Abstract
Studies of microbiomes are booming, along with the diversity of computational approaches to make sense out of the sequencing data and the volumes of accumulated microbial genotypes. A swift evaluation of newly published methods and their improvements against established tools is necessary to reduce the time between the methods' release and their adoption in microbiome analyses. The LEMMI platform offers a novel approach for benchmarking software dedicated to metagenome composition assessments based on read classification. It enables the integration of newly published methods in an independent and centralized benchmark designed to be continuously open to new submissions. This allows developers to be proactive regarding comparative evaluations and guarantees that any promising methods can be assessed side by side with established tools quickly after their release. Moreover, LEMMI enforces an effective distribution through software containers to ensure long-term availability of all methods. Here, we detail the LEMMI workflow and discuss the performances of some previously unevaluated tools. We see this platform eventually as a community-driven effort in which method developers can showcase novel approaches and get unbiased benchmarks for publications, and users can make informed choices and obtain standardized and easy-to-use tools.
Metagenomics has made possible the study of previously undiscovered, uncultured microorganisms. To probe the vast hidden microbial landscape we need effective bioinformatics tools, in particular taxonomic classifiers for binning the sequencing reads (i.e., grouping and labeling) and profiling the corresponding microbial community (i.e., defining the relative abundances of taxa). This is computationally challenging, requiring time and resources to query shotgun sequencing data against rapidly expanding genomic databases. Users have to choose a solution among the plethora of methods that are being developed in a quest for accuracy and efficiency, for instance, lowering the run time and memory usage by reducing the reference material while maintaining the diversity represented (Truong et al. 2015; Kim et al. 2016). To date, at least 100 published methods can be identified (https://microbe.land/97-metagenomics-classifiers/), and new developments that may bring innovations to the field cohabit with re-implementations of already-explored strategies, complicating method selection, fragmenting the community of users, and hindering experimental reproducibility. Papers describing these methods often fall in the “self-assessment trap” (Norel et al. 2011), in which all published methods are the best on carefully selected data. Such assessments give no indication to potential users about general performance, which should be a prerequisite before considering any case-specific improvements or novel features.
Independent comparative benchmarking studies (Peabody et al. 2015; Lindgreen et al. 2016; McIntyre et al. 2017; Gardner et al. 2019; Ye et al. 2019) and challenges (Sczyrba et al. 2017) are a major step toward a fair and efficient assessment of methods. They discuss the results and pinpoint advantages and weaknesses of different technical approaches, promote developments dedicated to specific technologies or problems (Bremges and McHardy 2018), and have introduced valuable benchmarking resources (Belmann et al. 2015; Meyer et al. 2018, 2019). Unfortunately, some tools have not been considered by any of the comparative studies published to date. For instance, MetaCache (Müller et al. 2017), despite being an alternative to well-adopted tools such as Kraken 2 (Wood et al. 2019), still lacks an independent confirmation of the favorable evaluation presented by the investigators in its accompanying paper. Although published benchmarking data sets can be reused in subsequent publications, follow-up studies comparing newly published methods under the same conditions are not the norm. More importantly, the true weakness of these studies is the absence of regularly scheduled publication of results, with possibly years between successive assessments, because no continuous monitoring strategy exists, perpetuating uncertainty about the true state of the art.
To offer a solution to this problem, we introduce A Live Evaluation of Computational Methods for Metagenome Investigation (LEMMI; https://lemmi.ezlab.org) (Fig. 1). It establishes continuous and generalizable comparisons of individual tools or heterogeneous multistep pipelines in a controlled environment to ensure published methods have a reliable benchmark on unified hardware. It hosts in its infrastructure a semiautomated evaluation and presents detailed results and rankings for multiple analytic objectives and their computational costs (e.g., accuracy vs. memory usage) in a web-based interface that allows users to assess the pros and cons of new entries with more flexibility than a static report would offer. The first available LEMMI component, which is the focus of this publication, assesses taxonomic profilers and binners. LEMMI follows several recommendations for efficient “omics” tool benchmarking (Capella-Gutierrez et al. 2017; Maier-Hein et al. 2018; Mangul et al. 2019a), namely, (1) using and distributing containers (i.e., isolated software packages) to allow reproducibility and long-term availability of tools (Mangul et al. 2019b); (2) reporting the consumption of computational resources, which is essential as available hardware limits the choice to otherwise less efficient methods; and (3) exploring parameters and avoiding rankings based on a single metric. In the particular case of taxonomic classification, a benchmark that uses the same reference for all methods is necessary to perform a valid evaluation of their respective algorithms, in addition to assessing the variety of available databases. Therefore, both strategies are implemented by LEMMI. With an expected delay of a few weeks between submission and appearance of the method on the website, our solution complements and supports analyses of specific cases discussed in self-evaluations or periodical comparative studies, bridging the time gap between them. We describe below the advantages of the LEMMI workflow for tool developers and users, and discuss the performances of some recently published methods.
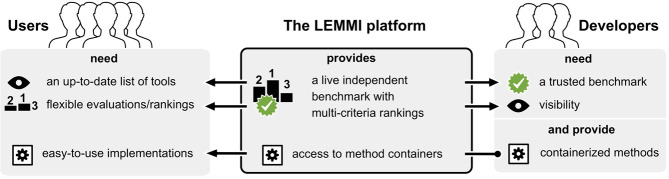
Figure 1.
Connecting users and developers through benchmarking. The LEMMI platform facilitates the access to up-to-date implementations of methods for taxonomic binning and profiling. LEMMI creates a link between developers and users as it provides independent and unbiased benchmarks that are valuable to both. Developers need comparative evaluations to keep improving the methodology and to get their work published in peer-reviewed journals. Method users need a resource that keeps track of new developments and provides a flexible assessment of their performances to match their experimental goals, resources, and expectations. The containerized approach of LEMMI guarantees the transfer of a stable and usable implementation of each method from the developer to the final user, as they appear in the evaluation.
