Abstract
Introduction
Over one in eight Canadians is affected by a foodborne infection annually; however, the long-term consequences, including the risks and costs of sequelae, are unclear. We aim to estimate the health burden and direct costs of 14 infections commonly transmitted by food, considering the acute illness and subsequent sequelae and mortality, for the population of British Columbia, Canada (~4.7 million).
Methods and analysis
We will conduct a population-based retrospective cohort study of the British Columbia provincial population, over a 10-year study period (1 January 2005 to 31 December 2014). Exposure is defined as a provincially reported illness caused by Clostridium botulinum, Campylobacter, Cryptosporidium, Cyclospora, Giardia, hepatitis A virus, Listeria, non-typhoidal Salmonella spp, Salmonella Typhi, Salmonella Paratyphi, Shiga toxin-producing Escherichia coli, Shigella, Vibrio parahaemolyticus or Yersinia (excluding pestis). We will link individual-level longitudinal data from eight province-wide administrative health and reportable disease databases that include physician visits, hospitalisations and day surgeries, deaths, stillbirths, prescription medications (except those to treat HIV) and reportable foodborne diseases. Using these linked databases, we will investigate the likelihood of various sequelae and death. Hazard models will be used to estimate the risk of outcomes and their association with the type of foodborne infection. Epidemiological analyses will be conducted to determine the progression of illness and the fraction of sequelae attributable to specific foodborne infections. Economic analyses will assess the consequent direct healthcare costs.
Ethics and dissemination
This study has been approved by a University of Waterloo Research Ethics Committee (no 30645), the University of British Columbia Behavioral Research Ethics Board (no H16-00021) and McGill University’s Institutional Review Board (no A03-M12-19A). Results will be disseminated via presentations to academics, public health practitioners and knowledge users, and publication in peer-reviewed journals. Where such publications are not open access, manuscripts will also be available via the University of Waterloo’s Institutional Repository (https://uwspace.uwaterloo.ca).
Keywords: epidemiology, public health, gastrointestinal infections
Strengths and limitations of this study.
This cohort is a near-complete set of individually linked administrative health and reportable foodborne infection data, covering the ~4.7 million residents of British Columbia, Canada, over 10 years (2005–2014).
To the best of our knowledge, the study described in this protocol will be the most comprehensive assessment of the risk of sequelae following foodborne infections across multiple pathogens to date.
Because all residents of British Columbia are covered by a mandatory, single provincial health insurance plan (with only a few exceptions, eg, members of the military), movement of individuals within the province or between employers does not create loss to follow-up.
Limitations include incomplete and lower quality (eg, misclassification, use of non-specific codes) information associated with administrative health data, and under-ascertainment of foodborne infections typical to reportable disease data.
Introduction
Infections commonly transmitted via food, such as Salmonella spp and Shiga toxin-producing Escherichia coli (STEC) are a global public health concern,1 and in Canada they affect over one in eight people annually.2 Beyond the acute stage of illness (typically characterised by diarrhoea and other gastrointestinal symptoms), these infections can cause severe and longer-term outcomes, such as spontaneous abortion, haemolytic uremic syndrome (HUS), Crohn’s disease and ulcerative colitis (inflammatory bowel disease), Guillain-Barré syndrome (GBS) and death.3–11
Estimates of the risk of sequelae following foodborne infection have come in part from prospective cohort studies conducted as follow-ups to outbreaks12–16 or to reports of sporadic cases (usually via laboratory testing and disease surveillance systems).4 17–23 While such prospective studies have the advantage of being able to tailor the data collection to address specific research questions, they have some important limitations. For example, outbreak follow-up studies are limited to specific strain(s) causing the outbreak and the specific population affected. Studies of sporadic cases often have short follow-up periods and rely on self-reported questionnaires to identify sequelae, both of which can lead to bias.
Retrospective, population-based cohort studies, in which administrative and registry data are analysed,10 11 24–32 offer several advantages that complement the prospective studies described above. They allow for a wider population to be covered, both sporadic and outbreak-associated infections (caused by the range of strains affecting the population) to be included, and the use of self-reports of event occurrence to be avoided. However, because they require population-wide, linked data on the exposures and outcomes of interest, they are less frequently conducted. To date, such studies have not been conducted in Canada.
Although infections such as Salmonella spp and STEC can be transmitted via several routes (eg, person-to-person, water), their transmission via food and their presence throughout the food system (eg, food animals as a reservoir for Campylobacter spp,33 food handlers shedding hepatitis A virus,34 the ability of Listeria monocytogenes to persist in food production equipment35) mean that these infections are often termed ‘foodborne’ although some fraction will not be transmitted via food directly. Here, we apply the term ‘foodborne infection’ to 14 infections that can be transmitted via food (Clostridium botulinum, Campylobacter, Cryptosporidium, Cyclospora, Giardia, hepatitis A virus, Listeria, non-typhoidal Salmonella spp, Salmonella Typhi, Salmonella Paratyphi, STEC, Shigella, Vibrio parahaemolyticus and Yersinia excluding pestis), recognising that not all result from direct foodborne transmission.
The overall goal of this study is to estimate the health burden and costs of these 14 infections, considering the acute illness and subsequent sequelae and associated mortality, for the population of British Columbia, Canada. Our specific objectives are to:
Determine the risk of developing sequelae following infection.
Describe the epidemiology and clinical progression across the range of outcomes, including acute illness, sequelae and death.
Quantify the direct healthcare costs due to these infections and their various outcomes.
Determine the risk of sequelae in the population attributable to these infections.
