Abstract
Despite the growing resources and tools for high-throughput characterization and analysis of genomic information, the discovery of the genetic elements that regulate complex traits remains a challenge. Systems genetics is an emerging field that aims to understand the flow of biological information that underlies complex traits from genotype to phenotype. In this study, we used a systems genetics approach to identify and evaluate regulators of the lignin biosynthesis pathway in Populus deltoides by combining genome, transcriptome, and phenotype data from a population of 268 unrelated individuals of P. deltoides. The discovery of lignin regulators began with the quantitative genetic analysis of the xylem transcriptome and resulted in the detection of 6706 and 4628 significant local- and distant-eQTL associations, respectively. Among the locally regulated genes, we identified the R2R3-MYB transcription factor MYB125 (Potri.003G114100) as a putative trans-regulator of the majority of genes in the lignin biosynthesis pathway. The expression of MYB125 in a diverse population positively correlated with lignin content. Furthermore, overexpression of MYB125 in transgenic poplar resulted in increased lignin content, as well as altered expression of genes in the lignin biosynthesis pathway. Altogether, our findings indicate that MYB125 is involved in the control of a transcriptional coexpression network of lignin biosynthesis genes during secondary cell wall formation in P. deltoides.
Uncovering the genetic elements that regulate complex traits is a critical but still challenging goal of biology. Genome-wide association (GWA) analysis has become the most common approach to unravel the relationship between genotype and phenotype. GWA studies have identified numerous DNA polymorphisms associated with complex traits in humans, plants, and animals (Porth et al. 2013; Sonah et al. 2015; Fahrenkrog et al. 2017a; Tieman et al. 2017; Turuspekov et al. 2017; Zhang et al. 2018; Furches et al. 2019; Guerra et al. 2019). These studies largely validated the early quantitative genetics theory that complex traits are influenced by many small-effect variants (Visscher et al. 2017) but also exposed the statistical power limitation of the approach. Furthermore, phenotype–DNA association studies do not account for many factors that may contribute to phenotypic variation by changes in gene expression (Vazquez et al. 2016). Integrating transcriptome data to the analysis of complex traits may contribute to a better understanding of what determines a phenotype, as well as targets to modify them.
Single-nucleotide polymorphisms (SNPs) associated with complex phenotypes are enriched for variants that affect gene expression (Nicolae et al. 2010). Gene expression regulation itself is largely genetically controlled and heritable, as first described in yeast (Brem 2002) and later in several plant species (Schadt et al. 2003; Kirst et al. 2005; West et al. 2007). Therefore, transcript abundance can be considered a complex, quantitative phenotype and can be subjected to association mapping to identify polymorphisms responsible for transcript abundance variation among individuals and gene expression regulation. In this context, polymorphisms associated with gene expression are referred to as expression quantitative trait loci (eQTL) and are classified as local (putative cis) and distant (putative trans), depending on their physical location relative to the gene whose expression is affected. To date, most eQTL studies in plants have been conducted on relatively small, closely related, and unreplicated populations. This limits the power to detect variants affecting gene expression in cis and, to a greater extent, in trans, because they usually have smaller effects (Holloway et al. 2011; Wang et al. 2014; Ranjan et al. 2016; Mähler et al. 2017). Hence, the use of a small sample size in eQTL studies has hampered the discovery of variants that contribute to gene expression regulation and further identification of potential master regulators of biosynthetic pathways and metabolic networks.
Integrative systems analysis, such as systems genetics, is an emerging field that aims to decipher biological networks and pathways involved in complex traits and map their genetic variants (Ayroles et al. 2009; Civelek and Lusis 2014; Moreno-Moral and Petretto 2016). The starting point of a systems genetics approach is the identification of polymorphisms modulating gene expression. Once genes that are under genetic control are identified, their respective gene regulatory networks are further explored to obtain a deeper understanding of the mechanisms underlying complex traits. One of the most common approaches is the use of genome-wide transcript abundances to construct gene coexpression networks, as coexpressed genes often function in the same pathway (Mackay 2014). By coupling information from coexpression networks and eQTL studies, it is possible to develop hypotheses regarding the regulatory function of genes in a given pathway. This leads to the systematic unraveling of key genetic drivers of biosynthetic pathways or metabolic networks underlying complex traits. Integration of information from different layers of omics data has been shown to improve the understanding of complex phenotypes in humans (Vazquez et al. 2016; Hasin et al. 2017; Bakker et al. 2018; Qin et al. 2019) and some plant species (Acharjee et al. 2016; Chauffour et al. 2019).
In this study, we used a systems genetics approach to identify regulators of the lignin biosynthesis pathway in Populus deltoides. Lignin is a complex phenolic polymer derived from the phenylpropanoid pathway that provides structural support to plants. Lignin is also the major contributor to the recalcitrance of biomass (Studer et al. 2011), which considerably increases its processing cost and, consequently, hinders biofuel production. Because of the vast industrial and biological importance of lignin, uncovering the elements of its biosynthesis pathway and regulation is critical for industrial and agricultural purposes. To date, the genes encoding for enzymes in the lignin biosynthesis pathway have been largely identified (Hefer et al. 2015; Wang et al. 2018). Studies in model systems have revealed that the expression of these genes is primarily under the coordinated control of secondary cell wall (SCW) NAC-mediated and R2R3-MYB–mediated transcriptional network (Xie et al. 2018). Several R2R3-MYB transcription factors playing important roles in regulating secondary wall thickening have been identified and well characterized in Arabidopsis (Xie et al. 2018). It is hypothesized that in perennial woody species, such as poplar, a similar but more complicated regulatory mechanism of the lignin biosynthesis pathway exists. For instance, the Populus genome has at least 192 annotated genes encoding R2R3-MYB transcription factors (Wilkins et al. 2009), yet only a few members of this family have been functionally characterized (Wilkins et al. 2009; McCarthy et al. 2010; Zhong et al. 2011; Tian et al. 2013; Zhong and Ye 2014; Li et al. 2015; Petzold et al. 2018a; Gui et al. 2019, 2020). Therefore, the complete genetic networks underlying the control of the lignin biosynthesis pathway for Populus and other commercially important woody species remain largely unknown.
Here we used a system genetics framework to uncover the gene expression network implicated in the control of lignin biosynthesis in Populus. First, we surveyed the genetic regulation of gene expression in differentiating xylem of a genetically unrelated population of P. deltoides using an eQTL approach. Next, we integrated information from eQTL and gene coexpression network to dissect the genetic regulation of the lignin trait.
Results
A large fraction of the xylem transcriptome of P. deltoides is highly heritable
To obtain a comprehensive understanding of the global profile of the P. deltoides xylem transcriptome, we sequenced paired-end, 150-bp RNA-seq libraries from 343 individuals. This population shows low levels of subpopulation differentiation (FST = 0.022–0.106), high genetic diversity (θW = 0.00100, Π = 0.00170), a large effective population size (Ne ≈ 32,900), and low to moderate levels of linkage disequilibrium (LD) (Fahrenkrog et al. 2017a). These individuals were previously genotyped with 68,885 SNP markers by targeted resequencing of 18,153 genes and 23,835 intergenic regions (Fahrenkrog et al. 2017b). In total, 23.1 billion reads were generated with an average of 23.4 million reads per sample. To be considered for further analysis, each genotype was required to have at least 15 million reads for at least two of the three biological replicates, resulting in 268 individuals being used for downstream analysis. Next, the raw reads were filtered and mapped to the Populus trichocarpa v3 reference genome. The mapped reads were independently assembled using alternative approaches (see Methods), and the transcripts constructed by each assembler were grouped into a single transcriptome file, which resulted in 18,207 genes composed of 52,708 transcripts expressed in the population. Over 92% of the genes expressed in the population are present in the P. trichocarpa v3 reference genome annotation. Therefore, 1709 transcriptional units represent potentially novel, unannotated open reading frames composed of 1914 unannotated transcripts.
We used mixed-effects models to estimate the broad-sense heritability (H2) of each of the 18,207 expressed genes (Supplemental Fig. S1). Heritability estimates ranged from 0.0 to 0.88 with a mean (±SD) of 0.15 (±0.13). The experimental design explained on average 12.3% of the variance in FPKM. However, the percentage of variance in gene expression varied among genes (10th percentile 3.5%, 90th 23.2%). In this data set, genetic structure is obvious in the first two principal components, which group the genotypes according to their geographic origin (Fahrenkrog et al. 2017b). The first five principal components accounted for 9.9% of the total sum of the eigenvalues of the genomic relationship matrix. The proportion of variance of gene expression accounted for by these five principal components was on average (i.e., across genes) 2.2%, with a 10th–90th percentile range of 0.5%–4.4%.
Permutation analyses showed that 12,579 genes had H2 estimates greater than what was expected by chance (P-value < 0.01) (Supplemental Fig. S1). Analysis of Gene Ontology (GO) of the genes with high heritability (H2 > 0.5) showed enrichment for GO categories such as programed cell death (PCD; GO: 0012501, P-value = 3.6 × 10−7) (Supplemental Table S1).
