Abstract
Objective
Coronavirus disease 2019 (COVID-19) is a fatal and fast-spreading viral infection. To date, the number of COVID-19 patients worldwide has crossed over six million with over three hundred and seventy thousand deaths (according to the data from World Health Organization; updated on 2 June 2020). Although COVID-19 can be rapidly diagnosed, efficient clinical treatment of COVID-19 remains unavailable, resulting in high fatality. Some clinical trials have identified vitamin C (VC) as a potent compound pneumonia management. In addition, glycyrrhizic acid (GA) is clinically as an anti-inflammatory medicine against pneumonia-induced inflammatory stress. We hypothesized that the combination of VC and GA is a potential option for treating COVID-19.
Methods
The aim of this study was to determine pharmacological targets and molecular mechanisms of VC + GA treatment for COVID-19, using bioinformational network pharmacology.
Results
We uncovered optimal targets, biological processes and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of VC + GA against COVID-19. Our findings suggested that combinatorial VC and GA treatment for COVID-19 was associated with elevation of immunity and suppression of inflammatory stress, including activation of the T cell receptor signaling pathway, regulation of Fc gamma R-mediated phagocytosis, ErbB signaling pathway and vascular endothelial growth factor signaling pathway. We also identified 17 core targets of VC + GA, which suggest as antimicrobial function.
Conclusions
For the first time, our study uncovered the pharmacological mechanism underlying combined VC and GA treatment for COVID-19. These results should benefit efforts to address the most pressing problem currently facing the world.
Keywords: vitamin C, glycyrrhizic acid, COVID-19, bioinformatics analysis, biotarget
Introduction
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and first reported from Wuhan, Hubei Province, China [1]. The virus spread across China before it could be restrained effectively, then subsequently infected populations worldwide, resulting in a pandemic with increasing death tolls [2]. Clinical manifestations of COVID-19 include acute pneumonia detected via computed tomography imaging or radiological examination. Systemic fever is another visible symptom [3]. SARS-CoV-2 has greater infectivity than severe acute respiratory syndrome-related coronavirus (SARS-CoV), because the former’s binding capacity to angiotensin-converting enzyme II is approximately 10–100-fold higher than the latter’s [4]. Patients infected with SARS-CoV-2 exhibit high mortality rate, especially if they are over 60 years of age [5]. Although the medical diagnosis of COVID-19 is rapid and efficient, effective treatments do not yet exist [6]. Thus, potential agents for treatment should be screened rapidly.
Vitamin C (VC) possesses antiaging, antiviral and anti-immunocompromise properties, making the compound useful for preventing various health conditions [7]. Some evidence suggests that VC supplements may enhance cellular immunological capacity for phagocyte induction, T lymphocyte activation and interferon release [8]. Moreover, clinical studies suggest that the antiviral effects of VC can eliminate virus-caused respiratory complications [9]. In addition, VC appears to be beneficial against the avian coronavirus, altering host susceptibility to viral infection [10]. Notably, VC supplements can reduce viral respiratory infections during influenza, suggesting anti-pneumonia potential [11]. Controlled clinical trials demonstrated that VC-supplemented patients have reduced pneumonia incidence, implying potential effectiveness against lower respiratory tract viral infections [12]. During the SARS epidemic, VC was recommended as preventive medication and for adjuvant therapy that significantly lowered the incidence of pneumonia [13, 14]. In China, clinical trials are being conducted with patients at Wuhan hospitals to determine whether high doses of VC can manage COVID-19 [ClinicalTrials.gov Identifier: NCT04264533]. However, VC overdose has unwanted side effects, including kidney stones and diarrhea [15]. Thus, combining VC with existing medicines or bioactive compounds may enhance the former’s therapeutic effectiveness against COVID-19 while minimizing side effects. Rhizoma Glycyrrhizae (Gancao) is common, multifunctional ingredient of traditional Chinese medicine, such as viral infection [16]. Glycyrrhizic acid (GA) is a major bioactive ingredient extracted from Rhizoma Glycyrrhizae. It has evident pharmacological properties, including detoxifying, cough-relieving, anti-inflammatory, antitumor and antibacterial capacity [17]. Furthermore, GA may inhibit propagation of the pseudorabies virus and porcine epidemic diarrhea virus, indicating potential antiviral action [18]. GA has been used in clinical treatment of inflammation-induced diseases [19]. Based on the potent pharmacological properties of both VC and GA, their combined use may effectively manage COVID-19. At this time, no empirical data supports the clinical efficacy of VC + GA treatment against COVID-19 has not been investigated.
However, we can perform bioinformatics analysis as a theoretical evaluation of potential anti-COVID-19 molecular mechanisms of VC + GA to support future clinical trials. Bioinformatics-based network pharmacology has emerged as a powerful tool to reveal active ingredients, biological targets and signaling pathways linked to certain diseases [20]. Our previous studies used network pharmacology to identify anti-disease targets and pathways of certain bioactive components [21, 22]. In this study, we aimed to establish a component-target-pathway network of VC + GA treatment against the novel coronavirus, thereby uncovering anti-COVID-19 processes and mechanisms in detail (Figure 1).
Figure 1.
A stepwise workflow showed the combined antiviral activity of VC and GA against COVID-19 through network pharmacology. We identified candidate biotargets of VC and GA, then determined and mapped their combined core biotargets against COVID-19. A PPI diagram of VC + GA against COVID-19 was generated. Our analysis revealed the pharmacological functions and molecular pathways of VC + GA action against COVID-19.
Methods
Screening potential targets of VC and GA
Potential targets were acquired after a thorough analysis of the Traditional Chinese Medicine Systems Pharmacology (TCMSP) [23], Drugbank [24], SuperPred, Swiss Target Prediction [25], ChemMapper [26], Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine (BATMAN TCM) [27] and SuperPred webserver [28] datasets. Functional targets of VC and GA in humans were selected from the Swiss-Prot and Uniprot databases [29]. Genecard [30] and Online Mendelian Inheritance in Man (OMIM) [31] databases were used to screen SARS-CoV-2 pathogenic targets. All shortlisted targets of VC, GA and SARS-CoV-2 were then subjected to intersection analysis via an online toolkit. Custom Venn diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn/) were created to show correlative targets of VC and GA against COVID-19.
Gene ontology (GO) and KEGG pathway enrichment analysis of correlative targets
R-language packages ‘ClusterProfiler,’ ‘ReactomePA,’ ‘org.Hs.eg.Db,’ and ‘GOplot’ were used for analysis and visualization of correlative targets. GO data were analyzed using ‘org.Hs.eg.Db,’ with cutoffs for enrichment being P < 0.05 and q < 0.05; outputs were bubble charts and Circos-circle charts. The ‘pathview’ package was used to create pathway diagrams for correlative targets of enriched KEGG pathways; this step allowed for further analysis of the results [32, 33].
Establishment of network relevance visualization
Cytoscape version 3.7.1 was used to construct visualizations of the component-target-pathway network, GO biological processes and KEGG pathways of VC and GA’s individual and combined effects against COVID-19 [34].
