Abstract
Chitinase 3–like-1 (Chi3l1) and IL-13 are both ligands of IL-13 receptor α2 (IL-13Rα2). The binding of the former activates mitogen-activated protein kinase, AKT, and Wnt/β-catenin signaling, and plays important roles in innate and adaptive immunity, cellular apoptosis, oxidative injury, allergic inflammation, tumor metastasis and wound healing, fibrosis, and repair in the lung. In contrast, the latter binding is largely a decoy event that diminishes the effects of IL-13. Here, we demonstrate that IL-13Rα2 N-glycosylation is a critical determinant of which ligand binds. Structure–function evaluations demonstrated that Chi3l1–IL-13Rα2 binding was increased when sites of N-glycosylation are mutated, and studies with tunicamycin and Peptide:N-glycosidase F (PNGase F) demonstrated that Chi3l1–IL-13Rα2 binding and signaling were increased when N-glycosylation was diminished. In contrast, structure–function experiments demonstrated that IL-13 binding to IL-13Rα2 was dependent on each of the four sites of N-glycosylation in IL-13Rα2, and experiments with tunicamycin and PNGase F demonstrated that IL-13–IL-13Rα2 binding was decreased when IL-13Rα2 N-glycosylation was diminished. Studies with primary lung epithelial cells also demonstrated that Chi3l1 inhibited, whereas IL-13 stimulated, N-glycosylation as evidenced by the ability of Chi3l1 to inhibit and IL-13 to stimulate the subunits of the oligosaccharide complex A and B (STT3A and STT3B). These studies demonstrate that N-glycosylation is a critical determinant of Chi3l1 and IL-13 binding to IL-13Rα2, and highlight the ability of Chi3l1 and IL-13 to alter key elements of the N-glycosylation apparatus in a manner that would augment their respective binding.
Keywords: IL-13, chitinase 3–like-1, oligosaccharyltransferase, IL-13 receptor α2, N-glycosylation
Clinical Relevance
This is the first study demonstrating the significance of N-glycosylation in Chi3l1 (chitinase 3–like-1) and IL-13 binding to IL-13Rα2. Because these molecules play an important role in regulating proinflammatory mechanisms under many different conditions, unraveling the determinants of this process is clinically and scientifically significant.
The GH 18 (18 glycosyl hydrolase) gene family contains true enzymatically active chitinases that degrade chitin polysaccharides and CLPs (chitinase-like proteins) that bind, but do not degrade, chitin (1, 2). Chitinase 3–like-1 (Chi3l1; also called YKL-40 in humans and breast regression protein-39 [BRP-39] in rodents) is the prototype of the CLP gene subfamily. The retention of the GH 18 moieties over species and evolutionary time (3) has led to the belief that they play essential roles in biology. In support of this speculation, recent studies from our laboratory demonstrated that Chi3l1 plays a major role in antipathogen, antigen- and oxidant-induced inflammatory, repair, and remodeling responses by regulating a variety of essential biologic processes, including oxidant injury, apoptosis, pyroptosis, inflammasome activation, T-helper cell type 1/-2 cytokine balance, M2 macrophage differentiation, TGF-β1 (transforming growth factor-β1) elaboration, dendritic cell accumulation and activation, fat accumulation, and the activation of mitogen-activated protein kinase (MAPK), Akt, and Wnt/β-catenin signaling in the lung (4–8). These studies also suggest that Chi3l1 is part of a primordial protective response based on its ability to decrease epithelial cell apoptosis, while simultaneously stimulating fibroproliferative repair (9). In keeping with these pleiotropic effects, studies from our laboratory and others also demonstrated that Chi3l1 is dysregulated in a variety of diseases characterized by injury, inflammation, and/or remodeling, including asthma, pulmonary fibrosis, pneumonia, visceral tumors, and obesity (1, 4, 9–16).
To further understand the biology of Chi3l1, we addressed the possibility that Chi3l1 drives its effector responses via a ligand–receptor mechanism. To do this, we used a variety of approaches to define the receptor proteins that bind to Chi3l1. These studies demonstrated that Chi3l1 binds to the known receptor for IL-13, IL-13 receptor α2 (IL-13Rα2) (8). They also demonstrated that Chi3l1 does not compete with IL-13 for binding to IL-13Rα2 and that YKL-40/Chi3l1/BRP-39, IL-13Rα2, and IL-13 interact to form a multimeric receptor complex called the chitosome (8). Lastly, they demonstrate that Chi3l1/YKL-40 activates MAPK, AKT/protein kinase B (PKB), and Wnt/β-catenin signaling pathways and regulates apoptosis, pyroptosis, inflammasome activation, oxidant injury, antibacterial responses, melanoma metastasis, and TGF-β1 elaboration in the cells derived from the lung or in vivo lungs via IL-13Rα2–dependent mechanisms (8). These studies demonstrate that ligand–receptor interactions of Chi3l1 and IL-13 to common receptor, IL-13Rα2, are an important factor modulating lung pathologies in which these two ligands are implicated. However, the mechanisms that control the binding of Chi3l1 and IL-13 to IL-13Rα2 have not been appropriately addressed.
Asparagine (N)-linked glycosylation is the most frequent modification of membrane and secretory proteins in eukaryotes (17, 18). This modification has a number of important biologic consequences, including the regulation of protein folding and sorting and the regulation of protein and cellular interactions with its environment (17). IL-13 is known to bind to IL-13Rα2 (8, 19). In this setting, IL-13Rα2 often acts as a decoy receptor that diminishes IL-13–induced cell and tissue responses (19). Studies by Kioi and colleagues (20) have demonstrated that N-glycosylation plays an essential role in IL-13 binding to IL-13Rα2 and IL-13Rα2 decoy responses. In contrast, the importance of N-glycosylation in Chi3l1–IL-13Rα2 binding and Chi3l1 signaling have not been investigated.
We hypothesized that N-glycosylation plays a critical role in the degree to which IL-13 and/or Chi3l1 bind to IL-13Rα2. To test this hypothesis, we defined the importance of N-glycosylation in the physical interactions of IL-13Rα2 and its ligands, IL-13 and Chi3l1. We also characterized the effects of N-glycosylation on Chi3l1-induced cell signaling and the effects of IL-13 and Chi3l1 on the N-glycosylation apparatus of primary cells. These studies demonstrate that, in contrast to the essential role of N-glycosylation in IL-13–IL-13Rα2 binding, Chi3l1 binds to IL-13Rα2 via a N-glycosylation–independent mechanism, and that Chi3l1–IL-13Rα2 binding is increased and Chi3l1-induced signaling is augmented by interventions that inhibit or enzymatically abrogate IL-13Rα2 N-glycosylation. They also highlight the ability of IL-13 and Chi3l1 to stimulate the cellular N-glycosylation apparatus, and thereby potentially contribute to the resolution of IL-13–induced cell and/or tissue responses.
Methods
For a full description of the methods, please refer to the data supplement.
Mice
C57BL/6 mice were purchased from the Jackson Laboratory and were housed at the Brown animal facility until they were used. Chi3l1/YKL-40–transgenic (Tg) mice (YKL-40 Tg) were generated and characterized in our laboratory as previously described (5, 21). IL-13Rα2–null mutant mice were generated and provided by Dr. M. Grusby of Harvard University School of Public Health (22), and the mice were back-crossed to a C57BL/6J background for more than 10 generations. All murine procedures were approved by the Institutional Animal Care and Use Committees at Brown University.
Yeast 2 Hybrid Assays
The screening and identification of specific molecular interaction was undertaken according to the procedures described by our laboratory (8).
Co-IP and Immunoblot
The co-IP and immunoblot (IB) with antibodies against IL-13Rα2, Chi3l1, or IL-13 as previously described by our laboratory (8, 19).
