Abstract
Polycomb group proteins are important for maintaining gene expression patterns and cell identity in metazoans. The mammalian Polycomb repressive deubiquitinase (PR-DUB) complexes catalyze removal of monoubiquitination on lysine 119 of histone H2A (H2AK119ub1) through a multiprotein core comprised of BAP1, HCFC1, FOXK1/2, and OGT in combination with either of ASXL1, 2, or 3. Mutations in PR-DUB components are frequent in cancer. However, mechanistic understanding of PR-DUB function in gene regulation is limited. Here, we show that BAP1 is dependent on the ASXL proteins and FOXK1/2 in facilitating gene activation across the genome. Although PR-DUB was previously shown to cooperate with PRC2, we observed minimal overlap and functional interaction between BAP1 and PRC2 in embryonic stem cells. Collectively, these results demonstrate that PR-DUB, by counteracting accumulation of H2AK119ub1, maintains chromatin in an optimal configuration ensuring expression of genes important for general functions such as cell metabolism and homeostasis.
The maintenance and dynamics of gene expression in metazoans is tightly controlled and is crucial for cell identity, differentiation, proper development, and cell fate. Site-specific DNA binding of transcription factors (TFs) plays a major role in instructing cell type–specific gene expression patterns (Niwa 2018; Stadhouders et al. 2019). In addition, chromatin undergoes covalent histone modifications and DNA methylation, which further recruit chromatin modifying factors to impart precise catalysis of transcription and enhance or repress gene expression (Yadav et al. 2018; Talbert et al. 2019).
One of the major families involved in gene regulation at the chromatin level is the Polycomb group family of proteins (PcG) that are essential for maintaining genes in a repressed state (Chittock et al. 2017; Schuettengruber et al. 2017). PcG proteins can be broadly divided into three subgroups, Polycomb Repressive Complex 1 (PRC1), Polycomb Repressive Complex 2 (PRC2), and Polycomb Repressive DeUBiquitinase complex (PR-DUB). These complexes catalyze certain covalent modifications of histones that in turn contribute to their negative impact on gene expression. PRC1 catalyzes monoubiquitination of H2AK119 (H2AK119ub1), whereas PRC2 catalyzes mono-, di- and trimethylation of histone H3 Lys 27 (H3K27me1, 2, and 3). Although PR-DUB has been reported to mediate gene repression, the complex acts to deubiquitinate H2AK119ub1, a chromatin modification associated with PRC1-mediated gene silencing (Scheuermann et al. 2010).
The PR-DUB complex was originally defined in Drosophila melanogaster to consist of the gene products of Asx and Calypso (Scheuermann et al. 2010). Asx and Calypso belong to the enhancer of Trithorax and Polycomb (ETP) group, and their deletion causes both posterior and anterior transformation of the body plan (Simon et al. 1992; Sinclair et al. 1992; de Ayala Alonso et al. 2007). Calypso is an ubiquitin carboxy-terminal hydrolase that deubiquitylates H2AK118ub1 (the analog of mammalian H2AK119ub1), and the PR-DUB complex binds to PcG target genes in Drosophila (Scheuermann et al. 2010). Mutations in both Asx and Calypso lead to an increase in the levels of H2AK118ub1, which correlates with derepression of PcG-targeted Hox genes (Scheuermann et al. 2010). Therefore, this complex was designated as Polycomb Repressive DeUBiquitinase complex. Although several models for the repressive function of the PR-DUB complex have been proposed (Scheuermann et al. 2010; Dey et al. 2012; Abdel-Wahab and Dey 2013; LaFave et al. 2015; Micol and Abdel-Wahab 2016; Campagne et al. 2019), the mechanism by which it mediates gene repression remains uncertain.
Three PR-DUB-like complexes exist in mammals that all contain BAP1 (the human homolog of Calypso), HCFC1, OGT, and FOXK1/2 and either ASXL1, ASXL2, or ASXL3 (the human homologs of ASX) (Fig. 1A; Micol and Abdel-Wahab 2016). BAP1 was originally identified as a binding partner for BRCA1 and, besides being the core enzymatic component of PR-DUB, it might play a role in DNA repair (Carbone et al. 2013; Murali et al. 2013). BAP1, ASXL1, ASXL2, and ASXL3 are frequently mutated in a variety of human cancers (Carbone et al. 2013; Murali et al. 2013; Micol and Abdel-Wahab 2016). BAP1 mutations are observed in mesothelioma and uveal melanoma and in familial melanoma (Carbone et al. 2013; Murali et al. 2013). ASXL1 is mutated with high frequency in hematological malignancies such as myelodysplastic syndrome and acute myeloid leukemia (Micol and Abdel-Wahab 2016). ASXL1 and ASXL3 mutations are also causal for the severe congenital disorders Bohring-Opitz and Bainbridge-Ropers syndromes, respectively (Hoischen et al. 2011; Bainbridge et al. 2013; Srivastava et al. 2016). In both cancers and congenital disorders, ASXL1/2/3 have been linked to repression of HOX genes (Abdel-Wahab et al. 2012; Inoue et al. 2013). In myeloid malignancies, ASXL1 has been demonstrated to bind directly to PRC2 and to be important for its recruitment to and repression of HOX genes (Abdel-Wahab et al. 2012. Inoue et al. 2013). Moreover, genome-wide analyses in murine myeloid cells and human cell lines have shown that ASXL1 and BAP1 can bind to CpG islands, which show a strong enrichment for ETS binding motifs (Dey et al. 2012; Abdel-Wahab et al. 2013). Studies of specific loci in tumor cell lines have also shown that members of the FOX transcription factor family can recruit BAP1 to chromatin (Ji et al. 2014; Okino et al. 2015).
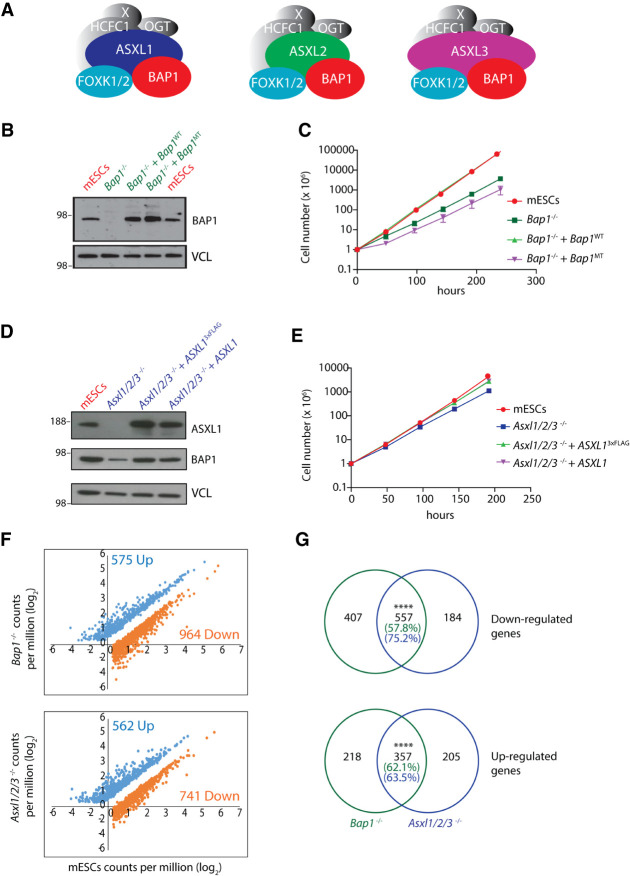
Figure 1.
BAP1 and ASXL1/2/3 are required for normal cell proliferation and for the expression of a common set of genes. (A) Illustration of the three different PR-DUB complexes and their components. (B) Western blot showing BAP1 levels in wild-type mESCs, Bap1−/− mESCs, Bap1−/− + Bap1WT, and Bap1−/− + Bap1MT mESCs. Vinculin (VCL) was used as a loading control. (C) Cell proliferation and growth curves of wild-type mESCs, Bap1−/− mESCs, Bap1−/− + Bap1WT, and Bap1−/− + Bap1MT mESCs were performed in three independent biological replicates per condition. (D) Western blots showing BAP1 and ASXL1 levels in wild-type mESCs, Asxl1/2/3−/− mESCs, Asxl1/2/3−/− + Asxl13xFLAG, and Asxl1/2/3−/− + hASXL1 mESCs. Vinculin was used as a loading control. (E) Cell proliferation of wild-type mESCs, Asxl1/2/3−/− mESCs, Asxl1/2/3−/− + Asxl13xFLAG, and Asxl1/2/3−/− + hASXL1 mESCs. The results present three independent biological replicates per condition. (F) Gene expression analysis of the indicated cell lines. Log2-normalized mean counts of mapped reads in Bap1−/− (top) and Asxl1/2/3−/− (bottom) mESCs versus wild-type mESCs. Only genes up- (blue) and down-regulated (orange) are shown. Down-regulated genes in Bap1−/− and Asxl1/2/3−/− mESCs were defined with the following criteria: log2 fold change ≤ −1, P-value ≤ 0.05, log2CPM in mESCs ≥ 0.5. Up-regulated genes in Bap1−/− and Asxl1/2/3−/− mESCs were defined with the following criteria: log2 fold change ≥ 1, P value ≤ 0.05, log2 CPM in KO ≥ 0.5. (G) Euler diagrams demonstrating down-regulated (top) and up-regulated (bottom) genes in common between Bap1−/− and Asxl1/2/3−/− mESCs versus wild-type mESCs. Fisher's exact test, (****) P-value < 0.0001.
