To the Editor:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel virus first identified in December 2019 in Wuhan, China, as causing coronavirus disease (COVID-19), with more than 7.5 million cases currently reported worldwide (1). ACE2 (angiotensin-converting enzyme 2) is the receptor for SARS-CoV-2 and has recently been identified as an IFN-stimulated gene (2). Rhinovirus (RV) infections are potent inducers of IFN-stimulated genes and subsequent cytokine production. RV infections are the most frequent virus identified in the common cold and are responsible for the majority of asthma exacerbations in children and adults (3). Young people with asthma have higher rates of COVID-19, accounting for 27% of hospitalized patients in the United States in the 18- to 49-year-old age group (4). We hypothesized that RV infections could increase expression of ACE2 and subsequently activate cytokine pathways associated with severe COVID-19 infections.
We developed air–liquid interface (ALI) cultures from nasal tissues biopsied from 30 adults with physician-diagnosed asthma. Subjects averaged 35 years of age, 60% were non-Hispanic white individuals, and subjects were evenly divided by sex. We infected ALI cultures with common RV strains RV-A16 (1 × 105 RNA copies/well), RV-C15 (1 × 105 RNA copies/well), or Dulbecco’s modified Eagle medium/F12 media (control) for 4 hours at 34°C, 5% CO2. RNA was then extracted from whole-cell lysates, sequenced using KAPA Stranded RNA-Seq libraries on an Illumina HiSeq 3000 for a 1 × 50 run, demultiplexed with Illumina Bcl2fastq2 (v2.17), and then mapped to the UCSC transcript set using Bowtie2 (v2.1.0). We processed the discovery (n = 22) and validation (n = 8) cohorts separately through the NOISeq library (5) to filter out genes with low counts (counts per million < 30), resulting in 7,474 and 7,905 unique genes in the discovery and validation cohorts. We then used the function “ARSyNseq” followed by “voomWithQualityWeights” (6) to process RNA counts for downstream statistical analysis with the linear model implemented in the LIMMA R library. We used the moderate t test for paired samples for statistical analyses to prioritize 402 differentially expressed genes (DEGs) adjusted by false discovery rate <1% and absolute log2 fold change >0.5.
When compared with controls, both RV-A16– and RV-C15–infected ALI cultures resulted in a greater than threefold increase in ACE2 expression in the discovery and validation cohorts (Figure 1). Interestingly, levels of TMPRSS2 (transmembrane serine protease 2), a protease that primes the SARS-CoV-2 virus for cellular entry, were not increased after either RV-A16 or RV-C15 infections. How could RV infections induce ACE2 expression? Ziegler and colleagues determined that stimulation of primary nasal epithelial cells with IFN increased ACE2 expression. They also identified four potential ACE2 transcription factors located within 2 kbp of the ACE2 start site: STAT1, STAT3, IRF8, and IRF1 (2). Of these four transcription factors, only IRF1 was reproducibly differentially expressed in our data set and showed a significant threefold increase in expression after RV-A and RV-C infections.
Figure 1.
ACE2 (angiotensin-converting enzyme 2) is overexpressed in human rhinovirus (RV)-infected human nasal tissue cultures. ACE2 fold change of expression varies from 3.2 to 3.6 in the discovery and validation cohorts after RV-A and RV-C infection. TMPRSS2 is not reproducibly altered by RV infections. The mRNA expression was calculated by normalization of voom counts. n.s. = not significant; TMPRSS2 = transmembrane serine protease 2.
Next, we sought to determine if the patterns observed in nasal cells among patients with asthma were also observed for other viruses in human bronchial epithelial cells unselected for asthma. We analyzed microarray data (GSE32140) to quantify gene expression changes after exposure to influenza A and respiratory syncytial virus in ALI cultures of human bronchial epithelial cells. Two hours after infection with influenza A or respiratory syncytial virus, ACE2 expression levels were sixfold higher whereas TMPRSS2 levels were not altered compared with control uninfected cells (data not shown).
