Abstract
This study aimed to determine the value of ARL9 expression or methylation as a biomarker for LGG survival. We investigated the expression, methylation, prognosis and immune significance of ARL9 through bioinformatics analysis. ARL9 is negatively regulated by ARL9 methylation, leading to its low expression in LGG tissues. Both low ARL9 expression and hypermethylation predicted favorable OS and PFS in LGG patients, according to the TCGA database. Cox regression demonstrated that low ARL9 expression and ARL9 hypermethylation were independent biomarkers for OS. Moreover, three other glioma databases were utilized to verify the prognostic role of ARL9 in LGG, and the similar results were reached. A meta-analysis revealed that low ARL9 expression was closely relevant to better OS. Finally, ARL9 expression exhibited a close correlation with some immune cells, especially CD8+ T cells. ARL9 could constitute a promising prognostic biomarker, and probably plays an important role in immune cell infiltration in LGG.
Keywords: Low-grade glioma, ARL9, Methylation, Survival, Immune cells
Abbreviations: LGG, Low-grade glioma; ARL9, ADP-ribosylation factor-like 9; OS, Overall survival; PFS, Progression-free survival; TCGA, the Cancer Genome Atlas; CGGA, Chinese Glioma Genome Atlas; Rembrandt, Repository for Molecular Brain Neoplasia Data; ROC, Receiver operating characteristic; GEPIA, Gene Expression Profiling Interactive Analysis; TIMER, Tumor Immune Estimation Resource
Highlights
-
•
This is the first study to report the clinical and prognostic significance of ARL9, a methylation-driven gene,in LGG.
-
•
Meta-analysis could be used for bioinformatics analysis to assess the overall effect of the gene from different datasets.
-
•
ARL9 probably plays a role in the infiltration of immune cells, and acts as a promising prognostic marker in LGG patients.
1. Introduction
Low-grade glioma (LGG) represents the most common primary malignancy occurred in the brain, and LGG exhibits great intrinsic heterogeneity with regard to tumor biological behavior [1]. Despite comprehensive therapy for LGG, including neurosurgical resection, chemotherapy and radiotherapy, therapeutic resistance and tumor recurrence appear to be inevitable [2,3]. Some patients with LGG have indolent outcomes, while others may rapidly progress to high grade glioblastoma (GBM), which usually represents an unfavorable outcome [4]. Hence, finding a novel biomarker with high accuracy for predicting prognosis of LGG patients is urgently needed.
The ADP-ribosylation factor (ARF) family generally belongs to the RAS superfamily which plays an oncogenic role in the pathogenesis and metastasis of glioma [5,6]. Both ADP-ribosylation factor-like (ARL) 2 and ARL3, as the classic members of the ARF family, were reported to expressed at low levels in glioma and their expression is inversely correlated with unfavorable prognosis among glioma patients [7,8]. ARL9 is a kind of novel GTP-binding protein, and is extremely conserved and ubiquitously expressed in eukaryotes [9]. However, as a novel member of the ARF family, the clinical and prognostic significance of ARL9 in glioma is still unknown, and its functional role in LGG has never been documented.
In this study, we first explored the differential expression of ARL9 mRNA in LGG tissues and normal tissues, analyzed the correlation between ARL9 expression and ARL9 DNA methylation in the LGG dataset of The Cancer Genome Atlas (TCGA), and assessed the prognostic significance of ARL9 expression and its DNA methylation. Then, we validated the prognostic role of ARL9 using data from the Chinese Glioma Genome Atlas (CGGA), Gravendeel and Repository for Molecular Brain Neoplasia Data (Rembrandt) databases. In addition, we performed a comprehensive meta-analysis to assess the overall prognostic significance of ARL9 using data from four public databases. Moreover, as substantial attention has been focused on the crucial role of the immune microenvironment in the progression of LGG [[10], [11], [12], [13]], we also mined Tumor Immune Estimation Resource (TIMER) database to evaluate the potential correlation between ARL9 and immune infiltration levels in LGG. Finally, we examined the biological processes of ARL9 in which ARL9 is involved through gene enrichment analysis, to study the functional mechanism of ARL9 in LGG.
2. Materials and methods
2.1. Data mining from public databases
First, we searched an online website Gene Expression Profiling Interactive Analysis (GEPIA) [14], which is available at http://gepia.cancer-pku.cn/index.html, to investigate the differential expression of ARL9 mRNA in LGG tissues and normal tissues. Then, we downloaded the clinical data, transcriptional along with methylation profiles of patients with LGG from TCGA database via the Cbioportal website [15] (https://www.cbioportal.org/). The inclusion criteria were (1) patients with WHO grade II or III and (2) patients with complete clinical and transcriptional data. Moreover, we further downloaded clinical data and ARL9 mRNA expression data from three other public databases, namely, CGGA [16] (http://www.cgga.org.cn/), Rembrandt (http://gliov is.bioin fo.cnio.es/) and Gravendeel ((http://gliov is.bioin fo.cnio.es/) to verify the prognostic role of ARL9 in LGG. A total of 1367 patients with confirmed LGG (TCGA:527 patients; CGGA:592 patients; Rembrandt:139 patients; Gravendeel:109 patients) were included in this analysis.
2.2. Meta-analysis
The PubMed, Web of Science and Embase databases were systematically searched to identify all published studies on the association between ARL9 and prognosis of LGG or glioma. As this is the first study to investigate the prognostic role of ARL9 in LGG, no previous studies were obtained from the databases. Therefore, we could utilize the meta-analysis to assess the overall prognostic significance of ARL9 in patients with LGG from 4 datasets. Combined HR and 95% CI were calculated to evaluate the correlation of ARL9 expression with prognosis of LGG patients. The heterogeneity across four datasets was assessed by the Q test (I2 statistics). A fixed-effects model would be selected for combination if no obvious heterogeneity (I2 < 50%). Otherwise, a random-effects model would be applied. The meta-analysis was completed using STATA 15.1 software.
2.3. TIMER database analysis
TIMER is a comprehensive website for the automatic analysis and visualization of association between immune infiltrate levels and a series of variables (https://cistrome.shinyapps.io/timer/) [17]. We assessed the correlation of ARL9 expression with the abundance of six kinds of immune cells (CD4 + T cells, CD8 + T cells, B cells, neutrophils, dendritic cells and macrophages) in LGG via the TIMER algorithm. Then, we also explored the prognostic value of ARL9 in LGG patients with different abundance of immune cells. In addition, we exploited the correlation module to estimate the correlation of ARL9 with the type markers of T cells (general), CD8 + T cells, B-cells, CD8 + T cells, neutrophils, monocytes, tumor-associated macrophages (TAM), M1 cells, M2 cells, dendritic cells, NK cells, Th1 cells, Th2 cells, Treg cells in LGG. These gene markers of immune cells were well illustrated in prior studies [[18], [19]].
