Abstract
Objective:
To assess the impact of somatic gene mutations on survival among patients undergoing resection of colorectal liver metastases (CLM).
Summary Background Data:
Patients undergoing CLM resection have heterogeneous outcomes, and accurate risk stratification is necessary to optimize patient selection for surgery.
Methods:
Next-generation sequencing of 50 cancer-related genes was performed from primary tumors and/or liver metastases in 401 patients undergoing CLM resection. Missense TP53 mutations were classified by the Evolutionary Action Score (EAp53), a novel approach that dichotomizes mutations as low or high risk.
Results:
The most frequent somatic gene mutations were TP53 (65.6%), followed by KRAS (48.1%) and APC (47.4%). Double mutation in RAS/TP53, identified in 31.4% of patients, was correlated with primary tumor location in the right colon (P = 0.006). On multivariable analysis, RAS/TP53 double mutation was an independent predictor of shorter overall survival (hazard ratio, 2.62; 95% confidence interval 1.41 to 4.87; P = 0.002). In patients with co-mutated RAS, EAp53 high risk mutations were associated with shorter 5-year overall survival of 12.2%, compared with 55.7% for TP53 wild-type (P < 0.001). The negative prognostic effects of RAS and TP53 mutations were limited to tumors harboring mutations in both genes.
Conclusions:
Concomitant RAS and TP53 mutations are associated with decreased survival after CLM resection. A high EAp53 predicts a subset of patients with worse prognosis. These preliminary analyses suggest that surgical resection of liver metastases should be carefully considered in this subset of patients.
INTRODUCTION
Colorectal cancer is the second leading cause of cancer mortality in the United States and is estimated to account for over 49,000 deaths in 2016.1 Most patients dying from colorectal cancer will have liver metastases. Surgical resection of colorectal liver metastases (CLM) is associated with a 5-year overall survival (OS) rate of 58% and is accepted as standard of care.2 Owing to advances in systemic therapy, radiology, and surgical technique, more patients are eligible for hepatic resection. However, colorectal cancer is a heterogeneous disease, and subsets of patients undergoing surgery will succumb early to disease recurrence.3 With systemic therapy alone, median OS rates exceeding 2 years are reported for metastatic colorectal cancer.4 Therefore, accurate prognostic markers are needed for risk stratification and optimization of patient selection for hepatic resection.
Currently, few biologic markers are used to guide therapy and prognostication in CLM. Defective DNA mismatch repair (MMR) is established as an important factor in colorectal cancer pathogenesis, treatment, and outcome.5 However, MMR-deficient tumors are found in a minority of patients undergoing CLM resection.6 Also uncommon, occurring in < 5% of patients, BRAF mutations are associated with particularly poor outcomes, with median overall survival (OS) of 22.6 months after CLM resection.7 To date, RAS mutations are the most prevalent predictive and prognostic genetic alterations in CLM, detected in 18%−52% of patients undergoing CLM resection.7, 8 In addition to predicting resistance to anti-epidermal growth factor receptor therapies, RAS mutations are associated with disease recurrence in the lungs, higher rate of positive margins, and worse survival after CLM resection.9, 10
As next-generation sequencing technology becomes widely available, multigene analysis is expected to inform clinical decisions in CLM beyond BRAF and RAS. TP53 is the most frequently mutated gene in metastatic colorectal cancer, but previous studies on TP53 mutations and colorectal cancer prognosis have yielded conflicting results.11–13 Preclinical studies have demonstrated that cooperation between mutated TP53 and RAS is critical for malignant transformation of colorectal cancer cells.14, 15 The malignant state of colorectal cancer cells depends upon genes controlled synergistically by loss of wild-type p53 function and RAS activation.14 In this article, we evaluated cooperativity between somatic mutations in RAS and TP53 among patients undergoing CLM resection. Since TP53 mutations are not all functionally equivalent, we categorized mutations according to the Evolutionary Action (EA) score, a novel computational approach to assess the functional impact of genetic mutations.16 We hypothesized that integrating the mutational status of RAS and TP53 provides information on patients’ prognosis after CLM resection that is complementary to standard clinicopathologic factors to guide treatment decisions.
METHODS
Patients
From 1151 patients undergoing CLM resection at the University of Texas MD Anderson Cancer Center between 2005 and 2015, 401 patients underwent multigene panel testing from their primary tumors and/or hepatic metastases. For patients undergoing initial CLM resection before 2012, genomic analysis was performed from archived tumor samples or repeat hepatectomy specimens. Computerized medical records were queried for data on clinicopathologic factors, disease recurrence, and patient survival. Synchronous metastases were defined as metastases diagnosed within 6 months of primary tumor diagnosis. Positive surgical margin was defined as tumor cells at the line of transection and included patients undergoing radiofrequency ablation at time of hepatic resection. The number and diameter of liver metastases were determined from surgical pathology specimens. Major hepatectomy was defined as resection of 3 or more contiguous Couinaud liver segments.17
Primary tumor location was classified as right colon if the vascular supply originated from the superior mesenteric artery, including the proximal transverse colon. Primary tumors in the left were defined by vascular supply from the inferior mesenteric artery, including distal transverse colon and rectal cancers. MMR status was determined as previously described.5 Primary tumor T stage was classified according to the American Joint Committee on Cancer, 7th edition.18
Preoperative and postoperative chemotherapy regimens were selected at the discretion of treating medical oncologists. Our institutional practice is to administer short-course preoperative chemotherapy to most patients before liver resection, followed by postoperative chemotherapy to complete 6 months total of systemic therapy. Preoperative chemotherapy was administered to 338 of the 401 patients (84.3%) and included regimens containing oxaliplatin (63.6%), irinotecan (12.0%), or both oxaliplatin and irinotecan (4.7%). Anti-epidermal growth factor receptor (EGFR) therapy was administered to 20 patients (5.0%) before liver resection and 16 patients (4.0%) after liver resection.
