Abstract
Purpose:
Machine-learning algorithms are a subset of artificial intelligence that have proven to enhance analytics in medicine across various platforms. Spine surgery has the potential to benefit from improved hardware placement utilizing algorithms that autonomously and accurately measure pedicle and vertebral body anatomy. The purpose of this study was to assess the accuracy of an autonomous convolutional neural network (CNN) in measuring vertebral body anatomy utilizing clinical lumbar computed tomography (CT) scans and automatically segment vertebral body anatomy.
Methods:
The CNN was trained utilizing 8000 manually segmented CT slices from 15 cadaveric specimens and 30 adult diagnostic scans. Validation was performed with twenty randomly selected patient datasets. Anatomic landmarks that were segmented included the pedicle, vertebral body, spinous process, transverse process, facet joint, and lamina. Morphometric measurement of the vertebral body was compared between manual measurements and automatic measurements.
Results:
Automatic segmentation was found to have a mean accuracy ranging from 96.38% to 98.96%. Coaxial distance from the lamina to the anterior cortex was 99.10% with pedicle angulation error of 3.47%.
Conclusion:
The CNN algorithm tested in this study provides an accurate means to automatically identify the vertebral body anatomy and provide measurements for implants and placement trajectories.
Keywords: Artificial intelligence, navigation, spine surgery
INTRODUCTION
Recent advances in surgical robotics and image-guided surgery have led to increased utilization of these systems in spine surgery.[1] The reported advantages include increased implant placement accuracy and decreased radiation exposure to the surgeon. Drawbacks of these systems include a significant surgeon involvement in preoperative planning, lack of system autonomy during surgery (i.e., master–slave approach), and the inability of the system to provide real-time intraoperative warnings and suggestions to the surgeon. With progress in the field of artificial intelligence and image recognition, it has become possible to develop a machine-learning algorithm for autonomously identifying and labeling anatomical landmarks on spine computed tomography (CT) scans.[2,3,4] A neural network trained in identifying vertebral anatomy would assist with presurgical planning and intraoperative surgical assistance and potentially lowered radiation exposure and accelerated operative times. Some of the advantages of such an application would include the ability to autonomously and preoperatively formulate a surgical plan with minimal surgeon input, select the correct implant sizes (allowing manufacturers to ship in only necessary implants and highly reducing costs), suggest implant placement trajectories (reducing human errors), and provide real-time warnings if deviations from the plan occurred intraoperatively (increasing surgical outcomes). Coupling this type of application to a robotic or image guidance platform would improve patient safety by reducing the chance for incorrect preoperative plan formulated by novice surgeons, poor implant selection, and incorrect implant trajectory selection. The purpose of this study is to validate the accuracy of the machine-learning software that was developed for teaching the computer how to autonomously identify bony anatomical landmarks of the lumbar spine on diagnostic and intraoperative grade CT scans.
METHODS
A convolutional neural network (CNN) was developed and trained[5](Holosurgical, Inc., Chicago, IL, USA) how to properly segment, identify, and label vertebral landmarks. Measurements included the pedicle, vertebral, spinous process, transverse process, facet joints, and lamina of the lumbar spine. The training phase consisted of 8000 manually segmented sliced of 24 intraoperative CT-scan datasets of 15 cadaveric lumbar spine specimens (192 slices each). The validation of the CNN algorithm was performed with twenty datasets randomly pulled from a pool of 2000 datasets with a total of 2200 validation images utilized. We hypothesized that the CNN algorithm would be able to measure vertebral anatomy with a high degree of accuracy from lumbar spine CT images.
The training procedure, as presented in Figure 1, comprises the following steps. First, a set of digital imaging and communications in medicine (DICOM) images are generated by a preoperative or an intraoperative CT, representing consecutive slices with visible tissues [Figure 1a]. Next, all slices are processed to perform automatic vertebral segmentation [Figure 1b]. Then, the original information obtained from DICOM images and the segmentation results are merged to obtain combined images comprising information about both the tissue density and its classification corresponding to each anatomy parts [Figure 1c].
Figure 1.