Heatmap construction of Biological Process (BP) biological process and KEGG enrichment pathway of VC and GA
We performed pairwise comparisons on bioinformational data of the top 20 biological processes and KEGG-enriched pathways involved in the separate therapeutic effects of VC and GA against COVID-19. We used -log10 (P.adjust) as the heatmap parameter and drew heatmaps in HemI 1.0 [35].
Optimal targets and construction of associated protein–protein interaction (PPI) network
We used common targets of VC and GA against SARS-CoV-19 as inputs to a STRING database (version 11.0) [36], creating target-to-target network interactions and a target interaction PPI network diagram. We then analyzed network topology parameters (e.g. median and maximum degrees of freedom) in the Tab Separated Values (TSV) data format using Network Analyzer in Cytoscape version 3.7.1; optimal targets were collected based on degree values. For filtering, the upper limit was maximum degree value in the topology data, and the lower limit was twice the median degree of freedom [37]. The degree of nodes (i.e. quantity of linking edges) indicates the probability distribution of degrees across the entire network [38, 39].
Results
Screening and identification of VC and GA targets
We screened Genecard and OMIM datasets to identify 426 COVID-19-associated genes. After data correction with Uniprot, we identified 338 VC- and 223 GA-pharmacological action genes. Venn diagrams revealed 34 VC-intersection (Figure 2A) and 28 GA-intersection (Figure 2B) targets against COVID-19.
Figure 2.
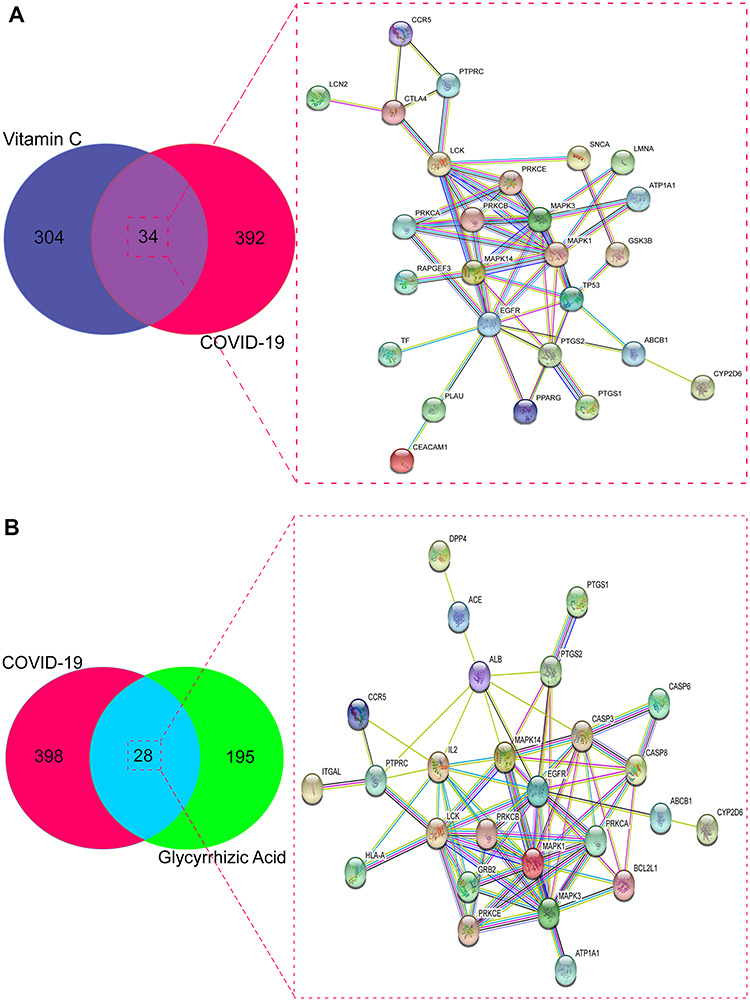
Targets of sole VC or sole GA against COVID-19. (A) Venn diagram and PPI network showed the 34 targets of VC against COVID-19. (B) Venn diagram and PPI network exhibited the 28 targets of GA against COVID-19.
GO biological process and KEGG pathway enrichment of VC and GA targets
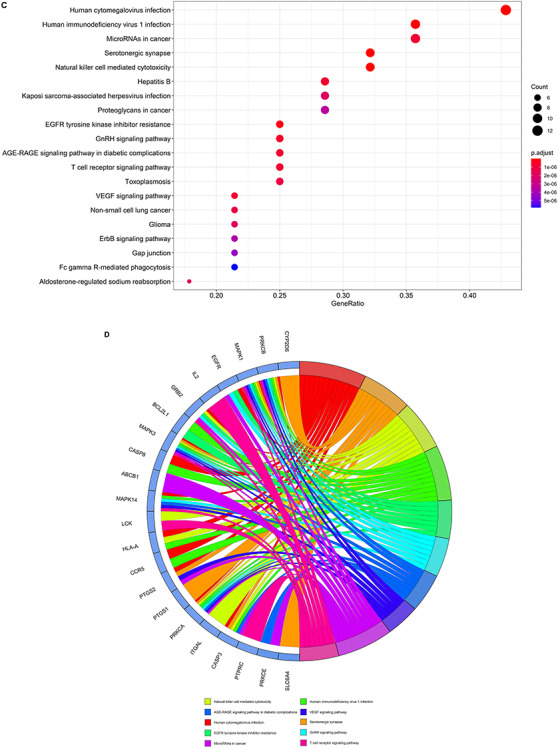
The results of GO analysis suggested that the biological processes of core VC targets against COVID-19 were mainly involved in peptidyl-serine phosphorylation, peptidyl-serine modification, regulation of cell–cell adhesion, cellular response to biotic stimulus, positive regulation of nucleocytoplasmic transport, regulation of leukocyte cell–cell adhesion, regulation of T cell activation, response to lipopolysaccharide, T cell activation, blood coagulation, leukocyte cell–cell adhesion, cellular response to external stimulus, hemostasis, coagulation, response to molecule of bacterial origin, platelet activation, fatty acid metabolic process, positive regulation of cell adhesion, regulation of nucleocytoplasmic transport and positive regulation of protein transport (Figure 3A and B and Supplementary Table 1, see Supplementary Data available online at https://academic.oup.com/bib). The 134 enriched KEGG pathways (P < 0.05) were involved in serotonergic synapse, Human cytomegalovirus infection, vascular endothelial growth factor (VEGF) signaling pathway, T cell receptor (TCR) signaling pathway, non-small cell lung cancer, microRNAs in cancer, aldosterone-regulated sodium reabsorption, sphingolipid signaling pathway, thyroid hormone signaling pathway, glioma, Kaposi sarcoma-associated herpesvirus infection, Epidermal Growth Factor Receptor (EGFR) tyrosine kinase inhibitor resistance, ErbB signaling pathway, proteoglycans in cancer, Fc gamma R-mediated phagocytosis, Gonadotropin-Releasing Hormone (GnRH) signaling pathway, interleukin (IL)-17 signaling pathway, prostate cancer, endocrine resistance and Advanced Glycation End Products-Receptor for Advanced Glycation End Products (AGE-RAGE) signaling pathway in diabetic complications (Figure 3C and D and Supplementary Table 2, see Supplementary Data available online at https://academic.oup.com/bib).