Methods and analysis
Study setting
British Columbia is Canada’s westernmost and third most populous province (~4.7 million circa 2014). The annual incidence of foodborne infections in British Columbia is comparable to annual incidences in Canada, the USA, Australia and Western Europe.2 36 37
All British Columbia residents (defined as citizens or permanent residents of Canada who are physically present in British Columbia for at least 6 months in a calendar year), their dependents and certain other individuals (eg, some holders of study or work permits) are covered by the province’s health insurance programme.38 Enrolment is mandatory, and this programme covers nearly all the British Columbia population (with the exception of members of the Canadian military, Royal Canadian Mounted Police and some First Nations individuals covered by federal insurance programmes). Physician visits, laboratory tests, hospitalisations, outpatient hospital services and drugs for certain populations are among the publicly funded benefits. The administrative datasets that contain these healthcare use data, along with vital statistics (VS) (eg, births, deaths), demographic data and other datasets, are held by the British Columbia Ministry of Health and are accessible to researchers via Population Data BC,39 a central repository and ‘multiuniversity, data and education resource’ that ‘support[s] research access to individual-level, deidentified longitudinal data on British Columbia’s 4.7 million residents’.40
The British Columbia Public Health Act mandates that reportable diseases, including several foodborne infections,41 be reported by health professionals and laboratories to the local and provincial public health authorities, and these data are managed provincially by the British Columbia Centre for Disease Control (BCCDC). These data are housed within Panorama, the provincial public health database of reportable diseases. In the Panorama database, as well as the administrative health and VS databases, individuals are recorded by their unique Personal Health Number (PHN), allowing information from these data sources to be linked by individual.
Study design, population and timeframe
This is a retrospective cohort study of the population of British Columbia, with additional descriptive, cost and population attributable risk analyses. Because this is a dynamic population in which exposure status of individuals changes over time, our study design assesses the risk and effect of exposure in terms of person-time. Thus, rather than using fixed cohorts of exposed versus unexposed individuals, we will track individuals over time and assign their person-time at risk to either ‘unexposed person-time’ (eg, prior to foodborne infection) or ‘exposed person-time’ (eg, after foodborne infection), as described further below.
The study population is all individuals in British Columbia registered with the provincial health insurance programme at any point during the study period, that is, all individuals with the following from 2005 to 2014 inclusive: one or more record in one or more of the Medical Services Plan (MSP), Discharge Abstracts Database (DAD), VS Deaths or PharmaNet; or record of coverage under the provincial insurance programme within the Consolidation File database (see table 1). The 10-year study period is 1 January 2005 to 31 December 2014, inclusive, with additional 2 year wash-in (1 January 2003 to 31 December 2004) and wash-out (1 January 2015 to 31 December 2016) periods. During these periods, we will identify occurrences of foodborne infections, sequelae and death. The 10-year study period was selected to more than encompass timeframes for initial sequelae development and ensuing healthcare use currently reflected in the literature (ie, days to years), although there is some evidence that sequelae can develop over longer timeframes (eg, over decades).42
Table 1.
Population-level administrative and reportable disease databases that will be used in this study (British Columbia, Canada)
| Database (reference) | Database description and summary of variables included for this study | Date range |
| Health Care and Health Services Data | ||
| Medical Services Plan (MSP) Payment Information File74 | Billing records for all medically necessary services provided by fee-for-service physicians. Includes PHN, service dates, up to five ICD-9/ICD-10 diagnostic codes, MSP-specific fee-item codes and physician specialties. | 01 January 2003 to 31 December 2016 |
| Discharge Abstracts Database (Hospital Separations)75 | Data on discharges, transfers and deaths of inpatients and day surgery patients from acute care hospitals in British Columbia; does not include emergency room visits. Includes PHN, admission and discharge dates, up to 25 ICD-10-CA diagnostic codes, service use and procedure codes, newborn and maternal data, discharge status and province issuing healthcare number. | 01 January 2003 to31 December 2016 |
| PharmaNet76 | All prescriptions (for drugs and medical supplies) dispensed from community pharmacies and from hospital outpatient pharmacies for patient use at home, in British Columbia. Includes PHN, date of dispensing, quantity, dose, costs/fees, the type of prescribing practitioner and the Drug Information Number (or Product Information Number) assigned by Health Canada. | 01 January 2003 to 31 December 2016 |
| Population and Vital Statistics Data | ||
| Vital Statistics Deaths77 | All deaths registered in British Columbia. Includes PHN, time and place data, and ICD-10 codes for the nature and causes of death. | 01 January 2005 to 31 December 2016 |
| Vital Statistics Stillbirths78 | All stillbirths registered in British Columbia. Includes the mother’s PHN, time and place data, number of stillborn and live born children in the event, gestation period and ICD-10 codes for the underlying cause of stillbirth. | 01 January 2005 to 31 December 2016 |
| Consolidation File79 | Population Data BC’s central demographics file for research requests, containing all individuals who are eligible to receive services in British Columbia. Includes PHN, age, sex, neighbourhood income deciles, and local health authority area, as well as the date and number of days registered in the provincial health insurance programme. | 01 January 2003 to 31 December 2016 |
| Statistics Canada Income Bands80 | 1000 income bands that contain information about the six-digit postal code area in which the individual resides. Includes the average and median equivalised disposable income (derived from Statistics Canada tax-filer data, and available for the years 1992, 2002 and 2006), and the number of families, adults and children in the area. | 2002, 2006 |
| Reportable Disease Data | ||
| Panorama Public Health Information System | All cases of the following 14 reportable diseases reported in British Columbia: Clostridium botulinum, Campylobacter, Cryptosporidium, Cyclospora, Giardia, hepatitis A virus, Listeria, non-typhoidal Salmonella spp, Salmonella Typhi, Salmonella Paratyphi, STEC, Shigella, Vibrio parahaemolyticus and Yersinia. Includes PHN, onset date, reported date, health authority and aetiological agent. | 01 January 2005 to 31 December 2014 |
BC, British Columbia; CA, Canada-specific modification of ICD-10; ICD, International Classification of Diseases; PHN, Personal Health Number; STEC, Shiga toxin-producing Escherichia coli.
We assume that enrolment in the provincial health insurance programme (ie, entry into the study population) and reasons for exit from the cohort (eg, moving away from British Columbia) are not related to the exposures nor the outcomes of interest.
Data sources and linkage
The study will use individually linked, longitudinal data from eight databases to investigate both acute and longer-term health outcomes following foodborne infection (table 1). In totality, these data contain information on 14 reportable foodborne infections, physician and hospital visits, prescription medications, VS and various demographic descriptors, for the British Columbia population across the study period. All data will be stored and analysed within Population Data BC’s virtual Secure Research Environment.