Distant-eQTLs explain a higher proportion of the genetic variance for gene expression
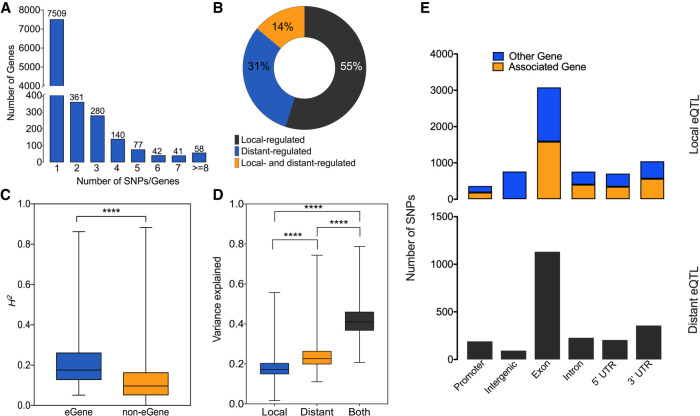
We decomposed the variation of gene expression in local and distant components to quantify the proportion of gene expression variance explained by each category. The local relationship matrix was built using an average of 404 SNPs, whereas the distant relationship matrix used an average of 68,480 SNPs per gene. The majority of the SNPs explained individually a small fraction of the phenotypic (gene-expression) variance, particularly for the distant SNPs (Supplemental Fig. S2A,B). However, the aggregate effect of the distant SNPs associated to each gene was larger than the local SNPs (mean 1.35-fold more variance explained, P-value < 0.0001) (Fig. 1A). These results indicate that distant SNPs contribute more to the heritability of gene expression, even though their individual effects are generally very small.
Figure 1.
Characterization of eQTL associations. (A) Expression variance explained by all SNPs for each gene. (B) Number of associated genes per SNP in significant eQTL associations. (C) The distribution of genes regulated by local and/or distant SNPs. (D) Broad sense heritability for genes with and without significant eQTL associations. (E) Genomic location of SNPs with significant eQTL associations. Promoters are defined as 500 bp upstream of the gene. Data were analyzed using Kruskal–Wallis test followed by Dunn's multiple comparison test. Significance is indicated by asterisks: (****) P-value < 0.0001.
To dissect the genetic regulation of gene expression, we performed an eQTL analysis of all genes with H2 estimates greater than expected by chance (P-value < 0.01). We detected a total of 11,334 eQTLs at a 5% empirical FDR. These significant eQTLs represent pairwise associations between the expression of 5533 genes and 8470 SNPs, with an average of 1.5 significant SNPs per gene (Fig. 1B). For >89% of the genes, a single SNP was detected in association with their expression. We classified the eQTL based on the physical distance between the SNP and the associated gene as local (if the SNP is located within the gene or up to 1 Mb upstream of or downstream from the transcript start site [TSS] of the gene), and distant otherwise. According to this classification, 6706 (59%) were local-eQTLs and 4628 (41%) were distant-eQTLs. Three thousand twenty genes had only distant-eQTLs, 1738 genes had only local-eQTLs, and 775 genes were regulated by both distant and local SNPs (Fig. 1C). On average, genes with significant association showed higher heritability than heritable genes for which no significant association was found (P < 0.0001) (Fig. 1D).
Next, we considered the genomic location of the SNPS with significant association. The distribution in the genome of SNPs (local and distant) with significant association and the distribution of all the SNPs were similar, with the majority of the SNPs located within the coding sequence of a gene (Fig. 1E; Supplemental Fig. S3; Supplemental Table S2). This result was expected because the majority of the probes (89%) used to genotype the population was designed to capture 18,153 genes (Fahrenkrog et al. 2017a). Moreover, we observed that local associations had a proportionally higher number of SNPs located in intergenic regions (21%) compared with distant associations and all SNPs (4.2% and 4.1%, respectively) (Supplemental Table S2). Finally, we observed that approximately half of the local SNPs were located within the associated gene, whereas the remaining were within an adjacent gene.
Coexpression network analysis shows two modules enriched for genes in the lignin biosynthesis pathway
Global coexpression networks were constructed from all the 18,207 genes expressed in the population using a weighted correlation network analysis (WGCNA) (Langfelder and Horvath 2008). Overall, we identified 28 coexpression modules with an average size of 650 genes (min = 56, max = 2316) (Supplemental Fig. S4). We performed KEGG pathway enrichment analysis in all the modules and found that two of them were enriched for genes involved in the lignin biosynthesis pathway (modules in black, containing 656 genes, and in red, containing 803 genes in Supplemental Fig. S5). Together, these modules included 20 of the 31 genes that are preferentially expressed in xylem and active in the phenylpropanoid biosynthetic pathway (Hefer et al. 2015). Moreover, we also found that six genes from the shikimate and methionine salvage pathways, which provide the precursors for the lignin biosynthesis pathway, were present in both modules (Supplemental Table S3).
Next, we investigated how the black and red modules relate to each other using their eigengene value (i.e., the first principal component of the module). After eigengenes were clustered (Supplemental Fig. S6), the modules black and red were found to be grouped together, which indicates that they have similar expression patterns (Supplemental Fig. S6). Thus, we constructed a new coexpression network with only the genes present in these two modules. This resulted in five coexpression modules with the number of genes varying from 16 to 1138 (Supplemental Fig. S7). The largest module (colored turquoise) contained 18 out of the 20 genes of the lignin biosynthesis pathway present originally in the black and red modules (Supplemental Table S4). An alternative approach to investigate whether the genes in modules red and black are indeed correlated is to adjust the mergeCutHeight parameter in the analysis. In fact, when we applied the mergeCloseModules function, genes in modules black and red were grouped.
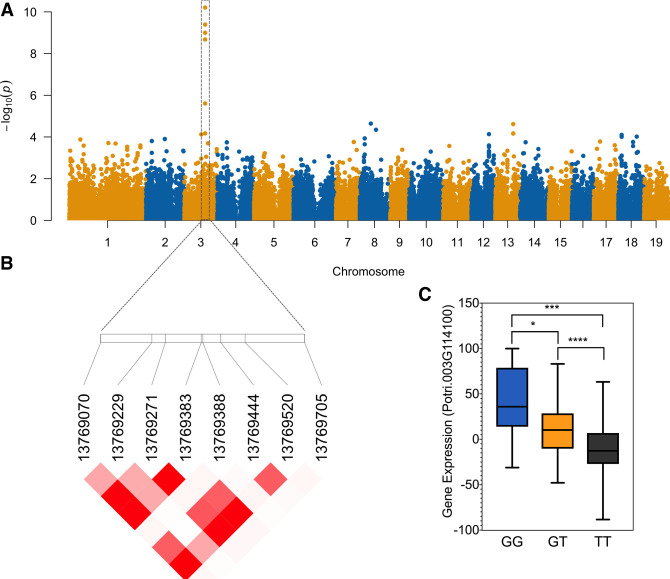
Following this, we selected the most connected genes of the turquoise module (Supplemental Fig. S7). These genes are expected to play a central role in this functional-based network. We considered genes with eigengene-based connectivities (kME) higher or equal to 0.7 to be the hub genes of the module and the most highly connected genes (Horvath 2011). By using this criterion, we identified 179 hub genes within the turquoise module (Supplemental Table S5). These included nine genes of the lignin biosynthesis pathway present in the turquoise module, as well as two genes from the shikimate pathway and two genes from the methionine salvage pathway. We further assessed if significant eQTL associations were detected for any of these genes and found that 10 of them had significant local-eQTL associations (Supplemental Table S5). Out of the 10 genes identified as being regulated by local SNPs, only MYB125 (Potri.003G114100) is annotated as a transcription factor. Because of its potential role in the expression regulation of other genes, we chose it for further analysis. We found five SNPs (Fig. 2A,B; Supplemental Table S6) to be significantly associated with the expression of MYB125 in cis (FDR < 0.05) (Fig. 2C; Supplemental Fig. S8). Out of the four SNPs located in the coding sequence, only one of them causes a nonsynonymous mutation, through a substitution of threonine (Thr) to alanine (Ala) in residue 255 of the protein (Supplemental Table S6). The pairwise LD pattern among the five SNPs shows that four of them (SNPs 1, 3, 4, and 5) are in almost complete LD (R2 > 0.97) (Supplemental Table S7). The fifth SNP (SNP 2) shows a lower LD (R2 varying from 0.26 to 0.28). The minor allele frequency (MAF) for this variant is 0.07, which is considerably lower compared with the MAF of the other four SNPs (MAF > 0.22) (Fig 2; Supplemental Table S6).
Figure 2.
Local SNPs regulate the expression of MYB125. (A) Manhattan plot displaying the eQTL results for MYB125. The significant SNPs associated with the expression of MYB125 are located within the coding and UTR sequences of MYB125. The x-axis shows the chromosome positions, and the y-axis shows the significance expressed as –log10. (B) Pattern of linkage disequilibrium in the region of MYB125 across the SNPs with significant association. Darker red indicates stronger correlation. (C) Boxplot for expression of MYB125 plotted as an effect of genotypes at the SNP with the smallest P-value for the eQTL association (for boxplot for expression of MYB125 plotted as an effect of genotypes of the other SNPs, see Supplemental Fig. S8). The horizontal line represents the median, and the vertical lines mark the range of the minimum and maximum values. Data were analyzed using ANOVA followed by Tukey's multiple comparison test. Significance is indicated by asterisks: (****) P-value < 0.0001; (***) P-value < 0.001; (*) P-value < 0.05.