Figure 3.
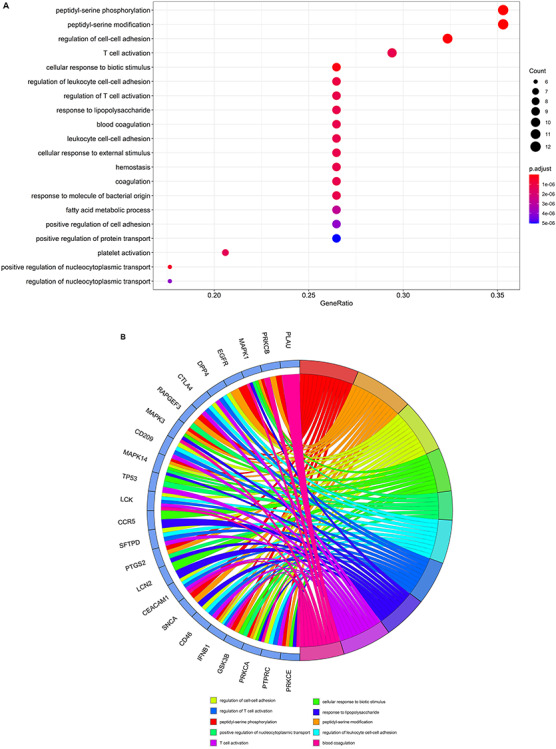
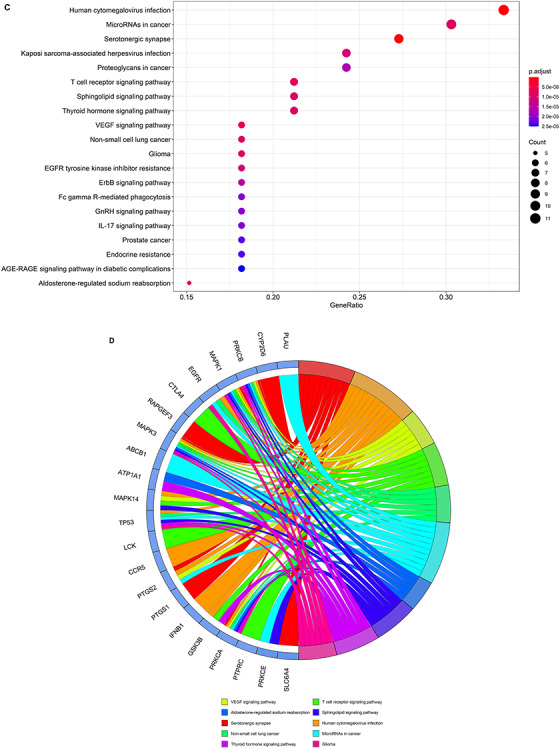
Biological processes and molecular pathways associated with core targets of VC against COVID-19. (A) Biological processes [from GO analysis] were presented by bubble diagrams with count algorithms and P-adjust values. (B) All core biotargets of VC against COVID-19 were linked to the top 10 most enriched GO terms in Circro diagrams. (C) Molecular pathways (from KEGG analysis) were presented by bubble diagrams based on count algorithms and P-adjust values. (D) Identified core biotargets of VC against COVID-19 were associated with the 10 most enriched KEGG terms in Circro diagrams.
The GO data indicated that functional processes of core GA targets against COVID-19 were associated with response to lipopolysaccharide, cellular response to abiotic stimulus, cellular response to environmental stimulus, response to molecule of bacterial origin, placenta development, peptidyl-serine phosphorylation, reproductive structure development, reproductive system development, peptidyl-serine modification, lipopolysaccharide-mediated signaling pathway, cellular response to external stimulus, platelet activation, regulation of cell–cell adhesion, cellular response to mechanical stimulus, positive regulation of establishment of protein localization, peptidyl-tyrosine autophosphorylation, T cell activation, cellular response to lipopolysaccharide, interaction with host and response to mechanical stimulus (Figure 4A and B and Supplementary Table 3, see Supplementary Data available online at https://academic.oup.com/bib). The 146 enriched KEGG pathways (P < 0.05) were mostly linked to human cytomegalovirus infection, serotonergic synapse, natural killer cell mediated cytotoxicity, human immunodeficiency virus (HIV)-1 infection, EGFR tyrosine kinase inhibitor resistance, GnRH signaling pathway, VEGF signaling pathway, AGE-RAGE signaling pathway in diabetic complications, TCR signaling pathway, Hepatitis B, microRNAs in cancer, non-small cell lung cancer, toxoplasmosis, aldosterone-regulated sodium reabsorption, Kaposi sarcoma-associated herpesvirus infection, glioma, proteoglycans in cancer, ErbB signaling pathway, gap junction, Fc gamma R-mediated phagocytosis (Figure 4C and D and Supplementary Table 4, see Supplementary Data available online at https://academic.oup.com/bib). We then used the ‘pathview’ package in R to generate network diagrams that highlighted core targets of enriched KEGG pathways for VC against COVID-19 (Supplementary Figure 1, see Supplementary Data available online at https://academic.oup.com/bib) and GA against COVID-19 (Supplementary Figure 2, see Supplementary Data available online at https://academic.oup.com/bib).
Figure 4.
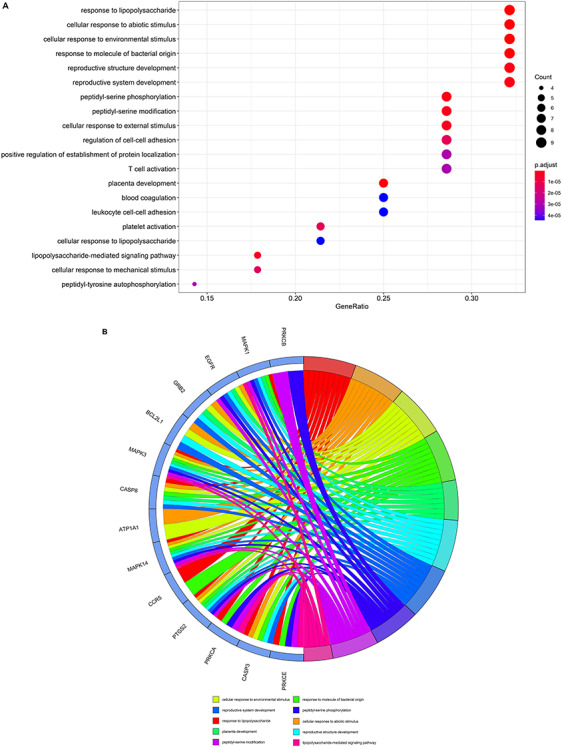
Biological processes and molecular pathways associated with core targets of GA against COVID-19. (A) Biological processes were presented by bubble diagrams generated through count algorithms and P-adjust calculation. (B) Core biotargets of GA against COVID-19 were related to the top 10 enriched GO terms in Circro diagrams. (C) Molecular pathways (from KEGG analysis) were presented by bubble diagrams based on count algorithms and P-adjust values. (D) Core biotargets of GA against COVID-19 were linked to the top 10 enriched KEGG terms in Circro diagrams.
