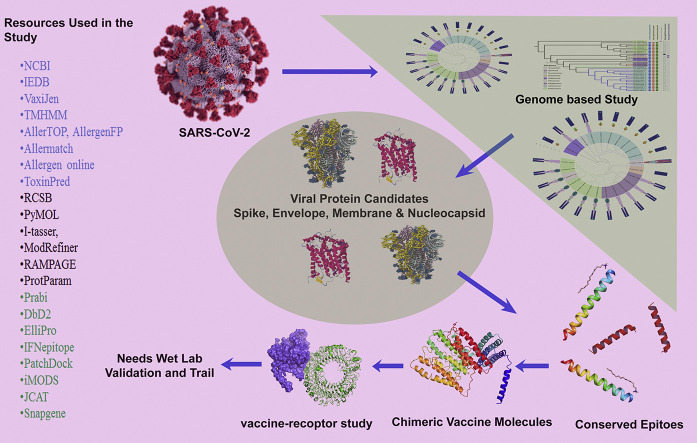
Abstract
The present study aimed to predict a novel chimeric vaccine by simultaneously targeting four major structural proteins via the establishment of ancestral relationship among different strains of coronaviruses. Conserved regions from the homologous protein sets of spike glycoprotein, membrane protein, envelope protein and nucleocapsid protein were identified through multiple sequence alignment. The phylogeny analyses of whole genome stated that four proteins reflected the close ancestral relation of SARS-CoV-2 to SARS-COV-1 and bat coronavirus. Numerous immunogenic epitopes (both T cell and B cell) were generated from the common fragments which were further ranked on the basis of antigenicity, transmembrane topology, conservancy level, toxicity and allergenicity pattern and population coverage analysis. Top putative epitopes were combined with appropriate adjuvants and linkers to construct a novel multiepitope subunit vaccine against COVID-19. The designed constructs were characterized based on physicochemical properties, allergenicity, antigenicity and solubility which revealed the superiority of construct V3 in terms safety and efficacy. Essential molecular dynamics and normal mode analysis confirmed minimal deformability of the refined model at molecular level. In addition, disulfide engineering was investigated to accelerate the stability of the protein. Molecular docking study ensured high binding affinity between construct V3 and HLA cells, as well as with different host receptors. Microbial expression and translational efficacy of the constructs were checked using pET28a(+) vector of E. coli strain K12. However, the in vivo and in vitro validation of suggested vaccine molecule might be ensured with wet lab trials using model animals for the implementation of the presented data.
Keywords: SARS-CoV-2, COVID-19, Chimeric vaccine, Evolutionary relationship, Normal mode analysis, Molecular docking, Restriction cloning
Graphical abstract
1. Introduction
Novel coronavirus named SARS-CoV-2/2019-nCoV was identified at the end of 2019 in Wuhan, a city in the Hubei province of China, causing severe pneumonia that leads to huge death cases (Wang et al., 2020). Gradually this virus emerged as a new threat to the whole world and affecting almost all parts of the world. To date, the pathogen has affected 198 countries, and thus becoming a global public health emergency. Global public health concern with pandemic notion of COVID-19 was declared on January 30th, 2020 by the World Health Organization (WHO), 2020a, World Health Organization (WHO), 2020b. Again, an adverse situation has also been announced on 13 March 2020 for increasing the infections of COVID-19 (Kunz and Minder, 2020). Till April 10, 2020, total virus affected people around the world exceeded 1,633,272 and more than 97,601 committed death, while 366,610 people fully recovered from the infection World Health Organization (WHO), 2020a, World Health Organization (WHO), 2020b). The alarming situation is that the number of confirmed cases worldwide has exceeded one million by this time. It took more than three months to reach the first 10,000 confirmed cases, while required only 12 days to detect the next 100,000 cases. The situation is getting worse in European region. Total death cases in Italy, Spain, USA, France, United Kingdom was 14,681,11,744,7847,6507 and 4313 respectively (till April 4, 2020) and this number is exacerbating day by day (World Health Organization (WHO), 2020a, World Health Organization (WHO), 2020b).
Some common clinical manifestations of COVID-19 are fever, sputum production and shortness in breath, cough, fatigue, sore throat and headache which leads to severe cases of pneumonia. A few patients also have gastrointestinal symptoms with diarrhea and vomiting (Guan et al., 2020). Though several early studies showed that the mortality rate for SARS-CoV-2 is not as high (2–3%), the latest global death rate for COVID-19 is 3.4% which indicates the increasing trends (Wu et al., 2020). The investigation of Chinese Center for Diseases Control and Prevention (2020) revealed that the prevalence of COVID-19 is more apparent in the people ages 50 years rather than the lower age groups (Jeong-ho et al., 2020). High fever and Lymphocytopenia were found more common in Covid-19, though the frequency of the patient without fever condition is also higher than in the earlier outbreaks caused by SARS-CoV (1%) and MERS-CoV (2%) (Huang et al., 2020; Chen et al., 2020).
SARS-CoV-2 is a beta-coronavirus that has a positive sense, 26–32 kb in length, single stranded RNA molecule as its genetic material and belongs to the family Coronaviridae, order Nidovirales (Hui et al., 2020). It shares genome similarity with SARS-CoV (79.5%) and bat coronavirus (96%) (Zhou et al., 2020a, Zhou et al., 2020b; Zhu et al., 2020). However, there are still obscured hypothesis regarding the vector or carrier of SARS-CoV-2, though its detection was primarily linked to Wuhan’s Huanan Seafood wholesale market (Lu et al., 2020; World Health Organization (WHO), 2020a, World Health Organization (WHO), 2020b). Though the species of SARS-CoV-1 and bat coronavirus shares sufficient sequence similarities with the COVID-19, the known way mechanism of infection to the host, and the death rate is quite different in case of the novel coronavirus. In addition, there is an evolutionary distance between SARS-CoV-1 and bat coronavirus as well as the COVID-19 (Hu et al., 2018; Wu et al., 2020; Wu 2020). Because of high sequence variability of the pathogen, many of the efforts that have been undertaken to develop vaccine against SARS-CoV remain unsuccessful (Graham et al. 2013). Therefore, there is an urgent need to develop vaccines for treatment of SARS-CoV-2 based on the understanding of actual evolutionary ancestral relationship. While some natural metabolites and traditional medication may come up with comfort and take the edge off few symptoms of COVID-19, there is no proof that existing treatment procedures can effectively combat against the diseased condition (World Health Organization (WHO), 2020a, World Health Organization (WHO), 2020b). However, inactivated or live-attenuated forms of pathogenic organisms are usually recommended for the initiation of antigen-specific responses that alleviate or reduce the possibility of host experience with secondary infections (Thompson and Staats 2011). Moreover, all of the proteins are not usually targeted for protective immunity, whereas only a few numbers of proteins are necessary depending on the microbes (Tesh et al., 2002, Li et al. 2014). Depending on sufficient antigen expression from experimental assays, traditional vaccine could take 15 years to develop, while sometimes can lead to undesirable consequences (Purcell et al. 2007; Petrovsky and Aguilar 2004).
Reverse vaccinology approach, on the other hand, is an effective way to develop vaccine against COVID-19. In this method, computation analysis towards genomic architecture of pathogenic candidate could predict the antigens of pathogens without the prerequisite to culture the pathogens in lab condition. Although, few pathogens that challenge to develop effective vaccines so far may become possible through such approach (Rappuoli 2000) which initiates a huge move in the development of vaccine against the deadly pathogens. The strategy included the comprehensive utilization of bioinformatics algorithm or tools to develop epitope based vaccine molecules, though further validation and experimental procedures are also needed (Moxon et al., 2019). In addition, peptide based subunit vaccines are biologically safer due to the absence of continuous in vitro culture during the production period, and also implies an appropriate activation of immune responses (Purcell et al. 2007; Dudek et al., 2010). Such immunoinformatic approaches have already been employed by the researchers to design vaccines against a number of deadly pathogens including Ebola virus (Khan et al. 2015), HIV (Pandey et al. 2018), Areanaviruses (Azim et al. 2019a), Marburgvirus (Hasan et al. 2019a), Norwalk virus (Azim et al. 2019b), Nipah virus (Saha et al. 2017), Influenza virus (Hasan et al. 2019b) and so on. At present, a suitable peptide vaccine to combat COVID-19 is urgently necessary that could efficiently generate enough immune response to destroy the virus. Hence, the study was designed to develop a chimeric recombinant vaccine against SARS-CoV-2 by targeting four major structural proteins of the pathogen, while revealing the evolutionary history of different species of coronavirus based on whole genome and protein domain-based phylogeny.
2. Materials and methods
2.1. Data acquisition
Complete Genomes of the COVID-19 and other coronaviruses were retrieved from the NCBI (https://www.ncbi.nlm.nih.gov/), using the keyword ‘coronavirus’ and the search option ‘nucleotide’. A total 61 complete genomes were retrieved, with unique identity (File S1). Protein sequence of the spike, envelope, membrane and nucleocapsid were also retrieved from the corresponding genome sequences found in NCBI (File S1).
2.2. Phylogeny construction and visualization
The complete genome sequences of coronaviruses and the sequences of viral proteins, such as spike glycoprotein, envelope, membrane and nucleocapsid were employed to construct different phylogenetic trees. Multiple sequence alignment (MSA) of the complete genome and protein sequences were performed using MAFFT v7.310 (Katoh and Standley 2013) tool. For the whole genome alignment, we used MAFFT Auto algorithm, while for the protein sequences alignment, MAFFT G-INS-I algorithm was used using default parameters. Next, alignment was visualized using the JalView-2.11 (Waterhouse et al. 2009). Alignment position with more than 50% gaps was pruned from coronavirus genome using Phyutility 2.2.6 program (Smith and Dunn 2008). Again, more than 20% gaps from the spike protein alignment were removed. PartitionFinder-2.1.1 (Lanfear et al., 2016) indicated the best fit substitution model of the completed genome sequences and the protein sequences. The phylogeny of the whole genome sequences of coronavirus was constructed using both the Maximum Likelihood Method and Bayesian Method. RAxML version 8.2.11 (Stamatakis 2014) with the substitution model GTRGAMMAI was used using 1000 rapid bootstrap replicates. MrBayes version 3.2.6 (Ronquist et al. 2012) with INVGAMMA model was used for the corona virus genomes. Phylogenetic analyses of four different protein sequences were performed by using RAxML-8.2.11 tool. For spike and nucleocapsid proteins, we found PROTGAMMAIWAG and PROTGAMMAIWAG as the best fit model, respectively. Again, PROTGAMMAWAG was the best fit model of evolution for both the membrane and envelope proteins. For the retrieval of the domain sequences of the stated protein sequences, InterPro database (https://www.ebi.ac.uk/interpro/) was utilized. Finally, the Interactive Tree of Life (iTOL; EMBL, Heidelberg, Germany) was used for the visualization of the phylogenetic trees. All the trees were rooted in the midpoint.
2.3. Identification of conserved regions as vaccine target
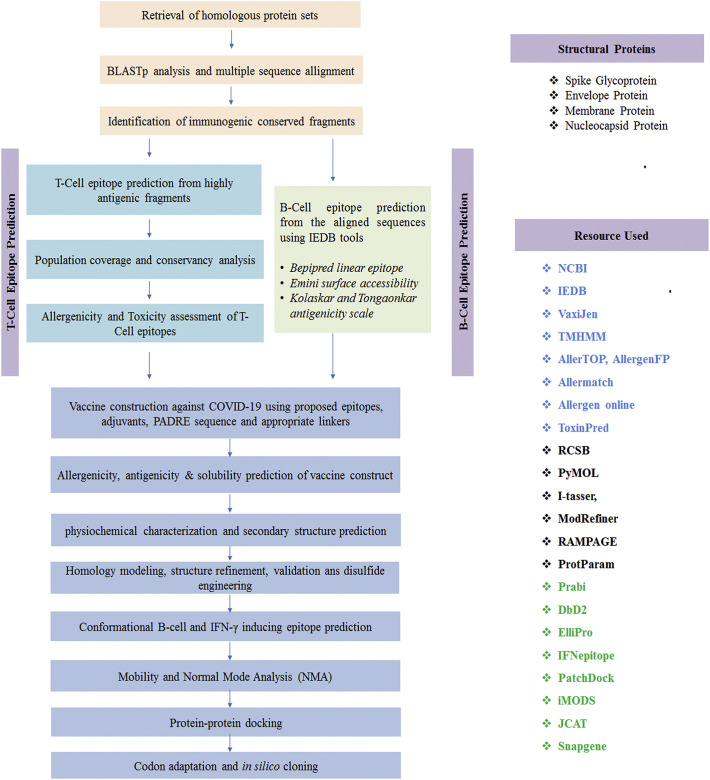
In the present study, reverse vaccinology technique was utilized to model a novel multiepitope subunit vaccine against 2019-nCoV. The scheme in Fig. 1 represents the complete methodology that has been adopted to develop the final vaccine construct. Among 496 proteins (available in the NCBI database) from different strains of novel corona virus, four structural proteins, i.e. spike glycoprotein, membrane glycoprotein, envelope protein and nucleocapsid protein, were prioritized for further investigation (File S2). After sequence retrieval from NCBI, the sequences were subjected to BLASTp analysis to find out the homologous protein sequences. Multiple sequence alignment was done by using Clustal Omega to identify the conserved regions (Sievers and Higgins 2014).The topology of each conserved regions were predicted by TMHMM Server v.2.0 (http://www.cbs.dtu.dk/services/TMHMM/), while the antigenicity of the conserved regions was determined by VaxiJen v2.0 (Doytchinova and Flower 2007a).
Fig. 1.
Flow chart summarizing the protocol of multi-epitope subunit vaccine design against SARS-CoV-2 through reverse vaccinology approach.
