Abstract
Coronavirus disease 2019 (COVID-19), which is caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was declared by the World Health Organization (WHO) as a global pandemic on March 11, 2020. SARS-CoV-2 targets the respiratory system, resulting in symptoms such as fever, headache, dry cough, dyspnea, and dizziness. These symptoms vary from person to person, ranging from mild to hypoxia with acute respiratory distress syndrome (ARDS) and sometimes death. Although not confirmed, phylogenetic analysis suggests that SARS-CoV-2 may have originated from bats; the intermediary facilitating its transfer from bats to humans is unknown. Owing to the rapid spread of infection and high number of deaths caused by SARS-CoV-2, most countries have enacted strict curfews and the practice of social distancing while awaiting the availability of effective U.S. Food and Drug Administration (FDA)-approved medications and/or vaccines. This review offers an overview of the various types of coronaviruses (CoVs), their targeted hosts and cellular receptors, a timeline of their emergence, and the roles of key elements of the immune system in fighting pathogen attacks, while focusing on SARS-CoV-2 and its genomic structure and pathogenesis. Furthermore, we review drugs targeting COVID-19 that are under investigation and in clinical trials, in addition to progress using mesenchymal stem cells to treat COVID-19. We conclude by reviewing the latest updates on COVID-19 vaccine development. Understanding the molecular mechanisms of how SARS-CoV-2 interacts with host cells and stimulates the immune response is extremely important, especially as scientists look for new strategies to guide their development of specific COVID-19 therapies and vaccines.
Keywords: Coronavirus, SARS-CoV-2, Investigational medications, Pandemic, Pathophysiology, Viral immune response
Abbreviations: ACE2, angiotensin-converting enzyme 2; AHFS, American Hospital Formula Service; ANGII, angiotensin II; APCs, antigen presenting cells; ARDS, acute respiratory distress syndrome; COVID-19, coronavirus disease; CoVs, coronaviruses; GVHD, graft versus host disease; HCoVs, human coronoaviruses; IBV, infectious bronchitis coronavirus; IFN-γ, interferon-gamma; ILCs, innate lymphoid cells; MERS-CoV, Middle East respiratory syndrome; nsps, nonstructural proteins; NKs, natural killer cells; ORFs, open reading frames; PAMPs, pathogen-associated molecular patterns; RdRp, RNA-dependent RNA polymerase; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SLE, systemic lupus erythematosus; TMPRSS2, transmembrane serine protease 2; WHO, World Health Organization
1. Introduction
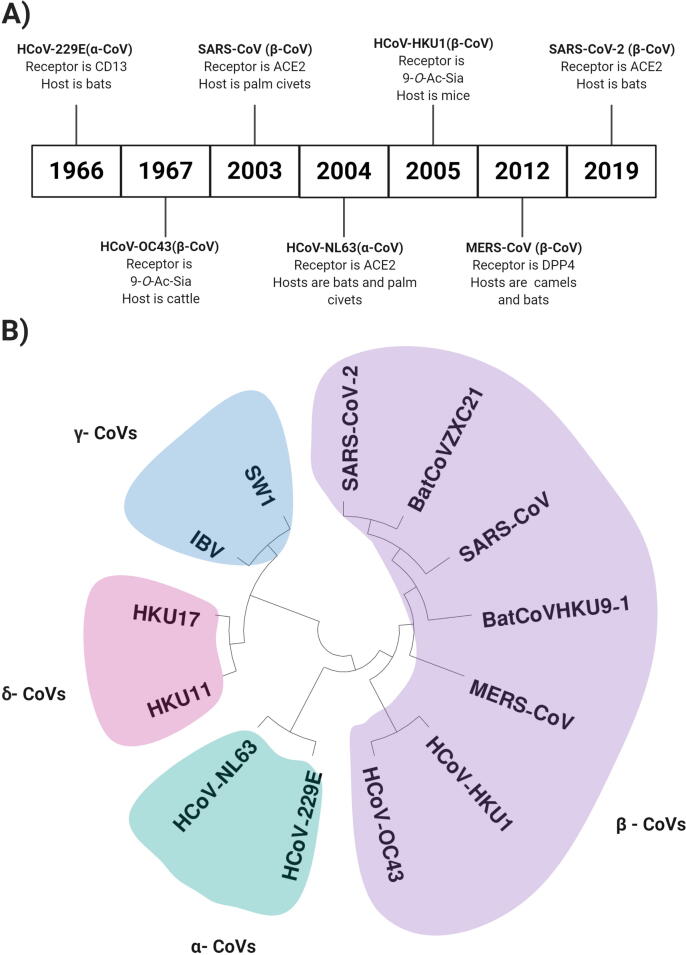
Coronaviruses (CoVs) can cause diseases in humans and other animals leading to the common cold, mild infections, or in some cases more serious lower respiratory system distress, targeting mainly the elderly (Saif et al., 2019). CoVs are named for the spike proteins that are shaped like crowns on the surface of the virus (Y. Chen et al., 2020). The avian infectious bronchitis coronavirus (IBV) was the earliest CoV isolated in 1937(Cavanagh, 2005); human CoVs (HCoVs) were first discovered in the 1960s (Hamre and Procknow, 1966) (Fig. 1A). CoVs belong to the very large Coronaviridae family of viruses, which is classified into four genera according to their genetic information as follows: Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV), and Deltacoronavirus (δ-CoV) (Forni et al., 2017). The α-CoVs include HCoV-229E and HCoV-NL63, whereas the β-CoVs include HCoV-OC43, HCoV-Hong Kong University 1 (HCoV-HKU1), Middle East respiratory syndrome (MERS-CoV), severe acute respiratory syndrome (SARS-CoV), and SARS-CoV-2 (Fig. 1B). Generally, α-CoVs and β-CoVs are limited to only infecting mammals, whereas γ-CoVs and δ-CoVs infect mostly birds but are capable of infecting mammals in some cases (Woo et al., 2012). To date, seven types of CoVs have been identified that have the ability to infect humans: HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, MERS-CoV, SARS-CoV, and most recentlySARS-CoV-2. The novel SARS-CoV-2 originated from Wuhan, China, with a subsequent international outburst leading to the current global pandemic (Pillaiyar et al., 2020) (Fig. 1A and B).
Fig. 1.
A) Timeline of the emergence of coronaviruses (CoVs) and their classification, functional receptor, and hosts. B) Phylogenetic analysis of 13 sequences from selected CoVs including α-CoVs: HCoV-229E (GenBank ID: KF514433.1) and HCoV-NL63 (KF530114); β-CoVs: HCoV-OC43 (AY391777), HCoV-HKU1 (NC_006577.2), MERS-CoV (NC_019843), SARS-CoV (NC_004718.3), SARS-CoV2 (NC_019843), bat-SL-CoVZXC21 (MG772934), and bat CoV HKU9-1 (EF065513); γ-CoVs: SW1 (NC_010646), and IBV (NC_001451.1); and δ-CoVs: HKU17 (YP_005352845) and HKU11 (YP_002308478.1).
The infectious lifecycle of CoVs begins when the virus binds to a particular receptor on the host cell surface, facilitating entry. The receptors and known or suspected host(s) of HCoVs are as follows: HCoV-229E binds to human aminopeptidase N (CD13) (Yeager et al., 1992) and is believed to have originated from bats; HCoV-NL63, SARS-CoV, and SARS-CoV-2 bind to angiotensin-converting enzyme 2 (ACE2) (Li et al., 2003, Wu et al., 2009, Zhang et al., 2020b) and are believed to have originated from bats; HCoV-OC43 and HCoV-HKU1 bind to 9-O-acetylated sialic acid (Butler et al., 2006, Huang et al., 2015) and originated from cattle and mice, respectively; and MERS-CoV binds to dipeptidyl peptidase 4 (van Doremalen et al., 2014) and originated from camels. Based on phylogenetic analysis (Fig. 1B), SARS-CoV-2 and hence COVID-19 might originate from bats, but this has not been confirmed. Moreover, the intermediary that facilitates the transfer of this mutated strain from bats to humans remains unknown (Yang et al., 2020).
CoVs can cross species barriers and in humans can cause illnesses ranging from the common cold to more severe diseases such as MERS and SARS (Lau and Chan, 2015). In December 2019, COVID-19 was first announced by the Chinese government and, subsequently, has taken the world by storm leading the World Health Organization (WHO) to issue a warning (Wang et al., 2020). To date, there is no verified cure for COVID-19; however, numerous studies are underway searching for and testing novel and repurposed drugs, and vaccines are currently under development and in initial testing phases. Most of these investigational medications are classified into two groups, those that target the viral pathophysiology and those that target the immune response (Crisci et al., 2020). Notably, scientists benefit from information already available on the therapeutic response of host cells infected by other CoVs (Crisci et al., 2020). Furthermore, the use of repurposed medications offers a practical approach, with many of the anti-viral medications under study having been shown to act by inhibiting viral replication leading to improved patient health (De Soto et al., 2020, Perricone et al., 2020). Thus, the current and future therapeutic drugs used against COVID-19 may in turn prove very promising against future viral infections.
Exploiting the molecular mechanisms of how SARS-CoV-2 interacts with host cells and stimulates the immune response may constitute an excellent strategy for developing new, specific COVID-19 therapies. This review will shed light on investigational medications that are undergoing clinical trials and are known and/or speculated to treat COVID-19.
2. SARS-CoV-2: Genome structure and pathophysiology
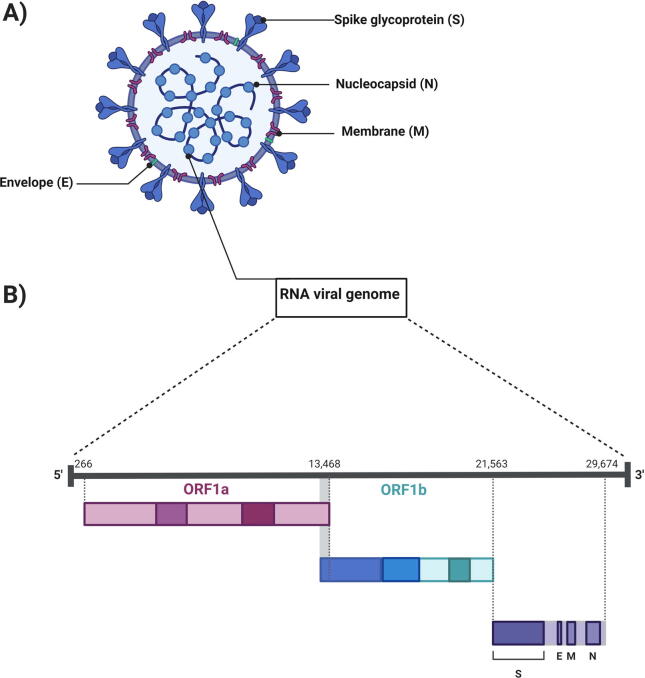
Finding suitable drugs to treat and cure COVID-19 is crucial; understanding the genome and pathophysiology of the causative virus can only help in this endeavor. CoVs are the largest RNA viruses with a genome size estimated to be 27–31 kb (Smith and Denison, 2013). The architecture of transcripts of every CoV includes a 5′ cap and a 3′ poly(A) tail (Y. Chen et al., 2020). When CoVs enter a host cell the viral genomic RNA (gRNA) acts as a transcript for gene 1, which consists of a pair of large open reading frames (ORFs), ORF1a and ORF1ab (Fig. 2). ORF1a (Cowley et al., 2000) is translated generating polypeptide1a (pp1a; 440–500 kDa); ORF1ab is translated generating polypeptide1ab (pp1ab; 740–810 kDa). These ORFs are then proteolytically cleaved into 16 nonstructural proteins (nsps) (Cong, 2019, Kim et al., 2020).
Fig. 2.
Schematic of SARS-CoV-2 virion and genomic structure. A) The SARS-CoV-2 virion includes four major structural proteins: spike glycoprotein (S), membrane protein (M), envelope protein (E), and nucleocapsid protein (N). B) The genomic single-stranded RNA can be considered in thirds: the first third encodes ORF1a, the second third encodes ORF1ab, and the last third encodes the four essential structural proteins and other accessory proteins.
Viral proteases facilitate the proteolytic cleavage process. One of the nsps generated in this process is the viral enzyme RNA-dependent RNA polymerase (RdRp), which is an essential part of the replication/transcription machinery of CoVs (Gao et al., 2020). The 3′ CoV gRNA encodes four essential structural proteins (sps): spike glycoprotein (S), integral membrane protein (M), small envelope protein (E), and nucleocapsid protein (N), along with several accessory proteins (Dhama et al., 2020) (Fig. 2).
The causative agent of COVID-19 is formally known as SARS-CoV-2, as named after SARS-CoV by the International Committee on Taxonomy of Viruses (ICTV) because of the ~80% similarity between the two viruses with regard to structure and pathogenicity (Wang et al., 2020). To understand the pathophysiology of SARS-CoV-2, its genomic structure needs to be considered (Guo et al., 2020). Microscopic images have revealed that SARS-CoV-2 is an enveloped virus with a linear, positive single-strand of RNA (+ssRNA) (Shereen et al., 2020). The sequenced SARS-CoV-2 genome has been published in many gene banks revealing that it consists of 29,891 nucleotides (nt) encoding approximately 9860 amino acids (Cascella et al., 2020, Oguh et al., 2020).
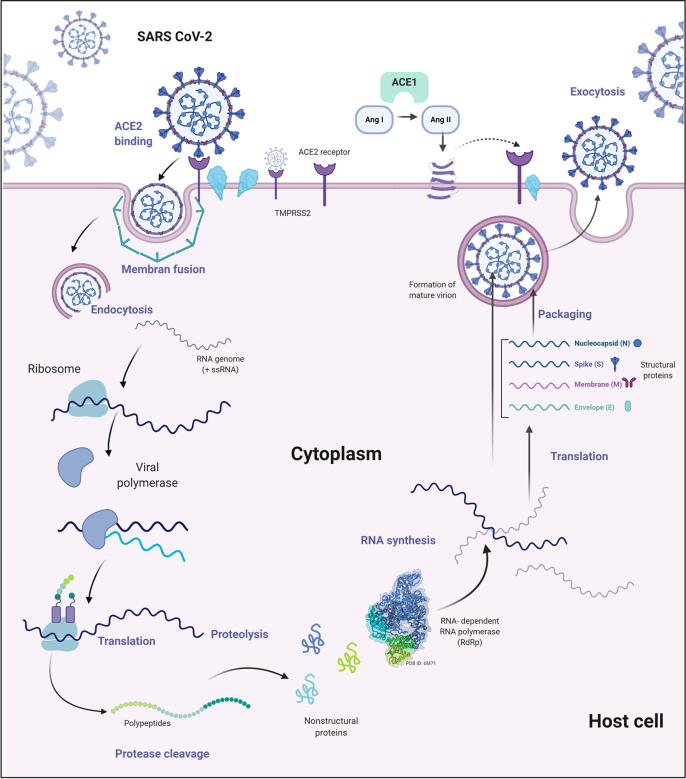
The pathophysiology of SARS-CoV-2 within the host epithelial cells begins with the attachment of the virus to the host cell through binding of its S glycoprotein to the ACE2 receptor (Fig. 3). Structural and functional examination of this binding process indicates that a conformational alteration facilitates the fusion and entry of the virus into the host cell; host transmembrane serine protease 2 (TMPRSS2) improves this process (Rothan and Byrareddy, 2020). After entry into the host cell, the virus is uncoated and releases its +ssRNA into the cytoplasm, whereby it acts as an mRNA and uses the translational machinery of the host cell to translate the viral mRNA into large polypeptides. These polypeptides are proteolytically cleaved by specific proteases into all of the protein components of SARS-CoV-2 (Chu et al., 2020). Meanwhile, the +ssRNA also uses its own RdRp to synthesize more copies of +ssRNA molecules (Davis et al., 2020). All four of the sps: S, M, E, and N, and the +ssRNA are assembled, matured, and packaged into new copies of the virus that can then exit the host cell via exocytosis (Ahn et al., 2020).
Fig. 3.
Schematic of SARS-CoV-2 pathophysiology. The infection cycle of SARS-COV-2 is initiated by binding of the virus through its spike (S) protein to an ACE2 receptor; the TMPRSS2 protease improves the binding process. When the virus enters the host cell it is un-coated and releases its +ssRNA into the host cell cytoplasm where it is translated into large polypeptides that are proteolytically cleaved to produce non-structural proteins including RNA-dependent RNA polymerase (RdRp). The viral +ssRNA also uses its own RdRp to synthesize a − ssRNA template to generate more copies of viral +ssRNA molecules. The four structural proteins S, M, E, and N, and the +ssRNA are assembled, packaged, and then matured to make several copies of the virus that exit the host cell via exocytosis.
3. Current anti-COVID-19 medications under investigation related to SARS-CoV-2 genome structure and pathophysiology
SARS-CoV-2 mainly targets the respiratory system resulting in COVID-19 with symptoms such as a fever, headache, dry cough, dyspnea, and dizziness (Rothan and Byrareddy, 2020). These symptoms vary from person to person, ranging from mild to hypoxia with acute respiratory distress syndrome (ARDS) and sometimes death (COVID, 2020, Negri et al., 2020).