Results
A fully containerized hosted evaluation
LEMMI inputs consist of methods wrapped in standardized containers resembling a previously suggested format, bioboxes (Belmann et al. 2015), which can be prepared by the LEMMI development teams; the vision is for method developers to contribute and submit to LEMMI themselves immediately on release of the tool. In the latter case, containers can be exchanged through public channels (e.g., https://hub.docker.com) to be loaded and run on the LEMMI platform (Supplemental Fig. S1A). Rules are well defined (https://www.ezlab.org/lemmi_documentation.html), and no human-made decisions and curations are needed once the container is submitted. This minimizes the time from submission to the publication of the results and also excludes any arbitrary interpretation. In addition, evaluated containers and their building sources can be downloaded by users from the LEMMI website, unless the method is commercialized and/or not under open source licensing. Thus, users have a simple access to the evaluated version to conduct their analyses. LEMMI is the first solution in its field to compel containerization to manage a benchmarking workflow instead of simply suggesting this solution. The benefits of such an approach regarding systematic reevaluation (Supplemental Fig. S1B) and reusability are likely greater in the long term than mere submission of results. As strategies that facilitate automated conversion between distribution channels are being explored, for instance from BioConda to biocontainers (da Veiga Leprevost et al. 2017), promoting benchmarking processes that rely exclusively on these channels to obtain all methods ensures that the otherwise subjective question of installability is addressed unambiguously.
A dynamic interface to explore multiple objectives
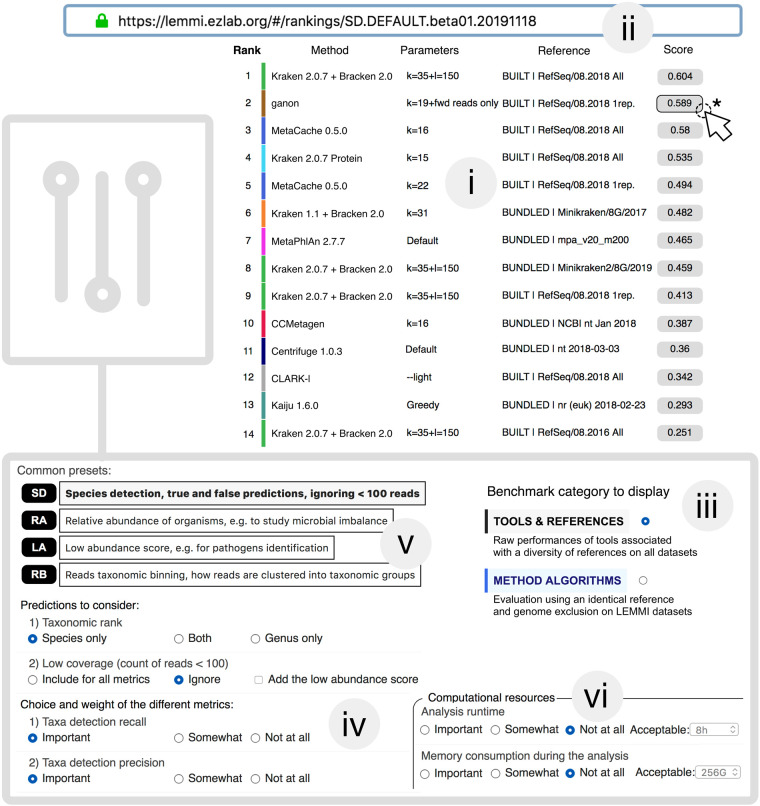
Making informed decisions when designing analyses or pipelines requires a thorough exploration of the parameter space of available algorithms. In LEMMI, multiple runs of a container enable such exploration as shown with the tool MetaCache, which was run with different k-mer lengths (Figs. 2, 3; Supplemental Figs. S2–S5, S7; Supplemental Table S1). On the other hand, users are sometimes looking for trade-offs between several features offered by a single tool or pipeline and its ability to complete multiple objectives with a single analysis (e.g., profiling and binning), given hardware limitations imposed by their environment. Thus, LEMMI does not segregate methods in categories with separate runs for the two objectives, because many tools provide both features or have extra scripts that blur the demarcation. For example, Kraken (Wood and Salzberg 2014; Wood et al. 2019) is a read classifier that can be associated with its companion tool Bracken (Lu et al. 2017) to produce a valid taxonomic abundance profile. LEMMI is “results-oriented,” the tools or pipelines wrapped in a container can output either abundance profiles, bins, or both in a single run. The metrics specified when exploring the results will define the dynamic ranking and determine whether the tool/pipeline meets the users’ needs without specifically asking for a profiler or a binner. These two-dimensional results can be visualized from different perspectives through a multicriteria ranking in which users can put the emphasis on metrics matching the objectives they recognize as important. These choices are algorithmically converted into scores, ordering methods to facilitate more detailed investigations (Figs. 2, 3; Supplemental Figs. S2–S8). In each displayed ranking, a green color highlights methods that score >90% of the theoretical maximum for the selected metrics, to give the user a sense of the room for improvement left to future developments.
Figure 2.