Population Data BC will link the eight databases directly by individual using PHN, with additional identifiers (eg, name, age, sex) used to validate linkages and link records probabilistically when PHNs are missing. Population Data BC’s detailed linkage process is fully described elsewhere.43 Note that because the Statistics Canada Income Bands database contains area-level data (whereas the other seven databases contain individual-level data), these data are first linked to individuals (and their PHNs) using their six-digit postal code. Once the linkage is complete, each individual is then assigned a unique study identifier. All individually linked, deidentified databases are provided by Population Data BC within their Secure Research Environment, a centralised online platform, accessible via virtual private network within Canada, for accessing and analysing research data, with security standards that meet Data Steward requirements.
Measuring exposure and outcomes
The exposures of interest are infections with the 14 foodborne pathogens. These 14 infections were selected because they (1) are considered a priority in terms of prevention potential and health impacts and (2) capture nearly all reportable foodborne infections in British Columbia. Note that brucellosis and paralytic shellfish poisoning were also reportable foodborne infections in British Columbia during the study period. However, since brucellosis is very rare and nearly always travel-related, and paralytic shellfish poisoning (also rare) is syndromic and diagnosis is uncertain, these two were not considered for inclusion in this study.
Individuals will be considered exposed when and if they have a laboratory-confirmed and provincially reported case of any of the following, recorded in the Panorama database during the study period: Clostridium botulinum, Campylobacter, Cryptosporidium, Cyclospora, Giardia, hepatitis A virus, Listeria, Salmonella spp (non-typhoidal, Typhi, Paratyphi), STEC, Shigella, Vibrio parahaemolyticus and Yersinia (excluding pestis). Case definitions for each of these infections are specified by the BCCDC.44 Individuals without a reported foodborne infection, but who have International Classification of Disease (ICD) codes either for one of our infections (eg, A02.0, ‘Salmonella enteritis’) or for non-specific gastroenteritis (eg, A08.4, ‘viral intestinal infection, unspecified’45) within the MSP and DAD databases will be considered potentially exposed. We will describe these individuals as a separate group in our descriptive, economic and population attributable fraction analyses, but will remove them from the main analyses of sequelae risk. We will, however, estimate the risk of sequelae among those who are potentially exposed as a secondary analysis.
It is possible for individuals to have more than one reported foodborne infection during the study period, either as a simultaneously occurring coinfection or as two or more distinct events. For these individuals, we will treat this as a complex exposure problem; sequelae will be associated with the most plausible explanatory infection, considering biology and timing, and we will adjust for the presence of concurrent foodborne infections if applicable.46 47
Our primary outcomes of interest are those sequelae for which the link to a given foodborne infection is either established or is possible (table 2). We selected sequelae (1) with evidence of an association with any of the 14 individual foodborne infections (eg, 55)48 and (2) that occur via direct effects of pathogens or their toxin, or via autoimmune or chronic inflammatory processes that can be triggered by the infection. We will classify individuals as having the sequelae via administrative case definitions, using ICD diagnostic codes within the MSP, DAD and VS data, with the exception of stillbirths which will be determined using recorded events in the VS-Stillbirths database. The ICD codes in the MSP data are generally considered accurate to the third digit.49 Although ICD codes in the VS and DAD data are ordered by most probable diagnosis, we will consider all codes, regardless of order.
Table 2.
Established (E) and possible (P) sequelae of foodborne infections that will be assessed in this study (British Columbia, Canada)
| Foodborne infection* | Acute kidney injury | Coeliac disease | Erythema nodosum | Graves’ disease | Guillain-Barré syndrome† | Haemolytic uremic syndrome | Inflammatory bowel disease | Irritable bowel syndrome | Neonatal listeriosis‡ | Stillbirth | Reactive arthritis§ | Thrombotic thrombocytopenic purpura¶ |
| Campylobacter | E | P | E | P | P | P | E | |||||
| Cryptosporidium | P | |||||||||||
| Cyclospora | P | P | ||||||||||
| Giardia | P | P | ||||||||||
| Hepatitis A virus | E | P | ||||||||||
| Listeria monocytogenes | E ¶ | E | ||||||||||
| Salmonella (non-typhoidal) | E | P | P | P | P | E | ||||||
| Salmonella Paratyphi | E | P | P | P | P | E | ||||||
| Salmonella Typhi | E | P | P | P | P | E | ||||||
| STEC | E | E | P | P | ||||||||
| Shigella | E | E | P | E | P | |||||||
| Yersinia (excluding pestis) | E | E | P | P | E |
*Clostridium botulinum and Vibrio parahaemolyticus do not have established or possible sequelae.
†This includes GBS variants such as Miller Fisher syndrome; other neurological conditions such as chronic inflammatory demyelinating polyneuropathy will also be assessed.
‡Considered here as a sequela of maternal Listeria infection.
§This includes associated diagnoses such as anterior uveitis and ankylosing spondylitis.
¶Shown not to be a sequela; retained to capture historical misdiagnosis of HUS.
GBS, Guillain-Barré syndrome; HUS, haemolytic uremic syndrome; STEC, Shiga toxin-producing Escherichia coli.
Validation of the ICD codes is currently in progress, via a literature review to identify administrative case definitions that have been validated in the Canadian context, medical expert consultation and for those sequelae without a relevant validated definition, a targeted chart review in British Columbia. In addition to the ICD codes, we may also use PharmaNet data to improve sequelae classification if needed by identifying pharmaceuticals given to patients (eg, tumour necrosis factor inhibitor use as an indication of reactive arthritis; intravenous immune globulin use for GBS).
Some of our sequelae of interest are lifelong (eg, Graves’ disease), and some are transient in that complete recovery is possible (eg, GBS, stillbirth). For lifelong sequelae, we will consider the individual as having the sequela on the earliest date they meet the administrative case definition for that sequela (with subsequent records considered as a continuation of the original event). For sequelae from which recovery and subsequent return to being at risk is possible, we will consider the individual as first having the sequela on the earliest date they meet the administrative case definition for that sequela; we will then apply a postsequela recovery time to determine the date on which the individual can be considered to be at risk for a new, subsequent occurrence of that sequela.