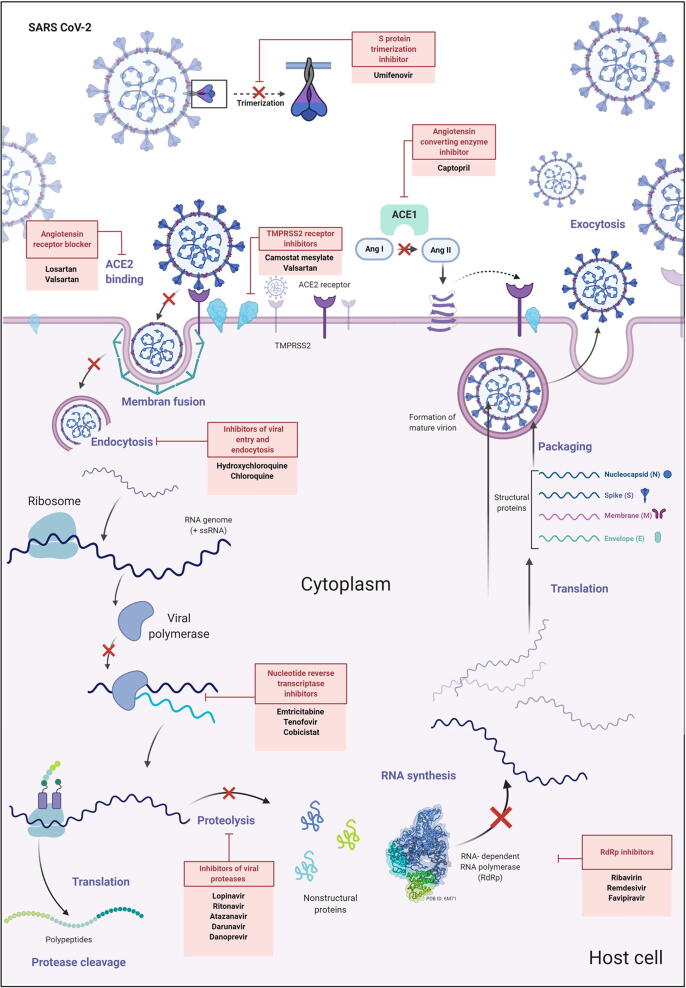
The fast rate of SARS-CoV-2 spread, together with the lack of effective vaccines or drugs that can be used to prevent and treat COVID-19, respectively, have made the pandemic situation worse. This has led to a very high demand for research and development of effective treatments on several fronts. Most agents used to treat COVID-19 at this time are not approved by the FDA for COVID-19; scientists worldwide are working diligently to develop an effective anti-COVID-19 treatment and are appealing for rapid approval for clinical use (Sanders et al., 2020). A wide spectrum of existing drugs whose general pharmacology, interactions, pharmacoeconomics, properties, spectra, and taxonomy are understood, might be beneficial and provide a head-start toward treating COVID-19 (Vellingiri et al., 2020). Here, we have reviewed some of these drugs that could potentially affect the pathophysiology of SARS-CoV-2, classified based on their mechanism of action (Table 1 and Fig. 4).
Table 1.
Current antiviral medications that could potentially affect the pathophysiology of COVID-19.
| Target | Mechanism of action | Drug | Drug bank ID | Chemical formula | AHFS | Clinical trial phase for the treatment of COVID-19 |
|---|---|---|---|---|---|---|
| ACE2 | Angiotensin receptor blocker | Losartan | DB00678 | C22H23ClN6O | Treat diabetic nephropathies (Pechlivanova et al., 2020), heart failure, hypertension (Corbett et al., 2020). | Phase 1 |
| Valsartan (Diovan) | DB00177 | C24H29N5O3 | Treat hypertension (Williams et al., 2017), heart failure (Sokos and Raina, 2020), post myocardial infarction (Gervais et al., 1999). | Phase 4 | ||
| Angiotensin converting enzyme inhibitor | Captopril | DB01197 | C9H15NO3S | Treat hypertension (Atkinson and Robertson, 1979). | Phase 2 | |
| TMPRSS2 | TMPRSS2 receptor inhibitor | Camostat mesylate | DB13729 | C20H22N4O5 | Treat chronic pancreatitis and drug-induced lung injury (Ota et al., 2016). | Phase 2 |
| Bromhexine | DB09019 | C14H20Br2N2 | An expectorant/mucolytic agent (Zanasi et al., 2017). | Phase 4 | ||
| S protein | Umifenovir | DB13609 | C22H25BrN2O3S | Treat influenza and other respiratory viral infections (Pshenichnaya et al., 2019). | Phase 4 | |
| Membranes fusion | Inhibitor of viral entry and endocytosis | Hydroxychloroquine | DB01611 | C18H26ClN3O | Treat uncomplicated malaria (Lim et al., 2009), rheumatoid arthritis (Furst, 1996), chronic discoid lupus erythematosus, and systemic lupus erythematosus (Fox, 1993). | Phase 4 |
| Chloroquine | DB00608 | C18H26ClN3 | Treat malaria (Mwanza et al., 2016), rheumatic diseases (Haładyj et al., 2018), and prophylaxis of Zika virus (Li et al., 2017). | Phase 2 | ||
| RdRp | RNA-dependent RNA polymerase (RdRp) inhibitor | Ribavirin | DB00811 | C8H12N4O5 | Treat chronic hepatitis C virus (HCV) infection (Martin and Jensen, 2008). | Phase 1 |
| Remdesivir | DB14761 | C27H35N6O8P | Treat Ebola virus (EBOV) (Warren et al., 2016). | Phase 2 | ||
| Favipiravir | DB12466 | C5H4FN3O2 | Treat influenza virus infections(Hayden and Shindo, 2019). | Phase 2 | ||
| Protease | Viral protease inhibitor (PI) | Lopinavir | DB01601 | C37H48N4O5 | Treat human immunodeficiency virus type 1 (HIV-1) infection (Reddy et al., 2007). | Phase 1 & 2 |
| Ritonavir | DB00503 | C37H48N6O5S2 | Treat HIV infections (Tigabu et al., 2020). | Not Applicable | ||
| Atazanavir | DB01072 | C38H52N6O7 | Treat HIV infections (Le Tiec et al., 2005). | Not Applicable | ||
| Darunavir | DB01264 | C27H37N3O7S | Treat HIV infections (Lombaard et al., 2018). | Phase 3 | ||
| Danoprevir | DB11779 | C35H46FN5O9S | Treat hepatitis C (Wei et al., 2019). | Phase 4 | ||
| Nucleoside reverse transcriptase | Nucleotide reverse transcriptase inhibitor (NRTI) | Emtricitabine | DB00879 | C8H10FN3O3S | Treat HIV infections (Sax et al., 2017). | Phase 3 |
| Tenofovir | DB14126 | C9H14N5O4P | Treat HIV infections (Klatt et al., 2017). | Phase 3 | ||
| Cobicistat | DB09065 | C40H53N7O5S2 | Treat HIV infections (Eron et al., 2018). | Phase 3 |
AHFS, American Hospital Formula Service.
Fig. 4.
Schematic of sites of action of drugs under study targeting SARS-CoV-2 viral pathophysiology and the development of COVID-19. The first group of drugs target functional ACE2 receptors and include: i) angiotensin receptor blockers such as valsartan and losartan, and ii) angiotensin converting enzyme inhibitors such as captopril. The second group blocks possible entry gateways of the virus into the host cell and includes: i) TMPRSS2 receptor inhibitors camostat mesylate and valsartan, and ii) viral entry and endocytosis inhibitors such as hydroxychloroquine and chloroquine, which block the release of viral genomic RNA into the host cell. The third group of drugs blocks transcriptional/translational machinery and includes: i) viral protease inhibitors such as lopinavir, ritonavir, atazanavir, darunavir, and danoprevir, ii) RNA-dependent RNA polymerase (RdRp) inhibitors such as ribavirin, remdesivir, and favipiravir, and iii) nucleotide reverse transcriptase inhibitors (NRTIs) such as emtricitabine, tenovir, and cobicistat.
The first group of anti-COVID-19 medications target the functional ACE2 receptor and include i) angiotensin receptor blockers such as valsartan and losartan (Marin, 2020) and ii) angiotensin converting enzyme inhibitors such as captopril (Zhang et al., 2020a, Zhang et al., 2020c). These medicines are usually prescribed to reduce high blood pressure and kidney and heart diseases, as well as being beneficial for the treatment of COVID-19.
The ACE2 receptor is a key element for activating the renin-angiotensin-aldosterone system pathway, which plays a crucial role in the inflammatory response (Patel et al., 2017). ACE2 is overexpressed in epithelial cells where it provides protection in the lungs, heart, kidneys, and blood vessels (Li et al., 2020). Type II pneumocytes (alveolar cells) that are present in lung alveoli are responsible for taking in oxygen and releasing carbon dioxide; these cells are found to significantly express ACE2 (Islam, 2017). ACE2 physiologically regulates the activity of angiotensin II (ANG II), a molecule that plays a critical role in causing hypertension and inducing inflammation. In normal cells ANG II is converted and neutralized by ACE2 to reduce its harmful effects (Liu et al., 2018).
In COVID-19, the ACE2 receptor has been confirmed to be the main factor in the entry of SARS-CoV-2 into the host alveolar cells, which are considered to be the main gate for virus entry into the lungs. When the cells are infected by SARS-CoV-2, the viral S protein binds to the ACE2 receptor resulting in inhibition of ACE2 regulation of ANG II, which leads to damaged cells and tissues. This explains one of the symptoms of COVID-19, lung injury (Hirano and Murakami, 2020). Furthermore, cells that overexpress ACE2 in the lungs and heart tissue also overexpress ACE1; these proteins normally work together to decrease the expression level of ANG II. However, with depressed ACE2 availability, ACE1 works in the opposite manner to elevate the expression level of ANG II (Gheblawi et al., 2020).
The second group of anti-COVID-19 medications block potential viral entry gateways into the host cell and include i) TMPRSS2 receptor inhibitors including camostat mesylate and bromhexine, which are widely investigated and undergoing clinical trials (Sonawane et al., 2020), ii) inhibitors of viral entry and endocytosis such as hydroxychloroquine and chloroquine which block the release of viral gRNA into the host cell (Colson et al., 2020), and iii) inhibitors that prevent S protein trimerization such as umifenovir (Costanzo et al., 2020) (Table 1, Fig. 4).
The serine protease TMPRSS2 is overexpressed in alveolar cells and enhances the binding between ACE2 and the S protein of SARS-CoV-2, mediating the endosomal and lysosomal entrance and processing of viral particles through host-viral membrane fusion (Sungnak et al., 2020). It has been clinically proven that TMPRSS2 inhibitors can block the entry of SARS-CoV-2 into host cells by inhibiting the acidic protease activity (Bittmann et al., 2020). This mechanism of action has drawn the attention of drug development designers targeting this process with chloroquine and the less toxic hydroxychloroquine (Blaess et al., 2020, Colson et al., 2020). The mechanism of action of both drugs is based on neutralizing the acidic pH of lysosomes by accumulating the drugs within the organelle and thus affecting protease activity leading to protein and mucopolysaccharide degradation (Ferner and Aronson, 2020). Hence, chloroquine and hydroxychloroquine could participate effectively in blocking and preventing SARS-CoV-2 from entering host cells via interference with the viral S protein and ACE2 receptor (Devaux et al., 2020, Zhou et al., 2020). As a result, patients with COVID-19 would benefit from these drugs only in an early stage of viral infection. However, the safety and effectiveness of both drugs are still being evaluated in clinical trials for treatment of patients with COVID-19.
Nevertheless, despite the lack of clinical evidence backing up the safety and efficiency of chloroquine and hydroxychloroquine, regulatory authorities nationwide are allowing the use of both drugs to treat patients with COVID-19 based on previous successful results in curing SARS-Cov-1 or MERS (Mpofu et al., 2020). Furthermore, these drugs are undergoing clinical trials in many countries as likely medications for use in treating COVID-19 (Chowdhury et al., 2020). However, numerous factors confounding the efficacy and clinical application of chloroquine and hydroxychloroquine have been recently reported, such as small sample size, intolerance to the medications, and possible dose-related adverse reactions including cardiotoxicity and mortality. These serious limitations have hindered the progress of the clinical trial-based advancement of hydroxychloroquine and chloroquine as therapeutic agents. Moreover, the report from a large retrospective observational study revealing a link between the use of these agents, ventricular fibrillation (VF), and the deaths of patients with COVID-19 in hospitals (Chorin et al., 2020, Szekely et al., 2020) has led several regulatory authorities in the majority of European countries such as the UK and France to directly pause or stop all clinical trials that involve examining chloroquine and hydroxychloroquine in patients with COVID-19 (White et al., 2020).
Alternatively, according to structural investigations, trimerization of the S protein can be targeted for blockage by the drug umifenovir (Arbidol®) (Vankadari, 2020). In vitro investigations have shown that umifenovir markedly affects replication of the SARS virus (Lian et al., 2020). The use of umifenovir to treat patients with COVID-19 is in phase 4 clinical trials.
A third group of anti-COVID-19 medications blocks transcriptional/translational machinery and includes i) viral protease inhibitors such as lopinavir, ritonavir, atazanavir, darunavir, and danoprevir (Mohamed et al., 2020), ii) RdRp inhibitors such as ribavirin, favipiravir, and remdesivir (Vafaei et al., 2019), and iii) nucleotide reverse transcriptase inhibitors such as emtricitabine, tenofovir, and cobicistat (Lythgoe and Middleton, 2020) (Table 1, Fig. 4).
The main viral protease, RdRp, is responsible for replicating the viral genome and synthesizing the essential viral functional proteins from their mRNA templates (Hillen et al., 2020). The activity of RdRp is stimulated by interacting with other trigger molecules at its active site. However, when the active site is bound to another molecule its activity is inhibited (Mothay and Ramesh, 2020); this provides a target for inhibition. Thus, discovering molecules that could inhibit RdRp of SARS-CoV2 may shed light on new therapeutic treatments (Gordon et al., 2020) and help defeat this global pandemic. Drugs with this type of mechanism have been widely used for years in HIV therapy and include lopinavir, ritonavir, atazanavir, darunavir, and danoprevir. Some RdRp inhibitors have also shown promising results in treating MERS and Ebola by delaying the replication process (J. Huang et al., 2020). Based on these previous success stories, these drugs have received considerable attention in attempts to develop a treatment for COVID-19. However, the safety and effectiveness of using this group of medications in patients with COVID-19 are unknown and currently undergoing clinical trials (Table 1).
An essential class of inhibitory drugs for curing CoVs is nucleoside reverse transcriptase inhibitors (NRTIs) (Fig. 4). These compounds are activated by phosphorylation inside the host cells to form triphosphates that compete with cellular substrates for the viral reverse transcriptase, thereby terminating transcription and inhibiting enzyme activity (Pau and George, 2014).
Overall, using these drugs alone or in combination with other drugs is not guaranteed to be effective against COVID-19 replication nor improve patient symptoms. In addition, side effects are very likely. The development of specific and effective drugs with no or low side effects for treating patients with COVID-19 is important. Furthermore, knowledge of the molecular mechanisms of how SARS-CoV-2 enters host cells and replicates can only enhance studies attempting to design new drugs.
4. Overview of the innate, adaptive and complement immune systems
The human immune system is comprised of cells, molecules, and processes that work together to provide protection to skin, respiratory passages, the gastrointestinal tract, and other parts of the body against foreign intruders such as bacteria, parasites, fungi, viruses, toxins, and cancer cells. The immune system can be simply viewed as two lines of defense: innate (non-specific) and adaptive (specific) immunity (Marshall et al., 2018). For the host immune system to contain viral spread and attenuate infection, it needs to employ both immune defenses; innate immunity can exert its effects through inflammatory cytokines and innate immune cells, whereas adaptive immunity clears the infection using T-helper cells CD4+, CD8+ cells to kill infected cells, and B cells to produce antibodies to neutralize and exterminate free virus (Christiaansen et al., 2015).
The innate immune system includes phagocytic cells (macrophages and neutrophils), mast cells, eosinophils, basophils, natural killer cells (NKs), dendritic cells (DCs), and lymphoid cells (Iwasaki and Medzhitov, 2010, Marshall et al., 2018). The innate immune system acts rapidly, within minutes of pathogen exposure. It is programmed to detect invariant microbial components shared by the majority of pathogen groups; moreover, it serves as the central player in activating subsequent responses of adaptive immunity (Iwasaki and Medzhitov, 2010, Turvey and Broide, 2010). Within innate immunity, macrophages and DCs both constitute phagocytic and antigen presenting cells (APCs), whereas mast cells (sentinel cells) and basophils are responsible for cytokine release and the initiation of acute inflammatory responses such as in asthma and allergies. In turn, eosinophils destroy parasites and NKs are responsible for the destruction of tumor cells and virus-infected cells by releasing granzymes and perforins, causing cell lysis. NKs are also an important source of interferon-gamma (IFN-γ), which mobilizes APCs and activates anti-viral immunity. Finally, innate lymphoid cells (ILCs) (including, ILC-1, ILC-2, and ILC-3) are responsible for cytokine-selective production such as IFN-γ and interleukins 4 and 17 (IL-4, IL-17) (Stone et al., 2010).