MYB125 is a major trans-acting regulator of the lignin biosynthesis pathway
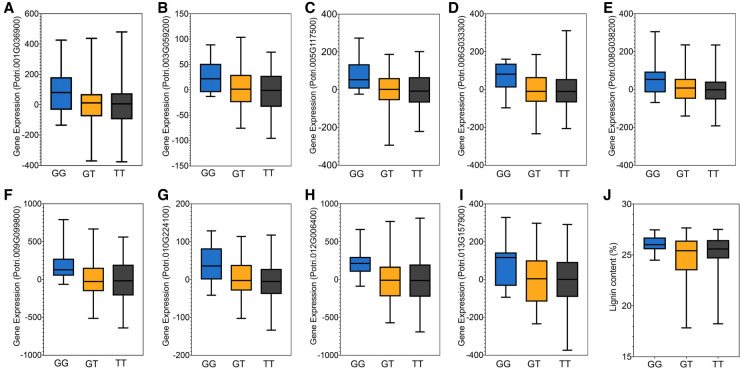
We used Cytoscape (Shannon 2003) to create a gene interaction network including all the genes present in the turquoise module (Supplemental Fig. S7), yielding a total of 713 nodes and 57,023 edges (Supplemental Fig. S9). MYB125 is directly connected to all nine genes that are part of the lignin biosynthesis pathway and identified as highly connected genes of the turquoise module (F5H indicates ferulic acid 5-hydrolase; C4H1, cinnamate-4-hydrolase 1; COMT1, O-methyltransferase 1; PAL4, phenylalanine ammonia-lyase 4; PAL2, phenylalanine ammonia-lyase 2; CCoACOMT1, caffeoyl-CoA 3-O-methyltransferase; C3′H, P-coumaroyl shikimate 3′-hydrolase; CSE, caffeoyl shikimate esterase; 4CL3, 4-coumarate:CoA ligase 3) (Supplemental Fig. S9). These results suggest that MYB125 may be a trans-acting regulator of these genes and, consequently, a key regulator of the lignin biosynthesis pathway. To investigate further, we individually tested the local-regulatory SNPs of MYB125 for trans-regulation of the nine genes of the lignin biosynthesis pathway that are directly connected to MYB125. The single marker regression analysis showed that all the nine genes are trans-regulated by the local-regulatory SNPs of MYB125 (P-value < 0.05) (Fig. 3A–I).
Figure 3.
MYB125 is a trans-acting regulator of genes from the lignin biosynthesis pathway. Single marker regression analysis between the local-regulatory SNP of MYB125 (with the smallest P-value) and the expression of (A) 4CL3 (P-value = 0.015); (B) CSE (P-value = 0.019); (C) F5H (P-value = 0.005); (D) C3′H (P-value = 0.003); (E) PAL2 (P-value = 0.014); (F) CCoAOMT1 (P-value = 0.015); (G) PAL4 (P-value = 0.003); (H) COMT1 (P-value = 0.003); and (I) C4H1 (P-value = 0.013). (J) Single marker regression analysis between the local-regulatory SNP of MYB125 (with the smallest P-value) and the lignin content percentage (P-value = 0.003). Boxplot for expression of genes from the lignin biosynthesis pathway (A–J) and lignin content percentage (I) plotted as an effect of genotypes at the SNP, with the smallest P-value, in the region of MYB125. The horizontal line represents the median, and the vertical lines mark the range of the minimum and maximum values.
Moreover, we found the expression of these nine genes to be positively correlated with the expression of MYB125 (R2 = 0.25–0.41, P-value < 0.0001) (Supplemental Fig. S10). We also tested the local-regulatory SNPs of MYB125 for trans-regulation of the other nine genes of the lignin biosynthesis pathway present in the turquoise module that are not directly connected to MYB125. Except for the gene cinnamyl alcohol dehydrogenase (Potri.009G095800), none of these other genes showed significant association with the local-regulatory SNPs of MYB125 (Supplemental Fig. S11). Finally, to validate the potential role of MYB125 as a key regulator of the lignin biosynthesis pathway, we performed a single marker regression between the local-regulatory SNPs of MYB125 and the lignin content percentage previously measured in this population (Fahrenkrog et al. 2017b). As expected, we observed a significant association between the local-regulatory SNPs of MYB125 and lignin content percentage (P-value = 0.003) (Fig. 3J). Accordingly, the lignin content percentage is also positively correlated with the expression of MYB125 (P-value = 0.03) (Supplemental Fig. S10).
MYB125 induces the expression of genes in the lignin biosynthesis pathway and increases the lignin content
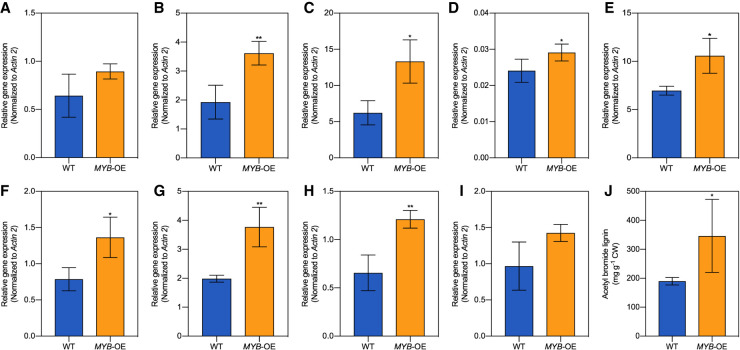
To verify the role of MYB125 in the regulation of lignin biosynthesis, we generated transgenic roots overexpressing MYB125 (Supplemental Fig. S12A). We performed qRT-PCR to determine the expression level of the genes in the lignin biosynthesis pathway shown to be possibly trans-regulated by MYB125 (Fig. 3, Supplemental Fig. S10). Of the nine genes, we confirmed eight genes (4CL3, CSE, F5H, C3′H, PAL2, CCoAOMT1, PAL4, and COMT1) to be significantly up-regulated in transgenic roots overexpressing MYB125, compared with WT roots (Fig. 4A–I).
Figure 4.
Overexpression of MYB125 activates the expression of genes in the lignin biosynthetic pathway. Relative expression levels of (A) C4H1; (B) CSE; (C) F5H; (D) C3′H; (E) PAL2; (F) CCoAOMT1; (G) PAL4; (H) COMT1; and (I) 4CL3. Relative transcript levels were quantified by RT-qPCR and normalized with the housekeeping gene Actin2. (J) Acetyl bromide lignin content of WT and transgenic roots overexpressing MYB125. Error bars, SD. One-tail Student's t-test was used to determine statistical significance; n = 3. Significance is indicated by asterisks: (*) P-value < 0.05; (**) P-value < 0.01.
To further confirm the role of MYB125 as a positive regulator of lignin biosynthesis, we measured the levels of acetyl bromide soluble lignin (ABSL) in both transgenic roots overexpressing MYB125 and WT roots. Indeed, all three biological replicates overexpressing MYB125 showed increased levels of ABSL content by 35% to 158% compared with WT roots (Fig. 4J). We observed a highly positive correlation (R2 = 0.83, P-value = 0.0118) between the expression of MYB125 and the lignin content (Supplemental Fig. S12B). These results support the hypothesis that MYB125 is a transcription activator of the lignin biosynthesis pathway during SCW formation.
Discussion
The genetic architecture of xylem gene expression in P. deltoides
In this study, we examined the xylem transcriptome of 268 genotypes from a population of unrelated individuals of P. deltoides that were previously genotyped (Fahrenkrog et al. 2017b). Our primary aim was to detect potential causal genes and regulatory networks of whole-plant traits, such as biomass productivity and wood quality. An initial analysis of the transcriptome showed that 18,207 genes were expressed; of which 1709 represent potentially novel, unannotated open reading frames. Transcription of a large proportion of these genes is heritable; 12,579 genes had heritabilities significantly higher than expected by chance. Among the genes with the highest heritability (H2 > 0.5, n = 172), we found significant enrichment for those involved in cell death. Xylogenesis, the formation of water-conducting vascular tissue, is a developmentally regulated process that begins with the division and expansion of cambial cells and concludes with their programmed cell death (Funada et al. 2016). This ontogenetic process is intimately connected with cambial cell differentiation and secondary genetic and physiological and self-destructive process (Danial and Korsmeyer 2004; Iakimova and Woltering 2017). Our data support that this developmental stage in xylogenesis is under significant genetic control in this population.
We assessed the genetic architecture of heritable gene expression in the P. deltoides population using an eQTL approach, in which we detected 11,334 significant associations. We identified more local- than distant-eQTLs, and an analysis of the genomic location of the SNPs with significant association showed that the majority of the SNPs were located within a gene. As mentioned previously, this result was expected because the majority of the probes used to genotype the population was designed to capture 18,153 genes (Fahrenkrog et al. 2017b). In addition, we showed that intergenic regions were the genomic features least represented in the analysis (in local SNPs, distant SNPs, and the whole set of SNPs), which again indicates a limitation of the genotyping method used to detect putative distant regulatory elements, such as enhancers. Although in lower number, we were still able to detect local-acting (21%) and distant-acting (4.2%) SNPs located in the intergenic regions. These results suggest a higher power in the detection of putative enhancers in cis-associations compared with the detection of enhancers that module the expression of a transcription factor, which in turn regulates the expression of another gene in trans. The large number of significant SNPs also supports the concept that gene expression is itself a complex trait. Furthermore, we found that distant-eQTLs explain significantly more of the variance of gene expression than local-eQTLs. Although in disagreement with previous studies (Keurentjes et al. 2007; Mähler et al. 2017), our results support the recent omnigenic model proposing that heritability of complex traits is primarily driven by weak distant-eQTLs, whose effects are mediated by peripheral genes that impact the expression of core genes (Liu et al. 2019). The disagreement between our findings and previous studies (Keurentjes et al. 2007; Mähler et al. 2017) is most likely owing to the population size. Most of the eQTL studies in plants have been performed on relatively small, closely related and unreplicated populations. This limits the power to detect distant-eQTLs, which usually have smaller effects. Based on our findings and in agreement with the omnigenic model (Liu et al. 2019), it appears that most of the heritability of gene expression is controlled by many trans (distant) eQTLs of small individual effects.
