To the Editor:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a coronavirus identified as the cause of an outbreak of coronavirus disease (COVID-19), which now causes death in over 6% of infected individuals worldwide (1–5). Patients with confirmed infection have reported respiratory illness, such as fever, cough, and shortness of breath (6). Once contacted with the human airway, the spike proteins of this virus can associate with the surface receptors of sensitive cells, which mediate the entrance of the virus into target cells for further replication. Xu and colleagues first modeled the spike protein to identify the receptor for SARS-CoV-2 and indicated that ACE2 (angiotensin-converting enzyme 2) could be the receptor for this virus (7). ACE2 is previously known as the receptor for severe acute respiratory syndrome coronavirus (SARS-CoV) and human coronavirus NL63 (HCoV-NL63) (8–10). Studies focusing on the genome sequence and structure of the receptor-binding domain of the spike proteins further confirmed that the new coronavirus can efficiently use ACE2 as a receptor for cellular entry, with an estimated 10- to 20-fold higher affinity to ACE2 than SARS-CoV (11, 12). Zhou and colleagues conducted virus infectivity studies and showed that ACE2 is essential for SARS-CoV-2 to enter HeLa cells (13). These data indicate that ACE2 is the receptor for SARS-CoV-2.
The tissue expression and distribution of the receptor decide the tropism of the virus infection, which has a major implication for understanding its pathogenesis and designing therapeutic strategies. Previous studies have investigated the RNA expression of ACE2 in 72 human tissues and demonstrated its expression in lung and other organs (14). The lung is a complex organ with multiple types of cells, so such real-time PCR RNA profiling based on bulk tissue could mask the ACE2 expression in each type of cell in the human lung. The ACE2 protein level was also investigated by immunostaining in lung and other organs (14, 15). These studies showed that in the normal human lung, ACE2 is mainly expressed by type II alveolar (AT2) and type I alveolar (AT1) epithelial cells. Endothelial cells were also reported to be ACE2 positive. Immunostaining detection is a reliable method for the identification of protein distribution, yet accurate quantification remains a challenge for such analysis. The recently developed single-cell RNA-sequencing technology enables us to study the ACE2 expression in each cell type and provides quantitative information at a single-cell resolution. Previous work has built up the online database for single-cell RNA-sequencing analysis of eight normal human lung transplant donors (16). In the current work, we used the updated bioinformatics tools to analyze the data. Some of the results of these studies have been previously reported in the form of a preprint (https://doi.org/10.1101/2020.01.26.919985) (16).
We analyzed 43,134 cells derived from the normal lung tissue of eight adult donors (Figure 1A). We performed unsupervised graph-based clustering (Seurat version 2.3.4), and for each individual, we identified 8–11 transcriptionally distinct cell clusters based on their marker gene expression profile. Typically, the clusters include AT2 cells, AT1 cells, airway epithelial cells (ciliated cells and club cells), fibroblasts, endothelial cells, and various types of immune cells. The cell cluster map of a representative donor (a 55-yr-old Asian man) was visualized using t-distributed stochastic neighbor embedding (tSNE), as shown in Figure 1B.
Figure 1.
