deletions in putative binding site 1 (dBS1), putative binding site 2 (dBS2), or in combination (dBS1 + 2). Relative Renilla luciferase activity following transfection of these constructs into HEK293T cells in the presence of control or miR760 mimics. (E) Basal Renilla luciferase activity of ATXN1 5′ UTR and dBS2 construct (without expression of miRNA constructs). In each case, Renilla luciferase activity was normalized to firefly luciferase activity. For each assay, a minimum of three replicates were performed. Simple comparisons used Student's t-test, whereas multigroup comparisons used one- or two-way ANOVAs. (*) P < 0.05; (****) P < 0.0001. All data are represented as means ± SEM.
To investigate which of the four sites miR760 preferentially binds, we first generated a transgenic DAOY cell line that only expresses ATXN1’s CDS fused to a Flag tag (Flag-ATXN1[82Q]). Transfection of miR760 mimics slightly, but significantly increased Flag-ATXN1 protein levels (Fig. 3B), suggesting that the negative regulatory effect of miR760 on ATXN1 requires either the 5′ UTR or the 3′ UTR. To determine whether miR760 binds to the 5′ UTR or the 3′ UTR, we then generated luciferase reporter constructs expressing luciferase alone, luciferase fused to ATXN1’s 5′ UTR, or luciferase fused to the region of ATXN1’s 3′ UTR containing miR760's predicted binding site and transfected them into HEK293T cells (Fig. 3C). Compared with luciferase alone, miR760 decreased luciferase levels only when luciferase was fused to ATXN1’s 5′ UTR, supporting our prediction that miR760's negative regulatory effect occurs via binding to the 5′ UTR (Fig. 3C).
Last, to determine which of the two predicted 5′ UTR binding sites miR760 is interacting with, we deleted three nucleotides from each site, either alone or in combination (Fig. 3D). Disruption of binding site 2 or binding sites 1 and 2 in combination abolished miR760's effect on luciferase activity, while deletion of binding site 1 alone had no effect (Fig. 3D). To ensure that this effect was not due to changes in mRNA structure, we used the Web Server mFold to predict the structure of ATXN1’s 5′ UTR with and without the 3-nt deletion in binding site 2 (Zuker 2003). We found that the overall structure remained unchanged upon binding site 2 deletion (Supplemental Fig. S6), indicating that the observed effect was due to abolishing miR760's binding on ATXN1's 5′ UTR and not due to alterations in the secondary structure of the 5′ UTR. Interestingly, in the absence of exogenously added miR760, the basal luciferase activity of the binding site 2 mutant was also increased (Fig. 3E), suggesting that endogenous miR760 is sufficient to regulate the 5′ UTR luciferase construct through this site.
These results show that miR760 negatively regulates ATXN1 expression by binding to the conserved binding site 2 in ATXN1’s 5′ UTR (chr6:16,328,355 to chr6: 16,328,361).
miR760 regulates ATXN1 expression via AGO2-mediated RNA degradation and translational inhibition
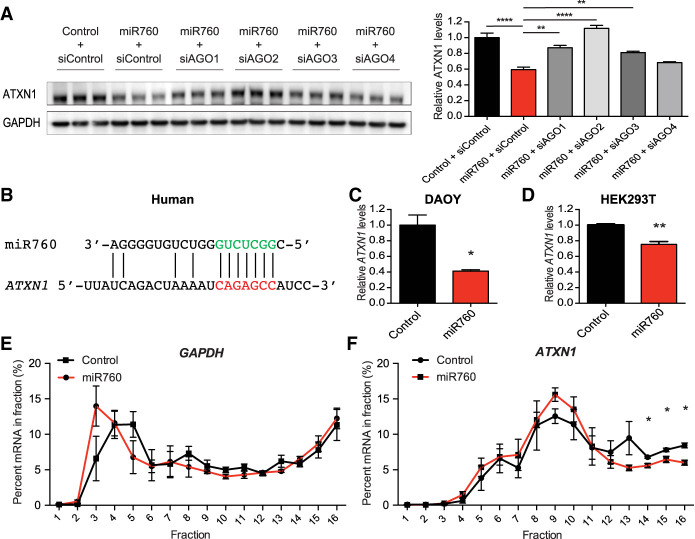
Most miRNAs are transcribed as primary transcripts (pri-miRNAs), which contain one or more hairpins. The pri-miRNAs then get processed by the nuclear microprocessor complex and the RNase III Drosha to generate a single hairpin, the precursor miRNA (pre-miRNA). After their export to the cytoplasm via exportin 5, Dicer cleaves off the loop of the hairpin, leaving a double-stranded RNA. One strand is then often degraded while the other, the mature miRNA, is loaded into the RNA-induced silencing complex (RISC), where it interacts with a member of the Argonaute (AGO) family (Meister 2013). In humans, the Argonaute family consists of eight members. The first four family members, Argonaute1-4 (AGO1-4), belong to the AGO subfamily and are ubiquitously expressed (Sasaki et al. 2003). The remaining four family members, the Piwi subfamily, are exclusively expressed in the germline (Sasaki et al. 2003; Siomi et al. 2011). While all four AGO subfamily members were shown to be involved in miRNA mediated silencing by inducing enhanced RNA degradation and/or translational inhibition, AGO2 is the only family member with catalytic activity, allowing it to also cleave its target mRNA (Meister 2013).
As previous studies suggested that certain miRNAs might preferentially bind to specific AGO proteins (Burroughs et al. 2011), we next investigated which of the four AGO proteins is required for the effect of miR760 on ATXN1 levels. To do so, we performed our miR760 mimic experiments in the presence of siRNAs against each AGO. Knockdown of AGO2 completely disrupted the effect of miR760 on ATXN1 protein levels (Fig. 4A). While AGO2 appears to be the primary effector of the 5′ UTR-mediated regulation of ATXN1 by miR760, knockdown of AGO1 and AGO3 also reduced miR760's effect on ATXN1 protein levels, indicating that AGO1 and AGO3 are also capable of mediating miR760's effect on ATXN1, albeit to a lesser extent (Fig. 4A).
Figure 4.
miR760 regulates ATXN1 expression via AGO2-mediated RNA degradation and translational inhibition. (A) Representative Western blot (left) and quantification (right) of ATXN1 levels in HEK293T cells treated with control or miR760 mimics and control siRNA or siRNAs targeting AGO1–4. ATXN1 levels were normalized to GAPDH. Statistical comparison of all groups with miR760 + siControl are shown. (B) Schematic showing the complementarity of miR760's seed region to its binding site in the 5′ UTR of human ATXN1. (C,D) Relative ATXN1 mRNA levels measured by qPCR in DAOY (C) and HEK293T (D) cells treated with control or miR760 mimics. ATXN1’s mRNA levels were normalized to GAPDH mRNA levels. (E,F) GAPDH (E) and ATXN1 (F) mRNA levels in single fractions following sucrose gradient fractionation. For each assay, a minimum of three replicates were performed. Simple comparisons used Student's t-test, whereas multigroup comparisons used one- or two-way ANOVAs. (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001. All data are represented as means ± SEM.