Selection criteria for liver resection included radiologic response or stable disease after preoperative chemotherapy, sufficient remnant liver volume, and technical resectability, as previously described.19 Patients with anticipated insufficient future liver remnant volume underwent portal vein embolization. Two-stage hepatectomy was performed for patients with bilateral disease that could not be safely resected in one stage. Patients with extrahepatic disease were considered for surgery if all sites of disease could be resected, or if patients had indeterminate or disappearing lung lesions.
This study was approved by the institutional review board at the University of Texas MD Anderson Cancer Center.
Genomic Analysis
Since 2012, metastatic colorectal cancer samples submitted for KRAS and/or BRAF testing are routinely analyzed by next-generation sequencing to detect somatic mutations in the coding sequence of 50 genes from DNA extracted from formalin-fixed paraffin-embedded tumor samples, as previously described.20
Sequence analysis of RAS included KRAS exons 2–4 (codons 5–66 and 114–150), NRAS exons 2–4 (codons 3–31, 43–69, and 124–150), and HRAS exons 2–3 (codons 5–35 and 42–82). For TP53, the exons (codons) tested were 2 (1–20), 4 (68–113), 5 (126–138), 5–6 (149–223), 7 (225–258), 8 (263–307), and 10 (332–367).
Mutation diagrams (lolliplots) were generated using the cBioPortal.21, 22
Classification of TP53 Mutations
Missense TP53 mutations were classified as low or high risk according to the Evolutionary Action (EAp53) score.16 EA measures the fitness effect of coding mutations. It models evolution as a mapping of genotypes (γ) to phenotypes (φ) in the fitness landscape via an evolutionary function f:γ→φ Assuming evolvability, f is differentiable, and following calculus, the action of a mutation dγ on fitness is dφ=f′(γ) dγ, where f′(γ) is the sensitivity of the mutated site. In practice, dγ is approximated with inverse amino acid substitution log-odds and f′(γ) with Evolutionary Trace ranks of importance. The fitness effect dφ, or EA, of coding mutations is then computable and correlates with experimental loss of function and morbidity. A training cohort of head and neck squamous cell cancer patients defined a threshold EAp53 value of > 75% that was associated with poor prognosis.23 Evolutionary Action and EAp53 are available for non-profit use at http://mammoth.bcm.tmc.edu/EvolutionaryAction and http://mammoth.bcm.tmc.edu/EAp53/, respectively.
Statistical Analysis
Group comparisons were performed using chi-square tests for categorical variables and Mann-Whitney for continuous variables. The Kaplan-Meier method was used to estimate probability of OS and recurrence-free survival (RFS) from date of liver resection. The log-rank test was used to compare OS and RFS between subgroups. Factors significant on univariable analysis were entered into a multivariable Cox analysis of OS. All P values were two-sided, and P < 0.05 was considered statistically significant. All analyses were performed with SPSS Statistics 23.0 (IBM Corp., Chicago, IL).
RESULTS
Clinical Characteristics
Baseline characteristics of 401 patients who underwent CLM resection are listed in Table 1. Among 381 patients who underwent evaluation of microsatellite instability (MSI), 376 (98.7%) had MSI-stable tumors. Overall, 155 (38.7%) patients underwent resection of a solitary hepatic metastasis, and 246 (61.3%) resection of multiple hepatic metastases. Median duration of follow-up was 35 months (range, 4–144 months).
Table 1.
Patient and Tumor Characteristics
| Characteristic | N | % |
|---|---|---|
| Median age, years (range) | 54 (23–79) | |
| Gender | ||
| Male | 223 | 55.6 |
| Female | 178 | 44.4 |
| Primary tumor location | ||
| Right colon | 99 | 24.7 |
| Left colon and rectum | 299 | 74.6 |
| Rectum only | 113 | 28.2 |
| NA | 3 | 0.8 |
| Primary tumor regional lymph node status | ||
| Positive | 274 | 68.3 |
| Negative | 114 | 28.4 |
| NA | 13 | 3.2 |
| Primary tumor T stage | ||
| T1–2 | 58 | 14.5 |
| T3–4 | 327 | 81.5 |
| NA | 16 | 4.0 |
| Microsatellite instability (MSI) status | ||
| MSI-stable | 376 | 93.8 |
| MSI-high | 5 | 1.2 |
| NA | 20 | 5.0 |
| Synchronous diagnosis of liver metastases | 278 | 69.3 |
| Chemotherapy before liver resection | 338 | 84.3 |
| Extrahepatic metastases | 73 | 18.2 |
| Median number of liver metastases (range) | 2 (1–40) | |
| Median size of largest liver metastasis, cm (range) | 2.6 (0.0–18.0) | |
| Surgical resection margin | ||
| R0 | 294 | 73.3 |
| R1 | 107 | 26.7 |
| Radiofrequency ablation at time of liver resection | 28 | 7.0 |
| Major hepatectomy | 185 | 46.1 |
| Two-stage hepatectomy | 36 | 9.0 |
| Postoperative chemotherapy | 204 | 50.9 |
| Disease recurrence | 333 | 83.0 |
Abbreviation: NA, data not available.