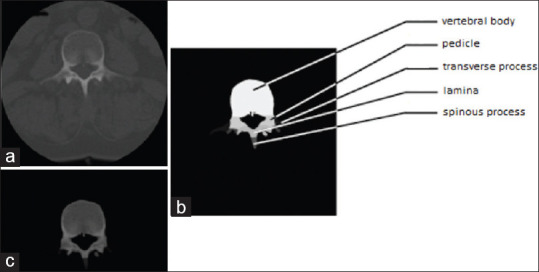
(a) Original digital imaging and communications in medicine image from intraoperative scanner. (b) Manual segmentation. (c) Segmentation used as a mask over the original image
Several morphometric measurements of both original DICOM and computer-segmented slices were performed by a board-certified orthopedic surgeon at various time points: anteroposterior vertebral depth (A), vertebral body width (B), spinous process height (C), pedicle angulation (D), pedicle diameter at the isthmus (E), and coaxial distance from facet cortex to the anterior vertebral cortex (F) [Figure 2]. Measurements of the neural network input (original) and output (computer-annotated) images [Figure 3] of the lumbar spine were recorded on separate datasheets, so that when entering the measurement results, the prior measurement results were not visible on the same sheet to avoid bias. The mean accuracy of each landmark measurement was recorded as a percentage by dividing the output measurements by the input measurements. The standard deviation of each landmark mean accuracy was reported as well as the error, which is expressed as the difference of the mean accuracy from 100%. All calculations were performed using SPSS software (IBM; Chicago, IL, USA).
Figure 2.
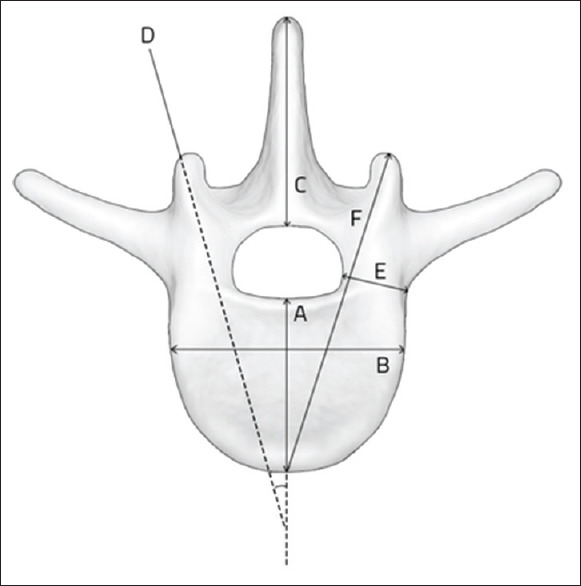
Measured anatomical landmarks
Figure 3.
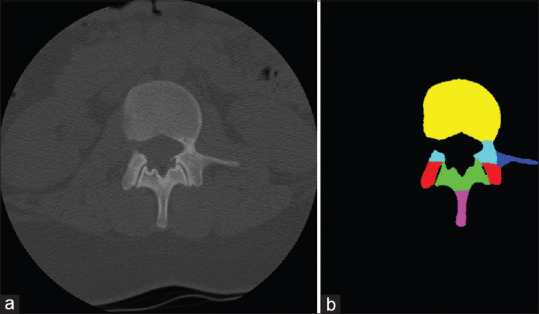
(a) Input and (b) output images of the neural network
RESULTS
A total of 37 pairs of original CT and computer-generated images were investigated.
The mean accuracy of each anatomical segmentation ranged between 96.38% and 98.96% [Table 1]. The measured mean error of all anatomical landmarks was 2.27%.
Table 1.
Accuracy of automated vertebral body measurements from computed tomography scans
| Anatomical landmarks | Accuracy |
Error (%) | |
|---|---|---|---|
| Mean (%) | SD (%) | ||
| A: AP vertebral depth | 97.65 | 1.77 | 2.35 |
| B: Vertebral body width | 98.38 | 0.99 | 1.62 |
| C: Spinous process height | 97.81 | 1.42 | 2.19 |
| D: Pedicle angulation | 96.53 | 3.84 | 3.47 |
| E: Pedicle diameter | 96.93 | 3.76 | 3.07 |
| F: Coaxial distance from lamina to anterior cortex | 99.10 | 0.87 | 0.90 |
| Average | 97.73 | 2.11 | 2.27 |
AP - Anteroposterior, SD - Standard deviation
Specifically, an error of 3.07% in the measurements of the anatomical landmark that determines pedicle screw selection (i.e., pedicle diameter) presented no statistically significant differences between the original DICOM and the computer-segmented image.