Bioinformatics analysis of potential VC + GA targets against COVID-19
We identified 17 intersecting targets of VC and GA against COVID-19: PRKCE, SLC6A4, CYP2D6, PRKCB, MAPK1, EGFR, DPP4, MAPK3, ABCB1, ATP1A1, MAPK14, LCK, CCR5, PTGS2, PTGS1, PRKCA and PTPRC (Figure 5). The top 10 GO terms of VC + GA against COVID-19 were cellular response to external stimulus (0071496), peptidyl-serine modification (0018209), peptidyl-serine phosphorylation (0018105), platelet activation (0030168), regulation of cell–cell adhesion (0022407), response to lipopolysaccharide (0032496), response to molecule of bacterial origin (0002237), T cell activation (0042110), blood coagulation (0007596) and leukocyte cell–cell adhesion (0007159) (Table 1). The top 15 KEGG signaling pathways of VC + GA against COVID-19 were AGE-RAGE signaling pathway in diabetic complications (hsa04933), Aldosterone-regulated sodium reabsorption (hsa04960), EGFR tyrosine kinase inhibitor resistance (hsa04960), ErbB signaling pathway (hsa04012), Fc gamma R-mediated phagocytosis (hsa04666), Glioma (hsa05214), GnRH signaling pathway (hsa04912), Human cytomegalovirus infection (hsa04912), Kaposi sarcoma-associated herpesvirus infection (hsa04912), MicroRNAs in cancer (hsa05206), Non-small cell lung cancer (hsa05206), Proteoglycans in cancer (hsa05205), Serotonergic synapse (hsa04726), TCR signaling pathway (hsa04660) and VEGF signaling pathway (hsa04370) (Table 2).
Figure 5.
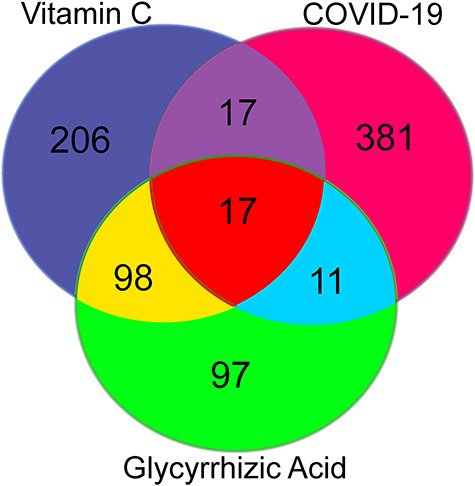
Targets of combined VC and GA against COVID-19. Venn diagram highlighted the intersecting targets of VC in combination with GA as a drug for COVID-19. Using online databases, we identified 19 shared biotargets of VC + GA against COVID-19.
Table 1.
Top 10 biological processes of VC in combination with GA against COVID-19
| ID | Description | Gene symbol | Number of gene |
|---|---|---|---|
| GO:0018105 | Peptidyl-serine phosphorylation | PRKCE, PRKCB, MAPK1, EGFR, MAPK3, MAPK14, PTGS2 | 7 |
| GO:0018209 | Peptidyl-serine modification | PRKCE, PRKCB, MAPK1, EGFR, MAPK3, MAPK14, PTGS2 | 7 |
| GO:0022407 | Regulation of cell–cell adhesion | DPP4, MAPK14, LCK | 3 |
| GO:0032496 | Response to lipopolysaccharide | PRKCE, MAPK1, MAPK3, MAPK14, CCR5, PTGS2, PRKCA | 7 |
| GO:0042110 | T cell activation | DPP4, LCK | 2 |
| GO:0007596 | Blood coagulation | PRKCE, PRKCB, MAPK1, MAPK3, LCK, PRKCA | 6 |
| GO:0007159 | Leukocyte cell–cell adhesion | DPP4, LCK, PTPRC | 3 |
| GO:0071496 | Cellular response to external stimulus | MAPK1, EGFR, MAPK3, ATP1A1, PTGS2, PTPRC | 6 |
| GO:0002237 | Response to molecule of bacterial origin | PRKCE, MAPK1, MAPK3, MAPK14, CCR5, PTGS2, PRKCA | 7 |
| GO:0030168 | Platelet activation | PRKCE, PRKCB, MAPK1, MAPK3, LCK, PRKCA | 6 |
Table 2.
KEGG pathway of VC in combination with GA against COVID-19
| ID | Description | Gene symbol | Number of gene |
|---|---|---|---|
| hsa05163 | Human cytomegalovirus infection | PRKCB, MAPK1, EGFR, MAPK3, MAPK14, CCR5, PTGS2, PRKCA | 8 |
| hsa04726 | Serotonergic synapse | SLC6A4, CYP2D6, PRKCB, MAPK1, MAPK3, PTGS2, PTGS1, PRKCA | 8 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | MAPK1, EGFR, MAPK3 | 3 |
| hsa04912 | GnRH signaling pathway | PRKCB, MAPK1, EGFR, MAPK3, MAPK14, PRKCA | 6 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | PRKCE, PRKCB, MAPK1, MAPK3, MAPK14, PRKCA | 6 |
| hsa04370 | VEGF signaling pathway | PRKCB, MAPK1, MAPK3, MAPK14, PTGS2, PRKCA | 6 |
| hsa05206 | MicroRNAs in cancer | PRKCE, PRKCB, MAPK1, EGFR, MAPK3, ABCB1, PTGS2, PRKCA | 8 |
| hsa04660 | TCR signaling pathway | MAPK1, MAPK3, MAPK14, LCK, PTPRC | 5 |
| hsa05223 | Non-small cell lung cancer | PRKCB, MAPK1, EGFR, MAPK3, PRKCA | 5 |
| hsa04960 | Aldosterone-regulated sodium reabsorption | PRKCB, MAPK1, MAPK3, ATP1A1, PRKCA | 5 |
| hsa05167 | Kaposi sarcoma-associated herpesvirus infection | MAPK1, MAPK3, MAPK14, CCR5, PTGS2 | 5 |
| hsa05214 | Glioma | PRKCB, MAPK1, EGFR, MAPK3, PRKCA | 5 |
| hsa05205 | Proteoglycans in cancer | PRKCB, MAPK1, EGFR, MAPK3, MAPK14, PRKCA | 6 |
| hsa04012 | ErbB signaling pathway | PRKCB, MAPK1, EGFR, MAPK3, PRKCA | 5 |
| hsa04666 | Fc gamma R-mediated phagocytosis | PRKCE, PRKCB, MAPK1, MAPK3, PRKCA, PTPRC | 6 |
Heatmaps of VC + GA against COVID-19
Respectively, Figure 6A and B shows the heatmaps depicting GO biological processes and KEGG signaling pathways of VC + GA against COVID-19, respectively. Detailed information regarding the raw data is listed in Supplementary Tables 5 and 6, see Supplementary Data available online at https://academic.oup.com/bib.