2.4. Gene ontology enrichment analysis
We applied GlioVis database (http://gliovis.bioinfo.cnio.es/) to complete the Gene ontology analysis. Patients with glioma from Rembrandt dataset were initially divided into high ARL9 expression and low ARL9 expression groups. Differentially expressed genes between the two groups were selected with the false discovery rate less than 0.05. Gene terms with |logFC| ≥ 1 combined with P value less than 0.05 were viewed as significant. Then we chose Gene ontology enrichment analysis and biological process to explore the functional role of ARL9 in glioma.
2.5. Statistical analysis
The data were analyzed using the Medcalc program (version 19.4) or GraphPad Prism (version 6.0). Low and high ARL9 expression groups were established based on the median ARL9 mRNA expression value in the separate datasets. Similarly, ARL9 hypomethylation and hypermethylation groups were established according to the median value of ARL9 DNA methylation in the TCGA-LGG dataset. The relationships between ARL9 expression or its DNA methylation and a series of categorical variables were analyzed by chi-square or Fisher exact-tests. The difference in continues indexes with normal distribution between two groups was determined by Student's t-test and continues indexes with skew distribution were inspected by a nonparametric test. The correlation of ARL9 expression with ARL9 DNA methylation level was measured by Pearson correlation coefficient. Moreover, we employed univariate along with multivariate Cox regression models to probe whether ARL9 expression was an independent prognostic index in patients with LGG. Kaplan-Meier curves were utilized to evaluate the prognostic significance of ARL9 expression along with ARL9 DNA methylation. Time dependent-receiver operating characteristic (td-ROC) analyses were utilized to assess the predictive performance of ARL9 in predicting OS. P-values less than 0.05 on both sides were statistically significant.
3. Results
3.1. The clinical and prognostic value of ARL9 expression and methylation according to TCGA database
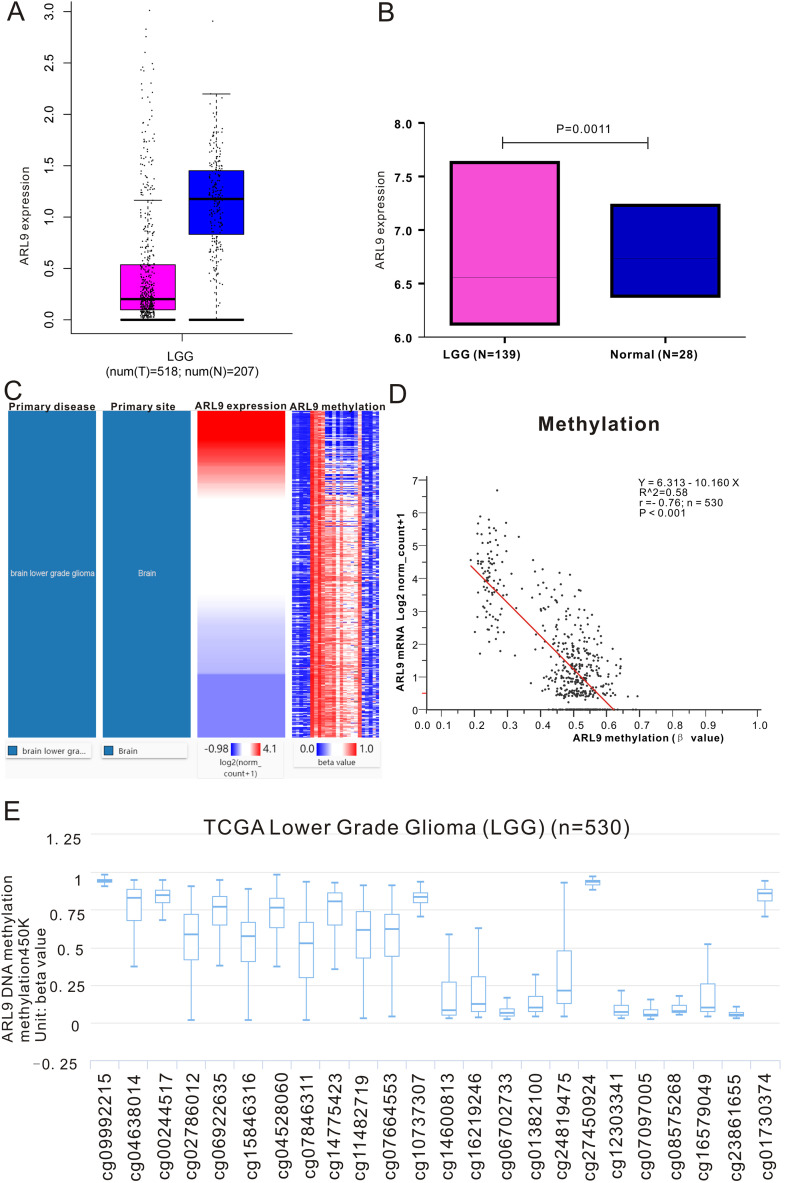
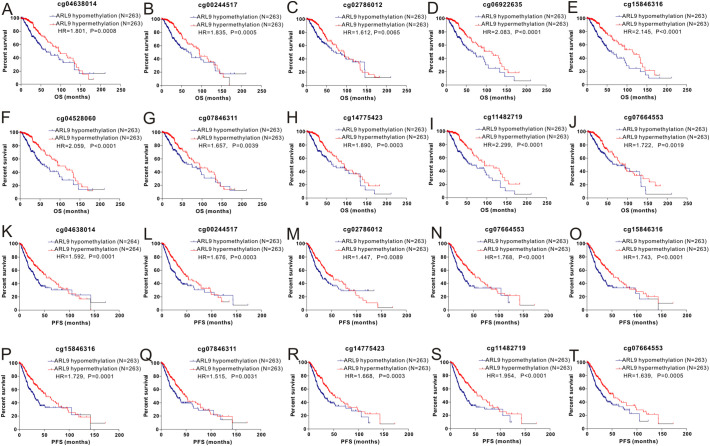
The RNA-sequencing data of 518 LGG tissues from TCGA and 207 normal samples from the GTEx project were analyzed with GEPIA, and we found that ARL9 mRNA was lowly expressed in LGG tissues, while highly expressed in normal tissues (Fig. 1A). This differential expression was also confirmed by data from the Rembrandt dataset (Fig. 1B). As exhibited in the heatmap (Fig. 1C), we could observe a strong negative correlation (r = −0.76, P < 0.0001) between ARL9 expression and ARL9 DNA methylation (Fig. 1D). The distribution of 24 ARL9 CpG sites was clearly exhibited in Fig. 1E. Then Pearson correlation analysis was exploited to identify the ARL9 CpG sites at which methylation was most strongly correlated with ARL9 mRNA expression. As listed in Table S1, except for 3 CpG sites (cg09992215, cg10737307 and cg01382100), methylation of the remaining CpG sites well correlated with the expression of ARL9. We selected the most relevant CpG sites (|r| > 0.7, P < 0.0001), as demonstrated in Fig. 2 , to investigate the prognostic values of these significant ARL9 DNA CpG sites in patients with LGG. Kaplan-Meier plots demonstrated that high levels of methylation of the selected CpG sites were correlated with not only more favorable OS (Fig. 2A-J) but also better PFS (Fig. 2K-T) among patients with LGG. Then, the LGG patients in TCGA were dichotomized into low or high subgroups when divided by the median value of ARL9 expression or ARL9 methylation. We implemented the chi-square test to study the detailed correlation of ARL9 expression as well as ARL9 methylation with a panel of clinical features. As shown in Table 1 , the expression of ARL9 was closely correlated with age (P < 0.0001), histological type (P = 0.0282), family history of cancer (P = 0.0473), molecular subtype (P < 0.0001), radiotherapy (P = 0.0015), living status (P = 0.002) and ARL9 methylation (P < 0.0001). Similarly, the level of ARL9 methylation was affected by gender (P = 0.0391), histological type (P = 0.0351), molecular subtype (P < 0.0001), radiotherapy (P = 0.0412), living status (P = 0.0225) and ARL9 expression (P < 0.0001).
