Abstract
Influenza viruses initiate infection in the upper respiratory tract (URT), but early viral tropism and the importance of cell-type-specific antiviral responses in this tissue remain incompletely understood. By infecting transgenic lox-stop-lox reporter mice with a Cre-recombinase-expressing influenza B virus, we identify olfactory sensory neurons (OSNs) as a major viral cell target in the URT. These cells become infected, then eliminate the virus and survive in the host post-resolution of infection. OSN responses to infection are characterized by a strong induction of interferon-stimulated genes and more rapid clearance of viral protein relative to other cells in the epithelium. We speculate that this cell-type-specific response likely serves to protect the central nervous system from infection. More broadly, these results highlight the importance of evaluating antiviral responses across different cell types, even those within the same tissue, to more fully understand the mechanisms of viral disease.
Keywords: olfactory sensory neurons, interferon stimulated genes, respiratory viruses, central nervous system, viral clearance, inflammation, epithelial cells
Graphical Abstract
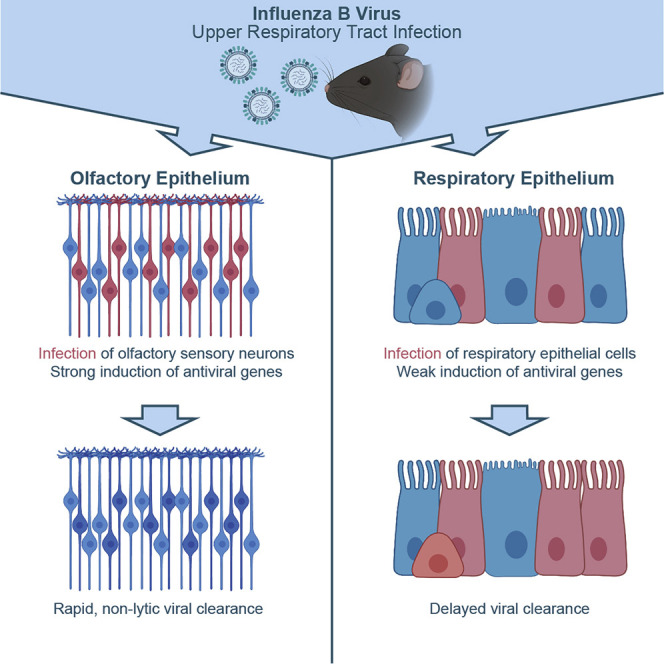
Highlights
-
•
Influenza B virus targets olfactory sensory neurons (OSNs) in the upper airways
-
•
OSNs exhibit a stronger and more rapid antiviral response compared to matched epithelial cells
-
•
Infected OSNs clear the virus non-lytically and persist in the host long term
-
•
The OSN antiviral response to infection may help prevent viral spread to the brain
In the upper respiratory tract, different cell types respond in different ways early after viral infection. Dumm et al. show that influenza B virus targets olfactory sensory neurons, but these neurons induce interferon-stimulated genes to rapidly clear the virus non-lytically and protect the CNS from disseminated infection.
Introduction
The upper respiratory tract (URT) is the primary site where respiratory pathogens, such as influenza viruses, initiate infection (Kuiken and Taubenberger, 2008; Tellier, 2006). Due to its location, the nature of its antiviral responses inherently affect the course of influenza disease (Reddy et al., 2016; van Riel et al., 2010). Failure to initially control infection in the URT can result in viral spread to other connected tissues, including the distal respiratory airways and central nervous system (CNS). Further, the cellular composition of URT is complex; cell types with unique and divergent functions populate functionally distinct regions such as the respiratory epithelium (RE) of the nasal concha, and the olfactory epithelium (OE) that mediates olfaction. Discrete cell types in the URT include subsets of epithelial cells responsible for mucous production and mucociliary clearance (Brooks and Wallingford, 2014; Spassky and Meunier, 2017), neuronal and neuroendocrine cells to transmit sensory and hormonal information (Schwob et al., 2017; Van Lommel, 2001), immune cells poised for incoming pathogens (Holtzman et al., 2002; Iwasaki et al., 2017), and connective cells to maintain tissue structure. Due to a lack of in vitro models that recapitulate this cellular diversity, as well as the technical challenges to studying the URT in vivo, interrogation of early viral tropism in this tissue has remained limited.
In addition to understanding which cells are infected, it is also important to understand their responses to infection. Different cell types can mount unique responses depending on their basal gene expression profiles, physiological function, and microenvironment (Bals and Hiemstra, 2004; Holtzman et al., 2002; Parker and Prince, 2011; Schleimer et al., 2007; Vareille et al., 2011; Whitsett and Alenghat, 2015). For example, inflammatory cytokines and chemokines produced by infected hematopoietic and epithelial cells can vary dramatically (Parker and Prince, 2011). Additionally, cell types are differentially permissive for infection, and even within a population of infectible cells, outcomes following infection vary. For cells that function primarily as barriers, high basal levels of antiviral effectors help to reduce viral infection rates and slow spread in the event of productive infection (Lazear et al., 2015; Wells and Coyne, 2018). Permissive infected cells can activate cell death pathways or be cleared by the immune system (Barber, 2001; Chen et al., 2018; Man et al., 2017; Orzalli and Kagan, 2017). Alternatively, some infected cells are able to non-lytically clear viral infection and survive beyond host viral clearance (Burdeinick-Kerr and Griffin, 2005; Chisari, 2000; Dumm et al., 2019; Fiege et al., 2019; Gomme et al., 2012; Heaton, 2017; Heaton et al., 2014; Reuther et al., 2015; Wheeler et al., 2017). Despite our understanding that cells can have a range of potential responses to infection, it remains generally unclear how differential cellular responses influence viral replication and host inflammation across disparate tissue types, including the URT.
In this study, our goal was to understand how cellular diversity in the URT may influence influenza virus infection. After surveying the URT for regions of infection, we observed that cells in both the RE and OE were productively infected. We defined the infected cells in the OE as predominantly olfactory sensory neurons (OSNs) and further showed that these infected cells non-lytically clear the virus and persist after viral clearance. Characterization of the antiviral responses of OSNs revealed higher expression of specific cell-intrinsic anti-influenza genes and more rapid viral clearance than those in other types of infected cells. These data highlight the importance of defining cell-type-specific antiviral responses even within a single tissue or organ.
Results
Characterization of Influenza B Virus Infection in the URT
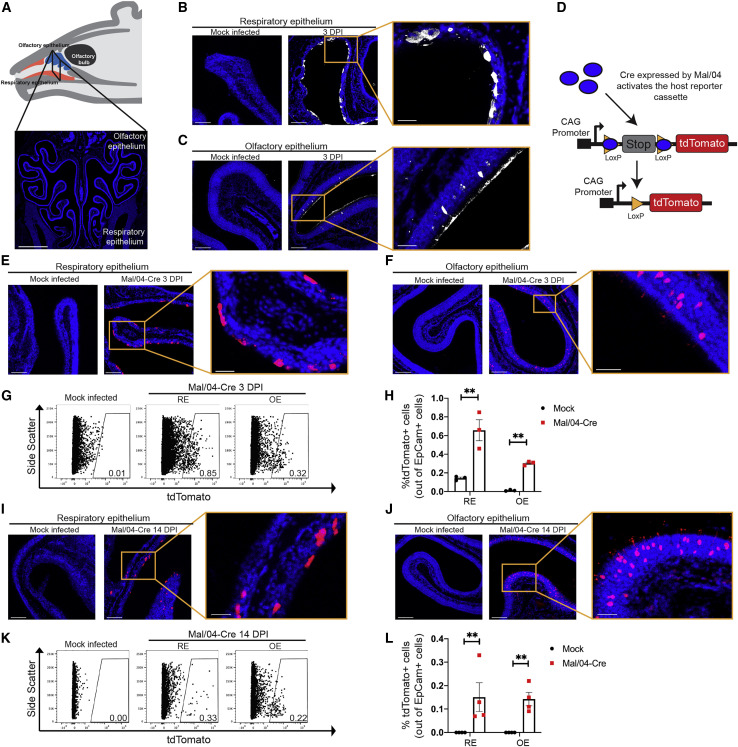
To model a human URT infection in mice, we chose a seasonal strain of influenza B virus (IBV; B/Malaysia/2506/2004-Mal/04), which preferentially replicates at the lower temperature of the URT (Bui et al., 2019; Laporte et al., 2019; Wang et al., 2007). We made use of a reporter Mal/04 variant, which expresses Cre recombinase after infection (Dumm et al., 2019); when target cells contain a lox-stop-lox-tdTomato cassette, Cre activity induces permanent fluorescent labeling, allowing us to identify actively infected cells and also cells that have cleared infection. To initially survey the infected cells in the URT (Figure 1 A), we infected mice and collected URT tissue at 3 days post-infection (DPI). Using polyclonal sera raised against Mal/04, we observed areas of the RE positive for viral protein (Figure 1B), with somewhat limited but still detectable levels of infection in the posterior OE (Figure 1C). These data are consistent with previous reports describing IBV replication primarily in the RE (Bui et al., 2019; Dando et al., 2014; Laporte et al., 2019; van Riel et al., 2015).