Despite the lack of antigen-specificity and immune memory in the innate immune system, it is capable of distinguishing between “self” components and “non-self“ invaders through molecular patterns present in pathogens but not in host cells. These molecules, known as pathogen-associated molecular patterns (PAMPs), are recognized by a finite number of germline-encoded pattern-recognition receptors (PRRs) (Silva et al., 2017). As a result, PAMP detection elicits the activation of redundant antiviral genes establishing a cellular antiviral state to enable cells to limit and/or clear infection (Brubaker et al., 2015). Toll-like receptors (TLRs) act as PRRs on the APCs and indirectly elicit adaptive immune responses by monitoring the expression of co-stimulatory molecules and the production of pro-inflammatory cytokines. Alternatively, damaged cell-mediated initiation of the immune response is amplified by endogenous molecules known as damage-associated molecular patterns (DAMPs). Both PAMPs and DAMPs initiate immune responses via the activation of PRRs, which include TLRs, NOD-like receptors (NLRs), C-type lectin receptors (CLRs), retinoid acid-inducible gene I (RIG-I), and myriad intracellular DNA sensors (Gong et al., 2020). The source of DAMPs is the mitochondria; these molecules are also therefore called mitochondrial DAMPs (MTDs) (Hu et al., 2015).
Additionally, the mitochondria also play a direct role in induction of the innate immune response as the outer membrane of the mitochondria contains mitochondrial antiviral signaling molecules (signalosomes) (MAVS). This molecule influences the function of RIG-1-like receptors (RLRs), which are specialized toward the recognition of viruses by innate immunity. RLR family members RIG-1, laboratory of genetic physiology 2 (LGP2), and melanoma differentiation-associated 5 (MDA5) sense the viral RNA in the perinuclear environment and activate the immune response by downstream signing of transcription factors including IFN regulatory factors (IRFs) 3 and 7, and NF-κB, which contribute to the abundant elevation of type 1 IFNs and pro-inflammatory cytokine production (Koshiba, 2013, Tiku et al., 2020).
Adaptive immune cells include T-lymphocytes, which mature in the thymus gland, and B-lymphocytes, which arise from the bone marrow and produce antibodies. T-lymphocytes play various roles including the elimination of pathogen-infected cells and functioning as helper cells through direct cellular contact or cytokine-dependent signaling to enhance the responses of B- and T- lymphocytes. They also activate mononuclear phagocytes, regulate immune responses, and limit tissue damage sustained by overall inflammatory responses. The largest group of T-cells in the body is CD4+, with most serving as helper cells (TH) cells (Bonilla and Oettgen, 2010). TH1 cells are differentiated from precursor naïve TH0 cells under the influence of IL-12 and INF-γ, also designated based on their ability to generate IL-2 and INF-γ cytokines, respectively. TH1 produces cytokines that mediate the activation of mononuclear phagocytes, NKs, and cytolytic T-cells to target and kill virally infected cells and intracellular microbes. Another group of helper cells, the TH17 cells, are protuberant in chronic inflammatory and allergic processes; they also produce five homologous molecules (IL-17A-F). IL-17A and IL-17F are proinflammatory cytokines that induce IL-6 and tumor necrosis factor (TNF) production, granulocyte recruitment, and tissue damage. IL-10 is produced by another type of T-cell known as regulatory T cells (TR1). The majority of circulating T-cells are CD8+ cells that eliminate pathogen-infected cells and transformed cells (tumor cells). Adaptive humoral immunity is mediated by antibodies produced from plasma cells developed from B-cells under the direction of T-cells and other cells such as, DCs (Bonilla and Oettgen, 2010).
The complement system contains 30 proteins found in blood-soluble form or as membrane-associated proteins. When this system is activated, it leads to initiation of different physiological responses ranging from chemoattraction to apoptosis. The roles played by the complement system can be seen in innate immunity by providing a robust and rapid response, in adaptive immunity through elimination of pathogens by T- and B-cell responses, and as immunologic memory maintenance to avoid re-invasion (Carroll, 2004, Sarma and Ward, 2011). Moreover, the complement system participates in tumor growth and tissue regeneration. Activation of the complement pathway occurs via PAMPs through three pathways: classical, lectin, and alternative. The most recognized structures involved in complement activation are mannan-binding lectin, C-reactive protein, ficolins, natural immunoglobulin M (IgM), and C1q. The complement system has been discussed in detail in previous reviews (Carroll, 2004, Sarma and Ward, 2011).
4.1. Cytokines targeted in immunomodulation
Cytokines are a group of small proteins secreted by several cells to establish communication and intercellular signaling. Cytokines target specific cells in an endocrine, paracrine, or autocrine manner. Moreover, they might have unrelated functions depending on the presence or absence of certain cytokines or the targeted cell (Tisoncik et al., 2012). By binding to certain receptors, specific cytokines trigger versatile responses such as cell differentiation, proliferation, angiogenesis, inflammation, and immune responses. These different responses comprise an overlapping network with varying degrees of redundancy involved in different and important modulations. Consideration should therefore be given to defining key steps of cytokine responses during an infection to mark specific cytokines as therapeutic intervention targets (Tisoncik et al., 2012).
In turn, a cytokine storm is the result of a combination of immune-activated molecules including IFNs, ILs, chemokines, TNF-α, and colony-stimulating factors (CSFs). All these elements are cytokine storm-initiating and will be generally reviewed below.
4.1.1. Interferons
IFNs are described as heterogenous soluble proteins possessing high antiviral activity. They are usually induced upon bacterial or viral infection and are classified into three types (I, II, and III) based on the structure of the cell membrane receptors in which they bind. IFNs have been used as therapeutic options against various cancers, autoimmune diseases, and viral infections such as hepatitis C virus (HCV) and hepatitis B virus (HBV) (Lin and Young, 2014). The type I IFN family contains homologues of IFNα, which consist of 13 subtypes of small proteins (cytokines) encoded by several genes in humans, a single IFN-β, and other types of IFNs produced by single genes that are not adequately characterized including IFN-ζ, IFN-ω, IFN-δ, IFN-ε, IFN-τ, and IFN-κ (Pestka et al., 2004). The type II IFN family is comprised of IFN-γ, which is a single gene product primarily produced by NKs and T-cells; IFN-γ can also affect a wide range of cells that contain IFN-γ receptors (IFN-γR) (Schoenborn and Wilson, 2007). The type III IFN family contains IFNλ1 (IL-29), IFNλ2 (IL-28A), IFNλ3(IL-28B), and IFNλ4 (O’Brien et al., 2014, Prokunina-Olsson et al., 2013). These IFNs are known for their ability to activate the antiviral state in virus-infected, non-infected, and bystander cells via the induction of a gene transcription program, which causes interference with different stages of the viral replication cycle by orchestrating various responses (Yan and Chen, 2012).
Upon viral or bacterial infection, cells secrete IFNs to launch protective activity via three major routes: i) activating cellular-intrinsic microbial states to limit the spread of infectious agents, especially viral agents during infection in both the infected and adjacent cells, ii) modulation of innate immunity responses by establishing balance and promoting the presentation of antigens and functions of natural NKs while preventing production of cytokines and pro-inflammatory pathways, and iii) activation of adaptive immunity responses to stimulate maturation of specific-antigen-high affinity B- and T- cell responses and create immunological memory (Ivashkiv and Donlin, 2014).
Most cells of hematopoietic origin produce IFNβ, whereas DCs (plasmacytoid) predominantly produce IFNα. Type I IFNs are induced after detecting microbial products via PRRs (Goubau et al., 2013, Iwasaki, 2012, Paludan and Bowie, 2013). Both IFNα and IFNβ bind to the IFNα receptor leading to activation of the receptor-associated protein tyrosine kinases Janus kinase 1 (JAK1) and tyrosine kinase 2 (TYK2), which activate the cytoplasmic transcription factors known as signal transducer and activator of transcription 1 and 2 (STAT1, STAT2). Moreover, the activation of IFN-γ is guided by the JAK1 and JAK2 phosphorylation of STAT1 (Levy and Darnell, 2002, Stark and Darnell, 2012). When STAT1 and STAT2 are tyrosine-phosphorylated, they dimerize and are then translocated to the nucleus where they bind to IFN-regulatory factor 9 (IRF9) leading to the formation of a trimolecular complex known as IFN-stimulated gene factor 3 (ISGF3). This factor binds to its analogous DNA sequence (TTTCNNTTTC), also known as IFN-stimulated response elements (ISREs), and activates transcription of ISGs, whereas other cytokines activate STAT homodimers leading to activation of distinguished gamma-activated sequence (GAS) (TTCNNNGAA). Eventually, several hundred ISRE-driven ISG subsets are induced by type1 IFN signaling to acquire the antiviral state (MacMicking, 2012, Rusinova et al., 2012, Schoggins et al., 2011, Stark and Darnell, 2012).
Proteins encoded by ISGs repress pathogens by different mechanisms such as restraining viral transcription, translation, replication, viral-nucleic acid destruction, and alteration of metabolism of cellular lipids (MacMicking, 2012, Saka and Valdivia, 2012). SARS-CoV-2 shows both genetic and clinical similarities with other CoV diseases including MERS and SARS, and both show presentations of elevated proinflammatory cytokines, indicating the potential in common therapeutic strategies for managing SARS-CoV-2 (Khan et al., 2020). Based on recent studies, it has been shown that SARS-CoV-2 is sensitive to INFα. This is due to the fact that SARS-CoV-2 loses its anti-IFN activity as the orf6 and orf3b proteins are truncated; these proteins are responsible for the inhibition of IRF3. This is unlike the ones found in SARS-CoV and MERS-CoV that are able to alter IFN responses. Therefore, INFβ1 can be used as a suitable COVID-19 treatment because SARS-CoV-2 has proven to be more sensitive to type I INFs than SARS-CoV or MERS-CoV. Moreover, assessment of the safety of type I INFs has been observed in several independent clinical trials (Sallard et al., 2020). In addition, both DAMPs and PAMPs lead to further activation of inflammatory surface receptors such as TLRs and triggering receptor expressed on myeloid cells 1 (TREM-1), with downstream signaling of transcription factors and kinases resulting in increased secretion of pro-inflammatory cytokines such as TNF-α, IL-1β, IL-6, and IFN-γ leading to ischemic events (Rai and Agrawal, 2017).
4.1.2. Interleukins
The immune system is regulated by a diverse family of ILs, which mainly function in the differentiation and activation of immune cells. These ILs can be either pro- or anti-inflammatory, and are able to generate a variety of responses similar to cytokines (Brocker et al., 2010). Proinflammatory cytokines ILα- and IL-β mediate infection-host responses using several mechanisms, both direct and indirect (Dinarello, 2009). The biological functions of ILs include acute phase signaling activation, immune cell trafficking to the primary site of infection, activation of epithelial cells, and the secondary production of cytokines. The production of IL-1β and IL-18 as defense mechanisms against certain pathogens occurs via inflammasomes, which are cytosolic macromolecular complexes that consist of members of the nucleotide binding domain and leucine-rich-repeat-containing receptor (NLR) family. The best characterized inflammasome is NLRP3, which produces both IL-1β and IL-18. The activity of both ILs is regulated by type I IFNs (Guarda et al., 2011). IL-6 is produced by a variety of cells; during cute inflammation it is produced by monocytes and macrophages but in chronic inflammation it is produced by T-cells, which are considered the main source of this particular cytokine (Dmitrieva et al., 2016). The acute phase response to an infection is a systemic response, proportional to the inflammatory stimulus severity and mediated by IL-1 and IL-6 (Gabay and Kushner, 1999).
Some diseases are characterized by the uncontrolled production of IL-1β secreted from innate immune cells; inhibiting this cytokine is a therapeutic target in inflammatory diseases (Schett et al., 2016). The pleiotropic immunomodulatory cytokine, IL-10, is produced by a variety of cells such as monocytes, DCs, macrophages, B- and T-lymphocytes, mast cells, epithelial cells, and keratinocytes. The key feature of IL-10 is its ability to work as an immunosuppressant against a wide range of cytokines and chemokines (Slobedman et al., 2009). Overexpressed IL-6 was first identified in cardiac myxoma tissue in 1987, where it contributed to the development of chronic inflammation and autoimmunity. Subsequently, IL-6 inhibition has been a successful therapeutic target in treating several chronic immune diseases and is expected to be used in future efforts to treat various diseases (Tanaka et al., 2018). Accordingly, the use of IL inhibitors such as IL-6 antagonists has been suggested as a treatment measure in treating patients with critical COVID-19 (Zhang et al., 2020a, Zhang et al., 2020c). In addition, IL-1 blockade is worthy of consideration in treating SARS-CoV-2 infection as it plays a role in coagulopathy reduction caused by endothelial dysfunction (Cavalli et al., 2020).
4.1.3. Chemokines
The chemokine super family is the largest family of cytokines with more than 40 members that are able to bind to one or more of the 21 G-protein-coupled receptors. This superfamily is comprised of small secreted proteins critical to a wide range of physiological and pathological processes such as embryogenesis and the immune system. They are recognized for their function in tumorigenesis, inflammatory responses, and cancer metastasis. Moreover, chemokines play a major role as extracellular mediators (chemo-attractants) of cellular migration, especially of immune cells (Comerford and McColl, 2011). Chemokines are classified based on the number and spacing between their N-terminal first two cysteine residues into four types: CXC, C, CC, and CX3C (Raman et al., 2011, Scanzello, 2017). Chemokine receptors are subdivided as well into two main classes based on their binding ability to CXC or CC. This enables them to be highly active during the inflammation response by promoting immune cell chemotactic migration to the inflammation site. However, chemokines can also produce undesirable inflammatory responses (Koelink et al., 2012). During microbial infection, an inflammatory response is elicited resulting in increased vascular permeability and leukocyte infiltration; specifically, neutrophils comprise the primary cells that infiltrate during acute inflammation. During chronic inflammation, monocytes and lymphocytes are the main infiltrating cells; however, in allergic reactions the predominant cells are eosinophils and lymphocytes. This selectivity of tissue infiltration is promoted by a combination of specific adhesion molecules and chemokine/chemokine receptors (Yoshie and Matsushima, 2017).
The chemokine ligand 5 (CCL5) C-C motif, also known as RANTES, is one of the associated-early expressed chemokines during viral infection. This is a result of degranulation from activated virus specific CD8+ T cells. RANTES is also released from CD4+ T lymphocytes, platelets, fibroblasts, and epithelial cells leading to the activation of NKs, T CD4+, monocytes, DCs, mast cells, and lymphocytes (Marques et al., 2013). CCR5 inhibitors are used to block HIV Env from binding to the CCR5 co-receptor via a competitive mechanism; this in turn prevents CCL5 chemotaxis elicitation and hence, inflammatory T cells and CCR5+ macrophages. This therapeutic approach is considered in the treatment of SARS-CoV-2 (Patterson et al., 2020).
4.1.4. Tumor necrosis factors
The TNF family includes TNFα, TNFβ, Fas ligand (FasL), TNF-related apoptosis inducing ligand (TRAIL), LIGHT (receptor expressed by T lymphocytes), and CD40 ligand (CD40L). TNFs are important cytokines involved in several physiological processes such as tumor lysis, systemic inflammation, acute phase reaction, and apoptosis (Chu, 2013). TNFs can stimulate their own secretion along with other inflammatory cytokines and chemokines; therefore, they are considered as key mediators in both acute and chronic inflammatory responses. They also play a role in the pathogenesis of autoimmune diseases such as rheumatoid arthritis, multiple sclerosis, Crohn's disease, ulcerative colitis, systemic sclerosis, and systemic lupus erythematosus (SLE). Moreover, TNFs play a major role in endotoxic-induced septic shock in animal models, whereas in patients with late-stage lung cancer TNFs can cause chemotherapy-induced septic shock (Chu, 2013). TNFα is a pyrogenic cytokine (causes heat/fever) usually secreted from immune cells during acute phase inflammation and infections, and is central in viral diseases. This cytokine is also associated with various autoimmune and chronic inflammatory diseases (Carswell et al., 1975). The use of anti-TNF treatment in treating patients with SARS-CoV-2 infection is being advocated based on observations of patients with COVID-19 suffering from inflammatory bowel diseases (IBDs) who are receiving anti-TNFs; these patients showed better outcomes and higher COVID-19 recovery rates than those who received other treatments (Feldmann et al., 2020).
4.1.5. Colony- stimulating factors
CSFs are proteins that include macrophage colony stimulating factor (M-CSF), granulocyte colony-stimulating factor (G-CSF), and granulocyte–macrophage colony-stimulating factor (GM-CSF). They function in stimulating the growth, proliferation, and differentiation of hematopoietic progenitor cells (Hamilton, 2008). CSFs are also associated with inflammation and are proposed to be part of a CSF network; their action was observed to be part of a pro-inflammatory cytokine network that includes TNF and IL-1. CSFs are thought to be involved in elevated macrophage-cytokine production at the inflammation site. Moreover, CSFs may play a role in amplification cascades leading to a perpetuated inflammatory response, which increases the likelihood of their involvement in the pathogenesis of several diseases. Hence, CSFs are considered as potential therapeutic targets (Hamilton, 2008). In spite of the downside of CSFs, it has been suggested that treatments stimulating GM-CSF should be used as they result in an elevation of leukocyte levels in the blood. Based on the finding of low white blood cells in patients with COVID-19, the use of such drugs is supported to increase leukocytes, especially phagocytes, which offer therapeutic advantage in fighting COVID-19 (Potter et al., 2020).
