Infection with the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can lead to severe pneumonia, lung function impairment, and multiple organ failure that can be fatal (1). There are currently no U.S. Food and Drug Administration–approved therapies across the spectrum for patients affected with coronavirus disease (COVID-19). However, several experimental approaches, including repurposing of the RNA polymerase–inhibiting antiviral agents, have improved the health outcomes among patients with COVID-19 (2). In Southeast Asia, a combination therapy of ribavirin, a nucleoside analog, together with two nonnucleosidic antivirals used to treat the human immunodeficiency virus (HIV) has shown some promise in mild-to-moderately ill patients (3), as did a study employing another nucleoside-based antiviral agent, favipiravir (4). In the United States, the most promising drug therapy thus far has been remdesivir (GS-441524). A multisite trial indicated that treatment with remdesivir was associated with speedy recovery among hospitalized patients infected with SARS-CoV-2, which prompted the U.S. Food and Drug Administration to allow emergency use access of the drug for COVID-19 treatment on May 1, 2020 (5). Despite these promising recent developments, strategies that could help clinicians predict which patients are most likely to respond effectively to a given therapeutic regimen remain perfunctory. Patient prioritization and treatment matching should be paramount in ensuring optimization of therapeutics to thwarting this pandemic.
Along these lines, we reported that patients who die from sepsis syndrome and acute respiratory failure initially present in the emergency department and the medical intensive care unit with a conspicuous metabolomic profile (6–9). Among the most striking changes were the increases in metabolites related to the de novo production of nicotinamide adenine dinucleotide (NAD; a key cofactor central to metabolism), mitochondrial function, and production of ATP as summarized in Table 1. In these patients, the normal endogenous precursors to NAD, as well as purine and pyrimidine nucleobases and nucleosides, were rerouted from their normal biosynthetic pathways. Furthermore, patients with poor outcomes presented with metabolomic dysfunction that appears to be irreversible as evidenced by the accumulation of unprocessed tricarboxylic acid cycle metabolites and carnitine esters. Together, these markers not only predict mortality but also suggest that nonsurvivors have an acute bioenergetic crisis likely attributable to severe decrements in mitochondrial function and metabolism that we have observed several days prior to death (6–9).
Table 1.
Changes in the Abundance of Selected Metabolites in Patients with Sepsis with Poor Outcomes, Altogether Markers of Metabolic Imbalance
| Pathway | Metabolite | Change in Nonsurvivors |
|---|---|---|
| NAD metabolism | N-acetyl-tryptophan | ↑↑↑ |
| Tryptophan | ↓ | |
| Kynurenine | ↑↑ | |
| 3-Hydroxy-kynurenine | ↑↑ | |
| Kynurenate | ↑↑ | |
| Picolinate | ↑↑ | |
| 2-Hydroxyadipate | ↑↑ | |
| Quinate | ↑ | |
| Quinolinate | ↑↑ | |
| 1-Methyl-nicotinamide | ↑↑ | |
| TCA β-oxidation | Succinate | ↑ |
| Succinylcarnitine | ↑↑ | |
| Acetylcarnitine | ↑↑ | |
| Glutarylcarnitine | ↑↑ | |
| 2-Methylbutyrylcarnitine | ↑↑ | |
| Nucleobases | 1,3-Dimethylurate | ↑↑ |
| 1-Methylxanthine | ↑↑ | |
| Adenine | ↑ | |
| Cytidine | ↑ | |
| Thymine | ↑↑ | |
| Uracil | ↑↑ | |
| Nucleosides | N2,N2-Dimethylguanosine | ↑↑ |
| N1-Methylguanosine | ↑↑ | |
| N1-Methyladenosine | ↑↑ | |
| N2-Methylguanosine | ↑↑ | |
| N6-Succinyladenosine | ↑↑ | |
| Uridine | ↓ | |
| 5-Methyluridine | ↓ | |
| Lipids | 1-Arachidonoyl-GPC | ↓↓ |
| 1-Palmitoyl-GPC | ↓↓ |
Definition of abbreviations: GPC = glycerophosphocholine; NAD = nicotinamide adenine dinucleotide; TCA = tricarboxylic acid.
Recent metabolomics and proteomics studies on patients with COVID-19 with associated severe respiratory distress demonstrated plasma metabolomic signatures similar to those described above for sepsis syndrome (10, 11). The results implicated dysregulation of macrophage function, platelet degranulation and complement system pathways, and metabolic suppression, similar to the acute bioenergetic crisis profile we previously observed in patients with sepsis with poor outcomes (6, 8).