A system genetics approach reveals MYB125 as a potential regulator of the lignin biosynthesis pathway
Gene networks can be used to infer the functions of genes based on their close network neighbors (guilt by association). Furthermore, by combining gene network with eQTL data one can also find genetic variants responsible for the coregulation of genes in that network. If multiple genes appear to be regulated by the same genetic variant through a trans-acting eQTL, it may be possible to determine the “master regulator” by identifying the cis-eQTL causing the trans-regulation. This is possible because genes under trans-regulation of specific phenotype-associated genetic variants are often functionally connected with the pathway associated with the corresponding phenotype. In plants and animals, the identification of such “master regulators” is critical for the manipulation of complex traits either through genetic engineering or traditional breeding, as they profoundly impact developmental and regulatory pathways.
We used a system genetics approach in which we combined eQTL analyses with data from coexpression networks generated by transcriptome analysis of 268 individuals. The network analysis revealed two modules enriched for genes involved in the lignin biosynthesis pathway. Together, these modules contained 20 of the 31 genes of the lignin biosynthesis pathway expressed in P. deltoides xylem. The global regulation of a coexpression network can be assigned to a specific locus by identifying a common eQTL that controls most members of that network (Moreno-Moral and Petretto 2016). Because MYB125 is directly connected to nine genes contained within modules enriched for the lignin biosynthesis pathway and is regulated by a local SNP, we considered it a potential trans-acting regulator of the lignin biosynthesis pathway. Although the trans-associations between the genetic variants of MYB125 and these nine genes of the lignin biosynthesis were not significant after multiple testing correction, the single marker regression showed that all the nine genes were identified as trans-regulated by the local-regulatory SNPs of MYB125 (P-value < 0.05). The proportion of the genetic variance in the expression of these nine genes in the lignin biosynthesis pathway explained by the cis-acting SNPs ranged from 4.83% to 8.71%; furthermore, the SNPs explained 5.79% of the variance in lignin content (Supplemental Table S8). It is important to highlight we performed 861,592,405 tests to identify distant-eQTL associations. It is widely known multiple testing correction when used in an association study, in which a large number of tests is performed, may lead to a very high rate of false negatives. In other words, multiple testing corrections control false positives at the expense of many more false negatives. Hence, in studies of genomics and statistical genetics, one must find a way to balance false-positive and false-negative results. In our study, the use of a system genetics approach allowed us to balance the false-positive and false-negative results because, through gene coexpression network analysis, we found that MYB125 is coexpressed and directly connected to 4CL3, CSE, F5H, C3′H, PAL2, CCoAOMT1, PAL4, COMT1, and C4H2. These results, together with the fact that MYB transcription factors are known to play a role in the regulation of lignin biosynthesis, led us to hypothesize that MYB125 is a potential “master regulator” of the lignin biosynthesis pathway in P. deltoides.
It is important to mention that the SNPs located in MYB125 that are associated with its expression were not identified as significantly correlated with lignin content in our previous GWA study (FDR = 0.42) (Fahrenkrog et al. 2017a). This is most likely owing to SNP effects not being of a sufficiently large magnitude to be detected. The current study was designed in part to address some of the shortcomings of our previous GWA study, in which we relied solely on the detection of direct associations between genotype and phenotype. Because in that study we had limited genotypic data in noncoding sequences, we pursued the approach of integrating transcriptome data to obtain additional supporting evidence to pursue the analysis of genes that regulate complex traits at the transcriptional level.
MYB125 is a transcriptional activator of the lignin biosynthetic pathway in P. deltoides
Wood is the most abundant biomass on earth, mainly composed of SCWs. The SCWs of xylem cells provide rigidity and mechanical strength and enable efficient water conduction through the vascular system. The SCW is composed of cellulose microfibrils embedded in a matrix of hemicellulose and lignin. The most abundant components of SCWs and terrestrial biopolymers are cellulose and lignin, accounting for ∼30% of the organic carbon present in the biosphere (Ralph et al. 2004). In addition, lignin is also the major contributor to the recalcitrance of biomass (Studer et al. 2011), which considerably increases its processing cost and, consequently, biofuel production. Because of the economic importance in pulp and biofuel production, understanding the molecular mechanisms regulating SCW deposition not only is an important topic in plant developmental biology but also is fundamental for providing molecular tools to manipulate wood composition for bioenergy use. Several studies have shown that SCW biosynthesis is controlled by a transcription factor network, including NAC and MYB transcription factors (Patzlaff et al. 2003; Schrader et al. 2004; Andersson-Gunnerås et al. 2006; Legay et al. 2007; Bomal et al. 2008; Zhong et al. 2008, 2011, 2013; Du et al. 2009; Wilkins et al. 2009; McCarthy et al. 2010; Lu et al. 2013; Tian et al. 2013; Li et al. 2015; Zhong and Ye 2015; Yang et al. 2017, 2019; Gui et al. 2019, 2020).
MYB transcription factors are at least one billion years old and represent one of the largest plant transcription factor families (Lipsick 1996). They are characterized by the presence of a highly conserved MYB domain (required for DNA binding) in the N-terminal of the protein and a modular and more diverse C-terminal (responsible for the protein's regulatory activity). The R2R3-MYBs are the most common type of MYB transcription factor in plants, and many of these are involved in plant secondary metabolism, including the phenylpropanoid pathway (Liu et al. 2015). To date, R2R3-MYB genes have been extensively described in species such as Arabidopsis (Stracke et al. 2001), maize (Du et al. 2012), wheat (Zhang et al. 2012), rice (Jiang et al. 2004), eucalyptus (Soler et al. 2015), and poplar (Wilkins et al. 2009). Among the 192 annotated genes encoding R2R3-MYB transcription factors in the Populus genome, only a small number involved in the regulation of SCW biosynthesis have been functionally characterized (Li et al. 2015).
Here we reported the role of MYB125 in activating the lignin biosynthetic pathway. MYB125 is a homolog of Arabidopsis MYB42 and MYB85, previously shown to be a positive regulator of SCW biosynthesis (Zhong et al. 2008). Overexpression of MYB125 in transgenic roots resulted in increase of lignin content and a significant up-regulation of 4CL3, CSE, F5H, C3′H, PAL2, CCoAOMT1, PAL4, and COMT1. Although MYB125 has not been associated with lignin regulation in poplar, it has been shown, in P. trichocarpa, to bind into the promoter and activate the expression of PAL4 and CCoAOMT1, Potri.010G224100 and Potri.009G099800, respectively (Petzold et al. 2018b). Consistent with our results, MYB92 (Potri.001G118800), a paralog of MYB125, was shown to activate the expression of the lignin biosynthetic genes and induce deposition of lignin in Populus tomentosa (Li et al. 2015). We evaluated the expression and segregation of MYB125 closely related homologs (MYB92, Potri.012G127700, and Potri.015G129100). Among the three closely related homologs, only MYB125 and MYB92 were expressed in xylem, and expression of MYB125 is on average 30 times higher than that of MYB92 in the population. We also evaluated the expression of MYB92 in our data and found that its expression is not as variable as MYB125 (Supplemental Fig. S13), showing limited segregation in the population. Finally, we found that the correlation between expression of MYB125 and MYB92 is relatively low (R2 = 0.22). Therefore, based on these findings, we can argue that MYB125, and not MYB92, is the cause of the effect observed in the phenotype (gene expression of trans-regulated genes and lignin content).
The regulation of C3′H, CSE, and 4CL by MYB125 suggests that this transcription factor could be a regulator of the lignin biosynthetic pathway branch in which CSE, in combination with 4CL, bypasses the second hydroxycinnamoyl transferase (HCT) reaction (Fig. 5). CSE was recently identified as an enzyme central to the lignin biosynthetic pathway in Arabidopsis (Vanholme et al. 2013). CSE catalyzes the conversion of caffeoyl shikimate into caffeate and its activity, combined with that of 4CL to produce caffeoyl-CoA, bypasses the second/reverse reaction of HCT.
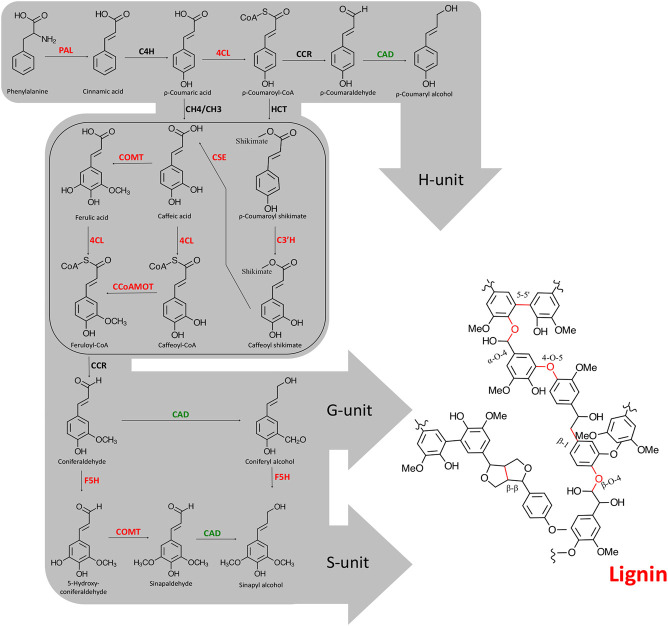
Figure 5.