Somatic DNA Mutations
Overall, 383 (95.5%) of 401 patients had at least one genetic mutation identified in their tumors. TP53 (65.6%) was the most frequently mutated gene, followed by KRAS (48.1%), APC (47.4%), PIK3CA (15.0%), and SMAD4 (11.7%) (Figure 1A). Three patients had mutations in both KRAS and NRAS. Among 16 (4.0%) patients with BRAF mutations, 6 had V600E mutations, and the remainder had mutations in BRAF codons 466, 469, 581, or 594.
Figure 1.
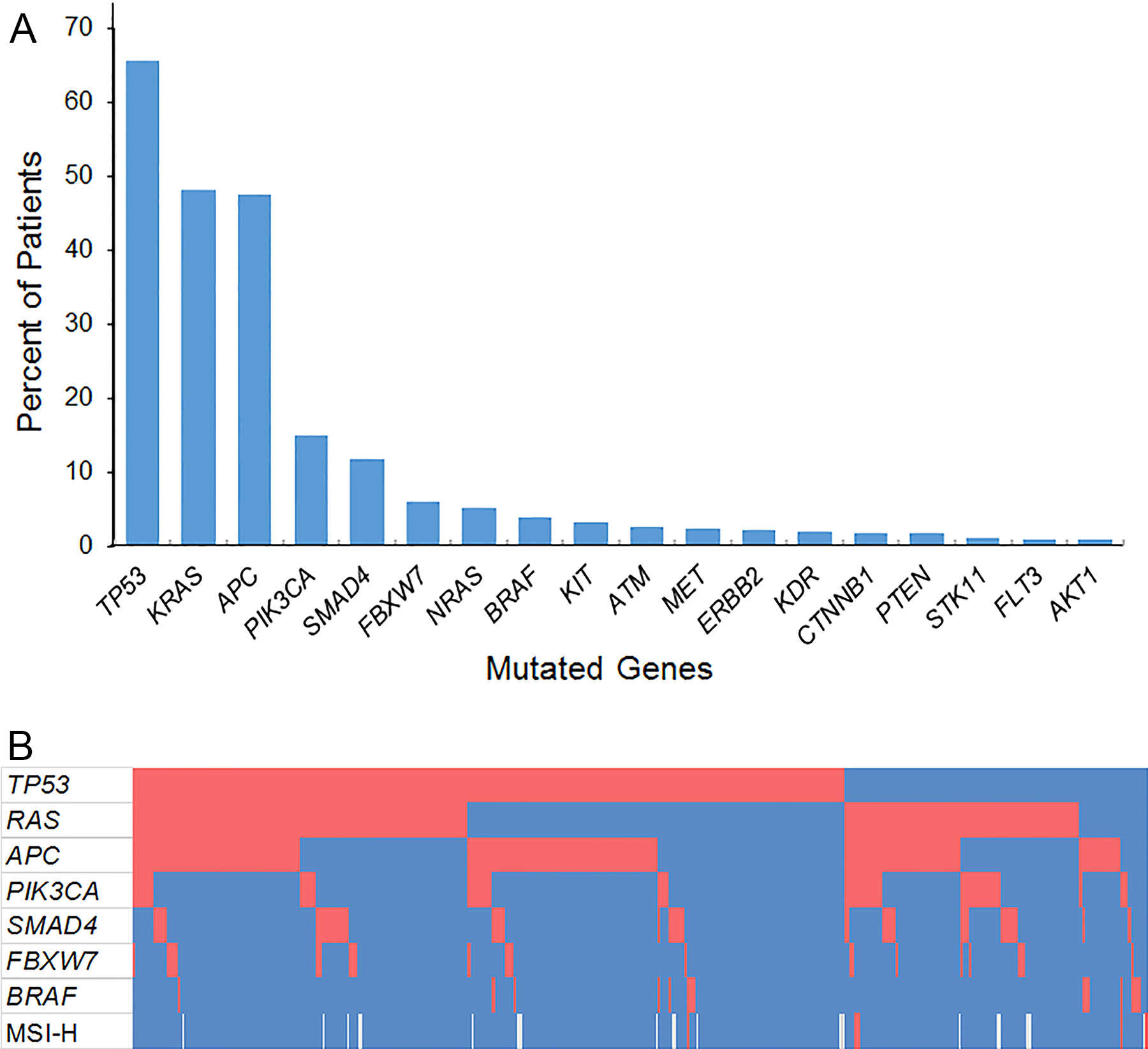
(A) Prevalence of somatic gene mutations among 401 patients undergoing resection of colorectal liver metastases. (B) Co-mutation plot showing patients in columns and genes in rows. Red blocks represent somatic mutations or deficient mismatch repair status. Blue blocks represent absence of somatic mutations or intact mismatch repair. Microsatellite instability status was not available for 20 patients, represented by white blocks. MSI-H, microsatellite instability-high.
Multigene panel testing was performed from the liver metastases in 173 patients, primary colorectal tumor in 144 patients, and lung metastases in 24 patients. Multigene panel testing from both the primary tumor and liver metastases was performed in 60 patients. Discordant results for KRAS were observed in 2 patients, NRAS in 1 patient, and TP53 in 1 patient.
The most frequent concurrent mutations with TP53 were RAS and APC (Figure 1B). Mutated TP53 and RAS were observed in 126 patients (31.4%), mutated TP53 and APC in 132 patients (32.9%), and triple mutation of TP53, RAS, and APC in 64 patients (16.0%).