RISC-mediated gene silencing can occur via multiple mechanisms: (1) direct cleavage of mRNA via AGO2, (2) enhanced mRNA degradation, and (3) translational silencing (Valencia-Sanchez et al. 2006). While AGO2 is the primary effector of miR760's effect on ATXN1 levels and the only known AGO to induce cleavage of its target mRNA, AGO-2 mediated mRNA cleavage remains unlikely as it requires extensive complementarity between miRNA and its target mRNA (Bartel 2009). As miR760 is only complementary at the seed region and a few additional bases on the 3′ end of the identified binding site in ATXN1’s 5′ UTR (Fig. 4B), we focused our efforts on the latter two possibilities.
To test for enhanced RNA degradation, we extracted RNA from cells treated with miR760 mimics and measured ATXN1’s mRNA levels by qPCR. In both DAOY and HEK293T cells, ATXN1’s mRNA levels were decreased, indicating an effect of miR760 on the stability of ATXN1’s mRNA (Fig. 4C,D). To investigate whether the mechanism also involves translational inhibition, we performed polysome profiling and assessed ATXN1’s translation efficiency in the presence of miR760. Cytoplasmic extracts of HEK293T cells transfected with either control or miR760 mimics were fractionated through a sucrose gradient. The lighter fractions consisted of small (40S) and large (60S) ribosomal subunits and monomers (80S), while higher fractions contained progressively larger polysomes (Supplemental Fig. S7). Global translation rates in control and miR760-transfected cells were unchanged (Supplemental Fig. S7). The distribution of ATXN1’s mRNA and the mRNA of housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were measured by qPCR analysis in all fractions. While the distribution of GAPDH mRNA was unaltered throughout the fractions in control and miR760 treated cells, significantly less ATXN1 mRNA sedimented in heavier polysome fractions 14–16 in the miR760 transfected cells, suggesting that transfection of miR760 alters the translation efficiency of ATXN1 mRNA (Fig. 4E,F).
Together, these data suggest that miR760 regulates ATXN1 expression via AGO2-mediated RNA degradation and decreased translation efficiency.
miR760 reduces ATXN1 protein levels in the mouse cerebellum
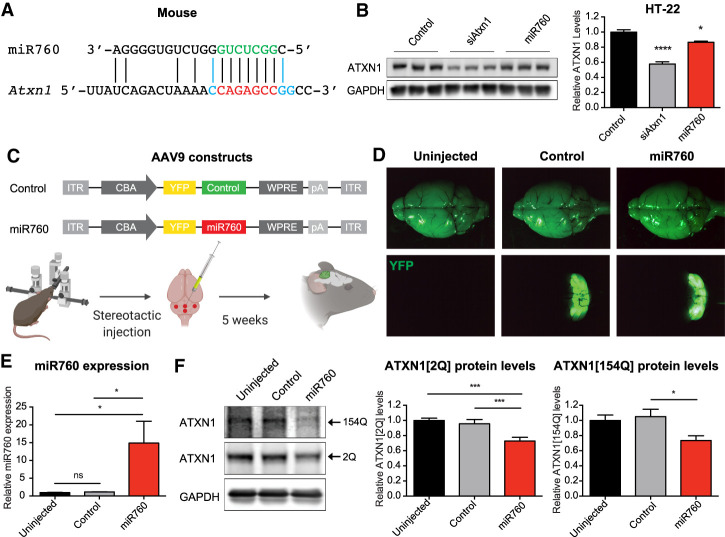
As miR760's sequence and its binding site in ATXN1's/Atxn1’s 5′ UTR are conserved between human and mouse (Fig. 5A), we wanted to investigate whether miR760's negative regulatory effect on Atxn1 is conserved in mice as well. Interestingly, the 5′ UTR of mouse Atxn1 contains several basepair substitutions, which increase miR760's complementarity to this region (Fig. 5A). To confirm our human cell data in a murine system, we transfected mouse hippocampal HT-22 cells with miR760 mimics. Consistent with our findings in human cells, we detected a reduction in ATXN1 protein levels in the presence of miR760 (Fig. 5B).
Figure 5.
miR760 regulates ATXN1 levels in the mouse brain. (A) Schematic showing miR760's complementarity to its seed region in mouse Atxn1. Nucleotides shown in blue are substitutions that differ between mouse Atxn1 and human ATXN1. (B) Representative Western blot (left) and quantification (right) of mouse hippocampal HT-22 cells transfected with control, siAtxn1, or miR760 mimics. ATXN1 levels were normalized to GAPDH. (C) Schematic overview of AAV9 constructs used for injections in the adult mouse cerebellum. (ITR) Inverted terminal repeat; (CBA) chicken β-actin promoter; (YFP) yellow fluorescent protein; (WPRE) woodchuck hepatitis virus posttranscriptional regulatory element; (pA) polyadenylation site. (D) Bright-field (top) and YFP fluorescent microscopy image (bottom) of brains from uninjected, AAV9 control, and AAV9 miR760-injected SCA1 mice. (E) qPCR of miR760 in cerebella in uninjected, AAV9 control-injected, and AAV9 miR760-injected SCA1 animals. miR760 expression was normalized to U6. (F) Representative Western blot (left) and quantification (right) of ATXN1[2Q] and [154Q] levels in cerebella of uninjected, AAV9 control-injected, and AAV9 miR760-injected SCA1 mice. ATXN1 levels were normalized to GAPDH. For each assay, a minimum of three replicates were performed. Simple comparisons used Student's t-test, whereas multigroup comparisons used one- or two-way ANOVAs. (*) P < 0.05; (***) P < 0.001. All data are represented as means ± SEM. Illustration was created with BioRender.com.
Given that transfection of miR760 mimics was able to decrease WT ATXN1 levels in a mouse cell line, we next determined whether increased miR760 expression could also reduce ATXN1 levels in the cerebellum of SCA1 (Atxn1154Q/2Q) mice, which express a WT (2Q) and a polyQ-expanded (154Q) Atxn1 allele (Watase et al. 2002). To this end, we cloned AAV9 constructs expressing either control or miR760 and yellow fluorescent protein (YFP) under the control of the chicken beta-actin (CBA) promoter (Fig. 5C). We performed stereotactic cerebellar injections on 5-wk-old SCA1 mice. Five weeks following injection, the virus was expressed in the cerebellum as indicated by YFP localization (Fig. 5D). Upon dissection of the cerebellum, we detected a 15-fold increase in miR760's expression (Fig. 5E). Despite interinjection variability in miR760 expression, we observed a decrease in polyQ-expanded and unexpanded ATXN1 levels in the micro-dissected YFP-positive cerebellar tissue (Fig. 5F). These data provide evidence that the negative regulatory effect of miR760 on ATXN1 levels observed in cells holds true in vivo and raises the question of whether overexpression of miR760 in the mouse cerebellum could mitigate pathogenesis in SCA1 mice.