LEMMI web interface. (i) The LEMMI users obtain a list of entries suited to their needs through the dynamic ranking interface that allows them to select and weight the criteria of interest. The ranking visible here shows the performances when identifying species while ignoring taxa under 100 reads, balancing precision and recall. The score assigned to each entry visible in LEMMI rankings averages selected metrics over all tested data sets. Underlying values can be seen by clicking on the score (*). (ii) A unique “fingerprint” allows custom rankings to be shared and restored at any time on the LEMMI platform through the address bar. (iii) The benchmark category can be selected in the dashboard. (iv) Prediction accuracy metrics can be chosen along with their importance (weight for “Important” is 3, “Somewhat” is 1, and “Not at all” is 0). This will cause the list to be updated with the corresponding scores. (v) Several presets corresponding to common expectations are available. (vi) Computational resources and the time required to complete the analysis can be included as an additional factor to rank the methods.
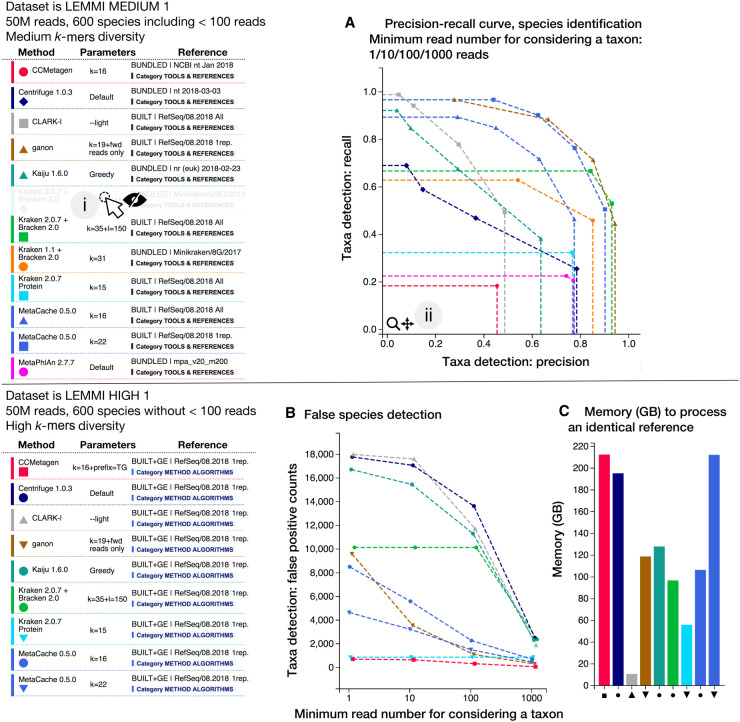
Figure 3.
“Detailed plots by data sets” interface. Plots for up to 17 metrics are available for each data set, and the LEMMI user can toggle each line representing a method associated with a reference and specific parameters individually (i). Plots can be zoomed in to disentangle overlapping points and permit focused interpretations (ii). The pages exist for all taxonomic ranks investigated, that is, genus and species in the release beta01. (A) The precision-recall curve in species identification, when filtering to only consider taxa represented by an increasing minimum of reads. If a curve contains fewer than four independent points (precision-recall at minimum 1/10/100/1000 reads), it indicates that filtering did not change the precision and recall at all thresholds, leading to overlaps. The entries are a mix of methods using references freely provided or built using the current maximal memory capacity of LEMMI of 245 GB. B and C show results when assessing methods using an identical built reference. (B) Illustration showing how LEMMI assesses the precision improvement enabled by filtering low abundance taxa at different thresholds. (C) Reporting the peak of memory necessary to construct the reference.
Prepackaged and built reference databases
Some method implementations provide prepackaged reference databases, and others provide scripts to generate them. LEMMI evaluates these prepackaged references to include tools whose value lies in providing a curated database, for example, marker genes (Truong et al. 2015), and to keep track of reference genome catalogs available to method users. However, because the use of different reference databases is likely a major source of result discrepancies, methods accepting any nucleotide or protein files to construct their database need to include the corresponding scripts in the container to create references as part of the benchmark. This enables LEMMI to report the resources (i.e., memory, run time) required to both build a reference and run the analysis, and then to assess the algorithm independently from its default reference database. To provide a continuous evaluation that is unaffected by the publication of new genomic sequences, LEMMI maintains an in-house repository currently based on all bacterial and archaeal RefSeq (O'Leary et al. 2016) assemblies. However, only entries having both nucleotide and corresponding protein files are kept to offer both types of sequences as a possible reference with equal representation (hereafter, the LEMMI/RefSeq repository). When conducting a benchmark run, LEMMI can subsample it by publication date, assembly states, or a fixed number of representatives per taxonomic identifier (taxid). As for parameters, this enables for each method the exploration of references, for instance, by restraining the source material to what was available at a given date or to specific criteria (e.g., assembly states being only “Complete Genome”). Not every method can process the 125,000 genomes included in LEMMI/RefSeq with the resources provided for benchmarking (245 GB of RAM, representing a medium-scale environment). It is therefore relevant to assess which subsets constitute good trade-offs in terms of run time, memory use, and accuracy of the predictions for each method, and to track the impact of continuous database growth on different methods (Supplemental Fig. S6; Nasko et al. 2018). For all methods allowing database creation, we first used as reference one representative per species taxid (entries labeled as RefSeq/08.2018/1rep.), which was defined empirically. This is suitable for most tools. Methods that could not build this reference owing to memory limitation were considered as being above the reasonable resource requirements to be useful to most users. They are listed on https://www.ezlab.org/lemmi_failed.html. On the other end of the range, memory-efficient methods that could process the complete set of reference genomes were also evaluated with the whole LEMMI/RefSeq repository to test whether this technical advantage translates into better predictions (entries labeled as RefSeq/08.2018/all) (Supplemental Table S1). Our results with Kraken 2 indicate that it is the case, with a cost in terms of extra memory that is relatively small with this tool. In the case of MetaCache, a trade-off had to be made to use more genomes, decreasing the k-mer length from 22 to 16. Results indicate it is an efficient choice with some configurations and data sets, as seen on Figure 2 and Supplemental Figure S5, in which MetaCache using k = 16 outperforms the use of k = 22 thanks to a larger reference (Supplemental Fig. S7). MetaCache with k = 16 is better ranked than with k = 22 in some configurations (Supplemental Fig. S2A,D), but it is not in others (Supplemental Fig. S2C) in which a larger k-mer value is more beneficial than a larger reference (Fig. 3).