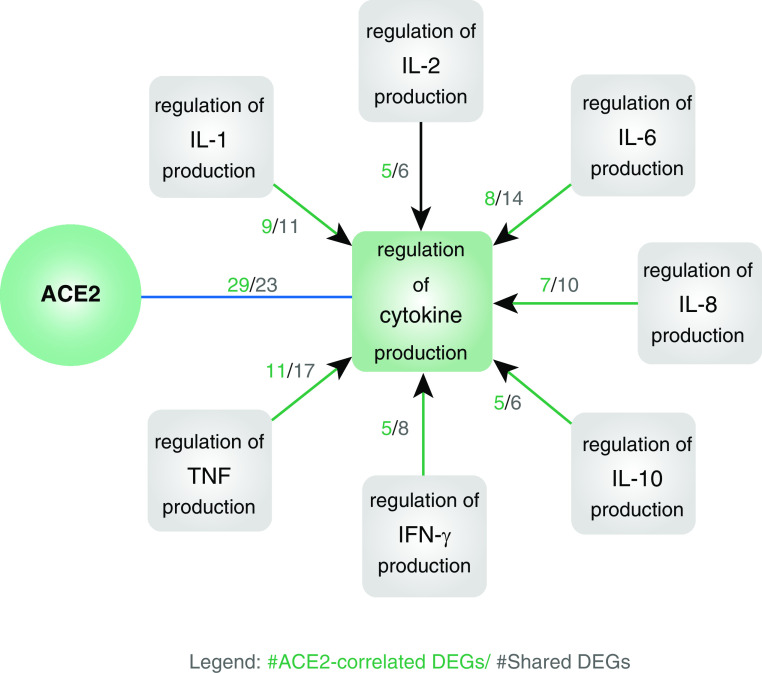
The role of ACE2 overexpression on the cytokine surge, which has been shown to be clinically relevant in the severity of COVID-19, is unknown. Huang and colleagues recently reported that critically ill patients with COVID-19 had high serum levels of IL-1β, IL-1RA, IL-2, IL-4, IL-7, IL-8, IL-9, IL-10, IL-13, IL-17, G-CSF, IFN-γ, IP-10, MCP-1, MIP-1A, and TNF-α (SARS-CoV-2–associated cytokine surge) (7). Using our in vitro model, we sought to identify DEGs associated with RV-induced ACE2 overexpression and with SARS-CoV-2 cytokine regulation. Sixty-three DEGs were correlated to RV-induced ACE2 overexpression and overrepresented in the “Regulation of cytokine production” gene ontology (GO) set (GO:0001817). We then identified 34 GO annotations correlated to the regulation and production of the SARS-CoV-2–associated cytokine surge (8, 9). Twenty-nine of these 63 DEGs were annotated in 7 GO annotations, and several of these genes have also been implicated in the aberrant antiviral response in asthma (Figure 2).
Figure 2.
Biomolecular mechanisms of response to rhinovirus (RV) infection in 30 asthmatic cultures that are both correlated with ACE2 (angiotensin-converting enzyme 2) overexpression and overrepresented in coronavirus disease (COVID-19) cytokine surge pathways. Twenty-nine of the 63 differentially expressed genes (DEGs) in response to RV-A and RV-C infections compared with controls (n = 22 patients with asthma in the discovery cohort, n = 8 in the validation cohort; adjusted with false discovery rate <1% in the discovery cohort and Bonferroni adjustment <5% in the validation cohort) were 1) reproducibly correlated with ACE2 expression and the gene ontology (GO) mechanism (GO:0001817): regulation of cytokine production (Bonferroni-adjusted P < 5%) (green square), and 2) also overrepresented in the GO mechanisms associated with the cytokine surge in ICU-admitted subjects with COVID-19 (gray squares). Twelve of these DEGs (CASP1, CEACAM1, EREG, GBP1, HLA-E, IFI16, ISG15, KLF4, MYD88, PML, TRIB2, and VTCN1) were associated with the regulation of a single cytokine, and the remainder of the genes (CD274, DDX58, F2R, FZD5, IDO1, IFIH1, IRAK3, JAK2, LGALS9, PRKD2, RIPK1, TICAM1, TLR2, TLR3, TNFAIP3, ZC3H12A, and ZFP36) were associated with the regulation of multiple cytokines. Genes in bold have been implicated in aberrant antiviral responses in asthma (references not shown).