Studies of Bap1 knockout mice have shown that loss of Bap1 leads to myeloid transformation and that BAP1 can deubiquitinate its complex partners OGT and HCFC1, thereby providing additional nonchromatin function to the protein (Dey et al. 2012). BAP1 has also been shown to stabilize ASXL2, whereas monoubiquitination of ASXLs led to stimulation of BAP1 enzymatic activity, indicating a complex role for BAP1 in regulating PR-DUB stability and function (Daou et al. 2018). Recently, a study showed a role for PR-DUB in CDKN2B gene activation (Wu et al. 2015), whereas two other studies showed that PR-DUB antagonizes PRC1-mediated H2AK119ub1 deposition and hence allows for target gene activation (Campagne et al. 2019; Kuznetsov et al. 2019). However, a general understanding of the steady state recruitment of PR-DUB in normal cells and its impact on transcription is still lacking.
Here, we address the role of PR-DUB in mouse embryonic stem cells (mESCs), how the complex is recruited to chromatin, and its functional relationship to PRC1/2.
Results
BAP1 and ASXL1/2/3 are required for normal cell proliferation and for the expression of a common set of genes
To dissect the role of PR-DUB and its core components, we deleted Bap1 in mESCs using CRISPR-Cas9 (Fig. 1B). Bap1 loss led to a significant reduction in cell growth rate and an increase of the doubling time of the mESCs from 15 to 22 hours (Fig. 1C; Supplemental Fig. S1A), consistent with a recent publication (He et al. 2019). We then ectopically expressed wild type (Bap1−/−+ Bap1WT) or a cancer-associated mutation (Bap1−/− + Bap1MT, A95D mutant) 3×FLAG-BAP1 in Bap1−/− mESCs and observed that wild-type BAP1 re-expression rescued the reduced cell growth, whereas the mutant did not. This suggests that reduced cell growth in BAP1 knockout mESCs is not a clonal artifact and that the catalytic activity of BAP1 contributes to the observed phenotype (Fig. 1B,C). Transcriptomic analysis revealed that loss of Bap1 led to deregulation of a large number of genes, and the expression of these genes was largely restored upon introduction of wild-type BAP1 (Supplemental Fig. S1B).
Next, we analyzed the effects of Asxl1/2/3 deletions on mESC cell proliferation by deleting the genes individually or in combination. (Fig. 1D). Although knockout of individual Asxl genes had no effect on cell proliferation, the deletion of all three genes led to a reduction of cell proliferation, albeit to a slightly lesser degree than observed for BAP1 knockout cells. (Fig. 1D,E). Similarly to BAP1, we observed a rescue of reduced cell growth rate by ectopic expression of wild-type ASXL1 (Asxl1/2/3−/−+ ASXL1). Collectively, this suggests a redundant role for the ASXL proteins in regulating PR-DUB activity in ES cells.
To analyze the effect of ASXL and BAP1 loss on gene expression, we performed RNA-seq on knockout clones and compared the expression changes. Principal component analysis of the biological replicates of Bap1−/− and Asxl1/2/3−/− mESCs revealed distinct clustering compared to parental mESCs as well as between them (Supplemental Fig. S1C). Differential gene expression analysis revealed that 964 and 741 genes were significantly down-regulated in Bap1−/− and Asxl1/2/3−/− mESCs, respectively, whereas 575 and 562 genes were significantly up-regulated in Bap1−/− and Asxl1/2/3−/− mESCs, respectively (Fig. 1F). The higher number of down-regulated genes could indicate that PR-DUB serves to promote gene expression on a wider level. We found that a large common set of genes to be significantly down- (557) or up-regulated (357) between Bap1−/− and Asxl1/2/3−/− mESCs (Fig. 1G), consistent with the idea that ASXL and BAP1 proteins affect a similar set of genes.
BAP1 binds to active genes involved in key cellular processes in an ASXL1/2/3-dependent manner
To obtain an understanding of how loss of PR-DUB leads to changes in gene expression, we mapped the genome-wide location of BAP1 in mESCs (Fig. 2A). We noticed that the enrichment of endogenous BAP1 on potential target genes was low using commercial and in-house produced antibodies. Therefore, we used two complementary strategies to map BAP1 binding sites genome-wide.
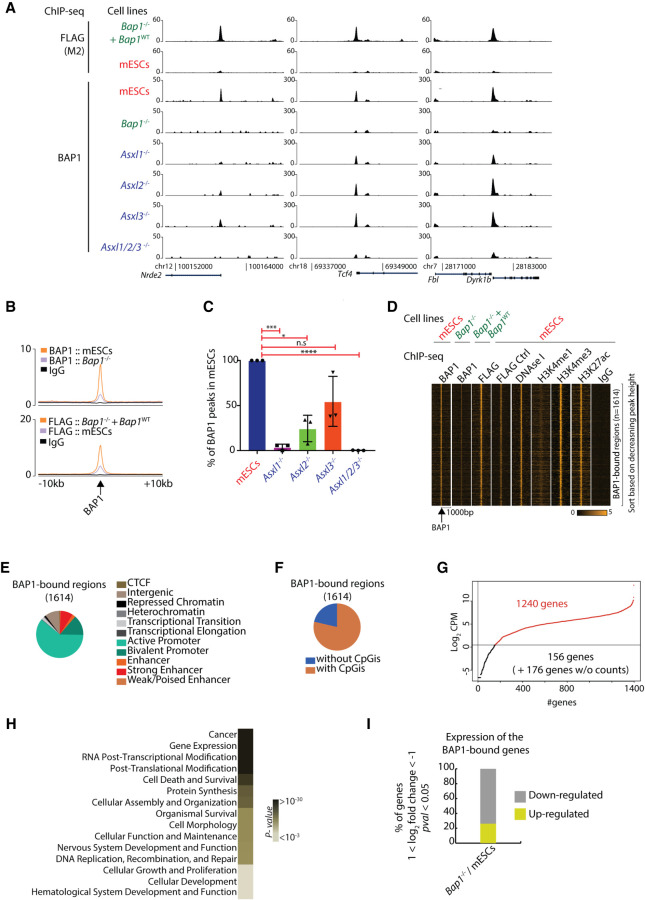
Figure 2.
BAP1 binds to active genes involved in key cellular processes in an ASXL1/2/3-dependent manner. (A) Screenshots for three representative loci showing ChIP-seq for 3×FLAG-BAP1 in Bap1−/−+ Bap1WT and wild-type mESCs (control; upper panel) and ChIP-seq for BAP1 in wild-type mESCs, Bap1−/−, Asxl1−/−, Asxl2−/−, Asxl3−/−, and Asxl1/2/3−/− mESCs. (B) Average profile of the BAP1 ChIP-seq in wild-type and Bap1−/− mESCs (upper) and 3×FLAG-BAP1 ChIP-seq in Bap1−/−+ Bap1WT and wild-type mESCs (bottom) signals in 20 kb around the BAP1-enriched regions. (C) Identification of the (percentage) BAP1 regions remaining in Asxl1−/−, Asxl2−/−, Asxl3−/−, and Asxl1/2/3−/− mESCs compared to wild-type mESCs (determined by three independent biological replicates). Unpaired t-test with Welch correction. (*) P-value = 0.0124; (***) P-value = 0.0004; (****) P-value < 0.0001. (D) Heat maps illustrating the signal of the indicated ChIP-seq profiles (BAP1, FLAG for BAP1, DNase I, H3K4me1, H3K4me3, H3K27ac, and IgG) in 2 kb around the 1614 identified BAP1-enriched regions. (E) Hidden Markov analysis for the 1614 BAP1 positions in mESCs and organization in the 11 indicated categories. (F) Identification of the enrichment of CpG islands (CpGi) for the 1614 BAP1 positions. (G) Log2-normalized mean counts of mapped reads for the 1572 BAP1-bound genes; 1240 genes are expressed and have an adequate number of mapped reads (log2 CPM ≥ 0.5). (H) BAP1 target genes as divided by the most significant GO terms. (I) Differential expression profile of the statistically significant BAP1-bound genes in Bap1−/− mESCs versus wild-type mESCs. The criteria for the selected genes are: 1 ≤ log2 fold change ≤ −1, P-value ≤ 0.05, log2 CPM in mESCs ≥ 0.5 (down-regulated genes) or log2 CPM in KO ≥ 0.5 (up-regulated genes).
First, we mapped the genome-wide localization of BAP1 using 3×FLAG-Bap1 expressing mESCs (Bap1−/−+ Bap1WT), where parental mESCs were used as a negative control using FLAG (M2) antibody (Fig. 2A top, B). Secondly, we complemented this data set with BAP1 profiles generated from an in-house-produced polyclonal antibody to increase sensitivity/specificity and to identify the location of endogenous BAP1 in mESCs, where Bap1−/− mESCs were used as a negative control (Fig. 2A bottom, B).
Using our FLAG-BAP1 data set, we observed that BAP1 localizes to 16,544 regions, identifying 8741 unique target genes. Meanwhile, mapping of endogenous BAP1 identified 1614 high-confidence regions (Fig. 2A,B). Given the specificity of both data sets, we concluded that mapping of endogenous BAP1 resulted in an underestimation of the absolute number of targets compared to the FLAG antibody in the transgenic mESCs. However, to avoid potential artifacts introduced by ectopic expression of BAP1 in further studies, we focused on the 1614 identified high-confidence regions and used the in-house-produced polyclonal antibody for further experiments.