In silico data sets and genome exclusion
LEMMI uses its repository not only as source material provided to each method to create their reference database, but also for sampling mock microbial communities to generate in silico paired-end short reads used to measure the accuracy of predictions made by each method. It implements a genome exclusion (Peabody et al. 2015) approach to simulate unknown organisms at various taxonomic ranks and to prevent overfitting by excluding the source of the analyzed reads from the reference (Supplemental Fig. S9; Methods). In most biological samples, it is expected that organisms unrepresented in databases constitute a fraction of the reads. A proper benchmark should assess the ability of methods to label unknown organisms at a given taxonomic level as being unknown. Simulating clades that have no corresponding organisms in the reference is therefore useful to spot methods producing an excess of false positive identifications under this situation (Fig. 3B). Data sets generated in-house by LEMMI contain randomly sampled bacteria and archaea, representing generic communities of variable complexity in terms of number of species, abundance distribution (i.e., absence or presence of taxa below 100 reads), and number of nonunique k-mers composing the reads. The description of all data sets is presented in Supplemental Table S2. Their detailed taxonomic compositions remain private as long as they are in use in the platform and will be published when LEMMI moves to its next major release (Methods). A demo data set is available to exemplify their typical composition and help with preparing a method container for evaluation. LEMMI data sets are complemented by others used in previously published benchmarking studies, namely CAMI1 (Sczyrba et al. 2017) and mockrobiota (Bokulich et al. 2016). They are useful because they enable comparisons with past benchmarks familiar to the users and introduce variety in terms of data sets tested, which permits the identification of unwanted patterns inherent in the benchmarking design (Supplemental Note S1). Detailed evaluation results using up to 17 metrics are presented individually in interactive plots for each data set and taxonomic rank, namely, genus and species (Fig. 3; Supplemental Figs. S3–S8; Supplemental Table S3).
Two benchmarking categories
Our platform offers evaluations conducted with preprocessed databases, meaning no possibility of genome exclusion, as well as evaluations conducted under an identical set of genomes absent from the source of in silico reads. Their results cannot be interpreted together, and LEMMI distinguishes two benchmarking categories (Fig. 2). When choosing “TOOLS & REFERENCES,” users can get an overview of all tools and references included in LEMMI and their respective abilities to capture the known microbial diversity. This is strongly influenced by the database sampling strategy and creation date, but reflects what practitioners will encounter when using ready-to-use solutions. The second category, “METHOD ALGORITHMS,” considers only methods and data sets that permit the creation of a reference that will be identical for each run. This is the best possible benchmark for developers to support their algorithm improvement claims and for advanced users interested in producing their own reference database after selecting the most efficient method.
Evaluation of recent methods
The release of the LEMMI benchmarking platform described here (beta01.20191118) includes the well-established methods Centrifuge (Kim et al. 2016), Kraken (Wood and Salzberg 2014) and Kraken 2 (Wood et al. 2019), along with Bracken (Lu et al. 2017), Kaiju (Menzel et al. 2016), and MetaPhlAn2 (Truong et al. 2015). They are a good representation of the field as it is today, because they stand among methods evaluated together recently (Ye et al. 2019). This allows LEMMI users to judge additional methods never before evaluated side by side with tools they are already familiar with. We highlight on Figures 2 and 3 and Supplemental Figures S2–S5 and S7 a glimpse of the performances of the novel algorithms implemented by ganon (Piro et al. 2020), MetaCache (Müller et al. 2017), and CCMetagen (Marcelino et al. 2020), which are described in their supporting publications but currently absent from comparative benchmarks. We report overall very good performances in recall and precision at the species level for MetaCache and ganon. These tools perform well when compared to other solutions using freely selected references (Figs. 2, 3A; Supplemental Fig. S2A) as well as when compared to other tools using an identical built reference (Fig. 3B; Supplemental Figs. S2E, S3). They are also top scorers when taxonomic binning objectives are considered (Supplemental Figs. S2D,F, S4A,C). CCMetagen minimizes false positives but suffers from poor recall (Fig. 3A,B), except when analyzing data sets having a lower number of species (Supplemental Fig. S5). As LEMMI minor releases are continuously produced, the current list of methods and databases (Supplemental Table S1) will rapidly be enriched. To appreciate the full extent of the results through detailed plots, we invite readers to visit the LEMMI web platform (https://lemmi.ezlab.org).
Evaluation of different references with Kraken 2
Taking advantage of the flexible reference selection in LEMMI, we investigated the improvement of the Kraken algorithm in its second version (Wood et al. 2019) regarding its ability to build large references without using a considerable amount of memory. Kraken 2 is among the most memory-efficient methods evaluated in LEMMI to date when building the medium-sized representative reference (RefSeq/08.2018/1rep.) used for all tools (Fig. 3C) or the largest reference (RefSeq/08.2018/all) (Supplemental Fig. S7). Kraken 2 combined with Bracken 2 remains among the best methods according to several metrics and is the top scorer in relative abundance estimation (Figs. 2, 3; Supplemental Fig. S2A,B,D). We then compared the most comprehensive reference built in LEMMI with one of Kraken's prebuilt databases: Minikraken. This 8 GB reference is intended for low memory environments and created by subsampling k-mers from RefSeq “Complete genomes” sequences only. The diversity of data sets offered by LEMMI shows that good performances with Minikraken depend almost entirely on the source of the in silico reads being complete genome sequences. Otherwise, Minikraken ignores the large part of the diversity proxied by NCBI species taxids that have no complete genome representatives, which is likely present in real samples. This can be observed when analyzing CAMI1 data sets, in which a large proportion of taxa is overlooked, whereas Kraken 2 can actually recover them when using the comprehensive LEMMI/RefSeq repository as reference (Supplemental Fig. S8).