Fig. 1.
The expression and methylation of ARL9 in LGG tissues and normal tissues revealed by bioinformatic analysis. ARL9 mRNA is lowly expressed in LGG tissues in TCGA dataset (A) and Rembrandt dataset (B). C. Heatmap of the correlation of ARL9 expression with methylation of ARL9 DNA CpG sites. D. The expression of ARL9 was negatively regulated by ARL9 DNA methylation. E. The distribution of 24 ARL9 DNA promoter CpG sites.
Fig. 2.
Kaplan-Meier curves of low and high ARL9 DNA promoter CpG sites in LGG patients. cg04638014 (A, K), cg00244517(B, L), cg02786012 (C, M), cg06922635 (D, N), cg15846316 (E, O), cg04528060 (F, P), cg07846311 (G, Q), cg14775423 (H, R), cg11482719 (I, S) and cg07664553 (J, T).
Table 1.
Correlation between ARL9 mRNA expression/methylation and clinicopathologic features in TCGA database.
| Clinical features | ARL9 expression |
P value | ARL9 methylation |
P value | |||
|---|---|---|---|---|---|---|---|
| Low (%) | High(%) | Low (%) | High(%) | ||||
| Age | ≤50 | 209(79.47) | 161(60.98) | <0.001 | 176(66.67) | 194(73.76) | 0.0749 |
| >50 | 54(20.53) | 103(39.02) | 88(33.33) | 69(26.24) | |||
| Gender | Female | 113(42.97) | 123(46.59) | 0.4027 | 130(49.24) | 106(40.30) | 0.0391 |
| Male | 150(57.03) | 141(53.41) | 134(50.76) | 157(59.70) | |||
| Racea | White/American | 248(96.12) | 239(92.64) | 0.2107 | 239(92.64) | 248(96.12) | 0.2107 |
| Black/Africa american | 7(2.71) | 15(5.81) | 15(5.81) | 7(2.71) | |||
| Asian | 3(1.16) | 4(1.55) | 4(1.55) | 3(1.16) | |||
| Histological type | Astrocytoma | 85(32.32) | 112(42.42) | 0.0282 | 109(41.29) | 88(33.46) | 0.0351 |
| Oligoastrocytoma | 67(25.48) | 67(25.38) | 71(26.89) | 63(23.95) | |||
| Oligodendroglioma | 111(42.21) | 85(32.20) | 84(31.82) | 112(42.59) | |||
| Family history of cancer | No | 120(67.04) | 100(56.82) | 0.0473 | 116(60.42) | 104(63.80) | 0.5124 |
| Yes | 59(32.96) | 76(43.18) | 76(39.58) | 59(36.20) | |||
| Family history of brain tumor | No | 171(95.00) | 174(97.21) | 0.2802 | 183(94.82) | 162(97.59) | 0.2805 |
| Yes | 9(5.00) | 5(2.79) | 10(5.18) | 4(2.41) | |||
| Molecular subtype | IDH mut-codel | 103(40.23) | 64(25.50) | <0.001 | 54(21.26) | 113(44.66) | <0.0001 |
| IDH mut-non-codel | 149(58.20) | 99(39.44) | 109(42.91) | 139(54.94) | |||
| IDH-wildtype | 4(1.56) | 88(35.06) | 91(35.83) | 1(0.40) | |||
| Lateralitya | Left | 131(50.58) | 124(47.15) | 0.6718 | 124(47.88) | 131(49.81) | 0.8331 |
| Midline | 4(1.54) | 3(1.14) | 3(1.16) | 4(1.52) | |||
| Right | 124(47.88) | 136(51.71) | 132(50.97) | 128(48.67) | |||
| WHO grade | II | 137(52.09) | 120(45.63) | 0.1381 | 124(46.97) | 133(50.76) | 0.3841 |
| III | 126(47.91) | 143(54.37) | 140(53.03) | 129(49.24) | |||
| Cancer status | With tumor | 90(38.63) | 93(39.41) | 0.8625 | 92(39.48) | 91(38.56) | 0.8372 |
| Tumor -free | 143(61.37) | 143(60.59) | 141(60.52) | 145(61.44) | |||
| KPSb | ≤80 | 42(26.42) | 54(36.49) | 0.0572 | 56(35.67) | 40(26.67) | 0.089 |
| >80 | 117(73.58) | 94(63.51) | 101(64.33) | 110(73.33) | |||
| Sample type | Primary | 258(97.73) | 256(96.97) | 0.5880 | 258(97.36) | 256(97.34) | 0.9885 |
| Recurrent | 6(2.27) | 8(3.03) | 7(2.64) | 7(2.66) | |||
| Radiotherapy | No | 113(45.02) | 75(31.12) | 0.0015 | 83(33.74) | 105(42.68) | 0.0412 |
| Yes | 138(54.98) | 166(68.88) | 163(66.26) | 141(57.32) | |||
| Seizure history | No | 95(38.93) | 87(34.94) | 0.3581 | 90(36.59) | 92(37.25) | 0.879 |
| Yes | 149(61.07) | 162(65.06) | 156(63.41) | 155(62.75) | |||
| Living status | Alive | 212(80.61) | 182(68.94) | 0.002 | 186(70.45) | 208(79.09) | 0.0225 |
| Dead | 51(19.39) | 82(31.06) | 78(29.55) | 55(20.91) | |||
| MTDc | ≤2 cm | 258(97.73) | 251(95.08) | 0.1019 | 253(95.47) | 256(97.34) | 0.2495 |
| >2 cm | 6(2.27) | 13(4.92) | 12(4.53) | 7(2.66) | |||
| ARL9 mRNA expression | Low | – | – | – | 89(33.58) | 175(66.54) | <0.0001 |
| High | – | – | 176(66.42) | 88(33.46) | |||
| ARL9 Methylation | Low | 89(33.71) | 176(66.67) | <0.001 | – | – | – |
| High | 175(66.29) | 88(33.33) | – | – | – | ||
Fisher's exact test.