Overexpression of miR760 ameliorates cerebellar SCA1 phenotypes
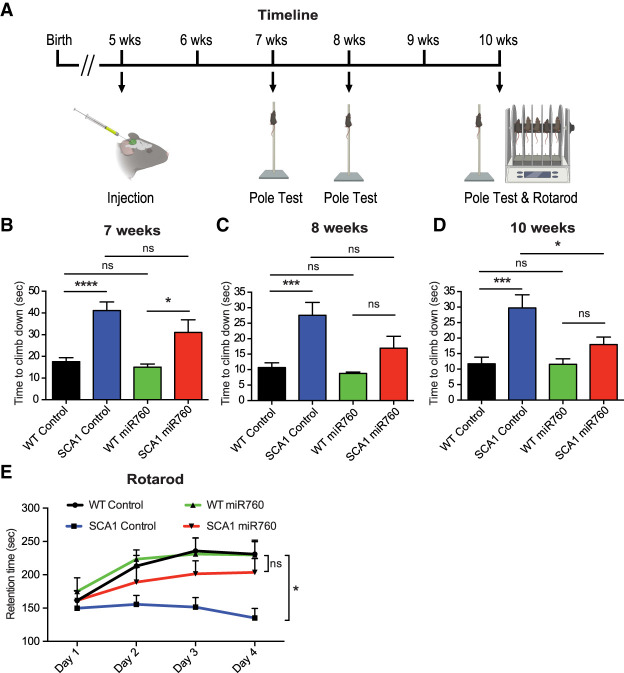
SCA1 mice recapitulate many pathological and behavioral characteristics of SCA1 including motor incoordination beginning at 5 wk of age (Watase et al. 2002). Moreover, reduction of ATXN1 levels has previously been shown to mitigate SCA1 disease progression in multiple SCA1 mouse models (Park et al. 2013; Keiser et al. 2014; Friedrich et al. 2018). To investigate whether lowering ATXN1 levels by overexpressing miR760 can similarly alleviate SCA1 phenotypes, we performed behavioral assays for motor coordination and motor learning upon injection of control and miR760 AAV9 in 5-wk-old WT and SCA1 animals (Fig. 6A).
Figure 6.
Injection of AAV9 expressing miR760 improves cerebellar phenotypes in SCA1 mice. (A) Time line of injection and behavioral experiments. (B–D) Time to climb down during the pole test for WT and SCA1 animals injected with AAV9 control (control) or AAV9 miR760 (miR760) at 7 wk (B), 8 wk (C), and 10 wk of age (D). (E) Retention time across four training days in the rotarod assay for WT and SCA1 animals injected with control or miR760. Each group contained 11–13 animals. Multigroup comparisons used one- and two-way ANOVAs. (*) P < 0.05; (***) P < 0.001; (****) P < 0.0001; (ns) P > 0.05. All data are represented as means ± SEM. Illustration was created with BioRender.com.
To track the progression of motor phenotypes in injected WT and SCA1 mice, we performed the pole test, an assay used to measure motor coordination, at 7, 8, and 10 wk of age. SCA1 animals that received either control or miR760-harboring AAV9s performed worse than WT control animals 2 wk following injection when YFP and miR760 are just starting to be expressed (Fig. 6B). At 3 wk after injection, SCA1 mice injected with the miR760-containing AAV9 showed a slight improvement (Fig. 6C). Five weeks after injection, SCA1 mice injected with the miR760-containing AAV9 showed a significant improvement in motor coordination compared with control-injected SCA1 mice (Fig. 6D).
Five weeks following injection, at 10 wk of age, the mice were subjected to a 4-d rotarod protocol to assess motor coordination and motor learning. As expected, the performance of WT control-injected mice improved over a 4-d period, whereas the performance of SCA1 control-injected mice failed to improve. Notably, miR760-injected SCA1 animals exhibited improvement over time and performed similarly to the control- and miR760-injected WT controls (Fig. 6E).
As weight could impact the performance on the rotarod and pole test, we compared the weight of the control and miR760-injected WT and SCA1 animals. We found that the weight was similar in control and miR760-injected SCA1 animals (Supplemental Fig. S8), indicating that the improvement on the pole test and rotarod was indeed due to rescue of cerebellar phenotypes.
Taken together, these data present evidence that overexpression of miR760 in the cerebellum of adult SCA1 mice can rescue their motor coordination and motor learning deficits.
Discussion
In this study, we identified a new mechanism regulating the levels of ATXN1, a dosage-sensitive gene that plays a critical role in the neurodegenerative disorder SCA1 (Banfi et al. 1994). As a dosage-sensitive gene, ATXN1 levels need to be under tight regulation. A slight increase in ATXN1 levels causes ataxia and motor coordination deficits (Gennarino et al. 2015), whereas a decrease of 50% or more of ATXN1 leads to an increased risk for developing Alzheimer's disease (Suh et al. 2019). Previous studies have focused on the importance of ATXN1’s 3′ UTR in regulating its mRNA stability through the recruitment of miRNAs and RNA-binding proteins (Lee et al. 2008; Gennarino et al. 2015). As such, miR101 was found to negatively regulate ATXN1 levels in cell culture, while the RNA-binding protein PUM1 was shown to have a negative regulatory effect on ATXN1 in the cerebellum and cerebrum (Lee et al. 2008; Gennarino et al. 2015). Here, we chose to investigate the regulatory role of ATXN1’s 5′ UTR. Previous studies have suggested that longer 5′ UTRs provide a greater opportunity for regulation, especially for genes requiring finely tuned expression such as growth factors, transcription factors, or proto-oncogenes (Kozak 1987; Mignone et al. 2002). Therefore, we were intrigued to find that ATXN1 contains one of the longest 5′ UTRs found in humans. We report that ATXN1’s long 5′ UTR negatively regulates its expression by decreasing ATXN1’s mRNA stability, a mechanism independent of the recently reported translational inhibition via upstream AUGs and alternative splicing in cerebellar and cortical tissues (Manek et al. 2019).