Here, we posit that success in reducing the viral burden in patients with SARS-CoV-2 using antiviral drugs that first require intracellular ATP-dependent activation will be contingent on the overall bioenergetic phenotype of the patient. All the nucleoside-based drugs currently considered for SARS-CoV-2 treatment (e.g., remdesivir, ribavirin, and favipiravir) require functional activation by host enzymes that employ endogenous ATP for their conversion to the active triphosphate species. For instance, remdesivir must be converted to its triphosphate form to become a substrate for the viral replicase-transcriptase and get integrated into the growing viral RNA chain to prevent the full replication of the virus (12). Ribavirin also needs ATP for activation, whereas favipiravir, a nucleobase analog, requires initial conversion to its nucleotide form via a mechanism that requires phosphoribosyl pyrophosphate, another high-energy intracellular biomolecule (13). These activation processes and their dependence on ATP levels may explain the limited success of some of the nucleoside-derived drugs targeted at the viral replicase-transcriptase. An impaired “energy status” of a patient, characterized by the decrement of high-energy metabolites such as ATP and phosphoribosyl pyrophosphate (14), may impede effective drug conversion and thus decrease efficacy against viral replication.
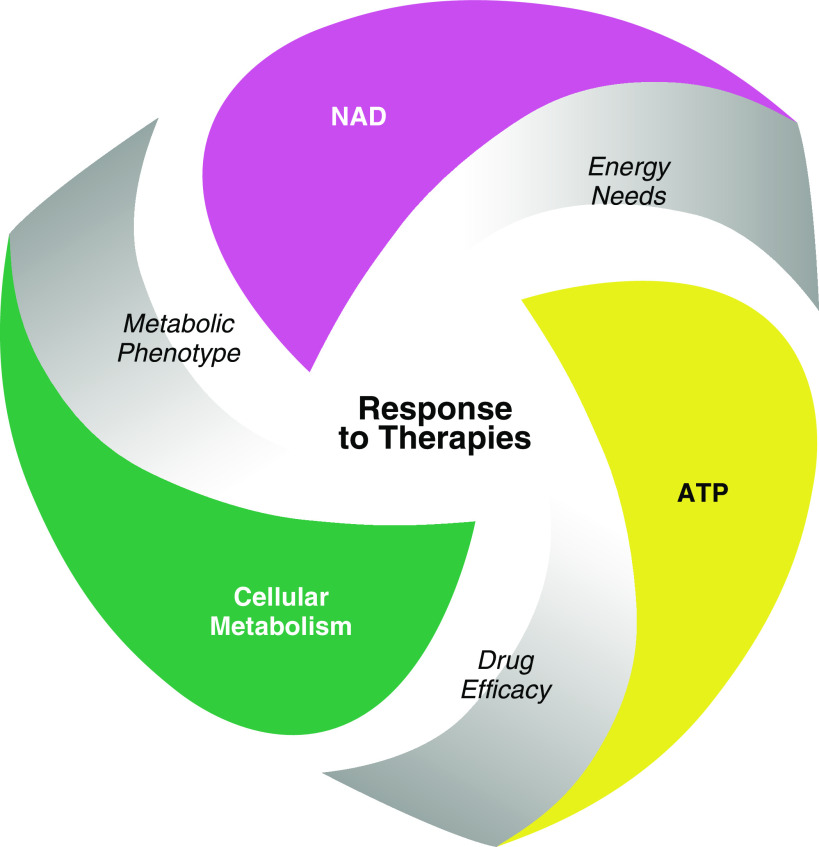
The implications of the present considerations (as outlined in Figure 1) offer opportunities for stratification of patients with COVID-19 based on their metabolic phenotype to maximize drug efficacy. Monitoring patients’ bioenergetics status might help rationalize why a given replicase-trancriptase inhibitor is successful in some patients and not in others. With this perspective, drugs with significant dependence on ATP will be less effective in patients presenting with advanced metabolic dysfunction. Therefore, we propose that ribavirin or favipiravir, drugs that require multiple-stage functionalization, would have a better chance of success in patients presenting with a near-normal metabolic profile. However, patients that present with a metabolomic phenotype of an acute bioenergetic crisis could be treated with drugs that require less energy, such as remdesivir, as it requires low ATP commitment for drug activation.
Figure 1.
Response to viral drug therapies in patients with severe acute respiratory distress syndrome coronavirus 2 (SARS-CoV-2) may be dependent on the metabolic status of the patient. A patient’s metabolomic phenotype can predict patient outcomes as well as the status of cellular metabolism. In particular, the function of nicotinamide adenine dinucleotide (NAD) is critical for cellular metabolism as well as energy production such as ATP. Because most viral transcriptase inhibitors are dependent on ATP for activation and incorporation with viral RNAs, cellular metabolism and energy production can critically affect the efficacy of certain antivirals. Monitoring the metabolomic phenotype in clinical trials that use antiviral drugs will be critical for optimization of drug efficacy.