Figure 1.
Influenza B Virus-Infected Cells in the Olfactory and Respiratory Epithelium Can Eliminate Viral Infection
(A) Schematic and a coronal section of the mouse URT. The respiratory epithelium (RE) is in the ventral region, while the olfactory epithelium (OE) is in the dorsal region. Scale bar: 1,000 μm. Blue represents DAPI.
(B and C) Microscopy of the RE (B) and OE (C) stained for viral protein from a Mock- and Mal/04-Cre-infected mouse at 3 days post-infection. Scale bar: 100 μm; zoom scale bar: 30 μm. Blue represents DAPI; white represents viral protein.
(D) Schematic of viral activation of the lox-stop-lox-tdTomato reporter cassette.
(E and F) Microscopy of the RE (E) and OE (F) from lox-stop-lox tdTomato transgenic mice, infected with Mock and Mal/04-Cre virus and imaged at 3 days post-infection. Scale bar: 100 μm. Zoom scale bar: 30 μm. Blue represents DAPI; red represents tdTomato.
(G) Flow cytometry of RE and OE cells expressing tdTomato from Mock- and Mal/04-Cre-infected mice at 3 days post-infection.
(H) Quantification of tdTomato+ cells out of total epithelial (EpCam+) cells from the RE and OE of a Mal04-Cre infected mouse. Three independent mice were used to collect matching RE and OE samples.
(I and J) Microscopy of the RE (I) and OE (J) from lox-stop-lox tdTomato transgenic mice that were infected with Mock and Mal/04-Cre virus and imaged at 14 days post-infection. Scale bar: 100 μm. Zoom scale bar: 30 μm. Blue represents DAPI; red represents tdTomato.
(K) Flow cytometry of RE and OE cells expressing tdTomato from a Mock- and Mal/04-Cre-infected mouse at 14 days post-infection.
(L) Quantification of tdTomato+ cells out of total epithelial (EpCam+) cells from the RE and OE of a Mal04-Cre infected mouse. Four independent mice were used to collect matching RE and OE samples.
For all graphs, error bars indicate the SEM. ∗∗p ≤ 0.001. All data are representative of at least two biological replicates.
To define whether some infected cells may be poorly detected by antibody staining, as well as assess differences in the ultimate fates of the infected cells, we performed microscopy for the virally Cre-activated tdTomato reporter (Figure 1D). As expected, we observed tdTomato+ cells in the RE and OE, but in contrast with the antibody staining, the numbers of tdTomato+ cells were similar (within ∼2-fold) between the two tissues (Figures 1E–1H; Figures S1A–S1C). We next looked for tdTomato+ in the URT after viral clearance (Figure S1D). We found that previously infected cells were present in both the RE and OE (Figures 1I and 1J) at similar rates (Figures 1K and 1L; Figure S2). We have previously identified the subtypes of cells in the RE that can survive influenza virus infection (Dumm et al., 2019; Heaton et al., 2014); however, the identity of the surviving cells in the OE was unknown.
IBV Infects OSNs and Results in Upregulation of Canonical Antiviral Genes
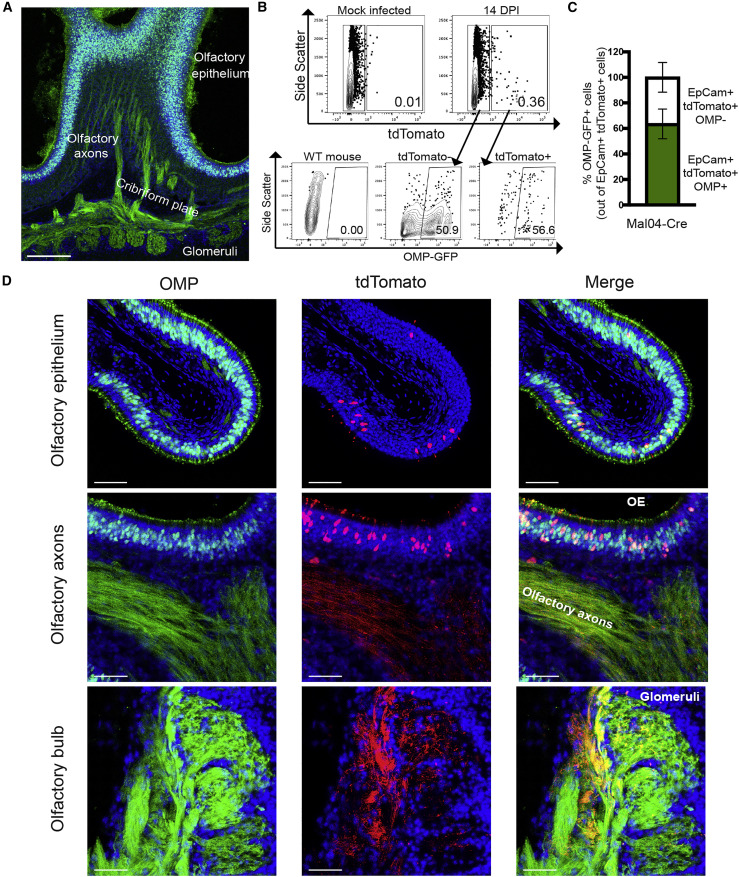
Closer analysis of the morphology of the tdTomato+ cells in the OE revealed a morphology reminiscent of OSNs, including dendritic knobs extending into the airspace (Figure 1J, inset). We therefore crossed a transgenic line expressing GFP under the control of the olfactory marker protein (OMP) promoter (Potter et al., 2001) to the lox-stop-lox-tdTomato mouse so that we could simultaneously visualize OSNs (Figure 2 A) and monitor infected/surviving cells. At 14 DPI, we found that on average ∼64% of tdTomato+ cells in the OE also express GFP, indicating that they are indeed OSNs (Figures 2B and 2C). The remainder of survivor cells that do not express OMP are likely either additional epithelial subtypes, contaminating RE cells, or immature OSNs with limited expression of OMP (Nickell et al., 2012). In addition to tdTomato/GFP colocalization in the OE, the axons of these cells were also co-labeled and can be seen terminating in the glomeruli of the olfactory bulb (Figure 2D). We failed to observe infection or replication in any other cell type in the olfactory bulb or other regions of the brain.
Figure 2.
Olfactory Sensory Neurons (OSNs) Can Survive Influenza B Virus Infection
(A) Microscopy of a sagittal section of the OE from OMP-GFP mice with axon tracks extending to the glomeruli of the olfactory bulb. Scale bar: 200 μm. Blue represents DAPI; green represents GFP.
(B) Flow cytometry of OE cells for tdTomato and OMP-GFP from mock or infected mice at 14 days post-infection.
(C) Quantification of OMP-GFP+ cells out of total survivor cells (tdTomato+) from a Mal04-Cre-infected mouse as measured via flow cytometry. n = 3 mice; error bars indicate the SEM.