4.2. Viral manipulation of immunity
Viruses can use cellular immune defense mechanisms to their favor to avoid viral clearance and achieve successful infection (Christiaansen et al., 2015). One of the most essential cytokines in this process is IL-10 as a result of the role it plays in suppression of inflammatory cytokines, impairment of maturation of DCs, inhibition of effector T-cells by down-regulating the major histocompatibility complex (MHC), and co-stimulation molecules on APCs (Couper et al., 2008, Wilson and Brooks, 2010). Host cellular IL-10 is stimulated by many viruses including HBV, HCV, and HIV. Alternatively, other viruses have the ability to encode IL-10 orthologs of their own, especially poxviruses and herpesviruses (Ouyang et al., 2014, Slobedman et al., 2009). The induction of IL-10 by the lymphocytic choriomeningitis virus in infected mice encourages virus persistence; therefore, blocking IL-10 signaling could facilitate viral clearance (Brooks et al., 2006).
Chemokines direct leukocyte movement by binding to their specific receptors and some viruses are able to alter chemokine signaling networks encouraging viral dissemination, modulation of host immune cell trafficking, inhibition of immune clearance, and creation of a suitable environment for viral replication (Berson et al., 1996, Feng et al., 1996). Other viruses can produce molecules that are able to “mimic” the chemokine system components by interacting with their receptors and interrupting the signaling leading to an altered immune response against the viral infection (Alcami, 2003). Antiviral response induced by IFN can be inhibited by some viruses by impairing the intracellular PRRs including intracellular RNA sensors and TLRs (Thiel and Weber, 2008). SARS-CoV is one of the viruses that can avoid the activation of IFN-induced cascades by the induction of double-membraned vesicles within the cytoplasm (perinuclear sites) where RNA is synthesized; this mechanism protects viral RNA replication from exposure to intracellular RNA sensors (Prentice et al., 2004, Snijder et al., 2006). Another mechanism used by SARS-CoV to inhibit IFN-induced cascade activation is by producing the ORF6 protein that inhibits the action of transcription factor-3 (IRF-3), a product of one of the IFN genes (Frieman et al., 2007, Kopecky-Bromberg et al., 2007).
Some viruses such as respiratory syncytial virus (RSV), human cytomegalovirus (HCMV), and Epstein-Barr virus (EBV) possess the ability to develop mechanisms that target inhibition of adaptive immune responses by altering CD4 T-cells, which produce TH1 cytokines including IFN-α and IFN-γ (Lukacs et al., 2006, Marshall et al., 2003, Mason et al., 2013, Moore et al., 2009, Voo et al., 2005). CD8 T-cells play a major role in controlling and clearing viral infection. Some viruses, including HCMV, possess tools that prevent MHC class 1 expression on virus-hosting cells, which blocks CD8 T-cell mediated cytolysis. This also targets the molecules for degradation (Besold et al., 2009, Wiertz et al., 1996). EBV down-modulates MHC class 1 expression (Griffin et al., 2013). B-cells produce antibodies that help in infection clearance and prevent future reinfection; however, some viruses have developed mechanisms to prevent B-cell mediated clearance, which include inhibiting B-cell maturation and residing within B-cells and hindering their survival. The influenza virus is able to directly infect antigen-specific B-cells in the lungs, which reduces the antibody response. However, infected individuals are still able to produce a neutralizing antibody response against the infecting strain. In comparison, antibodies produced specifically against RSV infection appear to be inadequate in preventing re-infection, mainly of the upper respiratory tract (Dougan et al., 2013, Habibi et al., 2015, Nothelfer et al., 2015).
Furthermore, a previous study has suggested that SARS-CoV-1 can manipulate the innate immune response presented by the mitochondria. The mechanism involves the ability of the virus to secrete a protein designated as ORF-9b; this protein causes the MAVS molecule and dynamin-like protein (DRP1), a host protein involved in mitochondrial fission, to degrade via direct and indirect events, which eventually leads to abnormal elongation of the mitochondria and suppression of the antiviral transcriptional response, resulting in mitochondrial dysfunction (Shi et al., 2014).
4.3. The cytokine storm
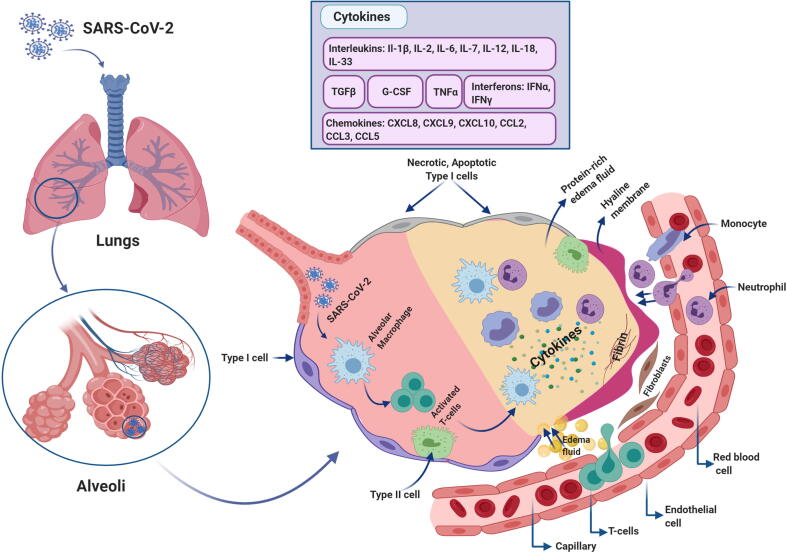
The term cytokine storm has gained attention since it was first introduced in the 1990s to describe the onset of events aimed at modulating graft-versus-host disease (GVHD) (Ferrara et al., 1993). Cytokine storm describes the role of the immune system in producing a generalized and uncontrolled inflammatory response (Tisoncik et al., 2012) and can be used in parallel with another term, cytokine release syndrome (CRS). Both terms reflect processes in which the same biomarker signatures and clinical phenotypes are presented; however, they possess distinctly different characteristics. CRS describes a spectrum of reactions observed post targeted therapy administration resulting in significant immune system activation; these therapies include chimeric antigen receptor (CAR) T-cell therapy and bispecific T-cell-engaging antibodies. In contrast, cytokine storm is an overwhelming activation of the immune system leading to systemic inflammation, which occurs independently from tumor targeting therapies (Porter et al., 2018). Some infectious and non-infectious diseases are associated with presentation of the cytokine storm; a drawback of some therapeutic intervention attempts is that this type of action is promoted (Suntharalingam et al., 2006). Cytokine storm has been associated with avian influenza virus (H5N1) infection (Yuen and Wong, 2005); clinical findings revealed that most infected patients presented with fever, dyspnea, dry cough, and lung bilateral ground-glass opacities on chest computed tomography (CT) scans (Guan et al., 2020). Early reports of COVID-19 indicated that one of the clinical aftereffects of SARS-CoV-2 infection appeared to be ARDS, which accounts for significant deaths among infected patients. Therefore, the process of cytokine storm should be considered as an immune-mediated hallmark of SARS-CoV-2 infection, as previously described for MERS-CoV and SARS-CoV (Xu et al., 2020). ARDS is defined by the presence of severe hypoxemia and bilateral lung infiltrates; it accounts for nearly 40% of mortality incidents and can be an unfortunate outcome of several clinical situations including pneumonia, blood transfusion, sepsis, and pancreatitis. The pathogenesis of ARDS results in alveolo-capillary membrane permeability owing to inflammatory injury, leading to protein-rich pulmonary fluid perfusion into the airspaces causing edema, which eventually leads to insufficient respiration (Bhatia et al., 2012). Increased levels of circulating pro-inflammatory cytokines (e.g., INFγ, IL-B1, IL-6, and IL-12) and chemokines (e.g., CXCL10 and CCL2) are features of pulmonary inflammation in patients with SARS and similar to MERS-CoV infection (Channappanavar and Perlman, 2017).
Recently, it was reported that patients diagnosed with COVID-19 were presenting with high levels of cytokines and chemokines (Huang et al., 2020). The increased levels of INFγ, IL-B1, CXCL10, and CCL2 strongly indicated activation of TH1 cell function; moreover, intensive care unit (ICU)-admitted patients with COVID-19 showed higher concentrations of CXCL10, CCL2, and TNFα compared to those of patients who did not require an ICU admission. Increased secretion of TH2 immune -oriented cytokines such as IL-4 and IL-10 was also observed; both suppress inflammation (Zhang et al., 2020a, Zhang et al., 2020c). According to the literature, in SARS-CoV infection the presence of ARDS is a result of a cytokine storm; the release of inflammatory cytokines (IFNγ, IFNα, TNFα, IL-1β, IL-6, IL-12, IL18, Il-33, and TGFβ) and chemokines (CCL2, CCl3, CCl5, CXCL8, CXCL9, and CXCL10) are participants in the divergent systemic inflammatory response (Cameron et al., 2008, Channappanavar and Perlman, 2017, Williams and Chambers, 2014, Zhang et al., 2020a, Zhang et al., 2020c). SARS-CoV-2 has been reported to initiate cytokine storms in the lungs by stimulating a variety of cytokines such as IL-2, IL-6, IL-7, TNFα, G-CSF, IP10 (CXCL10), MCP1 (CCL2), and MIP1A (CCL3) (Huang et al., 2020). The consequence of a cytokine storm is ARDS and organ failure as the immune system attacks the body, which eventually leads to death in the most severe cases of SARS-CoV-2 infection (Xu et al., 2020) (Fig. 5).
Fig. 5.
Cytokine storm pathogenesis. When SARS-CoV-2 infects the lungs, the immune cells including macrophages, neutrophils, and monocytes reach the infected site via chemotaxis, recognize the pathogen, and produce a large amount of cytokines and chemokines at the site of infection. This initiates a cycle of exaggerated inflammatory responses that eventually damage the lungs as a result of fibrin and scar tissue formation, which obstructs oxygen passage through the bloodstream. Owing to inflammation, the blood vessels weaken and become permeable leading to the build-up of protein-rich fluid and edema within the lung cavities. This eventually leads to acute respiratory distress syndrome (ARDS).
A recent study has suggested that the excessive buildup of cellular senescence observed in ageing or age-related diseases is a key player in chronic inflammation and tissue and organ dysfunction. The senescent cell secretome of the senescence-associated secretory phenotype (SASP), includes extracellular vesicles (EVs) containing miRNA and DNA, which spread to proximal and distant tissues resulting in catastrophic consequence in the organism as it promotes further inflammatory responses (cytokine storm). Senescent cells showed an association with mitochondria hyper-fusion resulting in abnormally elongated mitochondria, thereby increasing SASP. The mitochondrial dysfunction could exacerbate SASP via DAMPs in the elderly with SARS-CoV-2 infection (Malavolta et al., 2020).
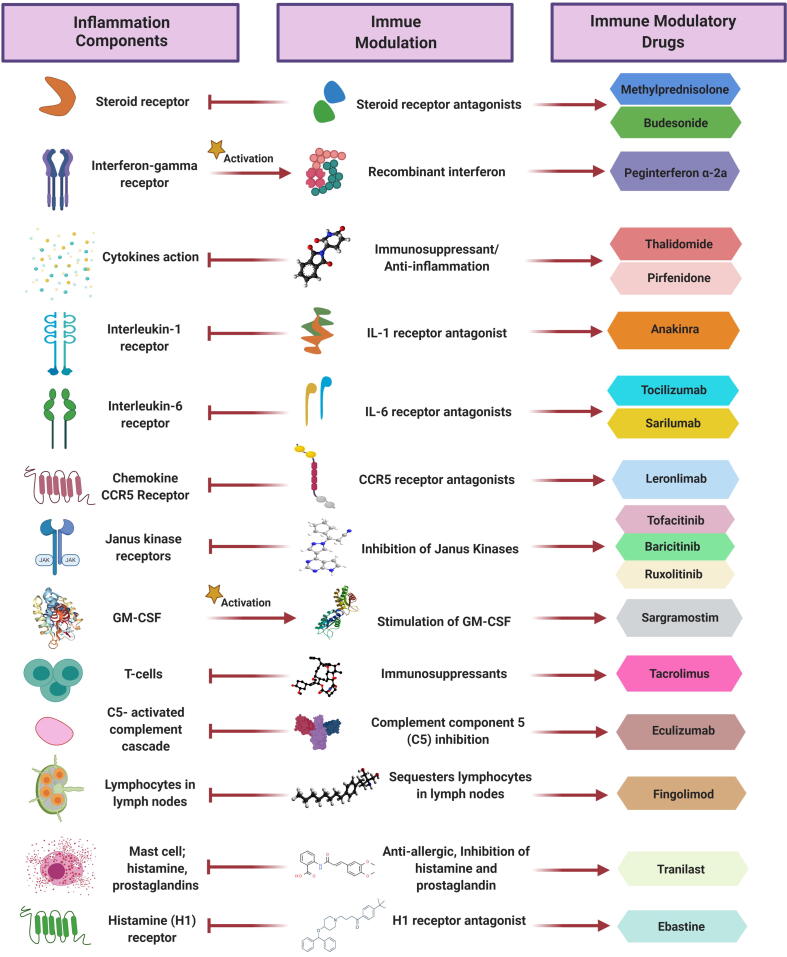
Notably, incorporating immunomodulators as part of the drug combinations used in treating SARS-CoV-2 infection has proven effective in helping to control the symptoms and complications resulting from COVID-19. Immunomodulators help attenuate the cytokine storm, leading to reduction in comorbidity of the affected population, mostly to ARDS. This is a result of targeting the activity of certain cytokines and/or by suppressing the exaggerated immune response. It is important to point out that the drugs are FDA-approved for treating diseases other than COVID-19 (Table 2; Fig. 6).
Table 2.
Immunomodulators engaged in COVID-19 clinical trials that balance and alleviate immune responses and cytokine storms.
| Target | Mechanism of action | Drug/ Drug bank ID | Chemical formula | AHFS | COVID-19 clinical trial phase |
|---|---|---|---|---|---|
| Inflammatory response | Immunosuppressant; steroid receptor antagonist | Methylprednisolone /(DB00959) | C22H30O5 | Treats acute phase relapse of multiple sclerosis (Sloka and Stefanelli, 2005), suppresses ARDS (Meduri et al., 2002). | Phase 2 NCT0424459 |
| Budesonide (DB01222) | C25H34O6 | Treats asthma (Brogden and McTavish, 1992) and active Crohn's disease (Greenberg et al., 1994). | Phase 2 NCT04331470 | ||
| Anti-viral state activation | Recombinant- interferon | Peginterferon α-2a (DB00008) | C860H1353N227O255S9 | Combined with imatinib treats chronic myeloid leukemia (Preudhomme et al., 2010). | Phase 4 ChiCTR2000030000 |
| Cytokine action | Immunosuppressive; anti-angiogenic activity | Thalidomide (DB01041) | C13H10N2O4 | Treats prurigo nodularis (refractory PN), multiple myeloma, leprosy complications, erythema nodosum leprosum (Ito et al., 2010, Taefehnorooz et al., 2011). | Phase 2 NCT04273529 |
| Down-regulation of multiple cytokines, collagen synthesis, and fibroblast proliferation | Pirfenidone (DB04951) | C12H11NO | Treats diabetic nephropathy (Sharma et al., 2011) and idiopathic pulmonary fibrosis (Noble et al., 2011). | Phase 3 NCT04282902 | |
| Interleukin-1 receptor | Inhibition of IL-1 receptor | Anakinra (DB00026) | C759H1186N208O232S10 | Treats rheumatoid arthritis (RA) (Mertens and Singh, 2009) and refractory pericarditis (Jain et al., 2015). | Phase 3 NCT04364009 |
| Interleukin-6 receptor | Inhibition of IL-6 receptor | Tocilizumab /(DB06273) | C6428H9976N1720O2018S42 | Treats RA (Oldfield et al., 2009). | Phase 2 NCT04335071 |
| Sarilumab/(DB11767) | C6388H9918N1718O1998S44 | Combined with methotrexate, treats RA (Genovese et al., 2015). | Phase 3 NCT04327388 | ||
| Chemokine CCR5 receptor | Anti-human CCR5 receptor | Leronlimab /(DB05941) | C6534 H10036 N1720 O2040 S42 | Anti-HIV (Promsote et al., 2020). | Phase 2 NCT04347239 |
| Janus kinase receptors | Inhibition of Janus kinases | Tofacitinib /(DB08895) | C16H20N6O | Treats ulcerative colitis and RA (Sandborn et al., 2017, van Vollenhoven et al., 2012) | Phase 2 NCT04412252 |
| Baricitinib/(DB11817) | C16H17N7O2S | Treats acute respiratory distress and RA (Richardson et al., 2020, Taylor et al., 2017). | Phase 2 NCT04358614 | ||
| Ruxolitinib/(DB08877) | C17H18N6 | Treats myelofibrosis and polycythemia vera (Harrison et al., 2012, Vannucchi et al., 2015). | Phase 3 NCT04348071 | ||
| GM-CSF | Stimulation of granulocytes and macrophage formation | Sargramostim/(DB00020) | C639H1006N168O196S8 | Induces remission in active Crohn's disease (Roth et al., 2011). | Phase 4 NCT04326920 |
| T-helper cells-dependent-B-cells responses, and IL-2 | Immunosuppressive | Tacrolimus/(DB00864) | C44H69NO12 | Treatment of atopic dermatitis and lupus nephritis (Cury Martins et al., 2015, Lee et al., 2011) | Phase 3 NCT04341038 |
| C5-activated complement cascade | Complement component 5 (C5) inhibition | Eculizumab/(DB01257) | C6442H9910N1694O2034S50 | Treatment of atypical hemolytic-uremic syndrome (Legendre et al., 2013). | Phase 2 NCT04346797 |
| Lymphocytes in lymph nodes | Sequestering lymphocytes in lymph nodes; hinder autoimmune reaction | Fingolimod/(DB08868) | C19H33NO2 | Treatment of multiple sclerosis (Cohen et al., 2010). | Phase 2 NCT04280588 |
| Mast cells | Anti-allergic, inhibition of histamine and prostaglandins from mast cells | Tranilast/(DB07615) | C18H17NO5 | Treats bronchial asthma, and vast array of inflammatory diseases, proliferation, some cancers such as, pancreatic, lung, prostate, uterine leiomyoma, neurofibroma, breast, bladder and desmoid tumor (Darakhshan and Pour, 2015). | Phase 4 ChiCTR2000030002 |
| Histamine (H1) receptor | Anti-allergic, H1 receptor antagonist | Ebastine/(DB11742) | C32H39NO2 | Treatment of chronic spontaneous urticaria (Godse, 2011). | Phase 4 ChiCTR2000030535 |
AHFS, American Hospital Formula Service.