Given studies that have demonstrated the ability of miRNAs to interact with 5′ UTRs to down-regulate the expression of their target genes (Lytle et al. 2007; Dewing et al. 2012; Zhou and Rigoutsos 2014) and ATXN1’s unusually long 5′ UTR, we focused our work on the regulation of ATXN1 via miRNAs as a potential mechanism of action. We identified miR760 as a miRNA that interacts with an evolutionarily conserved site in ATXN1’s 5′ UTR and leads to increased ATXN1 mRNA degradation and reduced translation rates. Overexpression of miR760 in the cerebellum of a SCA1 knock-in mouse model reduced ATXN1 levels and improved the cerebellar SCA1 motor coordination phenotypes, indicating that miR760's effect on ATXN1 is biologically relevant and disease-mitigating. As ATXN1 is negatively correlated with miR760 across multiple brain regions, it is likely that miR760 has a negative regulatory effect on ATXN1 in other brain regions as well. Future studies need to investigate whether the effects of miR760 on ATXN1 and the regulatory mechanisms are similar across multiple tissues and cell types.
It is worth noting that broad overexpression of endogenous miRNAs might not be advisable as they can regulate a multitude of target genes and cellular processes. As such, miR760 has been shown to be involved in the regulation of cell proliferation, migration, differentiation, and apoptosis (Liao et al. 2016; Yang et al. 2018; Tang et al. 2019). Alterations in miR760 expression, in both directions, up and down, have been associated with multiple forms of cancer, including ovarian, liver, and colorectal cancer (Liao et al. 2016; Li et al. 2017; Wang et al. 2019), making its systemic overexpression as long-term treatment for SCA1 patients challenging. However, having shown the benefit of local overexpression of miR760 gives hope that one can avoid systemic effects and warrants future studies that optimize its use to rescue cerebellar symptoms.
Importantly, we also found that inhibiting miR760 or mutating its binding site in the 5′ UTR of ATXN1 significantly increased ATXN1 protein levels. This finding, together with the fact that miR760 decreases ATXN1 by ∼25% in the cerebellum is important, as even a 20% increase in ATXN1 levels can lead to ataxia in both mice and humans (Gennarino et al. 2015, 2018). Currently, patients with ataxia are classified solely based on the presence of the polyQ expansion in either ATXN1 or other polyQ-encoding ATXNs, or the presence of known protein-coding mutations in other ataxia-associated genes, leaving a large number of patients unclassified (Alonso et al. 2007). We propose that some of these unclassified ataxia patients might contain mutations in regulatory elements, leading to changes in expression levels of ataxia-associated genes. In the case of ATXN1, a mutation in miR760's binding site within the 5′ UTR or mutations in the 3′ UTR binding sites of miRNAs and RNA-binding protein may increase ATXN1 expression, causing ataxia symptoms. The findings from this study add to the growing list of studies highlighting the critical need to identify and characterize regulatory regions of various ataxia-causing genes and to perform whole-genome sequencing in ataxia patients with unknown genetic etiologies in order to investigate potential disease-causing mutations in noncoding regulatory regions.
Materials and methods
Bioinformatic analysis
To perform the 5′ UTR analysis, the 5′ UTR length and exon number data for UCSC RefSeq (refGene) coding transcripts were downloaded from the UCSC Genome Browser database (http://genome.ucsc.edu). The 5′ UTR length of a gene was defined as the length of the longest 5′ UTR of all coding transcripts associated with the gene. The 5′ UTR exon number of a gene was defined as the largest 5′ UTR exon number of all the coding transcripts associated with the gene.
To identify potential miRNAs binding to ATXN1’s 5′ UTR, the custom prediction of the miRNA prediction tool miRDB (http://mirdb.org) was used (Wong and Wang 2015; Liu and Wang 2019). To calculate the correlation of miR760's expression to ATXN1, miR760 and ATXN1 expression data in the developing human brain (Kang et al. 2011) were obtained from the BrainSpan database (http://www.brainspan.org) and analyzed as described previously (Kim et al. 2016).
Cloning of ATXN1 5′ UTR and 3′ UTR luciferase constructs
For the cloning of ATXN1’s 5′ UTR upstream of luciferase, the pRL-TK vector was digested with EcoRI-HF and NheI-HF restriction enzymes (New England Biolabs). gBlocks containing part of the HSV TK promoter and human ATXN1’s 5′ UTR including the splice donor and acceptor of the first intron were purchased from Integrated DNA Technologies and digested with the same restriction enzymes. Both products were then ligated using T4 DNA ligase (New England Biolabs). Correct ligation was confirmed by Sanger sequencing.
For the cloning of ATXN1’s 3′ UTR downstream from luciferase, the pRL-TK vector was digested with XbaI and NotI-HF (New England Biolabs). gBlocks (Integrated DNA Technologies) containing the predicted miR760-binding site in ATXN1’s 3′ UTR and 500 bp upstream of and downstream from the predicted site were purchased from Integrated DNA Technologies and digested with the same restriction enzymes. Both products were then ligated using T4 DNA ligase (New England Biolabs). Correct ligation was confirmed by Sanger sequencing.
Site-directed mutagenesis of ATXN1 5′ UTR construct
To test the binding of miR760 to ATXN1’s 5′ UTR, both predicted binding sites of miR760 were mutated alone or in combination by deleting three nucleotides in the core binding region. To do so, the QuikChange Lightning multisite-directed mutagenesis kit (Agilent) was used according to the manufacturer's instructions using primers miR760_BS1 5′-CTCAGGAGCCCTCAGCCTGTGGGAAG-3′ and/or miR760_BS2 5′-GTGACTTTCCGTTTATCAGACTAAAATAGCCATCCAGACA-3′ (Sigma-Aldrich). The mutations were confirmed by Sanger sequencing.
Luciferase assay
All cells were cultured in DMEM supplemented with 10% FBS (Atlanta Biological) and 1% antibiotic/antimycotic (Invitrogen) at 37°C in 5% CO2. HEK293T cells were seeded at five 8 × 104 cells per well in 24-well plates. Twenty-four hours after seeding, the cells were transfected with either 12.5 ng of pRL-TK control, pRL-TK 5′ UTR construct, or pRL-TK 3′ UTR construct, and 50 ng of pGL3-control (Promega) using jetPRIME transfection reagent (Polyplus). For miR760 experiments, the cells were further transfected with either 30 pmol of mirVana miRNA mimic negative control #1 (Invitrogen 4464059) or 30 pmol of hsa-miR-760 mimic (Invitrogen 4464066 MC12767). Forty-eight hours after transfection, luciferase activities were measured using the dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions.