KPS represents karnofsky performance status.
MTD stands for maximum tumor diameter.
We utilized survival analyses to assess the association between ARL9 expression as well as ARL9 methylation and prognosis of patients with LGG. As depicted in Fig. S2, we discovered that patients with low ARL9 expression had an approximately a 2 fold longer OS than that with high ARL9 expression (HR = 0.4885, 95% CI = 0.3465–0.6889, P < 0.0001). Subgroup analyses were further implemented based on WHO grade, IDH1-mutation and histological type. We observed that low ARL9 expression was more closely correlated with favorable OS than high ARL9 expression in LGG patients with WHO grade III (P = 0.0002) and astrocytoma (P < 0.0001). Moreover, we also discovered that LGG patients with low ARL9 expression usually exhibited much better PFS than those with high ARL9 expression (HR = 0.6023, 95% CI = 0.4574–0.793, P = 0.0003, Fig. S2). Subgroup analyses revealed that low ARL9 expression was remarkably correlated with better PFS than high expression of ARL9 in LGG patients with WHO grade II (P = 0.0469), IDH1-wt (P = 0.0048), IDH1-mutation (P = 0.0362) and astrocytoma (P = 0.0006). Regarding the prognostic value of ARL9 methylation, patients with ARL9 hypermethylation exhibited longer OS (HR = 0.4681, 95%CI = 0.3298–0.6645, P < 0.0001) or PFS time (HR = 0.4945, 95% CI = 0.3726–0.6561, P < 0.0001) than those with ARL9 hypomethylation (Fig. S3). Subgroup analyses demonstrated that ARL9 hypermethylation was remarkably correlated with better OS or PFS than ARL9 hypomethylation in LGG patients with WHO grade II, grade III and astrocytoma. Furthermore, we conducted the univariate along with multivariate Cox regression analyses to probe the independent prognostic variables of patients with LGG, and we found that both low ARL9 expression (HR = 0.52, 95% CI = 0.283–0.957, P = 0.036) and ARL9 hypermethylation (HR = 0.40, 95% CI = 0.213–0.753, P = 0.004) were independent prognostic variables for favorable OS (Table S2). Similarly, ARL9 hypermethylation (HR = 0.45, 95% CI = 0.278–0.728, P = 0.001) was an independent prognostic marker for favorable PFS (Table S2).
3.2. The clinical and prognostic significance of ARL9 expression according to the CGGA database
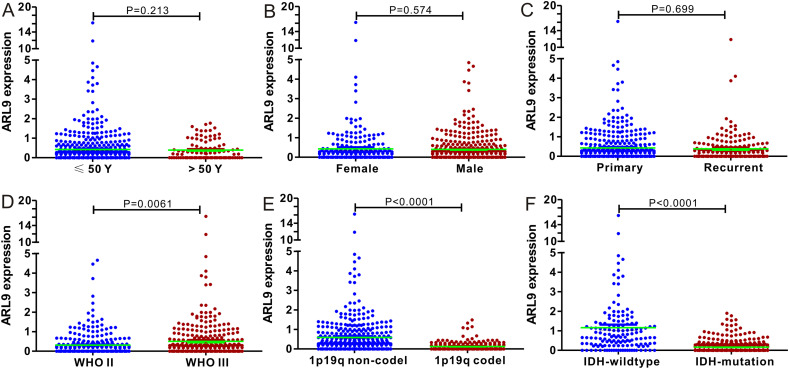
First, we used nonparametric test to compare the difference in ARL9 mRNA expression in groups divided by age, gender, cancer type, WHO grade, 1p19q codel and IDH1-mutation. As clearly exhibited in Fig. 3 , ARL9 mRNA expression was remarkably different in groups stratified by WHO grade (P = 0.0061), 1p19q codeletion (P < 0.001) and IDH1-mutation (P < 0.001), indicating the close correlation of ARL9 mRNA expression with a series of significant clinical parameters. While no association was found between ARL9 mRNA expression and age, gender and cancer type. In addition, we also confirmed the prognostic value of ARL9 expression in CGGA dataset by log rank test. LGG patients with low levels of ARL9 mRNA experienced a much longer OS time than LGG patients with high levels of ARL9 mRNA (HR = 0.6591, 95% CI = 0.5226–0.8313, P = 0.0004 Fig. S4). To investigate the impacts of clinical parameters on the prognostic significance of ARL9, we further implemented a series of subgroup analyses according to four crucial clinical indexes, including WHO grade, 1p19q codeletion, IDH1-mutation and radiotherapy. As revealed by a panel of Kaplan-Meier curves, the correlation of low ARL9 expression with better OS still existed in LGG patients with WHO III (P = 0.0002) and radiotherapy (P = 0.0001), while no correlation was observed among LGG patients with WHO II (P = 0.9487), 1p19q codeletion (P = 0.1913), 1p19q non-codeletion (P = 0.2245), IDH1-wt (P = 0.5633), IDH1-mutation (P = 0.0878) and no radiotherapy (P = 0.0693).
Fig. 3.
Correlation between ARL9 mRNA expression and clinical indexes of LGG patients from CGGA database. ARL9 mRNA expression is stratified by age (A), gender (B), sample type (C), WHO grade (D), 1p19q codeletion (E) and IDH1 mutation (F).