One could also consider the severity of the bioenergetic crisis in the context of cellular metabolism and its relationship to patient outcomes and survival. For example, antiviral agents may be infective in patients presenting with an advanced metabolic dysfunction. This may explain why remdesivir can improve duration of symptoms but has no statistical benefit on patient survival (11, 15). In such cases, targeted metabolic strategies or nutritional supplementation that include remediation of the NAD and ATP pools could be implemented to reduce the impact of the acute bioenergetic crisis on dysregulated immune and repair responses that lead to multiorgan failure. Moreover, correction of these nutritional deficiencies may be necessary to optimize drug responses.
In conclusion, metabolomic phenotyping may represent an important step toward personalized therapeutics in patients infected with COVID-19. First, it will help enhance the therapeutic efficacy of ATP-dependent replicase-trancriptase inhibitors currently under clinical investigations against COVID-19. Drugs with significant dependence on ATP to achieve functionality against the viral target might be less effective in patients presenting with advanced metabolic dysfunction. Second, this metabolomic phenotyping will also inform the need to integrate balanced metabolic and nutritional strategies within the treatment regimen to optimize patient recovery. Defining and modulating the bioenergetic state in a risk-stratified and personalized approach could have long-term impact in improving patient outcomes to SARS-CoV-2 infections.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank the study subjects, without whom this work could not be conducted, and Thomas Gagliano for graphic design.
Footnotes
Supported by 1KL2TR003097 (R.J.L.), R21AI144374 (H.S.C.), in part by Elysium Health (M.M.), and R01 HL113614, R01 GM127823, and UL1 TR001417 (M.N.G.). All grants are funded by the U.S. National Institutes of Health. The KL2 and UL1 grants are funded by the National Center for Advancing Translational Sciences.
Originally Published in Press as DOI: 10.1165/rcmb.2020-0206LE on June 23, 2020
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 3.Hung IF-N, Lung K-C, Tso EY-K, Liu R, Chung TW-H, Chu M-Y, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McKee DL, Sternberg A, Stange U, Laufer S, Naujokat C. Candidate drugs against SARS-CoV-2 and COVID-19. Pharmacol Res. 2020;157:104859. doi: 10.1016/j.phrs.2020.104859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, et al. ACTT-1 Study Group Members. Remdesivir for the treatment of covid-19 - preliminary report. N Engl J Med. [online ahead of print] 22 May 2020; DOI: 10.1056/NEJMoa2007764. [Google Scholar]
- 6.Langley RJ, Tsalik EL, van Velkinburgh JC, Glickman SW, Rice BJ, Wang C, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. 2013;5:195ra95. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers AJ, McGeachie M, Baron RM, Gazourian L, Haspel JA, Nakahira K, et al. Metabolomic derangements are associated with mortality in critically ill adult patients. PLoS One. 2014;9:e87538. doi: 10.1371/journal.pone.0087538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langley RJ, Tipper JL, Bruse S, Baron RM, Tsalik EL, Huntley J, et al. Integrative “omic” analysis of experimental bacteremia identifies a metabolic signature that distinguishes human sepsis from systemic inflammatory response syndromes. Am J Respir Crit Care Med. 2014;190:445–455. doi: 10.1164/rccm.201404-0624OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langley RJ, Flores L, Mohney R, Lovato J, Case L, Harrod KS, et al. Metabolomic analysis of the target cohort identifies serum signatures that outperform lactate in sepsis prognosis [abstract] Am J Respir Crit Care Med. 2015;191:A4007. [Google Scholar]
- 10.Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, et al. SARS-CoV-2 infected host cell proteomics reveal potential therapy targets. General Cell Biology & Physiology Virology. [online ahead of print] 22 Mar 2020; DOI: 10.21203/rs.3.rs-17218/v1. [Google Scholar]
- 11.Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, et al. Proteomic and metabolomic characterization of covid-19 patient sera. Cell. doi: 10.1016/j.cell.2020.05.032. [online ahead of print] 28 May 2020; DOI: S0092-8674(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naesens L, Guddat LW, Keough DT, van Kuilenburg AB, Meijer J, Vande Voorde J, et al. Role of human hypoxanthine guanine phosphoribosyltransferase in activation of the antiviral agent T-705 (favipiravir) Mol Pharmacol. 2013;84:615–629. doi: 10.1124/mol.113.087247. [DOI] [PubMed] [Google Scholar]
- 14.Frederick DW, Loro E, Liu L, Davila A, Jr, Chellappa K, Silverman IM, et al. Loss of nad homeostasis leads to progressive and reversible degeneration of skeletal muscle. Cell Metab. 2016;24:269–282. doi: 10.1016/j.cmet.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.