Potential Prognostic Factors by Mutational Status
Correlations between potential prognostic factors and double mutation in RAS/TP53 are presented in Table 2. Compared with patients lacking double mutations, those with mutated RAS and TP53 were more likely to have primary tumors in the right colon (P = 0.006). There was not a statistically significant association between double mutation in RAS/TP53 and site of disease recurrence.
Table 2.
Comparison of Potential Prognostic Factors by TP53 and RAS Mutations, N (%)
| Variable | RAS/TP53 Co-mutation N = 126 | Absence of RAS/TP53 Co-mutation N = 275 | P |
|---|---|---|---|
| Median age, years (range) | 54 (23–79) | 56 (26–79) | 0.55 |
| Male gender | 66 (52.4) | 157 (57.1) | 0.39 |
| Primary tumor location* | 0.006 | ||
| Right | 43 (34.1) | 56 (20.6) | |
| Left | 83 (65.9) | 216 (79.4) | |
| Node-positive primary tumor† | 95 (76.6) | 179 (67.8) | 0.094 |
| Primary tumor T stage‡ | 0.91 | ||
| T1–2 | 18 (14.8) | 40 (15.2) | |
| T3–4 | 104 (85.2) | 223 (84.8) | |
| Synchronous liver metastases | 81 (64.3) | 197 (71.6) | 0.16 |
| Extrahepatic metastases | 30 (23.8) | 43 (15.6) | 0.052 |
| Median number of liver metastases (range) | 2 (1–17) | 2 (1–40) | 0.75 |
| Median size of liver metastases, cm (range) | 2.4 (0.0–11.0) | 2.7 (0.0–18.0) | 0.10 |
| R1 resection margin | 31 (24.6) | 76 (27.6) | 0.55 |
| Major hepatectomy | 53 (42.1) | 132 (48.0) | 0.28 |
| Two-stage hepatectomy | 11 (8.7) | 25 (9.1) | 1.00 |
| Disease recurrence | 115 (91.3) | 220 (80) | 0.005 |
| Recurrence site | 0.30 | ||
| Liver | 38 (33.0) | 76 (34.5) | |
| Lungs | 40 (34.8) | 57 (25.9) | |
| Peritoneum | 6 (5.2) | 20 (9.1) | |
| Liver and lungs | 8 (7.0) | 15 (6.8) |
Data not available in 3 patients.
Data not available in 13 patients.
Data not available in 16 patients.
Survival by Mutational Status
RAS mutations were associated with worse median and 5-year OS rates of 48 months and 34.1%, compared with 71 months and 61.6% for RAS wild-type (P < 0.001, Figure 2A). When considered in isolation, TP53 mutations were not associated with OS, with median OS rates of 55 and 62 months, with and without TP53 mutations, respectively (P = 0.27, Figure 2B).
Figure 2.
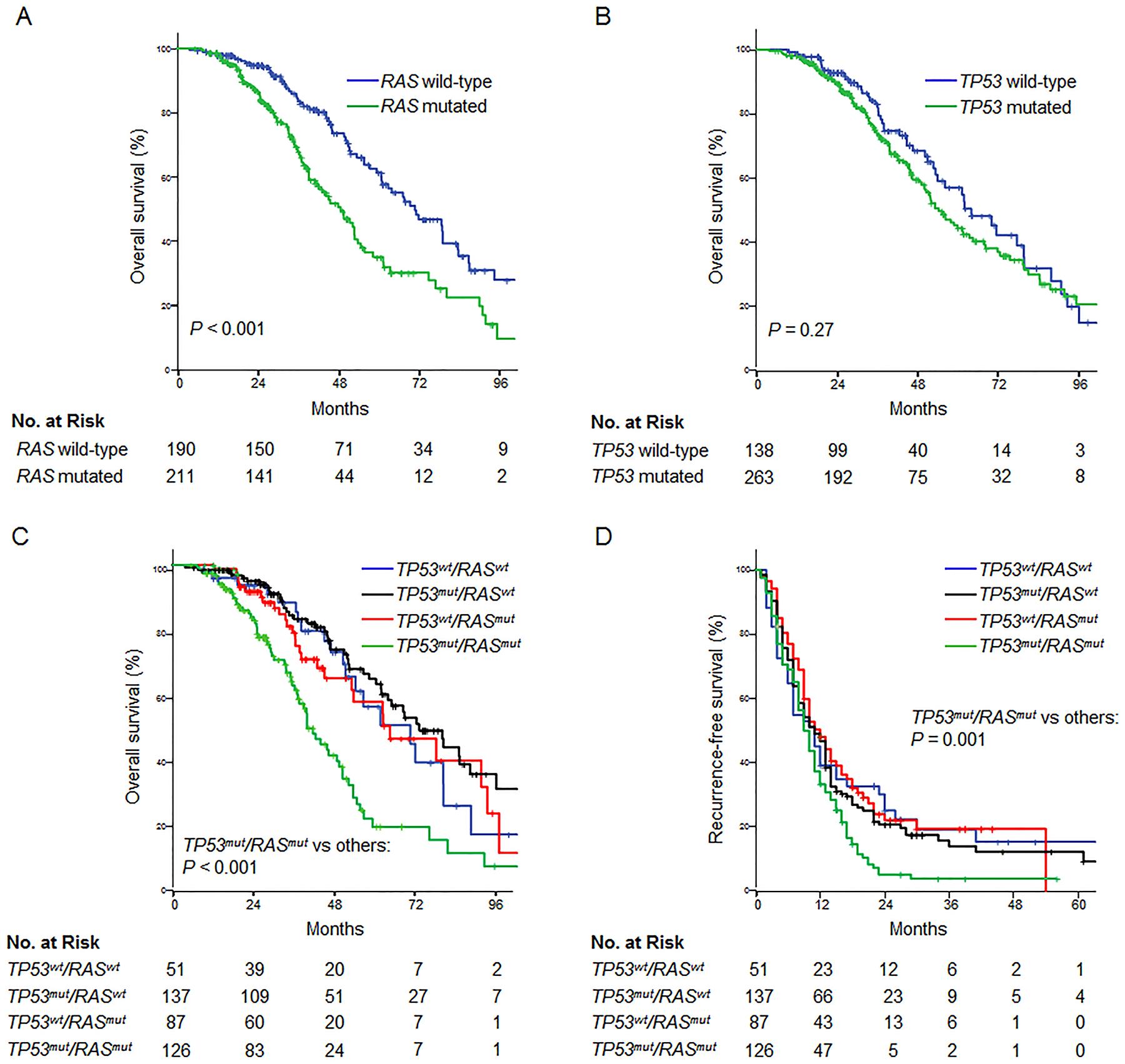
Relationship between RAS and TP53 mutations and overall survival after resection of colorectal liver metastases. Analysis of the entire patient cohort demonstrated a significant association between overall survival and RAS mutations (A) but not TP53 mutations (B). Stratification by both mutations revealed that TP53 mutations were significantly associated with overall survival among patients with concomitant RAS mutations (C). Similarly, the negative prognostic effect of RAS mutations was limited to patients with concurrent TP53 mutations. Recurrence-free survival was decreased in patients with double mutation in RAS/TP53 (D). Wt, wild-type; mut, mutated.