3.3. Validation of the prognostic value of ARL9 in two independent databases
To independently verify the prognostic significance of ARL9, we analyzed the results of microarray data of 139 LGG patients from the Rembrandt dataset and RNA sequencing data of 109 patients with LGG from the Gravendeel dataset. We employed Kaplan–Meier analysis to investigate the association between low ARL9 and OS in LGG patients (Fig. S5), and we found that low ARL9 was closely correlated with more favorable OS than high ARL9 both in the REMBRANDT dataset (HR = 0.5735, 95% CI: 0.3870–0.8499, P = 0.0059) and Gravendeel dataset (HR = 0.4272, 95% CI: 0.2723–0.6703, P = 0.0002). Subgroup analyses of Kaplan–Meier curves were conducted based on WHO grade, and we observed that low ARL9 was significantly correlated with much better OS than high ARL9 among patients with WHO III in both datasets. However, this association disappeared in patients with WHO II in Gravendeel dataset, perhaps due to the relatively small number of LGG patients with WHO II in this dataset.
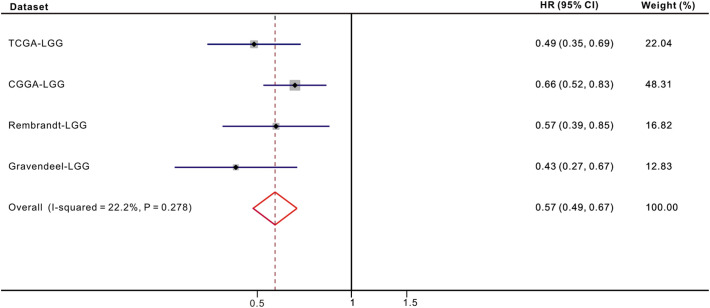
3.4. Meta-analysis and predictive performance of ARL9 expression
As no previous studies have reported an association between low ARL9 expression and OS among patients with LGG, we only included the results generated from four different datasets in meta-analysis. The pooled HR along with 95% CI for the association between low ARL9 expression and OS in 1367 cases of LGG patients was 0.57(0.49–0.67), and no significant heterogeneity among the 4 datasets was observed (I2 = 22.2%, P = 0.278, Fig. 4 ). Hence, we could confidently conclude that low ARL9 expression is a strong predictor of favorable OS among patients with LGG. Moreover, ROC curve analysis was applied to determine the predictive value of ARL9 expression for OS. The area under the curve (AUC) was calculated to predict the 1-year, 3-year and 5-year OS of LGG patients, and the ROC curves (Fig. S6) showed that ARL9 expression could predict 1-year, 3-year and 5-year OS with good predictability in LGG patients based on four datasets.
Fig. 4.
Forest plot of low ARL9 expression with better OS in LGG patients from four datasets.
3.5. Relationships of ARL9 with immune cells and PD-L1
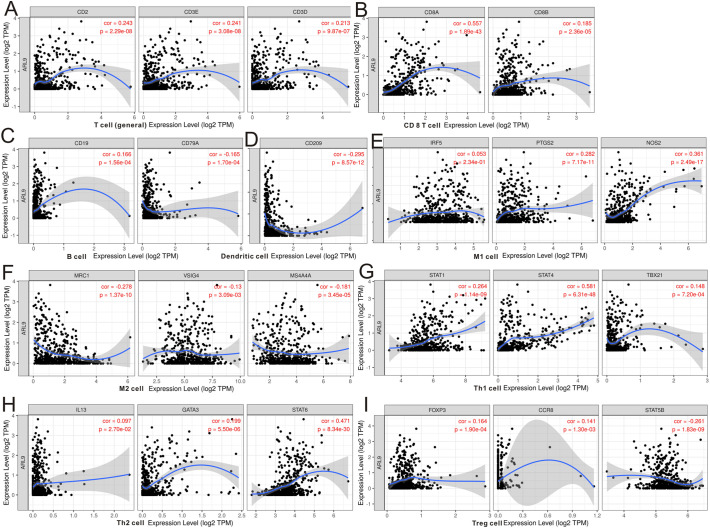
The TIMER database was searched to estimate the correlations of ARL9 mRNA expression with immune cell infiltration. As illustrated in the scatter plots (Fig. S7), the expression of ARL9 was negatively correlated with immune infiltration of CD4+ T cells (r = −0.115, P = 0.0118), and positively correlated with B cells (r = 0.128, P = 0.00492), CD8+ T cells (r = 0.357, P < 0.0001), and neutrophils (r = 0.156, P = 0.00065). No association was discovered between ARL9 expression and macrophage (r = 0.03, P = 0.513), or dendritic cell (r = 0.076, P = 0.0988) infiltration. In addition, we focused on the correlations of ARL9 expression with the markers of 14 immune cells in LGG using the TIMER database (Table S3). As shown in Fig. 5 , after adjusting for purity, the results demonstrated that ARL9 expression was remarkably correlated with most immune markers of T cells (general Fig. 5A), CD8 + T cells (Fig. 5B), B cells (Fig. 5C), dendritic cells(Fig. 5D), M1 cells (Fig. 5E), M2 cells (Fig. 5F), Th1 cells (Fig. 5G), Th2 cell (Fig. 5H) and Treg cells (Fig. 5I) in LGG. Moreover, due to the promising prospect of immunotherapy, we further determined the association between ARL9 expression and PD-1 (Fig. S7G) and PD-L1 (Fig. S7H). We also noticed a positive correlation of ARL9 with PD-1 (r = 0.14, P = 1.38e-3) and PD-L1 (r = 0.323, P = 5.08e-14).
Fig. 5.
The expression of ARL9 was related to a panel of gene markers of immune cells, including T cell (A), CD8+ T cell (B), B cell (C), dendritic cell (D), M1 cell (E), M2 cell (F), Th1 cell (G), Th2 cell (H) and Treg cell (I).
Given that expression of ARL9 was related to the immune infiltration in LGG, and that low ARL9 expression was also correlated with good OS in LGG, we speculated that ARL9 expression affected the prognosis in patients with LGG partly due to immune infiltration. Therefore, we further employed the Kaplan-Meier curves to validate our hypothesis (Fig. S8), and the results revealed that low ARL9 expression in enriched B cell (Fig. S8A), CD8+ T cells (Fig. S8B), CD4+ T cells (Fig. S8C), macrophages (Fig. S8D), neutrophils (Fig. S8E) and dendritic cells (Fig. S8F) was associated with more favorable prognosis in patients with LGG. Collectively, the above results indicated that low ARL9 expression may affect prognosis of patients with LGG partially due to immune infiltration.