Discussion
Here, we introduce the first-of-its-kind, continuous integration benchmarking platform for metagenomics classifiers to enable immediate and independent assessment of any newly published method using data sets of various compositions that can be generated or reused from previous effort. A key advantage of LEMMI is that the method and not the result of the computation is submitted for evaluation, enabling assessment under identical conditions and subsequent reevaluations on new data. It also proves that the tool can easily function outside the developer's environment. Thanks to this, LEMMI envisions a sustainable life cycle: It maintains a benchmark unchanged for a certain period of time to allow many methods to be integrated, and then migrates all the content to an updated version to meet the new expectations of the field. We are planning to produce a new major release of LEMMI every 2 yr to renew the data sets, for example, to support long read technologies, to integrate newly discovered taxa or covering currently unsupported clades, and above all to make the benchmark design evolve. For instance, although the initial release presented in this publication uses the NCBI taxonomy (Federhen 2012) and has put the focus on evaluating the lowest supported ranks, that is, genus and species, the pipeline is designed to integrate alternative ranks or taxonomies if future method implementations embrace them. There are calls to revise NCBI taxonomy (Parks et al. 2018, 2020) and future tools could be more agnostic regarding taxonomic systems (i.e., configuring the ranks and identifiers, instead of having the NCBI taxonomy entangled with the core classification algorithm). This would enable the exploration of new approaches meant to improve the classification resolution, for example, reaching bacterial strains or viral operational taxonomic units (Paez-Espino et al. 2019). Moreover, because LEMMI is an approach that avoids manual intervention, it encourages developers with different ideas to agree on comparable outputs besides standardized file formats. Post-processing and filtering procedures can be explored as parameters during the benchmarking. Feedback and exchanges within the community of method developers and users will be important aspects of the LEMMI evolution and will help to define new directions and rules. When a major LEMMI release occurs, methods that are still valuable will be reevaluated as they stand if their developers are no longer active, or will be replaced by an updated container otherwise (Supplemental Fig. S1B). Developers interested in submitting their tools can visit https://lemmi.ezlab.org, where documentation, support, a discussion board, and evaluated containers are available. Future extensions of LEMMI may also include a standalone version to allow private assessment and help with development before submitting to the public platform for an independent evaluation. At all times, LEMMI ensures a traceable archive of past rankings through the use of unique “fingerprints” to be reported in publications.
In addition to presenting the pipeline, we highlight the high potential of some recently published methods that had not previously been evaluated independently. MetaCache and ganon are efficient methods constituting an alternative to Kraken 2 combined with Bracken 2. When methods are evaluated using an identical reference of one representative per species, ganon is ranked first in accurately predicting the species composition, whereas MetaCache has the best binning performances. This makes them worth considering as alternatives to other solutions, especially if their specific innovations, for instance, the ability of ganon to incrementally update a reference database without full reconstruction, are beneficial to the user. However, Kraken 2 is definitely the most memory-efficient tool and, with the Bayesian reestimation offered by Bracken 2, remains the best solution among those compared for relative abundance estimation. We also show, by comparing Minikraken with other references, the importance of considering all assembly states issued from RefSeq to avoid missing most of the diversity available in public databases. Therefore, better subsampling strategies than simply keeping complete genome sequences should be explored by database creators to include the whole available genomic diversity while limiting the number of genomes to mitigate the constraints of insufficient computational resources. Moreover, to assess the true ability of methods to scale with the ever-growing diversity of sequenced taxa, future benchmarking efforts should not rely exclusively on complete genomes (Ye et al. 2019), because a substantial fraction of assemblies are deposited in databases as draft genomes (Supplemental Note S1).
Benchmarking has become a must-have requirement for publishing novel methods. To bring credibility and facilitate adoption by their target audience, it is essential that new methods appear rapidly side by side with established competitors in a trusted independent ranking. LEMMI encourages developers to consider biocontainers to disseminate their work and to standardize the results formats so users can obtain easy-to-use and stable implementations of up-to-date methods as they appear in the benchmark.
Methods
Release strategy
LEMMI rankings are identified by the major release version of the platform (e.g., beta01) and the minor release date (e.g., 20191118). Until the next major release, the content of the LEMMI/RefSeq repository, the version of the NCBI taxonomy, and the data sets are frozen to a specific version that guarantees exact comparisons. Continuous additions or withdrawals of an entry in the rankings generate a new minor release date, while previous rankings remain accessible by requesting old fingerprints (e.g., https://lemmi.ezlab.org/#/rankings/SD.DEFAULT.beta01.20191118).
Structure of the pipeline
An in-house Python3-based (McKinney 2010; Oliphant 2015) controller coordinates the many subtasks required to generate data sets, run the candidate containers, and compute the statistics. Snakemake 5.3.1 (Koster and Rahmann 2012) is used to supervise individual subtasks such as generating a data set or running one evaluation. The process is semiautomated through configuration files, designed to allow a potential full automation through a web application. To be easily deployable, the benchmarking pipeline itself is wrapped in a Docker container. The plots presented on the user interface are generated with the mpld3 library (https:// mpld3.github.io) (Hunter 2007).