To investigate if the ASXL proteins are involved in BAP1 recruitment, we performed ChIP-seq for BAP1 in Asxl1−/−, Asxl2−/−, Asxl3−/−, or Asxl1/2/3−/− mESCs (Fig. 2A, bottom). In general, loss of ASXL1 led to a bigger reduction in BAP1 binding than loss of ASXL2 or ASXL3. However, the simultaneous loss of all three ASXL proteins completely abolished BAP1 binding (Fig. 2C). We were not able to assess protein levels of ASXL2 or ASXL3 due to the lack of available antibodies. However, assuming equal stability and translation of transcribed Asxl1 and Asxl2 mRNAs (Supplemental Fig. S1D), our data suggest that ASXL1 is the most abundant ASXL protein in mESCs, which is consistent with our results showing it plays a more prominent role in recruiting BAP1 to chromatin than ASXL2 and ASXL3 in mESCs.
The majority of the BAP1-bound regions colocalized with chromatin signatures associated with transcriptionally active regions, such as H3K27ac, H3K4me1, H3K4me3, and DNase I hypersensitive sites (Fig. 2D). By performing a hidden Markov analysis for the 1614 high-confidence BAP1 peaks, we found that 61% of these peaks were located on active promoters, 14% on bivalent promoters, and 11% on enhancers (Fig. 2E). Overall, 78% of the BAP1-bound regions were also located on CpG islands (Fig. 2F). Relating BAP1 peaks to their target genes and their expression, we found that 1240 of the 1572 BAP1-bound genes were expressed in mESCs (Fig. 2G) and are involved in a variety of basic cellular processes (Fig. 2H). Furthermore, 74% of BAP1-bound genes were down-regulated upon BAP1 loss in mESCs (Fig. 2I). Taken together, we propose that, in normal steady state mESCs, BAP1 is dependent on stable ASXL expression to bind active regions and facilitate transcription of target genes, in contrast to the proposed repressive function of PR-DUB.
Site-specific association of BAP1 with chromatin is dependent on FOXK1 and FOXK2 and regulates H2AK119 ubiquitination
To identify potential TFs involved in PR-DUB recruitment to chromatin, we subjected the top 5000 BAP1 binding regions obtained from ChIP-seq analysis of the 3×FLAG-Bap1 transgenic mESCs (Bap1−/− + Bap1WT) to motif analysis. The strongest enrichment was found to match the consensus binding motif for the Forkhead (FOX) TF family, colocalizing with the apex of the BAP1 peaks (Fig. 3A). Since FOXK1/2 copurify with all three mammalian PR-DUB complexes and appear to be integral components of PR-DUB complexes (Yu et al. 2010; Dey et al. 2012; Ji et al. 2014), it prompted us to interrogate whether BAP1 binding to chromatin was dependent on FOXK1 and FOXK2.
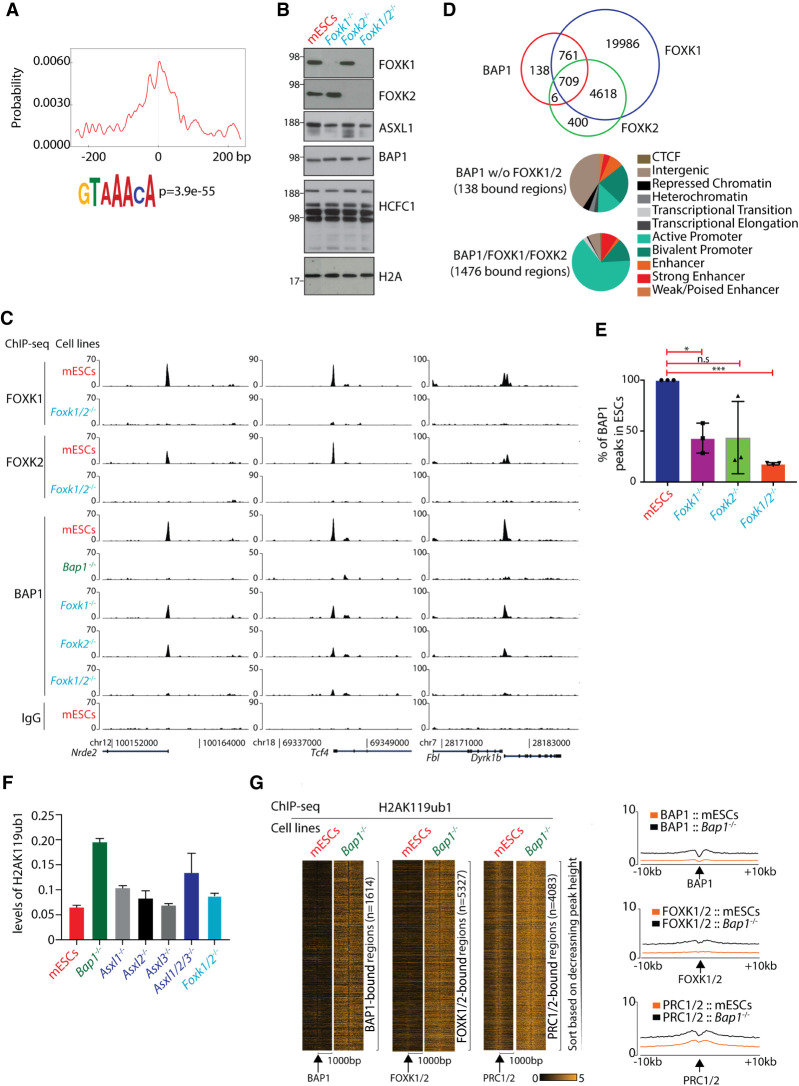
Figure 3.
Site-specific binding of BAP1 to chromatin is dependent on FOXK1 and FOXK2. (A) Motif analysis of the top 5000 3×FLAG-Bap1 binding regions revealed an enrichment for the FOX motifs centralized around the BAP1 peaks. (B) Western blots demonstrating the expression levels of FOXK1, FOXK2, ASXL1, BAP1, HCFC1, and H2AK119ub1 in wild-type mESCs, Foxk1−/−, Foxk2−/−, and Foxk1/2−/− mESCs. (C) ChIP-seq screenshot for three representative loci showing the binding of FOXK1 and FOXK2 in wild-type and Foxk1/2−/− mESCs and BAP1 in wild-type mESCs, Bap1−/−, Foxk1−/−, Foxk2−/−, and Foxk1/2−/− mESCs. (D) Euler diagrams showing the overlap between BAP1, FOXK1, and FOXK2 peaks, in wild-type mESCs showing that 1476 BAP1 regions colocalize with FOXK1/2 (top panel). Hidden Markov analysis for the 138 BAP1-bound not overlapping with FOXK1/2 binding in mESCs and the 1476 BAP1/FOXK1/FOXK2-bound regions and the organization of these regions into the 11 indicated categories (bottom panel). Based on hypergeometric test, the overlap between BAP1 and FOXK1 (P-value < 1.89 × −7) and the overlap between BAP1 and FOXK2 (P-value < 3.248 × −264) are statistically significant. (E) Identification of the (percentage) BAP1 peaks remaining in Foxk1−/−, Foxk2−/−, and Foxk1/2−/− mESCs compared to wild-type mESCs. Unpaired t-test with Welch correction. (*) P-value = 0.0215; (***) P-value < 0.0001. (F) The relative H2AK119ub1 levels in wild-type mESCs, Bap1−/−, Asxl1−/−, Asxl2−/−, Asxl3−/−, Asxl1/2/3−/−, and Foxk1/2−/− mESCs, as determined by mass spectrometry. (G) Heat maps illustrating enrichments of H2AK119ub1 in 2 kb around the BAP1, FOXK1/2, and PRC1/2 peaks in wild-type and Bap1−/− mESCs. The H2AK119ub1 ChIP-seqs were performed in triplicate and the first replicate is depicted. In the right panel, the average profiles of the H2AK119ub1 ChIP-seq signals in 20 kb around the BAP1, FOXK1/2, and PRC1/2 peaks are depicted.
First, we analyzed the genome-wide binding patterns of FOXK1 and FOXK2 in mESCs. For appropriate negative controls, we generated Foxk1−/−, Foxk2−/−, and Foxk1/2−/− mESCs using CRISPR-Cas9 (Fig. 3B). Loss of Foxk1/2 results in reduced proliferation of mESCs to levels similar to those observed in Bap1−/− and Asxl1/2/3−/− mESCs (Supplemental Fig. S2A), supporting the notion that FOXK1 and FOXK2 could be important components of the PR-DUB complex. Genome-wide ChIP-seq analysis of FOXK1 and FOXK2 in mESCs revealed 26,074 and 5733 binding regions, respectively, with 5327 regions in common, suggesting functional redundancy between FOXK1 and FOXK2 (Fig. 3C; Supplemental Fig. S2B). Approximately 70% of the FOXK1/FOXK2 binding sites were associated with active gene regulatory elements such as promoters and enhancers (Supplemental Fig. S2C). In agreement with the BAP1 binding pattern, GO term analysis of the FOXK1/2 target genes highlighted general functions in basic cellular processes and metabolism (Supplemental Fig. S2D).