The mean standard deviation of coaxial distance from the lamina to the anterior cortex, which determines screw length, was measured as only 0.8%, with an accuracy of 99.10%. The least accurate anatomical landmark was the pedicle angulation, with a 3.47% error.
DISCUSSION
Machine learning is a subset of artificial intelligence, which has a great potential to improve the practice of spine surgery. Prior studies have utilized machine-learning models to develop predictive algorithms for spinal tumor survival,[6,7] complications and sustained opioid use following anterior cervical discectomy and fusion surgery,[8,9] patient-reported outcomes and discharge disposition following lumbar spine surgery,[10,11] and blood transfusion in spinal deformity surgery.[12] All of these studies demonstrated high reliability in predictive power and proved superior to conventional statistical methods.
Recently, Ames et al. 2019 utilized a self-learning hierarchical clustering algorithm to propose a new classification system for adult spinal deformity.[13] The study analyzed a multisurgeon-multicenter database of 570 patients that underwent adult spinal deformity corrective surgery. Patients were clustered based on their curves' type and surgical interventions associated with optimal outcomes, which identified 12 unique classification groups. However, the classification system did not include specific radiographic curve morphology. This method of grouping spinal deformity proves to possibly strengthen classification systems that are already well accepted.[14,15]
Furthermore, these algorithms have been used to analyze preoperative and intraoperative radiographic data to improve implant placement in spine surgery. A proof of concept study by Esfandiari et al. tested pose estimation in 50,000 fluoroscopic images with pedicle screw random orientations.[16] The authors utilized a CNN and found a 93% and 83% accuracy in synthetic and clinically realistic radiographs, respectively. Although the results of this study were not extrapolated in vivo, they suggest machine-learning algorithms can assist surgeons with pedicle screw placement intraoperatively with negligible added operating room time. Moreover, Lafage et al. proposed that self-learning computers can aid in preoperative planning and can further be customized in treatment selection to surgeon-specific outcomes in spinal deformity corrective surgery.[17]
Combining artificial intelligence with robotic-assisted spine surgery has the potential to substantially benefit patients and surgeons. Recent literature has documented increased pedicle screw accuracy, decreased hospital stays, lower complications, and decreased radiation exposure with the use of robotics.[1] Artificial intelligence in conjunction with robotics can also decrease the registration time and decrease the time spent planning preoperatively. During spinal procedures, artificial intelligence would allow the robot to alert the surgeon about nuances in the procedure, such as precisely differentiating between bony, vascular, or nerve tissue and advising to redirect the implant trajectory in the desired direction. A key component for making this possible is automatic anatomical segmentation and landmark identification. For making surgical robotics practical and economical, it must rely on the images obtained from an intraoperative scanner at the time of surgery.
The neural network algorithm evaluated in this study automatically and autonomously analyzes DICOM spine scan data and then segments and identifies the various components of the vertebrae without any human intervention. The algorithm is able to recognize both normal and abnormal spine anatomy, as well as to identify and classify those components into the pedicle, vertebral body, lamina, transverse, or spinous process. This was done with a high degree of accuracy (97.73%) when compared with manually identified landmarks. Our findings are comparable to Burström et al., who reported a 95.4% accuracy of a machine-learning algorithm on 21 cadavers and 20 clinical cases using intraoperative quality CT scans.[2] This is concordant with prior work on automatic segmentation that has noted 90%–95% accuracy.[3,4,18,19] Manually segmented vertebrae on CT scans have been proven to be highly accurate with submillimeter accuracy and precision,[20] suggesting pedicle screw placement under machine-learned guidance to be well within the margin of safety by Gertzbein and Robbins.[21]
There are several immediate surgical applications for this algorithm. First, the system could perform a detailed analysis of each of the segmented parts, including pedicle length, diameter, volume, and angulation in real time. Once this information is available, the system could then be used to size and select an appropriate implant based on patient-specific anatomy. This feature, in turn, could be used to present the ideal trajectory for placing the implant. This trajectory can be displayed on a monitor for the surgeon or the information could be provided to the robotic platform, thus allowing the robot to autonomously target the pedicle without the need for surgeon input. The surgeons' role would be to ensure that the computer presents the proper trajectory and verify proper execution of the surgical plan.