Assessment of Effects of Tunicamycin and PNGase F
Preconditioned THP-1 cells or peritoneal macrophages were treated with tunicamycin (Sigma-Aldrich) in a dose of 2 μg/ml overnight before harvesting the cells. The Peptide:N-glycosidase F (PNGase F) (New England Biolabs) was treated on the THP-1 cell lysates with 20 μl (for 400 μg of cell lysates) of PNGase F (New England Biolabs) for 1 hour at 37°C in an incubator. Then the cell lysates were subjected to the Co-IP and IB evaluations to assess the effects of deglycosylation with specific antibodies against Chi3l1, IL-13, and IL-13Rα2.
Assessment of Chi3l1 Signaling
The activation of MAPK/Erk and Akt and nuclear accumulation of β-Catenin and c-fos were assessed by IB evaluations according to the procedures described previously by our laboratory (8).
Preparation and Stimulation of Peritoneal Macrophages
Peritoneal macrophages were isolated from 7-week-old IL-13Rα2–null and wild-type (WT) mice according to the previously described procedures (19). These cell populations were greater than 95% macrophages, as assessed by F4/80 antibody staining.
Ligand Regulation of STT3A, STT3B, and DAD1
MLE12 lung epithelial cells were stimulated with recombinant (r) Chi3l1 (rChi3l1) or rIL-13 for indicated doses and time points, then the cells were harvested for further analysis. The cell lysates were then subjected to qRT-PCR or Western blot evaluations for the assessment of mRNA and protein expression, respectively. The specific primers sequences (Table E1 in the data supplement) and antibodies against the catalytic subunits of the oligosaccharyltransferase complex A (STT3A; Millipore Sigma) and B (STT3B, Proteintech) and defender against apoptotic cell death 1 (DAD1) (Sigma-Aldrich) were used for these evaluations.
Assessment of the Effects of Chi3l1 on STT3A in the Murine Lung
Cell lysates from the lungs of WT and Chi3l1/YKL-40 Tg mice were generated and the levels of STT3A mRNA were quantified using qRT-PCR, as previously described by our laboratory (5, 21).
Statistical Analysis
Normally distributed data are expressed as mean (±SEM) and were assessed for significance by Student’s t test or ANOVA as appropriate. Statistical significance was defined at a P value less than 0.05. All statistical analyses were performed with SPSS version 13.0 (SPSS Inc.). Statistical significance was defined at a level of P less than 0.05.
Results
Effects of Mutation of IL-13Rα2 N-Glycosylation Sites in Chi3l1/YKL-40 Binding to IL-13Rα2
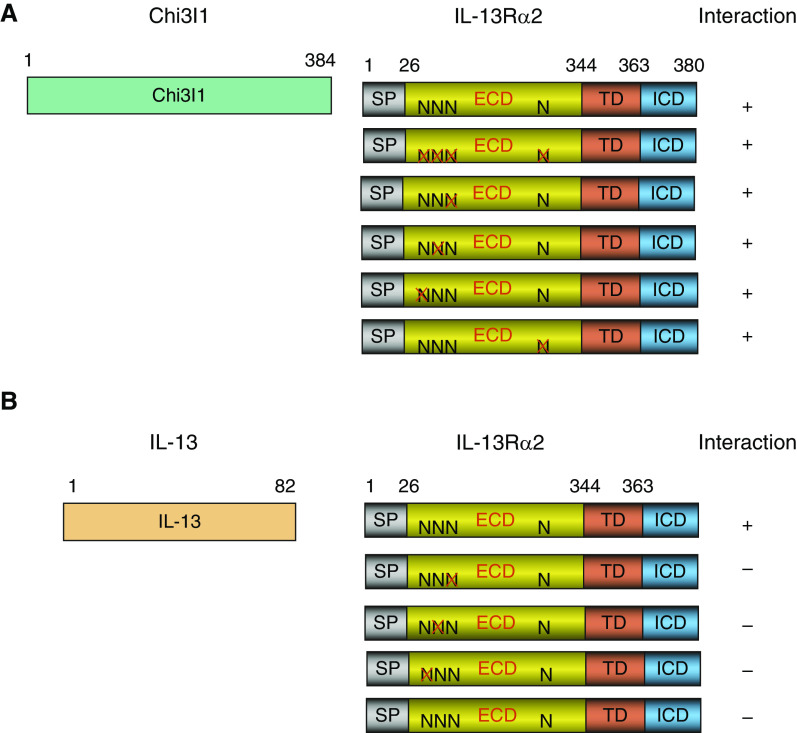
IL-13Rα2 has four N-glycosylation sites in its extracellular domain (20). To begin to understand the importance of these glycosylation sites in the binding of ligands, Y2H (yeast 2 hybrid) assays were undertaken. In these experiments, WT IL-13Rα2 and mutated IL-13Rα2 moieties that no longer contained N-glycosylation sites were used as bait, and binding to Chi3l1 was assessed. In accord with prior observations (8), Chi3l1 binding to WT IL-13Rα2 was readily appreciated (Figure 1A). Importantly, Chi3l1 binding to IL-13Rα2 was not abrogated by individual mutations or the simultaneous mutation of up to four N-glycosylation sites in these assays (Figure 1A). These studies demonstrate that Chi3l1/YKL-40 binding to IL-13Rα2 is not significantly diminished by the elimination of N-glycosylation sites of IL-13Rα2, suggesting that Chi3l1/YKL-40 binds to IL-13Rα2 via a mechanism(s) that is N-glycosylation independent.
Figure 1.
Yeast 2 hybrid characterization of human (h) Chi3l1 (Chitinase 3-like-1) and IL-13 interactions with IL-13 receptor α2 (IL-13Rα2) with alterations at sites of glycosylation. (A and B) Full-length Chi3l1 (composed of 384 amino acids [A]) or IL-13 (composed of 82 amino acids [B]) was used to evaluate the interactions with full-length IL-13Rα2 (380 amino acids) composed of signal peptide (SP), extracellular domain (ECD), transmembrane domain (TD), and C-terminal intracellular domain (ICD). The predicted N-glycosylation sites (N) in ECD were individually mutated (X), and the final interactions were indicated as positive (+) or negative (−). Each panel is representative of a minimum of three evaluations.
Effects of Mutation of IL-13Rα2 N-Glycosylation Sites in IL-13 Binding to IL-13Rα2
To begin to understand the importance of N-glycosylation in IL-13–IL-13Rα2 binding, a similar Y2H approach was employed. In these experiments, WT IL-13Rα2 and mutated IL-13Rα2 moieties that no longer contained N-glycosylation sites were used as bait, and binding to IL-13 was assessed. As can be seen in Figure 1B, IL-13 binding to IL-13Rα2 was readily appreciated when N-glycosylation of IL-13Rα2 was maintained. In contrast, IL-13 binding was markedly decreased when a single site of N-glycosylation was mutated (Figure 1B). These studies demonstrate that, in contrast to Chi3l1, IL-13 binds to IL-13Rα2 via a mechanism(s) that is N-glycosylation dependent.
Effects of Tunicamycin on Ligand–IL-13Rα2 Binding
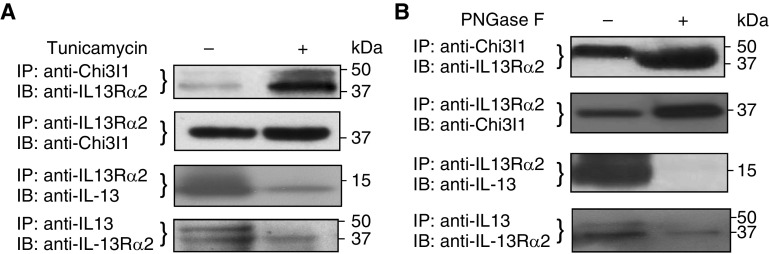
Studies were next undertaken to determine if inhibition of N-glycosylation with tunicamycin altered Chi3l1/YKL-40 or IL-13 binding to IL-13Rα2. In these experiments, THP-1 cells were preconditioned with rTNF-α and rIL-13 pretreatment (8), and the cells were exposed to tunicamycin (2 μg/ml) or vehicle (PBS) for 24 hours. Cell lysates were then prepared and IP was undertaken with antibodies against one of the ligands (either Chi3l1/YKL-40 or IL-13) or the receptor (IL-13Rα2). The composition of the precipitate was then evaluated using Western blotting with the antibodies against the moieties that were not targeted in the IP. In accord with prior observations (8), in the absence of tunicamycin, IP with anti–IL-13Rα2 simultaneously brought down Chi3l1 that could be detected by Western blotting. Similarly, IP with antibodies against Chi3l1 simultaneously brought down IL-13Rα2 (Figure 2A). Importantly, in the presence of tunicamycin, the binding of Chi3l1 to IL-13Rα2 was significantly enhanced (Figure 2A). Similarly, in the absence of tunicamycin, IP with anti–IL-13Rα2 simultaneously brought down IL-13 that could be detected by Western blotting, and IP with antibodies against IL-13 simultaneously brought down IL-13Rα2 (Figure 2A). Importantly, and in contrast to our findings with Chi3l1, IL-13 binding to IL-13Rα2 was markedly decreased after tunicamycin treatment. When viewed in combination, these studies demonstrate that IL-13Rα2 binding to Chi3l1 is augmented, whereas binding to IL-13 is diminished when N-glycosylation is diminished.