We performed RNA-seq to evaluate the effect of deleting Foxk1 and Foxk2 on gene expression and identified 593 and 146 statistically significant up-regulated and down-regulated genes, respectively, as compared to wild-type mESCs (Supplemental Fig. S2E). A higher fraction of genes down-regulated in Foxk1/2−/− showed an overlap with the commonly down-regulated genes by Bap1−/− and Asxl1/2/3−/− mESCs, as compared to the commonly up-regulated genes in the respective cell lineages (Supplemental Fig. S2F).
Analysis of the genome-wide occupancy of BAP1, FOXK1, and FOXK2 in mESCs revealed that they colocalize to DNase I hypersensitive site-positive region (Supplemental Fig. S2G,H). Ninety-one percent of the high-confidence 1614 BAP1-bound regions are co-occupied by either FOXK1 or FOXK2 or both (Fig. 3D), supporting a role for FOXK1/2 in the PR-DUB complex. Sixty-five percent of the BAP1/FOXK1/FOXK2-bound regions are located on active promoters, whereas only 14% of the BAP1-bound regions without FOXK1/2 are located on active promoters, suggesting a specific function of FOXK1/2 in maintaining transcription through the recruitment of BAP1 (Fig. 3D). We also analyzed the effect of deleting Asxl1/2/3 on the binding of FOXK1 and FOXK2. Although FOXK1 and FOXK2 binding to the BAP1/FOXK1/FOXK2-bound regions is slightly reduced, their specific binding to these regions is still retained and therefore appears independent of ASXL1/2/3 (Supplemental Fig. S2I).
To investigate whether FOXK1 or FOXK2 or both affect recruitment of BAP1, we determined the genome-wide BAP1-bound regions in Foxk1−/−, Foxk2−/−, and Foxk1/2−/− mESCs (Fig. 3C bottom). Approximately half of the BAP1-bound regions were lost in either Foxk1−/− or Foxk2−/− mESCs, whereas ∼75% of BAP1-bound regions were lost in Foxk1/2−/− mESCs (Fig. 3E). Overall, these results demonstrate that FOXK1 and FOXK2 are required for the association of PR-DUB with the majority of its binding sites in mESCs. These results are consistent with previous published data for cancer cell lines that showed the involvement of FOXK2 in the binding of BAP1 to specific genes in a lung cancer cell line (Okino et al. 2015) and an osteosarcoma cell line (Ji et al. 2014).
PR-DUB de-ubiquitinates H2AK119ub1, a chromatin mark imposed by PRC1. We analyzed the level of H2AK119ub1 in histones extracted from Bap1−/−, Asxl1−/−, Asxl2−/−, Asxl3−/−, Asxl1/2/3−/−, and Foxk1/2−/− mESCs using ArgC digestion followed by mass spectrometry analysis. Loss of BAP1 in mESCs increased the global levels of H2AK119ub1 by approximately threefold, and Asxl1/2/3−/− mESCs in parallel exhibited a similar but slightly lower increase of H2AK119ub1 (Fig. 3F). In contrast, the loss of ASXL1 alone or of FOXK1/2 only led to a minor increase in the global levels of H2AK119ub1 (Fig. 3F). This could be seen as a surprising result, in particular when taking into consideration the effect of deleting these genes on the site-specific localization of BAP1 (Figs. 2C, 3E). However, since H2AK119ub1 is associated with large parts of the genome in a non-sequence-specific manner and loss of BAP1 leads to a global increase in H2AK119ub1, this means that we are only able to identify a minor fraction of genomic sites where BAP1 is associated. In this perspective, it is not surprising that the deletion of Foxk1/2 does not lead to a strong global increase in H2AK119ub1, because these TFs “only” appear to be involved in the site-specific binding of PR-DUB (Fig. 3F; Supplemental Fig. S3A). Moreover, since ASXL2 and ASXL3 still can be expressed in the Asxl1−/− cells, these proteins can contribute to the global levels of PR-DUB activity.
To analyze the consequences of BAP1 loss on H2AK119ub1 on specific genomic regions, we performed ChIP-seq analysis for H2AK119ub1 in wild-type and Bap1−/− mESCs using Drosophila chromatin spike-in for normalization. Loss of BAP1 led to a local increase of H2AK119ub1 around the BAP1 or FOXK1/2 binding regions in mESCs, where the steady state levels of H2AK119ub1 in wild-type mESCs are overall lower than observed for bona fide PRC1/2 target genes (Fig. 3G; Supplemental Fig. S3B,C). Collectively, our results show that PR-DUB binding to target genes is dependent on FOXK1 and FOXK2 and that loss of the complex leads to increased H2AK119ub1 levels at the genes and their decreased expression.
PR-DUB and PRC1/2 share a limited set of target genes and operate independently during ESC differentiation
Previous studies have suggested that PR-DUB is required for PRC2 recruitment in mammalian cells and to colocalize with PRCs in Drosophila melanogaster (Scheuermann et al. 2010; Abdel-Wahab et al. 2012). To explore the potential interaction between PR-DUB and PRC1/2 in more detail, we compared the localization of components of the complexes in mESCs (Fig. 4A). Only 10% of the BAP1-bound regions (157 regions) colocalize with either SUZ12 (which is part of PRC2) or RING1B (which is part of PRC1), and the majority of these regions are located on bivalent promoters positive for H3K4me3 and H3K27me3 (Fig. 4A,B). In contrast, the majority of the BAP1-bound regions, not associated with PRC1/2, are located on active promoters defined by H3K4me3 and active transcription (Fig. 4B). The increased H2AK119ub1 levels in Bap1−/− mESCs are independent of detectable RING1B co-occupancy (Fig. 4C,D), suggesting that, in those genomic regions, RING1A may restore the H2AK119ub1 levels. In agreement with this, H2AK119ub1 and H3K27me3 levels were higher for the BAP1/PRC1/PRC2-bound regions compared to the BAP1-bound regions without PRC1/2.
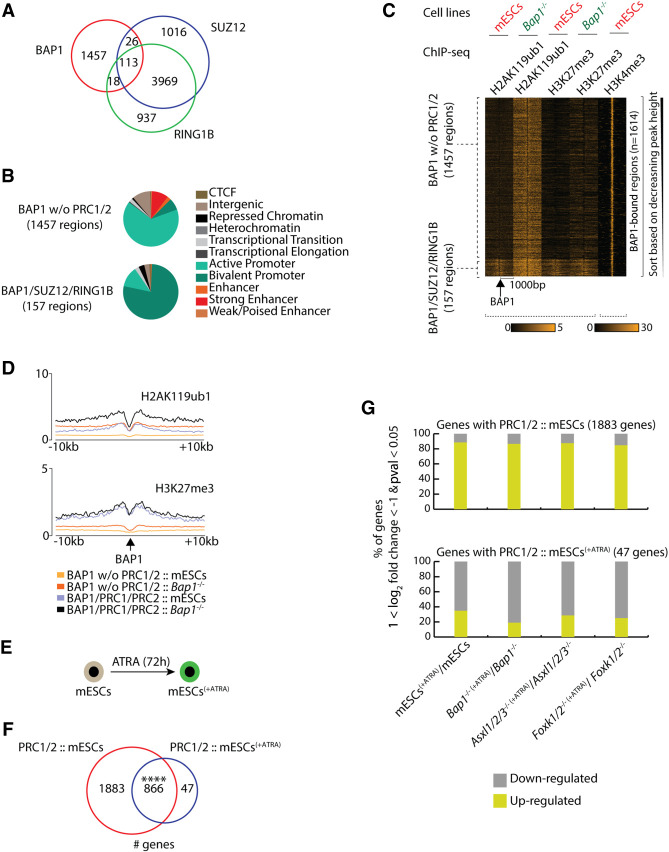
Figure 4.
PR-DUB and PRC1/2 share a limited set of bivalent target genes. (A) Euler diagram showing the overlap between BAP1, SUZ12, and RING1B binding regions in wild-type proliferating mESCs. (B) Hidden Markov analysis for the 1457 BAP1-bound regions positions not associated with SUZ12 or RING1B in mESCs, and for the 157 BAP1/SUZ12/RING1B-bound regions in wild-type mESCs. The distribution of the regions is shown in the 11 indicated categories. (C) Heat maps showing the depicted ChIP-seq profiles (H2AK119ub1, H3K27me3, and H3K4me3) in 2 kb centered around the BAP1-bound regions in wild-type and Bap1−/− mESCs. (D) The average signal of H2AK119ub1 and the H3K27me3 binding profile shown in a window of 20 kb around the BAP1-bound regions with or without SUZ12/RING1B in wild-type and Bap1−/− mESCs. (E) Schematic representation of mESCs subjected to all-trans retinoic acid (ATRA)-induced differentiation for 72 h (mESCs[+ATRA]). (F) Euler diagram showing the overlap of PRC1/2 target genes, identified in proliferating mESCs and in mESCs treated with ATRA for 72 h (mESCs[+ATRA]). Fisher's exact test, (****) P-value < 0.0001. (G) Expression changes of genes in response to ATRA-induced differentiation in wild-type, Bap1−/−, Asxl1/2/3−/−, and Foxk1/2−/− mESCs. Upper panel shows the % of the 1883 genes that are bound by PRC1/2 only in mESCs and are up- or down-regulated in response to ATRA. Lower panel shows the expression changes of the 47 genes that are only bound by PRC1/2 in mESCs treated with ATRA for 72 h (mESCs[+ATRA]).