There were several limitations of the current study that future work must address to make this technology more rigorous. First, CT modeling was only performed on the lumbar spine. Ideally, future projects would aim to develop software with accurate reconstruction of the thoracic and cervical spine to give further navigational capabilities. Another limitation unique to this type of analysis is that of the method used for image annotation and measurements performed by the orthopedic surgeon. Is the human examiner more accurate than the computer in making the measurements, or is it the other way around? Therefore, when analyzing the clinical significance of the reported error, this factor needs to take that into account. A further clinical comparison of the current studies' reconstruction software to intraoperative three-dimensional navigation would give further clarity to the ability of the device to streamline surgical spine procedures and increase surgical accuracy and efficiency.
CONCLUSION
Effective reconstruction and analytics of pre- and intraoperative imaging is paramount to the successful implementation of artificial intelligence applications and augmented reality devices in the surgical setting. The results of this study demonstrate that machine learning is able to produce highly reliable segmentation of bony anatomy based on preoperative CT scans, achieving an accuracy approaching 98%. To our knowledge, this is the first study that validates a machine-learning algorithm that is capable of autonomously and accurately segment and identify individual components of the bony spinal anatomy to this level of accuracy. These findings suggest CNN algorithms to have many powerful implications for further development of robotics and image-guided surgery.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Ghasem A, Sharma A, Greif DN, Alam M, Maaieh MA. The arrival of robotics in spine surgery: A review of the literature. Spine (Phila Pa 1976) 2018;43:1670–7. doi: 10.1097/BRS.0000000000002695. [DOI] [PubMed] [Google Scholar]
- 2.Burström G, Buerger C, Hoppenbrouwers J, Nachabe R, Lorenz C, Babic D, et al. Machine learning for automated 3-dimensional segmentation of the spine and suggested placement of pedicle screws based on intraoperative cone-beam computer tomography. Spine. 2019;31:147–54. doi: 10.3171/2018.12.SPINE181397. [doi: 10.3171/2018.12.SPINE181397] [DOI] [PubMed] [Google Scholar]
- 3.Ruikar DD, Santosh KC, Hegadi RS. Automated fractured bone segmentation and labeling from CT images. J Med Syst. 2019;43:60. doi: 10.1007/s10916-019-1176-x. [DOI] [PubMed] [Google Scholar]
- 4.Lessmann N, van Ginneken B, de Jong PA, Išgum I. Iterative fully convolutional neural networks for automatic vertebra segmentation and identification. Med Image Anal. 2019;53:142–55. doi: 10.1016/j.media.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Siemionow K, Luciano CJ, Kraft M. Automated Segmentation of Three Dimensional Bony Structure Images. 2017. Oct 10, [last accessed 2020 Mar 28]. Available from: https://patents.google.com/patent/US20190105009A1/en?inventor=siemionow+kris & oq=siemionow+kris .