Figure 2.
Effects of alterations in N-glycosylation on the interactions of Chi3l1 or IL-13 with IL-13Rα2. Preconditioned THP-1 cells expressing Chi3l1, IL-13, and IL-13Rα2 were subjected to these evaluations. (A) IP and IB assays on the cells treated with tunicamycin (2 μg/ml, overnight) before harvesting the cells. (B) IP and IB assays on the THP-1 cell lysates treated with Peptide:N-glycosidase F (PNGase F) (1 μl/20 μg of lysates) for 1 hour at 37°C. Each panel is representative of a minimum of three evaluations.
Effects of PNGase F on Ligand–IL-13Rα2 Binding
To determine if enzymatic abrogation of N-glycosylation altered ligand–IL-13Rα2 binding PNGase was employed. In these experiments, THP-1 cells were activated with rTNF-α and rIL-13 pretreatment, cell lysates were prepared and incubated in the presence or absence of PNGase for 1 hour, and IP/IB was undertaken as described previously here. As can be seen in Figure 2B, PNGase enhanced the binding of Chi3l1/YKL-40 to IL-13Rα2 (Figure 2B), while significantly decreasing IL-13 binding to IL-13Rα2 (Figure 2B).
Effects of Alterations in N-Glycosylation on Chi3l1 Signaling
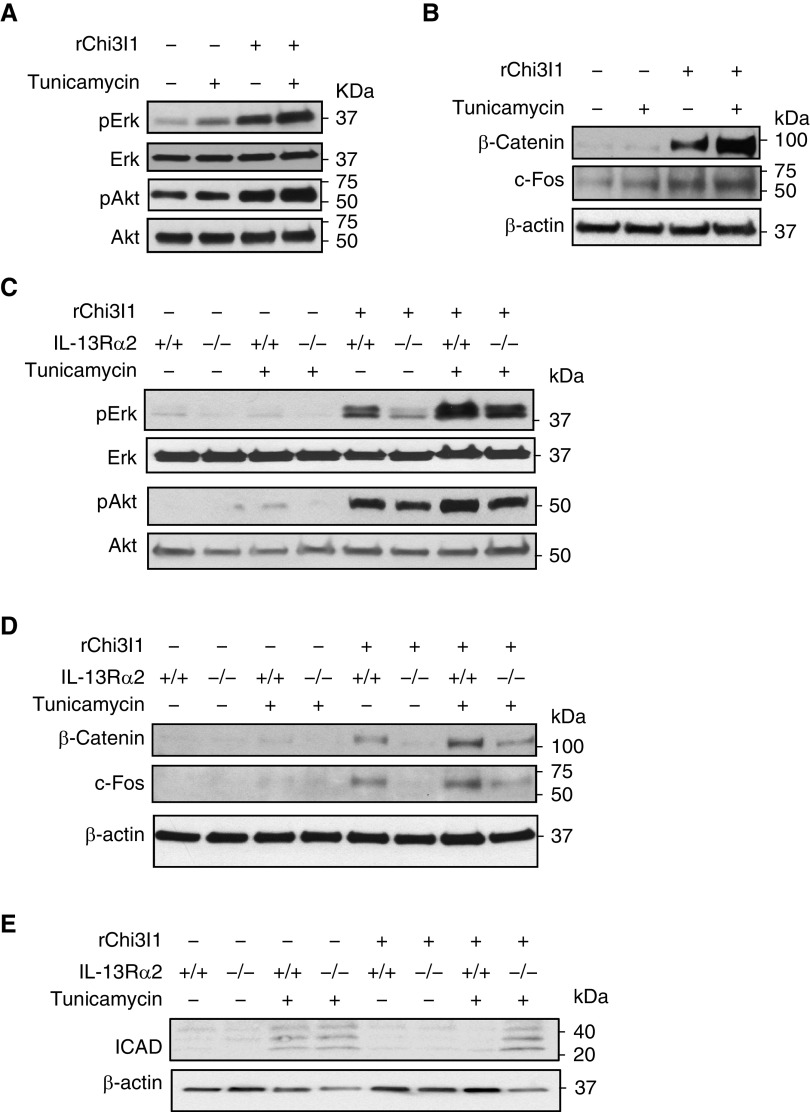
Studies were next undertaken to determine if the alterations in ligand–IL-13Rα2 binding noted previously here altered cell signaling responses. In these experiments, we initially determined if the ability of rChi3l1 to activate Erk, and Akt/PKB was altered by treatment with tunicamycin (2 μg/ml, overnight). As previously reported (8, 19), rChi3l1 was a potent stimulator of Erk and Akt phosphorylation in the absence of tunicamycin (Figure 3A). In keeping with the enhancement of Chi3l1–IL-13Rα2 binding that was seen when N-glycosylation was diminished, treatment with tunicamycin, augmented the ability of rChi3l1 to activate Erk and Akt (Figure 3A). In accord with these observations, tunicamycin treatment also increased rChi3l1 stimulation of β-catenin and c-fos (Figure 3B). Studies with cells from IL-13Rα2–null mice demonstrated that the enhanced MAPK, AKT, and Wnt/β-catenin signaling that was seen when tunicamycin was employed was mediated, to a great extent, via an IL-13Rα2–dependent mechanism(s) (Figures 3C and 3D). When viewed in combination, these studies demonstrate that the enhanced Chi3l1–IL-13Rα2 binding that is seen when N-glycosylation is decreased is associated with appropriate increases in Chi3l1-induced MAPK, AKT, and Wnt/β-catenin signaling.
Figure 3.
Effects of alterations in N-glycosylation on Chi3l1 signaling and tunicamycin-induced apoptosis. Peritoneal macrophages isolated from wild-type (WT) (+/+) and IL-13Rα2–null mutant mice (−/−) were stimulated with recombinant (r) Chi3l1 (rChi3l1) with and without tunicamycin treatment (2 μg/ml). (A) After overnight incubation, WT cells were harvested and total cell lysates were subjected to IB assays with phospho-specific antibodies against Erk (pErk), Akt (pAkt), together with antibodies detecting total forms of Erk and Akt. (B) Nuclear fractions of the cell lysates were used to detect the levels of β-catenin and c-Fos expression. (C and D) Cells isolated from WT and IL-13Rα2−/− mice were subjected to IB assays with antibodies used in A and B. (E) Western blot evaluations of inhibitor of caspase-activated DNase (ICAD) in cells from WT and IL-13Rα2–null mice after treatment of tunicamycin and rChi3l1. Each panel is representative of a minimum of three evaluations. Erk = extracellular regulated kinase.