To understand if PR-DUB can impact PRC1/2 binding, we mapped the genome-wide binding of RING1B and SUZ12 in wild-type and Bap1−/− mESCs by ChIP-seq. We found that the 4082 regions commonly bound by RING1B and SUZ12 (Fig. 4A) were only slightly reduced upon loss of BAP1 (Supplemental Fig. S4A), ASXL1/2/3, or FOXK1/2 (Supplemental Fig. S4B). These results demonstrate that PR-DUB binding does not influence the binding of PRC1/2 to chromatin in mESCs.
Next, we investigated whether PR-DUB is required for the regulation of PRC1/2 target genes during differentiation. We treated mESCs (wild type, Bap1−/−, Asxl1/2/3−/−, and Foxk1/2−/− mESCs) with all-trans retinoic acid (ATRA) and subjected them to a monolayer differentiation protocol for 72 h (Fig. 4E). To assess the impact of deleting components of the PR-DUB complex on gene expression during differentiation, we performed RNA-seq (Supplemental Fig. S4C) for untreated (0 h) and treated with ATRA mESCs (72 h). This analysis showed that the deletion of the PR-DUB encoded genes did not lead to detectable differences in the down-regulation of the pluripotent markers or the up-regulation of the neuronal markers during ATRA-induced differentiation, suggesting that the PR-DUB complex is not required for this differentiation (Supplemental Fig. S4D).
To understand how ATRA-induced differentiation would affect the binding of PR-DUB and PRC1/2, we performed ChIP-seq. This led to the identification of 2749 and 913 PRC1/2 target genes in untreated (0 h) and ATRA-treated mESCs (72 h), respectively, with 866 genes in common (Fig. 4F), suggesting that only a few genes acquire PRC1/2 using this differentiation protocol. During differentiation, 1883 PRC1/2 target genes lost their Polycomb status and RNA-seq demonstrated that these were mainly up-regulated, whereas 47 PcG negative genes expressed in mESCs gained Polycomb status and were mainly repressed upon differentiation (Fig. 4G). Importantly, the loss of BAP1, ASXL1/2/3, or FOXK1/2 did not affect the transcriptional activation of mESC-specific PRC1/2 targets or interfere with the repression of de novo PRC1/2 target genes during differentiation (Fig. 4G). The 866 genes that retained PRC1/2 binding upon differentiation were mainly up-regulated, even in the absence of BAP1, ASXL1/2/3, or FOXK1/2 (Supplemental Fig. S5A). Overall, these results suggest that PR-DUB does not have a major impact on PRC1/2 function or the regulation PcG target genes during ATRA-induced differentiation.
Subsequently, we investigated the H3K27me3 levels in untreated and ATRA-treated mESCs. In the treated cells, H3K27me3 was increased at genes (at the TSS and within their gene-bodies), which were down-regulated and were either de novo PRC1/2 target genes or genes which retained PRC1/2 in the presence of ATRA (Supplemental Fig. S5B,C). The specific PRC1/2 targets in untreated mESCs, which were up-regulated in response to ATRA treatment, had slightly higher levels of H3K27me3 in their gene-bodies in untreated cells (Supplemental Fig. S5B,C). The H3K27me3 levels, at genes bound by PRC1/2 at both untreated and treated mESCs, which were up-regulated by ATRA treatment, were slightly increased around their TSS in response to ATRA (Supplemental Fig. S5B,C). Taken together, we conclude that the H3K27me3 patterns correlate for most of the genes, as expected, with the changes in gene expression.
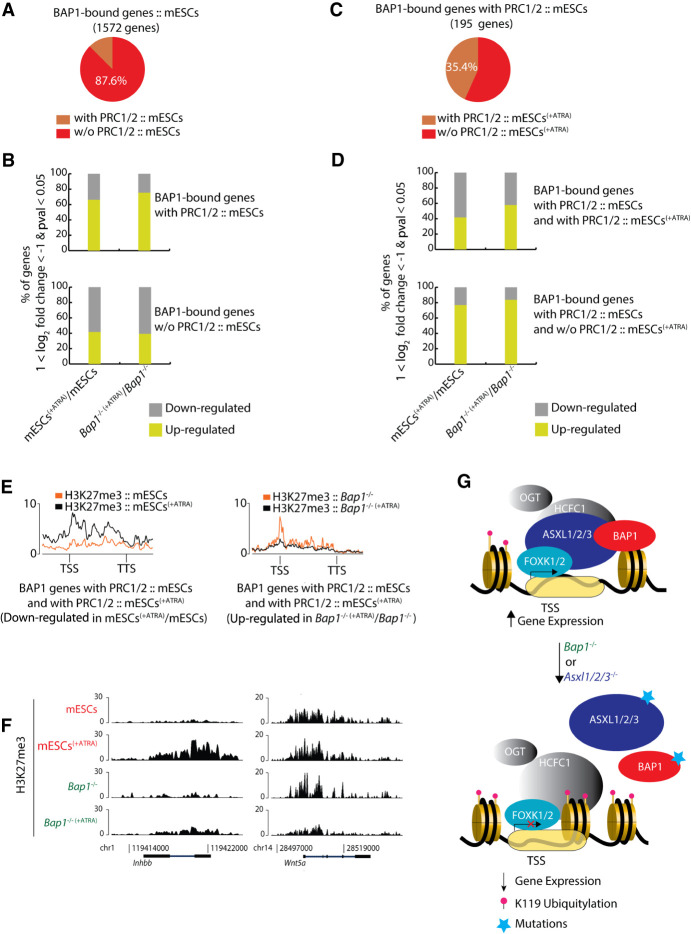
Subsequently, we focused on the BAP1 target genes with or without PRC1/2 binding, prior to the 72 h of ATRA-induced differentiation and the effect that loss of BAP1 had during differentiation (Fig. 5A). The majority of the genes bound only by BAP1 were repressed in response to ATRA, whereas the BAP1/PRC1/PRC2 cobound genes were activated, even in the absence of BAP1 (Fig. 5B). Almost none of the BAP1 target genes (99.7%) that were negative for PRC1/2 binding in mESCs acquired PRC1/2 binding upon differentiation, and their expression was not affected by loss of BAP1 in proliferating and differentiated mESCs (Supplemental Fig. S5D,E). The majority (64.6%) of the BAP1/PRC1/PRC2 target genes that lost Polycomb binding during differentiation were transcriptionally up-regulated, and the absence of BAP1 did not significantly change this (Fig. 5C,D). The remaining (35.4%) BAP1/PRC1/PRC2 target genes were repressed and retained their Polycomb status during differentiation, and loss of BAP1 only led to a minor change in their transcriptional response (Fig. 5C,D). Specifically, 58.3% of the BAP1/PRC1/PRC2 target genes in untreated mESCs, which retained PRC1/2 and were repressed in mESCs upon 72 h of ATRA-induced differentiation, exhibited higher levels of H3K27me3 in ATRA-treated mESCs compared to untreated mESCs (Fig. 5D–F). Moreover, 57.7% of the BAP1/PRC1/PRC2 target genes in untreated mESCs, which retained PRC1/2 and were up-regulated in Bap1−/− mESCs in response to ATRA, exhibited higher levels of H3K27me3 in untreated Bap1−/− mESCs compared to ATRA-treated Bap1−/− mESCs (Fig. 5D–F). Taken together, these data suggest that PR-DUB has a role in regulating gene expression, which could be either global (Fig. 4F,G) or local upon ATRA-induced differentiation (Fig. 5C,D). BAP1, as the core catalytic subunit of the PR-DUB complex, ensures that FOXK1/2-bound regions are kept free of H2AK119ub1 that can impede efficient expression of these target genes, which are required for cell growth and homeostasis.
Figure 5.
Correlation between PR-DUB target genes and PRC1/2 reveals the role of PR-DUB in gene expression. (A) Pie chart depicting the percentage of the 1572 BAP1-bound genes, which either colocalize or not with PRC1/2 in proliferating mESCs. (B) The upper panel shows the % of the BAP1-bound genes that are up- or down-regulated in response to ATRA and are also bound by PRC1/2 in proliferating (untreated) mESCs. The lower panel shows the % of the BAP1-bound genes that are not bound by PRC1/2 in proliferating (untreated) mESCs, which are up- or down-regulated in response to ATRA. (C) Pie chart depicting the percentage of the 195 BAP1/PRC1/PRC2-bound genes in proliferating (untreated) mESCs, illustrating the proportion that retain or lose PRC1/2 in response to 72 h ATRA treatment (mESCs[+ATRA]). (D) The upper panel shows the % of the BAP1/PRC1/PRC2-bound genes in mESCs, which retain PRC1/2 in mESCs(+ATRA) and are up- or down-regulated in response to ATRA-induced differentiation. The lower panel shows the % the BAP1/PRC1/PRC2-bound genes in untreated mESCs, which lose the PRC1/2 binding in mESCs(+ATRA) and are up- or down-regulated in response to 72 h of ATRA treatment. (E) The average signal (in metagenes) of H3K27me3 in the wild-type (left) or Bap1−/− (right) mESCs, in untreated (orange) and in response to ATRA-induced differentiation (black) for each cell type, for the BAP1/PRC1/PRC2 bound genes in mESCs which retain PRC1/2 in mESCs(+ATRA) and are either down-regulated (left) or up-regulated (right) upon 72 h of ATRA treatment. (F) ChIP-seq screenshot for two representative loci (based on Fig. 4E) showing the binding of H3K27me3 in untreated and ATRA-treated wild-type mESCs and Bap1−/− mESCs. (G) Model for the role of the PR-DUB complex in regulating histone H2AK119ub1 levels and transcription. BAP1 binding to chromatin depends on FOXK1/2 and the formation of a PR-DUB complex. The binding is required for retaining a chromatin environment that supports gene expression. Loss of BAP1, FOXK1/2, or ASXL1/2/3 leads to loss of PR-DUB from its target genes, the deposition of H2AK119ub1, and decrease in gene expression.