Figure 6.
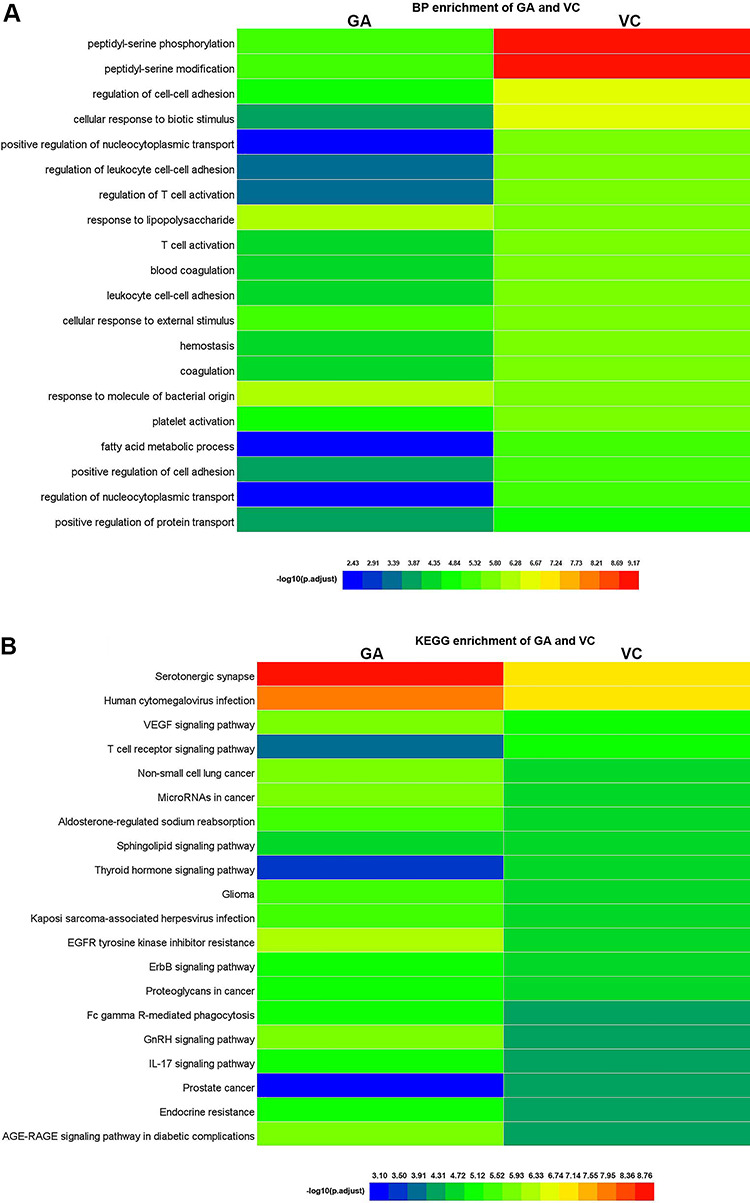
Biological processes and molecular pathways associated with core targets of combined VC + GA against COVID-19. (A) Heatmap showed GO biological processes of VC + GA against COVID-19. Optimal prioritization was determined via the -log10 (P.adjust) algorithm for visualization. (B) Heatmap uncovered KEGG signaling pathways of VC + GA against COVID-19. Different colors represented optimal prioritization of molecular pathways determined using the -log10 (P.adjust) algorithm.
Network diagrams of VC + GA against COVID-19
We created a network visualization of VC + GA-target-GO-KEGG-COVID-19 using the top 20 enriched GO terms and KEGG pathways (Figure 7). Our result showed that VC and GA shared 10 biological processes and 15 KEGG pathways, representing their combined anti-COVID-19 targets.
Figure 7.
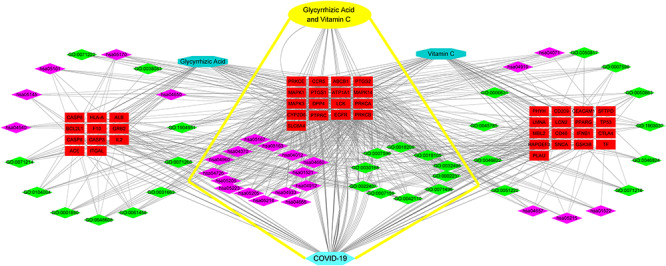
Interaction network of VC + GA target KEGG pathways against COVID-19. Detailed information on core biotargets, pharmacological functions and signaling pathways were presented.
Discussion
As the COVID-19 pandemic has spread worldwide, effective management and treatment strategies targeting SARS-CoV-2 are in urgent demand [40]. Although VC and GA individually have potent pharmacological efficacy against viral pneumonia [41, 42], no study has investigated their combined effects or potential mechanisms against COVID-19. Here, we used a potent strategy of network pharmacology and bioinformatics (including GO and KEGG) to uncover the integrative pharmacological mechanism of VC + GA against COVID-19. Our findings suggest that VC + GA may be able to suppress COVID-19 through their combined antioxidative, antiviral and anti-inflammatory effects, along with immune system activation. Specifically, notably enriched GO biological processes included T cell activation and leukocyte adhesion. Moreover, according to our KEGG pathway analysis, the pharmacological mechanisms of VC + GA against COVID-19 involved specific modulations of immune responses, such as Fc gamma R-mediated phagocytosis, TCR signaling pathway, VEGF signaling pathway and human cytomegalovirus infection.
Fc gamma R-mediated phagocytosis is an essential part of the innate immune response, when neutrophils and macrophages engulf IgG-coated particles [43]. TCR signaling has a key role in determining T cell fate [44]. Also, TCR is important to the differentiation, maintenance and function of regulatory T cells [45]. When VC + GA activates Fc gamma R-mediated phagocytosis and TCR, these processes may facilitate the host to initiate an immune response against COVID-19. VEGF signaling pathway is a major pathway that stimulates blood vessel formation [46]. Although most VEGF studies focused on cancer development, a mouse study demonstrated that exposure to recombinant VEGF dramatically reduced CD4+/CD8+ thymocytes [47]. Additionally, a glioblastoma rodent model showed that VEGF modulates the innate immune response through suppressing immunologic and pro-angiogenic functions of macrophages, suggesting VEGF is also essential for the adaptive immune system [48].