Fig. 6.
Immune modulatory drugs and their mechanism of action during inflammatory responses and cytokine storms. The drugs target several sites/actions within the cell to attenuate inflammation and manage the consequences, including preventing acute respiratory distress syndrome (ARDS).
5. Mesenchymal stem cells (MSCs) in COVID-19 treatment
MSCs have been broadly used in cell-based therapy basic research and in clinical trials and their safety has been reported in several clinical trials related to GVHD and SLE (Connick et al., 2012, Hashmi et al., 2016, Kamen et al., 2018, Wilson et al., 2015). MSC transplantation has been used to treat patients suffering from H7N9-induced ARDS; notably the results showed significant reduction in mortality rates (Chen et al., 2020a). Interesting findings from a recently conducted study on MSC-based treatment for SARS-CoV-2 suggest that MSCs lack SARS-CoV-2 infection-vital receptors (ACE2- and TMPRSS2-); therefore they are SARS-CoV-2 infection-free. Additionally the transplantation of MSCs in SARS-CoV-2-infected patients improved the outcomes owing to the exceptional immunosuppressant potential possessed by MSCs (Leng et al., 2020). Pro-inflammatory cytokines were dramatically reduced in the patient serum which led to less attraction of the leukocytes to the fragile lungs whereas IL-10 and vascular epidermal growth factor (VGEF) levels were notably elevated. Altogether this led to enhanced lung repair and the patients in critical condition were able to enter recovery (Leng et al., 2020). Other studies on umbilical cord-derived MSCs combined with other immunomodulator drugs also showed promising results in treating critically ill patients diagnosed with COVID-19. After receiving MSC-based treatments their health improved and they were able to leave the ICU (Liang et al., 2020). The curative properties of the MSCs against SARS-CoV-2 infection include: (i) direct induction of apoptosis via activated T-cells alleviating excessive immune responses; (ii) homeostasis maintenance and regeneration of specific lung injuries; (iii) release of cytokines to inhibit neutrophil intravasation and enhance macrophage differentiation which helps attenuate inflammation and also promotes the release of EVs which deliver microRNA mRNA DNA proteins and metabolites into host cells post lung injury which promotes repair regeneration and lung function restoration. Therefore MSCs should be considered as a potential treatment for critically ill patients with SARS-CoV-2 infection (Ji et al., 2020)
6. Update on COVID-19 vaccine development
The sequence of the SARS-CoV-2 RNA genome was published on January 11, 2020. This knowledge helped fuel global efforts in vaccine development using a wide range of technologies based on different platforms including nucleic acids (RNA and DNA), peptides, virus-like particles, recombinant proteins, viral vectors (replicating or non-replicating), and whole virus vaccines (live attenuated virus or inactivated virus) (Thanh Le et al., 2020). Several vaccine candidates are in various stages of development as summarized in Table 3.
Table 3.
Candidate COVID-19 vaccines.
| Vaccine candidate | Vaccine description | Association | Clinical trial phase | Reference |
|---|---|---|---|---|
| Viral vector (replicating/non-replicating) | Adenovirus-vectored using AdVac® and PER.C6® technology | Jonson &Johnson. New Brunswick, New Jersy, United States. | Pre-clinical (Phase 1, Sep 2020) | (Johnson and Johnson, 2020). |
| Adenovirus type 5 vector that expresses S protein (Ad5-nCoV) | CanSino Biologics. Tianjin, China. | Phase 1 | (Fengcai et al., 2020). | |
| Modified chimpanzee adenovirus (ChAdOx1) | Oxford's Jenner Institute. Oxford, United Kingdom. | Phase 1/2 | (Doremalen et al. (2020). | |
| Whole virus | Live-attenuated virus | Codagenix. Long Island, New York, United States.Serum Institute of India. Pune, India. | Pre-clinical | (Farmingdale, 2020). |
| Recombinant protein/ protein subunit | Recombinant nanoparticle technology | Novavax. Gaithersburg, Maryland, United States. | Pre-clinical | (Pharmaceutical Technology, 2020). |
| Protein-based vaccine using molecular clamp platform | University of Queensland. Brisbane, Australia./Coalition for Epidemic Preparedness Innovations (CEPI). Oslo, Norway. | Pre-clinical | (Hennessy, 2020). | |
| Coronavirus RBD protein-based vaccine | Baylor College of Medicine. Huston, Texas, United States. Fudan University. Shanghi, China. New York Blood Center. New York, United states. University of Texas Medical Branch. Texas, United States. | Pre-clinical | (Mukherjee, 2020). | |
| S-trimer recombinant protein using trimer-Tag technology | Clover Bipharmaceuticals. Chengdu, China. | Pre-clinical | (Clover Biopharmaceuticals, 2020). | |
| Oral recombinant protein vaccine using VAAST platform | Vaxart. San Francisco, California, United States. | Pre-clinical | (Vaxart, 2020). | |
| Nucleic Acid(DNA/RNA) | DNA vaccine (INO-4800, based on INO-4700 MERS vaccine) | INOVIO Pharmaceuticals. Pennsylvania, United States /Beijing Advaccine Biotechnology.Co. Beijin, China / CEPI | Phase 1 | (Yang, 2020). |
| mRNA vaccine | Moderna. Massachusettes, United States /CEPI/NIH. Bethesda, Maryland, United States. | Phase 2 | (ModernaTX, 2020). | |
| CEPI/ CureVac. Tubingen, Germany. | Pre-clinical | (Smith, 2020). | ||
| Vector-modified cells/ T-cell- based immunotherapy | LV-SMENP-DCmodified DCs with lentivirus vector to express minigene (SMENP) and immune modulatory genes | Shenzhen Geno-Immune Medical Institute. Shenzhen, Guandong, China. | Phase 2 | (Cheng, 2020a). |
| Modified pathogen-specific artificial antigen-presenting cells (aAPCs) by lentivirus vector to express immune modulatory and viral genes | Shenzhen Geno-Immune Medical institute | Phase 1 | (Cheng, 2020b). |
7. Conclusion
This review offers an overview of the various types of CoVs, focusing on SARS-CoV-2 and the disease it causes, COVID-19. A general understanding of viral structure, genomic sequence, and pathogenesis, and also the role of immune system elements in fighting pathogen attacks are key points to understanding the body's defense mechanisms against viral infections. Together, this information provides the cornerstone for the design of specific and effective therapeutic agents including identifying suitable drug combinations to fight CoVs. The drugs reviewed here are still investigational, in clinical trials, with all of the drugs in use being FDA approved to treat several diseases apart from COVID-19 including viral, inflammatory, and autoimmune diseases. The aim of these repurposed drugs is to inhibit SARS-CoV-2 entrance, multiplication, and manipulation of host cell activity, in addition to controlling the immune reaction via mitigation of cytokine activity. Drug combinations are mainly used to manage the symptoms and prevent patients from developing respiratory complications such as pneumonia and ARDS, which are the most observed consequence of severe SARS-CoV-2 infection. Moreover, cell-based therapy with MSCs alongside described drug treatments show promising results in managing and treating COVID-19 including decreasing mortality. Currently, several groups are racing against time to obtain a safe and suitable vaccine using various technologies. Some of these vaccines have already entered clinical trial stages targeting the viral activity on different levels along with the human immune system. Finally, since the outbreak of COVID-19 in China in late 2019, multiple efforts have been devoted to vanquishing SARS-CoV-2; however, at this point no approved vaccine is within reach and the pandemic continues.
Declaration of Competing Interest
None.
Acknowledgments
Acknowledgements
Declared none.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ahn D.-G., Shin H.-J., Kim M.-H., Lee S., Kim H.-S., Myoung J., Kim B.-T., Kim S.-J. Current Status of Epidemiology, Diagnosis, Therapeutics, and Vaccines for Novel Coronavirus Disease 2019 (COVID-19) J. Microbiol. Biotechnol. 2020;30:313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- Atkinson A., Robertson J.I. Captopril in the treatment of clinical hypertension and cardiac failure. Lancet. 1979;314:836–839. doi: 10.1016/S0140-6736(79)92186-X. [DOI] [PubMed] [Google Scholar]
- Berson J.F., Long D., Doranz B.J., Rucker J., Jirik F.R., Doms R.W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J. Virol. 1996;70:6288–6295. doi: 10.1128/JVI.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besold K., Wills M., Plachter B. Immune evasion proteins gpUS2 and gpUS11 of human cytomegalovirus incompletely protect infected cells from CD8 T cell recognition. Virology. 2009;391:5–19. doi: 10.1016/j.virol.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Bhatia M., Zemans R.L., Jeyaseelan S. Role of chemokines in the pathogenesis of acute lung injury. Am. J. Respir. Cell Mol. Biol. 2012;46:566–572. doi: 10.1165/rcmb.2011-0392TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittmann S., Weissenstein A., Villalon G., Moschuring-Alieva E., Luchter E. Simultaneous Treatment of COVID-19 With Serine Protease Inhibitor Camostat and/or Cathepsin L Inhibitor? J. Clin. Med. Res. 2020;12:320–322. doi: 10.14740/jocmr4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaess, M., Kaiser, L., Sauer, M., Deigner, H.-P., 2020. Lysosomotropic active compounds—hidden protection against COVID-19/SARS-CoV-2 infection? https://doi.org/10.20944/preprints202005.0061.v1
- Bonilla F.A., Oettgen H.C. Adaptive immunity. J. Allergy Clin. Immunol. 2010;125:S33–S40. doi: 10.1016/j.jaci.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Brocker C., Thompson D., Matsumoto A., Nebert D.W., Vasiliou V. Evolutionary divergence and functions of the human interleukin (IL) gene family. Hum. Genomics. 2010;5:30. doi: 10.1186/1479-7364-5-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden R.N., McTavish D. Budesonide: An Updated Review of its Pharmacological Properties, and Therapeutic Efficacy in Asthma and Rhinitis. Drugs. 1992;44:375–407. doi: 10.2165/00003495-199244030-00007. [DOI] [PubMed] [Google Scholar]
- Brooks D.G., Trifilo M.J., Edelmann K.H., Teyton L., McGavern D.B., Oldstone M.B.A. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker S.W., Bonham K.S., Zanoni I., Kagan J.C. Innate Immune Pattern Recognition: A Cell Biological Perspective. Annu. Rev. Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler N., Pewe L., Trandem K., Perlman S. Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated. Virology. 2006;347:410–421. doi: 10.1016/j.virol.2005.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., Bermejo-Martin J.F., Danesh A., Muller M.P., Kelvin D.J. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll M.C. The complement system in regulation of adaptive immunity. Nat. Immunol. 2004;5:981–986. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- Carswell E.A., Old L.J., Kassel R.L., Green S., Fiore N., Williamson B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S.C., Di Napoli, R., 2020. Features, evaluation and treatment coronavirus (COVID-19). In: Statpearls [Internet]. StatPearls Publishing. [PubMed]
- Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., Oltolini C., Castiglioni B., Tassan Din C., Boffini N., Tomelleri A., Farina N., Ruggeri A., Rovere-Querini P., Di Lucca G., Martinenghi S., Scotti R., Tresoldi M., Ciceri F., Landoni G., Zangrillo A., Scarpellini P., Dagna L. Interleukin-1 blockade with high-dose anakinra in patients with COVID-19, acute respiratory distress syndrome, and hyperinflammation: a retrospective cohort study. Lancet Rheumatol. 2020;9913:1–7. doi: 10.1016/S2665-9913(20)30127-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh D. Coronaviruses with Special Emphasis on First Insights Concerning SARS. Springer; 2005. Coronaviridae: a review of coronaviruses and toroviruses; pp. 1–54. [Google Scholar]
- Channappanavar, R., Perlman, S., 2017. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. https://doi.org/10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed]
- Chen J., Hu C., Chen Lijun, Tang L., Zhu Y., Xu X., Chen Lu., Gao H., Lu X., Yu L., Dai X., Xiang C., Li L. Clinical Study of Mesenchymal Stem Cell Treatment for Acute Respiratory Distress Syndrome Induced by Epidemic Influenza A (H7N9) Infection: A Hint for COVID-19 Treatment. Engineering. 2020 doi: 10.1016/j.eng.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L., 2020a. Immunity and Safety of Covid-19 Synthetic Minigene Vaccine [WWW Document]. ClinicalTrials.gov. URL https://clinicaltrials.gov/ct2/show/NCT04276896 (accessed 6.3.20).
- Cheng, L., 2020b. Safety and Immunity of Covid-19 aAPC Vaccine [WWW Document]. ClinicalTrials.gov. URL https://clinicaltrials.gov/ct2/show/NCT04299724 (accessed 6.3.20).
- Chorin, E., Wadhwani, L., Magnani, S., Dai, M., Shulman, E., Nadeau-Routhier, C., Knotts, R., Bar-Cohen, R., Kogan, E., Barbhaiya, C., Aizer, A., Holmes, D., Bernstein, S., Spinelli, M., Park, D.S., Stefano, C., Chinitz, L.A., Jankelson, L., 2020. QT interval prolongation and torsade de pointes in patients with COVID-19 treated with hydroxychloroquine/azithromycin. Hear. Rhythm. https://doi.org/10.1016/j.hrthm.2020.05.014. [DOI] [PMC free article] [PubMed]
- Chowdhury M.S., Rathod J., Gernsheimer J. A Rapid Systematic Review of Clinical Trials Utilizing Chloroquine and Hydroxychloroquine as a Treatment for COVID-19. Acad. Emerg. Med. 2020;27:493–504. doi: 10.1111/acem.14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaansen A., Varga S.M., Spencer J.V. Viral manipulation of the host immune response. Curr. Opin. Immunol. 2015;36:54–60. doi: 10.1016/j.coi.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H., Chan, J.F.-W., Wang, Y., Yuen, T.T.-T., Chai, Y., Hou, Y., Shuai, H., Yang, D., Hu, B., Huang, X., 2020. Comparative replication and immune activation profiles of SARS-CoV-2 and SARS-CoV in human lungs: an ex vivo study with implications for the pathogenesis of COVID-19. Clin. Infect. Dis. [DOI] [PMC free article] [PubMed]
- Chu W.-M. Tumor necrosis factor. Cancer Lett. 2013;328:222–225. doi: 10.1016/j.canlet.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clover Biopharmaceuticals, 2020. Clover Initiates Development of Recombinant Subunit-Trimer Vaccine for Wuhan Coronavirus (2019-nCoV) | Vaccines | News Channels [WWW Document]. PipelineReview.com. URL https://pipelinereview.com/index.php/2020012873644/Vaccines/Clover-Initiates-Development-of-Recombinant-Subunit-Trimer-Vaccine-for-Wuhan-Coronavirus-2019-nCoV.html (accessed 6.3.20).