2.4. T-cell epitope prediction, transmembrane topology screening and antigenicity analysis
Only the common fragments were used for T-Cell epitopes enumeration via T-Cell epitope prediction server of IEDB (http://tools.iedb.org/main/tcell/) (Vita et al., 2015). Again, TMHMM server was utilized for the prediction of transmembrane topology of predicted MHC-I and MHC-II binding peptides followed by antigenicity scoring via VaxiJen v2.0 server (Krogh et al. 2001; Doytchinova and Flower 2007b). The epitopes which have antigenic potency were picked and used for preceding analysis.
2.5. Conservancy analysis and toxicity profiling of the predicted epitopes
The level of conservancy scrutinizes the ability of epitope candidates to impart capacious spectrum immunity. Homologous sequence sets of the chosen antigenic proteins were retrieved from the NCBI database by utilizing BLASTp tool. Later, conservancy analysis tool (http://tools.iedb.org/conservancy/) in IEDB was used to demonstrate the conservancy level of the predicted epitopes among different viral strains. The toxicity of non-allergenic epitopes was enumerated by using ToxinPred server (Gupta, 2013).
2.6. Population coverage and allergenicity pattern of putative epitopes
Among different ethnic societies and geographic spaces, the HLA distribution varies around the world. Population coverage study was conducted by using IEDB population coverage calculation server (Vita et al., 2015). To check the allergenicity of the proposed epitopes, four distinct servers i.e. AllergenFP (Dimitrov et al. 2014b), AllerTOP (Dimitrov et al., 2014a, Dimitrov et al., 2014b), Allermatch (Fiers et al., and Allergen Online (http://www.allergenonline.org/) servers were utilized.
2.7. Identification of B-cell epitopes and conservancy analysis
Three different algorithms i.e. Bepipred Linear Epitope Prediction 2.0 (Jespersen et al. 2017), Emini surface accessibility prediction (Emini et al. 1985) and Kolaskar and Tongaonkar antigenicity scale (Kolaskar and Tongaonkar 1990) from IEDB predicted the potential B-Cell epitopes within conserved fragments of the chosen viral proteins. To cross check the epitope conservancy, we used 30 strains from 30 different countries using World Health Organization’s most infected countries’ list. We demonstrated the conservancy for both B-cell and T-cell epitopes used to design the final vaccine construct in this study.
2.8. Construction of vaccine molecules and prediction of allergenicity, antigenicity and solubility of the constructs
Top CTL, HTL and B cell epitopes were compiled to design the final vaccine constructs in the study. Each vaccine constructs commenced with an adjuvant followed by top CTL epitopes, HTL epitopes and BCL epitopes respectively. For construction of novel corona vaccine, the chosen adjuvants i.e. L7/L12 ribosomal protein, beta defensin (a 45 mer peptide) and HABA protein (M. tuberculosis, accession number: AGV15514.1) were used (Rana and Akhter, 2016). Several linkers such as EAAAK, GGGS, GPGPG and KK in association with PADRE sequence were incorporated to construct fruitful vaccine sequences against COVID-19. The constructed vaccines were then analyzed whether they are non-allergenic by utilizing the following tool named Algpred (Azim et al., 2019a, Azim et al., 2019b). The most potential vaccine among the three constructs was then determined by assessing the antigenicity and solubility of the vaccines via VaxiJen v2.0 (Doytchinova and Flower 2007b) and Proso II server (Smialowski et al., 2007), respectively.
2.9. Physicochemical characterization and secondary structure analysis
ProtParam tool (https://web.expasy.org/protparam/), provided by ExPASy server (Hasan et al. 2019c) was used to functionally characterize (Gasteiger et al., 2005) the vaccine constructs. The studied functional properties were isoelectric pH, molecular weight, aliphatic index, instability index, hydropathicity, estimated half-life, GRAVY values and other physicochemical characteristics. Alpha helix, beta sheet and coil structures of the vaccine constructs were analyzed through GOR4 secondary structure prediction method using Prabi (https://npsa-prabi.ibcp.fr/). In addition, Espript 3.0 (Robert and Gouet 2014) was also used to predict the secondary structure of the stated protein sequences.
2.10. Homology modeling, structure refinement, validation and disulfide engineering
Vaccine 3D model was generated on the basis of percentage similarity between target protein and available template structures from PDB by using I-TASSER (Peng and Xu 2011). The modelled structures were further refined via FG-MD refinement server. Structure validation was performed by Ramachandran plot assessment in RAMPAGE (Hasan et al. 2019b). By utilizing DbD2 server, probable disulfide bonds were designed for the anticipated vaccine constructs (Craig and Dombkowski, 2013). The value of energy was considered <2.5, while the chi3 value for the residue screening was chosen between −87 to +97 for the operation (Hasan et al. 2019b).
2.11. Conformational B-cell and IFN-α inducing epitopes prediction
The B-cell epitopes of putative vaccine molecules were predicted via ElliPro server (http://tools.iedb.org/ellipro/) with minimum score 0.5 and maximum distance of 7 Å (Ponomarenko et al., 2004). Moreover, IFN- inducing epitopes within the vaccine were predicted using IFNepitope with motif and SVM hybrid detection strategy (Hajighahramani et al., 2017).
2.12. Molecular dynamics and normal mode analysis (NMA)
Normal mode analysis (NMA) was performed to predict the stability and large scale mobility of the vaccine protein. The iMod server determined the stability of construct V3 by comparing the essential dynamics to the normal modes of protein (Aalten et al., 1997; Wüthrich et al., 1980). It is a recommended alternative to costly atomistic simulation (Tama and Brooks 2006; Cui and Bahar 2007) and shows much quicker and efficient assessments than the typical molecular dynamics (MD) simulations tools (Prabhakar et al. 2016; Awan et al. 2017). The main-chain deformability was also predicted by measuring the efficacy of target molecule to deform at each of its residues. The motion stiffness was represented via eigenvalue, while the covariance matrix and elastic network model was also analyzed.
2.13. Protein-protein interaction study
Patchdock server was prioritized for docking between different HLA alleles and the putative vaccine molecules (Azim et al. 2020). In addition, the superior construct was also docked with different human immune receptors such as ACE 3, APN, DPP4 and TLR-8. The 3D structures of these receptors were retrieved from RCSB protein data bank. Detection of highest binding affinity between the putative vaccine molecules and the receptor was experimented based on the lowest interaction energy of the docked structure.
2.14. Codon adaptation and in silico cloning
JCAT tool was utilized for codon adaptation in order to fasten the expression of vaccine construct V3 in E. coli strain K12. For this, some restriction enzymes (i.e. BglI and BglII), Rho independent transcription termination and prokaryote ribosome-binding site were put away from the work (Grote et al., 2005). After that, the mRNA sequence of constructed V3 vaccine was ligated within BglI (401) and BglII (2187) restriction site at the C-terminal and N-terminal sites respectively. SnapGene tool was utilized for in silico restriction cloning (Solanki and Tiwari 2018).
3. Results
3.1. COVID-19 exhibits close ancestral relation to SARS-CoV-1 and bat-coronavirus
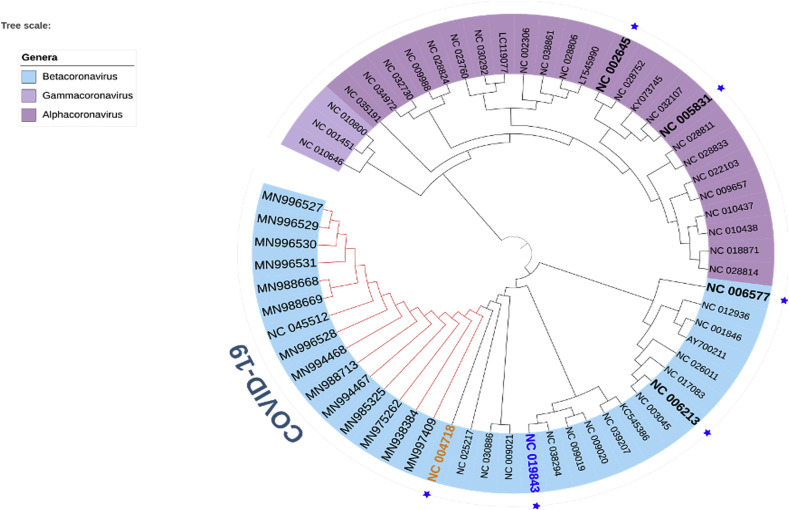
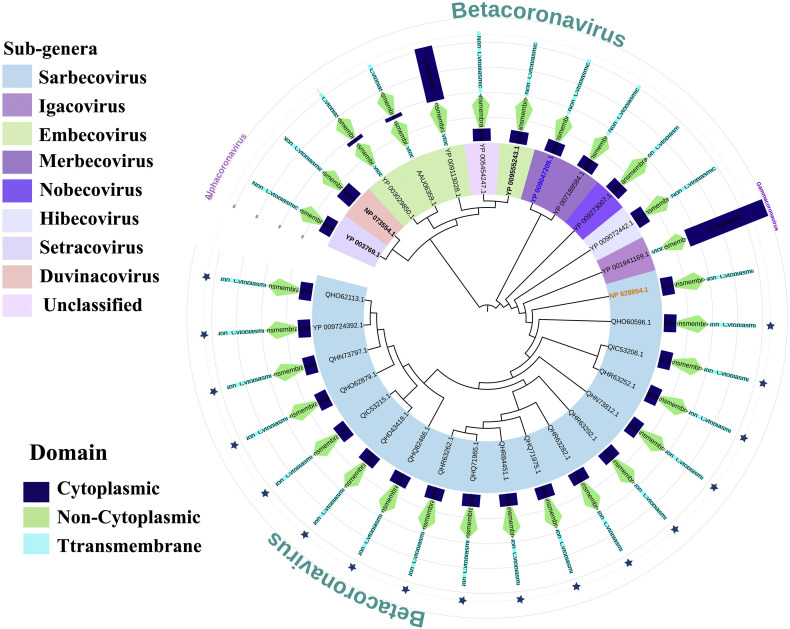
In the phylogenetic analysis, we introduced different coronavirus from three different genera: alpha-coronavirus, beta-coronavirus and gamma-coronavirus. Total 61 species of the corona virus covered 21 sub-genera (Table S1 and Fig. 2). These 61 species of corona viruses included 7 pathogenic species (Fig. 2), which are: COVID-19 or SARS-CoV-2, SARS-CoV-1 virus, MERS virus, HCoV-HKU1, HCoV-OC43, HCoV-NL63, HCoV-229E (Forni et al., 2017; Zhou et al., 2020a, Zhou et al., 2020b; Zumla et al. 2016). Among these, the first five species belong to the beta-coronavirus genera, while the last two belongs to the alpha-genera. Here, none of the members of Gammacoronavirus studied in this study were found to use human as potential host. Apart from the human coronaviruses, we introduced other coronaviruses which choose different species of bats, whale, turkey, rat, mink, ferret, swine, camel, rabbit, cow and others as host (Table S1).
Fig. 2.
Phylogeny of 61 species of coronaviruses. Seven pathogenic human coronaviruses have been represented by blue star and the IDs have been made bold. COVID-19 clade has been shown with red colour. SARS-CoV-1 and MERS virus have been represented by orange and blue colors, respectively. The genera have been represented on the left coloured labels. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The phylogeny of these species clearly revealed two broad clades (Fig. 2), where first large clade contains Gammacoronavirus and Alpha-coronavirus genera, while the other belongs to the beta-coronavirus. Within the beta-coronavirus clade, we found three clear divisions. HCoV-HKU1 and HCoV-OC43 have been placed in the first clade, while in the second clade we found MERS coronavirus. COVID-19 or SARS-COV-2 formed clade with SARS-COV-1 and bat beta-coronaviruses (Fig. 2), which is consistent with the previous finding (Ceraolo and Giorgi 2020). Though SARS-CoV-1 belongs to the same sub-genus as the COVID-19, the bat coronaviruses belong to two different sub-genera including Hibecovirus and Nobecovirus (File S1).
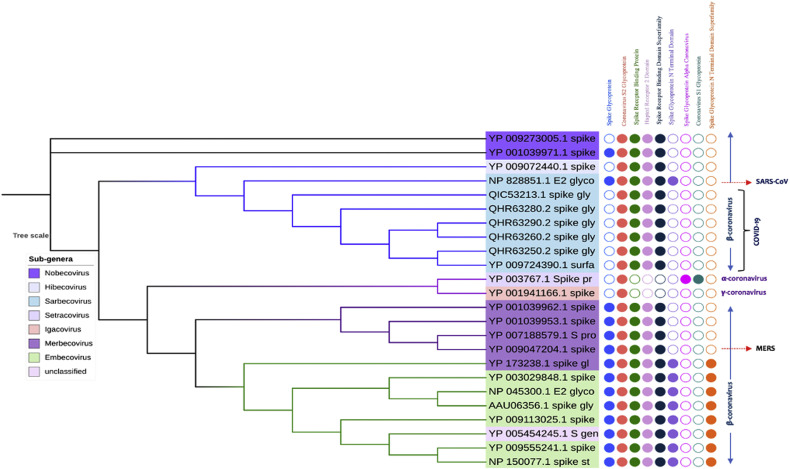
3.2. Evolution of spike proteins based on domain
Domain analysis of spike protein of coronaviruses reveals that they contain mainly one signature domains namely, coronavirus S2 glycoprotein (IPR002552), which is present in all the candidates. All other beta-coronavirus contains spike receptor binding protein (IPR018548), coronavirus spike glycoprotein hapted receptor 2 domain (IPR027400) and spike receptor binding domain superfamily (IPR036326). SARS-CoV-1 contains an extra domain, namely spike glycoprotein N-terminal domain (IPR032500), which is also present in some the sub-genera (Embecovirus) of beta-coronavirus, but not in COVID-19. One important finding in our study is that the COVID-19 candidates do not contain the domain spike glycoprotein (IPR042578), which is present in the SARS-CoV-1 (Fig. 3). The secondary structure prediction study shows a large numbers of cysteine residues which contribute to the formation of disulfide bonds within the spike protein. Most of them fall within the S1 spike protein, which is 654 amino acid long in SARS-CoV-1, while 672 amino acids long in COVID-19. The RGD motif which is conserved within the COVID-19 is present in the vicinity of the S1 protein. It exists as KGD that clearly demonstrates the mutation over the short time period. Again, the receptor binding domain and receptor binding motif analyses disclose variations within several region between the COVID19 and SARS-CoV-1 (File S2). The domain-based phylogenetic analysis reflects two main divisions, where the all the novel beta-coronavirus i.e., COVID19 form clade with the SARS-CoV-1; while other beta-coronavirus fall in another clade which further divide to give rise different sub-genera. This clearly shows that the COVID-19 exerts specific ancestral connection to the SARS-CoV-1 in terms of spike glycoproteins. Interestingly, our study also revealed close relatedness of both the SARS-CoV-1 and COVID-19 to the bat beta-coronavirus that belongs to the Hibecovirus sub-genus. However, in our study, the bat coronaviruses of Nobecovirus sub-genus did not fall into the same clade of novel coronaviruses. The phylogenetic study and MSA also revealed that, the functional portion of the spike glycoprotein domain and spike glycoprotein N-terminal domain might be lost from the COVID-19 during the course of evolution.