Our molecular functional analysis revealed that key effectors regulate the anti-COVID-19 properties just described. Effectors include protein kinase C (PKC) family (PRKCA, PRKCB and PRKCE), mitogen-activated protein kinase family (MAPK1, MAPK3 and MAPK14) and lymphocyte-specific protein tyrosine kinase (LCK). PKC alpha, PRKCB and PRKCE are central to immune biology [49, 50]. The first has an important function in different immune cell types. For instance, a PKCα-deficient mouse study showed that pkrc−/− cells were defective in IL-2 suppression because l17a promoter activity was reduced; this finding suggests that PRKCA participates in Th17 cell immune responses [51]. In addition, PRKCA is required for Fc receptors, which acts during IgA (FcalphaR) trafficking to major histocompatibility complex (MHC) class II compartments and FcalphaR-mediated antigen presentation [52]. Moreover, PRKCA promotes M1 macrophage polarization in colon cancer cells through the Mitogen-Activated Protein Kinase (MAPK) pathway [53]. Next, PRKCB is associated with phagocytosis function [54]. A mouse study demonstrated that PRKCB enhancement improves macrophage function in lupus patients with lipopolysaccharide (LPS) tolerance from chronic infection [55]. Also, PRKCB is a negative regulator of retinoic acid-inducible gene I antiviral signal transduction. This process is a key sensor of viral RNA in the cytosol and is therefore critical to establishing an interferon-mediated antiviral state [56]. Furthermore, PRKCB is integral to B cell survival and antigenic response, following B cell antigen receptor engagement of naïve B cells [57]. Finally, PRKCE is also necessary for innate immune responses [58]. A PKCɛ-knockout mouse study showed that prkce−/− macrophages produced fewer inflammatory cytokines, suggesting that the protein kinase is important for macrophage activation and defense against bacterial infection [59].
Our data implicated MAPK signaling as part of the mechanisms underlying VC + GA antiviral function. MAPK cascades are crucial signaling pathways in the regulation of host immune response to infection [60]. Specifically, MAPK signaling induces pro-inflammatory mediators and activation of anti-inflammatory pathways through controlling many downstream effectors [61]. For example, activator protein-1 is a downstream target of the MAPK signaling cascade and a critical regulator of nuclear gene expression during T cell activation [62]. In addition, MAPK pathway inhibition negatively affects T cell activity in breast cancer, suggesting that MAPK activity is required for T cell function [63].
The final protein kinase that appears to explain the action of VC + GA treatment against COVID-19 is LCK, a tyrosine kinase that contributes to intracellular signaling pathways of lymphocytes. These include transduction of TCR-mediated activation and cytokine production in T cells [64, 65]. Decreases in LCK activity facilitate viral infections such as HIV-1 in T cells [66].
It has been reported that COVID-19 patients with severe and fatal disease had significantly increase in white blood cell number and decreased lymphocyte and platelet counts [67, 68]. Our findings showed that the VC + GA treatment should exert potent antiviral and anti-inflammatory effects through the activation of immune system, suggesting the possible use of VC + GA as optional treatment of COVID-19. These attributes are consistent with other forms of combined modality therapy against clinical disease, which acts via regulating multiple targets and pathways. In upcoming clinical treatments for COVID-19, VC + GA supplementation may potentiate the therapeutic effectiveness of existing medicines such as Remdesivir. However, more studies are needed to confirm the action of VC + GA on COVID-19, as we currently do not know how the supplements target and influence the virus. Previous reports suggest that VC affects many cellular receptors, such as peroxisome proliferator-activated receptor α, fibroblast growth factor receptor 2 and angiotensin receptors [69–71]. Furthermore, considerable evidence exists to show that VC and GA can separately control some signaling pathways, including the three (MAPK, VEGF and PKC signaling), which we identified here using bioinformatics [72–76]. We thus find VC + GA, a promising supplementary treatment against COVID-19, which is worth further investigation.
Conclusion
In conclusion, network pharmacology revealed that anti-inflammation and immunity activation were primary target pathways of the VC + GA treatment against COVID-19. Hopefully, this combination can be applied to the development of effective SARS-CoV-2 treatments, based on the identified functional processes and pharmacological mechanisms.
Key Points
Bioinformatics analysis using network pharmacology is a valuable strategy for revealing candidate genes and molecular pathways of bioactive compounds effective against complex diseases.
Candidate targets and underlying mechanisms of the combined vitamin C (VC) and glycyrrhizic acid (GA) treatment against coronavirus disease 2019 (COVID-19) were identified and visualized.
Bioinformatics findings indicate that combined VC and GA can act as a potential therapeutic option for COVID-19 treatment.
Supplementary Material
Acknowledgement
K.P.L. is supported by Hong Kong SAR, Macao SAR and Taiwan Province Talent Young Scientist Program of Guangxi.
Rong Li is a researcher at Guilin Medical University. He plotted the figures based on network pharmacology, using online databases.
Ka Wu is a clinical pharmacist of The Second People’s Hospital of Nanning City and The Third Affiliated Hospital of Guangxi Medical University. He conducted data analysis and performed literature searches.
Yu Li is a master’s student of Guilin Medical University who contributed to data collection and figure plotting.
Xiao Liang is a master’s student of Guilin Medical University who worked on analyzing data and literature searches.
Professor Keng Po Lai is a researcher at Guilin Medical University. His research is focused on revealing cell signaling networks underlying complex diseases, through deep sequencing of transcriptomes, small RNA transcriptomes and methylomes, which is then integrated with data from antiviral bioactive agents.
Professor Jian Chen is a researcher at Guilin Medical University. His main interests lie in creating and analyzing network data and figures constructed using the framework of network pharmacology. His work contributes to improving our understanding of how bioactive compounds can treat complex diseases, including COVID-19.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Funding
This work was supported by the National Natural Science Foundation of China (81660610 and 81560134) and the National Natural Science Foundation of Guangxi Province (2019GXNSFBA185015, 2019GXNSFFA245001, 2017GXNSFDA198019 and 2018GXNSFAA281159).