Transfection of miRNA mimics, miRNA inhibitors, and siRNAs
All cell lines were cultured in DMEM supplemented with 10% FBS (Atlanta Biological) and 1% antibiotic/antimycotic (Invitrogen) at 37°C in 5% CO2. The number of cells seeded per well in a six-well plate varied based on the cell line used: 3 × 105 for HEK293T, 8 × 104 for DAOY, and 3 × 105 for HT-22. The following day, the cells were transfected with either 70 pmol of miRVana miRNA mimics (Invitrogen), 200 pmol of miRNA inhibitors (Invitrogen), and/or 40 pmol of siRNAs (Dharmacon or Invitrogen) using DharmaFECT 1 transfection reagent (Dharmacon) according to the manufacturer's instructions. The miRVana miRNA mimics used were mirVana miRNA mimic negative control #1 (Invitrogen 4464059) and miRVana hsa-miR-760 mimic (Invitrogen 4464066 MC12767) (see also Supplemental Table S1). The miRNA inhibitors used were mirVana miRNA inhibitor negative control #1 (Invitrogen 4464076) and miRVana hsa-miR-760 inhibitor (Invitrogen 4464084 MH12767) (see also Supplemental Table S2). The siRNAs used were ON-TARGETplus human ATXN1 siRNA (Dharmacon J-004510-06-0010), Silencer Select negative control #1 siRNA (Invitrogen 4390843), Silencer Select mouse Atxn1 (Invitrogen 4390771 s73331), Silencer Select siRNA AGO1 (Invitrogen 4427038 s25500), Silencer Select siRNA AGO2 (Invitrogen 4427037 s25931), Silencer Select siRNA AGO3 (Invitrogen 4427037 s46947), and Silencer Select siRNA AGO4 (Invitrogen 4427037 s46949). The cells were cultured for 72 h before harvesting for protein or mRNA analyses.
Generation of stable cell lines expressing Flag-tagged ATXN1
DAOY cells stably expressing ATXN1[82Q] fused with an N-terminal Flag tag were generated using the pINDUCER system (Meerbrey et al. 2011). For lentiviral production, 4 × 106 HEK239T cells were seeded in a 10-cm dish containing DMEM supplemented with 10% FBS (Atlanta Biological). The next day, 8 µg of pINDUCER_Flag_ATXN1[82Q] vector, 6 µg of psPAX2 vector (Addgene 12260), and 2 µg of pMD2.G vector (Addgene 12259) were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Twenty-four hours following transfection, an additional 5 mL of fresh medium was added to the dish and all media were collected the following day. For lentiviral transduction, 2 × 105 DAOY cells were seeded in six-well plates. Twenty-four hours after seeding, 50 µL of the lentiviral-containing medium was added to the cells. The next day, the medium was changed to selection medium containing DMEM supplemented with 10% FBS (Atlanta Biological), 1% antibiotic/antimycotic (Invitrogen), and 350 ng/mL Geneticin (Invitrogen). The cells were passaged for a minimum of three generations in geneticin-containing medium to ensure complete selection.
To test the effect of miR760 on ATXN1 levels in the pINDUCER_Flag_ATXN1[82Q] cell line, 8 × 104 DAOY cells were seeded in six-well plates with DMEM supplemented with 10% FBS (Atlanta Biological), 1% antibiotic/antimycotic (Invitrogen), and 300 ng/mL doxycycline hyclate (Sigma) at 37°C in 5% CO2 to induce ATXN1 expression. The next day, the cells were transfected as described above. Following transfection, cells were cultured for 72 h before harvesting for Western blot analyses.
Protein extraction and Western blot from cell culture experiments
Cells were scraped from the wells and homogenized in RIPA buffer (25 mM Tris-HCL at pH 7.6, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× protease inhibitor [Roche], 1× phosphatase inhibitor [Sigma]). After homogenization, the samples were placed for 30 min on ice at 4°C, sonicated five times, and spun down at 13,000 rpm for 15 min. Protein concentrations of the supernatant were measured using the Pierce BCA protein assay kit (Thermo Fisher Scientific) and samples were diluted and prepared in NuPAGE sample-reducing agent (Invitrogen) and NuPAGE LDS sample buffer (Invitrogen). The samples were boiled for 10 min and then run on NuPAGE 4%–12% Bis-Tris gel 1.5-mm 15-well gels (Invitrogen) or NuPAGE Nocex 4%–12% Bis-Tris Midi 26-well gels in MES running buffer (50 mM MES, 50 mM Tris base, 0.1% SDS, 1 mM EDTA). The proteins were subsequently transferred to Amersham Protran 0.2 NC nitrocellulose Western blotting membranes (GE Healthcare Life Sciences) using Tris glycine transfer buffer (25 mM Tris base, 192 mM glycine, 10% methanol) and blocked at room temperature for 1 h using 3% BSA in Tris-buffered saline (5 mM Tris at pH 7.5, 120 mM NaCl) with 0.1% Tween-20 (TBST). The membranes were probed overnight at 4°C with anti-ATXN1 (1:2000; 11750VII) or anti-GAPDH (1:10,000; 6C5; Sigma ) in 3% BSA in TBST. The next day, the membranes were washed three times with TBST and then incubated for 1 h at room temperature with either Immun-Star goat anti-rabbit HRP conjugate (1:10,000; Bio-Rad) to detect ATXN1 or Immun-Star goat anti-mouse HRP conjugate (1:10,000; Bio-Rad) to detect GAPDH. The membranes were then washed three times with TBST, incubated for 1 min in Amersham ECL Prime reagent (Fisher) and then imaged using an ImageQuant LAS 4000 imager (GE Healthcare Life Sciences).
RNA extraction and qPCR for cell culture experiments
Total RNA was isolated using the miRNeasy mini kit (Qiagen) following the manufacturer's instructions. For the luciferase RNA experiments, samples were treated with DNase using the Turbo DNA-free kit (Thermo Fisher Scientific) to remove the plasmid DNA. Random-primed cDNA was prepared from 1 µg of total RNA using M-MLV reverse transcriptase (Invitrogen). RT-qPCR was performed with PowerUp SYBR Green Master mix (Applied Biosystems), and samples were run on a real-time PCR detection system (Bio-Rad CFX96). All samples were analyzed in triplicate and expression levels were normalized to expression of the housekeeping gene GAPDH. Primers for GAPDH and ATXN1 were obtained from Sigma-Aldrich and were as follows: Renilla_For (GGAATTATAATGCTTATCTACGTGC), Renilla_Rev (CTTGCGAAAAATGAAGACCTTTTAC), Firefly_For (CTCACTGAGACTACATCAGC), Firefly_Rev (TCCAGATCCACAACCTTCGC), hATXN1_For (TCCAGCACCGTAGAGAGGAT), hATXN1_Rev (AGCCCTGTCCAAACACAAAA), hGAPDH_For (CGACCACTTTGTCAAGCTCA), and hGAPDH_Rev (TTACTCCTTGGAGGCCATGT).