Individuals may develop more than one sequela during the study period, either because they develop multiple different sequelae (eg, HUS and GBS) or because they develop multiple occurrences of a single sequela from which complete recovery is possible (eg, GBS, stillbirth). In all instances, the occurrence of multiple sequelae will be recorded and described. When individuals develop multiple different sequelae during the study period (eg, HUS and GBS), we will treat these as distinct outcomes in our risk estimates. When individuals develop multiple occurrences of the same sequela, we will treat these as distinct outcomes but account for recurrent events.47
Individuals with foodborne infections who develop a sequela listed in table 2, but for which there is no current evidence of an established or possible link to the specific pathogen (eg, Campylobacter and stillbirth), will be excluded from our estimates of sequelae risk (but included in sensitivity analyses).
For all 14 infections, the secondary outcome of interest is death which will be classified using recorded events in the VS-Mortality database. Finally, we are also interested in (1) the acute illnesses related to these infections (regardless of whether the individual develops sequelae or dies) and (2) additional outcomes following acute sequelae (eg, end-stage kidney disease and kidney transplant following HUS); these will be included only in our descriptive and economic analyses.
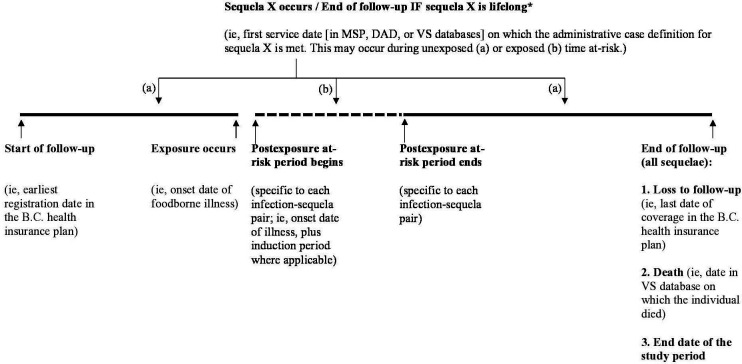
Measuring time at risk
For all individuals, time at risk for sequelae (figure 1) will be measured from the start of their entry into the study, which we define as the earliest registration date in the provincial health insurance programme (recorded in the Consolidation File). Individuals with foodborne infections may contribute to both the exposed time at risk (during the postinfection ‘at-risk’ period) and the unexposed time at risk (prior to, and after, the postinfection ‘at-risk’ period), while individuals without foodborne infections will only contribute to the unexposed time at risk. Time at risk for a specific sequela will be measured in days, from the date of entry into the study, until the development of that sequela, death, loss to follow-up or the end of the study. We define loss to follow-up as the last date of coverage in the provincial health insurance plan, calculated using the start day registered in the most recent year plus the total days registered in that year.
Figure 1.
Study follow-up period and time at risk for development of Sequela X (solid lines: unexposed time at risk; dashed lines: exposed time at risk. *For sequelae where recovery and return to being at risk is possible: this date + recovery time for Sequela X = date of return to being at risk for a subsequent occurrence of sequela X. B.C., British Columbia, Canada; DAD, Discharge Abstracts Database; MSP, Medical Services Plan; VS, Vital Statistics.
During the unexposed time at risk, we will treat all individuals as having the potential to develop any of the sequelae (with the exception of neonatal listeriosis and stillbirth, for which only those who are pregnant are at risk). For those who develop a foodborne infection, unexposed time at risk will end on the onset date of the infection. Infection onset date will be determined using the onset date reported in Panorama, and where this is missing, the date that the infection was reported minus the number of days between onset to reporting (eg, estimated using the Panorama data or from the literature).50
Exposed time at risk will be measured starting from the infection onset date, plus any additional induction periods (specific to each sequela and currently being determined via literature review and medical expert consultation). The end of the exposed time at risk period is currently being determined via literature review and medical expert consultation. During the exposed time at risk, individuals will be classified as having a sequela specific to their infection (table 2) on the date within the ‘at-risk’ period on which they meet the administrative case definition for that sequela (eg, the date of the physician visit or hospitalisation). After the postinfection at-risk period ends, individuals will revert to contributing to the unexposed time at risk.
These data are subject to censoring and truncation. Individuals will be censored for the sequela in the event of: death, last date of coverage in the provincial health insurance plan (ie, loss to follow-up, calculated as above) or the end of the study period, whichever comes first.51 52 In our descriptive and economic analyses, we will include all related healthcare use and prescription medication costs over the course of the infection and sequela(e), and in our estimates of mortality we will include any deaths recorded during the study period, following the sequela. We will determine whether healthcare use is related to infection and sequelae using ICD diagnosis codes, and we will determine whether prescription medication use is related via medical expert consultation.
Analysis plan
Data will be analysed and results reported following the STROBE (Strengthening the Reporting of Observational Studies in Epidemiology) and RECORD (REporting of studies Conducted using Observational Routinely-collected Data) guidelines.53 54 Analyses will be performed using SAS V.9.4 (SAS Institute, Cary, North Carolina, USA) and R.55 The nature and extent of missing data will be described. If imputation is used to complete missing data, specific methods and assumptions will be reported. We will emphasise estimation over tests of statistical significance by reporting relative measures of effect along with associated 95% CIs.
The datasets in table 1 contain the variable ‘sex’ (identified via government records) that denotes whether individuals are ‘male’ or ‘female’, thereby capturing a composite of sex and gender. To reflect for potential sex and gender differences, we will report and interpret findings stratified by this variable, in addition to overall findings.
Objective 1: To determine the risk of developing sequelae following foodborne infection, we will estimate hazard ratios using Cox regression models56 that adjust for confounders and comorbidities (see below), along with the possible effect-modifying role of age, sex, comorbidity and medication use (eg, antibiotics). We will include comorbidities as a composite score, using the revised Charlson Comorbidity Index and its associated coding algorithms.57–60 Following foodborne infection, we will compare the cumulative risk of first diagnosis of each infection-specific sequela using life table and Kaplan-Meier approaches.56 61 In the event an individual dies, we will use competing risk analysis.62 63 For those who experience more than one foodborne infection across the study period, we will explore the impacts of having multiple infections on the risk of sequelae and mortality. We will also estimate the likelihood of dying following foodborne infection using the same methods described above.
Objective 2: To describe the epidemiology and clinical progression across the range of outcomes, including acute illness (typically diarrhoea and other gastrointestinal symptoms), sequelae and death, for each foodborne infection we will calculate incidence rates; demographic, geographic and temporal distributions; timing and progression of outcomes; and case fatality rates, for both the acute stage, and sequelae associated to the foodborne infection.