Summary of the lignin biosynthetic pathway in higher plants. Enzymes are abbreviated as follows: (PAL) phenylalanine ammonia-lyase; (C4H) cinnamate 4-hydroxylase; (4CL) 4-coumarate:CoA ligase; (C3H) p-coumarate 3-hydroxylase; (C3′H) p-coumarate shikimate 3-hydroxylase; (HCT) shikimate/quinate hydroxycinnamoyl transferase; (CCR) cinnamoyl-CoA reductase; (CAD) cinnamyl alcohol dehydrogenase; (CSE) caffeoyl shikimate esterase; (COMT) caffeic acid O-methyltransferase; (CCoAOMT) caffeoyl-CoA O-methyltransferase; (F5H) ferulate 5-hydroxylase. Enzymes in red were found to be directly connected to MYB125 in the gene expression network and to be differentially expressed when MYB125 is overexpressed in poplar transgenic roots. The enzyme in green was not directly connected to MYB125 in the gene expression network, but single regression analysis showed significant association with the local-regulatory SNPs of MYB125. The box includes the most recent step discovered in the lignin biosynthesis pathway in plants.
Furthermore, the role of CSE in lignification was confirmed with a loss-of-function mutant showing reduced lignin content and an increased relative proportion of H units in the lignin polymer (Vanholme et al. 2013). Although it was initially suggested that this enzymatic step was conserved in plants, it was later shown that many unrelated species lack a bona fide CSE homolog. Thus, CSE might not be essential for lignification in all plants (Ha et al. 2016). In addition to Arabidopsis, the role of CSE in lignification was also shown in Medicago truncatula (Ha et al. 2016) and poplar (Saleme et al. 2017), for which cse plants showed lower lignin levels and preferential accumulation of H units.
In conclusion, we showed how the use of a system genetics approach allowed the identification of master regulators that could not be identified by an eQTL study owing to the high rate of false-negative results in genetic association studies. By combining eQTL study with gene coexpression network analysis, we identified MYB125 as a key regulator of the lignin biosynthetic pathway, likely acting in the lignin biosynthetic pathway branch where CSE together with 4CL bypasses the second HCT reaction. In addition, our study provided molecular evidence suggesting that MYB125 acts as a positive regulator of the lignin biosynthetic pathway in vascular tissue. Finally, besides surveying the genetic regulation of gene expression in differentiating xylem of a genetically unrelated population of P. deltoides, our study shows the importance and applicability of a large-scale system genetics approach for identifying candidate genes associated with complex traits.
Methods
Samples
The P. deltoides Bartr. ex Marsh (Eastern cottonwood) population composed of 579 individuals was sampled in 15 states in the central, southern, and eastern United States and maintained at the University of Florida. Out of the 425 unrelated individuals comprising the P. deltoides association population, we clonally propagated a set of 343 individuals from apical green cuttings in a greenhouse at the University of Florida as previously described (Fahrenkrog et al. 2017b). Half of the population was grown in 2015 and the other half in 2016 in the same greenhouse, under the same conditions and time of the year (from April to August). Both experiments were set with three biological replicates, using a row–column design. After 16 wk, we collected the xylem from three biological replicates of 343 individuals after removing bark and phloem. Xylem was immediately flash-frozen in liquid nitrogen and stored at −80°C until lyophilization and RNA extraction.
RNA isolation
Lyophilized samples were ground using stainless steel beads in a Geno/Grinder tissue homogenizer (SPEX SamplePrep) before RNA extraction. Total RNA was extracted according to the CTAB-LiCl protocol (Chang et al. 1993). DNA contamination was removed using TURBO DNase kit (Thermo Fisher Scientific), and the RNA concentration was measured using the Qubit RNA assay kit with a Qubit 2.0 fluorimeter (Thermo Fisher Scientific). RNA purity and integrity were evaluated by the plant RNA nano kit for Bioanalyzer (Agilent Technologies).
Library preparation and sequencing
NEBNext ultra directional RNA library prep kit for Illumina (NEB) was used to generate the RNA-seq libraries following the manufacturer's instructions. Briefly, mRNA was purified from 1 µg of total RNA using poly(T) oligo–attached magnetic beads. The mRNA was further fragmented, reverse transcribed, and amplified for 12 cycles using dual indexes primers. The PCR products were purified with an AMPure XP system (Beckman Coulter). The libraries were quantified using a Qubit 2.0 fluorimeter and assessed for quality and purity (Agilent Bioanalyzer 2100 system) before pooling in equimolar proportions. Ninety-six libraries were pooled, and each pool was sequenced in paired-end mode (2 × 150 bp) using the HiSeq 3000 sequencing system (Illumina). Raw data and normalized and filtered gene expression data have been deposited in the NCBI Gene Expression Omnibus (GEO) (Edgar 2002) and are accessible through GEO Series accession number GSE140232 (see Data access).
Sequencing data processing
RNA-seq FASTQ-files were quality and adapter trimmed using Trimmomatic v0.32 (Bolger et al. 2014) with the parameters TRAILING:3, SLIDINGWINDOW:4:20, MINLEN:20. The preprocessed reads were aligned to v3.0 of the Populus trichocarpa reference genome using STAR 2.5.2b (Dobin et al. 2013) using the following parameters: “‐‐outFilterMismatchNmax 8 ‐‐sjdbOverhang 100 ‐‐outSAMtype BAM SortedByCoordinate ‐‐outSJfilterReads Unique ‐‐outSAMmultNmax 1 ‐‐outSAMattrRGline ID: < genotype > _ < rep number > SM: < genotype > _ < rep number > -outFilterType BySJout.” Picard v1.115 (https://broadinstitute.github.io/picard/) was used to remove duplicate reads, and only unique alignments were kept for further analysis. At the time of the RNA-seq data analysis, only version 3.0 of the P. trichocarpa reference genome was available. Because an improved version of the genome assembly and annotation has since became available (v4.0), we assessed any impact that this may have had in the data presented here, namely, on the estimations of gene expression and definition of cis and trans status of SNPs. Estimates of gene expression detected by aligning reads to both versions of the genome resulted in transcript estimates that are highly correlated (94%). Alignment of the sequence of the genes discussed in this study to the version 4.0 of the P. trichocarpa genome and of the cis and trans regulatory status of SNPs remained unchanged in the newest version of the genome.
Transcriptome assembly and gene expression quantification
We used three transcript assembly platforms to maximize the detection of novel genes: (1) Cufflinks version 2.2.1 (Bolger et al. 2014) with parameters “‐‐library-type fr-firststrand –u -F 0.05 ‐‐max-intron-length 12000 ‐‐no-faux-reads”; (2) StringTie version 1.3.3 (Pertea et al. 2016) with parameters “-f 0.05 -j 2 –rf”; and (3) Trinity version 2.3.2 (Grabherr et al. 2011) in genome guided mode with parameters “‐‐genome_guided_bam < input bam file > ‐‐genome_guided_max_intron 12000 ‐‐full_cleanup ‐‐SS_lib_type RF ‐‐min_contig_length 50.” The resulting transcripts detected for each sample using Cufflinks and StringTie were merged with StringTie with parameters “-F 1 -f 0.05.” We used PASA 2.0.2 (Haas et al. 2003) to combine this merged assembly and the assembly generated by Trinity using parameters “-C –R -t < Trinity fasta output > ‐‐cufflinks_gtf < merged gtf file > -I 12000 ‐‐ALT_SPLICE ‐‐ALIGNER gmap,blat.” The final assemblies for each sample were merged with Cuffmerge to generate the master transcriptome representative of all transcripts in the population. The master transcriptome was reformatted and annotated using gffcompare version 0.9.9c (https://github.com/gpertea/gffread) and subjected to expression filtering to remove artifacts generated by merging assemblies such that each transcript in the assembly was required to be expressed at FPKM ≥ 3 in at least two of the three biological replicates of at least three individuals and 50 observations.
Gene expression was measured using Cufflinks version 2.2.1 (Bolger et al. 2014). Cufflinks was run with the –G option to disable novel transcript detection, and the master transcriptome described above was used as a reference to quantify against. The Cufflinks output GTF file was parsed to obtain the expression of each isoform of a gene, and the sum of all of the isoforms’ expression values were used to represent the expression of that gene.
Adjustment of gene expression by systematic effects of the experiments
We used a standard linear mixed effect model to correct the gene expression data for the systematic effects of the experiment. Separate models were fitted to each one of the genes. The response was abundance (FPKM), and the model included an intercept, the fixed effect of the experiment, and the random effect of the genotype, row, and column within the experiment. Models were fitted using the lmer function of the lme4 R-package (Bates et al. 2015). From the fitted model, we extracted the best linear unbiased estimates (BLUEs) of the FPKM for each genotype as the sum of the best linear unbiased predictor (BLUP) of the genotype effect plus the average residuals.
Heritability estimation
The mixed-effects model above described also rendered estimates of variance parameters for each of the random effects, including genotype , row , column (), and errors (). From those, we estimated gene-specific heritabilities using
The empirical distribution of the genes’ heritability is displayed in Supplemental Figure S1.
Subsequently, we conducted 10,000 permutations of the data, from which we approximated the distribution of the gene-specific heritabilities under the null hypothesis, . This was performed by permuting the genotype ID while maintaining the rest of the data structure unchanged.
For the subsequent analyses, we used only the genes with estimated heritability greater than the 99th percentile of the corresponding permutation distribution. A total of 12,579 satisfied that criteria.