LEMMI containers
The LEMMI containers are implemented for Docker 18.09.0-ce. They partially follow the design (Belmann et al. 2015) introduced by http://bioboxes.org/ as part of the first CAMI challenge effort. The required output files are compatible with the profiling and binning format created for the CAMI challenge. Two tasks have to be implemented to generate a reference and conduct an analysis (Supplemental Fig. S1A). To take part in the benchmark, a method developer has to build the container on their own environment, while ensuring that both tasks can be run by an unprivileged user and return the desired outputs. A tutorial is available on https://www.ezlab.org/lemmi_documentation.html. The containers or the sources to recreate them are made available to the users. The sources of all method containers presented as results in this study are available on GitHub (https://gitlab.com/ezlab/lemmi/tree/beta01.20191118/containers).
Computing resources
During the benchmarking process, the container is loaded on a dedicated server and given 245 GB of RAM and 32 cores. Reaching the memory limit will cause the container to be killed, ending the benchmarking process unsuccessfully. All inputs and outputs are written on a local disk, and the container is not given access to the Internet.
Taxonomy
The NCBI taxonomy is used to validate all entries throughout the process, and unknown taxids are ignored (unclassified). The framework etetoolkit (ETE3) (Huerta-Cepas et al. 2016) is used to query the taxonomy. The database used in beta01 was downloaded on March 9, 2018, and remains frozen to this version until a new major release of the LEMMI platform.
RefSeq repository
All RefSeq assemblies for bacteria, archaea, and viruses were downloaded from https://ftp.ncbi.nlm.nih.gov/refseq/release/ (download date for the beta01 release was August 2018) with the conditions that they contained both a protein and a nucleotide file and that their taxid has a corresponding entry in the ETE3 NCBI taxonomy database, for a total of 132,167 files of each sequence type. The taxonomic lineage for the seven main levels was extracted with ETE3 (superkingdom, phylum, class, order, family, genus, species). Viruses were not used for generating references and data sets in the current release, and the number of genomes in the reference RefSeq/08.2018/all is 124,289. To subset the repository and keep one representative per species as inputs for the reference construction, the list of bacterial and archaeal genomes was sorted according to the assembly states (1:Complete Genome, 2:Chromosome, 3:Scaffold, 4:Contig) and the first entry for each species taxid was retained, for a total of 18,907 files. When subsampling the repository in the genome exclusion mode (i.e., for the METHOD ALGORITHMS category) (Supplemental Fig. S9), if the entry to be selected was part of the reads, the next representative in the list was used instead when available.
LEMMI data sets
To sample the genomes included in the LEMMI data sets, a custom Python script was used to randomly select representative genomes in the LEMMI/RefSeq assembly repository, among bacterial and archaeal content (Supplemental Table S2). Their abundance was randomly defined following a lognormal distribution (mean = 1, standard deviation) (Supplemental Table S2). In the case of LEMMI_LOWDIV data sets, additional low coverage species (abundance corresponding to <100 reads) were manually defined, but in LEMMI_MEDDIV data sets, low coverage species were produced as part of the random sampling procedure. Each species abundance was normalized according to the species average genome size (as available in the LEMMI/RefSeq repository) to get closer to organisms’ abundances (considering one genome copy per cell), and the total was normalized to one to constitute a relative abundance profile. Therefore, both tools classifying all reads and those using marker genes can normalize their output to provide a unified answer. BEAR (Johnson et al. 2014) was used to generate paired-end reads, 2 × 150 bp, and DRISEE (Keegan et al. 2012) was used to extract an error profile from the NCBI Sequence Read Archive (SRA) entry ERX2528389 to be applied onto the generated reads. This approach, while biased toward that particular sample, was preferred to a simulation to model the artificial reads with an error pattern coming from a real experiment. The ground truth profile for the seven taxonomic ranks and taxonomic bins for species and genus were kept. The nonunique 50-mers and 31-mers diversity of the obtained reads were generated with Jellyfish 2.2.8 (Marçais and Kingsford 2011) on the concatenated pair of reads using the following parameters: jellyfish count -m 31 -s 3G ‐‐bf-size 5G -t 8 -L 1 reads.fq.
Additional data sets
The CAMI1 data sets were obtained from https://data.cami-challenge.org/ (accessed September 2018) along with the metadata describing their content, already in the expected file format. The binning details were reprocessed to obtain distinct lists at the species and genus rank. CAMI data sets are in silico sequencing reads sampled from mock metagenomes generated using real sequenced bacteria, archaea, viruses, plasmids, and other circular elements that were new at the time of the challenge and subsequently made publicly available (Sczyrba et al. 2017). The mockrobiota-17 data set (Kozich et al. 2013; Bokulich et al. 2016) was obtained through GitHub (https://github.com/caporaso-lab/mockrobiota) and reprocessed to obtain a taxonomic profile in the appropriate format. It is composed of even amounts of genomic DNA from 21 bacterial strains. No binning detail is available for this data set; therefore, no assessment of this aspect is based on this data set. Diversity of 50-mers and 31-mers were computed as detailed above.