Objective 3: To quantify the direct healthcare costs due to these infections and their various outcomes, we will determine health service use (ie, patterns of use by type, frequency, timing of physician visits and hospitalisations), for both the acute foodborne infection and any sequelae. We will estimate direct healthcare costs of out-of-hospital physician visits, hospitalisations and prescription medications. Out-of-hospital costs will be estimated using the MSP variables ‘Fee Item’ and ‘Paid Service’ and fee rates from the British Columbia fee schedule.64 Costs of inpatient and day-case hospitalisations will be calculated using established case-mix methodology (ie, using the ‘Resource Intensity Weight’ of each hospitalisation),65 and the British Columbia Ministry of Health unit costs for hospital stays.66 Total prescription medication costs will be calculated using the drug cost claimed by the pharmacist which includes the ingredient cost, professional dispensing fee and other special service fees (if applicable). Because these costs are captured directly in the PharmaNet data, they will be tallied directly. We will also apply these methods to determine the direct costs per sequelae, regardless of exposure. Costs will be adjusted for inflation using the Canadian Consumer Price Index.67 Results will be reported to allow comparability with other estimates (eg, 2010/2011 Canadian dollars and US dollars).
Objective 4: To determine the risk of sequelae in the population attributable to foodborne infections, we will calculate population attributable fractions using standard formulae.68 We will also describe the proportion of cases of each sequela with specific foodborne infections. Here, the total number of cases of each sequela occurring in British Columbia during the study period will be the denominator (eg, total number of cases of acute kidney injury), and the numerators will be the numbers of cases of each sequela occurring in those with specific foodborne infections (eg, total number of cases of hepatitis A virus, and of STEC, with acute kidney injury). We will also describe the proportion of individuals with sequela who do not have a foodborne infection, but who do have an ICD code for prior gastroenteritis, and use this to estimate the additional proportions of sequelae that may have an unidentified foodborne infection. We will calculate population attributable fractions and proportions for both established and possible sequelae, but clearly distinguish between the two when reporting findings.
Potential confounders and their adjustment
We will use propensity score matching and inclusion of potential confounders as covariates in our analyses, as our primary methods to adjust for confounding.69 The databases in table 1 include direct measurements of important known, strong confounders (eg, age, sex), as well as other potential factors (eg, use of protein pump inhibitors and antibiotics, disease severity). We will consider the following variables as potential confounders: age, sex, local health area, income band/area income (as a proxy for socioeconomic status), month/year, seasonality, immune status (eg, indication of immunosuppressant drugs, presence of conditions like cancer, pregnancy), use of medications like antibiotics and Charlson Comorbidity Index. Because none of our data sources include ethnicity nor race, we are unable to adjust, or conduct subanalyses, for these factors. Although ethnicity may impact healthcare-seeking behaviours, we anticipate these impacts will apply equally regardless of exposure, and thus we expect any bias in our findings to be negligible. Nevertheless, to assess residual confounding, we will conduct sensitivity analyses70 71 and perform indirect adjustments.72
Planned sensitivity analyses and study limitations
We are planning several sensitivity analyses to explore assumptions, methodological decisions, limitations in the data and robustness of results. We will explore the impact of propensity score matching on our sequelae and mortality risk estimates by also using (1) the whole unexposed population and (2) a random sample of unexposed individuals (matched on time), instead of propensity score-matched individuals. We may also explore additional matching and control strategies (eg, matching on age and sex). We will also explore the impacts of including individuals with foodborne infections who develop a sequela for which there is no current evidence of an established or possible link to the specific pathogen (eg, Campylobacter and stillbirth), in our estimates of sequelae risk. Our primary analyses will use a composite of all infections (ie, a report of any of the 14 foodborne infections) and their various sequelae. We will also analyse and present results for each of the individual foodborne infections and for each of the sequela.
A main recognised limitation of reportable disease data, such as the Panorama data in this study, is the under-ascertainment of foodborne infections. Here, this limitation means that individuals with foodborne infections who do not seek care, do not get tested, or who test negative will be misclassified as unexposed. We will assess the impacts of such potential misclassification via sensitivity analyses that illustrate how our findings could be impacted by different misclassification rates, using estimates of misclassification from the literature2 73 and from our data (eg, individuals with non-specific gastroenteritis). An additional limitation is that if sequelae develop over longer timeframes than our 10-year study (eg, over decades),42 our study cannot assess this scenario.
Ethics and dissemination
This study has received approval by a University of Waterloo Research Ethics Committee (no 30645), the University of British Columbia Behavioral Research Ethics Board (no H16-00021) and McGill University’s Institutional Review Board (no A03-M12-19A). In addition to conference presentations and dissemination to public health practitioners and other knowledge users, results will be published in peer-reviewed journals, and where such publications are not open access, they will also be stored on UWSpace, the University of Waterloo’s Institutional Repository (https://uwspace.uwaterloo.ca).
Supplementary Material
Acknowledgments
We thank Bei Yuan (Ethan) Zhang (University of British Columbia) and Dr Jenna Dixon (University of Waterloo) for helping in drafting early versions of this paper, and Dr Yuen Hung (University of Waterloo) for compiling relevant background literature. Study team members, Drs Jonathan Chan (University of British Columbia), Kristine Chapman (Vancouver General Hospital), Susan Horton (University of Waterloo), Douglas Matsell (British Columbia Children’s Hospital Research Institute) and Sara Pires (Technical University of Denmark), served as expert advisors and critically reviewed the study plan.
Footnotes
Contributors: SEM and EG are the co-principal investigators on this study. SEM, EG and MT conceived the study. The overall design was first developed by SEM, EG, MT and DP, with critical revisions from RJC, SE, STL, GGK and DMP. All coauthors developed the analysis plan, with specific statistical expertise provided by RJC, AC and MRG, and economic expertise by DP. SEM drafted the manuscript; all authors provided feedback on manuscript drafts and approved the final version to be published. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding: This work is supported by the Canadian Institutes of Health Research (CIHR), Project Grant programme, grant number 156385 (Principal Investigator (PI): SEM) and also received seed funding from the University of Waterloo’s Research Incentive Fund (PI: SEM) and the BCCDC Foundation for Public Health’s Open Award Programme (PI: EG).