Fig. 3.
Phylogeny of spike glycoprotein of coronavirus. The sub-genera have been labeled in the left table. The filled and unfilled circles show the presence and absence of the domains labeled on the top.
3.3. Domain architecture and ancestral state of envelope proteins
The envelope proteins of both beta-coronavirus and Alpha-coronavirus contain only one protein domain (IPR003873) namely, Nonstructural protein NS3 or small envelope protein E (NS3/E). This domain is well conserved in coronavirus and also found in murine hepatitis virus. On the other hands, the gamma coronavirus shows the exception, which possess (IPR005296) IBV3C protein domain, which thought to be expressed from the ORF3C gene of infectious bronchitis virus (Jia and Naqi 1997) (File S1, File S2). The length of the domains for the COVID-19, SARS-CoV and MARS virus are 75, 76 and 82 amino acids, respectively. on the contrary, the length of the gamma coronavirus candidate in our study which utilizes turkey as host is 99 amino acids.
The average length of the COVID-19 envelope proteins is 75 amino acids long. The NS3/E protein domains span the whole length of the protein and possess mainly one transmembrane domain, one non-cytoplasmic domain and a cytoplasmic domain. However, some species from the sub-genera of Embecovirus shows two transmembrane domains (Fig. 4, File S1, File S2). While in our in-silico study, we found 2 transmembrane domains in SARS-CoV-1 and 2–3 transmembrane domains in MERS virus, previous experiments proved that both contains only one α-helical transmembrane domains (Nieto-Torres et al. 2011; Surya et al. 2015). Though the computational analysis of CoVID-19 envelope protein secondary structure shows 2 transmembrane domains, our domain analysis shows only one such domain in their structures. Again, the Turkey corona viruses also possess the same transmembrane, non-cytoplasmic domain and cytoplasmic domain, with a variation in the orientation. The domain-based phylogeny of the envelope proteins of novel corona viruses reveals close ancestral relationship with the SARS-CoV-1 and bat coronaviruses (Fig. 4). In spite to the previous findings, where it was found that the envelope proteins of the MERS virus and SARS-CoV-1 exerted close proximity in terms of secondary structure and functions (Surya et al. 2015). Unlike to earlier finding, we got that gamma-coronavirus candidate in our study shows close connection with both SARS-CoV-1 and COVID-19 in terms of envelope proteins.
Fig. 4.
Phylogeny of envelope proteins of coronaviruses. The sub-genera under three different genera have been shown on the left labels. The star signs represent the COVID-19 virus. 5 different pathogenic human corona viruses have been shown in bold form, including the MERS virus (blue) and SARS-CoV-1 (yellow). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
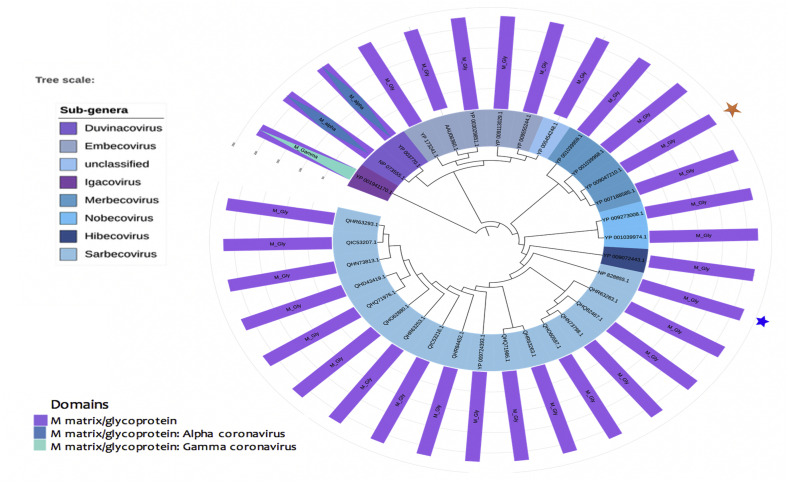
3.4. Domain architecture and phylogeny of membrane proteins
Membrane proteins of all the coronavirus mainly contain coronavirus M matrix/glycoprotein (IPR002574) domain family. However, the candidates of alpha- and gamma-coronaviruses contain M matrix/glycoprotein: Alpha-coronavirus (IPR042551) and M matrix/glycoprotein: gamma-coronavirus (IPR042550) domains, respectively. The next two domains belong to the M matrix/glycoprotein domain family. The membrane proteins of coronaviruses range from 221 to 230 amino acid long. Computational analysis of secondary structures shows some variations of COVID-19 with SARS-CoV-1 and MERS virus (File S3). SARS-CoV-2 possesses alpha helical structure in their structure, while the other two completely devoid of this structure. Again, both SARS-CoV-1 and MARS contain parallel beta sheets, while it is completely absent in the novel corona virus (File S3). The phylogenetic analysis of the membrane supports that the novel coronavirus is closely connected to the SARS-CoV-1 virus membrane protein. As well as it produced connections with bat coronaviruses of Hibecovirus and Nobecovirus sub-genus (Fig. 5).
Fig. 5.
Phylogeny of membrane proteins of coronavirus. The sub-genera under three different genera have been shown on the left labels. The blue and orange star represent the SARS-CoV1 and MERS virus, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
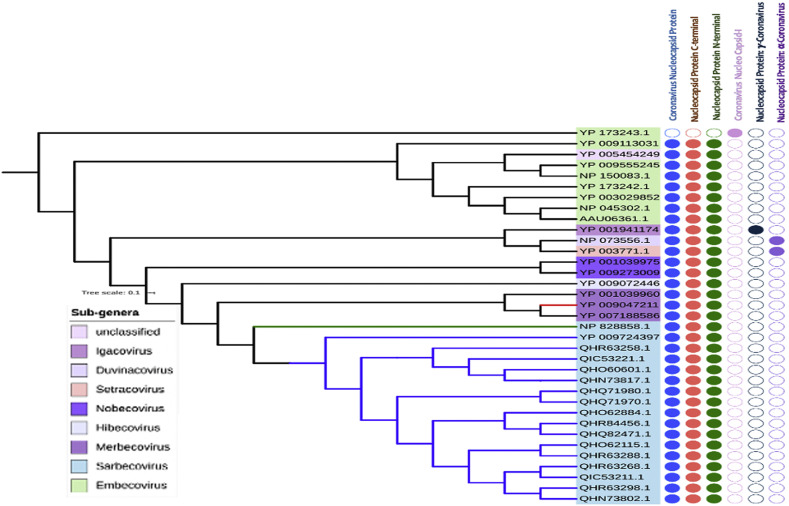
3.5. Domain-based phylogeny of nucleocapsid proteins
The length of nucleocapsid proteins of beta-coronavirus genus ranges from 410 to 450 amino acids. Three signature domains are mainly present in the nucleocapsid proteins, which are: Coronavirus Nucleocapsid protein (IPR001218), Nucleocapsid Proteins C-terminal (IPR037179) and Nucleocapsid Proteins N-terminal (IPR037195). However, in our experiment, we didn't find these domains in HCoV-HKU1 (Fig. 6); this virus belongs to Embecovirus sub-genus and contains only Coronavirus Nucleocapsid I (IPR004876) domain. The candidates of alpha- and gamma-coronavirus have their special domains. According to our domain-based phylogeny study of nucleocapsid proteins, we found the close approximation of the COVID-19 with the SARS-CoV-1, which is consistent with the findings of phylogeny of whole genome. Strikingly, unlike spike and membrane proteins, the closest homologs of COVID-19 are not only the SARS-COV-1 and bat coronaviruses (Sub-genus: Hibecovirus and Nobecovirus), but it also includes the MERS viruses and other proteins which is from the Merbecovirus sub-genus.
Fig. 6.
Phylogeny of nucleocapsid proteins of coronavirus. The sub-genera under three different genera have been shown on the left labels. The filled and unfilled circles show the presence and absence of the domains labeled on the top. COVID-19, SARS-CoV1 and MERS viruses are clades are labeled with blue, green and red colors, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Identification of conserved regions as vaccine target
A total 31, 24, 29 and 29 sequences of spike glycoprotein, membrane protein, envelope protein and nucleocapsid protein were retrieved from the NCBI database belonging to different strains of SARS-CoV-2. Following by BLASTp analysis and Multiple Sequence Alignment, two conserved regions were detected for membrane glycoprotein and nucleocapsid, while single fragments were identified for both spike glycoprotein and envelope protein (Table 1 ).Results showed that, all the conserved sequences except one from membrane glycoprotein met the criteria of default threshold standard in VaxiJen. Again, transmembrane topology scrutinizing showed that among the immunogenic conserved sequences from the corresponding proteins except spike glycoprotein met the criteria of desired exomembrane characteristics (Table 1).
Table 1.
Identified conserved regions among different homologous protein sets.
| Protein | Conserved region | Vaxijen score | Topology |
|---|---|---|---|
| Spike glycoprotein | NVYADSFVIRGDEVRQIAPGQTGKIADYNYKLPDD | 0.5471 | Inside |
| Membrane protein | LACFVLAAVYRINWITGGIAIAMACLVGLMWLSYFIASFRLFARTRSMWSFNPETNILL | 0.7352 | Outside |
| NVPLHGTILTRPLLESELVIGAVILRGHLRIAGHHLGRCDIKDLPKEI | 0.2688 | Outside | |
| Envelope protein | MYSFVSEETGTLIVNSVLLFLAFVVFLLVTLAILTALRLCAYCCNIVNVSLVKPSFYVYSRVKNLNSSRVPDLLV | 0.6025 | Outside |
| Nucleocapsid protein | MSDNGPQNQRNAPRITFGGPSDSTGSNQNGERSGARSKQRRPQGLPNNTASWFTALTQHGKEDLKFPRGQGVPINTNSSPDDQIGYYRRATRRIRGGDGKMKDLSPRWYFYYLGTGPEAGLPYGANKDGIIWVATEGALNTPKDHIGTRNPANNAAIVLQLPQGTTLPKGFYAEGSRGGSQASSRSSSRSRNS | 0.3985 | Outside |
| RNSTPGSSRGTSPARMAGNGGDAALALLLLDRLNQLESKMSGKGQQQQGQTVTKKSAAEASKKPRQKRTATKAYNVTQAFGRRGPEQTQGNFGDQELIRQGTDYKHWPQIAQFAPSASAFFGMSRIGMEVTPSGTWLTYTGAIKLDDKDPNFKDQVILLNKHIDAYKTFPPTEPKKDKKKKADETQALPQRQKKQQTVTLLPAADLDDFSKQLQQSMSSADSTQA | 0.5965 | Outside |
3.7. T-cell epitope prediction, transmembrane topology screening and antigenicity analysis
A plethora of immunogenic epitopes were generated from the conserved sequences that were able to bind with most noteworthy number of HLA cells (Table S1, Table S2, Table S3, Table S4). Top epitopes with exomembrane characteristics were ranked for each individual protein after investigating their antigenicity score and transmembrane topology (Table 2).
Table 2.