3.6. ARL9-related signaling pathways in glioma
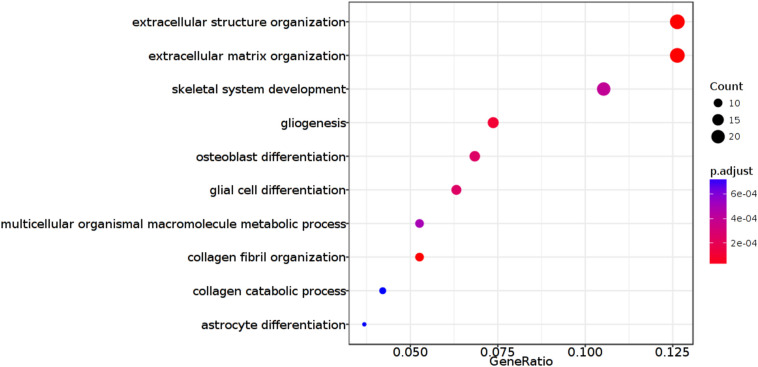
To gain insight into the functional role of ARL9 in glioma, we exploited the GlioVis database to determine the potential biological processes. The Rembrandt dataset was selected for Gene Ontology and the results are exhibited in Fig. 6 . ARL9 is involved in pathways of extracellular structure organization, extracellular matrix organization, skeletal system development, gliogenesis, osteoblast differentiation, and glial cell differentiation among others. Of note, ARL9 was implicated in gliogenesis and glial cell differentiation, and abnormal expression of ARL9 might lead to the occurrence and progression of glioma.
Fig. 6.
ARL9-related biological pathways in glioma.
4. Discussion
In our study, we analyzed the clinical and prognostic role of ARL9 mRNA expression as well as ARL9 methylation in LGG according to TCGA database. For the first time, we discovered low expression of ARL9 mRNA in LGG tissues, and a strongly negative association between ARL9 mRNA expression and ARL9 methylation. We found that both ARL9 expression and ARL9 methylation were closely associated with a series of significant features, including histological type and molecular type. Cox regression models established the critical role of low ARL9 expression and ARL9 hypermethylation in the favorable prognosis of patients with LGG. Moreover, we also verified the prognostic role of ARL9 expression in three other datasets, and the results all emphasized the promising prognostic value of ARL9 expression in LGG patients. Our meta-analysis containing 1367 LGG patients from 4 different databases further demonstrated that low ARL9 expression was an independent prognostic variable of OS in LGG patients. Finally, our analyses revealed that the levels of immune infiltration and a list of immune markers were significantly correlated with ARL9 expression in LGG. To our knowledge, our analyses provide novel insights into the prognostic role of ARL9 and potential role of ARL9 in the tumor immunology of LGG.
Some oncogenes implicated in the pathogenesis of gliomas have been identified and documented [20,21], but less is known about the clinical and prognostic role of ARL genes. Among the genes, the ARL9 has only been reported to be negatively correlated with malignant progression in prostate tumor [9], but has never studied in LGG. In this study, we systematically explored the association between low ARL9 expression and survival in four public databases. Encouragingly, low ARL9 expression was highly relevant with more favorable OS among LGG patients than high ARL9 expression. This conclusion was further confirmed by multivariate Cox regression which revealed that low ARL9 expression was a potent prognostic factor of OS in LGG patients. Finally, we adopted a meta-analysis to integrate four databases to assess the overall prognostic value of ARL9, and the pooled data of 1367 patients further proved that low ARL9 expression was truly associated with favorable OS among LGG patients. Collectively, our analyses emphasized that ARL9 is a promising biomarker for predicting prognosis of patients with LGG.
Previous studies [10,[22], [23], [24]] pointed out that tumor-infiltrating immune cells have emerged as a key regulator of tumor growth and progression in LGG. Tumor-infiltrating immune cells are part of the complex microenvironment and are associated with the biological behavior and patient survival in LGG [[25], [26], [27], [28], [29]]. In the present study, we observed that ARL9 expression was positively correlated (r = 0.357) with CD8+ T cells in LGG tissues. We further analyzed the cell markers (CD8A and CD8B) of T cells, and obtained consistent results. A recent study [22] reported that activated CD8+ T cells could secrete CCL4 which recruits microglia to secrete CCL5, a key growth factor for LGG stem cell survival. Although no study has provided insight into the correlation of ARL9 with immune cells, expression of ARL3, which belongs to the ARF family, also well correlated with the high abundance of CD8 + T cells, and these results were similar to the results from ARL9. Taken together, these results demonstrated that ARL9 was implicated in the tumor immune microenvironment mainly through the regulation of CD8+ T cells in LGG.
Increasing evidence [4,30] proves that abnormal DNA methylation occupies an essential role in the induction and progression of LGG. In our analysis, we first determined whether the ARL9 methylation status could influence ARL9 mRNA expression through Pearson coefficients. A potent negative correlation (r = −0.76) between ARL9 methylation and ARL9 mRNA expression existed in LGG tissues. This negative correlation could well explain the low expression of ARL9 in LGG tissues. Then, we further identified the specific CpG sites in the ARL9 DNA promoter at which methylation is significantly correlated with ARL9 mRNA expression. Surprisingly, almost all the CpG sites except for cg09992215, cg10737307 and cg01382100, showed significant associations with ARL9 expression. Among the remaining 21 significant CpG sites at which the ARL9 DNA promoter was methylated, methylation of 10 CpG sites showed very close correlation (|r| > 0.7) with ARL9 expression. In previous studies [[31], [32], [33]], the association between specific gene expression and its DNA methylation ranged from weak to moderate, and very few genes that are strongly regulated by DNA methylation were reported. In addition, we also investigated the prognostic significance of ARL9 DNA methylation and 10 selected CpG sites, and we found that ARL9 hypermethylation correlated well with favorable OS and PFS in patients with LGG, which was further proven by multivariate Cox regression. Taken together, ARL9 was negatively regulated by ARL9 methylation, and ARL9 methylation status might be a potent indicator of favorable OS and PFS.
It is worth noting that three inevitable limitations exist in our analysis. First, due to the limitations of CGGA, Rembrandt and Gravendeel databases, we could not validate the prognostic role of ARL9 methylation in LGG patients from the three databases. Then, only TCGA database contained PFS information, and the association of ARL9 expression and PFS could not be verified in three other database. Thus, it was unlikely to perform a meta-analysis of PFS. Finally, although we preliminarily explored the biological process of ARL9 in glioma through enrichment analysis, the detailed mechanism that links ARL9 expression and ARL9 methylation with LGG progression requires further biomedical experiments. Nevertheless, the current results are encouraging, and noteworthy in the field of identifying promising prognostic biomarkers for LGG.