Effects of Chi3l1–IL-13Rα2 Binding on Downstream Cellular Responses
Studies were next undertaken to determine if the enhanced Chi3l1 signaling responses that were seen when IL-13Rα2 N-glycosylation was decreased were associated with appropriate alterations in significant cellular responses that are downstream of these signaling events. These studies focused on inhibitor of caspase-activated DNase (ICAD), which is also known as DNA fragmentation factor 45 kD, and is encoded by alternatively spliced mRNAs (23). It was chosen because of the important role that it plays in apoptosis, where ICAD-null cells are often resistant to many inducers of apoptosis (24). It is also an important regulator of cellular differentiation (25). In these experiments, primary macrophages were obtained from WT and IL-13Rα2–null mice and incubated in the presence and absence of tunicamycin and/or Chi3l1. As previously reported (26, 27), tunicamycin treatment induced cellular apoptosis represented by cleaved ICAD (Figure 3E). Caspase 3 activation was also appreciated (Figure E1). Treatment with Chi3l1 reduced tunicamycin-induced ICAD cleavage. (Figure 3E). Importantly, this inhibitory effect required IL-13Rα2 (and Chi3l1–IL-13Rα2 binding), because null mutations of IL-13Rα2 abrogated this Chi3l1-induced inhibitory response, while augmenting caspase 3 cleavage (Figure 3E and Figure E1). When viewed in combination, these studies demonstrate that the enhanced Chi3l1–IL-13Rα2 binding and signaling that are seen when N-glycosylation is decreased are associated with appropriate increases in events that are downstream of these signaling responses, such as the regulation of critical apoptosis moieties like ICAD and caspase 3.
Effects of IL-13 and Chi3l1 on Catalytic Components of Oligosaccharyltransferase
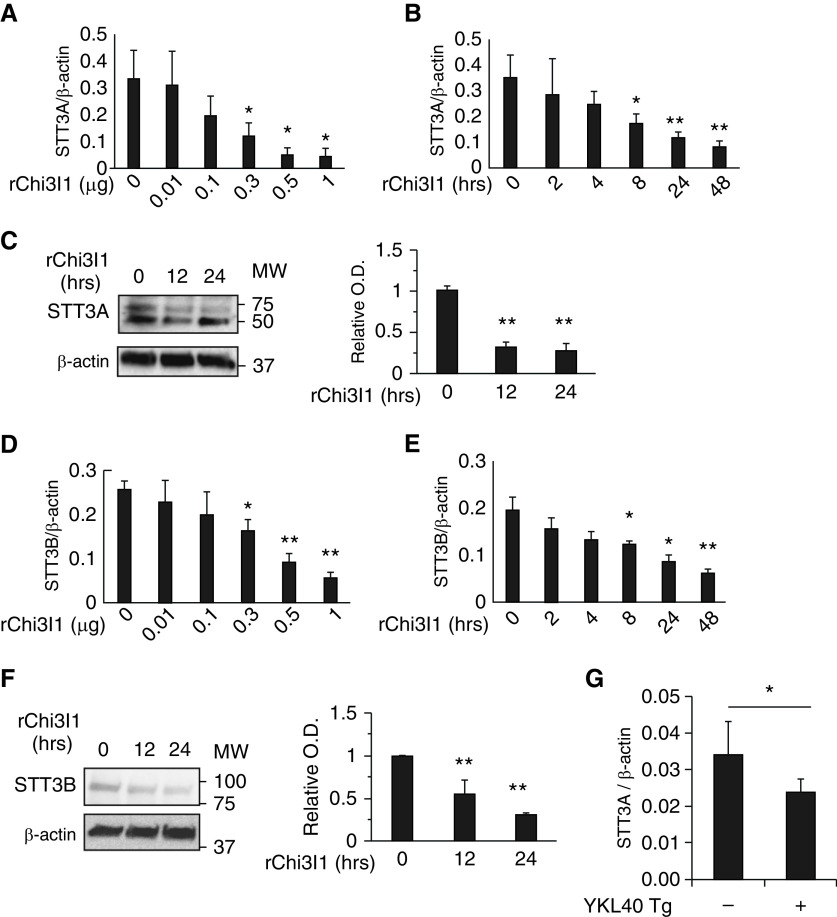
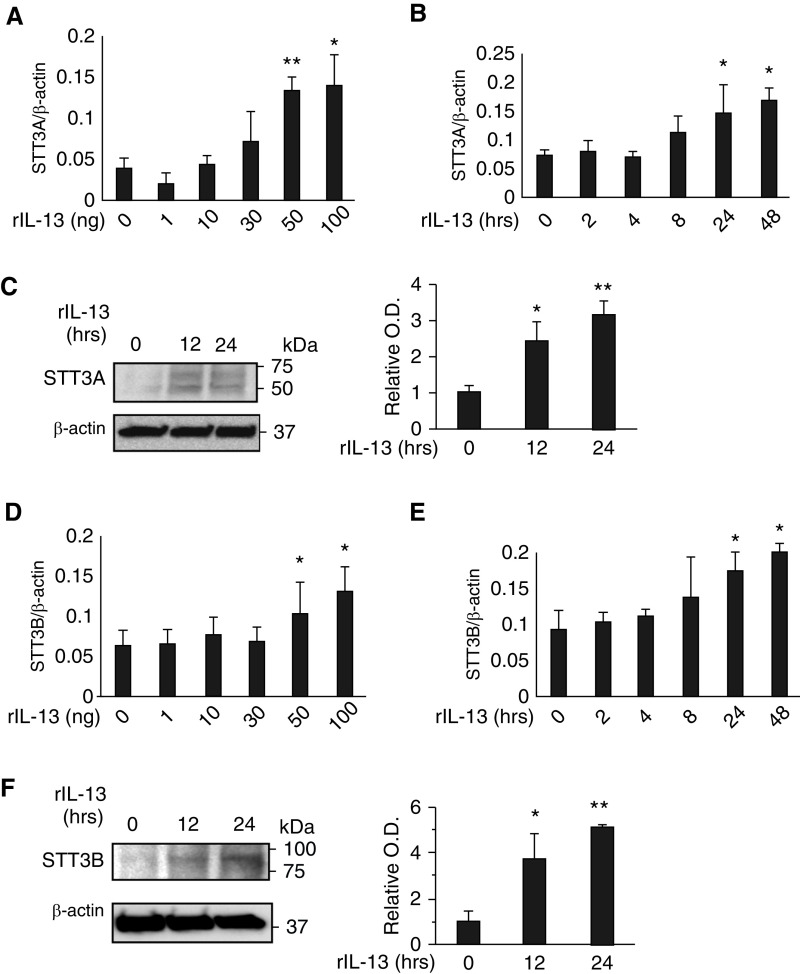
In the central reaction of the N-glycosylation pathway, oligosaccharyltransferase (OST) transfers a preassembled oligosaccharide to selected asparagine (N) residues (17). To further understand the biology of IL-13Rα2 and its ligands, studies were undertaken to define the effects of Chi3l1 and IL-13 on the expression and accumulation of major subunits of the OST complex. Our studies initially focused on STT3A and STT3B which are the major catalytic components of the complex (18, 28–30). Chi3l1 inhibited the expression and accumulation of STT3A and STT3B in lung epithelial cells (Figures 4A–4C). Chi3l1 also stimulated STT3B in a similar manner (Figures 4D–4F). Similar induction was noted in vivo in lungs from Chi3l1 overexpression Tg mice (Figure 4G). In contrast, IL-13 stimulated the expression and accumulation of STT3A and -B in lung epithelial cells in a dose- and time-dependent manner (Figure 5). In keeping with our demonstration that Chi3l1 and IL-13 binding to IL-13Rα2 are increased and decreased, respectively, when N-glycosylation is diminished, these studies demonstrate that Chi3l1 inhibits, and IL-13 stimulates, key catalytic aspects of the N-glycosylation apparatus in lung epithelial cells.
Figure 4.