Predicted T-cell (CTL and HTL) epitopes of Spike glycoprotein, membrane protein, envelope protein and nucleocapsid protein.
| Types | Proteins | Epitope | Start | End | Vaxijen score | No. of HLAs | Conservancy |
|---|---|---|---|---|---|---|---|
| CTL epitopes | Spike glycoprotein | ADYNYKLPD | 26 | 34 | 1.3382 | 81 | 100% (47/47) |
| Membrane protein | GIAIAMACL | 18 | 26 | 1.2059 | 54 | 10.6% (5/47) | |
| LACFVLAAV | 1 | 9 | 1.1825 | 27 | 93.6% (44/47) | ||
| LVGLMWLSY | 26 | 34 | 1.0633 | 54 | 8.51% (4/47) | ||
| LACFVLAAVY | 1 | 10 | 1.0354 | 27 | 91.5% (43/47) | ||
| CLVGLMWLSY | 25 | 34 | 1.0255 | 27 | 8.51% (4/47) | ||
| LVIGAVILR | 18 | 26 | 1.1027 | 54 | 8.51% (4/47) | ||
| ELVIGAVILR | 17 | 26 | 0.9998 | 27 | 8.51% (4/47) | ||
| ESELVIGAV | 15 | 23 | 0.9872 | 54 | 100% (47/47) | ||
| IGAVILRGH | 20 | 28 | 0.9127 | 27 | 8.51% (4/47) | ||
| LESELVIGA | 14 | 22 | 0.8597 | 27 | 23.4% (11/47) | ||
| Envelope protein | VKPSFYVYS | 52 | 60 | 1.0547 | 27 | 8.33% (3/36) | |
| FLLVTLAIL | 26 | 34 | 0.9645 | 81 | 88.9% (32/36) | ||
| VVFLLVTLA | 24 | 32 | 0.9374 | 27 | 83.3% (30/36) | ||
| LAILTALRL | 31 | 39 | 0.8872 | 54 | 91.7% (33/36) | ||
| LNSSRVPDL | 65 | 73 | 0.8553 | 54 | 5.56% (2/36) | ||
| LLFLAFVVF | 18 | 26 | 0.8144 | 81 | 63.9% (23/36) | ||
| VFLLVTLAI | 25 | 33 | 0.8134 | 54 | 88.9% (32/36) | ||
| LAFVVFLLV | 21 | 29 | 0.7976 | 54 | 72.2% (26/36) | ||
| VSLVKPSFY | 49 | 57 | 0.7476 | 81 | 11.11% (4/36) | ||
| FVVFLLVTL | 23 | 31 | 0.7403 | 81 | 83.3% (30/36) | ||
| Nucleocapsid | RSGARSKQR | 32 | 40 | 1.7874 | 81 | 6.41% (5/78) | |
| Protein | DLSPRWYFY | 103 | 111 | 1.7645 | 81 | 7.69% (6/78) | |
| DGKMKDLSP | 98 | 106 | 1.7554 | 27 | 7.69% (6/78) | ||
| KMKDLSPRW | 100 | 108 | 1.7462 | 54 | 7.69% (6/78) | ||
| TQHGKEDLKF | 57 | 66 | 1.646 | 54 | 7.69% (6/78) | ||
| KLDDKDPNF | 144 | 152 | 2.6591 | 27 | 93.6% (73/78) | ||
| AIKLDDKDP | 142 | 150 | 2.167 | 27 | 8.97% (7/78) | ||
| LDDKDPNFK | 145 | 153 | 1.9433 | 54 | 93.6% (73/78) | ||
| GAIKLDDKDP | 141 | 150 | 1.9075 | 27 | 39.7% (31/78) | ||
| TQGNFGDQE | 88 | 96 | 1.8694 | 27 | 8.97% (7/78) | ||
| Spike glycoprotein | SFVIRGDEVRQIAPG | 6 | 20 | 0.5882 | 27 | 8.70% (8/92) | |
| DSFVIRGDEVRQIAP | 5 | 19 | 0.1792 | 8.70% (8/92) | |||
| Membrane glycoprotein | LACFVLAAVYRINWI | 1 | 15 | 1.2905 | 27 | 8.51% (4/47) | |
| FVLAAVYRINWITGG | 4 | 18 | 1.0230 | 27 | 8.51% (4/47) | ||
| AIAMACLVGLMWLSY | 20 | 34 | 0.9526 | 27 | 8.51% (4/47) | ||
| IAIAMACLVGLMWLS | 19 | 33 | 0.9464 | 27 | 8.51% (4/47) | ||
| ITGGIAIAMACLVGL | 15 | 29 | 0.9310 | 27 | 6.38% (3/47) | ||
| LVIGAVILRGHLRIA | 18 | 32 | 0.8769 | 27 | 8.51% (4/47) | ||
| ELVIGAVILRGHLRI | 17 | 31 | 0.7972 | 27 | 8.51% (4/47) | ||
| PLLESELVIGAVILR | 12 | 26 | 0.7261 | 27 | 8.51% (4/47) | ||
| SELVIGAVILRGHLR | 16 | 30 | 0.6768 | 27 | 8.51% (4/47) | ||
| LESELVIGAVILRGH | 14 | 28 | 0.6528 | 27 | 8.51% (4/47) | ||
| Envelope protein | VTLAILTALRLCAYC | 29 | 43 | 0.8599 | 27 | 75.0% (27/36) | |
| LAFVVFLLVTLAILT | 21 | 35 | 0.8229 | 27 | 63.9% (23/36) | ||
| LLFLAFVVFLLVTLA | 18 | 32 | 0.8122 | 27 | 58.3% (21/36) | ||
| VSLVKPSFYVYSRVK | 49 | 63 | 0.7974 | 27 | 8.33% (3/36) | ||
| VVFLLVTLAILTALR | 24 | 38 | 0.7559 | 27 | 77.8% (28/36) | ||
| VNVSLVKPSFYVYSR | 47 | 61 | 0.7513 | 27 | 8.33% (3/36) | ||
| FLAFVVFLLVTLAIL | 20 | 34 | 0.7476 | 27 | 63.9% (23/36) | ||
| LFLAFVVFLLVTLAI | 19 | 33 | 0.7471 | 27 | 61.1% (22/36) | ||
| ILTALRLCAYCCNIV | 33 | 47 | 0.7427 | 27 | 83.3% (30/36) | ||
| LVKPSFYVYSRVKNL | 51 | 65 | 0.7311 | 27 | 8.33% (3/36) | ||
| Nucleocapsid protein | DLSPRWYFYYLGTGP | 103 | 117 | 1.5180 | 27 | 7.69% (6/78) | |
| LSPRWYFYYLGTGPE | 104 | 118 | 1.3086 | 27 | 100% (78/78) | ||
| KDLSPRWYFYYLGTG | 102 | 116 | 1.2051 | 27 | 7.69% (6/78) | ||
| GGDGKMKDLSPRWYF | 96 | 110 | 1.169 | 27 | 7.69% (6/78) | ||
| GDGKMKDLSPRWYFY | 97 | 11 | 1.1013 | 27 | 7.69% (6/78) | ||
| LDDKDPNFKDQVILL | 145 | 159 | 1.4829 | 27 | 8.97% (7/78) | ||
| WLTYTGAIKLDDKDP | 136 | 150 | 1.2787 | 27 | 6.41% (5/78) | ||
| DDKDPNFKDQVILLN | 146 | 160 | 1.2508 | 27 | 8.97% (7/78) | ||
| AFFGMSRIGMEVTPS | 119 | 133 | 1.1085 | 27 | 83.3% (65/78) | ||
| DPNFKDQVILLNKHI | 149 | 163 | 1.1072 | 27 | 8.97% (7/78) |
3.8. Conservancy analysis, toxicity profiling, population coverage and allergenicity pattern of the predicted epitopes
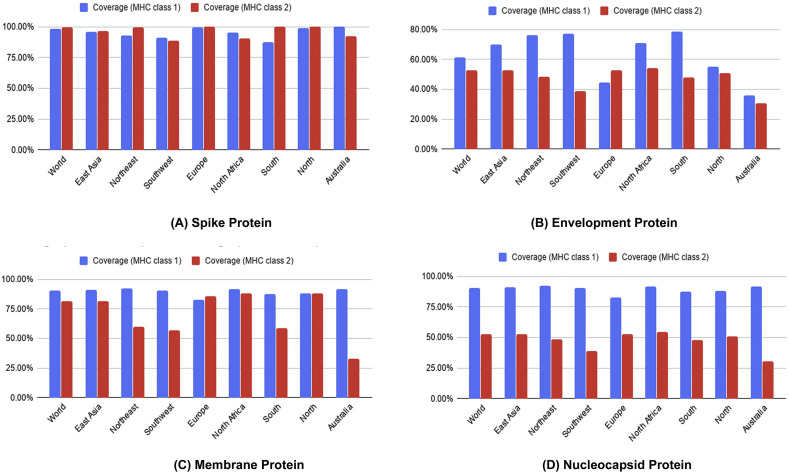
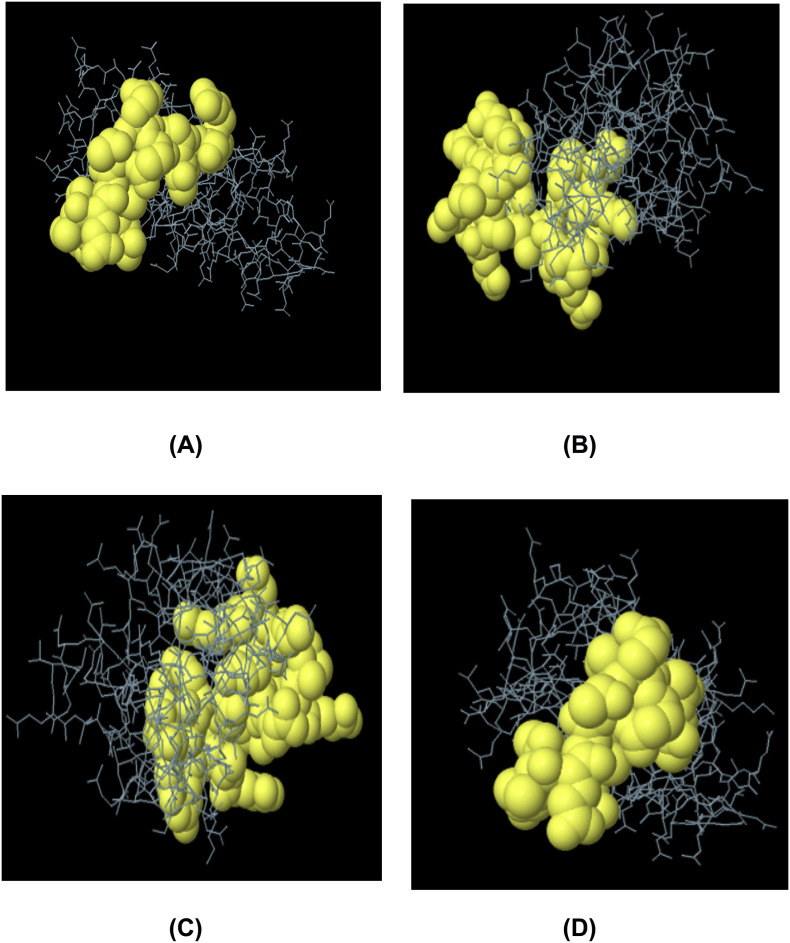
Epitopes from each protein showed high level of conservancy up to 100% (Table 2). ToxinPred server predicted the relative toxicity of each epitope which indicated that the top epitopes were non-toxin in nature (Table S5). Population coverage of four structures proteins were also done for the predicted CTL and HTL epitopes. From the screening, results showed that population of the various geographic regions could be covered by the predicted T-cell epitopes (Fig. 7).Finally, the allergenic epitopes were excluded from the list based on the evaluation of four allergenicity prediction server (Table S5).
Fig. 7.
Population coverage analysis of spike protein (A), envelope protein (B), membrane protein (C) and nucleocapsid proteins (D).
3.9. Identification of B cell epitopes and conservancy analysis
Top B-cell epitopes were predicted for Spike glycoprotein, membrane protein, envelope protein and nucleocapsid protein using 3 distinct algorithms (i.e. Bepipred Linear Epitope prediction, Emini Surface Accessibility, Kolaskar and Tongaonkar Antigenicity prediction) from IEDB. Epitopes were also allowed to analyze their vaxijen scoring and allergenicity (Table 3). Results revealed that top B-cell and T-cell epitopes were highly conserved (up to100%) among different strains of SARS-CoV-2 (Table 4), while 19 out of final 20 epitopes showed conservancy > 90%. Therefore, the designed vaccine construct is expected to confer broad range immunity in the host.
Table 3.
Allergenicity and antigenicity assessment of predicted B-cell epitopes.
| Protein | Algorithms | Top peptide sequence | Allergenicity | Vaxijen score |
|---|---|---|---|---|
| Spike glycoprotein | Bepipred Linear Epitope Prediction 2.0 | IRGDEVRQIAPGQTGKIADYNYK | Non-allergen | 1.0547 |
| Emini surface accessibility prediction | GDEVRQ | Non-allergen | 0.6701 | |
| Kolaskar and Tongaonkar antigenicity | VRQIAPG | Non-allergen | 1.2611 | |
| Membrane glycoprotein | Bepipred Linear Epitope Prediction 2.0 | MWSFNPETN | Non-allergen | 0.5509 |
| Emini surface accessibility prediction | LFARTRSMWSFNPET | Non-allergen | 0.9033 | |
| Kolaskar and Tongaonkar antigenicity | AMACLVGLM | Non-allergen | 0.6251 | |
| Envelope protein | Bepipred Linear Epitope Prediction 2.0 | YVYSRVKNLNSSRVP | Non-allergen | 0.4492 |
| Emini surface accessibility prediction | PSFYVYSRVKNLNSSRVP | Non-allergen | 0.5796 | |
| Kolaskar and Tongaonkar antigenicity | VNSVLLFLAFVVFLLVTLA | Non-allergen | 0.5893 | |
| Nucleocapsid protein | Bepipred Linear Epitope Prediction 2.0 | PGSSRGTSPARMAGNGG | Non-allergen | 0.3854 |
| Emini surface accessibility prediction | TEPKKDKKKKAD | Non-allergen | 0.2378 | |
| Kolaskar and Tongaonkar antigenicity | HWPQIAQFAPSASAF | Non-allergen | 0.4320 |
Table 4.