Analysis of the results
The profile and binning reports are processed with OPAL 0.2.8 (Meyer et al. 2019) and AMBER 0.7.0 (Meyer et al. 2018) against the ground truth to obtain a wide range of metrics. Binning reports are processed to obtain a file for each taxonomic rank (genus/species), moving reads up from lowest levels. The profiles of the candidate methods and the ground truth are filtered to discard low coverage taxa at different thresholds (below values corresponding to 1/10/100/1000 reads), and all metrics are computed for all values. When a container is not able to provide a profile as output, the LEMMI platform generates one using the proportion of reported reads. Taxa detection metrics are based on OPAL and thus on the profile output. Methods reporting a profile with a zero score for low abundance taxa despite being present in their binning files will shift the balance from recall to precision. The low abundance score takes into account both the profile and binning output and is a custom metric calculated separately to evaluate the ability of the method to correctly identify organisms represented with very low read coverage (<100 reads). However, it penalizes methods likely to recover these organisms by recurrent reporting of the same taxids owing to very poor precision. Because false positives always have a true abundance of zero, it is not possible in a given data set to filter low abundance false positives to compute them against low abundance true positives and obtain a precision restricted to these taxa. A different approach has to be used. The metric is computed by pairing two data sets to judge whether a prediction can be trusted. As illustrated with Supplemental Figure S10, the data sets D1 and D2 contain a subset of low abundance taxa (D1_low, D2_low, <100 reads coverage). When computing the metric on predictions made by a method on D1, all correctly identified taxa belonging to D1_low increase the low abundance score for that method. However, if one of these taxa is also predicted in D2 when it does not exist in D2, the method is not reliable in detecting this taxon, and this decreases the low abundance score for D1. This is a proxy for low abundance precision. The metric is also applied from the D2 perspective, D2_low being validated through the same process against D1. Thus, both paired data sets provide a low abundance score between 0.0 and 1.0. In the current release of LEMMI, this metric is only defined for the pair of LEMMI_LOWDIV and the pair of LEMMI_MEDDIV data sets (low abundance species: n = 10, n = 8, n = 98, and n = 138 for LEMMI_LOWDIV_001, LEMMI_LOWDIV_002, LEMMI_MEDDIV_001, and LEMMI_MEDDIV_002, respectively). The run time corresponds to the time in minutes during which the container is loaded. The memory is the peak value of total RSS memory reported when the container is loaded to complete one task. Methods unable to deliver part of the expected outputs are assigned 0.0 for the corresponding metrics (e.g., methods unable to provide a read binning report).
Ranking score calculation
Metrics are averaged over all data sets. Values that are not already between 0.0 and 1.0, with 1.0 being the best score, are transformed. The details of these transformations are presented in Supplemental Table S4. Any resulting metric below 0.0 or above 1.0 is set to be 0 (in practice, 10−5 to prevent divisions by zero) and 1.0, respectively. Each value is calculated for genus and species at 1, 10, and 100 reads low coverage filtering level and used or ignored according to the choice of the LEMMI user regarding these parameters. The final score displayed in the ranking is the harmonic mean of all retained metrics, taken into account 0, 1, or 3 times, depending on the weight assigned to the metric by the LEMMI user. Harmonic mean is preferred to arithmetic mean to combine quantities with different measures in the same way that the F1-score averages precision and recall.
Data access
All containers representing evaluated methods are available on https://lemmi.ezlab.org and as Supplemental Code. The LEMMI demo data set is available in the Zenodo repository (https://zenodo.org/record/2651062).
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank all members of the Zdobnov group for discussions and feedback and Maxime Arbez for his help. This work was supported by the Swiss National Science Foundation funding 31003A_166483 and 310030_189062 to E.M.Z.
Author contributions: M.S. and E.M.Z. conceived the study. M.S. coded the platform. M.S. and M.M. conducted the analyses. M.S. and M.M. wrote the documentation. M.S., M.M., and E.M.Z. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.260398.119.
Freely available online through the Genome Research Open Access option.
References
- Belmann P, Dröge J, Bremges A, McHardy A, Sczyrba A, Barton M. 2015. Bioboxes: standardised containers for interchangeable bioinformatics software. GigaSci 4: 47 10.1186/s13742-015-0087-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokulich N, Rideout J, Mercurio W, Shiffer A, Wolfe B, Maurice C, Dutton R, Turnbaugh P, Knight R, Caporaso J. 2016. mockrobiota: a public resource for microbiome bioinformatics benchmarking. mSystems 1: e00062-16 10.1128/mSystems.00062-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremges A, McHardy A. 2018. Critical assessment of metagenome interpretation enters the second round. mSystems 3: e00103-18 10.1128/mSystems.00103-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S, de la Iglesia D, Haas J, Lourenco A, Fernández J, Repchevsky D, Dessimoz C, Schwede T, Notredame C, Gelpi J, et al. 2017. Lessons learned: recommendations for establishing critical periodic scientific benchmarking. bioRxiv 10.1101/181677 [DOI]