(D) Microscopy of the OE, olfactory axons, and olfactory bulb from OMP-GFP; lox-stop-lox-tdTomato transgenic mice that were infected with Mal/04-Cre virus and imaged at 14 days post-infection. Scale bar: 30 μm. Blue represents DAPI; green represents GFP; red represents tdTomato. All data are representative of at least two biological replicates.
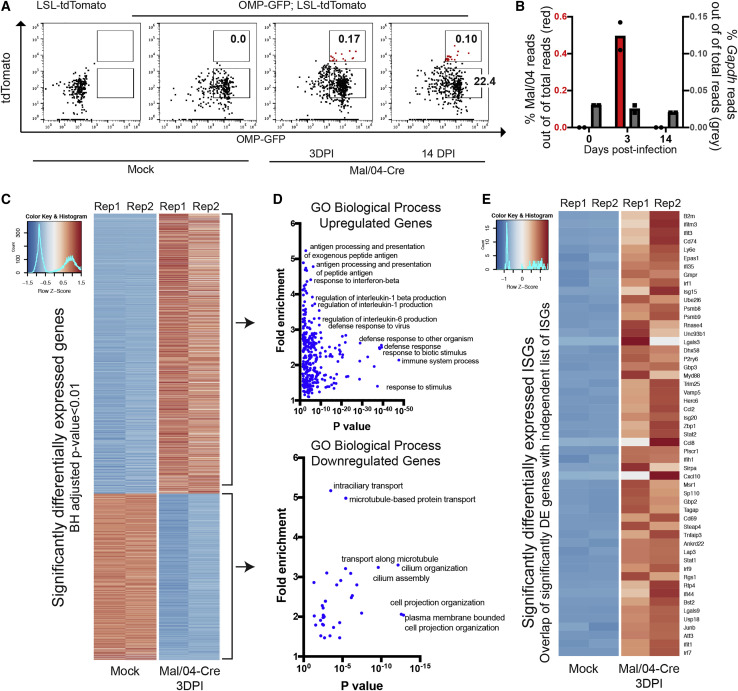
Neuropsychiatric complications of influenza infection are not restricted to neurotropic strains of the virus (Britton et al., 2017; Mizuguchi, 2013; Studahl, 2003), yet relatively little is known about infection or responses of neuronal cells to seasonal influenza strains (Studahl, 2003). We were therefore interested in understanding how OSNs, which link the brain to the URT, respond to viral infection and mediate non-lytic viral clearance. We sorted OMP+ OSNs from mock-infected mice and infected OMP+/tdTomato+ OSNs at 3 DPI and both OMP+/tdTomato− and OMP+/tdTomato+ cells 14 DPI. From these cells, we isolated mRNA and performed RNA sequencing (Figure 3 A; Data S1). Approximately 0.5% of total reads in the 3 DPI samples mapped to the Mal/04 genome, and as expected, viral RNA (vRNA) was cleared by 14 DPI (Figure 3B). High-level analysis of the host response revealed clusters of differentially induced genes between the uninfected, actively infected, and previously infected samples (Figure S3A).
Figure 3.
OSNs Induce Strong Antiviral Responses upon Infection
(A) Representative fluorescence-activated cell sorting (FACS) gating strategy to isolate live EpCam+ OMP+ OSNs from the OE of Mock- and Mal/04-Cre-infected mice is shown. For sequencing, tissues from three independent mice per time point were collected and pooled at 0, 3, and 14 days post-infection for mock, actively infected, and previously infected treatment groups, respectively.
(B) Reads mapped to the Mal/04 genome (shown in red) and Gapdh (shown in gray) and normalized to total reads in the Mock (0) and tdTomato+ cell population at 3 and 14 days post-infection.
(C) Heatmap of top significantly differentially expressed genes as ranked by a Benjamini-Hochberg adjusted p value. All “Olfr-” genes were excluded manually prior to analysis to account for sampling variability.
(D) Gene ontology analysis (PANTHER) of both upregulated and downregulated genes in infected OSNs.
(E) Heatmap of differentially expressed genes from an independent list (Schoggins et al., 2014) of interferon-stimulated antiviral genes. Rep, biological replicate.
Compared with mock-infected OSNs, actively infected OSNs at 3 DPI displayed the largest magnitude of gene expression changes (Figures 3C and 3D; Data S2). Gene Ontology (GO) analysis of upregulated genes revealed strong enrichment of expected pathways, such as immune responses against viral infection (Figure 3D; Data S2), which included key antiviral genes (Figure 3E). We also observed the downregulation of certain GO processes, such as cytoskeletal- and microtubule-dependent transport (Figure 3D; Data S2), pathways that we have previously observed to be suppressed after Mal/04 infection in other cell types (Dumm et al., 2019). At 14 DPI, we observed only minor transcriptional changes in bystander tdTomato− OSNs compared with OSNs from uninfected animals, indicating that the inflammatory environment induced by viral infection has limited effects on these cells (Figure S3B). For the surviving tdTomato+ OSNs at 14 DPI, we observed the same general gene expression patterns as the actively infected 3 DPI tdTomato+ OSNs; however, the magnitude of the transcriptional signature was lower (Figure S3B). Outside of antiviral response genes at 14 DPI, we did not observe major alterations to OSN gene expression patterns (Figure S3C; Data S3).
OSNs Express Higher Levels of Antiviral Effectors Than Do Similarly Infected Cells in the RE
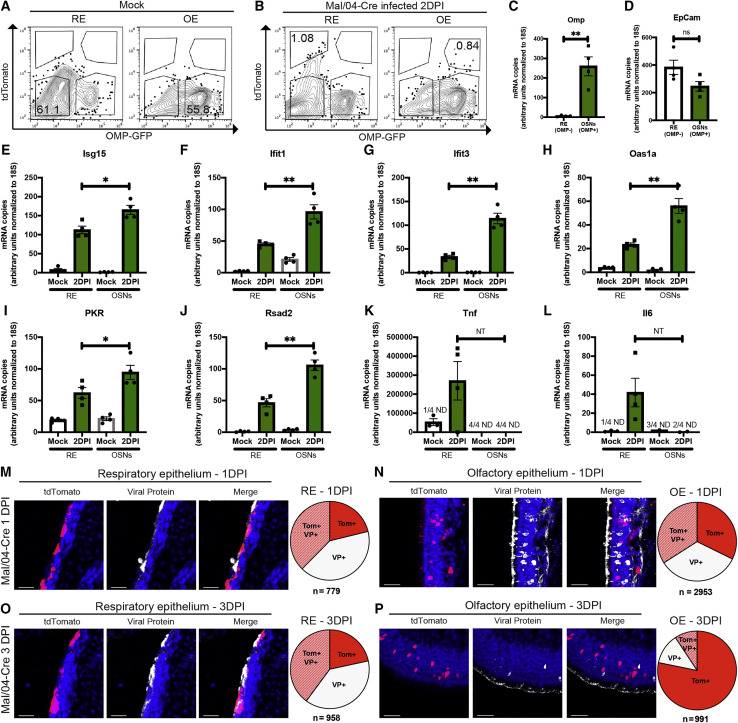
Because RNA sequencing identified a strong antiviral response in OSNs, we repeated our mouse infections and collected both OSNs and RE cells (along with corresponding tissues) to compare their early responses to infection (Figures 4 A–4D). qRT-PCR for the IBV genome, infectious virus titers, and tdTomato+OMP+ foci indicated productive infections of similar magnitude in both OSN and RE cell populations (Figures S4A–S4J). Quantification of anti-influenza interferon stimulated genes (ISGs) upregulated in the sequencing dataset (García-Sastre, 2011; Schoggins et al., 2014) revealed that many, including Isg15, Ifit1, Ifit3, Rsad2, Oas1a, and PKR, were expressed to a larger extent in OSNs compared with RE cells (Figures 4E–4J). This heightened antiviral response in OSNs did not extend to every antiviral gene that was tested as some genes, such as Ifit2, Ifitm3 (Figures S4K and S4L) and Tnf and Il6, were either induced to the same extent or more highly in RE cells (Figures 4K and 4L). Based on these data, we developed a model (Figure S4) in which initial viral burden is similar between the OE and RE, but the antiviral response of OSNs could shorten the duration of viral replication (Figure S4M).