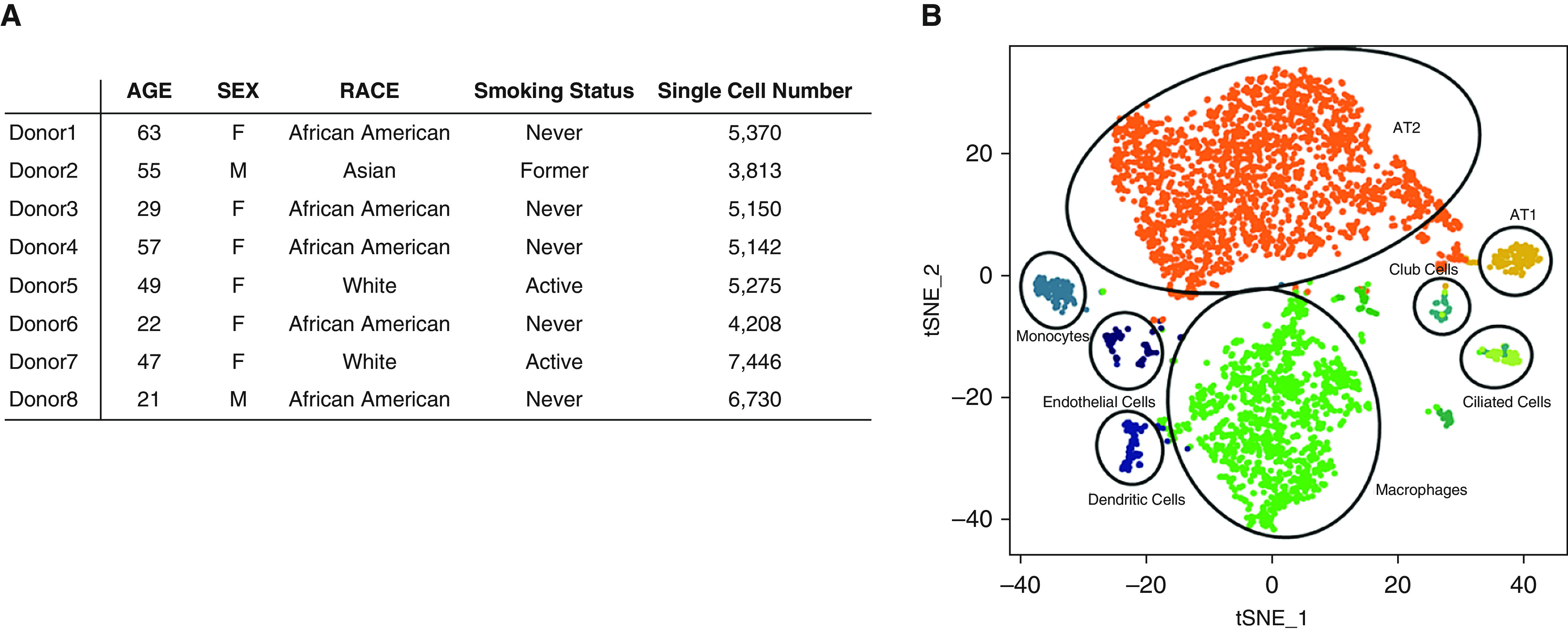
Single-cell RNA-sequencing analysis of normal human lungs. (A) Characteristics of lung transplant donors for single-cell RNA-sequencing analysis. (B) Cellular cluster map of the Asian male donor. All eight samples were analyzed using the Seurat R package. Cells were clustered using a graph-based, shared nearest-neighbor clustering approach and visualized using a t-distributed stochastic neighbor embedding plot. AT1 = type I alveolar; AT2 = type II alveolar; tSNE = t-distributed stochastic neighbor embedding.
Next, we analyzed the cell type–specific expression pattern of ACE2 in each individual. For all donors, ACE2 is expressed in 0.64% of all human lung cells. The majority of the ACE2-expressing cells (83% in average) are AT2 cells. Other ACE2-expressing cells include AT1 cells, airway epithelial cells, fibroblasts, endothelial cells, and macrophages. However, their ACE2-expressing cell ratio is relatively low and is variable among individuals. For the representative donor (Asian male, 55 yr old), the expressions of ACE2 and cell type–specific markers in each cluster are demonstrated in Figure 2A.
Figure 2.