SNP liftover from P. trichocarpa hybrid genome to P. trichocarpa V3 coordinates and SNP filtering
A collection of 358,809 high-quality SNPs identified by Fahrenkrog et al. (2017b) using a hybrid Populus genome assembly were converted to locations in the P. trichocarpa v3.0 reference genome assembly using a modified liftOver pipeline (http:// genomewiki.ucsc.edu/index.php/Minimal_Steps_For_LiftOver). This pipeline extracts the sequence flanking a SNP from the genome it was identified in and aligns the sequence to the genome one wishes to convert the SNP coordinates. In total, 313,036 of the 358,809 SNPs were converted to P. trichocarpa v3 reference genome coordinates using this pipeline. A stepwise walkthrough of this pipeline can be found at GitHub (https://github.com/jdLikesPlants/SNP-liftover). We also filtered out SNPs that departed from Hardy–Weinberg equilibrium by filtering out SNPs with a P-value <0.01 on a chi-square test. Finally, we further filtered the SNP set to remove SNPs with MAF below 0.05, mean sequencing depth in the population below 8, genotype quality below 20, and missing data percentage in the population >75%. This resulted in a total of 68,885 SNPs that were used for the analysis herein described.
Expression QTL analysis
The BLUEs for each gene expression were combined with the filtered SNPs and jointly analyzed to identify eQTLs. To avoid having an excess of false discoveries, the FPKM BLUES were further adjusted by the first five principal components, which were derived using scaled and centered SNP genotypes. Subsequently, we regressed the adjusted FPKM on each of the SNPs using an ordinal least squares (OLS) regression that had the adjusted FPKM as the response variable and a SNP as the predictor. From this regression, we extracted association P-values for all gene–SNP combinations. We used the p.adjust function of R 3.2.2 (R Core Team 2019) to obtain the FDR-adjusted P-values considering the method proposed by Benjamini and Hochberg (Benjamini and Hochberg 1995). Final inferences were thus based on FDR-adjusted P-values.
Partition of the gene variance into cis and trans components
We estimated the overall proportion of variance of FPKM explained by the SNPs and its cis and trans-regulation components for all mapped genes. To this end, for each gene, SNPs were classified as being in cis (covering the gene region plus 1 Mbp upstream of or downstream from the transcription start site of the tested gene) (Cubillos et al. 2012; Zan et al. 2016; Azodi et al. 2020) or trans (elsewhere); all the SNPs that passed the quality control were used in the analyses. Subsequently, for every gene we fitted a “GBLUP” model with adjusted FPKM as response and two random effects, one (uLi) capturing the total effect of the SNPs in cis (local) and another one (uDi) capturing the joint effects of the SNPs trans (distant), that is
The genomic effects of local and distant SNPs were assumed to follow a multivariate normal distribution (MVN) with a mean equal to zero and variance–covariance matrix proportional to genomic relationship matrices () derived from the SNPs local (GL) and distant (GD) to the gene. The genomic relationship matrices were computed using the getG function of the BGData R-package (Grueneberg and de los Campos 2019). Thus, the joint distribution of the data and the random effects were
The above-described model was implemented in a Bayesian setting using the BGLR R-package (Pérez and de los Campos 2014) using the default prior settings, 35,000 iterations, a burn-in of 5000, and a thinning of 5.
From the variance parameters, we estimated the proportion of the phenotypic variance of each gene explained by SNPs local and distant to the gene, that is,
and
respectively.
Weighted gene coexpression network analysis
Coexpression network analysis was performed using the R package WGCNA (Langfelder and Horvath 2008). Briefly, the gene coexpression network was constructed from the pairwise Pearson correlation coefficients for all gene–gene comparisons using an unsigned network and soft thresholding power of eight, which was the smallest threshold that resulted in a scale-free topology (R2> 0.9), a minimum module size of 30, a maximum block size equal to the total number of genes, and the other parameters set as default. The topological overlap matrix (TOM) was further generated using the TOMsimilarity function. A dissimilarity matrix based on TOM (1 − TOM) was used to identify network modules through a dynamic tree-cutting algorithm (Langfelder et al. 2008). Enrichment analyses for GO categories and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was performed using the Populus Genome Integrative Explorer (PopGenIE) platform in PlantGenIE.org (Sjödin et al. 2009). The enrichment analyzes were performed using all the genes as a background, in part because our data suggest that a large proportion of poplar tree genes are expressed in the differentiating xylem. Finally, the gene coexpression network was visualized and analyzed by Cytoscape 3.7.1 (Shannon 2003).
After the initial modules were generated using the default, the output was manually curated by inspecting each module for their eigengene values. In this step, we noticed that the modules black and red showed a similar expression pattern and showed KEGG enrichment for lignin biosynthesis genes. Thus, the genes presented in the modules black and red were used to construct a new coexpression network. The analyzes were performed as described above but using a soft thresholding power of seven.
Generation of transgenic poplar hairy roots
For transgenic hairy root generation, we followed a protocol previously described (Yoshida et al. 2015). Briefly, we excised leaves from in vitro grown P. tremula × P. alba clone INRA 717-1-B4 plants and placed them on preculture solid medium (0.25 g of MES, 0.1 g of myo-inositol, 30 g of sucrose, and 4.33 g of MS salts in 1 L at pH 5.7). The leaves were wounded by cutting the veins gently before transformation with Agrobacterium rhizogenes carrying a binary plasmid vector containing the coding sequence of MYB125 under the control of Actin2 promoter. The construct also contained a fluorescent marker (TdTomato) for rapid detection of transgenic roots. The binary vector construct was assembled using Golden Gate Modular Cloning (Engler et al. 2014). Hairy roots arising from transformed leaves were excised and maintained in the dark at 25°C in petri dishes on solid antibiotic-containing medium (cefotaxime 200 mg/mL, Timentin 200 mg/mL). Individual hairy roots clones were subcultured every 21 d. Screening for positive transformants was conducted by TdTomato detection under fluorescent microscope. Wild-type and transgenic roots were harvested and immediately frozen in liquid nitrogen for total RNA extraction and lignin content measurement.
RT-qPCR expression analysis of MYB125 and genes of lignin biosynthesis pathway in WT and transgenic roots of poplar
Five-to-eight WT and transgenic roots (∼3 g) were pooled to generate one biological replicate. Roots were ground in liquid nitrogen and divided in two samples containing ∼1.5 g of ground tissue each (one sample was used for RNA extraction and the second sample for cell wall isolation). Total RNA was extracted according to the CTAB-LiCl protocol (Chang et al. 1993) and treated with DNase to remove genomic DNA contamination. Quantitative reverse transcription–polymerase chain reaction (RT-qPCR) analyses were performed using a Luna universal probe one-step RT-qPCR kit (New England Biolabs) following the manufacturer's instructions, using 200 ng of total RNA. Gene expression levels were assessed in three independent biological replicates. The relative expression of each sample was determined after normalization to Actin2 using the relative standard curve method (Larionov et al. 2005). Actin2 was chosen based on a previous study that showed that it does not show significant variation in expression among different tissues in poplar and one of the smallest residue values on average among the housekeeping genes tested (Brunner et al. 2004). Means of different groups were compared and analyzed using a one-tail Student's t-test. Differences were reported as statistically significant when P-value < 0.05. Sequences of gene-specific primers used in the experiment are found in Supplemental Table S9.
Cell wall isolation and measurement of acetyl bromide lignin
Cell wall material (alcohol insoluble residue [AIR]) was obtained from three independent biological replicates of WT and transgenic roots as previously described (Barnes and Anderson 2017). Briefly, ground tissue (1.5 g) from each sample was freeze-dried, and 100 mg of dried sample was washed with 70% (v/v) ethanol followed by washing with chloroform/methanol (1:1 v/v) and 100% acetone. The pellet was air dried to obtain the AIR. To remove starch from the sample, the AIR pellet was resuspended in 0.1 M of sodium acetate (pH 5.0) and heated for 20 min at 80°C . The ice-cooled samples were further digested with α-amylase (0.5 U/mg AIR, from Bacillus species, Sigma-Aldrich) and pullulanase (0.3 U/mg AIR, from Bacillus acidopullulyticus, Sigma-Aldrich) overnight at 37°C under gentle shaking. The pellet was washed three times with water and once with acetone. The pellet was air dried, and AIR samples (2 mg) were incubated with 100 µL of freshly made acetyl bromide solution (25% v/v acetyl bromide in glacial acetic acid) for 4 h with vortexing every 15 min at 50°C. After complete digestion, samples were cooled down, and 400 µL of 2M NaOH, 70 µL of freshly prepared 0.5 M hydroxylamine hydrochloride, and glacial acetic acid (to complete 2 mL) were added to into the samples. After inverting the tubes several times to mix, 200 µL of the solution was transferred into a UV-specific 96-well plate. The ABSL was calculated from the UV absorbance at 280 nm using a molar extinction coefficient of 18.21 g−1 L cm−1 and a 0.539-cm path length, according to the following formula:
Means of different groups were compared and analyzed using a one-tail Student's t-test. Differences were reported as statistically significant when P-value < 0.05.
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) (Edgar 2002) under accession number GSE140232. All significant associations are available at Dryad Data set (https://doi.org/10.5061/dryad.12jm63xt6). Custom scripts are available in the Supplemental Material.
Competing interest statement
The authors declare no competing interests.
Supplementary Material
Acknowledgments
This study was funded by the U.S. National Science Foundation Plant Genome Research Program (grant NSF IOS-1444543).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.261438.120.