Discussion
Different components of the PR-DUB complex are mutated with high frequency in a variety of cancers and some rare congenital diseases. However, the mechanism by which PR-DUB regulates transcription has been unclear, and the complex has been reported to have both transcriptional activating and repressive properties. Here, we have systematically analyzed the regulatory function of PR-DUB in mouse embryonic stem cells and the mechanism by which the complex is recruited to specific sites. We have shown that the FOXK1 and FOXK2 transcription factors are essential for recruiting PR-DUB to high-confidence PR-DUB binding sites (Fig. 3). Furthermore, we have demonstrated that the three ASXL proteins also are required for the recruitment of BAP1 to these sites (Fig. 2). PR-DUB binds to a broad range of genes involved in basic cellular processes. Our results show that PR-DUB binds to active genes and is involved in maintaining their activity (Fig. 2), which is in agreement with previous reports (Wu et al. 2015; Campagne et al. 2019). Thus, PR-DUB is involved in transcriptional activation. We propose that PR-DUB, by preventing ubiquitination of H2AK119, acts to safeguard active genes from repression and to maintain chromatin in an optimal configuration, ensuring the full potential transcriptional output and response to transcriptional cues.
We and others have previously shown that FOXK1 and FOXK2 copurify with ASXL1/2/3 and BAP1 (Yu et al. 2010; Baymaz et al. 2014; Ji et al. 2014; Wu et al. 2015). Purification of proteins associated with FOXK1 and FOXK2 also shows robust enrichment of PR-DUB, strongly supporting that they are integral components of PR-DUBs (Ji et al. 2014; Okino et al. 2015). FOXK1 and FOXK2 also copurify with other chromatin modifying complexes (Liu et al. 2019; Nestal de Moraes et al. 2019), suggesting that these TFs target a variety of enzymatic activities to chromatin to regulate transcription. Although other genome-wide studies have found strong enrichment of ETS motifs for BAP1 and ASXL1 binding regions (Abdel-Wahab et al. 2012; Dey et al. 2012), our analysis of BAP1 binding regions showed that the strongest enrichment of the consensus motif for members was the FOX family. In agreement with this, we found that FOXK1 and FOXK2 are essential for site-specific recruitment of BAP1 in mESCs and that they appear to have overlapping function (Fig. 3). Our data suggest that FOXK1 in addition to FOXK2 (Ji et al. 2012) could act as pioneering TFs, similar to the FOXA TFs (Iwafuchi-Doi et al. 2016). The observation that FOXK1/2 target genes are, in general, enriched for genes regulating basic cellular processes and cell metabolism (Supplemental Fig. S2) is in agreement with recent studies that implicate these TFs in the regulation of glycolysis and mitochondrial metabolism (He et al. 2018; Sukonina et al. 2019).
The genes bound by PR-DUB/FOXK1/FOXK2 are, in general, transcriptionally active and are therefore typically not bound by the PRC1 and PRC2 complexes. However, we have observed that low levels of H2AK119ub1 are distributed over the entire mESC genome. Although the widespread distribution of low levels of H2AK119ub1 may have a role in transcriptional regulation, it could also be the default state of chromatin. In contrast, high levels of H2AK119ub1 are observed at Polycomb target genes (Figs. 3, 4), which are transcriptionally repressed. Since, H2AK119 is not ubiquitinated around the TSSs of the expressed genes, the targeted binding of PR-DUB by FOXK1/2 could be one of the mechanisms by which chromatin is kept in a transcriptionally permissive state. Similar mechanisms of action have been suggested for histone lysine demethylases and methylcytosine hydroxylases that are, by default, recruited to CpG islands either by binding to H3K4me3 or directly to CpG-rich sequences (Blackledge et al. 2010; Schmitz et al. 2011; Williams et al. 2011). However, the major difference between PR-DUB and such factors is that PR-DUB depends on a TF-based recruitment mechanism.
Although PR-DUB might also contribute to gene repression by eliminating H2AK119ub1, allowing the efficient interaction of repressive factors with chromatin, it appears that the major role of FOXK1/2 is to recruit PR-DUB to active genes (Fig. 3). In fact, we observed a limited overlap between PR-DUB and PRC1/2 in mESCs, restricted to bivalent promoters (Fig. 4). In addition, we only observed a minor effect on PRC1/2-regulated genes upon PR-DUB loss in mESCs (Figs. 4, 5). Thus, we conclude that the major site-specific function of PR-DUB is to ensure the expression of genes bound by FOXK1 and FOXK2.
We found that BAP1 targeting to chromatin is also dependent on the ASXL proteins, suggesting that, in disease, mutations in the ASXL genes decouple BAP1 from chromatin (Fig. 5G). ASXL1 is frequently found mutated in clonal hematopoiesis, with high frequency in myelodysplastic syndromes (MDS), and various forms of myeloproliferative disease (MPD), including acute myeloid leukemia (AML) (Micol and Abdel-Wahab 2016; Bowman et al. 2018). Moreover, truncation of ASXL1 in myeloid leukemias dysregulates BAP1, enhancing its activity (Balasubramani et al. 2015; Asada et al. 2018). Taken together, this suggests that initially a variety of genes involved in basic cellular processes and metabolism are deregulated due to accumulation of H2AK119ub1 resulting in poor cell growth, homeostasis, and differentiation. This agrees with the clinical features observed of MDS patients harboring mutations in ASXL1 (Micol and Abdel-Wahab 2016). Progression of MDS to MPD/AML then requires further mutations leading to oncogene activation, alleviating the proliferative defects imposed by ASXL1 mutation for progression to MPD. Mutations of ASXLs are often heterozygous (Micol and Abdel-Wahab 2016), and the frequency of ASXL mutations found in different tissues may reflect ASXL expression patterns. Thus, ASXL mutations may be less detrimental than BAP1 mutations due both to the type of mutation and to the redundancy of ASXL expression. In support of this notion is the recent finding that BAP1 mutations induce apoptosis in a number of tissues (He et al. 2019).
Collectively, we present a comprehensive analysis of the PR-DUB complexes in mouse embryonic stem cells. We show that BAP1 is recruited to site-specific regions of the genome by FOXK1/2 and that this recruitment is also dependent on the ASXL proteins. Furthermore, our results demonstrate that PR-DUB plays a role in ensuring a chromatin environment that is permissive for transcriptional activation of genes important for general cellular functions such as cell metabolism and homeostasis (Fig. 5G) and, in agreement with this, that PR-DUB-deficient cells grow slower.
Methods
Cell lines
Mouse embryonic stem cells were cultured on gelatin-coated dishes in 2i/LIF-containing medium, supplemented with 1× Pen-Strep (Gibco), 50 µM β-mercaptoethanol (Gibco), N2 + B27 (Thermo Fisher Scientific), GSK3i (CHIR99021), MEKi (PD0325901), and Leukemia Inhibitory Factor (LIF; produced in the lab). Cells were passaged every 2–3 d by removing the medium, washing cells with PBS, dissociating cells with either trypsin-EDTA (Gibco) or Accutase (Sigma-Aldrich), resuspending cells in medium, and pelleting by centrifugation. Finally, the cells were resuspended at a density of ∼106 cells/10 cm dish. All-trans retinoic acid treatment was used at 1 µM to induce monolayer differentiation of mESCs for 72 h. The CAG promoter present in the Piggy-BAC vector directed the expression of 3×FLAG-Bap1 in the transgenic mESCs.
CRISPR-Cas9
Knockout mESC lines were generated by CRISPR-Cas9 targeting in E14 cells. Single guide (sg) RNAs were cloned into pX458 and transfected into E14 cells using Lipofectamine 2000 (Thermo Fisher Scientific). The sequences of sgRNA encoding oligonucleotides were: Bap1 exon 5 (CCCACGCTGAGCCGAATGA), Asxl1 exon 3 (TGAAAAGACTAATGCGGCC), Asxl2 exon 3 (TATACACGCCCATTCTACC), Asxl3 exon 6 (CGTAAGGCTGCAATCTCGAG), Foxk1 exon 1 (GCTCGGAGCTGCGCGAACA), and Foxk2 exon 1 (GCAGGCGATGGC GGCCGCG). Single cell clones were isolated by GFP sorting using FACS Aria III into 0.2% gelatin-coated 96-well plates containing 2i/LIF and expanded. Knockout clones were validated by Sanger sequencing of genomic DNA and western blotting, when a specific antibody was available.
Antibodies
Monoclonal antibodies to BAP1 (M2, available from Diagenode as C15200212) and ASXL1 (MAb32) were generated using standard protocols following immunization of BALB/c mice with the full-length BAP1 and ASXL1 human proteins, respectively. These antibodies were used in western blotting. Polyclonal mouse antisera from mice hyperimmunized by human BAP1 protein were used for ChIP. In addition, the following antibodies were used for western blotting and/or ChIP assays as indicated: FOXK1 (Abcam, ab18196), FOXK2 (Abcam, ab83286), HCFC1 (Bethyl, A301-399), H2A (Abcam, ab18255), H2AK119ub1 (Cell Signaling Technology, 8240), H3K27me3 (Cell Signaling Technology, 9733), RING1B (Cell Signaling Technology, 5694), SUZ12 (Cell Signaling Technology, 3737), anti-FLAG M2 (Merck, F1804), and Vinculin (Merck, V4505).