Conservancy of the final epitopes among different SARS-CoV-2 strains (retrieved from the list of World Health Organization for top 30 infected regions).
| Protein | Epitope | Conservancy |
|---|---|---|
| Spike glycoprotein | ADYNYKLPD | 100.00% (31/31) |
| SFVIRGDEVRQIAPG | 100.00% (31/31) | |
| IRGDEVRQIAPGQTGKIADYNYK | 100.00% (31/31) | |
| GDEVRQ | 100.00% (31/31) | |
| VRQIAPG | 100.00% (31/31) | |
| Envelope protein | FVVFLLVTL | 96.30% (26/27) |
| LCAYCCNIV | 92.59% (25/27) | |
| VTLAILTALRLCAYC | 96.30% (26/27) | |
| YVYSRVKNLNSSRVP | 92.59% (25/27) | |
| PSFYVYSRVKNLNSSRVP | 92.59% (25/27) | |
| Membrane glycoprotein | LVIGAVILR | 96.67% (29/30) |
| LACFVLAAVYRINWI | 100.00% (30/30) | |
| MWSFNPETN | 100.00% (30/30) | |
| LFARTRSMWSFNPET | 100.00% (30/30) | |
| AMACLVGLM | 100.00% (30/30) | |
| Nucleocapsid protein | AIKLDDKDP | 93.33% (28/30) |
| TEPKKDKKKKAD | 93.33% (28/30) | |
| HWPQIAQFAPSASAF | 93.33% (28/30) | |
| DLSPRWYFYYLGTGP | 93.33% (28/30) | |
| PGSSRGTSPARMAGNGG | 76.67% (23/30) |
3.10. Construction of vaccine molecules and prediction of allergenicity, antigenicity and solubility of the constructs
Three putative vaccine molecules (i.e. V1, V2 and V3) were constructed, each comprising a protein adjuvant, eight T-cell epitopes, twelve B-cell epitopes and respective linkers (Table S6). PADRE sequence was included to extend the efficacy and potency of the constructed vaccine. The putative vaccine constructs, V1, V2 and V3 were 397, 481 and 510 residues long respectively. However, allergenicity score of V3 (−0.89886723) revealed that it was superior among the three constructs in terms safety and efficacy. V3 also had a solubility score (0.60) (Fig. 8E) and antigenicity (0.58) over threshold value (Table 5).
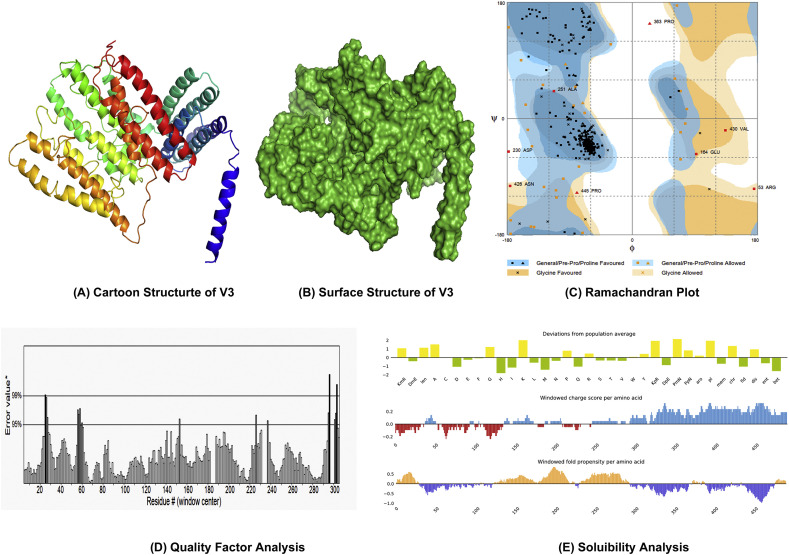
Fig. 8.
Homology modeling, structure validation and solubility prediction of construct V3 (A: Cartoon structure, B: Surface structure, C: Ramachandran Plot analysis, D: Quality factor analysis, E: Solubility analysis).
Table 5.
Allergenicity, antigenicity and solubility prediction of designed vaccine constructs.
| Vaccine constructs | Composition | Complete sequence of vaccine construct | Allergenecity (Threshold −0.4) | Antigenicit (Threshold 0.4) | Solubility (Threshold 0.45) |
|---|---|---|---|---|---|
| V1 | Predicted CTL,HTL and BCL epitopes with defensin adjuvant and PADRE sequence | EAAAKGIINTLQKYYCRVRGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKKEAAAKAKFVAAWTLKAAAGGGSADYNYKLPDGGGSLVIGAVILRGGGSFVVFLLVTLGGGSAIKLDDKDPGPGPGSFVIRGDEVRQIAPGGPGPGLACFVLAAVYRINWIGPGPGVTLAILTALRLCAYCGPGPGDLSPRWYFYYLGTGPKKIRGDEVRQIAPGQTGKIADYNYKKKGDEVRQKKVRQIAPGKKMWSFNPETNKKLFARTRSMWSFNPETKKAMACLVGLMKKYVYSRVKNLNSSRVPKKPSFYVYSRVKNLNSSRVPKKLCAYCCNIVKKPGSSRGTSPARMAGGGKKTEPKKDKKKKADKKHWPQIAQFAPSASAFKKAKFVAAWTLKAAAGGGS | −0.17698947 | 0.60 | 0.61 |
| V2 | Predicted CTL, HTL and BCL epitopes with L7/L12 ribosomal protein adjuvant and PADRE sequence | EAAKMAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPAGAAVEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGAPKPLLEKVAKEAADEAKAKLEAAGATVTVKEAAAKAKFVAAWTLKAAAGGGSADYNYKLPDGGGSLVIGAVILRGGGSFVVFLLVTLGGGSAIKLDDKDPGPGPGSFVIRGDEVRQIAPGGPGPGLACFVLAAVYRINWIGPGPGVTLAILTALRLCAYCGPGPGDLSPRWYFYYLGTGPKKIRGDEVRQIAPGQTGKIADYNYKKKGDEVRQKKVRQIAPGKKMWSFNPETNKKLFARTRSMWSFNPETKKAMACLVGLMKKYVYSRVKNLNSSRVPKKPSFYVYSRVKNLNSSRVPKKLCAYCCNIVKKPGSSRGTSPARMAGGGKKTEPKKDKKKKADKKHWPQIAQFAPSASAFKKAKFVAAWTLKAAAGGGS | 0.13269078 | 0.55 | 0.64 |
| V3 | Predicted CTL, HTL and BCL epitopes with HABA adjuvant and PADRE sequence | EAAKMAENPNIDDLPAPLLAALGAADLALATVNDLIANLRERAEETRAETRTRVEERRARLTKFQEDLPEQFIELRDKFTTEELRKAAEGYLEAATNRYNELVERGEAALQRLRSQTAFEDASARAEGYVDQAVELTQEALGTVASQTRAVGERAAKLVGIELEAAAKAKFVAAWTLKAAAGGGSADYNYKLPDGGGSLVIGAVILRGGGSFVVFLLVTLGGGSAIKLDDKDPGPGPGSFVIRGDEVRQIAPGGPGPGLACFVLAAVYRINWIGPGPGVTLAILTALRLCAYCGPGPGDLSPRWYFYYLGTGPKKIRGDEVRQIAPGQTGKIADYNYKKKGDEVRQKKVRQIAPGKKMWSFNPETNKKLFARTRSMWSFNPETKKAMACLVGLMKKYVYSRVKNLNSSRVPKKPSFYVYSRVKNLNSSRVPKKLCAYCCNIVKKPGSSRGTSPARMAGGGKKTEPKKDKKKKADKKHWPQIAQFAPSASAFKKAKFVAAWTLKAAAGGGS | −0.89886723 | 0.58 | 0.60 |
The bold sequences are linker sequences
3.11. Physicochemical characterization and secondary structure analysis of the construct
ProtParam tool was employed to analyze the physicochemical properties of V3. Molecular weight of 3 was scored as 55.181 kDa. The extinction coefficient of V3 was calculated as 63,830 at 0.1% absorption. It had been found that the protein would have net negative charge which was higher than the recommended pI 9.81. Aliphatic index and GRAVY value were found 77.80 and −0.383 respectively, which could express the thermostability and hydrophilic status of the V3 vaccine construct. Around sixty minutes in vitro half-life stability in mammalian reticulocytes was predicted for V3. The computed instability index (II) 36.98 classified the protein as a stable one. In contrast, Secondary structure of V3 exhibited to have 46.47% alpha helix, 15.00% sheet and 38.63% coil structure (Fig. S1).
Fig. S1.
Secondary structure prediction of constructed vaccine protein V3.
3.12. Homology modeling, structure refinement, validation and disulfide engineering
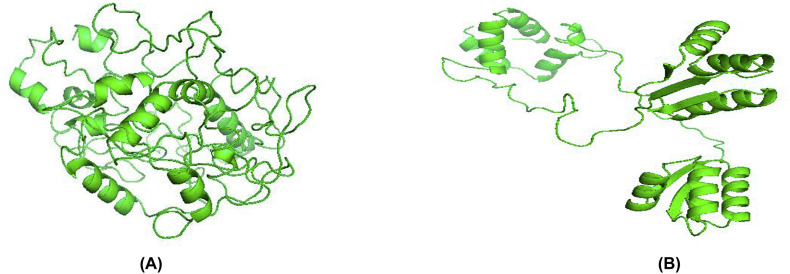
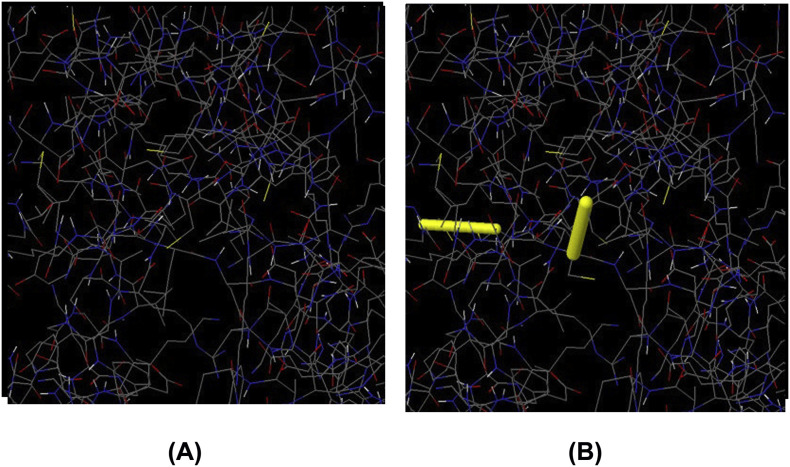
Tertiary structure of the putative vaccine construct V3 was generated using I-TASSER server (Fig. 8A and B). The server used 10 best templates with highest significant (measured via Z-score) from the LOMETS threading program to model the 3D structure. After refinement, Ramachandran plot analysis revealed that 92.7% and 5.7% residues were in the favored and allowed regions respectively, while only 8 residues (1.6%) occupied in the outlier region (Fig. 8C). The overall quality factor determined by ERRAT server was 91.56% (Fig. 8D). 3D modelled structure of V1 and V2 are shown in Fig. S2. DbD2 server recognized 33 pairs of amino acid residue with the potentiality to create disulfide bond between them. After analysis chi3 and B-factor parameter of residue pairs on the basis of energy, only 2 pairs (PRO 277-THR 329 and LEU 425-CYS 435) met the criteria for disulfide bond formation which were changed with cysteine (Fig. S3).
Fig. S2.
3D modelled structure of vaccine protein V1 and V2.
Fig. S3.
Disulfide engineering of vaccine protein V3 (A: Initial form, B: Mutant form).
3.13. Conformational B-cell and IFN- inducing epitopes prediction
Ellipro server predicted a total 6 conformational B-cell epitopes from the 3D structure of the construct V3. Epitopes No. 1 were considered as the broadest conformational B cell epitopes with 25 amino acid residues (Fig. 9 and Table S7). Results also revealed that predicted linear epitopes from 76 to 101, 131–143 and 48–55 were included in the conformational B-cell epitopes. Moreover, the sequence of the final vaccine was scanned for 15-mer IFN-inducing epitopes. Results showed that there were 292 positive IFN- inducing epitopes from which 20 had a score ≥ 5 (Table S8). Residues of 194–209 regions (GGGSLVIGAVILRGG) in the vaccine showed highest score of 17 (Table S8).
Fig. 9.
Predicted conformational epitopes (A and B) and linear epitopes (C and D) within construct V3.
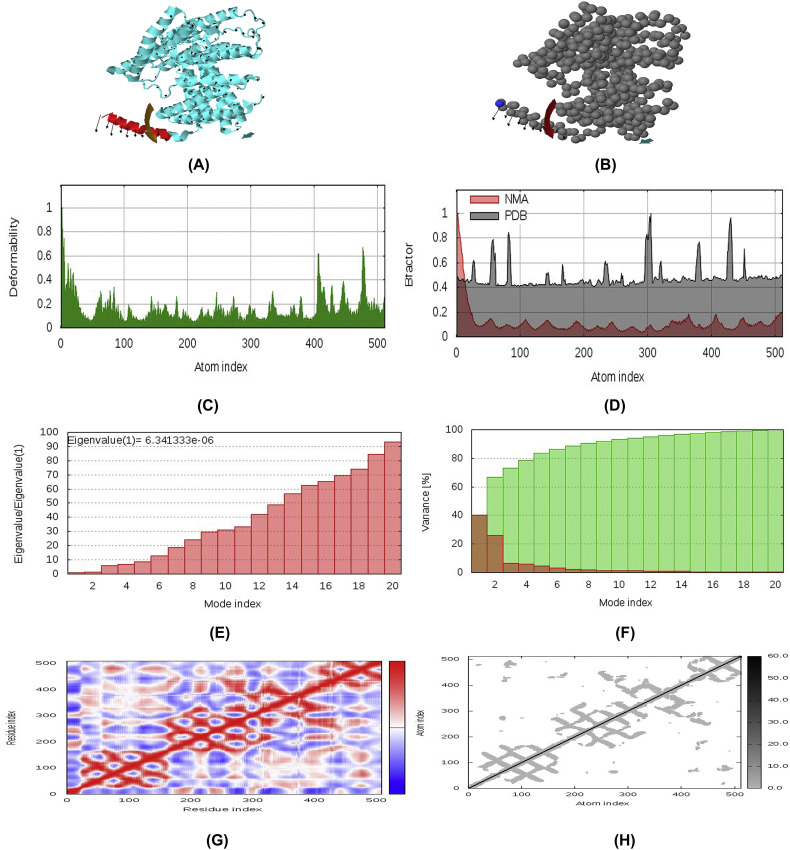
3.14. Molecular dynamics and normal mode analysis
Stability of the vaccine construct V3 was investigated through mobility analysis (Fig. 10A and B), B-factor, eigenvalue and deformability analysis, covariance map and recommended elastic network model. Results revealed that the placements of hinges in the chain was insignificant (Fig. 10C) and the B-factor column gave an averaged RMS (Fig. 10D). The estimated higher eigenvalue 6.341333e−06 (Fig. 10E) indicated low chance of deformation of vaccine protein V3. The correlation matrix and elasticity of the construct have been shown in Fig. 10G and Fig. 10H, respectively.