Figure 4.
High-Magnitude Antiviral Responses Are Correlated with Rapid Viral Clearance from Infected OSNs
(A and B) FACS was used to isolate live CD31−CD45−CD326+ cells from the RE and OE of Mock- (A) and Mal/04-Cre-infected (B) mice at 2 days post-infection. Infected OSNs were identified as CD31−CD45−CD326+ OMP+tdTomato+ cells, and infected RE cells were identified as CD31−CD45−CD326+ OMP−tdTomato+ cells.
(C–L) qRT-PCR on transcripts from RE and OSN cells was used to quantify transcripts for (C) Omp, (D) EpCam, (E) Isg15, (F) Ifit1, (G) Ifit3, (H) Oas1a, (I) PKR, (J) Rsad2, (K) Tnf, and (L) Il6. All genes were quantified based on a standard curve and normalized to endogenous 18S expression either in the same sample or samples run on the same plate. Four independent mice were used to collect matching RE and OE samples; a one-way ANOVA with Tukey’s multiple test comparison was used to determine significance. For all graphs, error bars indicate the SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.001. ND, not detected; ns, not significant; NT, not tested due to undetected samples. Data are representative of at least two independent experiments.
(M and N) Microscopy of the RE (M) and OE (N) stained for viral protein from Mal/04-Cre-infected mice at 1 day post-infection. Scale bar: 30 μm. Blue represents DAPI; white represents viral protein; red represents tdTomato. Pie charts represent the number of cells positive for either viral protein only (VP+), viral protein and tdTomato (Tom+ VP+), or tdTomato only (Tom+) collected from 16 images from two independent mice.
(O and P) Microscopy of the RE (O) and OE (P) stained for viral protein from Mal/04-Cre-infected mice at 3 days post-infection. Scale bar: 30 μm. Blue represents DAPI; white represents viral protein; red represents tdTomato. Pie charts represent the number of cells for the indicated groups collected from 24 images from three independent mice.
After Infection, OSNs Clear Viral Protein More Quickly Than RE Cells Do
In order to directly test viral clearance kinetics in different URT cell types, we utilized our Cre-based labeling system in conjunction with viral protein staining, allowing us to categorize infected cells as being either early or late in viral infection, or post-viral clearance. At 1 DPI, in both the RE and OE, we observed roughly equal percentages of cells expressing only viral protein, cells co-expressing both viral protein and tdTomato, and cells expressing only tdTomato (Figures 4M and 4N). Similar analyses at 3 DPI showed that although the RE looked similar to the 1 DPI time point (Figure 4O), there were striking differences in the OE. Only 12.5% of cells maintained viral protein expression in the OE, whereas 9.3% of cells were co-labeled and 78.2% of cells had completely cleared viral infection and expressed tdTomato alone (Figure 4P). These data are consistent with more rapid intrinsic viral clearance in OSNs compared with cells in the RE and are further supported by an overall more rapid decrease in numbers of cells positive for viral protein in the OE compared with RE (Figures S4N and S4O).
Discussion
In this study, we demonstrate involvement of the OE and OSNs during infection with a seasonal strain of IBV. OSN responses to IBV are characterized by a strong induction of antiviral gene responses, which led to more rapid clearance of virus when compared with RE cells. Despite regional heterogeneity across the URT, the entire tissue must function as an effective barrier to protect more vulnerable tissues from any airborne pathogens (Diamond et al., 2000; Parker and Prince, 2011). Earlier studies have reported that OSNs undergo apoptosis following influenza infection and suggested this as a neuroprotective mechanism (Mori et al., 2002). However, our study shows that OSNs can be infected and go on to survive. Understanding the frequency of apoptosis and non-lytic clearance, as well as how that may affect barrier function/viral dissemination, is an important area of future study.
Unlike neighboring areas of the RE within the URT, the OE is not specialized for barrier function per se, but rather chemosensation. To rapidly convey environmental odorant information, OSNs position their dendrite in the airway, while also extending an axon to the olfactory bulb of the CNS. This arrangement means that OSNs provide a direct single-cell route from the outside world to the CNS. Viruses that enter and infect OSNs can bypass conventional protective barriers of the CNS (blood-brain barrier, blood-cerebrospinal fluid barrier) and travel intracellularly from the airway into the CNS with only the antiviral capacity of OSNs standing in opposition. The data presented in this work suggest that OSNs are equipped with specialized antiviral responses that prevent both infected cell death and viral dissemination.
Although our findings indicate that OSNs have a robust antiviral program, case reports of human influenza encephalopathy during seasonal influenza infection suggest these antiviral programs can fail (Lee et al., 2010; Mastrolia et al., 2019; Meijer et al., 2016). Despite CNS involvement in some human influenza cases, evidence of viral RNA or infectious viral particles are uncommon (Lee et al., 2010; Steininger et al., 2003). Congenital or transient variation in antiviral responsiveness and antiviral gene expression within OSNs may lead to enhanced viral replication and either increased release of inflammatory mediators or direct viral transmission into the CNS. We therefore propose that variations in OSN antiviral responses could contribute to why certain individuals present with overt neurological complications after IBV infection (McCullers et al., 1999; Newland et al., 2003; Popescu et al., 2017; Reyes et al., 2019). Not surprisingly, vaccination against influenza appears to reduce the frequency of influenza-associated encephalopathy, indicating that pre-existing adaptive immunity can lessen the burden of OSN innate antiviral capacity for CNS protection (Ono et al., 2003). Although not specifically addressed in our study, it will also be important to understand how viral infection and/or survival of OSNs affect CNS inflammation.
Our data also suggest influenza infection of the OE and OSNs is likely a common occurrence in humans, and that the OSN can respond with strong cell-intrinsic mechanisms but likely secrete few inflammatory mediators. Because OSN responses initiated via infection in the OE could still mediate distal effects within the CNS, follow-up studies are needed to assess potential long-term changes in immune readiness within the CNS. In addition, URT viral infections are clinically associated with an impairment in sense of smell, a disorder known as post-viral olfactory disorder (PVOD) (Seiden, 2004; Temmel et al., 2002). Recently, loss of smell has been identified as an early symptom of SARS-CoV-2 infection (Whitcroft and Hummel, 2020), suggesting that the virus disrupts olfactory neuronal sensory function. Whether this is due to direct infection remains unclear, but it is possible that SARS-CoV-2 neurological manifestations may be the result of individual variations in OSN antiviral responses. In future studies, it will be important to define how viral infection affects the ability of these individual surviving cells to participate in olfaction and whether there is long-term loss of olfactory capacity (Choi and Goldstein, 2018).