Effects of Chi3l1 on catalytic components of oligosaccharyltransferase (OST). MLE12 lung epithelial cells were stimulated with rChi3l1 to determine Chi3l1 regulation of the catalytic subunits of the oligosaccharyltransferase complex A (STT3A) and B (STT3B), the major catalytic components of OST. (A and B) The mRNA expression of STT3A gene detected by qRT-PCR in the cells stimulated with different doses (A) and time points (B) of rChi3l1. (C) Representative IBs detecting STT3A protein expression in the MLE12 cells at different time points of rChi3l1 (0.5 μg/ml) stimulation. (D and E) The mRNA expression of STT3B gene detected by qRT-PCR in the cells stimulated with different doses (A) and time points (B) of rChi3l1. (F) A representative IB detecting STT3B protein expression in the MLE12 cells with different time points of rChi3l1 stimulation. (G) STT3A expression in the lungs of 6- to 8-week-old WT and Chi3l1/YKL-40 Tg mice with doxycycline (0.5 g/L) transgene induction for 2 weeks (n = 4 mice/group). The values in A, B, D, E, and G represent mean ± SEM in triplicated samples in a minimum of two separate experiments. (C and F) Representative of a minimum of three evaluations. The bar graphs to the right of IBs in C and F illustrate the relative quantities of STT3A and STT3B from three experiments measured by densitometric image analysis. *P < 0.05 and **P < 0.01 compared with vehicle controls. MLE12 = mouse lung epithelial cells; MW = molecular weight; Tg = transgenic; O.D. = optical density.
Figure 5.
Effects of IL-13 on catalytic components of OST. MLE12 lung epithelial cells were stimulated with rIL-13 to determine IL-13 regulation of STT3A and STT3B. (A and B) The mRNA expression of STT3A gene detected by qRT-PCR in the cells stimulated with different doses (A) and time points (B) of rIL-13. (C) Representative IBs detecting STT3A protein expression in the MLE12 cells at different time points of rIL-13 (50 ng/ml) stimulation. (D and E) The mRNA expression of STT3B gene detected by qRT-PCR in the cells stimulated with different doses (A) and time points (B) of rIL13. (F) A representative IB detecting STT3B protein expression in MLE12 cells with different time points of rIL-13 (50 ng/ml) stimulation. The values in A, B, D, and E represent mean ± SEM in triplicated samples in a minimum of two separate experiments. (C and F) Representative of a minimum of three evaluations. The bar graphs to the right of the IBs in C and F illustrate the relative quantities of STT3A and STT3B from three experiments measured by densitometric image analysis. *P < 0.05 and **P < 0.01 compared with vehicle controls.
Effects of Chi3l1 and IL-13 on DAD1
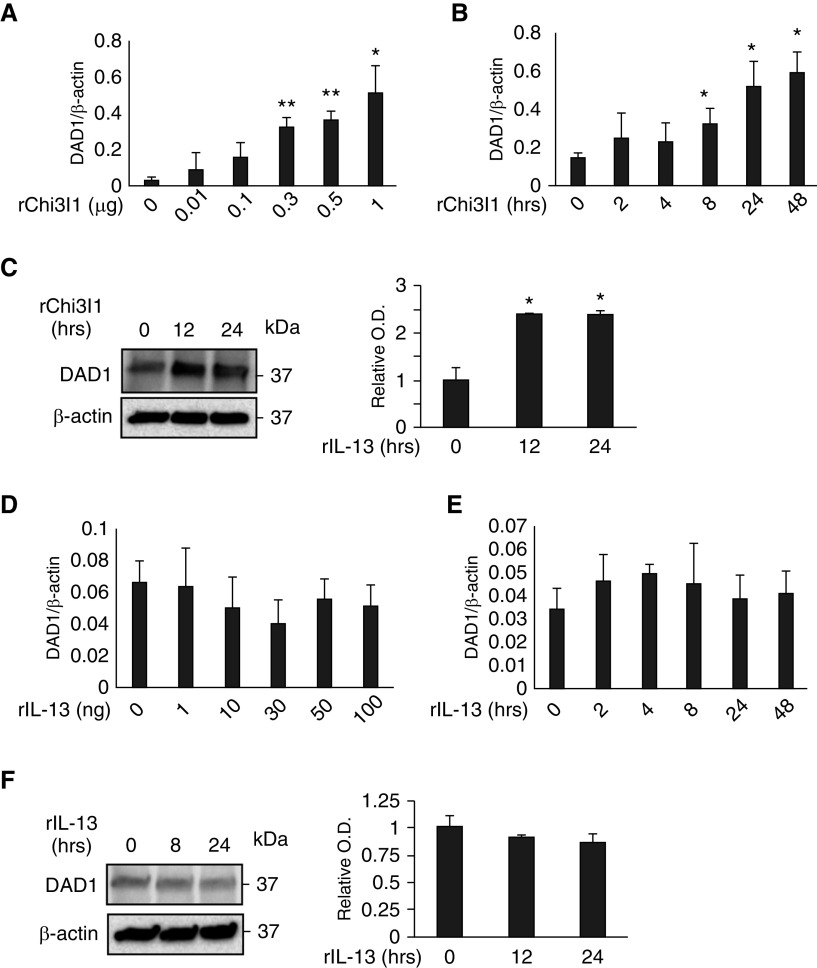
In addition to its catalytic components, the OST complex contains accessory protein that participates in N-glycosylation (18, 28–30). To further our understanding of the relationships between Chi3l1, IL-13, and IL-13Rα2, studies were undertaken to see if Chi3l1 or IL-13 regulated key OST regulatory proteins. These studies focused on DAD1, which is known to play a key role in the inhibition of apoptosis and induction of N-glycosylation (31, 32). As can be seen in Figures 6A–6C, Chi3l1 was a potent stimulator of DAD1 expression and accumulation in primary epithelial cells. In contrast, IL-13 did not stimulate DAD1 in a similar manner (Figures 6D–6F).
Figure 6.
Effects of Chi3l1 and IL-13 on DAD1 (defender against apoptotic cell death 1). MLE12 lung epithelial cells were stimulated with rChi3l1 and rIL-13, then the expression of DAD1 was detected by qRT-PCR and IB assays. (A and B) The mRNA expression of DAD1 gene detected by qRT-PCR in the cells stimulated with different doses (A) and time points (B) of rChi3l1. (C) Representative IBs detecting DAD1 protein expression in the MLE12 cells at different time points of rChi3l1 stimulation. (D and E) The mRNA expression of DAD1 gene detected by qRT-PCR in the cells stimulated with different doses (A) and time points (B) of rIL-13. (F) A representative IB detecting DAD1 protein expression in MLE12 cells with different time points of rIL-13 stimulation. The values in A, B, D, and E represent mean ± SEM in triplicated samples in a minimum of two separate experiments. (C and F) Representative of a minimum of three evaluations. The bar graphs to the right of the IBs in C and F show the relative quantities of DAD1 from three experiments measured by densitometric image analysis. *P < 0.05 and **P < 0.01, compared with vehicle controls.
Discussion
IL-13Rα2 was described as a high-affinity receptor for IL-13 that is distinct from the IL-13Rα1–IL-4Rα receptor dimer that IL-13 shares with IL-4 (33, 34). It was initially believed to be a decoy receptor, because it only contains a 17–amino acid cytoplasmic domain (35), and early studies highlighted its ability to diminish IL-13 responses (22, 33, 36). However, more recent studies have demonstrated that IL-13 also signals and regulates a variety of cellular and tissue responses via IL-13Rα2 (34, 37–43). The complexity of IL-13Rα2 was further reinforced when it was appreciated that it also binds and transmits signals from Chi3l1, and that IL-13, Chi3l1, and IL-13Rα2 participate in a multimeric receptor complex called the chitosome (8). N-glycosylation is required for IL-13 binding to IL-13Rα2 and the inhibition of IL-13 effector responses (20). The present studies add to our understanding of IL-13, Chi3l1, IL-13Rα2, and the chitosome by demonstrating that, in contrast to IL-13, which binds to IL-13Rα2 with higher affinity than IL-13Rα1 in the presence of N-glycosylation, Chi3l1 binding to IL-13Rα2 and Chi3l1-induced IL-13Rα2–mediated signaling are augmented when N-glycosylation is diminished. When viewed in combination, these studies highlight a mechanism by which protein glycosylation can determine the degree to which IL-13 or Chi3l can bind to and mediate responses via IL-13Rα2.