Disclaimer: All inferences, opinions and conclusions drawn in this research protocol are those of the authors and do not reflect the opinions or policies of the Data Steward(s).
Competing interests: SEM and EG report funding for this study as per the funding statement. SEM reports other relationships; she is an associate editor at Epidemiology and Infection (for which she receives a small honorarium); she has served as a paid expert on behalf of the Attorney General of Canada in legal proceedings, providing evidence on the public health risks and benefits of unpasteurised milk, and she is an expert on the Joint FAO/WHO Expert Meetings on Microbiological Risk Assessment (JEMRA) Roster of Experts. GGK reports honoraria for speaking or consultancy from Abbvie, Janssen, Pfizer and Takeda. He has received research support from Ferring, Janssen, Abbvie, GlaxoSmith Kline, Merck and Shire. He shares ownership of a patent: TREATMENT OF INFLAMMATORY DISORDERS, AUTOIMMUNE DISEASE, AND PBC. UTI Limited Partnership, assignee. Patent WO2019046959A1. PCT/CA2018/051098. 7 September 2018. EG’ spouse works for QHR Technologies, a Canadian medical records company; these records were not used in this study. All other authors have nothing to disclose.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Havelaar AH, Kirk MD, Torgerson PR, et al. World Health organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med 2015;12:e1001923. 10.1371/journal.pmed.1001923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas MK, Murray R, Flockhart L, et al. Estimates of the burden of foodborne illness in Canada for 30 specified pathogens and unspecified agents, circa 2006. Foodborne Pathog Dis 2013;10:639–48. 10.1089/fpd.2012.1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awofisayo A, Amar C, Ruggles R, et al. Pregnancy-Associated listeriosis in England and Wales. Epidemiol Infect 2015;143:249–56. 10.1017/S0950268814000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlier C, Perrodeau Élodie, Leclercq A, et al. Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect Dis 2017;17:510–9. 10.1016/S1473-3099(16)30521-7 [DOI] [PubMed] [Google Scholar]
- 5.Karmali MA, Petric M, Lim C, et al. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J Infect Dis 1985;151:775–82. 10.1093/infdis/151.5.775 [DOI] [PubMed] [Google Scholar]
- 6.Gould LH, Demma L, Jones TF, et al. Hemolytic uremic syndrome and death in persons with Escherichia coli O157:H7 infection, foodborne diseases active surveillance network sites, 2000-2006. Clin Infect Dis 2009;49:1480–5. 10.1086/644621 [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez LA, Ruigómez A, Panés J. Acute gastroenteritis is followed by an increased risk of inflammatory bowel disease. Gastroenterol 2006;130:1588–94. [DOI] [PubMed] [Google Scholar]
- 8.Small CL, Xing L, McPhee JB, et al. Acute infectious gastroenteritis potentiates a Crohn's disease pathobiont to fuel ongoing inflammation in the post-infectious period. PLoS Pathog 2016;12:e1005907. 10.1371/journal.ppat.1005907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruzante JM, Majowicz SE, Fazil A, et al. Hospitalization and deaths for select enteric illnesses and associated sequelae in Canada, 2001-2004. Epidemiol Infect 2011;139:937–45. 10.1017/S0950268810001883 [DOI] [PubMed] [Google Scholar]
- 10.Tam CC, Rodrigues LC, Petersen I, et al. Incidence of Guillain-Barré syndrome among patients with Campylobacter infection: a general practice research database study. J Infect Dis 2006;194:95–7. 10.1086/504294 [DOI] [PubMed] [Google Scholar]
- 11.Helms M, Vastrup P, Gerner-Smidt P, et al. Short and long term mortality associated with foodborne bacterial infections: registry-based study. Br Med J 2003;326:1–5. [PMC free article] [PubMed] [Google Scholar]
- 12.Bremell T, Bjelle A, Svedhem A. Rheumatic symptoms following an outbreak of Campylobacter enteritis: a five year follow up. Ann Rheum Dis 1991;50:934–8. 10.1136/ard.50.12.934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall JK, Thabane M, Garg AX, et al. Incidence and epidemiology of irritable bowel syndrome after a large waterborne outbreak of bacterial dysentery. Gastroenterology 2006;131:445–50. 10.1053/j.gastro.2006.05.053 [DOI] [PubMed] [Google Scholar]
- 14.Garg AX, Pope JE, Thiessen-Philbrook H, et al. Arthritis risk after acute bacterial gastroenteritis. Rheumatology 2008;47:200–4. 10.1093/rheumatology/kem339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanevik K, Wensaas K-A, Rortveit G, et al. Irritable bowel syndrome and chronic fatigue 6 years after Giardia infection: a controlled prospective cohort study. Clin Infect Dis 2014;59:1394–400. 10.1093/cid/ciu629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehn M, Wallensten A, Widerström M, et al. Post-infection symptoms following two large waterborne outbreaks of Cryptosporidium hominis in northern Sweden, 2010-2011. BMC Public Health 2015;15:529. 10.1186/s12889-015-1871-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannu T, Mattila L, Rautelin H, et al. Campylobacter‐triggered reactive arthritis: a population‐based study. Rheumatol 2002;41:312–8. [DOI] [PubMed] [Google Scholar]
- 18.Hunter PR, Hughes S, Woodhouse S, et al. Health sequelae of human cryptosporidiosis in immunocompetent patients. Clin Infect Dis 2004;39:504–10. 10.1086/422649 [DOI] [PubMed] [Google Scholar]