Traditionally, influenza is thought of as an exclusively respiratory pathogen, except in rare cases (such as with some highly pathogenic avian strains) that can replicate in the CNS (Aronsson et al., 2003; Jang et al., 2009). Despite this, the most commonly reported extra-respiratory symptom of influenza disease is CNS related, and the mechanisms underlying these symptoms are unclear (Goenka et al., 2014; Lee et al., 2010). By using an indelible viral labeling strategy, we were able to uncover robust infection of cells that directly link the airway to the CNS that may be at least partially responsible for these phenotypes. Specialized antiviral responses in this tissue likely developed to protect the CNS, and we suspect that analogous selective pressures drove the evolution of other tissue-specific immune responses (Hu and Pasare, 2013; Moutsopoulos and Konkel, 2018). This work and future studies defining viral/host interactions at the sub-tissue or even cell-type level will be important to fully define the mechanisms of viral pathogenesis and may identify mechanisms that can be targeted to alleviate influenza disease.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| EpCam/CD326 | BD Biosciences | Cat# 563478; RRID: AB_2738234 |
| CD45 | BD Biosciences | Cat# 557659; RRID: AB_396774 |
| CD31 | Biolegend | Cat# 102410; RRID: AB_312905 |
| Polyclonal sera from B/Mal/04 infected mice | Dumm et al., 2019 | N/A |
| Bacterial and Virus Strains | ||
| B/Malaysia/2506/2004 PB1-Cre | Dumm et al., 2019 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase D | Sigma | Cat# 11088858001 |
| DNase | Sigma | Cat# D4527 |
| Dispase | Corning | Cat# 354235 |
| Critical Commercial Assays | ||
| SMART-Seq v4 Ultra Low Input RNA Kit | Takara | Cat# 634889 |
| Nextera XT Index Kit | Illumina | Cat# FC-131-1024 |
| Deposited Data | ||
| Bulk RNA sequencing data | This paper | GSE145110 |
| Experimental Models: Cell Lines | ||
| MDCK | ATCC | CCL-34 |
| Experimental Models: Organisms/Strains | ||
| B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (lsl-tdTomato) | Jackson Laboratories | 007914 |
| OMP-GFP mice (Potter et al., 2001) | Dorian McGavern (NINDS, NIH) | N/A |
| Oligonucleotides | ||
| Primers, see Supplemental Dataset 8 | This paper | N/A |
| Software and Algorithms | ||
| Imaris x64 7.4.2 | Bitplane | N/A |
| Bowtie | Illumina BaseSpace | N/A |
| DESeq2 | Love et al., 2014 | N/A |
| Panther Gene Ontology | Mi et al., 2019 | N/A |
Resource Availability
Lead Contact
Further information and requests for reagents should be directed to Dr. Nicholas Heaton (nicholas.heaton@duke.edu).
Materials Availability
Data and reagents are available from the lead contact at request.
Data and Code Availability
The raw and analyzed for RNA-sequencing is accessible at NCBI GEO under the accession number GSE145110.
Experimental Model and Subject Details
Mouse lines
B6.Cg-Gt(ROSA)26Sor tm14(CAG-tdTomato)Hze/J (lsl-tdTomato) were purchased from Jackson Laboratory. OMP-GFP mice (Potter et al., 2001) on a C57BL/6 background were kindly provided by Dorian McGavern (NINDS, NIH). All colonies of homozygous and OMP-GFP; lsl-tdTomato heterozygous mice were bred and maintained in Duke Facilities. No blinding was performed in animal experiments. Both male and female animals were used at 6-12 weeks of age and randomly assigned to mock and infected treatment groups. All experiments involving animals were conducted in accordance with Duke University Animal Care and Use Committee.
Viruses and infections
Influenza B viruses (B/Malaysia/2506/2004) were used for these experiments. We utilized a recombinant strain expressing a Cre recombinase in the PB1 segment of the Mal/04 genome (Dumm et al., 2019). All viral stocks were grown in embryonated chicken eggs in a humidified incubator at 33°C. Mice were anesthetized with 0.1 mL of ketamine/xylazine and subsequently administered between 10-40 μL of virus diluted in pharmaceutical grade PBS. Infected animals in Figures 1 and 2 were carried out with mice infected with 100,000 PFU in 40 uL. Subsequent experiments shown in Figures 3 and 4 were infected with 500,000 PFU in 10 uL to target the upper respiratory tract. Infection doses ranged from 100,000-500,000 PFU/mouse. Mice were monitored for weight loss as a sign of morbidity and 80% of their starting bodyweight was used as a humane endpoint, however, we used primarily sublethal doses of virus for these experiments.
Method Details
Plaque assays
Upper respiratory tract tissues were collected from mice at 48 h post-infection and minced. After filtration, samples were plaqued on MDCK cells (ATCC, CCL-34) and incubated at 33C for 72 h. The cultures were then fixed and stained with polyclonal serum to recognize viral proteins and plaque-forming units were quantified.
Microscopy
Mice were collected at the appropriate time point, injected intraperitoneally with 500 μL of chloral hydrate (4 g/mL), and then systemically perfused with 2.5% buffered formalin in PBS. Mouse heads were dissected and skin and fur removed before incubation overnight in fixative. The tissue was then decalcified by incubation in 0.5M EDTA for at least 5 days at 4°C before mounting in tissue freezing medium (General Data #TFM-5) and sectioning. The frozen tissue was sectioned by a cryostat at −20°C, and each 30-micron section was adhered to a treated microscope slide (Fisher #12-550-15). Each slide was incubated with 400 μL of primary antibody stain overnight at 4°C. Slides were washed with PBS and incubated for 2 hr at room temperature with 400 μL of secondary antibody. Slides were washed with PBS and coverslipped with Mowiol mounting media. Primary antibody stain: DAPI 1 mg/mL (1:800) and anti-Mal/04 sera (1:260,000). Secondary antibody stain: AlexaFluor 647 goat anti-mouse IgG 1.5 mg/mL (1:1250) (Jackson ImmunoResearch #115-605-003). All images were captured with a Leica SP8 Upright Confocal microscope. Image analysis was performed with Imaris x64 7.4.2.
Flow cytometry
Mice were collected at the appropriate time point, euthanized, and then systemically perfused with PBS. Mouse heads were dissected to remove connective tissue then the brain was removed to access the olfactory epithelium in the posterior region of the nasal cavity and respiratory epithelium in the anterior section of the snout. Olfactory epithelial tissue was then digested in Hibernate A (Thermofisher) containing 0.5 mg/mL Collagenase D (Sigma 11088858001) and 0.5 mg/mL DNase (Sigma D4527). Respiratory epithelium was digested in DMEM containing 20 U/mL Dispase (Corning 354235). Following 30 min of digestion, the tissue was minced, washed 2X with PBS and filtered using a 70 μM filter (Corning #431751) to generate a single-cell suspension. Cells were then stained with Live/Dead (ThermoFisher L34964) EpCam (BD Biosciences #563478), CD45 (BD Biosciences 557659), CD31 (Biolegend 102410), or polyclonal serum from previously infected mice which will react to multiple viral proteins. All flow cytometry experiments were run on a BD Fortessa.
Prep for RNA sequencing
OMP;lox-stop-lox tdTomato mice were collected at 0 and 3 DPI for Mock and actively infected time points and olfactory tissue was digested as outlined above for flow cytometry. Olfactory epithelial cells were then stained using Live/Dead, EpCam and identified as OMP-GFP+ using tdTomato as a marker for infection. Cells were sorted on a SONY SH800S Cell Sorter using a 100 μM microfluidic sorting chip. cDNA libraries were then prepped using the SMART-Seq v4 Ultra Low Input RNA Kit for sequencing directly from sorted cells. Samples were barcoded for Illumina sequencing using Nextera XT Index Kit and sequenced on a single lane of HiSeq with 150 bp paired end reads. Data can be accessed at NCBI GEO under the accession number (GSE145110). Reads were mapped to the mm10 mouse transcriptome and the B/Mal04 genome using Bowtie on Basespace (Illumina).
RNA sequencing analysis using DeSeq2
Prior to analysis, we manually removed all olfactory sensory neuronal receptor genes (“Olfr-“) to minimize the sampling variability from sequencing a limited number of OSNs. Mapped reads were imported into R then scaled using the default “DESeq2” function using a log transformation and normalizing to capture moderately expressed genes (Love et al., 2014). Variance estimates were calculated across samples and using to normalize samples for heatmaps. The default “Results” function was used to compare the expression of Mock and actively infected samples and the top significantly differentially expressed genes (as defined by Benjamini-Hochberg adjusted p value < 0.01 were then selected to plot in a heatmap. These genes were then sorted as upregulated or downregulated by fold change and used as input to PANTHER Gene Ontology gene function analysis (Mi et al., 2019). Finally, all significantly differentially expressed genes were compared with an independent list of interferon-stimulated genes upregulated by influenza viruses (Schoggins et al., 2014) and the overlapping genes were plotted on a heatmap.