Among patients with co-mutated RAS, TP53 mutations were associated with median and 5-year OS rates of 41 months and 20.6%, compared with 62 months and 55.7% for TP53 wild-type (P = 0.003, Figure 2C). The negative prognostic effect of mutated RAS was restricted to patients with concurrent TP53 mutations. RFS was also shorter in patients with co-mutations in RAS and TP53, with median and 2-year RFS rates of 9 months and 5.2%, compared with 11 months and 23.6% without co-mutations (P = 0.001, Figure 2D).
After excluding the 20 patients (5.0%) who received anti-EGFR therapy before or after liver resection, the poor prognostic effect of concurrent mutations in RAS and TP53 remained significant for overall survival, with P-value of < 0.001.
In multivariable analysis, factors independently associated with OS were size of liver metastases ≥ 3 cm and double mutation in RAS/TP53 (Table 3).
Table 3.
Multivariable Overall Survival (OS) Analysis of 401 Patients Undergoing Resection of Colorectal Liver Metastases (CLM)
| Variable | Median OS (months) | 5-year OS (%) | Univariable P | Multivariable P | HR (95% CI) |
|---|---|---|---|---|---|
| Age, years | ‒ | ‒ | 0.79 | ||
| Gender | 0.037 | 0.42 | |||
| Female | 51 | 41.3 | |||
| Male | 63 | 54.9 | |||
| Primary tumor location* | 0.004 | 0.27 | |||
| Right | 45 | 28.9 | |||
| Left | 62 | 54.0 | |||
| Primary tumor lymph node status† | 0.009 | 0.084 | |||
| Positive | 58 | 49.6 | |||
| Negative | 72 | 54.2 | |||
| Primary tumor T stage‡ | 0.006 | 0.10 | |||
| T1–2 | 80 | 72.9 | |||
| T3–4 | 56 | 46.3 | |||
| Synchronous CLM | 0.90 | ||||
| Yes | 59 | 49.9 | |||
| No | 56 | 47.5 | |||
| Extrahepatic metastases | 0.76 | ||||
| Yes | 53 | 41.4 | |||
| No | 60 | 50.1 | |||
| Number of CLM | 0.005 | 0.30 | |||
| Solitary | 77 | 58.9 | |||
| Multiple | 51 | 43.0 | |||
| Size of CLM | 0.012 | 0.006 | 1.66 (1.16‒2.38) | ||
| ≥ 3 cm | 50 | 45.2 | |||
| < 3 cm | 60 | 50.4 | |||
| Surgical margin | 0.005 | 0.060 | |||
| Positive | 50 | 38.9 | |||
| Negative | 61 | 52.4 | |||
| Major hepatectomy | 0.40 | ||||
| Yes | 56 | 47.7 | |||
| No | 59 | 49.9 | |||
| Two-stage hepatectomy | 0.61 | ||||
| Yes | 50 | 33.5 | |||
| No | 59 | 50.0 | |||
| RAS and TP53 status | < 0.001 | ||||
| RASwt/TP53wt | 70 | 56.9 | Ref | Ref | |
| RASmut/TP53mut | 41 | 20.6 | 0.002 | 2.62 (1.41–4.87) | |
| RASmut/TP53wt | 62 | 55.7 | 0.79 | ||
| RASwt/TP53mut | 72 | 64.0 | 0.88 | ||
| Double mutation in APC and PIK3CA | 0.033 | 0.073 | |||
| Yes (N = 32) | 48 | 12.6 | |||
| No (N = 369) | 61 | 50.7 |
Abbreviations: HR, hazard ratio; CI, confidence interval; wt, wild-type; mut, mutated; Ref, reference variable.
Data not available in 3 patients.
Data not available in 13 patients.
Data not available in 16 patients.
Distribution and Classification of TP53 Mutations
Among 263 patients with TP53 mutations, 9 patients harbored two distinct TP53 mutations. Overall, 202 missense, 68 truncating (nonsense, splice site, or frameshift), and 2 in-frame deletion TP53 mutations were identified. Among the missense mutations, 121 mutations (59.9%) resulted in amino acid substitutions at six residues established in the literature as hotspots most frequently mutated in human cancers, R175, G245, R248, R249, R273, and R282 (Figure 3A).