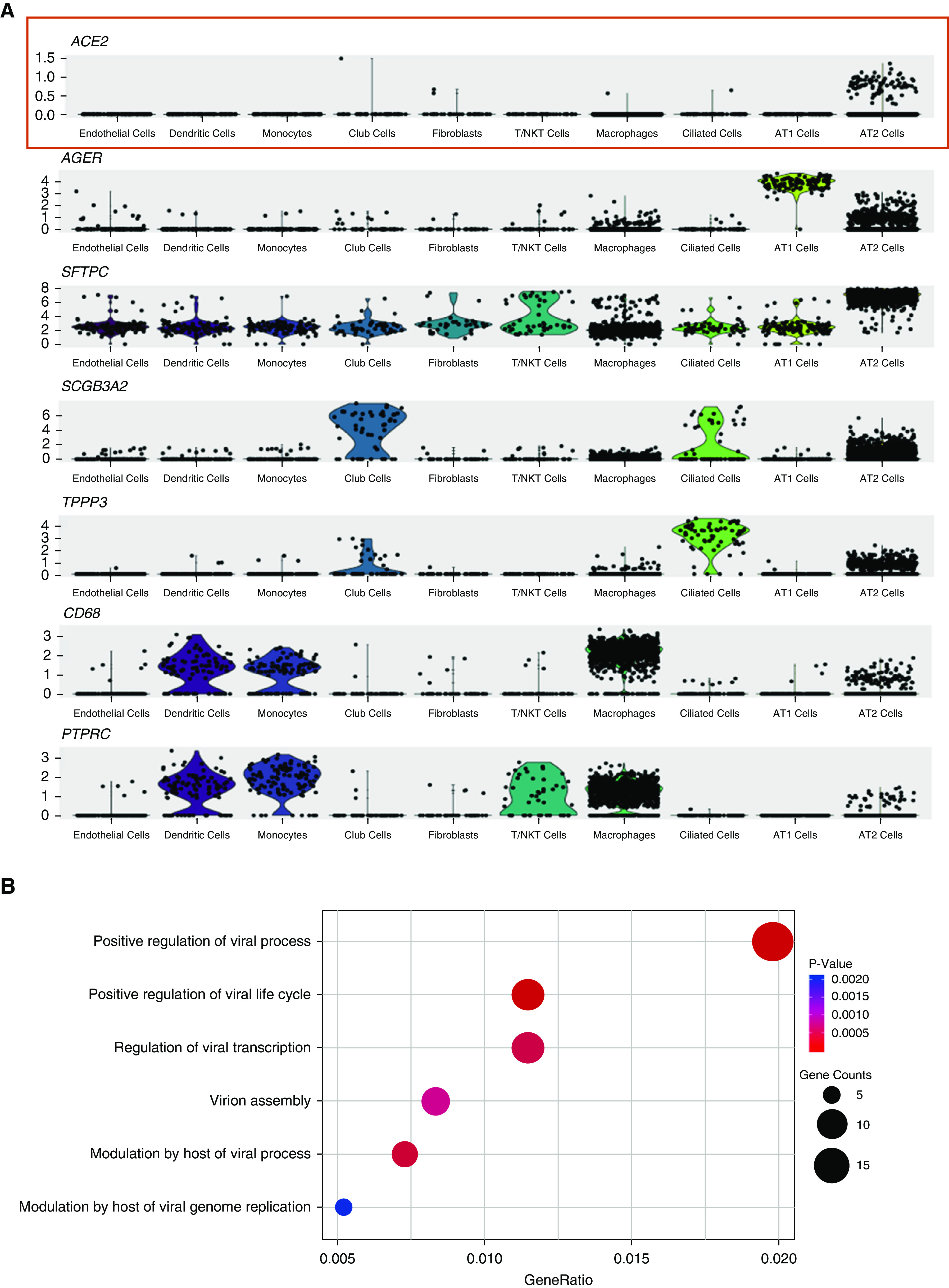
Gene expression analysis in ACE2 (angiotensin-converting enzyme 2)-expressing type II alveolar (AT2) cell population. (A) Violin plots of expression for ACE2 and select cell type–specific marker genes significantly upregulated in distinct lung cell clusters from an Asian male donor. AGER is a type I alveolar cell marker, SFTPC (SPC) is an AT2 cell marker, SCGB3A2 is a club cell marker, TPPP3 is a ciliated cell marker, CD68 is a macrophage marker, and PTPRC (CD45) is a panimmune cell marker. (B) Dot plot of gene ontology enrichment analysis demonstrating enriched virus-related biological processes in the ACE2-expressing AT2 population. AT1 = type I alveolar; NK = natural killer.
There are 1.4 ± 0.4% of AT2 cells expressing ACE2. To further understand the special population of ACE2-expressing AT2, we performed a gene ontology (GO) enrichment analysis to study which biological processes are involved with this cell population by comparing them with the AT2 cells not expressing ACE2. Surprisingly, we found that multiple viral life cycle–related functions are significantly overrepresented in ACE2-expressing AT2 cells, including those relevant to viral replication and transmission (Figure 2B). We found an upregulation of CAV2 and ITGB6 genes in ACE2-expressing AT2. These genes are components of caveolae, which is a special subcellular structure on the plasma membrane critical to the internalization of various viruses, including SARS-CoV (17–19). We also found an enrichment of multiple ESCRT (endosomal sorting complex required for transport) machinery gene members (including CHMP3, CHMP5, CHMP1A, and VPS37B) in ACE2-expressing AT2 cells that were related to virus budding and release (20, 21). These data showed that this small population of ACE2-expressing AT2 cells is particularly prone to SARS-CoV-2 infection.