Quantitative PCR
OMP;lox-stop-lox tdTomato mice were collected at 2 DPI for an actively infected time point and respiratory and olfactory tissue was digested as outlined above for flow cytometry. Samples were stained using Live/Dead, CD326 (Epcam), CD31 and CD45. RE cells were defined as CD326+CD31-CD45-OMP- from respiratory epithelial digests. OE cells were defined as CD326+CD31-CD45-OMP+ from olfactory epithelial tissue digests. Endogenous tdTomato was used to distinguish infected cells from uninfected cells. At least 200 cells were collected from 4 mice for each RE and OE samples and samples prepared using Smart-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Cat# 634889). Samples were amplified using primers from this kit to generate cDNA libraries. Quantitative PCR was performed using TaqMan Universal PCR Master Mix (ThermoFisher Scientific, Cat# 4304437) and using 18S as an endogenous control 18S (ThermoFisher Scientific 4319413E). qPCR was performed using the following assays: Isg15 (Mm01705338_s1), Ifit1 (Mm00515153_m1, Ifit2 (Mm00492606_m1), Ifit3 (Mm01704846_s1), PKR (Mm01235643_m1), Oas1a (Mn_00836412_m1), Rsad2 (Mm00491265_m1), EpCam (Mm00493214_m1), OMP (Mm00448081_s1), and Ifitm3 (Mm00847057_s1). Gene expression was plotted on an arbitrary standard curve then normalized to 18S either in the same sample or from the same sample on the same plate as an endogenous control. 4 independent mice were used to collect matching RE and OE samples, a one-way ANOVA with Tukey’s multiple test comparison was used to determine significance. Quantification of viral reads was performed with the following custom primers and probes (IDT): PB2 – F: CACTGAAGGCTCAGTTTCTTCTA, R: CTGTACTGACCAGCCATCTTC, P: /56-FAM/AGAGGACAT/ZEN/GTTCCAATGGGATGCA/3IABkFQ/, PB1 – F: GCAACCGCTGGAATACAAATC, R: CATTTGACAGTTTGGCCTTCTT, P: /56-FAM/CCCACGGGC/ZEN/AAACCACTTTGTTC/3IABkFQ/, PA – F: GCAAGGATGTCTCCCTTAGTATC, R: CTTCTGGTAGCTCATGGTTGT, P: /56-FAM/AGGACCTAA/ZEN/GACCAATAGGGCCTCA/3IABkFQ/, HA – F: CCCAGAAGTTCACCTCATCTG, R: GTAGTCCTCCGTCTTCTGTTTG, P: /56-FAM/CCAACGGAG/ZEN/TGACCACACATTACGT/3IABkFQ/, NP – F: GAGTTGGACTTGACCCTTCAT, R: CACTAGAGTTCCACCTCCTTTG, P: /56-FAM/TACCTTTGC/ZEN/AGGAAGCACACTCCC/3IABkFQ/, NA – F: AGGCATCAAGGGAGGATTTG, R: GGGTCTCCATCATACTTGACATAC, P: /56-FAM/TGGAAGGTG/ZEN/GTACTCTCGAACGATGT/3IABkFQ/, M – F: CTAGGAACGCTCTGTGCTTTAT, R: TAGCTGAGACCATCTGCATTTC, P: /56-FAM/AGCATCACA/ZEN/TTCACACAGGGCTCA/3IABkFQ/, NS – F: CTCACTCTTCGAGCGTCTTAAT, R: CCAGTCTAATTGTCTCCCTCTTC, P: /56-FAM/TAAGACTCC/ZEN/CACCGCAGTTTCAGC/3IABkFQ/. Any fold changes described in the text were calculated from the average of two independent qPCR replicates.
Quantification and Statistical Analysis
For all tests involving a single comparison between two samples, statistical analysis was done using a two-tailed Student’s t test. For all analyses involving comparisons between multiple samples, a one-way ANOVA was done followed by a Tukey’s multiple test comparison to determine significance. Standard error of the mean (SEM) was plotted to indicate variability around the mean for tests samples. All experiments included the number of biological replicates indicated in the figure legend and were reproduced at least twice. Sample sizes were chosen based on the anticipated magnitude of the measured phenotype. Statistical analysis was not done on any comparisons where values were lower than the limit of detection, and this was noted in the figure legend. No samples were excluded from statistical analysis for any comparisons with significance noted. Analysis was performed using Prism 7 software (Graphpad). For all comparisons, ∗p ≤ 0.05, ∗∗p ≤ 0.001, ns not significant.
Acknowledgments
We would like to acknowledge assistance from Mike Cook and the Duke Cancer Institute Flow Cytometry Core, as well as the Duke Light Microscopy Core Facility. We would like to thank David Sachs for development of the bioinformatic applications to map sequencing reads to the mouse transcriptome and the influenza genome. We would also like to thank Ben Chambers, Brook Heaton, and Heather Froggatt for critical reading of this manuscript and Hiroaki Matsunami (as well as members of his laboratory) for helpful discussions. N.S.H. is partially supported by grants from the National Heart, Lung, and Blood Institute (R01-HL142985) and the National Institute of Allergy and Infectious Diseases (R01-AI137031). E.A.M. is partially supported by the Duke School of Medicine Whitehead Family Scholarship.
Author Contributions
R.E.D., E.A.M., and N.S.H. designed the study. R.E.D., E.A.M., and S.A.W. performed experiments. R.E.D., S.A.W., E.A.M., and N.S.H. performed data analysis and interpretation. R.E.D., E.A.M., and N.S.H. wrote the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: September 1, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108103.
Supplemental Information
Tissues from three independent mice were pooled prior to cell isolation, the replicates represent biological replicates. All ‘Olfr-’ genes were excluded manually prior to subsequent analysis to account for stochastic isolation of olfactory receptor genes due to sampling variability. Related to Figure 3.
Tab 1: Top variably expressed genes between Mock and 3 DPI (actively infected) time points, as displayed in the heatmap in Figure 3C. Genes are ranked by fold change difference in expression. Tab 2: Gene ontology of overrepresented Biological Process categories using upregulated genes between Mock and 3 DPI (actively infected) time points. This analysis was done using PANTHER Biological Process and displayed in the Figure 3D. Tab 3: Gene ontology of overrepresented Biological Process categories using downregulated genes between Mock and 3 DPI (actively infected) time points. This analysis was done using PANTHER Biological Process and displayed in the Figure 3D. Related to Figure 3.
Tab 1: Top variably expressed genes between 14 tdTomato- and 14 tdTomato+ DPI (survivor cell) populations. Tab 2: Gene ontology of overrepresented Biological Process categories using upregulated genes between 14 tdTomato- and 14 tdTomato+ DPI (survivor cell) populations. This analysis was done using PANTHER Biological Process and displayed in Figure S3C. Tab 3: Gene ontology of overrepresented Biological Process categories using downregulated genes between 14 tdTomato- and 14 tdTomato+ DPI (survivor cell) populations. This analysis was done using Panther Biological Process and displayed in Figure S3C. Related to Figure 3.