Mass spectrometry
Histone extraction was performed as described previously with minor modifications (Sidoli et al. 2016). Briefly, cell pellets were resuspended directly in 100 µL of 0.2 M sulfuric acid for 2 h at 4°C, skipping the nuclei isolation step to minimize de-ubiquitinylation. After centrifugation at 3400g for 5 min, the supernatant was mixed with 33% trichloroacetic acid (by adding 50 µL of 100% trichloroacetic acid to the previous solution) and incubated for 1 h on ice. After centrifugation at 3400g for 5 min, the supernatant was discarded, and the pellet was washed twice with acetone to remove acid in excess. The analysis was performed with a nanoHPLC Dionex Ultimate 3000 RSLC coupled online with an Orbitrap Fusion Lumos (both Thermo Fisher Scientific). Peptides were loaded onto an in-house-packed nanocolumn (25 cm length, 75 µm ID, 3 µm C18-AQ particles) and separated using a 45-min gradient from 2% to 30% buffer B (Buffer A: 0.1% formic acid; Buffer B: 80% acetonitrile, 0.1% formic acid). Electrospray voltage was set at 2.5 kV and capillary temperature was set at 275°C. Samples were acquired using a data-dependent acquisition (DDA) and a data-independent acquisition (DIA) method. DDA and DIA acquisitions were performed as previously described (Sidoli et al. 2015), except for the DIA MS/MS resolution, which was set at 15,000. Higher-energy collisional dissociation (HCD) was set to 27. The spectra obtained from the DDA analysis were searched using Proteome Discoverer (v2.4, Thermo Fisher Scientific). These searches were performed using a database of histone proteins (UniProt), specifying “ArgC” as the digestion enzyme and including the GlyGly dynamic modification on lysine residues as a possible match. GlyGly corresponds to the mass of two glycine residues covalently bound to the lysine side chain, which is the short ubiquitin C terminus remaining attached to the histone peptide after ArgC digestion (as the C terminus of ubiquitin is RGG). After identifying the H2A peptide with the GlyGly group and its respective unmodified counterpart (VTIAQGGVLPNIQAVLLPKKTESHHK), the extracted ion chromatogram was performed with Skyline (MacLean et al. 2010). The area of the chromatogram of the ubiquitinylated peptide was divided by the sum of the unmodified + the ubiquitinylated to obtain the relative abundance in percentage. This percentage is not to be intended as exact stoichiometry, as the two peptides might have minimal differences in ionization efficiency. However, since the extra GlyGly group is a minor mass compared to the peptide sequence, the calculated percentage is a relatively accurate approximation of the relative abundance of ubiquitinylation on chromatin.
RNA-seq and data analysis
RNA was isolated from cells and prepared for sequencing based on standard kits (TrueSeq RNA library prep kit v2, Illumina) and the manufacturer's instructions and sequenced on Illumina NextSeq 500. Reads were aligned to the mouse reference genome (mm10) using HISAT2 with standard parameters (Kim et al. 2019). Mapped reads were counted with the iRNA-seq pipeline using the “gene” option for analysis of differential gene expression (Madsen et al. 2015). In all cases, up- and down-regulated genes were selected to have at least ±1 log2 fold-change in RNA levels and CPM > 0.5. These genes were used to for performing Gene Ontology analysis (http://www.metascape.org/ and Ingenuity Pathway analysis [Krämer et al. 2014; Zhou et al. 2019]).
ChIP-seq and data analysis
ChIP assays were performed according to the standard protocols. Chromatin was crosslinked with 1% formaldehyde (Sigma-Aldrich) directly in the dish, and DNA was sonicated to 200 to 400 bp fragments (Bioruptor, Diagenode). Fifty to 1000 ug of chromatin of mESCs were incubated with the indicated antibodies. Rabbit IgG was used as the negative control. ChIP-seq libraries were prepared according to standard protocols (NEB and Illumina). ChIP-seq for H2AK119ub1 and H3K27me3 were spiked-in with 5% Drosophila chromatin and normalized accordingly. Raw reads were aligned to the mouse reference genome (mm10) using Bowtie 2, discarding reads mapping to multiple genomic locations (Langmead and Salzberg 2012). Data processing and analysis were mainly performed as previously described (Soler et al. 2011; Stadhouders et al. 2015; Kolovos et al. 2016). Peaks were called based on standard criteria as stated for each ChIP-seq in the results section. Peaks for the BAP1 FLAG ChIP-seq were defined with the following criteria: peak ≥ 25, FDR ≤ 0.001, fold change over M2 control ≥ 3. The polyclonal BAP1 antibody high-confidence regions were defined with the following criteria: peak ≥ 20, FDR ≤ 0.001, fold change over Bap1−/− control ≥ 3. FOXK1 and FOXK2 regions were defined with the following criteria: peak ≥ 25, FDR ≤ 0.001, fold change over Foxk1/2−/− control ≥ 3. The identification of the target genes of the peaks and the overlap between binding sites were identified using the iRanges package and the “findOverlaps” function from GenomicRanges (Lawrence et al. 2013). Heat maps and average profiles were created using the EaSeq suite (Lerdrup et al. 2016). Motif analysis was performed using the MEME suite (Bailey et al. 2009). Hidden Markov analysis was performed based on already defined classes of ChromHMM states and as described previously (Pintacuda et al. 2017). ChIP-seqs for H3K4me1 (GSM1000121), H3K4me3 (GSM1000124), and DNase I (GSM1014154) in mESCs were obtained from the ENCODE consortium (Yue et al. 2014) and analyzed as described above.
Statistics
Fisher's exact test was performed in GraphPad (https://www.graphpad.com/quickcalcs/). Growth curves were plotted in Prism, where three independent replicates were plotted for each condition, depicting also their standard deviation error. Hypergeometric tests for the Euler diagrams were plotted in R using the appropriate functions (R Core Team 2018).
Data access
All raw mass spectrometry files generated in this study have been submitted to the free repository Chorus (https://chorusproject.org/) under project no. 1657. All raw sequencing data (ChIP-seq and RNA-seq) generated in this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP221249.
Competing interest statement
K.H. has been a consultant for Novo Holdings A/S. All other authors declare no competing interests.
Supplementary Material
Acknowledgments
We thank the members of the Helin lab for discussion, technical advice, and support. We thank Ulla Toftegaard for excellent technical assistance. P.K. was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (Rubicon fellowship; 019.162LW.011) and a Marie Curie Individual Fellowship (H2020-MSCA-IF-2017). K.N. was supported by Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers, Japan Society for the Promotion of Science (S2704). The work in the Helin laboratory was supported by the Danish Cancer Society (R167-A10877), the Danish National Research Foundation (DNRF82), the Independent Research Fund Denmark (6153-000005; 7016-00067), The Neye Foundation, through a center grant from the Novo Nordisk Fonden (NNF) to the NNF Center for Stem Cell Biology (NNF17CC0027852), and through the Memorial Sloan Kettering Cancer Center Support Grant (National Institutes of Health P30 CA008748).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.261016.120.