References
- Acharjee A, Kloosterman B, Visser RGF, Maliepaard C. 2016. Integration of multi-omics data for prediction of phenotypic traits using random forest. BMC Bioinformatics 17: 185 10.1186/s12859-016-1043-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson-Gunnerås S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B. 2006. Biosynthesis of cellulose-enriched tension wood in Populus: Global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45: 144–165. 10.1111/j.1365-313X.2005.02584.x [DOI] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, Rollmann SM, Duncan H, Lawrence F, Anholt RRH, et al. 2009. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet 41: 299–307. 10.1038/ng.332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azodi CB, Pardo J, VanBuren R, de los Campos G, Shiu S-H. 2020. Transcriptome-based prediction of complex traits in maize. Plant Cell 32: 139–151. 10.1105/tpc.19.00332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker OB, Aguirre-Gamboa R, Sanna S, Oosting M, Smeekens SP, Jaeger M, Zorro M, Võsa U, Withoff S, Netea-Maier RT, et al. 2018. Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses. Nat Immunol 19: 776–786. 10.1038/s41590-018-0121-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W, Anderson C. 2017. Acetyl bromide soluble lignin (ABSL) assay for total lignin quantification from plant biomass. Bio Protoc 7: 2149 10.21769/BioProtoc.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw 67: 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57: 289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JEK, Blais S, Tremblay L, Morency M-J, Pavy N, et al. 2008. Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: a comparative in planta analysis. J Exp Bot 59: 3925–3939. 10.1093/jxb/ern234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem RB. 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755. 10.1126/science.1069516 [DOI] [PubMed] [Google Scholar]
- Brunner AM, Yakovlev IA, Strauss SH. 2004. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4: 14 10.1186/1471-2229-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11: 113–116. 10.1007/BF02670468 [DOI] [Google Scholar]
- Chauffour F, Bailly M, Perreau F, Cueff G, Suzuki H, Collet B, Frey A, Clément G, Soubigou-Taconnat L, Balliau T, et al. 2019. Multi-omics analysis reveals sequential roles for ABA during seed maturation. Plant Physiol 180: 1198–1218. 10.1104/pp.19.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. 2014. Systems genetics approaches to understand complex traits. Nat Rev Gen 15: 34–48. 10.1038/nrg3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, Yansouni J, Khalili H, Balzergue S, Elftieh S, Martin-Magniette M-L, Serrand Y, Lepiniec L, Baud S, Dubreucq B, et al. 2012. Expression variation in connected recombinant populations of Arabidopsis thaliana highlights distinct transcriptome architectures. BMC Genomics 13: 117 10.1186/1471-2164-13-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. 2004. Cell death. Cell 116: 205–219. 10.1016/S0092-8674(04)00046-7 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhang L, Liu L, Tang X-F, Yang W-J, Wu Y-M, Huang Y-B, Tang Y-X. 2009. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry (Moscow) 74: 1–11. 10.1134/S0006297909010015 [DOI] [PubMed] [Google Scholar]
- Du H, Feng B-R, Yang S-S, Huang Y-B, Tang Y-X. 2012. The R2R3-MYB transcription factor gene family in maize. PLoS One 7: e37463 10.1371/journal.pone.0037463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert T-M, Werner S, Jones JDG, Patron NJ, Marillonnet S. 2014. A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843. 10.1021/sb4001504 [DOI] [PubMed] [Google Scholar]
- Fahrenkrog AM, Neves LG, Resende MFR Jr, Dervinis C, Davenport R, Barbazuk WB, Kirst M. 2017a. Population genomics of the eastern cottonwood (Populus deltoides). Ecol Evol 7: 9426–9440. 10.1002/ece3.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog AM, Neves LG, Resende MFR, Vazquez AI, de los Campos G, Dervinis C, Sykes R, Davis M, Davenport R, Barbazuk WB, et al. 2017b. Genome-wide association study reveals putative regulators of bioenergy traits in Populus deltoides. New Phytologist 213: 799–811. 10.1111/nph.14154 [DOI] [PubMed] [Google Scholar]
- Funada R, Yamagishi Y, Begum S, Kudo K, Nabeshima E, Nugroho WD, Hasnat R, Oribe Y, and Nakaba S. 2016. Xylogenesis in trees: from cambial cell division to cell death. In Secondary xylem biology (ed. Yoon SK, et al. ), pp. 25–43. Academic Press, San Diego: https://linkinghub.elsevier.com/retrieve/pii/B9780128021859000024. [Google Scholar]
- Furches A, Kainer D, Weighill D, Large A, Jones P, Walker AM, Romero J, Gazolla JGFM, Joubert W, Shah M, et al. 2019. Finding new cell wall regulatory genes in Populus trichocarpa using multiple lines of evidence. Front Plant Sci 10: 1249 10.3389/fpls.2019.01249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueneberg A, de los Campos G. 2019. BGData: a suite of R packages for genomic analysis with big data. G3: Genes, Genomes, Genet 9: 1377–1383. 10.1534/g3.119.400018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra FP, Suren H, Holliday J, Richards JH, Fiehn O, Famula R, Stanton BJ, Shuren R, Sykes R, Davis MF, et al. 2019. Exome resequencing and GWAS for growth, ecophysiology, and chemical and metabolomic composition of wood of Populus trichocarpa. BMC Genomics 20: 875 10.1186/s12864-019-6160-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui J, Luo L, Zhong Y, Sun J, Umezawa T, Li L. 2019. Phosphorylation of LTF1, an MYB transcription factor in Populus, acts as a sensory switch regulating lignin biosynthesis in wood cells. Mol Plant 12: 1325–1337. 10.1016/j.molp.2019.05.008 [DOI] [PubMed] [Google Scholar]
- Gui J, Lam PY, Tobimatsu Y, Sun J, Huang C, Cao S, Zhong Y, Umezawa T, Li L. 2020. Fibre-specific regulation of lignin biosynthesis improves biomass quality in Populus. New Phytologist 226: 1074–1087. 10.1111/nph.16411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Escamilla-Trevino L, Yarce JCS, Kim H, Ralph J, Chen F, Dixon RA. 2016. An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J 86: 363–375. 10.1111/tpj.13177 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666. 10.1093/nar/gkg770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin Y, Seldin M, Lusis A. 2017. Multi-omics approaches to disease. Genome Biol 18: 83 10.1186/s13059-017-1215-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hefer CA, Mizrachi E, Myburg AA, Douglas CJ, Mansfield SD. 2015. Comparative interrogation of the developing xylem transcriptomes of two wood-forming species: Populus trichocarpa and Eucalyptus grandis. New Phytologist 206: 1391–1405. 10.1111/nph.13277 [DOI] [PubMed] [Google Scholar]
- Holloway B, Luck S, Beatty M, Rafalski J-A, Li B. 2011. Genome-wide expression quantitative trait loci (eQTL) analysis in maize. BMC Genomics 12: 336 10.1186/1471-2164-12-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2011. Weighted network analysis. Springer New York, New York: http://link.springer.com/10.1007/978-1-4419-8819-5. [Google Scholar]
- Iakimova ET, Woltering EJ. 2017. Xylogenesis in zinnia (Zinnia elegans) cell cultures: unravelling the regulatory steps in a complex developmental programmed cell death event. Planta 245: 681–705. 10.1007/s00425-017-2656-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Gu X, Peterson T. 2004. Identification of conserved gene structures and carboxy-terminal motifs in the Myb gene family of Arabidopsis and Oryza sativa L. ssp. indica. Genome Biol 5: R46 10.1186/gb-2004-5-7-r46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu J, Terpstra IR, Garcia JM, van den Ackerveken G, Snoek LB, Peeters AJM, Vreugdenhil D, Koornneef M, Jansen RC. 2007. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci 104: 1708–1713. 10.1073/pnas.0610429104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirst M, Basten CJ, Myburg AA, Zeng Z-B, Sederoff RR. 2005. Genetic architecture of transcript-level variation in differentiating xylem of a eucalyptus hybrid. Genetics 169: 2295–2303. 10.1534/genetics.104.039198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Zhang B, Horvath S. 2008. Defining clusters from a hierarchical cluster tree: the dynamic tree cut package for R. Bioinformatics 24: 719–720. 10.1093/bioinformatics/btm563 [DOI] [PubMed] [Google Scholar]
- Larionov A, Krause A, Miller W. 2005. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics 6: 62 10.1186/1471-2105-6-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legay S, Lacombe E, Goicoechea M, Brière C, Séguin A, Mackay J, Grima-Pettenati J. 2007. Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci 173: 542–549. 10.1016/j.plantsci.2007.08.007 [DOI] [Google Scholar]
- Li C, Wang X, Ran L, Tian Q, Fan D, Luo K. 2015. PtoMYB92 is a transcriptional activator of the lignin biosynthetic pathway during secondary cell wall formation in Populus tomentosa. Plant Cell Physiol 56: 2436–2446. 10.1093/pcp/pcv157 [DOI] [PubMed] [Google Scholar]
- Lipsick JS. 1996. One billion years of Myb. Oncogene 13: 223–235. [PubMed] [Google Scholar]
- Liu J, Osbourn A, Ma P. 2015. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8: 689–708. 10.1016/j.molp.2015.03.012 [DOI] [PubMed] [Google Scholar]
- Liu X, Li YI, Pritchard JK. 2019. Trans effects on gene expression can drive omnigenic inheritance. Cell 177: 1022–1034.e6. 10.1016/j.cell.2019.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Li Q, Wei H, Chang M-J, Tunlaya-Anukit S, Kim H, Liu J, Song J., Sun Y-H, Yuan L, et al. 2013. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci 110: 10848–10853. 10.1073/pnas.1308936110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC. 2014. Epistasis and quantitative traits: using model organisms to study gene–gene interactions. Nat Rev Genet 15: 22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähler N, Wang J, Terebieniec BK, Ingvarsson PK, Street NR, Hvidsten TR. 2017. Gene co-expression network connectivity is an important determinant of selective constraint. PLoS Genet 13: e1006402 10.1371/journal.pgen.1006402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy RL, Zhong R, Fowler S, Lyskowski D, Piyasena H, Carleton K, Spicer C, Ye Z-H. 2010. The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol 51: 1084–1090. 10.1093/pcp/pcq064 [DOI] [PubMed] [Google Scholar]