Glycation is a form of cotranslational and post-translational protein modification that adds glycosylphosphatidylinositol anchors that link proteins to lipids through glycan linkages (44). All N-linked carbohydrates are linked through N-acetylglucosamine and the amino acid, asparagine. They occur at sites with amino acid sequences Asp-X-serine/threonine, where the X can be anything but proline (17, 18). The addition of N-linked carbohydrates can affect protein folding, the sorting of proteins in the endoplasmic reticulum, and multiple protein–protein interactions and cellular responses in the protein’s environment (17). The importance of N-glycosylation of IL-13Rα2 is highlighted in these studies, which demonstrate that it is essential in IL-13 binding to IL-13Rα2 and IL-13Rα2–induced decoy responses, and that it diminishes Chi3l1 binding to IL-13Rα2 and IL-13Rα2–mediated Chi3l1 signaling. Unfortunately, we cannot determine if N-glycosylation is a widely used mechanism to control IL-13 receptor–ligand binding, because the importance of N-glycosylation in IL-13 binding to its major receptor (the IL-13Rα1/IL-4 Rα heterodimer) has not been defined.
N-linked protein glycosylation is a covalent protein modification that occurs across all three domains of life: Bacteria, Archaea, and Eukarya (29). Proper N-linked glycosylation plays a critical role in protein folding, as well as stability, biosynthetic quality control, intracellular and extracellular trafficking, and physiologic function (45). In keeping with the importance of N-glycosylation, we used two different approaches to address its importance in IL-13 and Chi3l1 binding and signaling. Early studies used tunicamycin, which inhibits N-glycosylation by blocking the transfer of N-acetylglucosamine (GlcNAc)-P from uridine diphosphate-GlcNAc to dolichol (46). Because tunicamycin can also cause abnormal protein folding and decrease protein incorporation into cellular membranes (47), we also compared the results obtained with tunicamycin to those induced with the deglycosylase PNGase. Importantly, the enhanced ability of Chi3l1 and decreased ability of IL-13 to bind to IL-13Rα2, respectively, was seen with both interventions. These similar results establish the importance of N-glycosylation in the noted protein–protein interactions. They are also in accord with the yeast hybrid studies that highlight the importance of N-glycosylation site in the physical binding of IL-13Rα2 to IL-13, and its ability to diminish Chi3l1 binding to IL-13Rα2.
Glycans are involved in fundamental molecular and cell biology processes occurring in cancer, including cell signaling and communication, tumor cell dissociation and invasion, cell matrix interactions, tumor angiogenesis, immune modulation, and metastasis formation (48). Studies from our laboratory and others have highlighted the frequent dysregulation of Chi3l1 in a wide variety of visceral tumors (1, 10–12). They have also highlighted the critical role that Chi3l1 plays in the pathogenesis of pulmonary metastasis (11, 12, 49). These studies also highlighted the importance of IL-13Rα2 and the chitosome in these responses (8, 19). When viewed in combination with the present studies, one can see how the degree of glycosylation of IL-13Rα2 could play a major role in these responses, especially in diseases like glioblastoma, which are associated with the exaggerated expression of Chi3l1 and IL-13Rα2 (50).
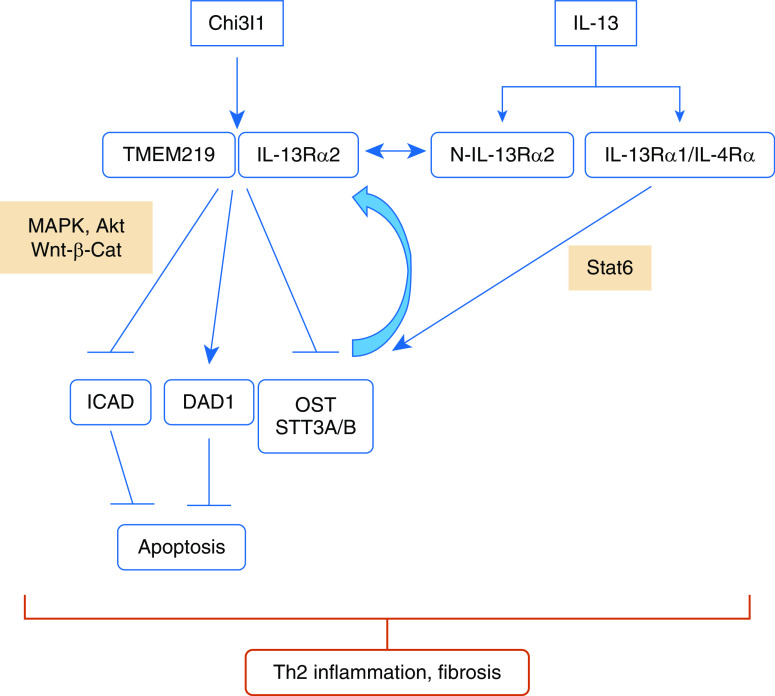
OST is a multisubunit enzyme complex that N-glycosylates proteins in the secretory pathway (18, 28–30). Mammalian OSTs are hetero-oligomeric membrane complexes, and contain one of two separately encoded catalytic subunits, STT3A or STT3B (28). STT3A is associated with the translocon, regulates cotranslational glycosylation, and is responsible for the majority of N-linked glycosylation in mammalian cells (28, 45). In contrast, STT3B glycosylates a smaller number of sequons, carries out post-translational glycosylation, and maximizes sequon occupancy by glycosylating sites that are skipped by STT3A (28, 45). OST is largely considered to be constitutive and unregulated (28, 45). Importantly, our studies suggest that this may not be the case. Specifically, our studies demonstrate that Chi3l1 is a potent inhibitor, and IL-13 is a potent stimulator, of STT3A and STT3B in epithelial cells in vitro and in vivo in the murine lung. When these findings are combined with our demonstration that Chi3l1 binding to and signaling via IL-13Rα2 are increased when N-glycosylation is decreased, whereas IL-13 binding to IL-13Rα2 is dependent on and abrogated when N-glycosylation is decreased, one can envision feedback loops that regulate these and other important processes. These feedback mechanisms may be particularly important for type 2 inflammatory and immune responses, which we previously demonstrated are augmented by Chi3l1–IL-13Rα2 binding (which augments the survival of type 2 inflammatory cells) and decreased by IL-13 binding to the decoy IL-13Rα2 receptor (5, 19). Specifically, our studies demonstrate that, in settings where Chi3l1 predominates, N-glycosylation is decreased, which increases Chi3l1–IL-13Rα2 binding and signaling, which further decreases STT3A and -B and N-glycosylation. This would augment type 2 inflammatory and fibrotic responses by augmenting Chi3l1–IL-13Rα2 responses and decreasing IL-13 binding to IL-13Rα2, which would otherwise act as an IL-13 decoy receptor. In contrast, in settings where IL-13 predominates N-glycosylation is increased, this would decrease type 2 responses by decreasing Chi3l1–IL-13Rα2 binding and increasing IL-13 binding to the IL-13Rα2 decoy receptor. In combination, these findings highlight the importance of IL-13Rα2 N-glycosylation in determining IL-13Rα2–ligand binding and its subsequent importance in the abnormal inflammatory and tissue responses noted in the various lung diseases that are associated with the dysregulation of Chi3l1 and/or IL-13. The envisioned role(s) of N-glycosylation is summarized in Figure 7.
Figure 7.
Schematic illustration of the role(s) of N-glycosylation in IL-13 and Chi3l1 binding to IL-13Rα2. IL-13Rα2 may be N-glycosylated (N-IL-13Rα2) or may not be (IL-13Rα2). Chi3l1 binding to IL-13Rα2 that is not N-glycosylated is augmented and, in conjunction with the β subunit of the chitosome transmembrane protein 219 (TMEM219), activates mitogen-activated protein kinase (MAPK), Akt, and Wnt-β-catenin signaling. This inhibits OST while inducing DAD1 and inhibiting ICAD. The inhibition of the STT3A and -B subunits of OST decreases N-glycosylation, which further increases Chi31–IL-13Rα2 binding, whereas the induction of DAD1 and inhibition of ICAD inhibit apoptosis. In contrast, IL-13 binds to both IL-13Rα2 and the IL-13Rα1/IL-4Rα heterodimer. The former requires N-glycosylation and diminishes IL-13 effector responses. In contrast, the latter induces IL-13 effector responses, largely via the activation of STAT6 (signal transducer and activator of transcription 6). This includes the induction of STT3A and –B, which feeds back to further augment IL-13Rα2 N-glycosylation and IL-13 binding to IL-13Rα2, and dampens IL-13 effector responses, such as those seen in T-helper cell type 2 (Th2) inflammation and tissue fibrosis.