Fig. 10.
Normal Mode Analysis (NMA) of vaccine protein V3. The directions of each residues are given by arrows and the length of the line represented the degree of mobility in the 3D model (A and B). The main-chain deformability derived from high deformability regions indicated by hinges in the chain which are negligible (C). The experimental B-factor was taken from the corresponding PDB field and calculated from NMA (D). The eigenvalue represents the motion stiffness and directly related to the energy required to deform the structure (E). The variance associated to each normal mode is inversely related to the eigenvalue. Coloured bars show the individual (red) and cummulative (green) variances (F). The covariance matrix indicates coupling between pairs of residues, where they may be associated with correlated, uncorrelated or anti-correlated motions, indicated by red, white and blue colors respectively (G). The elastic network model identifies the pairs of atoms connected via springs. Each dot in the diagram is coloured based on extent of stiffness between the corresponding pair of atoms. The darker the greys, the stiffer the springs (H). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.15. Protein-protein docking
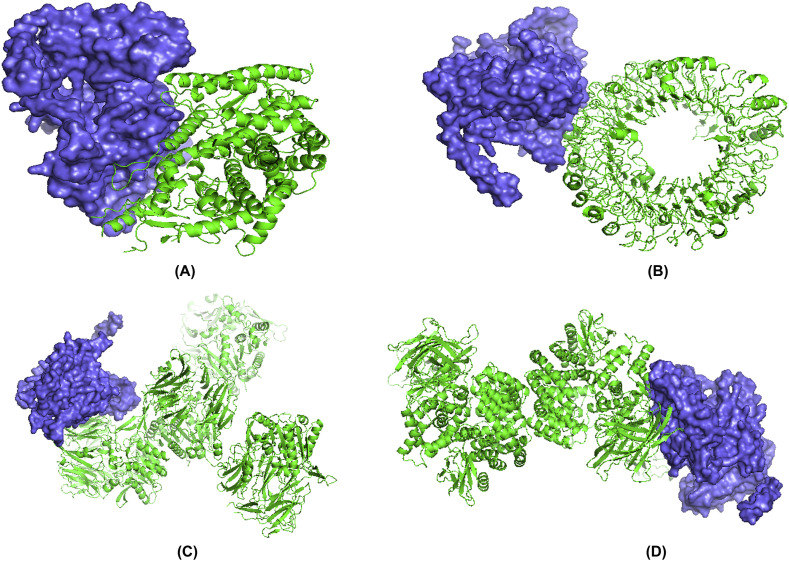
The structural interaction between HLA alleles and the designed vaccines were investigated by molecular docking approach. The server detected the complexed structure by focusing on complementary score, ACE (Atomic Contact Energy) and estimated interface area of the compound (Table 6 ). The molecular affinity between the putative vaccine molecules V3 and several immune receptors were also experimented. The result showed that construct V3 interacted with each receptor with significantly lower binding energy (Fig. 11 ).
Table 6.
Docking score of vaccine construct V3 with different HLA alleles and human immune receptors.
| Vaccine construct | PDB ID's HLAs | Global energy | Hydrogen bond energy | ACE | Score | Area |
|---|---|---|---|---|---|---|
| V1 | 1a6a | −11.07 | −2.61 | 8.85 | 17,640 | 2211.50 |
| 1h15 | −17.51 | −1.51 | 4.35 | 15,862 | 2302.20 | |
| 2fse | −5.38 | 0.00 | 5.61 | 16,720 | 2493.50 | |
| 2q6w | −10.48 | −0.39 | −1.45 | 15,348 | 2289.00 | |
| 2seb | 9.03 | 0.00 | 0.20 | 17,640 | 2903.00 | |
| 3c5j | −25.18 | −4.60 | 8.20 | 16,804 | 3022.80 | |
| V2 | 1a6a | −37.24 | −3.11 | −9.54 | 19,666 | 2703.90 |
| 1h15 | −9.33 | 0.00 | 4.33 | 16,132 | 2312.70 | |
| 2fse | −0.33 | 0.00 | −0.60 | 16,702 | 2956.10 | |
| 2q6w | −10.85 | −3.06 | 1.20 | 15,062 | 1924.40 | |
| 2seb | 1.38 | 0.00 | −0.79 | 18,008 | 3034.30 | |
| 3c5j | 10.58 | −1.56 | 5.54 | 16,158 | 2385.30 | |
| V3 | 1a6a | −4.14 | −1.49 | 12.24 | 14,358 | 2208.30 |
| 1h15 | −3.15 | −4.45 | 10.93 | 13,908 | 1637.70 | |
| 2fse | −8.68 | −2.76 | 9.98 | 18,340 | 3024.40 | |
| 2q6w | −21.02 | −2.41 | 5.05 | 17,060 | 2626.90 | |
| 2seb | −17.54 | −5.91 | 3.87 | 16,094 | 2946.80 | |
| 3c5j | −2.30 | 0.00 | −2.30 | 18,550 | 3041.00 |
Fig. 11.
Docked complex of vaccine construct V3 with human ACE 2 (A), TLR- (B), DPP4 (C) and APN (D).
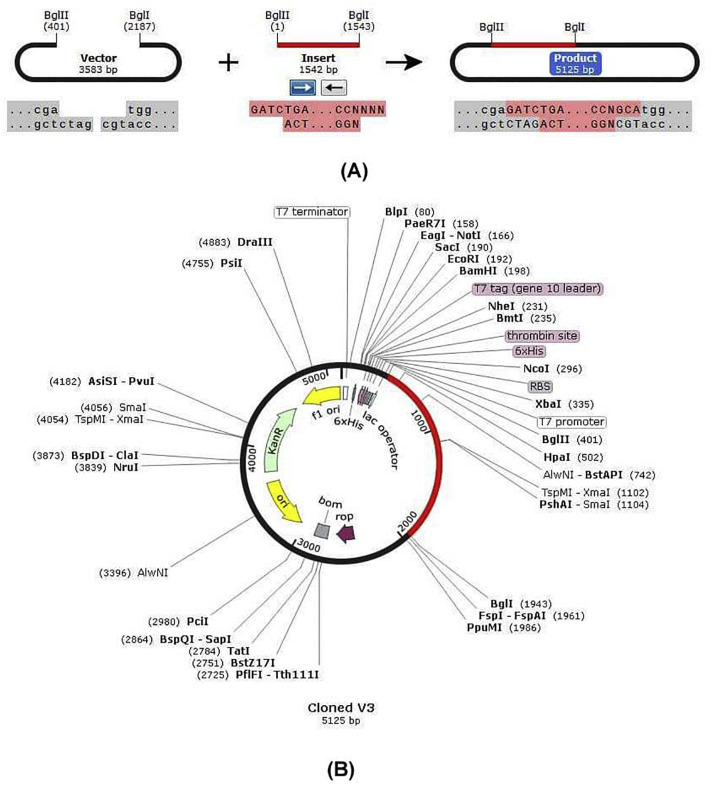
3.16. Codon adaptation and in silico cloning
The Codon Adaptation Index (CAI) and GC content for the predicted codons of the putative vaccine constructs V1 were demonstrated as 1.0 and 51.56% respectively. An insert of 1542 bp was found which lacked the restriction sites for BglI and BglII, thus providing comfort zone for cloning. The codons were inserted into pET28a(+) vector alongside two restriction sites (BglI and BglII) and a clone of 5125 base pair was generated (Fig. 12 ).
Fig. 12.
In silico restriction cloning of the gene sequence of final vaccine construct V3 into pET28a(+) expression vector (A: Restriction digestion of the vector pET28a(+) and construct V3 with BglII and BglI, B: Inserted desired fragment (V3 Construct) between BglII (401) and BglI (1943) indicated in red colour. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In December 2019, a new coronavirus prevalence flourished in Wuhan, China, causing clutter among the medical community, as well as to the rest of the world (Sun et al., 2020a, Sun et al., 2020b). The new species has been renamed as 2019-nCoV or, SARS-CoV-2, already causing considerable number infections and deaths in China, Italy, Spain, Iran, USA and to a growing degree throughout the world. The major outbreak and spread of SARS-CoV-2 in 2020 forced the scientific community to make considerable investment and research activity for developing a vaccine against the pathogen. However, owing to high infectivity and pathogenicity, the culture of SARS-CoV-2 needs biosafety level 3 conditions, which may obstructed the rapid development of any vaccine or therapeutics. It had been found that about 35 companies and academic institutions are engaged in such works (Spinney et al., 2020, Ziady, 2020). Among the potential SARS-CoV-2 vaccines in the pipeline, four have nucleic acid based designs, four involve non-replicating viruses or protein constructs, two contain live attenuated virus and one involves a viral vector (Pang et al. 2020), while only one, called mRNA-1273 (developed by NIAID collaboration with Moderna, Inc.), has confirmed to start phase-1 trial (NIH News Event, 2020). However, in this study we emphasized on a different approaches by prioritizing the advantages of different genome and proteome database using the immunoinformatic approach. Computational vaccine predictions were adopted by the researchers to design vaccines against both MERS-CoV (Sudhakar et al. 2013; Fernando et al., 2013) and SARS-CoV-1 (; Oany et al., 2014), targeting the outer membrane or functional proteins (Sharmin and Islam 2014). Several in silico strategies have also been employed to predict potential T cell and B cell epitopes against SARS-CoV-2, either emphasizing on spike glycoprotein or envelope proteins (Behbahani 2020; Rasheed et al., 2020). None of the studies, however, focused on other structural proteins. Moreover, random genetic changes and mutations in the protein sequences (Yin, 2020) may obstruct the development of effective vaccines and therapeutics against human coronavirus in the future. Hence, the present study was employed to identify the similarity and divergence among the close relatives of the target pathogen and develop a novel chimeric recombinant vaccine considering all major structural proteins i.e. spike glycoprotein, membrane glycoprotein, envelope protein and nucleocapsid protein simultaneously.
Structural proteins are often chosen as attractive therapeutic targets by the researchers due to their surface exposure. Spike (S) protein is currently the most promising antigen formulation for SARS-CoV-2 vaccine research which is able to be directly recognized by host immune system (Wrapp et al. 2020) and already been used for vaccine development to prevent Severe Acute Respiratory Syndrome (SARS) and Middle-East Respiratory Syndrome (MERS) (Zhou et al. 2018; Du et al. 2009). However, Membrane (M) protein, the most abundant one on the virus surface (Neuman et al. 2011), was also reported to stimulate efficient neutralizing antibodies in SARS patients upon immunization (Pang et al. 2004). Moreover, structural investigation revealed the existence of T-cell epitope cluster on the trans-membrane domain of M protein with the ability to induce a strong cellular immune response (Liu et al. 2010). Thus, it can be utilized as a candidate antigen for developing SARS-CoV-2 vaccine. Researchers have already considered Envelope (E) protein for vaccine candidacy against COVID-19 due to high antigenicity (Abdelmageed et al. 2020). It was also investigated against SARS-CoV in 2003 and more recently, against MERS-CoV (Schoeman and Fielding 2019). Nucleocapsid (N) protein, on the other hand, is believed to be more conserved than spike or membrane glycoprotein with the potential to elicit broad-based cellular responses (Zhao et al. 2005). Previously, cellular response against the N protein of some animal coronaviruses enhanced recovery from viral infection (Glansbeek et al. 2002; Wasmoen et al., 1995; Wesseling et al. 1993). A study showed that N protein of SARS-CoV act as an important B cell immunogen, indicating its potential value in vaccine development against SARS (Zhao et al. 2005). All these data suggest that S, E, M and N protein can induce both humoral and cellular immune responses against SARS-CoV-2.
Phylogenetic analysis is used for fundamental as well as applied issues of virus research and it includes the development of antiviral drugs and vaccines (Gorbalenya and Lauber 2017). Here, we used phylogeny data to perform similarity analysis between different coronavirus subfamiles to finalize which structural protein to be used as vaccine target in this study. We wanted to target the proteins which are most common in each subfamily and exhibit significant similarities to design a broad and universally effective vaccine candidate. The results revealed that each of the four major structural proteins possess highly conserved domains among alpha-, beta- and gamma-coronaviruses. Therefore we considered all major structural proteins to design a multiepitope subunit vaccine which is expected to stimulate more effective and broad response in the host. The topology of the phylogenetic trees of the whole genome and the stated four proteins sequences from different species of coronaviruses reveal that SARS-CoV-1 and bat coronaviruses are the closest homologs of the novel coronaviruses. Our results infer a significant level of similarities within the COVID-19 and SARS-CoV-1 which was also aligned with the previous findings (Jaimes et al., 2020; Sun et al., 2020a, Sun et al., 2020b; Wu 2020). The sequence similarities between the SARS-CoV, bat coronaviruses and the COVID-19 from the reported studies (Hu et al. 2018; Wu et al. 2020; Wu 2020) suggests that those are distantly related, in spite those are capable of infecting the humans and therefore possess the adaptive convergent evolution. Interestingly, the COVID-19 envelope proteins form clade with the Turkey coronavirus which belongs to gamma-coronavirus genus. So, in terms of envelope proteins, the envelope gene of turkey coronavirus might contribute to the convergence process, which needs further analysis. In addition, from the domain-based phylogeny of nucleocapsid proteins, it can be deduced that this protein might have originated in bats and was transmitted to camels and then later on choose human as the potential host. Overall, the COVID-19 might go through complex adaptation strategies in order to be transmitted into the human via different animals.