References
- Abdel-Wahab O, Dey A. 2013. The ASXL–BAP1 axis: new factors in myelopoiesis, cancer and epigenetics. Leukemia 27: 10–15. 10.1038/leu.2012.288 [DOI] [PubMed] [Google Scholar]
- Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, Pandey S, Patel JP, Chung YR, Koche R, et al. 2012. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 22: 180–193. 10.1016/j.ccr.2012.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Gao J, Adli M, Dey A, Trimarchi T, Chung YR, Kuscu C, Hricik T, Ndiaye-Lobry D, Lafave LM, et al. 2013. Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J Exp Med 210: 2641–2659. 10.1084/jem.20131141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada S, Goyama S, Inoue D, Shikata S, Takeda R, Fukushima T, Yonezawa T, Fujino T, Hayashi Y, Kawabata KC, et al. 2018. Mutant ASXL1 cooperates with BAP1 to promote myeloid leukaemogenesis. Nat Commun 9: 2733 10.1038/s41467-018-05085-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge MN, Hu H, Muzny DM, Musante L, Lupski JR, Graham BH, Chen W, Gripp KW, Jenny K, Wienker TF, et al. 2013. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med 5: 11 10.1186/gm415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramani A, Larjo A, Bassein JA, Chang X, Hastie RB, Togher SM, Lähdesmaki H, Rao A. 2015. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1–BAP1 complex. Nat Commun 6: 7307 10.1038/ncomms8307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baymaz HI, Fournier A, Laget S, Ji Z, Jansen PW, Smits AH, Ferry L, Mensinga A, Poser I, Sharrocks A, et al. 2014. MBD5 and MBD6 interact with the human PR-DUB complex through their methyl-CpG-binding domain. Proteomics 14: 2179–2189. 10.1002/pmic.201400013 [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190. 10.1016/j.molcel.2010.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman RL, Busque L, Levine RL. 2018. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell 22: 157–170. 10.1016/j.stem.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne A, Lee MK, Zielinski D, Michaud A, Le Corre S, Dingli F, Chen H, Shahidian LZ, Vassilev I, Servant N, et al. 2019. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat Commun 10: 348 10.1038/s41467-018-08255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. 2013. BAP1 and cancer. Nat Rev Cancer 13: 153–159. 10.1038/nrc3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittock EC, Latwiel S, Miller TC, Müller CW. 2017. Molecular architecture of polycomb repressive complexes. Biochem Soc Trans 45: 193–205. 10.1042/BST20160173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou S, Barbour H, Ahmed O, Masclef L, Baril C, Sen Nkwe N, Tchelougou D, Uriarte M, Bonneil E, Ceccarelli D, et al. 2018. Monoubiquitination of ASXLs controls the deubiquitinase activity of the tumor suppressor BAP1. Nat Commun 9: 4385 10.1038/s41467-018-06854-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ayala Alonso AG, Gutiérrez L, Fritsch C, Papp B, Beuchle D, Müller J. 2007. A genetic screen identifies novel Polycomb group genes in Drosophila. Genetics 176: 2099–2108. 10.1534/genetics.107.075739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Seshasayee D, Noubade R, French DM, Liu J, Chaurushiya MS, Kirkpatrick DS, Pham VC, Lill JR, Bakalarski CE, et al. 2012. Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337: 1541–1546. 10.1126/science.1221711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Gomes AP, Wang X, Yoon SO, Lee G, Nagiec MJ, Cho S, Chavez A, Islam T, Yu Y, et al. 2018. mTORC1 promotes metabolic reprogramming by the suppression of GSK3-dependent Foxk1 phosphorylation. Mol Cell 70: 949–960.e4. 10.1016/j.molcel.2018.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Chaurushiya MS, Webster JD, Kummerfeld S, Reja R, Chaudhuri S, Chen YJ, Modrusan Z, Haley B, Dugger DL, et al. 2019. Intrinsic apoptosis shapes the tumor spectrum linked to inactivation of the deubiquitinase BAP1. Science 364: 283–285. 10.1126/science.aav4902 [DOI] [PubMed] [Google Scholar]
- Hoischen A, van Bon BW, Rodríguez-Santiago B, Gilissen C, Vissers LE, de Vries P, Janssen I, van Lier B, Hastings R, Smithson SF, et al. 2011. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat Genet 43: 729–731. 10.1038/ng.868 [DOI] [PubMed] [Google Scholar]
- Inoue D, Kitaura J, Togami K, Nishimura K, Enomoto Y, Uchida T, Kagiyama Y, Kawabata KC, Nakahara F, Izawa K, et al. 2013. Myelodysplastic syndromes are induced by histone methylation–altering ASXL1 mutations. J Clin Invest 123: 4627–4640. 10.1172/JCI70739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. 2016. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell 62: 79–91. 10.1016/j.molcel.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Donaldson IJ, Liu J, Hayes A, Zeef LA, Sharrocks AD. 2012. The forkhead transcription factor FOXK2 promotes AP-1-mediated transcriptional regulation. Mol Cell Biol 32: 385–398. 10.1128/MCB.05504-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Mohammed H, Webber A, Ridsdale J, Han N, Carroll JS, Sharrocks AD. 2014. The forkhead transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. Nucleic Acids Res 42: 6232–6242. 10.1093/nar/gku274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37: 907–915. 10.1038/s41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolovos P, Georgomanolis T, Koeferle A, Larkin JD, Brant L, Nikolicc M, Gusmao EG, Zirkel A, Knoch TA, van Ijcken WF, et al. 2016. Binding of nuclear factor κB to noncanonical consensus sites reveals its multimodal role during the early inflammatory response. Genome Res 26: 1478–1489. 10.1101/gr.210005.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A, Green J, Pollard J Jr, Tugendreich S. 2014. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics 30: 523–530. 10.1093/bioinformatics/btt703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov JN, Aguero TH, Owens DA, Kurtenbach S, Field MG, Durante MA, Rodriguez DA, King ML, Harbour JW. 2019. BAP1 regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Sci Adv 5: eaax1738 10.1126/sciadv.aax1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFave LM, Beguelin W, Koche R, Teater M, Spitzer B, Chramiec A, Papalexi E, Keller MD, Hricik T, Konstantinoff K, et al. 2015. Loss of BAP1 function leads to EZH2-dependent transformation. Nat Med 21: 1344–1349. 10.1038/nm.3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol 9: e1003118 10.1371/journal.pcbi.1003118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. 2016. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol 23: 349–357. 10.1038/nsmb.3180 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ding W, Ge H, Ponnusamy M, Wang Q, Hao X, Wu W, Zhang Y, Yu W, Ao X, et al. 2019. FOXK transcription factors: regulation and critical role in cancer. Cancer Lett 458: 1–12. 10.1016/j.canlet.2019.05.030 [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968. 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen JG, Schmidt SF, Larsen BD, Loft A, Nielsen R, Mandrup S. 2015. iRNA-seq: computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res 43: e40 10.1093/nar/gku1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micol JB, Abdel-Wahab O. 2016. The role of additional sex combs-like proteins in cancer. Cold Spring Harb Perspect Med 6: e026526 10.1101/cshperspect.a026526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murali R, Wiesner T, Scolyer RA. 2013. Tumours associated with BAP1 mutations. Pathology 45: 116–126. 10.1097/PAT.0b013e32835d0efb [DOI] [PubMed] [Google Scholar]
- Nestal de Moraes G, Carneiro LDT, Maia RC, Lam EW, Sharrocks AD. 2019. FOXK2 transcription factor and its emerging roles in cancer. Cancers (Basel) 11: 393 10.3390/cancers11030393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. 2018. The principles that govern transcription factor network functions in stem cells. Development 145: dev157420 10.1242/dev.157420 [DOI] [PubMed] [Google Scholar]
- Okino Y, Machida Y, Frankland-Searby S, Machida YJ. 2015. BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the ubiquitin-mediated activation of FoxK2 target genes. J Biol Chem 290: 1580–1591. 10.1074/jbc.M114.609834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, et al. 2017. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol Cell 68: 955–969.10. 10.1016/j.molcel.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247. 10.1038/nature08966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K. 2011. Jarid1b targets genes regulating development and is involved in neural differentiation. EMBO J 30: 4586–4600. 10.1038/emboj.2011.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. 2017. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171: 34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Sidoli S, Simithy J, Karch KR, Kulej K, Garcia BA. 2015. Low resolution data-independent acquisition in an LTQ-orbitrap allows for simplified and fully untargeted analysis of histone modifications. Anal Chem 87: 11448–11454. 10.1021/acs.analchem.5b03009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidoli S, Bhanu NV, Karch KR, Wang X, Garcia BA. 2016. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J Vis Exp 111: 54112 10.3791/54112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W. 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114: 493–505. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Campbell RB, Nicholls F, Slade E, Brock HW. 1992. Genetic analysis of the additional sex combs locus of Drosophila melanogaster. Genetics 130: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, Boer E, Bryne JC, Thongjuea S, Rijkers E, Demmers J, van IJcken W, Grosveld F. 2011. A systems approach to analyze transcription factors in mammalian cells. Methods 53: 151–162. 10.1016/j.ymeth.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Srivastava A, Ritesh KC, Tsan YC, Liao R, Su F, Cao X, Hannibal MC, Keegan CE, Chinnaiyan AM, Martin DM, et al. 2016. De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge–Ropers syndrome. Hum Mol Genet 25: 597–608. 10.1093/hmg/ddv499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Cico A, Stephen T, Thongjuea S, Kolovos P, Baymaz HI, Yu X, Demmers J, Bezstarosti K, Maas A, et al. 2015. Control of developmentally primed erythroid genes by combinatorial co-repressor actions. Nat Commun 6: 8893 10.1038/ncomms9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadhouders R, Filion GJ, Graf T. 2019. Transcription factors and 3D genome conformation in cell-fate decisions. Nature 569: 345–354. 10.1038/s41586-019-1182-7 [DOI] [PubMed] [Google Scholar]
- Sukonina V, Ma H, Zhang W, Bartesaghi S, Subhash S, Heglind M, Foyn H, Betz MJ, Nilsson D, Lidell ME, et al. 2019. FOXK1 and FOXK2 regulate aerobic glycolysis. Nature 566: 279–283. 10.1038/s41586-019-0900-5 [DOI] [PubMed] [Google Scholar]
- Talbert PB, Meers MP, Henikoff S. 2019. Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat Rev Genet 20: 283–297. 10.1038/s41576-019-0105-7 [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348. 10.1038/nature10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bekker-Jensen IH, Christensen J, Rasmussen KD, Sidoli S, Qi Y, Kong Y, Wang X, Cui Y, Xiao Z, et al. 2015. Tumor suppressor ASXL1 is essential for the activation of INK4B expression in response to oncogene activity and anti-proliferative signals. Cell Res 25: 1205–1218. 10.1038/cr.2015.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav T, Quivy JP, Almouzni G. 2018. Chromatin plasticity: a versatile landscape that underlies cell fate and identity. Science 361: 1332–1336. 10.1126/science.aat8950 [DOI] [PubMed] [Google Scholar]
- Yu H, Mashtalir N, Daou S, Hammond-Martel I, Ross J, Sui G, Hart GW, Rauscher FJ III, Drobetsky E, Milot E, et al. 2010. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of gene expression. Mol Cell Biol 30: 5071–5085. 10.1128/MCB.00396-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. 10.1038/nature13992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. 2019. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10: 1523 10.1038/s41467-019-09234-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.