Polysome profiling
HEK293T cells (8 × 106) were seeded in 15-cm dishes. Twenty-four hours following seeding the cells were transfected with 750 pmol of miRVana miRNA mimic negative control #1 (Invitrogen 4464059) or 750 pmol of miRVana hsa-miR-760 mimics (Invitrogen 4464066 MC12767). The next day, the cells were washed with PBS containing 100 µg/mL cycloheximide (Sigma-Aldrich) before being lysed in polysome lysis buffer (10 mM HEPES-KOH, 150 mM KCL, 5 mM MgCl2 in RNase-free water, supplemented with 0.5 mM DTT, 100 µg/mL cycloheximide [Sigma-Aldrich], RNase inhibitors [Invitrogen], and Xpert protease inhibitor cocktail [GenDEPOT]) using dounce homogenization. The samples were spun down at 2000 rcf for 10 min at 4°C. The supernatant was transferred to a prechilled Eppendorf tube and NP-40 (at a final concentration of 0.5%) was added. The samples were incubated for 5 min on ice and centrifuged again at 20,000 rcf for 10 min at 4°C. The samples were incubated for 20 min on ice and then transferred onto a 17%–50% sucrose gradient. The gradient was spun at 35,000 rcf for 2 h at 4°C. After centrifugation, the fractions were obtained using a BR-184 tube piercer and syringe pump (Brande). Absorbance at 254 nm was monitored using a UA-6 UV detector (Teledyne ISCO) and a DI-158U USB data acquisition device (DATQ Instruments).
RNA was extracted from each fraction using Trizol LS according to the manufacturer's instructions. Random-primed cDNA was prepared using M-MLV reverse transcriptase (Invitrogen). RT-qPCR was performed with PowerUp SYBR Green Master mix Applied Biosystems), and samples were run on a real-time PCR detection system (Bio-Rad CFX96). All samples were analyzed in triplicate. Primers for GAPDH and ATXN1 were obtained from Sigma-Aldrich (Table 4).
Generation of AAV9 viruses expressing miR760
To generate an AAV vector expressing mouse miR760, a previously described AAV vector (Rousseaux et al. 2018b) containing both YFP and a miRE cassette-containing control shRNA under the control of the chicken β-actin promoter was modified. The vector was digested using NotI-HF and HindIII-HF (New England Biolabs) to remove the control shRNA and replace it with the mouse miR760 sequence. To do so, gBlocks (Integrated DNA Technologies) containing miR760 pri-miRNA sequence were purchased and digested with the same restriction enzymes. Both products were then ligated using T4 DNA ligase (New England Biolabs). To generate AAV9 viruses, the vectors were packaged by the Baylor College of Medicine Gene Vector Core.
Mouse husbandry
All mice were housed and maintained in the animal facilities at Baylor College of Medicine. Atxn1154Q/2Q mice, whose generation and characterization were described previously (Watase et al. 2002) were backcrossed to C57BL/6J background for a minimum of 10 generations. The mice were kept on a 12-h light/dark cycle, and behavioral tests were always conducted during the light phase of the cycle. The mice had access to food and water ad libitum except during tests. All mice were age- and sex-matched within experiments. Equal or similar numbers of male and female mice were used for all experiments and no differences between sexes were observed. Mice were randomly assigned to treatment and experimental conditions. All experiments used littermates as controls and were carried out and analyzed with the experimenter blinded to genotype and treatment. Animal care and experimental procedures were approved by the institutional animal care and use committee of Baylor College of Medicine, according to U.S. National Institutes of Health Guidelines.
Intracerebellar AAV9 injections
AAV9-YFP-control and AAV9-YFP-miR760 were injected at four cerebellar sites in 5-wk-old mice at a rate of between 0.1 and 0.15 μL/min. A total volume of 2 μL of each virus (4.0 × 1013 –8.0 × 1013 g.c/mL) was injected at each site. After the injection, the syringe was left in place for a minimum of 10 min before it was removed. The coordinates used for each site were anterior–posterior (AP): 6.2 mm, medial–lateral (ML): 0 mm, and dorsal–ventral (DV): 1.5 mm; AP: 6.96 mm, ML: 0 mm, DV: 1.5 mm; AP: 6.2 mm, ML: 1.8 mm, and DV: 2.2 mm; and AP: 6.2 mm, ML: 1.8 mm, and DV: 2.2 mm. Correct injection of the AAV was confirmed by fluorescent imaging of whole brains using a Leica M80HD. Two mice were excluded from the behavioral studies as the injections were mistargeted and resulted in strong expression of the virus in the brain stem.
RNA extraction and qPCR from AAV transduced cerebella
For the AAV injection experiments, cerebella from 10-wk-old mice were dissected. Total RNA was isolated using the miRNeasy mini kit from Qiagen following the manufacturer's instructions. cDNA synthesis was performed using the TaqMan microRNA reverse transcription kit (Invitrogen) according to the manufacturer's instructions. RT-qPCR was performed with TaqMan Universal PCR Master mix (Invitrogen) using specific primers for miR760 (Invitrogen 4427975, ID: 002328) and U6 (Invitrogen 4427975, ID: 001973). All samples were analyzed in triplicate and expression levels were normalized to U6.
Protein extraction and Western blot from AAV transduced cerebella
Cerebellar tissues were dissected at 10 wk of age. Cerebellar homogenates from each genotype were prepared by dounce homogenization in NETN buffer (100 mM NaCl, 20 mM Tris-HCl at pH 8.0, 0.5 mM EDTA, 1.5% NP-40, 1× protease inhibitor [Roche], 1× phosphatase inhibitor [Sigma]). Samples were incubated for 30 min at 4°C, sonicated a total of 10 times, and spun at 13,000 rpm for 15 min at 4°C. Supernatants were prepared with NuPAGE sample-reducing agent (Invitrogen) and NuPAGE LDS sample buffer (Invitrogen), boiled for 10 min, and run on NuPAGE 4%–12% Bis-Tris gel 1.5-mm 15-well gels (Invitrogen) in MES running buffer (50 mM MES, 50 mM Tris base, 0.1% SDS, 1 mM EDTA). The proteins were subsequently transferred to 0.45-µm Immobilon-FL PVDF membranes (Invitrogen) using Tris glycine transfer buffer (25 mM Tris base, 192 mM glycine, 10% methanol) and dried for a minimum of 1 h. Membranes were then blocked for 1 h at room temperature with 1:1 Odyssey TBS blocking buffer (LI-COR) in TBS and probed overnight at 4°C with anti-ATXN1 (1:2000; 11750VII) and anti-GAPDH (1:10,000; 6C5; Sigma) in 1:1 Odyssey TBS blocking buffer (LI-COR) in TBST. The next day, the membranes were washed three times with TBST and then incubated for 1 h at room temperature with either rabbit IgG (H&L) antibody DyLight 680 conjugate (1:10,000; Rockland Immunochemicals) to detect anti-ATXN1 or mouse IgG (H&L) antibody DyLight 800 conjugate (1:10,000; Rockland Immunochemicals) to detect anti-GAPDH. The membranes were washed three times with TBST and then imaged using the Odyssey CLx imaging system (LI-COR).