References
- [1]. Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun 2020;109:102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Heymann DL, Shindo N; WHO Scientific and Technical Advisory Group for Infectious Hazards. COVID-19: what is next for public health? Lancet 2020;395:542–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. AJR Am J Roentgenol 2020;1–7. [DOI] [PubMed] [Google Scholar]
- [4]. Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020;367:1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. Livingston E, Bucher K. Coronavirus disease 2019 (COVID-19) in Italy. JAMA 2020;323:1335. [DOI] [PubMed] [Google Scholar]
- [6]. Freund Y. The challenge of emergency medicine facing the COVID-19 outbreak. Eur J Emerg Med 2020;27:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Righi NC, Schuch FB, De Nardi AT, et al. Effects of vitamin C on oxidative stress, inflammation, muscle soreness, and strength following acute exercise: meta-analyses of randomized clinical trials. Eur J Nutr 2020. [DOI] [PubMed] [Google Scholar]
- [8]. Carr AC, Maggini S. Vitamin C and immune function. Nutrients 2017;9:1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Vorilhon P, Arpajou B, Vaillant Roussel H, et al. Efficacy of vitamin C for the prevention and treatment of upper respiratory tract infection. A meta-analysis in children. Eur J Clin Pharmacol 2019;75:303–11. [DOI] [PubMed] [Google Scholar]
- [10]. Chand N, Naz S, Khan A, et al. Performance traits and immune response of broiler chicks treated with zinc and ascorbic acid supplementation during cyclic heat stress. Int J Biometeorol 2014;58:2153–7. [DOI] [PubMed] [Google Scholar]
- [11]. Kim Y, Kim H, Bae S, et al. Vitamin C is an essential factor on the anti-viral immune responses through the production of interferon-α/β at the initial stage of influenza a virus (H3N2) infection. Immune Netw 2013;13:70–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Hemilä H, Louhiala P. Vitamin C for preventing and treating pneumonia. Cochrane Database Syst Rev 2013;(8). doi: 10.1002/14651858.CD005532.pub3. [DOI] [PubMed] [Google Scholar]
- [13]. Hemilä H. Vitamin C and SARS coronavirus. J Antimicrob Chemother 2003;52:1049–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Hemilä H. Vitamin C intake and susceptibility to pneumonia. Pediatr Infect Dis J 1997;16:836–7. [DOI] [PubMed] [Google Scholar]
- [15]. Marosz A, Chlubek D. The risk of abuse of vitamin supplements. Ann Acad Med Stetin 2014;60:60–4. [PubMed] [Google Scholar]
- [16]. Li N, Zhou T, Wu F, et al. Pharmacokinetic mechanisms underlying the detoxification effect of Glycyrrhizae Radix et Rhizoma (Gancao): drug metabolizing enzymes, transporters, and beyond. Expert Opin Drug Metab Toxicol 2019;15:167–77. [DOI] [PubMed] [Google Scholar]
- [17]. Chen K, Yang R, Shen F, et al. Advances in pharmacological activities and mechanisms of glycyrrhizic acid. Curr Med Chem 2019. [DOI] [PubMed] [Google Scholar]
- [18]. Sun ZG, Zhao TT, Lu N, et al. Research progress of glycyrrhizic acid on antiviral activity. Mini Rev Med Chem 2019;19:826–32. [DOI] [PubMed] [Google Scholar]
- [19]. Cao ZY, Liu YZ, Li JM, et al. Glycyrrhizic acid as an adjunctive treatment for depression through anti-inflammation: a randomized placebo-controlled clinical trial. J Affect Disord 2020;265:247–54. [DOI] [PubMed] [Google Scholar]
- [20]. Su M, Guo C, Liu M, et al. Therapeutic targets of vitamin C on liver injury and associated biological mechanisms: a study of network pharmacology. Int Immunopharmacol 2019;66:383–7. [DOI] [PubMed] [Google Scholar]
- [21]. Zhou R, Wu K, Su M, et al. Bioinformatic and experimental data decipher the pharmacological targets and mechanisms of plumbagin against hepatocellular carcinoma. Environ Toxicol Pharmacol 2019;70:103200. [DOI] [PubMed] [Google Scholar]
- [22]. Wu K, Wei P, Liu M, et al. To reveal pharmacological targets and molecular mechanisms of curcumol against interstitial cystitis. J Adv Res 2019;20:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Chem 2014;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46:1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47:357–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26]. Gong J, Cai C, Liu X, et al. ChemMapper: a versatile web server for exploring pharmacology and chemical structure association based on molecular 3D similarity method. Bioinformatics 2013;29:1827–9. [DOI] [PubMed] [Google Scholar]
- [27]. Liu Z, Guo F, Wang Y, et al. BATMAN-TCM: a Bioinformatics Analysis Tool for Molecular mechANism of Traditional Chinese Medicine. Sci Rep 2016;6:21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Nickel J, Gohlke BO, Erehman J, et al. SuperPred: update on drug classification and target prediction. Nucleic Acids Res 2014;42:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res 2018;46:2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Fishilevich S, Nudel R, Rappaport N, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017:bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Amberger JS, Bocchini CA, Schiettecatte F, et al. Online Mendelian Inheritance in Man (OMIM), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43:789–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32]. Li R, Ma X, Song Y, et al. Anti-colorectal cancer targets of resveratrol and biological molecular mechanism: analyses of network pharmacology, human and experimental data. J Cell Biochem 2019;120:11265–73. [DOI] [PubMed] [Google Scholar]
- [33]. Zhou R, Wu K, Su M, et al. Bioinformatic and experimental data decipher the pharmacological targets and mechanisms of plumbagin against hepatocellular carcinoma. Environ Toxicol Pharmacol 2019;70:103200. [DOI] [PubMed] [Google Scholar]
- [34]. Wu K, Wei P, Liu M, et al. To reveal pharmacological targets and molecular mechanisms of curcumol against interstitial cystitis. J Adv Res 2019;20:43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Li R, Song Y, Ji Z, et al. Pharmacological biotargets and the molecular mechanisms of oxyresveratrol treating colorectalcancer: network and experimental analyses. Biofactors 2020;46:158–67. [DOI] [PubMed] [Google Scholar]
- [36]. Deng W, Wang Y, Liu Z, et al. HemI: a toolkit for illustrating heatmaps. PLoS One 2014;9:e111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Ge B, Guo C, Liang Y, et al. Network analysis, and human and animal studies disclose the anticystitis glandularis effects of vitamin C. Biofactors 2019;45:912–9. [DOI] [PubMed] [Google Scholar]
- [39]. Li J, Guo C, Lu X, et al. Anti-colorectal cancer biotargets and biological mechanisms of puerarin: study of molecular networks. Eur J Pharmacol 2019;858:172483. [DOI] [PubMed] [Google Scholar]
- [40]. Watkins J. Preventing a covid-19 pandemic. BMJ 2020;368:810. [DOI] [PubMed] [Google Scholar]
- [41]. Hemilä H, Vitamin C, Infections . Nutrients. 2017;9: pii: E339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42]. Smirnov VS, Garshinina AV, Shtro AA, et al. Anti-viral activity of a complex of the glycyrrhizic acid-alpha-glutamyltryptophan against the experimental lethal influenza infection in white mice caused by the oseltamivir-resistant strain of the virus. Vopr Virusol 2014;59:31–8. [PubMed] [Google Scholar]