The homologous protein sets for four structural proteins of Coronavirus were sorted to identify conserved regions through BLASTp analysis and MSA. Only the conserved sequences were utilized to identify potential B-cell and T-cell epitopes for each individual protein (Table 1). Thus, our constructs are expected to stimulate a broad-spectrum immunity in host upon administration. Cytotoxic CD8 + T lymphocytes (CTL) play a crucial role to control the spread of pathogens by recognizing and killing diseased cells or by means of antiviral cytokine secretion (Garcia et al. 1999). Thus, T cell epitope-based vaccination is a unique process to confer defensive response against pathogenic candidates (Shrestha and Diamond, 2004). Approximately 800 MHC-I peptides (CTL epitopes) and 600 MHC-II peptides (HTL epitopes) were predicted via IEDB server, from which we screened the top ones through analyzing the antigenicity score, trans-membrane topology, conservancy level and other important physiochemical parameters employing a number of bioinformatics tools (Table 2). The top 10 epitopes from each protein was further assessed by investigating the toxicity profile and allergenicity pattern. Different servers rely on different parameters to predict the allergenic nature of small peptides. Therefore, we used 4 distinct servers for such assessment and the epitopes predicted as non-allergen at least via 3 servers were retained for further analysis (Table S5). Vaccine initiates the generation of effective antibodies that are usually produced by B cells and plays effector functions by targeting specifically to a foreign particles (Cooper and Nemerow 1984). The potential B cell epitopes were generated by three different algorithms (Bepipred linear epitope prediction 2.0, Kolaskar and Tongaonkar antigenicity prediction and Emini surface accessibility prediction) from IEDB database (Table 3).
Suitable linkers and adjuvants were used to combine top finalized epitopes from each protein that led to develop multiepitope vaccine molecules (Table S6). As PADRE sequence was usually recommended to lessen the polymorphism of HLA molecules in the population (Ghaffari-Nazari et al. 2015), it was also considered to construct the final vaccine molecule. Here, adjuvants would enhance the immunogenicity of the vaccine constructs and appropriate separation of epitopes in the host environment would be ensured by the linker (Yang et al. 2015). Allergenicity, physiochemical properties, antigenicity and three-dimensional structure of vaccine constructs were characterized, and it was concluded that V3 was superior to V1 and V2 vaccine constructs due to its highly negative allergenicity score (−0.89886723). Therefore, it is expected to be less likely to cause adverse side effects or hypersensitivity. V3 also had a solubility score (0.60) and antigenicity (0.58) over threshold value (Table 5). Hence, construct V3 was proposed as the most suitable and safe vaccine protein for further analysis.
The final construct also occupied by several interferon-α producing epitopes (Table S8). The vaccine protein (V3) was subjected to di-sulfide engineering to enhance its stability. Analysis of the normal modes in internal coordinates by iMODS was employed to investigate the collective motion of vaccine molecules (López-Blanco et al., 2014). Negligible chance of deformability at molecular level was analyzed for the putative vaccine construct V3, thereby strengthening our prediction. Moreover, molecular docking was investigated to analyze the molecular affinity of the vaccine with different HLA molecules i.e. DRB1*0101, DRB5*0101, DRB3*0202, DRB1*0401, DRB3*0101 and DRB1*0301 (Table 6). It had been reported that a specific receptor-binding domain of CoV spike protein usually recognizes its host receptor ACE2 (angiotensin-converting enzyme 2) (Li et al., 2003; Li 2015). Previous studies also identified dipeptidyl peptidase 4 (DPP4) as a functional receptor for human coronavirus (Raj et al. 2013). Therefore, we performed another docking study prioritizing these immune receptors to strengthen our prediction (Fig. 11). Results showed that the designed construct bound with the selected receptors with minimum binding energy which was biologically significant. Finally, in-silico restriction cloning was adopted to check the suitability of construct V3 for entry into pET28a (+) vector and expression in E. coli strain K12 (Fig. 12).
Traditional ways to vaccine development are time consuming and laborious. Moreover, the result may not be always as expected or fruitful (Stratton et al., 2003; Hasan et al., 2019a, Hasan et al., 2019b). In silico prediction and prescreening methods, on the contrary, offer some advantages while saving time and cost for production. Therefore, the present study may aid in the development of preventive strategies and novel vaccines to combat infections caused by 2019-nCoV. However, further wet lab trials involving model organism needs to be experimented for validating our findings.
The following are the supplementary data related to this article
Predicted CTL and HTL epitopes of spike glycoprotein.
Predicted CTL and HTL epitopes of membrane.
Predicted CTL and HTL epitopes of envelope protein
Predicted CTL and HTL epitopes of nucleocapsid protein.
Allergenicity pattern and toxicity profile of top T cell epitopes.
Proposed CTL and HTL epitopes for vaccine construction.
Predicted conformational epitopes within construct V3.
Predicted IFN alpha producing epitopes in the vaccine construct V3.
NCBI IDs of the complete genome, spike glycoprotein, envelope protein, membrane protein and nucleocapsid protein of coronavirus with Genera and Sub-Genera.
Retrieved protein sequences of major structural proteins of COVID-19.
Secondary structure and domain analysis of spike glycoprotein, envelope protein, membrane protein and nucleocapsid proteins.
Credit author statement
Mst Rubaiat Nazneen Akhand: Conceptualization, Methodology, Data curation, Formal analysis, Software and Writing - original draft. Kazi Faizul Azim: Conceptualization, Methodology, Software, Writing - original draft Syeda Farjana Hoque: Software, Data Curation and Writing - original draft Mahmuda Akther Moli: Software, Data Curation and Writing - original draft Bijit Das Joy: Software, Validation, and Visualization Hafsa Akter: Software, Validation and Visualization Ibrahim Khalil Afif: Software and Validation Nadim Ahmed: Software and Validation Mahmudul Hasan: Conceptualization, Supervision, Project administration, Resources and Writing - review & editing.
Funding information
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
Authors declare that they have no conflict of interests.
Acknowledgements
Authors would like to acknowledge the Department of Biochemistry and Chemistry and Department of Pharmaceuticals and Industrial Biotechnology of Sylhet Agricultural University for the technical support of the project.
References
- Dudek N.L., Perlmutter P., Aguilar I., Croft N.P., Purcell A.W. Epitope discovery and their use in peptide based vaccines. Curr. Pharm. Des. 2010;16(28):3149–3157. doi: 10.2174/138161210793292447. [DOI] [PubMed] [Google Scholar]
- Spinney Laura. The Guardian; 2020. When Will A Coronavirus Vaccine Be Ready? (Retrieved 18 March 2020) [Google Scholar]
- World Health Organization (WHO) World Health Organization; 2020. Infection Prevention and Control During Health Care When COVID-19 Is Suspected: Interim Guidance; p. 2020. [Google Scholar]
- Ziady Hanna. CNN; 2020. Biotech Company Moderna Says Its Coronavirus Vaccine is Ready For First Tests. Archived from the original on 28 February 2020. (Retrieved 2 March 2020) [Google Scholar]
- Dimitrov I., Bangov I., Flower D.R., Doytchinova I. AllerTOP v.2 - a server for in silico prediction of allergens. J. Mol. Model. 2014;14(6):S4. doi: 10.1007/s00894-014-2278-5. [DOI] [PubMed] [Google Scholar]
- NIH News Event NIH Clinical Trial of Investigational Vaccine For COVID-19 Begins. Study Enrolling Seattle-Based Healthy Adult Volunteers. 2020. https://www.nih.gov/news-events/news-releases/nih-clinical-trial-investigational-vaccine-covid-19-begins (Accessed on 1st April 2020)
- World Health Organization (WHO) Coronavirus Disease (COVID-19) Outbreak. 2020. https://www.who.int (opens in new tab)
- Aalten D.M.F., Groot B.L., Findlay J.B.C., Berendsen H.J.C., Amadei A. A comparison of techniques for calculating protein essential dynamics. J. Comput. Chem. 1997;18(2):169–181. doi: 10.1002/(SICI)1096-987X. [DOI] [Google Scholar]
- Abdelmageed M.I., Abdelmoneim A.H., Mustafa M.I., Elfadol N.M., Murshed N.S., Shantier S.W., Makhawi A.M. Design of a multiepitope-based peptide vaccine against the E protein of human COVID-19: an immunoinformatics approach. Biomed. Res. Int. 2020;2020 doi: 10.1155/2020/2683286. (Jan 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awan F.M., Obaid A., Ikram A., Janjua H.A. Mutation-structure function relationship 893 based integrated strategy reveals the potential impact of deleterious missense mutations in 894 autophagy related proteins on hepatocellular carcinoma (HCC): a comprehensive 895 informatics approach. Int. J. Mol. Sci. 2017;18(1):139. doi: 10.3390/ijms18010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim K.F., Lasker T., Akter R., Hia M.M., Bhuiyan OF, Hasan M., et al. Conglomeration of highly antigenic nucleoproteins to inaugurate a heterosubtypic next generation vaccine candidate against Arenaviridae family. bioRxiv. 2019 doi: 10.1101/2019.12.29.885731. (Jan 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim K.F., Hasan M., Hossain M.N., Somana S.R., Hoque S.F., Bappy M.N., et al. Immunoinformatics approaches for designing a novel multi epitope peptide vaccine against human norovirus (Norwalk virus) Infect. Genet. Evol. 2019;74:103936. doi: 10.1016/j.meegid.2019.103936. (Oct 1) [DOI] [PubMed] [Google Scholar]
- Azim K.F., Ahmed S.R., Banik A., Khan M.M., Deb A., Somana S.R. Screening and druggability analysis of some plant metabolites against SARS-CoV-2: an integrative computational approach. Inform. Med. Unlocked. 2020;20:100367. doi: 10.1016/j.imu.2020.100367. (Jun 9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behbahani M. bioRxiv; 2020. In Silico Design of Novel Multi-Epitope Recombinant Vaccine Based on Coronavirus Spike Glycoprotein. [DOI] [Google Scholar]
- Ceraolo C., Giorgi F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020;92(5):522–528. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N.R., Nemerow G.R. The role of antibody and complement in the control of viral infections. J. Invest. Dermatol. 1984;83(1 Suppl):121–127. doi: 10.1038/jid.1984.33. [DOI] [PubMed] [Google Scholar]
- Craig D.B., Dombkowski A.A. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinformatics. 2013;14(1):346. doi: 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Bahar I. Normal mode analysis theoretical and applications to biological and chemical systems. Brief. Bioinform. 2007;8(5):378–379. doi: 10.1201/9781420035070. [DOI] [Google Scholar]
- Dimitrov I., Naneva L., Doytchinova I., Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014;30(6):846–851. doi: 10.1093/bioinformatics/btt619. (Mar 15) [DOI] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. Identifying candidate subunit vaccines using an alignment-independent method based on principal amino acid properties. Vaccine. 2007;25(5):856–866. doi: 10.1016/j.vaccine.2006.09.032. (Jan 15) [DOI] [PubMed] [Google Scholar]
- Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinformatics. 2007;8(1):4. doi: 10.1186/1471-2105-8-4. (Dec 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., He Y., Zhou Y., Liu S., Zheng B.J., Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7(3):226–236. doi: 10.1038/nrmicro2090. (Mar) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emini E.A., Hughes J.V., Perlow D., Boger J. Induction of hepatitis a virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985;55(3):836–839. doi: 10.1128/jvi.55.3.836-839.1985. (Sep 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando A., Marta L., Isabel S., Sonia Z., Jose L., Silvia L.J., et al. Engineering a replication-competent, propagation defective middle east respiratory syndrome coronavirus as a vaccine candidate. mBio. 2013;4(5) doi: 10.1128/mbio.00650-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia K.C., Teyton L., Wilson I.A. Structural basis of T cell recognition. Annu. Rev. Immunol. 1999;17(1):369–397. doi: 10.1146/annurev.immunol.17.1.369. (Apr) [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Wilkins M.R., Appel R.D., Bairoch A. The Proteomics Protocols Handbook. Humana Press; 2005. Protein identification and analysis tools on the ExPASy server; pp. 571–607. [DOI] [Google Scholar]
- Ghaffari-Nazari H., Tavakkol-Afshari J., Jaafari M.R., Tahaghoghi-Hajghorbani S., Masoumi E., Jalali S.A. Improving multi-epitope long peptide vaccine potency by using a strategy that enhances CD4+ T help in BALB/c mice. PLoS One. 2015;10(11) doi: 10.1371/journal.pone.0142563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glansbeek H.L., Haagmans B.L., te Lintelo E.G., Egberink H.F., Duquesne V., Aubert A., Horzinek M.C., Rottier P.J. Adverse effects of feline IL-12 during DNA vaccination against feline infectious peritonitis virus. J. Gen. Virol. 2002;83(1):1–10. doi: 10.1099/0022-1317-83-1-1. (Jan 1) [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E., Lauber C. Reference Module in Biomedical Sciences. 2017. Phylogeny of viruses. [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11(12):836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote A., Hiller K., Scheer M., Münch R., Nörtemann B., Hempel D.C., et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33(Suppl. 2):W526–W531. doi: 10.1093/nar/gki376. (Jul 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan M., Azim K.F., Begum A., Khan N.A., Shammi T.S., Imran A.S., et al. Vaccinomics strategy for developing a unique multi-epitope monovalent vaccine against Marburg marburg virus. Infect. Genet. Evol. 2019;70:140–157. doi: 10.1016/j.meegid.2019.03.003. (Jun 1) [DOI] [PubMed] [Google Scholar]