We further analyzed each donor and their ACE2-expressing patterns. As the sample size was very small, no significant association was detected between the ACE2-expressing cell number and any characteristics of the individual donors. But we did notice that one donor had a five-fold higher ACE2-expressing cell ratio than average. The observation on this case suggested that ACE2-expressing profile heterogeneity might exist between individuals, which could make some individuals more vulnerable to SARS-CoV-2 than others. However, these data need to be interpreted very cautiously because of the very small sample size of the current dataset, and a larger cohort study is necessary to draw conclusions.
Altogether, in the current study, we report the RNA expression profile of ACE2 in the human lung at single-cell resolution. Our analysis suggested that the expression of ACE2 is concentrated in a special small population of AT2 cells, which also expresses many other genes favoring the viral infection process. It seems that SARS-CoV-2 has cleverly evolved to hijack this population of AT2 cells for its reproduction and transmission. Targeting AT2 cells explained the severe alveolar damage and minimal upper airway symptoms after infection by SARS-CoV-2. The demonstration of the distinct number and distribution of the ACE2-expressing cell population in different cohorts can potentially help to identify the susceptible population in the future. The shortcomings of the study are the small sample number and the fact that the current technique can only analyze the RNA level and not the protein level of single cells. Furthermore, although previous studies reported abundant ACE2 expression in pulmonary endothelial cells (14, 22), we did not observe high ACE2 RNA levels in this population. This inconsistency may be partly due to the fact that the cell number and portion of endothelial cells in the current dataset is relatively smaller than expected. Indeed, because the limitation of sample collection and processing, the analyzed cells in this study may not fully represent the whole lung cell population. Future quantitative analysis at the transcriptomic and proteomic level in a larger total population of cells is needed to further dissect the ACE2 expression profile, which could eventually lead to novel anti-infective strategies, such as ACE2 receptor blockade (23, 24), ACE2 protein competition (25), or ACE2-expressing cell ablation.
Methods
Public datasets (Gene Expression Omnibus GSE122960) were used for bioinformatics analysis. First, Seurat (version 2.3.4) was used to read a combined gene-barcode matrix of all samples. Low-quality cells with less than 200 or more than 6,000 detected genes were removed; cells were also removed if their mitochondrial gene content was <10%. Only genes found to be expressed in more than three cells were retained. For normalization, the combined gene-barcode matrix was scaled by the total unique molecular identifier counts, multiplied by 10,000, and transformed to log space. The highly variable genes were identified using the function FindVariableGenes. Variants arising from number of unique molecular identifiers and the percentage of mitochondrial genes were regressed out by specifying the vars.to.regress argument in Seurat function ScaleData. The expression level of highly variable genes in the cells was scaled, centered along each gene, and conducted to principal component (PC) analysis.
Then the number of PCs to be included in downstream analysis was assessed by 1) plotting the cumulative SDs accounted for each PC using the function PCElbowPlot in Seurat to identify the “knee” point at a PC number after which successive PCs explain the diminishing degrees of variance and 2) by exploring primary sources of heterogeneity in the datasets using the PC Heatmap function in Seurat. Based on these two methods, the first top significant PCs were selected for two-dimensional tSNE, which was implemented by the Seurat software with the default parameters. FindClusters was used in Seurat to identify cell clusters for each sample. After clustering and visualization with tSNE, the initial clusters were subjected to inspection and merging based on the similarity of marker genes and a function for measuring phylogenetic identity using BuildClusterTree in Seurat. The identification of cell clusters was performed on the final aligned object, guided by marker genes. To identify the marker genes, differential expression analysis was performed by the function FindAllMarkers in Seurat with the Wilcoxon rank sum test. Differentially expressed genes that were expressed at least in 25% of cells within the cluster and with a fold change of >0.25 (log scale) were considered to be marker genes. tSNE plots and violin plots were generated using Seurat.
For GO enrichment analysis, differentially expressed genes of the ACE2-expressing AT2 cells were calculated for each donor when they were expressed in at least 25% of cells within the cluster and had a fold change of > 0.25 (log scale) compared with all AT2 cells. All differentially expressed genes were combined to a gene list for GO analysis by the ClusterProfiler R package. GO terms with a corrected P value of less than 0.05 were considered significantly enriched by differentially expressed genes. Dot plots were used to visualize enriched terms by the enrichplot R package.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Alexander Misharin’s group for sharing their original single-cell RNA-sequencing dataset with the public. They also thank the medical workers who gave their lives in the fight against the COVID-19 pandemic.