References
- Aronsson F., Robertson B., Ljunggren H.G., Kristensson K. Invasion and persistence of the neuroadapted influenza virus A/WSN/33 in the mouse olfactory system. Viral Immunol. 2003;16:415–423. doi: 10.1089/088282403322396208. [DOI] [PubMed] [Google Scholar]
- Bals R., Hiemstra P.S. Innate immunity in the lung: how epithelial cells fight against respiratory pathogens. Eur. Respir. J. 2004;23:327–333. doi: 10.1183/09031936.03.00098803. [DOI] [PubMed] [Google Scholar]
- Barber G.N. Host defense, viruses and apoptosis. Cell Death Differ. 2001;8:113–126. doi: 10.1038/sj.cdd.4400823. [DOI] [PubMed] [Google Scholar]
- Britton P.N., Blyth C.C., Macartney K., Dale R.C., Li-Kim-Moy J., Khandaker G., Crawford N.W., Marshall H., Clark J.E., Elliott E.J., Australian Childhood Encephalitis (ACE) Study Investigators, Influenza Complications Alert Network (FluCAN) Investigators, and Paediatric Active Enhanced Disease Surveillance (PAEDS) Network The Spectrum and Burden of Influenza-Associated Neurological Disease in Children: Combined Encephalitis and Influenza Sentinel Site Surveillance From Australia, 2013-2015. Clin. Infect. Dis. 2017;65:653–660. doi: 10.1093/cid/cix412. [DOI] [PubMed] [Google Scholar]
- Brooks E.R., Wallingford J.B. Multiciliated cells. Curr. Biol. 2014;24:R973–R982. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui C.H.T., Chan R.W.Y., Ng M.M.T., Cheung M.C., Ng K.C., Chan M.P.K., Chan L.L.Y., Fong J.H.M., Nicholls J.M., Peiris J.S.M., Chan M.C.W. Tropism of influenza B viruses in human respiratory tract explants and airway organoids. Eur. Respir. J. 2019;54:1900008. doi: 10.1183/13993003.00008-2019. [DOI] [PubMed] [Google Scholar]
- Burdeinick-Kerr R., Griffin D.E. Gamma interferon-dependent, noncytolytic clearance of sindbis virus infection from neurons in vitro. J. Virol. 2005;79:5374–5385. doi: 10.1128/JVI.79.9.5374-5385.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu S., Goraya M.U., Maarouf M., Huang S., Chen J.L. Host Immune Response to Influenza A Virus Infection. Front. Immunol. 2018;9:320. doi: 10.3389/fimmu.2018.00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari F.V. Rous-Whipple Award Lecture. Viruses, immunity, and cancer: lessons from hepatitis B. Am. J. Pathol. 2000;156:1117–1132. doi: 10.1016/s0002-9440(10)64980-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi R., Goldstein B.J. Olfactory epithelium: Cells, clinical disorders, and insights from an adult stem cell niche. Laryngoscope Investig. Otolaryngol. 2018;3:35–42. doi: 10.1002/lio2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dando S.J., Mackay-Sim A., Norton R., Currie B.J., St John J.A., Ekberg J.A., Batzloff M., Ulett G.C., Beacham I.R. Pathogens penetrating the central nervous system: infection pathways and the cellular and molecular mechanisms of invasion. Clin. Microbiol. Rev. 2014;27:691–726. doi: 10.1128/CMR.00118-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond G., Legarda D., Ryan L.K. The innate immune response of the respiratory epithelium. Immunol. Rev. 2000;173:27–38. doi: 10.1034/j.1600-065x.2000.917304.x. [DOI] [PubMed] [Google Scholar]
- Dumm R.E., Fiege J.K., Waring B.M., Kuo C.T., Langlois R.A., Heaton N.S. Non-lytic clearance of influenza B virus from infected cells preserves epithelial barrier function. Nat. Commun. 2019;10:779. doi: 10.1038/s41467-019-08617-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiege J.K., Stone I.A., Dumm R.E., Waring B.M., Fife B.T., Agudo J., Brown B.D., Heaton N.S., Langlois R.A. Long-term surviving influenza infected cells evade CD8+ T cell mediated clearance. PLoS Pathog. 2019;15:e1008077. doi: 10.1371/journal.ppat.1008077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 2011;162:12–18. doi: 10.1016/j.virusres.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka A., Michael B.D., Ledger E., Hart I.J., Absoud M., Chow G., Lilleker J., Lunn M., McKee D., Peake D. Neurological manifestations of influenza infection in children and adults: results of a National British Surveillance Study. Clin. Infect. Dis. 2014;58:775–784. doi: 10.1093/cid/cit922. [DOI] [PubMed] [Google Scholar]
- Gomme E.A., Wirblich C., Addya S., Rall G.F., Schnell M.J. Immune clearance of attenuated rabies virus results in neuronal survival with altered gene expression. PLoS Pathog. 2012;8:e1002971. doi: 10.1371/journal.ppat.1002971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton N.S. Revisiting the concept of a cytopathic viral infection. PLoS Pathog. 2017;13:e1006409. doi: 10.1371/journal.ppat.1006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton N.S., Langlois R.A., Sachs D., Lim J.K., Palese P., tenOever B.R. Long-term survival of influenza virus infected club cells drives immunopathology. J. Exp. Med. 2014;211:1707–1714. doi: 10.1084/jem.20140488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman M.J., Morton J.D., Shornick L.P., Tyner J.W., O’Sullivan M.P., Antao A., Lo M., Castro M., Walter M.J. Immunity, inflammation, and remodeling in the airway epithelial barrier: epithelial-viral-allergic paradigm. Physiol. Rev. 2002;82:19–46. doi: 10.1152/physrev.00020.2001. [DOI] [PubMed] [Google Scholar]
- Hu W., Pasare C. Location, location, location: tissue-specific regulation of immune responses. J. Leukoc. Biol. 2013;94:409–421. doi: 10.1189/jlb.0413207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Foxman E.F., Molony R.D. Early local immune defences in the respiratory tract. Nat. Rev. Immunol. 2017;17:7–20. doi: 10.1038/nri.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H., Boltz D., Sturm-Ramirez K., Shepherd K.R., Jiao Y., Webster R., Smeyne R.J. Highly pathogenic H5N1 influenza virus can enter the central nervous system and induce neuroinflammation and neurodegeneration. Proc. Natl. Acad. Sci. USA. 2009;106:14063–14068. doi: 10.1073/pnas.0900096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiken T., Taubenberger J.K. Pathology of human influenza revisited. Vaccine. 2008;26(Suppl 4):D59–D66. doi: 10.1016/j.vaccine.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte M., Stevaert A., Raeymaekers V., Boogaerts T., Nehlmeier I., Chiu W., Benkheil M., Vanaudenaerde B., Pöhlmann S., Naesens L. Hemagglutinin Cleavability, Acid Stability, and Temperature Dependence Optimize Influenza B Virus for Replication in Human Airways. J. Virol. 2019;94:e01430-19. doi: 10.1128/JVI.01430-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Nice T.J., Diamond M.S. Interferon-λ: Immune Functions at Barrier Surfaces and Beyond. Immunity. 2015;43:15–28. doi: 10.1016/j.immuni.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Wong C.K., Chan P.K., Lindegardh N., White N.J., Hayden F.G., Wong E.H., Wong K.S., Cockram C.S., Sung J.J., Hui D.S. Acute encephalopathy associated with influenza A infection in adults. Emerg. Infect. Dis. 2010;16:139–142. doi: 10.3201/eid1601.090077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Karki R., Kanneganti T.D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol. Rev. 2017;277:61–75. doi: 10.1111/imr.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrolia M.V., Rubino C., Resti M., Trapani S., Galli L. Characteristics and outcome of influenza-associated encephalopathy/encephalitis among children in a tertiary pediatric hospital in Italy, 2017-2019. BMC Infect. Dis. 2019;19:1012. doi: 10.1186/s12879-019-4636-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J.A., Facchini S., Chesney P.J., Webster R.G. Influenza B virus encephalitis. Clin. Infect. Dis. 1999;28:898–900. doi: 10.1086/515214. [DOI] [PubMed] [Google Scholar]
- Meijer W.J., Linn F.H., Wensing A.M., Leavis H.L., van Riel D., GeurtsvanKessel C.H., Wattjes M.P., Murk J.L. Acute influenza virus-associated encephalitis and encephalopathy in adults: a challenging diagnosis. JMM Case Rep. 2016;3:e005076. doi: 10.1099/jmmcr.0.005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi M. Influenza encephalopathy and related neuropsychiatric syndromes. Influenza Other Respir. Viruses. 2013;7(Suppl 3):67–71. doi: 10.1111/irv.12177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori I., Goshima F., Imai Y., Kohsaka S., Sugiyama T., Yoshida T., Yokochi T., Nishiyama Y., Kimura Y. Olfactory receptor neurons prevent dissemination of neurovirulent influenza A virus into the brain by undergoing virus-induced apoptosis. J. Gen. Virol. 2002;83:2109–2116. doi: 10.1099/0022-1317-83-9-2109. [DOI] [PubMed] [Google Scholar]