The OST complexes described previously here also share accessory subunits, such as ribophorins, DAD1, and OST 48 (28). DAD1 was identified as a mammalian cell death suppressor that may act downstream of Bcl-2 (31). Interestingly, the protein sequence of DAD1 is 40% identical to yeast Ost2 protein, the 16-kD subunit of yeast OST (31). In addition, DAD1 is an integral subunit of OST, and loss of DAD1 causes a defect in N-glycosylation (32). Recent studies from our laboratory demonstrated that Chi3l1 is a potent inhibitor of apoptosis and pyroptosis (4, 5), and the present studies demonstrate that Chi3l1 is a potent stimulator of DAD1, whereas IL-13 does not have the same effect. This allows for the interesting speculation that the anti–cell death effects of Chi3l1 are mediated, at least in part, by DAD1, and support the concept that N-linked glycosylation is an essential event in eukaryotes (31).
Our studies highlight the importance of N-glycosylation in Chi3l1 and IL-13 binding to IL-13Rα2 using two cell lines, primary murine macrophages, and multiple different approaches that alter N-glycosylation. The cell lines included THP-1 cells, which are human in origin, and MLE12 cells, which are murine in origin. We also used primary macrophages from WT and genetically modified mice. In all comparisons (human vs. murine and primary vs. transformed), these studies highlighted the important ability of interventions that decrease N-glycosylation to increase Chi3l1–IL-13Rα2 binding and effector responses, and the importance of N-glycosylation to allow IL-13–IL-13Rα2 binding. These studies suggest that N-glycosylation may also be a regulator of IL-13Rα2-ligand binding in vivo and in clinical conditions. It is important to point out, however, that this hypothesis is difficult to assess, because the tools and reagents that are required for these complex glycobiology experiments have not been generated. Specifically, reagents that can detect and differentiate N-glycosylated versus nonglycosylated IL-13Rα2 in vivo have not been described. In addition, murine knockin mice that contain versions of IL-13Rα2 that cannot be N-glycosylated have not been produced. Efforts are underway to generate these needed reagents and genetically modified mice. Hopefully, they will hopefully allow these important issues to be appropriately addressed.
In summary, these studies demonstrate that the levels of N-glycosylation play a major role in determining if Chi3l1 or IL-13 binds to IL-13Rα2, with Chi3l1 binding and signaling being increased and IL-13 binding being decreased when N-glycosylation is inhibited. They also demonstrate that Chi3l1 and IL-13 modulate N-glycosylation in a manner that augments their respective binding with Chi3l1 inhibiting, and IL-13 stimulating, the major catalytic subunits of OST (STT3A and STT3B). Lastly, they also provide an interesting link between N-glycosylation of IL-13Rα2 and the antiapoptotic effects of Chi3l1 by demonstrating that Chi3l1 augments, whereas IL-13 does not alter, the expression of the OST accessory protein and apoptosis inhibitor, DAD1. These findings allow for interesting speculations regarding the mechanisms by which Chi3l1, IL-13, and IL-13Rα2 regulate inflammatory and cell death responses. Additional investigation of the importance and roles of Chi3l1, IL-13, IL-13Rα2, and epigenetic N-glycosylation in health and disease is warranted.
Supplementary Material
Footnotes
Supported by U.S. National Institutes of Health, National Heart, Lung, and Blood Institute grants U01 HL108638 (J.A.E.), PO1 HL114501 (J.A.E. and A.M.K.C.), and R01 HL115813 (C.G.L.), and Department of Defense grant USAMRMC W81XWH-17-1-0196 (J.A.E.).
Author Contributions: C.H.H. and J.A.E. conceived project and developed working hypothesis. C.H.H., C.G.L., and J.A.E. designed experiments. C.H.H., B.M., and S.K. performed the experiments and analyzed the data. B.M. and S.K. provided technical assistance. C.G.L. and J.A.E. prepared the manuscript with input from all other authors. A.M.K.C. and J.A.E. supervised all steps of the project and reviewed the manuscript.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2019-0446OC on May 13, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Lee CG, Da Silva CA, Dela Cruz CS, Ahangari F, Ma B, Kang MJ, et al. Role of chitin and chitinase/chitinase-like proteins in inflammation, tissue remodeling, and injury. Annu Rev Physiol. 2011;73:479–501. doi: 10.1146/annurev-physiol-012110-142250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee CG. Chitin, chitinases and chitinase-like proteins in allergic inflammation and tissue remodeling. Yonsei Med J. 2009;50:22–30. doi: 10.3349/ymj.2009.50.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Funkhouser JD, Aronson NN., Jr Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol Biol. 2007;7:96. doi: 10.1186/1471-2148-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dela Cruz CS, Liu W, He CH, Jacoby A, Gornitsky A, Ma B, et al. Chitinase 3–like-1 promotes Streptococcus pneumoniae killing and augments host tolerance to lung antibacterial responses. Cell Host Microbe. 2012;12:34–46. doi: 10.1016/j.chom.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CG, Hartl D, Lee GR, Koller B, Matsuura H, Da Silva CA, et al. Role of breast regression protein 39 (BRP-39)/chitinase 3–like-1 in Th2 and IL-13–induced tissue responses and apoptosis. J Exp Med. 2009;206:1149–1166. doi: 10.1084/jem.20081271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sohn MH, Kang MJ, Matsuura H, Bhandari V, Chen NY, Lee CG, et al. The chitinase-like proteins breast regression protein-39 and YKL-40 regulate hyperoxia-induced acute lung injury. Am J Respir Crit Care Med. 2010;182:918–928. doi: 10.1164/rccm.200912-1793OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanneganti M, Mino-Kenudson M, Mizoguchi E. Animal models of colitis-associated carcinogenesis. J Biomed Biotechnol. 2011;2011:342637. doi: 10.1155/2011/342637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He CH, Lee CG, Dela Cruz CS, Lee CM, Zhou Y, Ahangari F, et al. Chitinase 3–like 1 regulates cellular and tissue responses via IL-13 receptor α2. Cell Rep. 2013;4:830–841. doi: 10.1016/j.celrep.2013.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou Y, He CH, Herzog EL, Peng X, Lee C-M, Nguyen TH, et al. Chitinase 3–like-1 and its receptors in Hermansky-Pudlak syndrome-associated lung disease. J Clin Invest. 2015;125:3178–3192. doi: 10.1172/JCI79792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffman FD. Chitinase 3–like-1 (CHI3L1): a putative disease marker at the interface of proteomics and glycomics. Crit Rev Clin Lab Sci. 2008;45:531–562. doi: 10.1080/10408360802334743. [DOI] [PubMed] [Google Scholar]