- 19.Wang L-H, Fang X-C, Pan G-Z. Bacillary dysentery as a causative factor of irritable bowel syndrome and its pathogenesis. Gut 2004;53:1096–101. 10.1136/gut.2003.021154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hannu T, Mattila L, Siitonen A, et al. Reactive arthritis attributable to Shigella infection: a clinical and epidemiological nationwide study. Ann Rheum Dis 2005;64:594–8. 10.1136/ard.2004.027524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saps M, Pensabene L, Di Martino L, et al. Post-Infectious functional gastrointestinal disorders in children. J Pediatr 2008;152:812–6. 10.1016/j.jpeds.2007.11.042 [DOI] [PubMed] [Google Scholar]
- 22.Rosner BM, Werber D, Höhle M, et al. Clinical aspects and self-reported symptoms of sequelae of Yersinia enterocolitica infections in a population-based study, Germany 2009-2010. BMC Infect Dis 2013;13:236. 10.1186/1471-2334-13-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litleskare S, Rortveit G, Eide GE, et al. Prevalence of irritable bowel syndrome and chronic fatigue 10 years after Giardia infection. Clin Gastroenterol Hepatol 2018;16:1064–72. 10.1016/j.cgh.2018.01.022 [DOI] [PubMed] [Google Scholar]
- 24.Helms M, Simonsen J, Mølbak K. Foodborne bacterial infection and hospitalization: a registry-based study. Clin Infect Dis 2006;42:498–506. 10.1086/499813 [DOI] [PubMed] [Google Scholar]
- 25.García Rodríguez LA, Ruigómez A, Panés J. Acute gastroenteritis is followed by an increased risk of inflammatory bowel disease. Gastroenterology 2006;130:1588–94. 10.1053/j.gastro.2006.02.004 [DOI] [PubMed] [Google Scholar]
- 26.Ternhag A, Törner A, Svensson A, et al. Short- and long-term effects of bacterial gastrointestinal infections. Emerg Infect Dis 2008;14:143–8. 10.3201/eid1401.070524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gradel KO, Nielsen HL, Schønheyder HC, et al. Increased short-and long-term risk of inflammatory bowel disease after Salmonella or Campylobacter gastroenteritis. Gastroenterol 2009;137:495–501. [DOI] [PubMed] [Google Scholar]
- 28.Jess T, Simonsen J, Nielsen NM, et al. Enteric Salmonella or Campylobacter infections and the risk of inflammatory bowel disease. Gut 2011;60:318–24. 10.1136/gut.2010.223396 [DOI] [PubMed] [Google Scholar]
- 29.Porter CK, Choi D, Cash B, et al. Pathogen-Specific risk of chronic gastrointestinal disorders following bacterial causes of foodborne illness. BMC Gastroenterol 2013;13:46. 10.1186/1471-230X-13-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riddle MS, Murray JA, Cash BD, et al. Pathogen-Specific risk of celiac disease following bacterial causes of foodborne illness: a retrospective cohort study. Dig Dis Sci 2013;58:3242–5. 10.1007/s10620-013-2733-7 [DOI] [PubMed] [Google Scholar]
- 31.Nakao JH, Collier SA, Gargano JW. Giardiasis and subsequent irritable bowel syndrome: a longitudinal cohort study using health insurance data. J Infect Dis 2017;215:798–805. 10.1093/infdis/jiw621 [DOI] [PubMed] [Google Scholar]
- 32.Painter JE, Collier SA, Gargano JW. Association between Giardia and arthritis or joint pain in a large health insurance cohort: could it be reactive arthritis? Epidemiol Infect 2017;145:471–7. 10.1017/S0950268816002120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, et al. Global epidemiology of Campylobacter infection. Clin Microbiol Rev 2015;28:687–720. 10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tricco AC, Pham Ba', Duval B, et al. A review of interventions triggered by hepatitis A infected food-handlers in Canada. BMC Health Serv Res 2006;6:157. 10.1186/1472-6963-6-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpentier B, Cerf O. Persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol 2011;145:1–8. 10.1016/j.ijfoodmicro.2011.01.005 [DOI] [PubMed] [Google Scholar]
- 36.British Columbia Centre for Disease Control British Columbia annual summary of reportable diseases, 2017. Available: http://www.bccdc.ca/resource-gallery/Documents/Statistics%20and%20Research/Statistics%20and%20Reports/Epid/Annual%20Reports/2017CDAnnualReportFinal.pdf [Accessed 12 Nov 2019].
- 37.Scallan E, Majowicz SE, Hall G, et al. Prevalence of diarrhoea in the community in Australia, Canada, Ireland, and the United States. Int J Epidemiol 2005;34:454–60. 10.1093/ije/dyh413 [DOI] [PubMed] [Google Scholar]
- 38.Government of British Columbia Available: https://www2.gov.bc.ca/gov/content/health/health-drug-coverage/msp/bc-residents/eligibility-and-enrolment/are-you-eligible [Accessed 12 Nov 2019].
- 39.Ark TK, Kesselring S, Hills B, et al. Population Data BC: supporting population data science in British Columbia. Int J Pop Data Sci 2020;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.About PopData Population Data BC. Available: https://www.popdata.bc.ca/aboutus [Accessed 12 Nov 2019].
- 41.Government of British Columbia Public Health Act [SBC 2008] Chapter 28. Available: http://www.bclaws.ca/civix/document/id/complete/statreg/08028_01 [Accessed 12 Nov 2019].
- 42.Moorin RE, Heyworth JS, Forbes GM, et al. Long-Term health risks for children and young adults after infective gastroenteritis. Emerg Infect Dis 2010;16:1440. 10.3201/eid1609.081665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Population Data BC Data linkage. Available: https://www.popdata.bc.ca/datalinkage [Accessed 12 Nov 2019].
- 44.British Columbia Centre for Disease Control Case definitions. Available: http://www.bccdc.ca/health-professionals/clinical-resources/case-definitions [Accessed 12 Nov 2019].
- 45.World Health Organization ICD-10 version, 2016. Available: https://icd.who.int/browse10/2016/en [Accessed 12 Nov 2019].
- 46.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer Science & Business Media 2013.