- da Veiga Leprevost F, Grüning B, Alves Aflitos S, Röst H, Uszkoreit J, Barsnes H, Vaudel M, Moreno P, Gatto L, Weber J, et al. 2017. BioContainers: an open-source and community-driven framework for software standardization. Bioinformatics 33: 2580–2582. 10.1093/bioinformatics/btx192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federhen S. 2012. The NCBI Taxonomy database. Nucleic Acids Res 40: D136–D143. 10.1093/nar/gkr1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P, Watson R, Morgan X, Draper J, Finn R, Morales S, Stott M. 2019. Identifying accurate metagenome and amplicon software via a meta-analysis of sequence to taxonomy benchmarking studies. PeerJ 7: e6160 10.7717/peerj.6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33: 1635–1638. 10.1093/molbev/msw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng 9: 90–95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Johnson S, Trost B, Long J, Pittet V, Kusalik A. 2014. A better sequence-read simulator program for metagenomics. BMC Bioinformatics 15: S14 10.1186/1471-2105-15-S9-S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan K, Trimble W, Wilkening J, Wilke A, Harrison T, D'Souza M, Meyer F. 2012. A platform-independent method for detecting errors in metagenomic sequencing data: DRISEE. PLoS Comput Biol 8: e1002541 10.1371/journal.pcbi.1002541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Song L, Breitwieser F, Salzberg S. 2016. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res 26: 1721–1729. 10.1101/gr.210641.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J, Rahmann S. 2012. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Kozich J, Westcott S, Baxter N, Highlander S, Schloss P. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79: 5112–5120. 10.1128/AEM.01043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen S, Adair K, Gardner P. 2016. An evaluation of the accuracy and speed of metagenome analysis tools. Sci Rep 6: 19233 10.1038/srep19233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Breitwieser F, Thielen P, Salzberg S. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci 3: e104 10.7717/peerj-cs.104 [DOI] [Google Scholar]
- Maier-Hein L, Eisenmann M, Reinke A, Onogur S, Stankovic M, Scholz P, Arbel T, Bogunovic H, Bradley A, Carass A, et al. 2018. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat Commun 9: 5217 10.1038/s41467-018-07619-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangul S, Martin L, Hill B, Lam A, Distler M, Zelikovsky A, Eskin E, Flint J. 2019a. Systematic benchmarking of omics computational tools. Nat Commun 10: 1393 10.1038/s41467-019-09406-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangul S, Martin L, Eskin E, Blekhman R. 2019b. Improving the usability and archival stability of bioinformatics software. Genome Biol 20: 47 10.1186/s13059-019-1649-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27: 764–770. 10.1093/bioinformatics/btr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino VR, Clausen PTLC, Buchmann JP, Wille M, Iredell JR, Meyer W, Lund O, Sorrell TC, Holmes EC. 2020. CCMetagen: comprehensive and accurate identification of eukaryotes and prokaryotes in metagenomic data. Genome Biol 21: 103 10.1186/s13059-020-02014-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre A, Ounit R, Afshinnekoo E, Prill R, Hénaff E, Alexander N, Minot S, Danko D, Foox J, Ahsanuddin S, et al. 2017. Comprehensive benchmarking and ensemble approaches for metagenomic classifiers. Genome Biol 18: 182 10.1186/s13059-017-1299-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney W. 2010. Data structures for statistical computing in Python. In Proceedings of the Ninth Python in Science Conference (ed. van der Walt S, et al.), pp. 56–61. SciPy 2010, Austin, TX. 10.25080/Majora-92bf1922-00a [DOI] [Google Scholar]
- Menzel P, Ng K, Krogh A. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7: 11257 10.1038/ncomms11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Hofmann P, Belmann P, Garrido-Oter R, Fritz A, Sczyrba A, McHardy A. 2018. AMBER: Assessment of Metagenome BinnERs. Gigascience 7: giy069 10.1093/gigascience/giy069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Bremges A, Belmann P, Janssen S, McHardy A, Koslicki D. 2019. Assessing taxonomic metagenome profilers with OPAL. Genome Biol 20: 51 10.1186/s13059-019-1646-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A, Hundt C, Hildebrandt A, Hankeln T, Schmidt B. 2017. MetaCache: context-aware classification of metagenomic reads using minhashing. Bioinformatics 33: 3740–3748. 10.1093/bioinformatics/btx520 [DOI] [PubMed] [Google Scholar]
- Nasko D, Koren S, Phillippy A, Treangen T. 2018. RefSeq database growth influences the accuracy of k-mer-based lowest common ancestor species identification. Genome Biol 19: 165 10.1186/s13059-018-1554-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norel R, Rice J, Stolovitzky G. 2011. The self-assessment trap: Can we all be better than average? Mol Syst Biol 7: 537 10.1038/msb.2011.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary N, Wright M, Brister J, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T. 2015. Guide to NumPy, 2nd ed. CreateSpace Independent Publishing Platform, Scotts Valley, CA. [Google Scholar]
- Paez-Espino D, Roux S, Chen I, Palaniappan K, Ratner A, Chu K, Huntemann M, Reddy T, Pons J, Llabrés M, et al. 2019. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res 47: D678–D686. 10.1093/nar/gky1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D, Chuvochina M, Waite D, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36: 996–1004. 10.1038/nbt.4229 [DOI] [PubMed] [Google Scholar]
- Parks D, Chuvochina M, Chaumeil PA, Rinke C, Mussig A, Hugenholtz P. 2020. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat Biotechnol. 10.1038/s41587-020-0501-8 [DOI] [PubMed] [Google Scholar]
- Peabody M, Van Rossum T, Lo R, Brinkman F. 2015. Evaluation of shotgun metagenomics sequence classification methods using in silico and in vitro simulated communities. BMC Bioinformatics 16: 362 10.1186/s12859-015-0788-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piro VC, Dadi TH, Seiler E, Reinert K, Renard BY. 2020. ganon: precise metagenomics classification against large and up-to-date sets of reference sequences. Bioinformatics 36: i12–i20. 10.1093/bioinformatics/btaa458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczyrba A, Hofmann P, Belmann P, Koslicki D, Janssen S, Dröge J, Gregor I, Majda S, Fiedler J, Dahms E, et al. 2017. Critical Assessment of Metagenome Interpretation—a benchmark of metagenomics software. Nat Methods 14: 1063–1071. 10.1038/nmeth.4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong D, Franzosa E, Tickle T, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12: 902–903. 10.1038/nmeth.3589 [DOI] [PubMed] [Google Scholar]
- Wood D, Salzberg S. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15: R46 10.1186/gb-2014-15-3-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20: 257 10.1186/s13059-019-1891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Siddle K, Park D, Sabeti P. 2019. Benchmarking metagenomics tools for taxonomic classification. Cell 178: 779–794. 10.1016/j.cell.2019.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.