Figure 3.
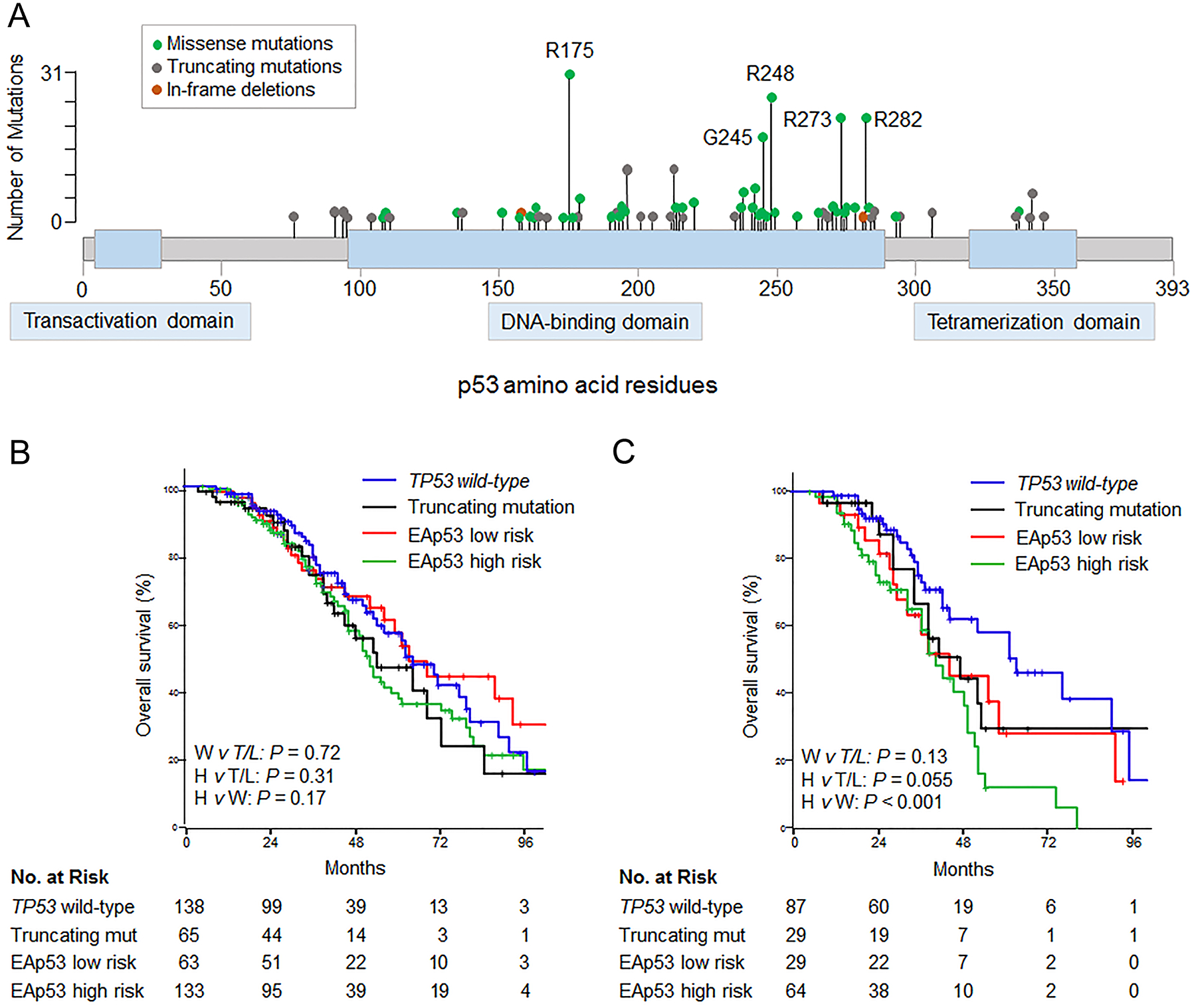
(A) Lolliplots showing distribution of somatic mutations in TP53 in 263 patients undergoing resection of colorectal liver metastases. (B) Overall survival by type of TP53 mutation (n = 399). Two patients with TP53 in-frame deletions were omitted from analysis. (C) Overall survival among RAS mutated patients, by type of TP53 mutation (n = 209). EAp53, Evolutionary Action of p53; W, TP53 wild-type; T/L, truncating or EAp53 low risk mutation; H, EAp53 high risk mutation; mut, mutation.
Survival analysis was performed after classifying TP53 mutations as truncating or missense, and further stratifying missense mutations as high or low risk according to the Evolutionary Action score (EAp53). Median OS rates were not significantly different between TP53 wild-type, truncating, EAp53 low, and EAp53 high risk mutations (wild-type: 62 months vs truncating: 54 months vs low risk: 63 months vs high risk: 52 months; Figure 3B). In patients with co-mutated RAS, OS was shorter with EAp53 high risk mutations, with median and 5-year OS rates of 41 months and 12.2%, compared with 62 months and 55.7% for TP53 wild-type (P < 0.001, Figure 3C). Median and 5-year OS rates were similar between EAp53 low risk (45 months and 28.4%) and truncating mutations (42 months and 33.0%, P = 0.71).