Behavioral tests
Rotarod
The rotarod test was performed at 10 wk of age to evaluate coordination and motor skill acquisition (type 7650; Ugo Basile). For each test, the mice were habituated for 30 min in the test room before testing. The mice were placed on the rotating rod (3-cm diameter, 30 cm long) for four trials every day for a period of 4 d. Each trial lasted a maximum of 10 min. The rod accelerated from 4 to 40 rpm in 5 min, and remained at 40 rpm for the remaining 5 min. The time the mice took to fall was recorded. Two subsequent rotations around the rod were also counted as a fall.
Pole test
The pole test was performed at 7, 8, and 10 wk of age. All mice were habituated for 30 min in the test room each day before testing. Each mouse was then placed head-upward at the top of a vertical threaded metal pole. Time of descent was measured with a 60-sec cutoff time. Falls from the metal pole were counted as a 60-sec descent. In case the mouse fell off the pole within the first 10 sec, the trial was not counted, and the mouse was placed on the pole again to assure proper grip. The test was performed in triplicate for each animal.
Statistical analysis
Statistical tests were performed in accordance with the experimental design. Simple comparisons used Student's t-test, whereas multigroup comparisons used one- or two-way ANOVAs. In each case, a single asterisk denotes P < 0.05, double asterisks denote P < 0.01, triple asterisks denote P < 0.001, quadruple asterisks denote P < 0.0001, and ns denotes P > 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Joel Neilson for advice on AAV vector cloning, Dr. Hyung-Kyoung Lee and Dr. Mauro Costa-Mattioli for equipment access, and Dr. James Orengo, Dr. Sameer Bajikar, Manar Zaghlula, and Dany El-Najjar for their critical comments on the manuscript. This work was funded by National Institute of Neurological Disorders and Stroke grant 4R37NS027699 (HYZ), the Howard Hughes Medical Institute (HYZ), Baylor Research Advocates for Student Scientists (LN), the Young Investigator Research Grant by the National Ataxia Foundation (VAG), and Intellectual and Developmental Disabilities Research Center at Baylor College of Medicine grant 1U54HD083092 (Administrative and Behavioral Cores) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the National Institutes of Health. This work was also supported by the Gene Vector Core at Baylor College of Medicine.
Author contributions: L.N. and H.Y.Z. conceived the study, designed experiments, and analyzed and interpreted data. L.N. performed molecular and behavioral assays and wrote the manuscript. L.N. and H.Y.Z. edited the manuscript. A.T. performed stereotaxic injections. S.L.C., E.X., J.L.J., and F.A.B. helped with molecular assays. V.A.G. performed COMETA analysis, and K.P. and Z.L. performed bioinformatic analysis. All authors reviewed the manuscript and provided input.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.339317.120.
References
- Alonso E, Martínez-Ruano L, De Biase I, Mader C, Ochoa A, Yescas P, Gutiérrez R, White M, Ashizawa T, Bidichandani S, et al. 2007. Distinct distribution of autosomal dominant spinocerebellar ataxia in the Mexican population. Mov Disord 22: 1050–1053. 10.1002/mds.21470 [DOI] [PubMed] [Google Scholar]
- Banfi S, Servadio A, Chung My, Kwiatkowski TJ, McCall AE, Duvick LA, Shen Y, Roth EJ, Orr HT, Zoghbi HY. 1994. Identification and characterization of the gene causing type 1 spinocerebellar ataxia. Nat Genet 7: 513–520. 10.1038/ng0894-513 [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, de Hoon ML, Tomaru Y, Suzuki H, Hayashizaki Y, Daub CO. 2011. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol 8: 158–177. 10.4161/rna.8.1.14300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, Levecque C, Larvor L, Andrieux J, Hulihan M, et al. 2004. α-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet 364: 1167–1169. 10.1016/S0140-6736(04)17103-1 [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Reinstein E, Sun Y, Antalffy B, Jiang YH, Ciechanover A, Orr HT, Beaudet AL, Zoghbi HY. 1999. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron 24: 879–892. 10.1016/S0896-6273(00)81035-1 [DOI] [PubMed] [Google Scholar]
- Dewing AST, Rueli RH, Robles MJ, Nguyen-Wu ED, Zeyda T, Berry MJ, Bellinger FP. 2012. Expression and regulation of mouse selenoprotein P transcript variants differing in non-coding RNA. RNA Biol 9: 1361–1369. 10.4161/rna.22290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J, Kordasiewicz HB, O'Callaghan B, Handler HP, Wagener C, Duvick L, Swayze EE, Rainwater O, Hofstra B, Benneyworth M, et al. 2018. Antisense oligonucleotide-mediated ataxin-1 reduction prolongs survival in SCA1 mice and reveals disease-associated transcriptome profiles. JCI insight 3 10.1172/jci.insight.123193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer JD, Yu P, Kang H, Mandel-brehm C, Carter AN, Crespo-barreto J, Gao Y, Flora A, Shaw C, Orr HT. 2011. Exercise and genetic rescue of SCA1 via the transcriptional repressor capicua. Science 334: 690–693. 10.1126/science.1212673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, D'Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, et al. 2012. Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res 22: 1163–1172. 10.1101/gr.130435.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Singh RK, White JJ, De Maio A, Han K, Kim JY, Jafar-Nejad P, Di Ronza A, Kang H, Sayegh LS, et al. 2015. Pumilio1 haploinsufficiency leads to SCA1-like neurodegeneration by increasing wild-type Ataxin1 levels. Cell 160: 1087–1098. 10.1016/j.cell.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennarino VA, Palmer EE, McDonell LM, Wang L, Adamski CJ, Koire A, See L, Chen CA, Schaaf CP, Rosenfeld JA, et al. 2018. A mild PUM1 mutation is associated with adult-onset ataxia, whereas haploinsufficiency causes developmental delay and seizures. Cell 172: 924–936.e11. 10.1016/j.cell.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AMM, Pletikos M, Meyer KA, Sedmak G, et al. 2011. Spatio-temporal transcriptome of the human brain. Nature 478: 483–489. 10.1038/nature10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsnelson A, De Strooper B, Zoghbi HY. 2016. Neurodegeneration: from cellular concepts to clinical applications. Sci Transl Med 8: 364ps18 10.1126/scitranslmed.aal2074 [DOI] [PubMed] [Google Scholar]