5. Conclusion
ARL9 is downregulated in LGG and negatively regulated by DNA methylation in LGG. Either low ARL9 expression or ARL9 hypermethylation predicts favorable prognosis in LGG patients. Moreover, ARL9 expression potentially contributes to the regulation of T cells, B cells and macrophage cells. Hence, ARL9, which is negatively regulated by DNA methylation, probably plays an important role in the infiltration of immune cells, and could act as a promising prognosing biomarker in LGG patients.
Author statement
Author contribution: Kai Shu, Hongquan Niu and Ting Lei designed this study. Yutang Tan analyzed the data and wrote the manuscript. Weihua Liu, Kuan Huang, and Weidong Tian collected the data. Junwen Wang, Kai Zhao and Qungen Xiao analyzed the data. Suojun Zhang revised the manuscript. All authors approved the final version for submission.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
Not applicable.
References
- 1.Ostrom Q.T., Gittleman H., Farah P., Ondracek A., Chen Y., Wolinsky Y., Stroup N.E., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl. 2) doi: 10.1093/neuonc/not151. (ii1–56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayes J., Yu Y., Jalbert L.E., Mazor T., Jones L.E., Wood M.D., Walsh K.M., Bengtsson H., Hong C., Oberndorfer S., Roetzer T., Smirnov I.V., Clarke J.L., Aghi M.K., Chang S.M., Nelson S.J., Woehrer A., Phillips J.J., Solomon D.A., Costello J.F. Genomic analysis of the origins and evolution of multicentric diffuse lower-grade gliomas. Neuro-Oncology. 2018;20:632–641. doi: 10.1093/neuonc/nox205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brat D.J., Verhaak R.G., Aldape K.D., Yung W.K., Salama S.R., Cooper L.A., Rheinbay E., Miller C.R., Vitucci M., Morozova O., Robertson A.G., Noushmehr H., Laird P.W., Cherniack A.D., Akbani R., Huse J.T., Ciriello G., Poisson L.M., Barnholtz-Sloan J.S., Berger M.S., Brennan C., Colen R.R., Colman H., Flanders A.E., Giannini C., Grifford M., Iavarone A., Jain R., Joseph I., Kim J., Kasaian K., Mikkelsen T., Murray B.A., O’Neill B.P., Pachter L., Parsons D.W., Sougnez C., Sulman E.P., Vandenberg S.R., Van Meir E.G., von Deimling A., Zhang H., Crain D., Lau K., Mallery D., Morris S., Paulauskis J., Penny R., Shelton T., Sherman M., Yena P., Black A., Bowen J., Dicostanzo K., Gastier-Foster J., Leraas K.M., Lichtenberg T.M., Pierson C.R., Ramirez N.C., Taylor C., Weaver S., Wise L., Zmuda E., Davidsen T., Demchok J.A., Eley G., Ferguson M.L., Hutter C.M., Mills S.K., Ozenberger B.A., Sheth M., Sofia H.J., Tarnuzzer R., Wang Z., Yang L., Zenklusen J.C., Ayala B., Baboud J., Chudamani S., Jensen M.A., Liu J., Pihl T., Raman R., Wan Y., Wu Y., Ally A., Auman J.T., Balasundaram M., Balu S., Baylin S.B., Beroukhim R., Bootwalla M.S., Bowlby R., Bristow C.A., Brooks D., Butterfield Y., Carlsen R., Carter S., Chin L., Chu A. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y.A., Zhou Y., Luo X., Song K., Ma X., Sathe A., Girard L., Xiao G., Gazdar A.F. SHOX2 is a potent independent biomarker to predict survival of WHO grade II-III diffuse gliomas. EBioMedicine. 2016;13:80–89. doi: 10.1016/j.ebiom.2016.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong C., Shu M., Ye J., Wang X., Chen X., Liu Z., Zhao W., Zhao B., Zheng Z., Yin Z., Gao M., Zhao H., Wang K., Zhao S. Oncogenic Ras is downregulated by ARHI and induces autophagy by Ras/AKT/mTOR pathway in glioblastoma. BMC Cancer. 2019;19:441. doi: 10.1186/s12885-019-5643-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan Y., Jiang Y. RACK1 affects glioma cell growth and differentiation through the CNTN2-mediated RTK/Ras/MAPK pathway. Int. J. Mol. Med. 2016;37:251–257. doi: 10.3892/ijmm.2015.2421. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y., Guan G., Cheng W., Jiang Y., Shan F., Wu A., Cheng P., Guo Z. ARL2 overexpression inhibits glioma proliferation and tumorigenicity via down-regulating AXL. BMC Cancer. 2018;18:599. doi: 10.1186/s12885-018-4517-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Zhao W., Liu X., Guan G., Zhuang M. ARL3 is downregulated and acts as a prognostic biomarker in glioma. J. Transl. Med. 2019;17:210. doi: 10.1186/s12967-019-1914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Louro R., Nakaya H.I., Paquola A.C., Martins E.A., Da S.A., Verjovski-Almeida S., Reis E.M. RASL11A, member of a novel small monomeric GTPase gene family, is down-regulated in prostate tumors. Biochem. Biophys. Res. Commun. 2004;316:618–627. doi: 10.1016/j.bbrc.2004.02.091. [DOI] [PubMed] [Google Scholar]
- 10.Wu F., Li G.Z., Liu H.J., Zhao Z., Chai R.C., Liu Y.Q., Jiang H.Y., Zhai Y., Feng Y.M., Li R.P., Zhang W. Molecular subtyping reveals immune alterations in IDH wild-type lower-grade diffuse glioma. J. Pathol. 2020;251(3):272–283. doi: 10.1002/path.5468. [DOI] [PubMed] [Google Scholar]
- 11.Wu F., Wang Z.L., Wang K.Y., Li G.Z., Chai R.C., Liu Y.Q., Jiang H.Y., Zhai Y., Feng Y.M., Zhao Z., Zhang W. Classification of diffuse lower-grade glioma based on immunological profiling. Mol. Oncol. 2020 doi: 10.1002/1878-0261.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang M., Wang X., Chen X., Zhang Q., Hong J. Novel immune-related gene signature for risk stratification and prognosis of survival in lower-grade glioma. Front. Genet. 2020;11:363. doi: 10.3389/fgene.2020.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng X., Lin D., Zhang X., Shen X., Yang Z., Yang L., Lu X., Yu L., Zhang N., Lin J. Profiles of immune-related genes and immune cell infiltration in the tumor microenvironment of diffuse lower-grade gliomas. J. Cell. Physiol. 2020;235(10):7321–7331. doi: 10.1002/jcp.29633. [DOI] [PubMed] [Google Scholar]