DISCUSSION
High-throughput genomic analysis is elucidating the molecular complexity underlying heterogeneous outcomes of patients with metastatic colorectal cancer. MMR status, KRAS, and BRAF are currently used as predictive and prognostic biomarkers.5, 7, 10 In this study, we analyzed results of next-generation sequencing of 50 cancer-related genes in patients undergoing CLM resection. TP53 mutations were the most frequent somatic gene mutations and associated with poor OS and RFS in the subset of patients with co-mutated RAS. Similarly, RAS mutations correlated with worse prognosis only in patients with co-occurring TP53 mutations. Three-quarters of TP53 mutations were missense, and when stratified by the EA score, high risk mutations were associated with 5-year OS of only 12.2% after liver resection.
According to the current study, the most frequent somatic gene mutation in patients with CLM is TP53, followed by KRAS and APC. These results are consistent with the distribution of molecular alterations identified in metastatic colorectal cancer patients screened for targeted therapy trials.24 Whole genome sequencing of primary colorectal cancers by The Cancer Genome Atlas identified 32 somatic recurrently mutated genes, including RAS and TP53.25 Only 1.3% of patients undergoing CLM resection had MSI-high tumors, reflecting the low frequency of MSI in metastatic colorectal cancer. Similarly, only 4.0% had BRAF-mutated tumors, and less than half were V600E mutations. As shown by previous reports, BRAF-mutated tumors are associated with peritoneal metastases and rarely liver-only metastases.26
On multivariable analysis, double mutation in RAS/TP53 was independently associated with worse OS, with a HR of 2.62. Prior studies have shown discordant results on the impact of TP53 mutations on colorectal cancer prognosis and response to systemic therapy.12, 27, 28 However, these studies did not analyze TP53 in the context of RAS. In this report, the negative prognostic effects of RAS and TP53 mutations were limited to tumors harboring both mutations, suggesting cooperativity between the two mutated genes. Cooperation between co-occurring genetic events is an important factor in cancer progression. Concurrent mutations in RAS and TP53 have been shown to drive carcinogenesis in vitro and in animal models.14, 15 In locally advanced rectal cancer, combined KRAS and TP53 mutations promote resistance to neoadjuvant chemoradiation.29 We previously showed that double mutation of APC and PIK3CA predicted poor survival in patients with CLM.30 In the present study, double APC-PIK3CA mutations were significantly associated with overall survival on univariable but not multivariable analysis. Thirty-two patients had double APC-PIK3CA mutations, compared to 126 patients with double RAS-TP53 mutations.
Co-mutations in RAS and TP53 were associated with primary tumor location in the right colon, suggesting their potential biologic significance. The embryologic origin of colorectal cancers from midgut or hindgut has emerged as an important biologic and prognostic factor.31, 32 Co-mutations in RAS and TP53 were associated with a trend toward higher rate of extrahepatic metastases (23.8% with co-mutations vs 15.6% without, P = 0.052), which may have not reached statistical significance due to the small number of patients with extrahepatic metastases. Given the heterogeneity of systemic therapy regimens administered, we did not evaluate the effect of co-mutations on response and resistance to systemic therapy. All of the patients in this study underwent CLM resection, and our practice is to consider liver resection for patients who do not progress on chemotherapy.
This report demonstrates that in patients undergoing CLM resection, most TP53 mutations are missense and occur at established TP53 hotspots, consistent with previously published studies.33 To identify potentially non-deleterious TP53 mutations, we classified mutations as truncating and missense, and further stratified missense mutations by EAp53, which is based upon the evolutionary importance of a mutation site and the relative odds of an amino acid substitution.16 EAp53 was previously shown to stratify outcomes in head and neck cancer.23 The current report demonstrates that EAp53 is also prognostic in CLM, highlighting the applicability of the EA model across tumor types. Patients with co-occurring RAS and EAp53 high risk, but not low risk, mutations had significantly worse OS than patients with TP53 wild-type tumors.
This study has several limitations. Patients undergoing CLM resection before 2012 did not initially undergo multigene panel testing, which was performed in these earlier patients from archived tumor or repeat hepatectomy specimens. In most patients, mutations were determined from DNA sequencing of either primary tumor or metastases samples. However, among 60 patients who underwent DNA sequencing from both the primary tumor and liver metastases, a high mutational concordance rate of 93.3% was observed for RAS and TP53, consistent with prior studies.34 These results suggest that in most patients, multigene panel testing from either the primary tumor or metastases is sufficient, and additional biopsies for molecular analyses are not necessary.