- 6.Karhade AV, Thio Q, Ogink P, Kim J, Lozano-Calderon S, Raskin K, et al. Development of machine learning algorithms for prediction of 5-year spinal chordoma survival. World Neurosurg. 2018;119:e842–7. doi: 10.1016/j.wneu.2018.07.276. [DOI] [PubMed] [Google Scholar]
- 7.Ryu SM, Lee SH, Kim ES, Eoh W. Predicting survival of patients with spinal ependymoma using machine learning algorithms with the SEER database. World Neurosurg. 2019:pii: S1878-8750(18)32914-0. doi: 10.1016/j.wneu.2018.12.091. [DOI] [PubMed] [Google Scholar]
- 8.Arvind V, Kim JS, Oermann EK, Kaji D, Cho SK. Predicting surgical complications in adult patients undergoing anterior cervical discectomy and fusion using machine learning. Neurospine. 2018;15:329–37. doi: 10.14245/ns.1836248.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karhade AV, Ogink PT, Thio QC, Broekman ML, Cha TD, Hershman SH, et al. Machine learning for prediction of sustained opioid prescription after anterior cervical discectomy and fusion. Spine J. 2019;19:976–83. doi: 10.1016/j.spinee.2019.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Staartjes VE, de Wispelaere MP, Vandertop WP, Schröder ML. Deep learning-based preoperative predictive analytics for patient-reported outcomes following lumbar diskectomy: Feasibility of center-specific modeling. Spine J. 2018;19:853–61. doi: 10.1016/j.spinee.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Karhade AV, Ogink P, Thio Q, Broekman M, Cha T, Gormley WB, et al. Development of machine learning algorithms for prediction of discharge disposition after elective inpatient surgery for lumbar degenerative disc disorders. Neurosurg Focus. 2018;45:E6. doi: 10.3171/2018.8.FOCUS18340. [DOI] [PubMed] [Google Scholar]
- 12.Durand WM, DePasse JM, Daniels AH. Predictive modeling for blood transfusion after adult spinal deformity surgery: A tree-based machine learning approach. Spine (Phila Pa 1976) 2018;43:1058–66. doi: 10.1097/BRS.0000000000002515. [DOI] [PubMed] [Google Scholar]
- 13.Ames CP, Smith JS, Pellisé F, Kelly M, Alanay A, Acaroǧlu E, et al. Artificial intelligence based hierarchical clustering of patient types and intervention categories in adult spinal deformity surgery: Towards a new classification scheme that predicts quality and value. Spine. 2019;44:915–26. doi: 10.1097/BRS.0000000000002974. [DOI] [PubMed] [Google Scholar]
- 14.Lenke LG, Betz RR, Harms J, Bridwell KH, Clements DH, Lowe TG, et al. Adolescent idiopathic scoliosis: A new classification to determine extent of spinal arthrodesis. J Bone Joint Surg Am. 2001;83-A:1169–81. [PubMed] [Google Scholar]
- 15.King HA, Moe JH, Bradford DS, Winter RB. The selection of fusion levels in thoracic idiopathic scoliosis. J Bone Joint Surg Am. 1983;65:1302–13. [PubMed] [Google Scholar]
- 16.Esfandiari H, Newell R, Anglin C, Street J, Hodgson AJ. A deep learning framework for segmentation and pose estimation of pedicle screw implants based on C-arm fluoroscopy. Int J Comput Assist Radiol Surg. 2018;13:1269–82. doi: 10.1007/s11548-018-1776-9. [DOI] [PubMed] [Google Scholar]
- 17.Lafage R, Pesenti S, Lafage V, Schwab FJ. Self-learning computers for surgical planning and prediction of postoperative alignment. Eur Spine J. 2018;27:123–8. doi: 10.1007/s00586-018-5497-0. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Liang W, Zhang Y, Tan J. Automatic global level set approach for lumbar vertebrae CT image segmentation. Biomed Res Int. 2018;2018:6319879. doi: 10.1155/2018/6319879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu C, Belavý DL, Armbrecht G, Bansmann M, Felsenberg D, Zheng G. Fully automatic localization and segmentation of 3D vertebral bodies from CT/MR images via a learning-based method. Pham D, ed. PLoS One. 2015;10:e0143327. doi: 10.1371/journal.pone.0143327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook DJ, Gladowski DA, Acuff HN, Yeager MS, Cheng BC. Variability of manual lumbar spine segmentation. Int J Spine Surg. 2012;6:167–73. doi: 10.1016/j.ijsp.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gertzbein SD, Robbins SE. Accuracy of pedicular screw placement in vivo. Spine (Phila Pa 1976) 1990;15:11–4. doi: 10.1097/00007632-199001000-00004. [DOI] [PubMed] [Google Scholar]