- 11.Ma B, Herzog EL, Lee CG, Peng X, Lee CM, Chen X, et al. Role of chitinase 3–like-1 and semaphorin 7a in pulmonary melanoma metastasis. Cancer Res. 2015;75:487–496. doi: 10.1158/0008-5472.CAN-13-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma B, Herzog EL, Moore M, Lee CM, Na SH, Lee CG, et al. RIG-like helicase regulation of chitinase 3–like 1 axis and pulmonary metastasis. Sci Rep. 2016;6:26299. doi: 10.1038/srep26299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Peng H, Sun H, Peng X, Tang C, Gan Y, et al. Chitinase 3–like 1 suppresses injury and promotes fibroproliferative responses in mammalian lung fibrosis. Sci Transl Med. 2014;6:240ra76. doi: 10.1126/scitranslmed.3007096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahangari F, Sood A, Ma B, Takyar S, Schuyler M, Qualls C, et al. Chitinase 3–like-1 regulates both visceral fat accumulation and asthma-like Th2 inflammation. Am J Respir Crit Care Med. 2015;191:746–757. doi: 10.1164/rccm.201405-0796OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ober C, Tan Z, Sun Y, Possick JD, Pan L, Nicolae R, et al. Effect of variation in CHI3L1 on serum YKL-40 level, risk of asthma, and lung function. N Engl J Med. 2008;358:1682–1691. doi: 10.1056/NEJMoa0708801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, He S, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–2027. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 17.Mohorko E, Glockshuber R, Aebi M. Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J Inherit Metab Dis. 2011;34:869–878. doi: 10.1007/s10545-011-9337-1. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21:576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Lee CM, He CH, Nour AM, Zhou Y, Ma B, Park JW, et al. IL-13Rα2 uses TMEM219 in chitinase 3–like-1-induced signalling and effector responses. Nat Commun. 2016;7:12752. doi: 10.1038/ncomms12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kioi M, Seetharam S, Puri RK. N-linked glycosylation of IL-13R alpha2 is essential for optimal IL-13 inhibitory activity. FASEB J. 2006;20:2378–2380. doi: 10.1096/fj.06-5995fje. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, et al. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood N, Whitters MJ, Jacobson BA, Witek J, Sypek JP, Kasaian M, et al. Enhanced interleukin (IL)-13 responses in mice lacking IL-13 receptor alpha 2. J Exp Med. 2003;197:703–709. doi: 10.1084/jem.20020906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Wang X, Bove KE, Xu M. DNA fragmentation factor 45–deficient cells are more resistant to apoptosis and exhibit different dying morphology than wild-type control cells. J Biol Chem. 1999;274:37450–37454. doi: 10.1074/jbc.274.52.37450. [DOI] [PubMed] [Google Scholar]
- 25.Larsen BD, Rampalli S, Burns LE, Brunette S, Dilworth FJ, Megeney LA. Caspase 3/caspase-activated DNase promote cell differentiation by inducing DNA strand breaks. Proc Natl Acad Sci USA. 2010;107:4230–4235. doi: 10.1073/pnas.0913089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guha P, Kaptan E, Gade P, Kalvakolanu DV, Ahmed H. Tunicamycin induced endoplasmic reticulum stress promotes apoptosis of prostate cancer cells by activating mTORC1. Oncotarget. 2017;8:68191–68207. doi: 10.18632/oncotarget.19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshino H, Kumai Y, Kashiwakura I. Effects of endoplasmic reticulum stress on apoptosis induction in radioresistant macrophages. Mol Med Rep. 2017;15:2867–2872. doi: 10.3892/mmr.2017.6298. [DOI] [PubMed] [Google Scholar]
- 28.Rinis N, Golden JE, Marceau CD, Carette JE, Van Zandt MC, Gilmore R, et al. Editing N-glycan site occupancy with small-molecule oligosaccharyltransferase inhibitors. Cell Chem Biol. 2018;25:1231–1241.e4. doi: 10.1016/j.chembiol.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aebi M. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 2013;1833:2430–2437. doi: 10.1016/j.bbamcr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: recognition and processing in the ER. Trends Biochem Sci. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Kelleher DJ, Gilmore R. DAD1, the defender against apoptotic cell death, is a subunit of the mammalian oligosaccharyltransferase. Proc Natl Acad Sci USA. 1997;94:4994–4999. doi: 10.1073/pnas.94.10.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakashima T, Sekiguchi T, Kuraoka A, Fukushima K, Shibata Y, Komiyama S, et al. Molecular cloning of a human cDNA encoding a novel protein, DAD1, whose defect causes apoptotic cell death in hamster BHK21 cells. Mol Cell Biol. 1993;13:6367–6374. doi: 10.1128/mcb.13.10.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lupardus PJ, Birnbaum ME, Garcia KC. Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Ralpha2. Structure. 2010;18:332–342. doi: 10.1016/j.str.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strober W, Kitani A, Fichtner-Feigl S, Fuss IJ. The signaling function of the IL-13Rα2 receptor in the development of gastrointestinal fibrosis and cancer surveillance. Curr Mol Med. 2009;9:740–750. doi: 10.2174/156652409788970652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konstantinidis AK, Puddicombe SM, Mochizuki A, Sheth PD, Yang IA, Yoshisue H, et al. Cellular localization of interleukin 13 receptor alpha2 in human primary bronchial epithelial cells and fibroblasts. J Investig Allergol Clin Immunol. 2008;18:174–180. [PubMed] [Google Scholar]
- 36.Chiaramonte MG, Mentink-Kane M, Jacobson BA, Cheever AW, Whitters MJ, Goad ME, et al. Regulation and function of the interleukin 13 receptor alpha 2 during a T helper cell type 2–dominant immune response. J Exp Med. 2003;197:687–701. doi: 10.1084/jem.20020903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daines MO, Tabata Y, Walker BA, Chen W, Warrier MR, Basu S, et al. Level of expression of IL-13R alpha 2 impacts receptor distribution and IL-13 signaling. J Immunol. 2006;176:7495–7501. doi: 10.4049/jimmunol.176.12.7495. [DOI] [PubMed] [Google Scholar]
- 38.Fichtner-Feigl S, Fuss IJ, Young CA, Watanabe T, Geissler EK, Schlitt HJ, et al. Induction of IL-13 triggers TGF-beta1–dependent tissue fibrosis in chronic 2,4,6-trinitrobenzene sulfonic acid colitis. J Immunol. 2007;178:5859–5870. doi: 10.4049/jimmunol.178.9.5859. [DOI] [PubMed] [Google Scholar]
- 39.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med. 2006;12:99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- 40.Fichtner-Feigl S, Strober W, Geissler EK, Schlitt H-J. Cytokines mediating the induction of chronic colitis and colitis-associated fibrosis. Mucosal Immunol. 2008;1:S24–S27. doi: 10.1038/mi.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fichtner-Feigl S, Terabe M, Kitani A, Young CA, Fuss I, Geissler EK, et al. Restoration of tumor immunosurveillance via targeting of interleukin-13 receptor-alpha 2. Cancer Res. 2008;68:3467–3475. doi: 10.1158/0008-5472.CAN-07-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang JS, Allahverdian S, Singhera GK, MacRedmond RE, Dorscheid DR. IL-13Ralpha2/AP-1 complex signalling mediates airway epithelial repair without effects on remodeling pathways. Allergy Asthma Clin Immunol. 2010;6:P27–P28. [Google Scholar]
- 43.Yang JSY, Wadsworth SJ, Singhera GK, Dorscheid D. The regulation of IL-13ralpha1 and IL-13ralpha2 expression and distribution in airway epithelial repair [abstract] Am J Respir Crit Care Med. 2011;183:A2812. [Google Scholar]
- 44.Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 45.Shrimal S, Cherepanova NA, Gilmore R. Cotranslational and posttranslocational N-glycosylation of proteins in the endoplasmic reticulum. Semin Cell Dev Biol. 2015;41:71–78. doi: 10.1016/j.semcdb.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esko JD, Bertozzi C, Schnaar RL.Chemical tools for inhibiting glycosylation Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, et al.editors. Essentials of glycobiology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2015701–712. [PubMed] [Google Scholar]
- 47.Ferris SP, Kodali VK, Kaufman RJ. Glycoprotein folding and quality-control mechanisms in protein-folding diseases. Dis Model Mech. 2014;7:331–341. doi: 10.1242/dmm.014589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 49.Kim DH, Park HJ, Lim S, Koo JH, Lee HG, Choi JO, et al. Regulation of chitinase-3–like-1 in T cell elicits Th1 and cytotoxic responses to inhibit lung metastasis. Nat Commun. 2018;9:503. doi: 10.1038/s41467-017-02731-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman JP, Wang GY, Arima K, Guan SP, Waters MR, Cavenee WK, et al. Interleukin-13 receptor alpha 2 cooperates with EGFRvIII signaling to promote glioblastoma multiforme. Nat Commun. 2017;8:1913. doi: 10.1038/s41467-017-01392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.