- Cohen J.A., Barkhof F., Comi G., Hartung H.-P., Khatri B.O., Montalban X., Pelletier J., Capra R., Gallo P., Izquierdo G., Tiel-Wilck K., de Vera A., Jin J., Stites T., Wu S., Aradhye S., Kappos L. Oral Fingolimod or Intramuscular Interferon for Relapsing Multiple Sclerosis. N. Engl. J. Med. 2010;362:402–415. doi: 10.1056/NEJMoa0907839. [DOI] [PubMed] [Google Scholar]
- Colson P., Rolain J.-M., Lagier J.-C., Brouqui P., Raoult D. Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int. J. Antimicrob. Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105932. 105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford I., McColl S.R. Mini-review series: Focus on chemokines. Immunol. Cell Biol. 2011;89:183–184. doi: 10.1038/icb.2010.164. [DOI] [PubMed] [Google Scholar]
- Cong Y.-Y. University of Groningen; 2019. Molecular insights into viral respiratory infections. [Google Scholar]
- Connick P., Kolappan M., Crawley C., Webber D.J., Patani R., Michell A.W., Du M.Q., Luan S.L., Altmann D.R., Thompson A.J., Compston A., Scott M.A., Miller D.H., Chandran S. Autologous mesenchymal stem cells for the treatment of secondary progressive multiple sclerosis: An open-label phase 2a proof-of-concept study. Lancet Neurol. 2012;11:150–156. doi: 10.1016/S1474-4422(11)70305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett J.A., Naqvi S.Y., Bajwa F.A. Hypertension, Left Ventricular Hypertrophy, and Heart Failure. Hypertension. 2020;6 [Google Scholar]
- Costanzo M., De Giglio M.A.R., Roviello G.N. SARS-CoV-2: recent reports on antiviral therapies based on lopinavir/ritonavir, darunavir/umifenovir, hydroxychloroquine, remdesivir, favipiravir and other drugs for the treatment of the new coronavirus. Curr. Med Chem. 2020 doi: 10.2174/0929867327666200416131117. [DOI] [PubMed] [Google Scholar]
- Couper K.N., Blount D.G., Riley E.M. IL-10: the master regulator of immunity to infection. J. Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- COVID, C.D.C., Team, R., 2020. Severe outcomes among patients with coronavirus disease 2019 (COVID-19)—United States, February 12–March 16, 2020. MMWR Morb Mortal Wkly Rep 69, 343–346. [DOI] [PMC free article] [PubMed]
- Cowley J.A., Dimmock C.M., Spann K.M., Walker P.J. Gill-associated virus of Penaeus monodon prawns: an invertebrate virus with ORF1a and ORF1b genes related to arteri-and coronaviruses. J. Gen. Virol. 2000;81:1473–1484. doi: 10.1099/0022-1317-81-6-1473. [DOI] [PubMed] [Google Scholar]
- Crisci, C.D., Ardusso, L.R.F., Mossuz, A., Müller, L., 2020. A Precision Medicine Approach to SARS-CoV-2 Pandemic Management. Curr. Treat. Options Allergy 1. [DOI] [PMC free article] [PubMed]
- Cury Martins J., Martins C., Aoki V., Gois A.F.T., Ishii H.A., da Silva E.M.K. Topical tacrolimus for atopic dermatitis. Cochrane Database Syst. Rev. 2015;2015 doi: 10.1002/14651858.CD009864.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darakhshan S., Pour A.B. Tranilast: A review of its therapeutic applications. Pharmacol. Res. 2015;91:15–28. doi: 10.1016/j.phrs.2014.10.009. [DOI] [PubMed] [Google Scholar]
- Davis J.S., Ferreira D., Denholm J.T., Tong S.Y.C. Med. J; Aust: 2020. Clinical trials for the prevention and treatment of coronavirus disease 2019 (COVID-19): The current state of play; p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Soto, J.A., DSSc, F., Hakim, S.T., Foust, D., 2020. Complementary Pharmacological Treatment and Therapeutic Prospects for COVID-19. https://doi.org/10.31219/osf.io/rm5sz.
- Devaux C.A., Rolain J.-M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob Agents. 2020:105938. doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama K., Khan S., Tiwari R., Sircar S., Bhat S., Malik Y.S., Singh K.P., Chaicumpa W., Bonilla-Aldana D.K., Rodriguez-Morales A.J. Coronavirus Disease 2019–COVID-19. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello, C.A., 2009. Immunological and Inflammatory Functions of the Interleukin-1 Family. https://doi.org/10.1146/annurev.immunol.021908.132612. [DOI] [PubMed]
- Dmitrieva O.S., Shilovskiy I.P., Khaitov M.R., Grivennikov S.I. Interleukins 1 and 6 as main mediators of inflammation and cancer. Biochem. 2016;81:80–90. doi: 10.1134/S0006297916020024. [DOI] [PubMed] [Google Scholar]
- Doremalen, N. van, Lambe, T., Spencer, A., Belij-Rammerstorfer, S., Purushotham, J.N., Port, J.R., Avanzato, V., Bushmaker, T., Flaxman, A., Ulaszewska, M., Feldmann, F., Allen, E.R., Sharpe, H., Schulz, J., Holbrook, M., Okumura, A., Meade-White, K., Pérez-Pérez, L., Bissett, C., Gilbride, C., Williamson, B.N., Rosenke, R., Long, D., Ishwarbhai, A., Kailath, R., Rose, L., Morris, S., Powers, C., Lovaglio, J., Hanley, P.W., Scott, D., Saturday, G., Wit, E. de, Gilbert, S.C., Munster, V.J., 2020. ChAdOx1 nCoV-19 vaccination prevents SARS-CoV-2 pneumonia in rhesus macaques. bioRxiv 2020.05.13.093195. https://doi.org/10.1101/2020.05.13.093195.
- Dougan S.K., Ashour J., Karssemeijer R.A., Popp M.W., Avalos A.M., Barisa M., Altenburg A.F., Ingram J.R., Cragnolini J.J., Guo C., Alt F.W., Jaenisch R., Ploegh H.L. Antigen-specific B-cell receptor sensitizes B cells to infection by influenza virus. Nature. 2013;503:406–409. doi: 10.1038/nature12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eron J.J., Orkin C., Gallant J., Molina J.-M., Negredo E., Antinori A., Mills A., Reynes J., Van Landuyt E., Lathouwers E. A week-48 randomized phase-3 trial of darunavir/cobicistat/emtricitabine/tenofovir alafenamide in treatment-naive HIV-1 patients. AIDS. 2018;32:1431. doi: 10.1097/QAD.0000000000001817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmingdale, N.Y., 2020. Codagenix and Serum Institute of India Initiate Co-Development of a Scalable, Live-Attenuated Vaccine Against the 2019 Novel Coronavirus, COVID-19 | BioSpace [WWW Document]. Codagenix, Inc. URL https://www.biospace.com/article/releases/codagenix-and-serum-institute-of-india-initiate-co-development-of-a-scalable-live-attenuated-vaccine-against-the-2019-novel-coronavirus-covid-19/ (accessed 6.3.20).
- Feldmann M., Maini R.N., Woody J.N., Holgate S.T., Winter G., Rowland M., Richards D., Hussell T. Trials of anti-tumour necrosis factor therapy for COVID-19 are urgently needed. Lancet. 2020;395:1407–1409. doi: 10.1016/S0140-6736(20)30858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Broder C.C., Kennedy P.E., Berger E.A. HIV-1 Entry Cofactor: Functional cDNA Cloning of a Seven-Transmembrane, G Protein-Coupled Receptor. Science (80-.) 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- Fengcai, zhu, Xuhan, G., Wei, W., 2020. Phase I Clinical Trial of a COVID-19 Vaccine in 18-60 Healthy Adults [WWW Document]. URL https://clinicaltrials.gov/ct2/show/NCT04313127 (accessed 6.3.20).
- Ferner, R.E., Aronson, J.K., 2020. Chloroquine and hydroxychloroquine in covid-19. [DOI] [PubMed]
- Ferrara J.L.M., Abhyankar S., Gilliland D.G. Cytokine storm of graft-versus-host disease: A critical effector role for interleukin-1. Transpl. Proc. 1993:1216–1217. [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox R.I. Mechanism of action of hydroxychloroquine as an antirheumatic drug. Semin Arthritis Rheum. 1993;23:82–91. doi: 10.1016/s0049-0172(10)80012-5. [DOI] [PubMed] [Google Scholar]
- Frieman M., Yount B., Heise M., Kopecky-Bromberg S.A., Palese P., Baric R.S. Severe Acute Respiratory Syndrome Coronavirus ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the Rough Endoplasmic Reticulum/Golgi Membrane. J. Virol. 2007;81:9812–9824. doi: 10.1128/jvi.01012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst D.E. Pharmacokinetics of hydroxychloroquine and chloroquine during treatment of rheumatic diseases. Lupus. 1996;5(Suppl 1):S11–S15. [PubMed] [Google Scholar]
- Gabay C., Kushner I. Acute-Phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat L.W., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (80-.) 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese M.C., Fleischmann R., Kivitz A.J., Rell-Bakalarska M., Martincova R., Fiore S., Rohane P., Van Hoogstraten H., Garg A., Fan C., Van Adelsberg J., Weinstein S.P., Graham N.M.H., Stahl N., Yancopoulos G.D., Huizinga T.W.J., Van Der Heijde D. Sarilumab plus methotrexate in patients with active rheumatoid arthritis and inadequate response to methotrexate: Results of a phase III study. Arthritis Rheumatol. 2015;67:1424–1437. doi: 10.1002/art.39093. [DOI] [PubMed] [Google Scholar]
- Gervais M., Richer C., Fornes P., De Gasparo M., Giudicelli J. Valsartan and coronary haemodynamics in early post-myocardial infarction in rats. Fundam. Clin. Pharmacol. 1999;13:635–645. doi: 10.1111/j.1472-8206.1999.tb00374.x. [DOI] [PubMed] [Google Scholar]
- Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.-C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020;126:1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godse K. Ebastine in chronic spontaneous urticaria in higher doses. Indian J. Dermatol. 2011;56:597. doi: 10.4103/0019-5154.87168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong T., Liu L., Jiang W., Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020;20:95–112. doi: 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubau, D., Deddouche, S., Reis e Sousa, C., 2013. Cytosolic Sensing of Viruses. Immunity 38, 855–869. https://doi.org/10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed]
- Greenberg G.R., Feagan B.G., Martin F., Sutherland L.R., Thomson A., Williams C.N., Nilsson L.-G., Persson T. Oral Budesonide for Active Crohn’s Disease. N. Engl. J. Med. 1994;331:836–841. doi: 10.1056/NEJM199409293311303. [DOI] [PubMed] [Google Scholar]
- Griffin B.D., Gram A.M., Mulder A., Van Leeuwen D., Claas F.H.J., Wang F., Ressing M.E., Wiertz E. EBV BILF1 Evolved To Downregulate Cell Surface Display of a Wide Range of HLA Class I Molecules through Their Cytoplasmic Tail. J. Immunol. 2013;190:1672–1684. doi: 10.4049/jimmunol.1102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W., Ni Z., Hu Yu., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang Jin-lin, Liang Z., Peng Y., Wei L., Liu Y., Hu Ya-hua., Peng P., Wang Jian-ming, Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda G., Braun M., Staehli F., Tardivel A., Mattmann C., Förster I., Farlik M., Decker T., Du Pasquier R.A., Romero P., Tschopp J. Type I Interferon Inhibits Interleukin-1 Production and Inflammasome Activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habibi M.S., Jozwik A., Makris S., Dunning J., Paras A., DeVincenzo J.P., de Haan C.A.M., Wrammert J., Openshaw P.J.M., Chiu C. Impaired Antibody-mediated Protection and Defective IgA B-Cell Memory in Experimental Infection of Adults with Respiratory Syncytial Virus. Am. J. Respir. Crit. Care Med. 2015;191:1040–1049. doi: 10.1164/rccm.201412-2256OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haładyj E., Sikora M., Felis-Giemza A., Olesińska M. Antimalarials–are they effective and safe in rheumatic diseases? Reumatologia. 2018;56:164. doi: 10.5114/reum.2018.76904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J.A. Colony-stimulating factors in inflammation and autoimmunity. Nat. Rev. Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- Hamre D., Procknow J.J. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- Harrison C., Kiladjian J.-J., Al-Ali H.K., Gisslinger H., Waltzman R., Stalbovskaya V., McQuitty M., Hunter D.S., Levy R., Knoops L., Cervantes F., Vannucchi A.M., Barbui T., Barosi G. JAK Inhibition with Ruxolitinib versus Best Available Therapy for Myelofibrosis. N. Engl. J. Med. 2012;366:787–798. doi: 10.1056/NEJMoa1110556. [DOI] [PubMed] [Google Scholar]
- Hashmi S., Ahmed M., Murad M.H., Litzow M.R., Adams R.H., Ball L.M., Prasad V.K., Kebriaei P., Ringden O. Survival after mesenchymal stromal cell therapy in steroid-refractory acute graft-versus-host disease: systematic review and meta-analysis. Lancet Haematol. 2016;3:e45–e52. doi: 10.1016/S2352-3026(15)00224-0. [DOI] [PubMed] [Google Scholar]
- Hayden F.G., Shindo N. Influenza virus polymerase inhibitors in clinical development. Curr Opin Infect Dis. 2019;32:176–186. doi: 10.1097/qco.0000000000000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy, J., 2020. Australia’s Been Asked to Make a Coronavirus Vaccine at “Unprecedented Speed” [WWW Document]. Scince alert. URL https://www.sciencealert.com/australian-scientists-asked-to-make-coronavirus-vaccine-at-unprecedented-speed (accessed 6.3.20).
- Hillen, H.S., Kokic, G., Farnung, L., Dienemann, C., Tegunov, D., Cramer, P., 2020. Structure of replicating SARS-CoV-2 polymerase. bioRxiv. [DOI] [PubMed]
- Hirano T., Murakami M. COVID-19: A new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020 doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Wood C.R., Cimen S., Venkatachalam A.B., Alwayn I.P.J. Mitochondrial Damage-Associated Molecular Patterns (MTDs) Are Released during Hepatic Ischemia Reperfusion and Induce Inflammatory Responses. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0140105. e0140105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Song W., Huang H., Sun Q. Pharmacological Therapeutics Targeting RNA-Dependent RNA Polymerase, Proteinase and Spike Protein: From Mechanistic Studies to Clinical Trials for COVID-19. J. Clin. Med. 2020;9:1131. doi: 10.3390/jcm9041131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Dong W., Milewska A., Golda A., Qi Y., Zhu Q.K., Marasco W.A., Baric R.S., Sims A.C., Pyrc K. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J. Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, D., 2017. Lung microenvironment plays a key role on the protective vs. detrimental effects of mesenchymal stromal cell therapy in acute lung injury.
- Ito T., Ando H., Suzuki T., Ogura T., Hotta K., Imamura Y., Yamaguchi Y., Handa H. Identification of a Primary Target of Thalidomide Teratogenicity. Science (80-.) 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type i interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, A., 2012. A Virological View of Innate Immune Recognition. https://doi.org/10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed]
- Iwasaki A., Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science (80-.) 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Thongprayoon C., Espinosa R.E., Hayes S.N., Klarich K.W., Cooper L.T., Moder K.G., Anavekar N.S., Oh J.K., Matteson E.L. Effectiveness and Safety of Anakinra for Management of Refractory Pericarditis. Am. J. Cardiol. 2015;116:1277–1279. doi: 10.1016/j.amjcard.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Ji F., Li L., Li Z., Jin Y., Liu W. Mesenchymal stem cells as a potential treatment for critically ill patients with coronavirus disease 2019. Stem Cells Transl. Med. 2020;9:813–814. doi: 10.1002/sctm.20-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson & Johnson, 2020. Johnson & Johnson Announces a Lead Vaccine Candidate for COVID-19; Landmark New Partnership with U.S. Department of Health & Human Services; and Commitment to Supply One Billion Vaccines Worldwide for Emergency Pandemic Use | Johnson & Johnson [WWW Document]. URL https://bit.ly/36Sf5fc (accessed 6.3.20).
- Kamen, D.L., Nietert, P.J., Wang, H., Duke, T., Cloud, C., Robinson, A., Gilkeson, G.S., 2018. CT-04 Safety and efficacy of allogeneic umbilical cord-derived mesenchymal stem cells (MSCs) in patients with systemic lupus erythematosus: results of an open-label phase I study, in: Clinical Trials. Lupus Foundation of America, p. A46.2-A47. https://doi.org/10.1136/lupus-2018-lsm.76.
- Khan, F., Fabbri, L., Stewart, I., Robinson, K., Smyth, A.R., Jenkins, G., 2020. A systematic review of Anakinra, Tocilizumab, Sarilumab and Siltuximab for coronavirus-related infections. medRxiv. https://doi.org/10.1101/2020.04.23.20076612.