- 14.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:11. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu H., Mu Q., Bao Z., Chen Y., Liu Y., Chen J., Wang K., Wang Z., Nam Y., Jiang B., Sa J.K., Cho H.J., Her N.G., Zhang C., Zhao Z., Zhang Y., Zeng F., Wu F., Kang X., Liu Y., Qian Z., Wang Z., Huang R., Wang Q., Zhang W., Qiu X., Li W., Nam D.H., Fan X., Wang J., Jiang T. Mutational landscape of secondary glioblastoma guides MET-targeted trial in brain tumor. Cell. 2018;175:1665–1678. doi: 10.1016/j.cell.2018.09.038. (e18) [DOI] [PubMed] [Google Scholar]
- 17.Li T., Fan J., Wang B., Traugh N., Chen Q., Liu J.S., Li B., Liu X.S. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Danaher P., Warren S., Dennis L., D’Amico L., White A., Disis M.L., Geller M.A., Odunsi K., Beechem J., Fling S.P. Gene expression markers of tumor infiltrating leukocytes. J. Immunother. Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J., Li H., Hu S., Zhou Y. ACE2 correlated with immune infiltration serves as a prognostic biomarker in endometrial carcinoma and renal papillary cell carcinoma: implication for COVID-19. Aging (Albany NY) 2020;12:6518–6535. doi: 10.18632/aging.103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L.M., Li Z., Piao Y.S., Cai Y.N., Zhang L.Y., Ge H.J., Xu W.W., Lu D.H. Clinico-neuropathological features of isocitrate dehydrogenase 2 gene mutations in lower-grade gliomas. Chin. Med. J. 2019;132:2920–2926. doi: 10.1097/CM9.0000000000000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu F., Zhao Z., Chai R.C., Liu Y.Q., Li G.Z., Jiang H.Y., Jiang T. Prognostic power of a lipid metabolism gene panel for diffuse gliomas. J. Cell. Mol. Med. 2019;23:7741–7748. doi: 10.1111/jcmm.14647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X., Pan Y., Xiong M., Sanapala S., Anastasaki C., Cobb O., Dahiya S., Gutmann D.H. Midkine activation of CD8(+) T cells establishes a neuron-immune-cancer axis responsible for low-grade glioma growth. Nat. Commun. 2020;11:2177. doi: 10.1038/s41467-020-15770-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pi C.D., Jose-Lopez R., Fernandez F.F., Rabanal P.R., Mandara M.T., Arus C., Pumarola B.M. Expression of FOXP3 in canine gliomas: immunohistochemical study of tumor-infiltrating regulatory lymphocytes. J. Neuropathol. Exp. Neurol. 2020;79:184–193. doi: 10.1093/jnen/nlz120. [DOI] [PubMed] [Google Scholar]
- 24.Weenink B., Draaisma K., Ooi H.Z., Kros J.M., Sillevis S.P., Debets R., French P.J. Low-grade glioma harbors few CD8 T cells, which is accompanied by decreased expression of chemo-attractants, not immunogenic antigens. Sci. Rep. 2019;9:14643. doi: 10.1038/s41598-019-51063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weenink B., van Brakel M., Wijers R., Sillevis S.P., French P.J., Debets R. Lack of B and T cell reactivity towards IDH1(R132H) in blood and tumor tissue from LGG patients. J. Neuro-Oncol. 2019;144:79–87. doi: 10.1007/s11060-019-03228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klemm F., Maas R.R., Bowman R.L., Kornete M., Soukup K., Nassiri S., Brouland J.P., Iacobuzio-Donahue C.A., Brennan C., Tabar V., Gutin P.H., Daniel R.T., Hegi M.E., Joyce J.A. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell. 2020;181:1643–1660. doi: 10.1016/j.cell.2020.05.007. (e17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo X., Pan Y., Gutmann D.H. Genetic and genomic alterations differentially dictate low-grade glioma growth through cancer stem cell-specific chemokine recruitment of T cells and microglia. Neuro-Oncology. 2019;21:1250–1262. doi: 10.1093/neuonc/noz080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plant A.S., Koyama S., Sinai C., Solomon I.H., Griffin G.K., Ligon K.L., Bandopadhayay P., Betensky R., Emerson R., Dranoff G., Kieran M.W., Ritz J. Immunophenotyping of pediatric brain tumors: correlating immune infiltrate with histology, mutational load, and survival and assessing clonal T cell response. J. Neuro-Oncol. 2018;137:269–278. doi: 10.1007/s11060-017-2737-9. [DOI] [PubMed] [Google Scholar]
- 29.Han S., Zhang C., Li Q., Dong J., Liu Y., Huang Y., Jiang T., Wu A. Tumour-infiltrating CD4(+) and CD8(+) lymphocytes as predictors of clinical outcome in glioma. Br. J. Cancer. 2014;110:2560–2568. doi: 10.1038/bjc.2014.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathur R., Zhang Y., Grimmer M.R., Hong C., Zhang M., Bollam S., Petrecca K., Clarke J., Berger M.S., Phillips J.J., Oberheim-Bush N.A., Molinaro A.M., Chang S.M., Costello J.F. MGMT promoter methylation level in newly diagnosed low-grade glioma is a predictor of hypermutation at recurrence. Neuro-Oncology. 2020 doi: 10.1093/neuonc/noaa059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Zhang L., Tang Y., Wang C., Chen Y., Shu J., Zhang K. Systemic screening identifies GABRD, a subunit gene of GABAA receptor as a prognostic marker in adult IDH wild-type diffuse low-grade glioma. Biomed. Pharmacother. 2019;118:109215. doi: 10.1016/j.biopha.2019.109215. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Wang Z., Zhang C., Liu X., Li G., Liu S., Sun L., Liang J., Hu H., Liu Y., Zhang W., Jiang T. Genetic and clinical characterization of B7-H3 (CD276) expression and epigenetic regulation in diffuse brain glioma. Cancer Sci. 2018;109:2697–2705. doi: 10.1111/cas.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma X., Shang F., Zhu W., Lin Q. CXCR4 expression varies significantly among different subtypes of glioblastoma multiforme (GBM) and its low expression or hypermethylation might predict favorable overall survival. Expert. Rev. Neurother. 2017;17:941–946. doi: 10.1080/14737175.2017.1351299. [DOI] [PubMed] [Google Scholar]