Footnotes
Supported by the National Key Research and Development Program of China (2017YFA0104600 to W.Z.), the National Science Foundation of China (81770073 to W.Z.), the Shanghai Science and Technology Talents Program (19QB1403100 to W.Z.), the Youth 1000 Talent Plan of China (to W.Z.), Tongji University (Basic Scientific Research Interdisciplinary Fund and 985 grant to W.Z.), and Guangzhou Medical University (annual grant to W.Z.).
Author Contributions: W.Z. designed the project. Y. Zhao, Z.Z., Y.W., Y. Zhou, and Y.M. performed the analysis. Y.W. and W.Z. drafted the manuscript.
Originally Published in Press as DOI: 10.1164/rccm.202001-0179LE on July 14, 2020
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.Huang CL, Wang YM, Li XW, Ren LL, Zhao JP, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv. 2020 Available from: https://www.medrxiv.org/content/10.1101/2020.02.06.20020974v1.
- 4.World Health Organization. Coronavirus disease 2019 (COVID-19) situation report-89. Geneva, Switzerland: World Health Organization; 2020. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200418-sitrep-89-covid-19.pdf?sfvrsn=3643dd38_2.
- 5.Bedford J, Enria D, Giesecke J, Heymann DL, Ihekweazu C, Kobinger G, et al. WHO Strategic and Technical Advisory Group for Infectious Hazards. COVID-19: towards controlling of a pandemic. Lancet. 2020;395:1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlos WG, Dela Cruz CS, Cao B, Pasnick S, Jamil S. Novel Wuhan (2019-nCoV) coronavirus. Am J Respir Crit Care Med. 2020;201:P7–P8. doi: 10.1164/rccm.2014P7. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, Chen P, Wang J, Feng J, Zhou H, Li X, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Sui J, Huang IC, Kuhn JH, Radoshitzky SR, Marasco WA, et al. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology. 2007;367:367–374. doi: 10.1016/j.virol.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu K, Li W, Peng G, Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci USA. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L, Ding Y, Zhang Q, Che X, He Y, Shen H, et al. Expression of elevated levels of pro-inflammatory cytokines in SARS-CoV-infected ACE2+ cells in SARS patients: relation to the acute lung injury and pathogenesis of SARS. J Pathol. 2006;210:288–297. doi: 10.1002/path.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus: a first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JK, Lin SS, Ji XJ, Guo LM. Binding of SARS coronavirus to its receptor damages islets and causes acute diabetes. Acta Diabetol. 2010;47:193–199. doi: 10.1007/s00592-009-0109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyfman P, Walter J, Joshi N, Anekalla K, McQuattie-Pimentel A, Chiu S, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Embong AK, Duffney P, Thatcher TH, Sime PJ. Cigarette smoke increases uptake of influenza A virus by lung epithelial cells by increasing expression of caveolin-1 [abstract] Am J Respir Crit Care Med. 2020;201:A4051. [Google Scholar]
- 18.Lu Y, Liu DX, Tam JP. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem Biophys Res Commun. 2008;369:344–349. doi: 10.1016/j.bbrc.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salanueva IJ, Cerezo A, Guadamillas MC, del Pozo MA. Integrin regulation of caveolin function. J Cell Mol Med. 2007;11:969–980. doi: 10.1111/j.1582-4934.2007.00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pincetic A, Leis J. The mechanism of budding of retroviruses from cell membranes. Adv Virol. 2009;2009:6239691–6239699. doi: 10.1155/2009/623969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Dong J, Martin M, He M, Gongol B, Marin TL, et al. AMP-activated protein kinase phosphorylation of angiotensin-converting enzyme 2 in endothelium mitigates pulmonary hypertension. Am J Respir Crit Care Med. 2018;198:509–520. doi: 10.1164/rccm.201712-2570OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug Dev Res. doi: 10.1002/ddr.21656. [online ahead of print] 4 Mar 2020; DOI: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng Y, Ling Y, Bai T, Xie Y, Huang J, Li J, et al. COVID-19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med. 2020;201:1380–1388. doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monteil V, Kwon H, Prado P, Hagelkrüys A, Wimmer RA, Stahl M, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913, e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