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt N.R., Cheu R., Birse K., Zevin A.S., Perner M., Noël-Romas L., Grobler A., Westmacott G., Xie I.Y., Butler J. Vaginal bacteria modify HIV tenofovir microbicide efficacy in African women. Science (80-.) 2017;356:938–945. doi: 10.1126/science.aai9383. [DOI] [PubMed] [Google Scholar]
- Koelink P.J., Overbeek S.A., Braber S., De Kruijf P., Folkerts G., Smit M.J., Kraneveld A.D. Targeting chemokine receptors in chronic inflammatory diseases: An extensive review. Pharmacol. Ther. 2012;133:1–18. doi: 10.1016/j.pharmthera.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martínez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/jvi.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T. Mitochondrial-mediated antiviral immunity. Biochim. Biophys. Acta - Mol. Cell Res. 2013;1833:225–232. doi: 10.1016/j.bbamcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Lau, S.K.P., Chan, J.F.W., 2015. Coronaviruses: emerging and re-emerging pathogens in humans and animals. [DOI] [PMC free article] [PubMed]
- Le Tiec C., Barrail A., Goujard C., Taburet A.M. Clinical pharmacokinetics and summary of efficacy and tolerability of atazanavir. Clin Pharmacokinet. 2005;44:1035–1050. doi: 10.2165/00003088-200544100-00003. [DOI] [PubMed] [Google Scholar]
- Lee Y., Lee H.-S., Choi S., Ji J.D., Song G. Efficacy and safety of tacrolimus therapy for lupus nephritis: a systematic review of clinical trials. Lupus. 2011;20:636–640. doi: 10.1177/0961203310389486. [DOI] [PubMed] [Google Scholar]
- Legendre C.M., Licht C., Muus P., Greenbaum L.A., Babu S., Bedrosian C., Bingham C., Cohen D.J., Delmas Y., Douglas K., Eitner F., Feldkamp T., Fouque D., Furman R.R., Gaber O., Herthelius M., Hourmant M., Karpman D., Lebranchu Y., Mariat C., Menne J., Moulin B., Nürnberger J., Ogawa M., Remuzzi G., Richard T., Sberro-Soussan R., Severino B., Sheerin N.S., Trivelli A., Zimmerhackl L.B., Goodship T., Loirat C. Terminal complement inhibitor eculizumab in atypical hemolytic-uremic syndrome. N. Engl. J. Med. 2013;368:2169–2181. doi: 10.1056/NEJMoa1208981. [DOI] [PubMed] [Google Scholar]
- Leng Z., Zhu R., Hou W., Feng Y., Yang Y., Han Q., Shan G., Meng F., Du D., Wang S., Fan J., Wang W., Deng L., Shi H., Li H., Hu Z., Zhang F., Gao J., Liu H., Li X., Zhao Y., Yin K., He X., Gao Z., Wang Y., Yang B., Jin R., Stambler I., Lim L.W., Su H., Moskalev A., Cano A., Chakrabarti S., Min K.-J., Ellison-Hughes G., Caruso C., Jin K., Zhao R.C. Transplantation of ACE2- Mesenchymal Stem Cells Improves the Outcome of Patients with COVID-19 Pneumonia. Aging Dis. 2020;11:216. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E., Darnell J.E. STATs: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Li C., Zhu X., Ji X., Quanquin N., Deng Y.Q., Tian M., Aliyari R., Zuo X., Yuan L., Afridi S.K., Li X.F., Jung J.U., Nielsen-Saines K., Qin F.X., Qin C.F., Xu Z., Cheng G. Chloroquine, a FDA-approved Drug, Prevents Zika Virus Infection and its Associated Congenital Microcephaly in Mice. EBioMedicine. 2017;24:189–194. doi: 10.1016/j.ebiom.2017.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zhou W., Yang L., You R. Physiological and pathological regulation of ACE2, the SARS-CoV-2 receptor. Pharmacol. Res. 2020;157:104833. doi: 10.1016/j.phrs.2020.104833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian N., Xie H., Lin S., Huang J., Zhao J., Lin Q. Clin. Microbiol Infect; 2020. Umifenovir treatment is not associated with improved outcomes in patients with coronavirus disease 2019: A retrospective study. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, S., Jiao, H.L., Chi, L.K., Shi, X.Y., Liang, A.M., Tian, Y., Han, J.L., Ma, S.S., Yang, B., Guan, F.X., 2020. Clinical remission of a critically ill COVID-19 patient treated by human umbilical cord mesenchymal stem cells. Chinese J. Tissue Eng. Res. 16, 9179–9185. https://doi.org/202002.00084v1.
- Lim H.S., Im J.S., Cho J.Y., Bae K.S., Klein T.A., Yeom J.S., Kim T.S., Choi J.S., Jang I.J., Park J.W. Pharmacokinetics of hydroxychloroquine and its clinical implications in chemoprophylaxis against malaria caused by Plasmodium vivax. Antimicrob Agents Chemother. 2009;53:1468–1475. doi: 10.1128/aac.00339-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, F. ching, Young, H.A., 2014. Interferons: Success in anti-viral immunotherapy. Cytokine Growth Factor Rev. 25, 369–376. https://doi.org/10.1016/j.cytogfr.2014.07.015. [DOI] [PMC free article] [PubMed]
- Liu P., Wysocki J., Souma T., Ye M., Ramirez V., Zhou B., Wilsbacher L.D., Quaggin S.E., Batlle D., Jin J. Novel ACE2-Fc chimeric fusion provides long-lasting hypertension control and organ protection in mouse models of systemic renin angiotensin system activation. Kidney Int. 2018;94:114–125. doi: 10.1016/j.kint.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Lombaard, J., DeJesus, E., Sklar, P., Nguyen, B.-Y., Hanna, G.J., Cahn, P.E., Lopardo, G.D., Porteiro, N., Bloch, M.T., Baker, D.A., 2018. Doravirine versus ritonavir-boosted darunavir in antiretroviral-naive adults with HIV-1 (DRIVE-FORWARD): 48-week results of a randomised, double-blind, phase 3, non-inferiority trial. [DOI] [PubMed]
- Lukacs N.W., Moore M.L., Rudd B.D., Berlin A.A., Collins R.D., Olson S.J., Ho S.B., Peebles R.S. Differential Immune Responses and Pulmonary Pathophysiology Are Induced by Two Different Strains of Respiratory Syncytial Virus. Am. J. Pathol. 2006;169:977–986. doi: 10.2353/ajpath.2006.051055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol Sci. 2020 doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J.D. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat. Rev. Immunol. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavolta M., Giacconi R., Brunetti D., Provinciali M., Maggi F. Exploring the Relevance of Senotherapeutics for the Current SARS-CoV-2 Emergency and Similar Future Global Health Threats. Cells. 2020;9:909. doi: 10.3390/cells9040909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin G.H. Facts and reflections on COVID-19 and anti-hypertensives drugs. Drug Discov. Ther. 2020;14:105–106. doi: 10.5582/ddt.2020.01017. [DOI] [PubMed] [Google Scholar]
- Marques R.E., Guabiraba R., Russo R.C., Teixeira M.M. Targeting CCL5 in inflammation. Expert Opin. Ther. Targets. 2013;17:1439–1460. doi: 10.1517/14728222.2013.837886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J.S., Warrington R., Watson W., Kim H.L. An introduction to immunology and immunopathology. Allergy, Asthma. Clin. Immunol. 2018;14:49. doi: 10.1186/s13223-018-0278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N.A., Vickers M.A., Barker R.N. Regulatory T Cells Secreting IL-10 Dominate the Immune Response to EBV Latent Membrane Protein 1. J. Immunol. 2003;170:6183–6189. doi: 10.4049/jimmunol.170.12.6183. [DOI] [PubMed] [Google Scholar]
- Martin P., Jensen D.M. Ribavirin in the treatment of chronic hepatitis C. J. Gastroenterol. Hepatol. 2008;23:844–855. doi: 10.1111/j.1440-1746.2008.05398.x. [DOI] [PubMed] [Google Scholar]
- Mason G.M., Jackson S., Okecha G., Poole E., Sissons J.G.P., Sinclair J., Wills M.R. Human Cytomegalovirus Latency-Associated Proteins Elicit Immune-Suppressive IL-10 Producing CD4+ T Cells. PLoS Pathog. 2013;9:e1003635. doi: 10.1371/journal.ppat.1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meduri G.U., Tolley E.A., Chrousos G.P., Stentz F. Prolonged Methylprednisolone Treatment Suppresses Systemic Inflammation in Patients with Unresolving Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2002;165:983–991. doi: 10.1164/ajrccm.165.7.2106014. [DOI] [PubMed] [Google Scholar]
- Mertens M., Singh J. Anakinra for Rheumatoid Arthritis: A Systematic Review. J. Rheumatol. 2009;36:1118–1125. doi: 10.3899/jrheum.090074. [DOI] [PubMed] [Google Scholar]
- Moderna, T.X., 2020. Dose-Confirmation Study to Evaluate the Safety, Reactogenicity, and Immunogenicity of mRNA-1273 COVID-19 Vaccine in Adults Aged 18 Years and Older - Full Text View - ClinicalTrials.gov [WWW Document]. ClinicalTrials.gov. URL https://clinicaltrials.gov/ct2/show/NCT04405076 (accessed 6.20.20).
- Mohamed, K., Yazdanpanah, N., Saghazadeh, A., Rezaei, N., 2020. Computational Drug Discovery and Repurposing for the Treatment of COVID-19: A Systematic Review. Available SSRN 3583748. [DOI] [PMC free article] [PubMed]
- Moore M.L., Chi M.H., Luongo C., Lukacs N.W., Polosukhin V.V., Huckabee M.M., Newcomb D.C., Buchholz U.J., Crowe J.E., Goleniewska K., Williams J.V., Collins P.L., Peebles R.S. A Chimeric A2 Strain of Respiratory Syncytial Virus (RSV) with the Fusion Protein of RSV Strain Line 19 Exhibits Enhanced Viral Load, Mucus, and Airway Dysfunction. J. Virol. 2009;83:4185–4194. doi: 10.1128/JVI.01853-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothay D., Ramesh K.V. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. VirusDisease. 2020;1 doi: 10.1007/s13337-020-00585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpofu, A.R., Banda, C., Gunter, H., Mondleki, E., Tatz, G., Sinxadi, P., 2020. Current evidence on chloroquine and hydroxychloroquine and their role in the treatment and prevention of COVID-19 0–22. https://doi.org/10.14293/111.000/000008.v1.
- Mukherjee, S., 2020. The first coronavirus drug candidate is set for testing in China | Fortune [WWW Document]. Fortune. URL https://fortune.com/2020/02/03/coronavirus-vaccine-testing-in-china/ (accessed 6.3.20).
- Mwanza S., Joshi S., Nambozi M., Chileshe J., Malunga P., Kabuya J.-B.B., Hachizovu S., Manyando C., Mulenga M., Laufer M. The return of chloroquine-susceptible Plasmodium falciparum malaria in Zambia. Malar. J. 2016;15:584. doi: 10.1186/s12936-016-1637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri, E.M., Piloto, B., Morinaga, L.K., Jardim, C.V.P., Lamy, S.A.E.-D., Ferreira, M.A., D’Amico, E.A., Deheinzelin, D., 2020. Heparin therapy improving hypoxia in COVID-19 patients-a case series. medRxiv. [DOI] [PMC free article] [PubMed]
- Noble P.W., Albera C., Bradford W.Z., Costabel U., Glassberg M.K., Kardatzke D., King T.E., Lancaster L., Sahn S.A., Szwarcberg J., Valeyre D., du Bois R.M. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet. 2011;377:1760–1769. doi: 10.1016/S0140-6736(11)60405-4. [DOI] [PubMed] [Google Scholar]
- Nothelfer K., Sansonetti P.J., Phalipon A. Pathogen manipulation of B cells: the best defence is a good offence. Nat. Rev. Microbiol. 2015;13:173–184. doi: 10.1038/nrmicro3415. [DOI] [PubMed] [Google Scholar]
- O’Brien T.R., Prokunina-Olsson L., Donnelly R.P. IFN-λ4: The paradoxical new member of the interferon lambda family. J. Interf. Cytokine Res. 2014;34:829–838. doi: 10.1089/jir.2013.0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguh C.E., Obiwulu E.N.O., Oniwon W.O., Okekeaji U., Ugwu C.V., Umezinwa O.J., Osuji C.A. Structure and Function of COVID-19 Encode Proteins in the Transcription and Replication Mechanism with Its Preventive Measures and Propose Efficacy Treatments: A Critical Systematic Review. Asian. J. Immunol. 2020:15–29. [Google Scholar]
- Oldfield, V., Dhillon, S., Plosker, G.L., 2009. Tocilizumab. Drugs 69, 609–632. https://doi.org/10.2165/00003495-200969050-00007. [DOI] [PubMed]
- Ota S., Hara Y., Kanoh S., Shinoda M., Kawano S., Fujikura Y., Kawana A., Shinkai M. Acute eosinophilic pneumonia caused by camostat mesilate: The first case report. Respir Med Case Rep. 2016;19:21–23. doi: 10.1016/j.rmcr.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang P., Rakus K., van Beurden S.J., Westphal A.H., Davison A.J., Gatherer D., Vanderplasschen A.F. IL-10 encoded by viruses: a remarkable example of independent acquisition of a cellular gene by viruses and its subsequent evolution in the viral genome. J. Gen. Virol. 2014;95:245–262. doi: 10.1099/vir.0.058966-0. [DOI] [PubMed] [Google Scholar]
- Paludan S.R., Bowie A.G. Immune Sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Rauf A., Khan H., Abu-Izneid T. Renin-angiotensin-aldosterone (RAAS): The ubiquitous system for homeostasis and pathologies. Biomed. Pharmacother. 2017;94:317–325. doi: 10.1016/j.biopha.2017.07.091. [DOI] [PubMed] [Google Scholar]
- Patterson, B.K., Seethamraju, H., Dhody, K., Corley, M.J., 2020. Disruption of the CCL5 / RANTES-CCR5 Pathway Restores Immune Homeostasis and Reduces Plasma Viral Load in Critical COVID-19. https://doi.org/10.1101/2020.05.02.20084673.
- Pau A.K., George J.M. Antiretroviral therapy: current drugs. Infect. Dis. Clin. 2014;28:371–402. doi: 10.1016/j.idc.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechlivanova D., Krumova E., Kostadinova N., Mitreva-Staleva J., Grozdanov P., Stoynev A. Protective effects of losartan on some type 2 diabetes mellitus-induced complications in Wistar and spontaneously hypertensive rats. Metab. Brain Dis. 2020:1–12. doi: 10.1007/s11011-020-00534-1. [DOI] [PubMed] [Google Scholar]
- Perricone C., Triggianese P., Bartoloni E., Cafaro G., Bonifacio A.F., Bursi R., Perricone R., Gerli R. The anti-viral facet of anti-rheumatic drugs: Lessons from covid-19. J. Autoimmun. 2020;102468 doi: 10.1016/j.jaut.2020.102468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka, S., Krause, C.D., Walter, M.R., 2004. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. https://doi.org/10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed]
- Pharmaceutical Technology, 2020. Coronavirus: Vir Biotechnology and Novavax announce vaccine plans [WWW Document]. URL https://www.pharmaceutical-technology.com/news/coronavirus-vir-biotechnology-novavax-vaccine/ (accessed 6.3.20).
- Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D., Frey N., Wood P.A., Weng Y., Grupp S.A. Grading of cytokine release syndrome associated with the CAR T cell therapy tisagenlecleucel. J. Hematol. Oncol. 2018;11:1–12. doi: 10.1186/s13045-018-0571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, H., Tyler, K.L., Boyd, T.D., Clarke, P., Pelak, V.S., 2020. Recruiting the innate immune system with GM-CSF to fight viral diseases , including West Nile Virus encephalitis and COVID-19 [ version 1 ; peer review : awaiting peer review ] 1–8. https://doi.org/10.12688/f1000research.23729.1. [DOI] [PMC free article] [PubMed]
- Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preudhomme, C., Guilhot, J., Nicolini, E., Guerci-Bresler, A., Rigal-Huguet, F., Maloisel, F., Coiteux, V., Gardembas, M., Berthou, C., Vekhoff, A., Rea, D., Jourdan, E., Allard, C., Delmer, A., Rousselot, P., Legros, L., Berger, M., Etienne, G., Roche-Lestienne, C., Eclache, V., Mahon, F.-X., Guilhot, F., 2010. Imatinib plus Peginterferon Alfa-2a in Chronic Myeloid Leukemia A BS T R AC T, N Engl J Med. [DOI] [PubMed]
- Prokunina-Olsson L., Muchmore B., Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I., Chen S., Brand N., Tarway M., Liu L., Sheikh F., Astemborski J., Bonkovsky H.L., Edlin B.R., Howell C.D., Morgan T.R., Thomas D.L., Rehermann B., Donnelly R.P., O’Brien T.R. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promsote W., DeMouth M.E., Almasri C.G., Pegu A. Anti-HIV-1 Antibodies: An Update. BioDrugs. 2020;34:121–132. doi: 10.1007/s40259-020-00413-2. [DOI] [PubMed] [Google Scholar]
- Pshenichnaya N., Bulgakova V., Selkova E., Maleyev V., Lvov N., Leneva I., Grekova A., Shestakova I. Umifenovir in treatment of influenza and acute respiratory viral infections in outpatient care. Int. J. Infect. Dis. 2019;79:103. [Google Scholar]
- Rai V., Agrawal D.K. The role of damage- and pathogen-associated molecular patterns in inflammation-mediated vulnerability of atherosclerotic plaques. Can. J. Physiol. Pharmacol. 2017;95:1245–1253. doi: 10.1139/cjpp-2016-0664. [DOI] [PubMed] [Google Scholar]
- Raman D., Sobolik-Delmaire T., Richmond A. Chemokines in health and disease. Exp Cell Res. 2011;317:575–589. doi: 10.1016/j.yexcr.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G.S., Ali A., Nalam M.N., Anjum S.G., Cao H., Nathans R.S., Schiffer C.A., Rana T.M. Design and synthesis of HIV-1 protease inhibitors incorporating oxazolidinones as P2/P2’ ligands in pseudosymmetric dipeptide isosteres. J. Med. Chem. 2007;50:4316–4328. doi: 10.1021/jm070284z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020 doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, L., MacDonald, J.K., McDonald, J.W., Chande, N., 2011. Sargramostim (GM-CSF) for induction of remission in Crohn’s disease. Cochrane Database Syst. Rev. https://doi.org/10.1002/14651858.cd008538.pub2. [DOI] [PubMed]
- Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;102433 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., Hertzog P.J. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2012;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saif, L.J., Wang, Q., Vlasova, A.N., Jung, K., Xiao, S., 2019. Coronaviruses. In: Zimmerman, J.J., Karriker, L.A., Ramirez, A., Schwartz, K.J., Stevenson, G.W., Zhang, J. (Eds.), Diseases of Swine. Wiley, pp. 488–523. https://doi.org/10.1002/9781119350927.ch31.