Here, we present novel findings suggesting that 1) RV infections are potential mechanisms of ACE2 overexpression in patients with asthma and 2) ACE2 activation regulates multiple cytokine antiviral responses. These results suggest that viral infections associated with asthma exacerbations exhibit synergistic biomolecular interactions with SARS-CoV-2 infection. Therefore, coinfections with RV and SARS-CoV-2 may pose significant risks for patients with asthma. One limitation of this study was that we did not evaluate the surface protein expression of ACE2 after RV infection. Unfortunately, testing of current available ACE2 antibodies has been nonspecific or inconclusive (2). We also were unable to directly infect our ALI cultures with SARS-CoV-2 owing to safety concerns. However, the recent availability of pseudotyped viral models expressing the SARS-CoV-2 spike protein will be invaluable to assess differences in SARS-CoV-2 binding in correlation to ACE2 expression. Although we used RV infection as a model of ACE2 activation and cytokine induction, it is not known if similar findings are found in the cytokine surge in severe SARS-CoV-2 infections seen in ICU patients. Are there potential therapies that could downregulate ACE2 expression to decrease SARS-CoV-2 susceptibility? Zaheer and colleagues found that knockdown of IRF-1 abrogated the production of antiviral cytokines after RV infections (3). Further studies are required to determine if IRF-1 blockade also affects ACE2 expression. Peters and colleagues also identified that the use of inhaled corticosteroids in individuals with asthma was associated with lower ACE2 expression levels, suggesting that nasal or inhaled corticosteroid use could be a potential therapy in ACE2 downregulation (10). Our study suggests that common viral infections may prime the host to respond excessively to COVID-19 infections and potentially correspond to an increase in disease severity when multiple respiratory viruses are circulating.
Supplementary Material
Footnotes
Supported in part by The University of Arizona Health Sciences Center for Biomedical Informatics and Biostatistics, the BIO5 Institute, and the NIH (HL132532, HL147187, and AI146131). The University of Arizona Health Sciences Center for Biomedical Informatics and Biostatistics supported in part the salaries of N.P. and Y.A.L. This article did not receive sponsorship for publication.
Author Contributions: Conception and design: E.H.C., Y.A.L., and F.D.M. Analysis and interpretation: E.H.C., A.L.W., C.E.R., D.A.C., N.P., J.L., Y.A.L., and F.D.M. Drafting the manuscript for important intellectual content: E.H.C., A.L.W., C.E.R., D.A.C., N.P., J.L., Y.A.L., and F.D.M.
Originally Published in Press as DOI: 10.1164/rccm.202004-1343LE on July 10, 2020
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.World Health Organization. Coronavirus disease 2019 (COVID-19): situation report-145. [accessed 2020 Jun 13]. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200613-covid-19-sitrep-145.pdf?sfvrsn=bb7c1dc9_4.
- 2.Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. HCA Lung Biological Network. Electronic address: lung-network@humancellatlas.org; HCA Lung Biological Network. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035, e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zaheer RS, Proud D. Human rhinovirus-induced epithelial production of CXCL10 is dependent upon IFN regulatory factor-1. Am J Respir Cell Mol Biol. 2010;43:413–421. doi: 10.1165/rcmb.2009-0203OC. [DOI] [PubMed] [Google Scholar]
- 4.Garg S, Kim L, Whitaker M, O’Halloran A, Cummings C, Holstein R, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019 - COVID-NET, 14 States, March 1-30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:458–464. doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tarazona S, Furió-Tarí P, Turrà D, Pietro AD, Nueda MJ, Ferrer A, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43:e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Y, Sam L, Li J, Friedman C, Lussier YA. Information theory applied to the sparse gene ontology annotation network to predict novel gene function. Bioinformatics. 2007;23:i529–i538. doi: 10.1093/bioinformatics/btm195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Li J, Lee Y, Lussier YA. GO-Module: functional synthesis and improved interpretation of Gene Ontology patterns. Bioinformatics. 2011;27:1444–1446. doi: 10.1093/bioinformatics/btr142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peters MC, Sajuthi S, Deford P, Christenson S, Rios CL, Montgomery MT, et al. COVID-19-related genes in sputum cells in asthma: relationship to demographic features and corticosteroids. Am J Respir Crit Care Med. 2020;202:83–90. doi: 10.1164/rccm.202003-0821OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.