- 47.Cook RJ, Lawless JF. The statistical analysis of recurrent events. New York, NY: Springer-Verlag, 2007. [Google Scholar]
- 48.Batz MB, Henke E, Kowalcyk B. Long-Term consequences of foodborne infections. Infect Dis Clin North Am 2013;27:599–616. 10.1016/j.idc.2013.05.003 [DOI] [PubMed] [Google Scholar]
- 49.Hu W. Diagnostic codes in MSP claim data, summary report. Victoria: Medical Services Plan, 1996. [Google Scholar]
- 50.Galanis E, Taylor M, Romanowski K, et al. Evaluating the timeliness of enteric disease surveillance in British Columbia, Canada, 2012-13. Can J Infect Dis Med Microbiol 2017;2017:1–7. 10.1155/2017/9854103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klein JP, Moeschberger ML. Survival analysis: techniques for censored and truncated data. 2nd edn New York, NY: Springer-Verlag New York Inc, 2003. [Google Scholar]
- 52.Qin J, Shen Y. Statistical methods for analyzing right-censored length-biased data under COX model. Biometrics 2010;66:382–92. 10.1111/j.1541-0420.2009.01287.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.von Elm E, Altman DG, Egger M, et al. Strengthening the reporting of observational studies in epidemiology (STROBE) statement: guidelines for reporting observational studies. BMJ 2007;335:806–8. 10.1136/bmj.39335.541782.AD [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benchimol EI, Smeeth L, Guttmann A, et al. The reporting of studies conducted using observational Routinely-collected health data (record) statement. PLoS Med 2015;12:e1001885 https://doi.org/ 10.1371/journal.pmed.1001885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R Core Team R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2019. http://www.R-project.org/ [Google Scholar]
- 56.DRJ C. Regression models and life-tables (with discussion). J R Stat Soc, B 1972;34:187–220. [Google Scholar]
- 57.Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis 1987;40:373–83. 10.1016/0021-9681(87)90171-8 [DOI] [PubMed] [Google Scholar]
- 58.Charlson M, Szatrowski TP, Peterson J, et al. Validation of a combined comorbidity index. J Clin Epidemiol 1884;47:1245–51. [DOI] [PubMed] [Google Scholar]
- 59.Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care 2005;43:1130–9. 10.1097/01.mlr.0000182534.19832.83 [DOI] [PubMed] [Google Scholar]
- 60.Quan H, Li B, Couris CM, et al. Updating and validating the Charlson comorbidity index and score for risk adjustment in hospital discharge Abstracts using data from 6 countries. Am J Epidemiol 2011;173:676–82. 10.1093/aje/kwq433 [DOI] [PubMed] [Google Scholar]
- 61.Kaplan EL, Meier PJ. Nonparametric estimation for incomplete observations. J Am Stat Assoc 1958;53:457–81. [Google Scholar]
- 62.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 1999;94:496–509. [Google Scholar]
- 63.Lau B, Cole SR, Gange SJ. Competing risk regression models for epidemiologic data. Am J Epidemiol 2009;170:244–56. 10.1093/aje/kwp107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Government of British Columbia Medical services Commission payment schedule, 2015. Available: https://www2.gov.bc.ca/assets/gov/health/practitioner-pro/medical-services-plan/msc_payment_schedule.pdf [Accessed 12 Nov 2019].
- 65.Poole B, Robinson S, MacKinnon M. Resource intensity weights and Canadian hospital costs: some preliminary data. Healthc Manage Forum 1998;11:22–6. 10.1016/S0840-4704(10)61000-9 [DOI] [PubMed] [Google Scholar]
- 66.Canadian Institute for Health Information Patient cost estimator: methodological notes and glossary. Ottawa, ON: CIHI, 2016. https://www.cihi.ca/en/pce_methodology_notes_en.pdf [Google Scholar]
- 67.Statistics Canada Consumer price index (CPI). Available: http://www23.statcan.gc.ca/imdb/p2SV.pl?Function=getSurvey&SDDS=2301 [Accessed 12 Nov 2019].
- 68.Mansournia MA, Altman DG. Population attributable fraction. BMJ 2018;360:k757. 10.1136/bmj.k757 [DOI] [PubMed] [Google Scholar]
- 69.Holmes WM. Using propensity scores in quasi-experimental designs. Los Angeles, Calif: Sage Publishing, 2014. [Google Scholar]
- 70.Stürmer T, Wyss R, Glynn RJ, et al. Propensity scores for confounder adjustment when assessing the effects of medical interventions using nonexperimental study designs. J Intern Med 2014;275:570–80. 10.1111/joim.12197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brookhart MA, Schneeweiss S, Rothman KJ, et al. Variable selection for propensity score models. Am J Epidemiol 2006;163:1149–56. 10.1093/aje/kwj149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuenzig ME, Barnabe C, Seow CH, et al. Asthma Is Associated With Subsequent Development of Inflammatory Bowel Disease: A Population-based Case-Control Study. Clin Gastroenterol Hepatol 2017;15:1405–12. 10.1016/j.cgh.2017.02.042 [DOI] [PubMed] [Google Scholar]
- 73.MacDougall L, Majowicz S, Doré K, et al. Under-Reporting of infectious gastrointestinal illness in British Columbia, Canada: who is counted in provincial communicable disease statistics? Epidemiol Infect 2008;136:248–56. 10.1017/S0950268807008461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.British Columbia Ministry of Health [creator] Medical Services Plan (MSP) Payment Information File. Population Data BC [publisher]. Data Extract. MOH, 2018. Available: http://www.popdata.bc.ca/data
- 75.Canadian Institute for Health Information [creator] Discharge Abstract Database (Hospital Separations). Population Data BC [publisher]. Data Extract. MOH, 2019. Available: http://www.popdata.bc.ca/data
- 76.British Columbia Ministry of Health [creator] PharmaNet. BC Ministry of Health [publisher]. Data Extract. Data Stewardship Committee, 2019. Available: http://www.popdata.bc.ca/data
- 77.British Columbia Vital Statistics Agency [creator] Vital Statistics Deaths. Population Data BC [publisher]. Data Extract. BC Vital Statistics Agency, 2019. Available: http://www.popdata.bc.ca/data
- 78.British Columbia Vital Statistics Agency [creator] Vital Statistics Stillbirths. Population Data BC [publisher] Data Extract. BC Vital Statistics Agency, 2019. Available: http://www.popdata.bc.ca/data
- 79.British Columbia Ministry of Health [creator] Consolidation File (MSP Registration & Premium Billing). Population Data BC [publisher]. Data Extract. MOH, 2019. Available: http://www.popdata.bc.ca/data
- 80.Statistics Canada [creator] Statistics Canada Income Band Data. Catalogue Number: 13C0016. Population Data BC [publisher]. Data Extract. Population Data BC (2018), 2009. Available: http://www.popdata.bc.ca/data
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.