- Keiser MS, Boudreau RL, Davidson BL. 2014. Broad therapeutic benefit after RNAi expression vector delivery to deep cerebellar nuclei: implications for spinocerebellar ataxia type 1 therapy. Mol Ther 22: 588–595. 10.1038/mt.2013.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Zhang Y, Pang K, Kang H, Park H, Lee Y, Lee B, Lee HJ, Kim WK, Geum D, et al. 2016. Bipolar disorder associated microRNA, miR- 1908-5p, regulates the expression of genes functioning in neuronal glutamatergic synapses. Exp Neurobiol 25: 296–306. 10.5607/en.2016.25.6.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger rNAS. Nucleic Acids Res 15: 8125–8148. 10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Samaco RC, Gatchel JR, Thaller C, Orr HT, Zoghbi HY. 2008. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat Neurosci 11: 1137–1139. 10.1038/nn.2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ding Y, Liu N, Sun Q, Zhang J. 2017. MicroRNA-760 inhibits cell proliferation and invasion of colorectal cancer by targeting the SP1-mediated PTEN/AKT signalling pathway. Mol Med Rep 16: 9692–9700. 10.3892/mmr.2017.7814 [DOI] [PubMed] [Google Scholar]
- Liao Y, Deng Y, Liu J, Ye Z, You Z, Yao S, He S. 2016. MiR-760 overexpression promotes proliferation in ovarian cancer by downregulation of PHLPP2 expression. Gynecol Oncol 143: 655–663. 10.1016/j.ygyno.2016.09.010 [DOI] [PubMed] [Google Scholar]
- Liu W, Wang X. 2019. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol 20: 18 10.1186/s13059-019-1629-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. 2007. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci 104: 9667–9672. 10.1073/pnas.0703820104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manek R, Nelson T, Tseng E, Rodriguez-Lebron E. 2019. 5′UTR-mediated regulation of Ataxin-1 expression. Neurobiol Dis 134: 104564 10.1016/j.nbd.2019.104564 [DOI] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, et al. 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci 108: 3665–3670. 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G. 2013. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet 14: 447–459. 10.1038/nrg3462 [DOI] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G. 2002. Untranslated regions of mRNAs. Genome Biol 3: reviews0004.1–rereviews0004.10. 10.1186/gb-2002-3-3-reviews0004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Thermann R, Hentze MW. 2010. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA 16: 2493–2502. 10.1261/rna.2384610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom UA, Nielsen FC, Lund AH. 2008. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 30: 460–471. 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- Orr H, Chung M, Banfi S, Kwiatkowski J, Servadio A, Beaudet A, McCall A, Duvick L, Ranum L, Zoghbi H. 1993. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet 4: 221–226. 10.1038/ng0793-221 [DOI] [PubMed] [Google Scholar]
- Park J, Al-Ramahi I, Tan Q, Mollema N, Diaz-Garcia JR, Gallego-Flores T, Lu HC, Lagalwar S, Duvick L, Kang H, et al. 2013. RAS–MAPK–MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature 498: 325–331. 10.1038/nature12204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux MWC, Tschumperlin T, Lu HC, Lackey EP, Bondar VV, Wan YW, Tan Q, Adamski CJ, Friedrich J, Twaroski K, et al. 2018a. ATXN1–CIC complex is the primary driver of cerebellar pathology in spinocerebellar ataxia type 1 through a gain-of-function mechanism. Neuron 97: 1235–1243.e5. 10.1016/j.neuron.2018.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux MWC, Vázquez-Vélez GE, Al-Ramahi I, Jeong HH, Bajić A, Revelli JP, Ye H, Phan ET, Deger JM, Perez AM, et al. 2018b. A druggable genome screen identifies modifiers of α-synuclein levels via a tiered cross-species validation approach. J Neurosci 38: 9286–9301. 10.1523/JNEUROSCI.0254-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerrière A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, et al. 2006. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 38: 24–26. 10.1038/ng1718 [DOI] [PubMed] [Google Scholar]
- Rumble B, Retallack R, Hilbich C, Simms G, Multhaup G, Martins R, Hockey A, Montgomery P, Beyreuther K, Masters CL. 1989. Amyloid A4 protein and its precursor in down's syndrome and alzheimer's disease. N Engl J Med 320: 1446–1452. 10.1056/NEJM198906013202203 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Shiohama A, Minoshima S, Shimizu N. 2003. Identification of eight members of the Argonaute family in the human genome. Genomics 82: 323–330. 10.1016/S0888-7543(03)00129-0 [DOI] [PubMed] [Google Scholar]
- Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, et al. 2003. α-Synuclein locus triplication causes Parkinson's disease. Science 302: 841 10.1126/science.1090278 [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258. 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, Del-Favero J, Cruts M, Van Duijn CM, Van Broeckhoven C. 2006. APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain 129: 2977–2983. 10.1093/brain/awl203 [DOI] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, Jaenisch R. 2016. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533: 95–99. 10.1038/nature17939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Romano DM, Nitschke L, Herrick SP, DiMarzio BA, Dzhala V, Bae J-S, Oram MK, Zheng Y, Hooli B, et al. 2019. Loss of Ataxin-1 potentiates Alzheimer's pathogenesis by elevating cerebral BACE1 transcription. Cell 178: 1159–1175.e17. 10.1016/j.cell.2019.07.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Wang J, Zhou S, Zhou J, Jia G, Wang H, Xin C, Fu G, Zhang J. 2019. MiR-760 regulates skeletal muscle proliferation in rheumatoid arthritis by targeting Myo18b. Mol Med Rep 20: 4843–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns J, Brouwers N, Engelborghs S, Sleegers K, Bogaerts V, Corsmit E, De Pooter T, Van Duijn CM, De Deyn PP, Van Broeckhoven C. 2006. Promoter mutations that increase amyloid precursor-protein expression are associated with Alzheimer disease. Am J Hum Genet 78: 936–946. 10.1086/504044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524. 10.1101/gad.1399806 [DOI] [PubMed] [Google Scholar]
- Wang ZC, Yang S, Chen MQ, Wu SS, Lv HH, Jin WX. 2019. LINC01296 promotes the proliferation and invasion by regulating microRNA-760 expression and predicts poor prognosis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 23: 9848–9856. [DOI] [PubMed] [Google Scholar]
- Watase K, Weeber EJ, Xu B, Antalffy B, Yuva-Paylor L, Hashimoto K, Kano M, Atkinson R, Sun Y, Armstrong DL, et al. 2002. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron 34: 905–919. 10.1016/S0896-6273(02)00733-X [DOI] [PubMed] [Google Scholar]
- Wong N, Wang X. 2015. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43: D146–D152. 10.1093/nar/gku1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YZ, Zhang YF, Yang L, Xu J, Mo XM, Peng W. 2018. MiR-760 mediates hypoxia-induced proliferation and apoptosis of human pulmonary artery smooth muscle cells via targeting TLR4. Int J Mol Med 42: 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhou H, Rigoutsos I. 2014. MiR-103a-3p targets the 5′ UTR of GPRC5A in pancreatic cells. RNA 20: 1431–1439. 10.1261/rna.045757.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. 1995. Spinocerebellar ataxia type 1 (SCAl). Cell Biol 1: 29–35. [DOI] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.