- Moutsopoulos N.M., Konkel J.E. Tissue-Specific Immunity at the Oral Mucosal Barrier. Trends Immunol. 2018;39:276–287. doi: 10.1016/j.it.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newland J.G., Romero J.R., Varman M., Drake C., Holst A., Safranek T., Subbarao K. Encephalitis associated with influenza B virus infection in 2 children and a review of the literature. Clin. Infect. Dis. 2003;36:e87–e95. doi: 10.1086/368184. [DOI] [PubMed] [Google Scholar]
- Nickell M.D., Breheny P., Stromberg A.J., McClintock T.S. Genomics of mature and immature olfactory sensory neurons. J. Comp. Neurol. 2012;520:2608–2629. doi: 10.1002/cne.23052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Kudo M., Aoki K., Ezaki F., Misumi J. Effect of mass immunization against influenza encephalopathy on mortality rates in children. Pediatr. Int. 2003;45:680–687. doi: 10.1111/j.1442-200x.2003.01813.x. [DOI] [PubMed] [Google Scholar]
- Orzalli M.H., Kagan J.C. Apoptosis and Necroptosis as Host Defense Strategies to Prevent Viral Infection. Trends Cell Biol. 2017;27:800–809. doi: 10.1016/j.tcb.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker D., Prince A. Innate immunity in the respiratory epithelium. Am. J. Respir. Cell Mol. Biol. 2011;45:189–201. doi: 10.1165/rcmb.2011-0011RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu C.P., Florescu S.A., Lupulescu E., Zaharia M., Tardei G., Lazar M., Ceausu E., Ruta S.M. Neurologic Complications of Influenza B Virus Infection in Adults, Romania. Emerg. Infect. Dis. 2017;23:574–581. doi: 10.3201/eid2304.161317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S.M., Zheng C., Koos D.S., Feinstein P., Fraser S.E., Mombaerts P. Structure and emergence of specific olfactory glomeruli in the mouse. J. Neurosci. 2001;21:9713–9723. doi: 10.1523/JNEUROSCI.21-24-09713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K.P., Bajwa E.K., Parker R.A., Onderdonk A.B., Walensky R.P. Relationship Between Upper Respiratory Tract Influenza Test Result and Clinical Outcomes Among Critically Ill Influenza Patients. Open Forum Infect. Dis. 2016;3:ofw023. doi: 10.1093/ofid/ofw023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuther P., Göpfert K., Dudek A.H., Heiner M., Herold S., Schwemmle M. Generation of a variety of stable Influenza A reporter viruses by genetic engineering of the NS gene segment. Sci. Rep. 2015;5:11346. doi: 10.1038/srep11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes C., Miranda S., Fica A., Navarrete M. [Encephalitis caused by type B influenza virus in an adult. Report of one case] Rev. Med. Chil. 2019;147:922–927. doi: 10.4067/S0034-98872019000700922. [DOI] [PubMed] [Google Scholar]
- Schleimer R.P., Kato A., Kern R., Kuperman D., Avila P.C. Epithelium: at the interface of innate and adaptive immune responses. J. Allergy Clin. Immunol. 2007;120:1279–1284. doi: 10.1016/j.jaci.2007.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., MacDuff D.A., Imanaka N., Gainey M.D., Shrestha B., Eitson J.L., Mar K.B., Richardson R.B., Ratushny A.V., Litvak V. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwob J.E., Jang W., Holbrook E.H., Lin B., Herrick D.B., Peterson J.N., Hewitt Coleman J. Stem and progenitor cells of the mammalian olfactory epithelium: Taking poietic license. J. Comp. Neurol. 2017;525:1034–1054. doi: 10.1002/cne.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiden A.M. Postviral olfactory loss. Otolaryngol. Clin. North Am. 2004;37:1159–1166. doi: 10.1016/j.otc.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Spassky N., Meunier A. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 2017;18:423–436. doi: 10.1038/nrm.2017.21. [DOI] [PubMed] [Google Scholar]
- Steininger C., Popow-Kraupp T., Laferl H., Seiser A., Gödl I., Djamshidian S., Puchhammer-Stöckl E. Acute encephalopathy associated with influenza A virus infection. Clin. Infect. Dis. 2003;36:567–574. doi: 10.1086/367623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studahl M. Influenza virus and CNS manifestations. J. Clin. Virol. 2003;28:225–232. doi: 10.1016/s1386-6532(03)00119-7. [DOI] [PubMed] [Google Scholar]
- Tellier R. Review of aerosol transmission of influenza A virus. Emerg. Infect. Dis. 2006;12:1657–1662. doi: 10.3201/eid1211.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmel A.F., Quint C., Schickinger-Fischer B., Klimek L., Stoller E., Hummel T. Characteristics of olfactory disorders in relation to major causes of olfactory loss. Arch. Otolaryngol. Head Neck Surg. 2002;128:635–641. doi: 10.1001/archotol.128.6.635. [DOI] [PubMed] [Google Scholar]
- Van Lommel A. Pulmonary neuroendocrine cells (PNEC) and neuroepithelial bodies (NEB): chemoreceptors and regulators of lung development. Paediatr. Respir. Rev. 2001;2:171–176. doi: 10.1053/prrv.2000.0126. [DOI] [PubMed] [Google Scholar]
- van Riel D., den Bakker M.A., Leijten L.M., Chutinimitkul S., Munster V.J., de Wit E., Rimmelzwaan G.F., Fouchier R.A., Osterhaus A.D., Kuiken T. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am. J. Pathol. 2010;176:1614–1618. doi: 10.2353/ajpath.2010.090949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D., Verdijk R., Kuiken T. The olfactory nerve: a shortcut for influenza and other viral diseases into the central nervous system. J. Pathol. 2015;235:277–287. doi: 10.1002/path.4461. [DOI] [PubMed] [Google Scholar]
- Vareille M., Kieninger E., Edwards M.R., Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin. Microbiol. Rev. 2011;24:210–229. doi: 10.1128/CMR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Tian X., Chen X., Ma J. Structural basis for receptor specificity of influenza B virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 2007;104:16874–16879. doi: 10.1073/pnas.0708363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells A.I., Coyne C.B. Type III Interferons in Antiviral Defenses at Barrier Surfaces. Trends Immunol. 2018;39:848–858. doi: 10.1016/j.it.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler D.L., Athmer J., Meyerholz D.K., Perlman S. Murine Olfactory Bulb Interneurons Survive Infection with a Neurotropic Coronavirus. J. Virol. 2017;91:e01099-17. doi: 10.1128/JVI.01099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcroft K.L., Hummel T. Olfactory Dysfunction in COVID-19: Diagnosis and Management. JAMA. 2020;323:2512–2514. doi: 10.1001/jama.2020.8391. [DOI] [PubMed] [Google Scholar]
- Whitsett J.A., Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 2015;16:27–35. doi: 10.1038/ni.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tissues from three independent mice were pooled prior to cell isolation, the replicates represent biological replicates. All ‘Olfr-’ genes were excluded manually prior to subsequent analysis to account for stochastic isolation of olfactory receptor genes due to sampling variability. Related to Figure 3.
Tab 1: Top variably expressed genes between Mock and 3 DPI (actively infected) time points, as displayed in the heatmap in Figure 3C. Genes are ranked by fold change difference in expression. Tab 2: Gene ontology of overrepresented Biological Process categories using upregulated genes between Mock and 3 DPI (actively infected) time points. This analysis was done using PANTHER Biological Process and displayed in the Figure 3D. Tab 3: Gene ontology of overrepresented Biological Process categories using downregulated genes between Mock and 3 DPI (actively infected) time points. This analysis was done using PANTHER Biological Process and displayed in the Figure 3D. Related to Figure 3.
Tab 1: Top variably expressed genes between 14 tdTomato- and 14 tdTomato+ DPI (survivor cell) populations. Tab 2: Gene ontology of overrepresented Biological Process categories using upregulated genes between 14 tdTomato- and 14 tdTomato+ DPI (survivor cell) populations. This analysis was done using PANTHER Biological Process and displayed in Figure S3C. Tab 3: Gene ontology of overrepresented Biological Process categories using downregulated genes between 14 tdTomato- and 14 tdTomato+ DPI (survivor cell) populations. This analysis was done using Panther Biological Process and displayed in Figure S3C. Related to Figure 3.
Data Availability Statement
The raw and analyzed for RNA-sequencing is accessible at NCBI GEO under the accession number GSE145110.