- Saka H.A., Valdivia R. Emerging Roles for Lipid Droplets in Immunity and Host-Pathogen Interactions. Annu. Rev. Cell Dev. Biol. 2012;28:411–437. doi: 10.1146/annurev-cellbio-092910-153958. [DOI] [PubMed] [Google Scholar]
- Sallard E., Lescure F., Yazdanpanah Y., Mentre F., Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antiviral Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandborn W.J., Su C., Sands B.E., D’Haens G.R., Vermeire S., Schreiber S., Danese S., Feagan B.G., Reinisch W., Niezychowski W., Friedman G., Lawendy N., Yu D., Woodworth D., Mukherjee A., Zhang H., Healey P., Panés J. Tofacitinib as Induction and Maintenance Therapy for Ulcerative Colitis. N. Engl. J. Med. 2017;376:1723–1736. doi: 10.1056/NEJMoa1606910. [DOI] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Sarma J.V., Ward P.A. The complement system. Cell Tissue Res. 2011;343:227–235. doi: 10.1007/s00441-010-1034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax P.E., Pozniak A., Montes M.L., Koenig E., DeJesus E., Stellbrink H.-J., Antinori A., Workowski K., Slim J., Reynes J. Coformulated bictegravir, emtricitabine, and tenofovir alafenamide versus dolutegravir with emtricitabine and tenofovir alafenamide, for initial treatment of HIV-1 infection (GS-US-380–1490): a randomised, double-blind, multicentre, phase 3, non-inferiori. Lancet. 2017;390:2073–2082. doi: 10.1016/S0140-6736(17)32340-1. [DOI] [PubMed] [Google Scholar]
- Scanzello C.R. Chemokines and inflammation in osteoarthritis: Insights from patients and animal models. J. Orthop. Res. 2017;35:735–739. doi: 10.1002/jor.23471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schett G., Dayer J.-M., Manger B. Interleukin-1 function and role in rheumatic disease. Nat. Rev. Rheumatol. 2016;12:14–24. doi: 10.1038/nrrheum.2016.166. [DOI] [PubMed] [Google Scholar]
- Schoenborn, J.R., Wilson, C.B., 2007. Regulation of Interferon-γ During Innate and Adaptive Immune Responses. Adv. Immunol. https://doi.org/10.1016/S0065-2776(07)96002-2. [DOI] [PubMed]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Ix J.H., Mathew A.V., Cho M., Pflueger A., Dunn S.R., Francos B., Sharma S., Falkner B., McGowan T.A., Donohue M., RamachandraRao S., Xu R., Fervenza F.C., Kopp J.B. Pirfenidone for Diabetic Nephropathy. J. Am. Soc. Nephrol. 2011;22:1144–1151. doi: 10.1681/ASN.2010101049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-S., Qi H.-Y., Boularan C., Huang N.-N., Abu-Asab M., Shelhamer J.H., Kehrl J.H. SARS-Coronavirus Open Reading Frame-9b Suppresses Innate Immunity by Targeting Mitochondria and the MAVS/TRAF3/TRAF6 Signalosome. J. Immunol. 2014;193:3080–3089. doi: 10.4049/jimmunol.1303196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva A.L., Peres C., Conniot J., Matos A.I., Moura L., Carreira B., Sainz V., Scomparin A., Satchi-Fainaro R., Préat V., Florindo H.F. Nanoparticle impact on innate immune cell pattern-recognition receptors and inflammasomes activation. Semin. Immunol. 2017;34:3–24. doi: 10.1016/j.smim.2017.09.003. [DOI] [PubMed] [Google Scholar]
- Slobedman B., Barry P.A., Spencer J.V., Avdic S., Abendroth A. Virus-Encoded Homologs of Cellular Interleukin-10 and Their Control of Host Immune Function. J. Virol. 2009;83:9618–9629. doi: 10.1128/jvi.01098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloka J.S., Stefanelli M. The mechanism of action of methylprednisolone in the treatment of multiple sclerosis. Mult. Scler. J. 2005;11:425–432. doi: 10.1191/1352458505ms1190oa. [DOI] [PubMed] [Google Scholar]
- Smith E.C., Denison M.R. Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity. PLoS Pathog. 2013;9:e1003760. doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J., 2020. CureVac Bids to Develop First mRNA Coronavirus Vaccine [WWW Document]. Labiotech.eu. URL https://www.labiotech.eu/medical/curevac-coronavirus-outbreak-cepi/ (accessed 6.3.20).
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J.M., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and Origin of Membrane Vesicles Associated with the Severe Acute Respiratory Syndrome Coronavirus Replication Complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokos G.G., Raina A. Understanding the early mortality benefit observed in the PARADIGM-HF trial: considerations for the management of heart failure with sacubitril/valsartan. Vasc. Health Risk Manag. 2020;16:41. doi: 10.2147/VHRM.S197291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonawane, K.D., Barale, S.S., Dhanavade, M.J., Waghmare, S.R., Nadaf, N.H., Kamble, S.A., Mohammed, A.A., Makandar, A.M., Fandilolu, P.M., Unit, S.B., 2020. Homology modeling and docking studies of TMPRSS2 with experimentally known inhibitors Camostat mesylate, Nafamostat and Bromhexine hydrochloride to control SARS-Coronavirus-2. ChemRxiv. https://doi.org/10.26434/chemrxiv.12162360.v1. [DOI] [PMC free article] [PubMed]
- Stark G.R., Darnell J.E. The JAK-STAT Pathway at Twenty. Immunity. 2012;36:503–514. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone K.D., Prussin C., Metcalfe D.D. IgE, mast cells, basophils, and eosinophils. J. Allergy Clin. Immunol. 2010;125:S73–S80. doi: 10.1016/j.jaci.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020:1–7. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam G., Perry M.R., Ward S., Brett S.J., Castello-Cortes A., Brunner M.D., Panoskaltsis N. Cytokine Storm in a Phase 1 Trial of the Anti-CD28 Monoclonal Antibody TGN1412. N. Engl. J. Med. 2006;355:1018–1028. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]
- Szekely Y., Lichter Y., Shrkihe B.A., Bruck H., Oster H.S., Viskin S. Chloroquine-induced torsades de pointes in a patient with coronavirus disease 2019. Hear. Rhythm. 2020;395:1315. doi: 10.1016/j.hrthm.2020.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taefehnorooz H., Truchetet F., Barbaud A., Schmutz J.L., Bursztejn A.C. Efficacy of thalidomide in the treatment of prurigo nodularis. Acta Derm. Venereol. 2011;91:344–345. doi: 10.2340/00015555-0997. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Narazaki M., Kishimoto T. Interleukin (IL-6) Immunotherapy. Cold Spring Harb. Perspect. Biol. 2018;10:a028456. doi: 10.1101/cshperspect.a028456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.C., Keystone E.C., van der Heijde D., Weinblatt M.E., del Carmen Morales L., Reyes Gonzaga J., Yakushin S., Ishii T., Emoto K., Beattie S., Arora V., Gaich C., Rooney T., Schlichting D., Macias W.L., de Bono S., Tanaka Y. Baricitinib versus Placebo or Adalimumab in Rheumatoid Arthritis. N. Engl. J. Med. 2017;376:652–662. doi: 10.1056/NEJMoa1608345. [DOI] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- Thiel V., Weber F. Interferon and cytokine responses to SARS-coronavirus infection. Cytokine Growth Factor Rev. 2008;19:121–132. doi: 10.1016/j.cytogfr.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigabu B.M., Agide F.D., Mohraz M., Nikfar S. Atazanavir/ritonavir versus Lopinavir/ritonavir-based combined antiretroviral therapy (cART) for HIV-1 infection: a systematic review and meta-analysis. Afr. Health Sci. 2020;20:91–101. doi: 10.4314/ahs.v20i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiku V., Tan M.-W., Dikic I. Mitochondrial Functions in Infection and Immunity. Trends Cell Biol. 2020;30:263–275. doi: 10.1016/j.tcb.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisoncik J.R., Korth M.J., Simmons C.P., Farrar J., Martin T.R., Katze M.G. Into the Eye of the Cytokine Storm. Microbiol. Mol. Biol. Rev. 2012;76:16–32. doi: 10.1128/mmbr.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turvey S.E., Broide D.H. Innate immunity. J. Allergy Clin. Immunol. 2010;125:S24–S32. doi: 10.1016/j.jaci.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafaei, S., Razmi, M., Mansoori, M., Asadi-Lari, M., Madjd, Z., 2019. Spotlight of Remdesivir in Comparison with Ribavirin, Favipiravir, Oseltamivir and Umifenovir in Coronavirus Disease 2019 (COVID-19) Pandemic. Favipiravir, Oseltamivir Umifenovir Coronavirus Dis.
- van Doremalen N., Miazgowicz K.L., Milne-Price S., Bushmaker T., Robertson S., Scott D., Kinne J., McLellan J.S., Zhu J., Munster V.J. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J. Virol. 2014;88:9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vollenhoven R.F., Fleischmann R., Cohen S., Lee E.B., García Meijide J.A., Wagner S., Forejtova S., Zwillich S.H., Gruben D., Koncz T., Wallenstein G.V., Krishnaswami S., Bradley J.D., Wilkinson B. Tofacitinib or Adalimumab versus Placebo in Rheumatoid Arthritis. N. Engl. J. Med. 2012;367:508–519. doi: 10.1056/NEJMoa1112072. [DOI] [PubMed] [Google Scholar]
- Vankadari N. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking the trimerization of viral spike glycoprotein? Int. J. Antimicrob Agents. 2020:105998. doi: 10.1016/j.ijantimicag.2020.105998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannucchi A.M., Kiladjian J.J., Griesshammer M., Masszi T., Durrant S., Passamonti F., Harrison C.N., Pane F., Zachee P., Mesa R., He S., Jones M.M., Garrett W., Li J., Pirron U., Habr D., Verstovsek S. Ruxolitinib versus Standard Therapy for the Treatment of Polycythemia Vera. N. Engl. J. Med. 2015;372:426–435. doi: 10.1056/NEJMoa1409002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaxart, 2020. Vaxart Announces Initiation of Coronavirus Vaccine Program | Vaccines | News Channels [WWW Document]. PipelineReview.com. URL https://pipelinereview.com/index.php/2020020273689/Vaccines/Vaxart-Announces-Initiation-of-Coronavirus-Vaccine-Program.html (accessed 6.3.20).
- Vellingiri, B., Jayaramayya, K., Iyer, M., Narayanasamy, A., Govindasamy, V., Giridharan, B., Ganesan, S., Venugopal, A., Venkatesan, D., Ganesan, H., 2020. COVID-19: A promising cure for the global panic. Sci. Total Environ. 138277. [DOI] [PMC free article] [PubMed]
- Voo K.S., Peng G., Guo Z., Fu T., Li Y., Frappier L., Wang R.-F. Functional Characterization of EBV-Encoded Nuclear Antigen 1–Specific CD4 + Helper and Regulatory T Cells Elicited by In vitro Peptide Stimulation. Cancer Res. 2005;65:1577–1586. doi: 10.1158/0008-5472.CAN-04-2552. [DOI] [PubMed] [Google Scholar]
- Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int. J. Antimicrob. Agents. 2020;55:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., Siegel D., Perron M., Bannister R., Hui H.C., Larson N., Strickley R., Wells J., Stuthman K.S., Van Tongeren S.A., Garza N.L., Donnelly G., Shurtleff A.C., Retterer C.J., Gharaibeh D., Zamani R., Kenny T., Eaton B.P., Grimes E., Welch L.S., Gomba L., Wilhelmsen C.L., Nichols D.K., Nuss J.E., Nagle E.R., Kugelman J.R., Palacios G., Doerffler E., Neville S., Carra E., Clarke M.O., Zhang L., Lew W., Ross B., Wang Q., Chun K., Wolfe L., Babusis D., Park Y., Stray K.M., Trancheva I., Feng J.Y., Barauskas O., Xu Y., Wong P., Braun M.R., Flint M., McMullan L.K., Chen S.S., Fearns R., Swaminathan S., Mayers D.L., Spiropoulou C.F., Lee W.A., Nichol S.T., Cihlar T., Bavari S. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Shang J., Ma Y., Xu X., Huang Y., Guan Y., Duan Z., Zhang W., Gao Z., Zhang M., Li J., Jia J., Yang Y., Wen X., Wang M., Jia Z., Ning B., Chen Y., Qi Y., Du J., Jiang J., Tong L., Xie Y., Wu J.J. Efficacy and Safety of 12-week Interferon-based Danoprevir Regimen in Patients with Genotype 1 Chronic Hepatitis C. J. Clin. Transl. Hepatol. 2019;7:1–5. doi: 10.14218/jcth.2019.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, N.J., Watson, J.A., Hoglund, R.M., Chan, X.H.S., Cheah, P.Y., Tarning, J., 2020. COVID-19 prevention and treatment : a critical analysis of chloroquine and hydroxychloroquine clinical pharmacology 1–34. [DOI] [PMC free article] [PubMed]
- Wiertz E.J.H.J., Tortorella D., Bogyo M., Yu J., Mothes W., Jones T.R., Rapoport T.A., Ploegh H.L. Sec6l-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- Williams A.E., Chambers R.C. The mercurial nature of neutrophils: still an enigma in ARDS? Am. J. Physiol. Cell. Mol. Physiol. 2014;306:L217–L230. doi: 10.1152/ajplung.00311.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B., Cockcroft J.R., Kario K., Zappe D.H., Brunel P.C., Wang Q., Guo W. Effects of sacubitril/valsartan versus olmesartan on central hemodynamics in the elderly with systolic hypertension: the PARAMETER study. Hypertension. 2017;69:411–420. doi: 10.1161/HYPERTENSIONAHA.116.08556. [DOI] [PubMed] [Google Scholar]
- Wilson, E.B., Brooks, D.G., 2010. The Role of IL-10 in Regulating Immunity to Persistent Viral Infections, in: Assessment & Evaluation in Higher Education. pp. 39–65. https://doi.org/10.1007/82_2010_96. [DOI] [PMC free article] [PubMed]
- Wilson J.G., Liu K.D., Zhuo H., Caballero L., McMillan M., Fang X., Cosgrove K., Vojnik R., Calfee C.S., Lee J.-W., Rogers A.J., Levitt J., Wiener-Kronish J., Bajwa E.K., Leavitt A., McKenna D., Thompson B.T., Matthay M.A. Mesenchymal stem (stromal) cells for treatment of ARDS: a phase 1 clinical trial. Lancet Respir. Med. 2015;3:24–32. doi: 10.1016/S2213-2600(14)70291-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N., Bai R., Teng J.L.L., Tsang C.C.C., Wang M. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavi. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Li W., Peng G., Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc. Natl. Acad. Sci. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L., Tai Y., Bai C., Gao T., Song J., Xia P., Dong J., Zhao J., Wang F.-S. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N., Chen Z.J. Intrinsic antiviral immunity. Nat. Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S., 2020. Safety, Tolerability and Immunogenicity of INO-4800 for COVID-19 in Healthy Volunteers [WWW Document]. ClinicalTrials.gov. URL https://clinicaltrials.gov/ct2/show/study/NCT04336410 (accessed 6.3.20).
- Yang Y., Peng F., Wang R., Guan K., Jiang T., Xu G., Sun J., Chang C. The deadly coronaviruses: The 2003 SARS pandemic and the 2020 novel coronavirus epidemic in China. J. Autoimmun. 2020;109:102434. doi: 10.1016/j.jaut.2020.102434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshie, O., Matsushima, K., 2017. Chemokines and Chemotaxis. In: Inflammation - From Molecular and Cellular Mechanisms to the Clinic. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany, pp. 619–650. https://doi.org/10.1002/9783527692156.ch25.
- Yuen, K., Wong, S., 2005. Human infection by avian influenza A H5N1. [PubMed]
- Zanasi A., Mazzolini M., Kantar A. A reappraisal of the mucoactive activity and clinical efficacy of bromhexine. Multidiscip. Respir. Med. 2017;12:7. doi: 10.1186/s40248-017-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wu Z., Li J.-W., Zhao H., Wang G.-Q. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int. J. Antimicrob. Agents. 2020;105954 doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020:1–5. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zhu L., Cai J., Lei F., Qin J.-J., Xie J., Liu Y.-M., Zhao Y.-C., Huang Xuewei, Lin L., Xia M., Chen M.-M., Cheng X., Zhang X., Guo D., Peng Y., Ji Y.-X., Chen J., She Z.-G., Wang Y., Xu Q., Tan R., Wang H., Lin J., Luo P., Fu S., Cai H., Ye P., Xiao B., Mao W., Liu L., Yan Y., Liu M., Chen M., Zhang X.-J., Wang X., Touyz R.M., Xia J., Zhang B.-H., Huang Xiaodong, Yuan Y., Rohit L., Liu P.P., Li H. Association of Inpatient Use of Angiotensin-Converting Enzyme Inhibitors and Angiotensin II Receptor Blockers With Mortality Among Patients With Hypertension Hospitalized With COVID-19. Circ. Res. 2020;126:1671–1681. doi: 10.1161/CIRCRESAHA.120.317134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, D., Dai, S.-M., Tong, Q., 2020. COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J. Antimicrob. Chemother. [DOI] [PMC free article] [PubMed]