- Moreno-Moral A, Petretto E. 2016. From integrative genomics to systems genetics in the rat to link genotypes to phenotypes. Dis Models Mech 9: 1097–1110. 10.1242/dmm.026104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, Gamazon E, Zhang W, Duan S, Eileen Dolan M, Cox NJ. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6: e1000888 10.1371/journal.pgen.1000888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patzlaff A, McInnis S, Courtenay A, Surman C, Newman LJ, Smith C, Bevan MW, Mansfield S, Whetten RW, Sederoff RR, et al. 2003. Characterisation of a pine MYB that regulates lignification. Plant J 36: 743–754. 10.1046/j.1365-313X.2003.01916.x [DOI] [PubMed] [Google Scholar]
- Pérez P, de los Campos G. 2014. Genome-wide regression and prediction with the BGLR statistical package. Genetics 198: 483–495. 10.1534/genetics.114.164442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 11: 1650–1667. 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold HE, Chanda B, Zhao C, Rigoulot SB, Beers EP, Brunner AM. 2018a. Divaricata and radialis interacting factor (DRIF) also interacts with WOX and KNOX proteins associated with wood formation in Populus trichocarpa. Plant J 93: 1076–1087. 10.1111/tpj.13831 [DOI] [PubMed] [Google Scholar]
- Petzold HE, Rigoulot SB, Zhao C, Chanda B, Sheng X, Zhao M, Jia X, Dickerman AW, Beers EP, Brunner AM. 2018b. Identification of new protein–protein and protein–DNA interactions linked with wood formation in Populus trichocarpa. Tree Physiol 38: 362–377. 10.1093/treephys/tpx121 [DOI] [PubMed] [Google Scholar]
- Porth I, Klapšte J, Skyba O, Hannemann J, McKown AD, Guy RD, DiFazio SP, Muchero W, Ranjan P, Tuskan GA., et al. 2013. Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytologist 200: 710–726. 10.1111/nph.12422 [DOI] [PubMed] [Google Scholar]
- Qin H, Niu T, Zhao J. 2019. Identifying multi-omics causers and causal pathways for complex traits. Front Genet 10: 110 10.3389/fgene.2019.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. 2004. Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60. 10.1023/B:PHYT.0000047809.65444.a4 [DOI] [Google Scholar]
- Ranjan A, Budke JM, Rowland SD, Chitwood DH, Kumar R, Carriedo L, Ichihashi Y, Zumstein K, Maloof JN, Sinha NR. 2016. eQTL regulating transcript levels associated with diverse biological processes in tomato. Plant Physiol 172: 328–340. 10.1104/pp.16.00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2019. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/bin/windows/base/old/3.2.2/ [Google Scholar]
- Saleme MdLS, Cesarino I, Vargas L, Kim H, Vanholme R, Goeminne G, Van Acker R, de Assis Fonseca FC, Pallidis A, Voorend W, et al. 2017. Silencing CAFFEOYL SHIKIMATE ESTERASE affects lignification and improves saccharification in poplar. Plant Physiol 175: 1040–1057. 10.1104/pp.17.00920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. 10.1038/nature01434 [DOI] [PubMed] [Google Scholar]
- Schrader J, Nilsson J, Mellerowicz E, Berglund A, Nilsson P, Hertzberg M, Sandberg G. 2004. A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell 16: 2278–2292. 10.1105/tpc.104.024190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjödin A, Street NR, Sandberg G, Gustafsson P, Jansson S. 2009. The Populus genome integrative explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytologist 182: 1013–1025. 10.1111/j.1469-8137.2009.02807.x [DOI] [PubMed] [Google Scholar]
- Soler M, Camargo ELO, Carocha V, Cassan-Wang H, Clemente HS, Savelli B, Hefer CA, Paiva JAP, Myburg AA, Grima-Pettenati J. 2015. The Eucalyptus grandis R2R3-MYB transcription factor family: evidence for woody growth-related evolution and function. New Phytologist 206: 1364–1377. 10.1111/nph.13039 [DOI] [PubMed] [Google Scholar]
- Sonah H, O'Donoughue L, Cober E, Rajcan I, Belzile F. 2015. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol J 13: 211–221. 10.1111/pbi.12249 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. 2001. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456. 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Studer MH, DeMartini JD, Davis MF, Sykes RW, Davison B, Keller M, Tuskan GA, Wyman CE. 2011. Lignin content in natural Populus variants affects sugar release. Proc Natl Acad Scis 108: 6300–6305. 10.1073/pnas.1009252108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Wang X, Li C, Lu W, Yang L, Jiang Y, Luo K. 2013. Functional characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation. PLoS One 8: e76369 10.1371/journal.pone.0076369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D, Zhu G, Resende MFR, Lin T, Nguyen C, Bies D, Rambla JL, Beltran KSO, Taylor M, Zhang B, et al. 2017. A chemical genetic roadmap to improved tomato flavor. Science 355: 391–394. 10.1126/science.aal1556 [DOI] [PubMed] [Google Scholar]
- Turuspekov Y, Baibulatova A, Yermekbayev K, Tokhetova L, Chudinov V, Sereda G, Ganal M, Griffiths S, Abugalieva S. 2017. GWAS for plant growth stages and yield components in spring wheat (Triticum aestivum L.) harvested in three regions of Kazakhstan. BMC Plant Biol 17: 190 10.1186/s12870-017-1131-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan GA, Muchero W, Tschaplinski TJ, Ragauskas AJ. 2019. Population-level approaches reveal novel aspects of lignin biosynthesis, content, composition and structure. Curr Opin Biotechnol 56: 250–257. 10.1016/j.copbio.2019.02.017 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J., Morreel K, Araujo P, et al. 2013. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106. 10.1126/science.1241602 [DOI] [PubMed] [Google Scholar]
- Vazquez AI, Veturi Y, Behring M, Shrestha S, Kirst M, Resende MFR, de Los Campos G. 2016. Increased proportion of variance explained and prediction accuracy of survival of breast cancer patients with use of whole-genome multiomic profiles. Genetics 203: 1425–1438. 10.1534/genetics.115.185181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 2017. Ten years of GWAS discovery: biology, function, and translation. Am J Human Genet 101: 5–22. 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Han Y, Teng W, Zhao X, Li Y, Wu L, Li D, Li W. 2014. Expression quantitative trait loci infer the regulation of isoflavone accumulation in soybean (Glycine max L. Merr.) seed. BMC Genomics 15: 680 10.1186/1471-2164-15-680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JP, Matthews ML, Williams CM, Shi R, Yang C, Tunlaya-Anukit S, Chen H-C, Li Q, Liu J, Lin C-Y, et al. 2018. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat Commun 9: 1579 10.1038/s41467-018-03863-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St. Clair DA. 2007. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450. 10.1534/genetics.106.064972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. 2009. Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149: 981–993. 10.1104/pp.108.132795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Zhang J, Tschaplinski TJ, Tuskan GA, Chen J-G, Muchero W. 2018. Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front Plant Sci 9: 1427 10.3389/fpls.2018.01427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhao X, Ran L, Chaofeng Li DF, Luo K. 2017. PtoMYB156 is involved in negative regulation of phenylpropanoid metabolism and secondary cell wall biosynthesis during wood formation in poplar. Sci Rep 7: 41209 10.1038/srep41209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yoo CG, Rottmann W, Winkeler KA, Collins CM, Gunter LE, Jawdy SS, Yang X, Pu Y, Ragauskas AJ, et al. 2019. PdWND3A, a wood-associated NAC domain-containing protein, affects lignin biosynthesis and composition in Populus. BMC Plant Biol 19: 486 10.1186/s12870-019-2111-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Ma D, Peter Constabel C. 2015. The MYB182 protein down-regulates proanthocyanidin and anthocyanin biosynthesis in poplar by repressing both structural and regulatory flavonoid genes. Plant Physiol 167: 693–710. 10.1104/pp.114.253674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan Y, Shen X, Forsberg SKG, Carlborg Ö. 2016. Genetic regulation of transcriptional variation in natural Arabidopsis thaliana accessions. G3: Genes, Genomes, Genet 6: 2319–2328. 10.1534/g3.116.030874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhao G, Jia J, Liu X, Kong X. 2012. Molecular characterization of 60 isolated wheat MYB genes and analysis of their expression during abiotic stress. J Exp Bot 63: 203–214. 10.1093/jxb/err264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen X, Lu C, Ye J, Zou M, Lu K, Feng S, Pei J, Liu C, Zhou X, et al. 2018. Genome-wide association studies of 11 agronomic traits in cassava (Manihot esculenta Crantz). Front Plant Sci 9: 503 10.3389/fpls.2018.00503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H. 2014. Complexity of the transcriptional network controlling secondary wall biosynthesis. Plant Sci 229: 193–207. 10.1016/j.plantsci.2014.09.009 [DOI] [PubMed] [Google Scholar]
- Zhong R, Ye Z-H. 2015. Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56: 195–214. 10.1093/pcp/pcu140 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye Z-H. 2008. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20: 2763–2782. 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, McCarthy RL, Lee C, Ye Z-H. 2011. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol 157: 1452–1468. 10.1104/pp.111.181354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, McCarthy RL, Haghighat M, Ye Z-H. 2013. The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS One 8: e69219 10.1371/journal.pone.0069219 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.