In conclusion, double mutation in RAS/TP53 is a strong independent predictor of worse survival after CLM resection. Five-year OS after resection of CLM with concurrent RAS and high EAp53 mutations is only 12.2%, and future studies are needed to validate these results. On the other hand, patients with traditionally poor prognostic factors, such as multiple liver metastases and extrahepatic disease, can be considered for surgery if their mutational status is favorable. Further investigations are needed to determine the mechanistic and downstream effects of co-occurring RAS and TP53 mutations in CLM.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2.Abdalla EK, Vauthey JN, Ellis LM, et al. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg 2004; 239(6):818–25; discussion 825–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passot G, Denbo JW, Yamashita S, et al. Is hepatectomy justified for patients with RAS mutant colorectal liver metastases? An analysis of 524 patients undergoing curative liver resection. Surgery 2017; 161(2):332–340. [DOI] [PubMed] [Google Scholar]
- 4.Loupakis F, Cremolini C, Masi G, et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med 2014; 371(17):1609–18. [DOI] [PubMed] [Google Scholar]
- 5.de Rosa N, Rodriguez-Bigas MA, Chang GJ, et al. DNA Mismatch Repair Deficiency in Rectal Cancer: Benchmarking Its Impact on Prognosis, Neoadjuvant Response Prediction, and Clinical Cancer Genetics. J Clin Oncol 2016; 34(25):3039–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haddad R, Ogilvie RT, Croitoru M, et al. Microsatellite instability as a prognostic factor in resected colorectal cancer liver metastases. Ann Surg Oncol 2004; 11(11):977–82. [DOI] [PubMed] [Google Scholar]
- 7.Schirripa M, Bergamo F, Cremolini C, et al. BRAF and RAS mutations as prognostic factors in metastatic colorectal cancer patients undergoing liver resection. Br J Cancer 2015; 112(12):1921–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vauthey JN, Zimmitti G, Kopetz SE, et al. RAS mutation status predicts survival and patterns of recurrence in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg 2013; 258(4):619–26; discussion 626–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kemeny NE, Chou JF, Capanu M, et al. KRAS mutation influences recurrence patterns in patients undergoing hepatic resection of colorectal metastases. Cancer 2014; 120(24):3965–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brudvik KW, Mise Y, Chung MH, et al. RAS Mutation Predicts Positive Resection Margins and Narrower Resection Margins in Patients Undergoing Resection of Colorectal Liver Metastases. Ann Surg Oncol 2016; 23(8):2635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conlin A, Smith G, Carey FA, et al. The prognostic significance of K-ras, p53, and APC mutations in colorectal carcinoma. Gut 2005; 54(9):1283–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosty C, Chazal M, Etienne MC, et al. Determination of microsatellite instability, p53 and K-RAS mutations in hepatic metastases from patients with colorectal cancer: relationship with response to 5-fluorouracil and survival. Int J Cancer 2001; 95(3):162–7. [DOI] [PubMed] [Google Scholar]
- 13.Pilat N, Grunberger T, Langle F, et al. Assessing the TP53 marker type in patients treated with or without neoadjuvant chemotherapy for resectable colorectal liver metastases: a p53 Research Group study. Eur J Surg Oncol 2015; 41(5):683–9. [DOI] [PubMed] [Google Scholar]
- 14.McMurray HR, Sampson ER, Compitello G, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature 2008; 453(7198):1112–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parada LF, Land H, Weinberg RA, et al. Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature 1984; 312(5995):649–51. [DOI] [PubMed] [Google Scholar]
- 16.Katsonis P, Lichtarge O. A formal perturbation equation between genotype and phenotype determines the Evolutionary Action of protein-coding variations on fitness. Genome Res 2014; 24(12):2050–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couinaud C [Liver lobes and segments: notes on the anatomical architecture and surgery of the liver]. Presse Med 1954; 62(33):709–12. [PubMed] [Google Scholar]
- 18.Edge SB, American Joint Committee on Cancer., American Cancer Society. AJCC cancer staging handbook : from the AJCC cancer staging manual. 7th ed New York: Springer, 2010. [DOI] [PubMed] [Google Scholar]
- 19.Charnsangavej C, Clary B, Fong Y, et al. Selection of patients for resection of hepatic colorectal metastases: expert consensus statement. Ann Surg Oncol 2006; 13(10):1261–8. [DOI] [PubMed] [Google Scholar]
- 20.Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol 2015; 33(25):2753–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2(5):401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neskey DM, Osman AA, Ow TJ, et al. Evolutionary Action Score of TP53 Identifies High-Risk Mutations Associated with Decreased Survival and Increased Distant Metastases in Head and Neck Cancer. Cancer Res 2015; 75(7):1527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overman MJ, Morris V, Kee B, et al. Utility of a molecular prescreening program in advanced colorectal cancer for enrollment on biomarker-selected clinical trials. Ann Oncol 2016; 27(6):1068–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas N Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487(7407):330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yaeger R, Cercek A, Chou JF, et al. BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer 2014; 120(15):2316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bazan V, Agnese V, Corsale S, et al. Specific TP53 and/or Ki-ras mutations as independent predictors of clinical outcome in sporadic colorectal adenocarcinomas: results of a 5-year Gruppo Oncologico dell’Italia Meridionale (GOIM) prospective study. Ann Oncol 2005; 16 Suppl 4:iv50–55. [DOI] [PubMed] [Google Scholar]
- 28.Ince WL, Jubb AM, Holden SN, et al. Association of k-ras, b-raf, and p53 status with the treatment effect of bevacizumab. J Natl Cancer Inst 2005; 97(13):981–9. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Aguilar J, Chen Z, Smith DD, et al. Identification of a biomarker profile associated with resistance to neoadjuvant chemoradiation therapy in rectal cancer. Ann Surg 2011; 254(3):486–92; discussion 492–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamashita S, Chun YS, Kopetz SE, et al. APC and PIK3CA Mutational Cooperativity Predicts Pathologic Response and Survival in Patients Undergoing Resection for Colorectal Liver Metastases. Ann Surg 2017. [DOI] [PubMed] [Google Scholar]
- 31.Loupakis F, Yang D, Yau L, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst 2015; 107(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita S, Brudvik KW, Kopetz SE, et al. Embryonic Origin of Primary Colon Cancer Predicts Pathologic Response and Survival in Patients Undergoing Resection for Colon Cancer Liver Metastases. Ann Surg 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naccarati A, Polakova V, Pardini B, et al. Mutations and polymorphisms in TP53 gene--an overview on the role in colorectal cancer. Mutagenesis 2012; 27(2):211–8. [DOI] [PubMed] [Google Scholar]
- 34.Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol 2012; 30(24):2956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]