- Hasan M., Ghosh P.P., Azim K.F., Mukta S., Abir R.A., Nahar J., et al. Reverse vaccinology approach to design a novel multi-epitope subunit vaccine against avian influenza A (H7N9) virus. Microb. Pathog. 2019;130:19–37. doi: 10.1016/j.micpath.2019.02.023. [DOI] [PubMed] [Google Scholar]
- Hasan M., Islam S., Chakraborty S., Mustafa A.H., Azim K.F., Joy Z.F., et al. Contriving a chimeric polyvalent vaccine to prevent infections caused by herpes simplex virus (Type-1 and Type-2): an exploratory immunoinformatic approach. J. Biomol. Struct. Dyn. 2019;9:1–8. doi: 10.1080/07391102.2019.1647286. (Aug) [DOI] [PubMed] [Google Scholar]
- Hu D., Zhu C., Ai L., He T., Wang Y., Ye F., et al. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg. Microbes Infect. 2018;7(1):1–10. doi: 10.1038/s41426-018-0155-5. (Dec 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/s0140-6736(20)30183-5. (Feb 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., I Azhar E, Madani TA, Ntoumi F, Kock R, Dar O, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health - the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes J.A., André N.M., Millet J.K., Whittaker G.R. Structural modeling of 2019-novel coronavirus (nCoV) spike protein reveals a proteolytically-sensitive activation loop as a distinguishing feature compared to SARS-CoV and related SARS-like coronaviruses. ArXiv. 2020;2002(06196) doi: 10.1101/2020.02.10.942185. [DOI] [Google Scholar]
- Jespersen M.C., Peters B., Nielsen M., Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45(W1):W24–W29. doi: 10.1093/nar/gkx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W., Naqi S.A. Sequence analysis of gene 3, gene 4 and gene 5 of avian infectious bronchitis virus strain CU-T2. Gene. 1997;189(2):189–193. doi: 10.1016/s0378-1119(96)00847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A., Hossain M.U., Rakib-Uz-Zaman S.M., Morshed M.N. Epitope-based peptide vaccine design and target site depiction against Ebola viruses: an immunoinformatics study. Scand. J. Immunol. 2015;82(1):25–34. doi: 10.1111/sji.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaskar A.S., Tongaonkar P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990;276(1–2):172–174. doi: 10.1016/0014-5793(90)80535-q. [DOI] [PubMed] [Google Scholar]
- Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kunz R., Minder M. COVID-19 pandemic: palliative care for elderly and frail patients at home and in residential and nursing homes. Swiss Med. Wkly. 2020;150(1314) doi: 10.4414/smw.2020.20235. [DOI] [PubMed] [Google Scholar]
- Lanfear R., Frandsen P.B., Wright A.M., Senfeld T., Calcott B. Partitionfinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016;34(3):772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89(4):1954–1964. doi: 10.1128/jvi.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Joshi M., Singhania S., Ramsey K., Murthy A. Peptide vaccine: progress and challenges. Vaccines. 2014;2(3):515–536. doi: 10.3390/vaccines2030515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sun Y., Qi J., Chu F., Wu H., Gao F., Li T., Yan J., Gao G.F. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010;202(8):1171–1180. doi: 10.1086/656315. (Oct 15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Blanco J.R., Aliaga J.I., Quintana-Ortí E.S., Chacón P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(W1):W271–W276. doi: 10.1093/nar/gku339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Stratton C.W., Tang Y. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J. Med. Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon R., Reche P.A., Rappuoli R. Editorial: reverse vaccinology. Front. Immunol. 2019:2776. doi: 10.3389/fimmu.2019.02776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman B.W., Kiss G., Kunding A.H., Bhella D., Baksh M.F., Connelly S., Droese B., Klaus J.P., Makino S., Sawicki S.G., Siddell S.G. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174(1):11–22. doi: 10.1016/j.jsb.2010.11.021. (Apr 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Torres J.L., DeDiego M.L., Álvarez E., Jiménez-Guardeño J.M., Regla-Nava J.A., Llorente M., et al. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology. 2011;415(2):69–82. doi: 10.1016/j.virol.2011.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oany A.R., Emran A.A., Jyoti T.P. Design of an epitope-based peptide vaccine against spike protein of human coronavirus: an in silico approach. Drug Des. Devel. Ther. 2014;8:1139. doi: 10.2147/dddt.s67861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R.K., Ojha R., Aathmanathan V.S., Krishnan M., Prajapati V.K. Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection. Vaccine. 2018;36(17):2262–2272. doi: 10.1016/j.vaccine.2018.03.042. [DOI] [PubMed] [Google Scholar]
- Pang H., Liu Y., Han X., Xu Y., Jiang F., Wu D., Kong X., Bartlam M., Rao Z. Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J. Gen. Virol. 2004;85(10):3109–3113. doi: 10.1099/vir.0.80111-0. (Oct 1) [DOI] [PubMed] [Google Scholar]
- Pang J., Wang M.X., Ang I.Y.H., Tan S.H.X., Lewis R.F., Chen J.I.-P., et al. Potential rapid diagnostics, vaccine and therapeutics for 2019 novel coronavirus (2019-nCoV): a systematic review. J. Clin. Med. 2020;9(3):623. doi: 10.3390/jcm9030623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Xu J. Raptorx: exploiting structure information for protein alignment by statistical inference. Proteins. 2011;79(S10):161–171. doi: 10.1002/prot.23175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovsky N., Aguilar J.C. Vaccine adjuvants: current state and future trends. Immunol. Cell Biol. 2004;82(5):488–496. doi: 10.1111/j.0818-9641.2004.01272.x. [DOI] [PubMed] [Google Scholar]
- Prabhakar P.K., Srivastava A., Rao K.K., Balaji P.V. Monomerization alters the dynamics 897 of the lid region in campylobacter jejuni CstII: an MD simulation study. J. Biomol. Struct. Dyn. 2016;34(4):778–779. doi: 10.1080/07391102.2015.1054430. [DOI] [PubMed] [Google Scholar]
- Purcell A.W., McCluskey J., Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 2007;6(5):404–414. doi: 10.1038/nrd2224. [DOI] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495(7440):251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R. Reverse vaccinology. Curr. Opin. Microbiol. 2000;3(5):445–450. doi: 10.1016/s1369-5274(00)00119-3. [DOI] [PubMed] [Google Scholar]
- Rasheed M.A., Raza S., Zohaib A., Yaqub T., Rabbani M., Riaz M.I., et al. In silico identification of novel B cell and T cell epitopes of Wuhan coronavirus (2019-nCoV) for effective multi epitope-based peptide vaccine production. Preprints. 2020 doi: 10.20944/preprints202002.0359.v1. [DOI] [Google Scholar]
- Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42(W1):320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. (May 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha C.K., Hasan M.M., Hossain M.S., Jahan M.A., Azad A.K. In silico identification and characterization of common epitope-based peptide vaccine for Nipah and Hendra viruses. Asian Pac J Trop Med. 2017;10(6):529–538. doi: 10.1016/j.apjtm.2017.06.016. (Jun 1) [DOI] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16(1):1–22. doi: 10.1186/s12985-019-1182-0. (Dec) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmin R., Islam A.B. A highly conserved WDYPKCDRA epitope in the RNA directed RNA polymerase of human coronaviruses can be used as epitope-based universal vaccine design. BMC Bioinformatics. 2014;15(1):161. doi: 10.1186/1471-2105-15-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha B., Diamond M.S. Role of CD8+ T cells in control of West Nile virus infection. J. Virol. 2004;78(15):8312–8321. doi: 10.1128/jvi.78.15.8312-8321.2004. (Aug 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Higgins D.G. Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 2014;1079:105–116. doi: 10.1007/978-1-62703-646-7_6. [DOI] [PubMed] [Google Scholar]
- Smialowski P., Martin-Galiano A.J., Mikolajka A., Girschick T., Holak T.A., Frishman D. Protein solubility: sequence based prediction and experimental verification. Bioinformatics. 2007;23(19):2536–2542. doi: 10.1093/bioinformatics/btl623. (Oct 1) [DOI] [PubMed] [Google Scholar]
- Smith S.A., Dunn C.W. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 2008;24(5):715–716. doi: 10.1093/bioinformatics/btm619. [DOI] [PubMed] [Google Scholar]
- Solanki V., Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci. Rep. 2018;8(1):1–9. doi: 10.1038/s41598-018-26689-7. (Jun 13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton K., Almario D.A., Wizemann T.M., McCormick M.C. National Academies Press; 2003. Immunization Safety Review: Vaccinations And Sudden Unexpected Death In Infancy, Institute of Medicine (US) [DOI] [PubMed] [Google Scholar]
- Sudhakar A., Robin G., Boyd L., Agnihothram S., Gopal R., Yount B.L., Donaldson E.F., et al. Platform strategies for rapid response against emerging coronaviruses: MERS-CoV serologic and antigenic relationships in vaccine design. J. Infect. Dis. 2013;209(7):995–1006. doi: 10.1093/infdis/jit609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., He W.T., Wang L., Lai A., Ji X., Zhai X., et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol. Med. 2020 doi: 10.1016/j.molmed.2020.02.008. (Mar 21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z., Thilakavathy K., Kumar S.S., He G., Liu S.V. Potential factors influencing repeated SARS outbreaks in China. Int. J. Environ. Res. Public Health. 2020;17(5):1633. doi: 10.3390/ijerph17051633. (Jan) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surya W., Li Y., Verdià-Bàguena C., Aguilella V.M., Torres J. MERS coronavirus envelope protein has a single transmembrane domain that forms pentameric ion channels. Virus Res. 2015;201:61–66. doi: 10.1016/j.virusres.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tama F., Brooks C.L. Symmetry, form, and shape: guiding principles for robustness in macromolecular machines. Annu. Rev. Biophys. Biomol. Struct. 2006;35(1):115–133. doi: 10.1146/annurev.biophys.35.040405.102010. [DOI] [PubMed] [Google Scholar]
- Tesh R.B., Arroyo J., da Rosa A.P., Guzman H., Xiao S.Y., Monath T.P. Efficacy of killed virus vaccine, live attenuated chimeric virus vaccine, and passive immunization for prevention of West Nile virus encephalitis in hamster model. Emerg. Infect. Dis. 2002;8(12):1392–1397. doi: 10.3201/eid0812.020229. (Dec) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A.L., Staats H.F. Cytokines: the future of intranasal vaccine adjuvants. Clin. Dev. Immunol. 2011;2011:1–7. doi: 10.1155/2011/289597. (Jul 31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vita R., Overton J.A., Greenbaum J.A., Ponomarenko J., Clark J.D., Cantrell J.R., Wheeler D.K., Gabbard J.L., Hix D., Sette A., Peters B. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(D1):D405–D412. doi: 10.1093/nar/gkq888. (Jan 28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/s0140-6736(20)30185-9. (Internet] Elsevier BV. (Feb) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmoen T.L., Kadakia N.P., Unfer R.C., Fickbohm B.L., Cook C.P., Chu H.J., Acree W.M. Corona And Related Viruses. Springer; Boston, MA: 1995. Protection of cats from infectious peritonitis by vaccination with a recombinant raccoon poxvirus expressing the nucleocapsid gene of feline infectious peritonitis virus; pp. 221–228. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling J.G., Godeke G.J., Schijns V.E., Prevec L., Graham F.L., Horzinek M.C., Rottier P.J. Mouse hepatitis virus spike and nucleocapsid proteins expressed by adenovirus vectors protect mice against a lethal infection. J. Gen. Virol. 1993;74(10):2061–2069. doi: 10.1099/0022-1317-74-10-2061. (Oct 1) [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. (Mar 13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. Strong evolutionary convergence of receptor-binding protein spike between COVID-19 and SARS-related coronaviruses. bioRxiv. 2020 doi: 10.1101/2020.03.04.975995. [DOI] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K., Wagner G., Richarz R., Braun W. Correlations between internal mobility and stability of globular proteins. Biophys. J. 1980;32(1):549–560. doi: 10.1016/s0006-3495(80)84989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sun W., Guo J., Zhao G., Sun S., Yu H., et al. In silico design of a DNA-based HIV-1 multi-epitope vaccine for Chinese populations. Hum. Vaccin. Immunother. 2015;11(3):795–805. doi: 10.1080/21645515.2015.1012017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C. Genotyping coronavirus SARS-CoV-2: methods and implications. ArXiv. 2020;2003(10965) doi: 10.1016/j.ygeno.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Cao J., Zhao L.J., Qin Z.L., Ke J.S., Pan W., Ren H., Yu J.G., Qi Z.T. Immune responses against SARS-coronavirus nucleocapsid protein induced by DNA vaccine. Virology. 2005;331(1):128–135. doi: 10.1016/j.virol.2004.10.016. (Jan 5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong-ho Lee, Zheng William, Zhou Laura. South China Morning Post; 2020. Chinese Scientists Race To Develop Vaccine As Coronavirus Death Toll Jumps. Archived from the original on 26 January 2020. (Retrieved 28 January 2020) [Google Scholar]
- Zhou Y., Jiang S., Du L. Prospects for a MERS-CoV spike vaccine. Expert Rev. Vaccines. 2018;17(8):677–686. doi: 10.1080/14760584.2018.1506702. (Aug 3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6(1):1–18. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.-Y. Coronaviruses-drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Predicted CTL and HTL epitopes of spike glycoprotein.
Predicted CTL and HTL epitopes of membrane.
Predicted CTL and HTL epitopes of envelope protein
Predicted CTL and HTL epitopes of nucleocapsid protein.
Allergenicity pattern and toxicity profile of top T cell epitopes.
Proposed CTL and HTL epitopes for vaccine construction.
Predicted conformational epitopes within construct V3.
Predicted IFN alpha producing epitopes in the vaccine construct V3.
NCBI IDs of the complete genome, spike glycoprotein, envelope protein, membrane protein and nucleocapsid protein of coronavirus with Genera and Sub-Genera.
Retrieved protein sequences of major structural proteins of COVID-19.
Secondary structure and domain analysis of spike glycoprotein, envelope protein, membrane protein and nucleocapsid proteins.