- [43]. Botelho RJ, Tapper H, Furuya W, et al. Fc gamma R-mediated phagocytosis stimulates localized pinocytosis in human neutrophils. J Immunol 2002;169:4423–9. [DOI] [PubMed] [Google Scholar]
- [44]. Feng Y, van der Veeken J, Shugay M, et al. A mechanism for expansion of regulatory T-cell repertoire and its role in self-tolerance. Nature 2015;528:132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Li MO, Rudensky AY. T cell receptor signalling in the control of regulatory T cell differentiation and function. Nat Rev Immunol 2016;16:220–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell 2019;176:1248–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Ohm JE, Gabrilovich DI, Sempowski GD, et al. VEGF inhibits T-cell development and may contribute to tumor-induced immune suppression. Blood 2003;101:4878–86. [DOI] [PubMed] [Google Scholar]
- [48]. Turkowski K, Brandenburg S, Mueller A, et al. VEGF as a modulator of the innate immune response in glioblastoma. Glia 2018;66:161–74. [DOI] [PubMed] [Google Scholar]
- [49]. Isakov N, Altman A. Regulation of immune system cell functions by protein kinase C. Front Immunol 2013;4:384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50]. Altman A, Kong KF. Protein kinase C inhibitors for immune disorders. Drug Discov Today 2014;19:1217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Meisel M, Hermann-Kleiter N, Hinterleitner R, et al. The kinase PKCα selectively upregulates interleukin-17A during Th17 cell immune responses. Immunity 2013;38:41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Chen YW, Lang ML, Wade WF. Protein kinase C-alpha and -delta are required for FcalphaR (CD89) trafficking to MHC class II compartments and FcalphaR-mediated antigen presentation. Traffic 2004;5:577–94. [DOI] [PubMed] [Google Scholar]
- [53]. Cheng Y, Zhu Y, Xu W, et al. PKCα in colon cancer cells promotes M1 macrophage polarization via MKK3/6-P38 MAPK pathway. Mol Carcinog 2018;57:1017–29. [DOI] [PubMed] [Google Scholar]
- [54]. Ondee T, Jaroonwitchawan T, Pisitkun T, et al. Decreased protein kinase C-β type II associated with the prominent endotoxin exhaustion in the macrophage of FcGRIIb−/− lupus prone mice is revealed by phosphoproteomic analysis. Int J Mol Sci 2019;20: pii: E1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Ondee T, Jaroonwitchawan T, Pisitkun T, et al. Decreased protein kinase C-β type II associated with the prominent endotoxin exhaustion in the macrophage of FcGRIIb−/− lupus prone mice is revealed by phosphoproteomic analysis. Int J Mol Sci 2019;20: pii: E1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56]. Maharaj NP, Wies E, Stoll A, et al. Conventional protein kinase C-α (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction. J Virol 2012;86:1358–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Maharaj NP, Wies E, Stoll A, et al. Conventional protein kinase C-α (PKC-α) and PKC-β negatively regulate RIG-I antiviral signal transduction. J Virol 2012;86:1358–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58]. Blair D, Dufort FJ, Chiles TC. Protein kinase Cβ is critical for the metabolic switch to glycolysis following B-cell antigen receptor engagement. Biochem J 2012;448:165–9. [DOI] [PubMed] [Google Scholar]
- [59]. Castrillo A, Pennington DJ, Otto F, et al. Protein kinase Cepsilon is required for macrophage activation and defense against bacterial infection. J Exp Med 2001;194(9):1231–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60]. Kirk SG, Samavati L, Liu Y. MAP kinase phosphatase-1, a gatekeeper of the acute innate immune response. Life Sci 2020;241:117157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol 2013;13:679–92. [DOI] [PubMed] [Google Scholar]
- [62]. Atsaves V, Leventaki V, Rassidakis GZ, et al. AP-1 transcription factors as regulators of immune responses in cancer. Cancers (Basel) 2019;11:E1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63]. Dushyanthen S, Teo ZL, Caramia F, et al. Agonist immunotherapy restores T cell function following MEK inhibition improving efficacy in breast cancer. Nat Commun 2017;8:606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64]. Germani A, Malherbe S, Rouer E. The exon 7-spliced Lck isoform in T lymphocytes: a potential regulator of p56lck signaling pathways. Biochem Biophys Res Commun 2003;301:680–5. [DOI] [PubMed] [Google Scholar]
- [65]. Zhou J, Zhang Q, Henriquez JE, et al. Lymphocyte-specific protein tyrosine kinase (LCK) is involved in the aryl hydrocarbon receptor-mediated impairment of immunoglobulin secretion in human primary B cells. Toxicol Sci 2018;165:322–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Yousefi S, Ma XZ, Singla R, et al. HIV-1 infection is facilitated in T cells by decreasing p56lck protein tyrosine kinase activity. Clin Exp Immunol 2003;133:78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67]. Chen H, Ai L, Lu H, et al. Clinical and imaging features of COVID-19, Radiol Infect Dis 2020. [Online ahead of print]. [DOI] [PMC free article] [PubMed]
- [68]. Henry BM, de Oliveira MHS, Benoit S, et al. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med 2020. [Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- [69]. Lee H, Ahn J, Shin SS, et al. Ascorbic acid inhibits visceral obesity and nonalcoholic fatty liver disease by activating peroxisome proliferator-activated receptor α in high-fat-diet-fed C57BL/6J mice. Int J Obes (Lond) 2019;43:1620–30. [DOI] [PubMed] [Google Scholar]
- [70]. Gu X, Luo X, Wang Y, et al. Ascorbic acid attenuates cell stress by activating the fibroblast growth factor 21/fibroblast growth factor receptor 2/adiponectin pathway in HepG2 cells. Mol Med Rep 2019;20:2450–8. [DOI] [PubMed] [Google Scholar]
- [71]. Leclerc PC, Proulx CD, Arguin G, et al. Ascorbic acid decreases the binding affinity of the AT1 receptor for angiotensin II. Am J Hypertens 2008;21:67–71. [DOI] [PubMed] [Google Scholar]
- [72]. Cárcamo JM, Pedraza A, Bórquez-Ojeda O, et al. Vitamin C is a kinase inhibitor: dehydroascorbic acid inhibits IkappaBalpha kinase beta. Mol Cell Biol 2004;24:6645–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Nespereira B, Pérez-Ilzarbe M, Fernández P, et al. Vitamins C and E downregulate vascular VEGF and VEGFR-2 expression in apolipoprotein-E-deficient mice. Atherosclerosis 2003;171:67–73. [DOI] [PubMed] [Google Scholar]
- [74]. Baek MW, Cho HS, Kim SH, et al. Ascorbic acid induces necrosis in human laryngeal squamous cell carcinoma via ROS, PKC, and calcium signaling. J Cell Physiol 2017;232:417–25. [DOI] [PubMed] [Google Scholar]
- [75]. Li W, Guo F, Jiang X, et al. Compound ammonium glycyrrhizin protects hepatocytes from injury induced by lipopolysaccharide/florfenicol through oxidative stress and a MAPK pathway. Comp Biochem Physiol C Toxicol Pharmacol 2019;225:108585. [DOI] [PubMed] [Google Scholar]
- [76]. Khan R, Rehman MU, Khan AQ, et al. Glycyrrhizic acid suppresses 1,2-dimethylhydrazine-induced colon tumorigenesis in Wistar rats: alleviation of inflammatory, proliferation, angiogenic, and apoptotic markers. Environ Toxicol 2018;33:1272–83. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.


