Abstract
Viral infectious diseases have resulted in millions of deaths throughout history and have created a significant public healthcare burden. Tremendous efforts have been placed by the scientific communities, health officials and government organizations to detect, treat, and prevent viral infection. However, the complicated life cycle and rapid genetic mutations of viruses demand continuous development of novel medicines with high efficacy and safety profiles. Peptides provide a promising outlook as a tool to combat the spread and re-emergence of viral infection. This article provides an overview of five viral infectious diseases with high global prevalence: influenza, chronic hepatitis B, acquired immunodeficiency syndrome, severe acute respiratory syndrome, and coronavirus disease 2019. The current and potential peptide-based therapies, vaccines, and diagnostics for each disease are discussed.
Abbreviations: Flu, influenza; HBV, hepatitis B virus; CHB, chronic hepatitis B; AIDS, acquired immunodeficiency syndrome; HIV, human immunodeficiency virus; SARS, severe acute respiratory syndrome; SARS-CoV-1, severe acute respiratory syndrome coronavirus 1; COVID-19, coronavirus disease 2019; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; AMP, antimicrobial peptides; bnAbs, broadly naturalizing antibodies; HA, Hemagglutinin; NA, Neuraminidase; M2, matrix-2; FDA, U.S. Food and Drug Administration; MDCK, Madin-Darby Canine Kidney cells; PLpro, Papain-like protease; 3CLpro, 3C-like protease; Mpro, main protease; S, Spike protein; E, Envelope protein; M, Membrane glycoprotein; N, Nucleocapsid protein; HCV, hepatitis C virus; RSV, Respiratory syncytial viral, ACE2, Angiotensin-converting enzyme; CPE, Cytopathic effect; MOE, Molecular Operating Environment; CSSE, Center for Systems Science and Engineering; RBD, Receptor binding domain; PPI, Protein-protein interaction; PEG, Polyethylene glycol (PEG); NTCP, Sodium taurocholate cotransporting polypeptide; Env, Envelope; HR2, Heptad repeat 2; 6HB, six-helical bundle; ECL2, extracellular loop; NHR, N-terminal heptad repeat; CHR, C-terminal heptad repeat; WHO, World Health Organization; PCR, polymerase chain reaction; qRT-PCR, quantitative real-time PCR
Keywords: Peptides, Influenza, Chronic hepatitis B, Acquired immunodeficiency syndrome, Severe acute respiratory syndrome, Coronavirus disease 2019
1. Introduction
Viral infectious diseases have had a catastrophic impact in the past century. The “Spanish flu” pandemic of 1918, caused by the H1N1 influenza A virus, claimed the lives of 50 million people. The acquired immunodeficiency syndrome (AIDS) pandemic was started in the early 1980s, yet over 37 million people are still living with the disease in 2020 [200]. Health organizations have put together countless initiatives, including treatment, prevention, and detection strategies to combat the spread and re-emergence of viral infectious diseases [1]. Despite these efforts, fighting viral diseases has remained a formidable task. Presently, the coronavirus disease-2019 (COVID-19) pandemic has impacted nearly every continent within a few months, infecting millions of people, and causing hundreds of thousands of mortalities [2].
Viruses propagate only inside the living cell and their lifecycles include five stages: attachment to the host cell, fusion/cell entry, replication inside the host cell, assembly, and release from the host cell [131]. A significant number of antiviral agents have been developed to target host cell receptors and/or stages of the viral lifecycle. These antiviral agents can be divided into six classes, including nucleoside and non-nucleoside inhibitors, integrase inhibitors, protease inhibitors, fusion inhibitors, and coreceptor antagonists [131]. While current drugs have shown to be effective in reducing morbidity and mortality associated with the viral infection, the rapid mutation rate in the viral genome enables viruses to quickly adapt and develop drug resistance [98]. This challenge emphasizes the need for the discovery and development of novel molecules, such as peptide-based agents.
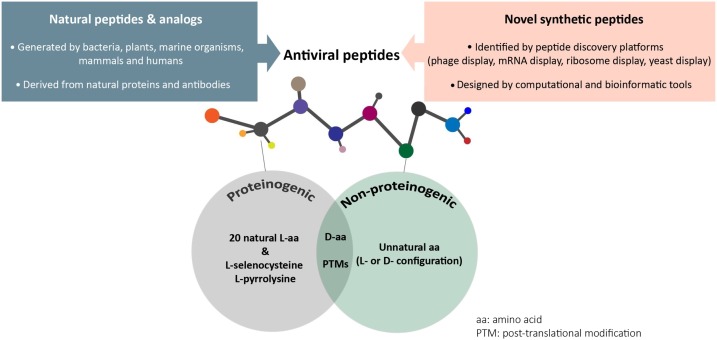
Peptides can be divided into two groups: natural and synthetic. Natural peptides, such as antimicrobial peptides (AMPs), have been shown to present antiviral and immunosuppressant activity and are reviewed extensively [134,41]. Other examples of natural peptides include hormones, neurotransmitters, and immunomodulators [226,194]. Alternatively, synthetic peptides are de novo peptides that are conceptualized by rational designs or discovered by the use of in vitro peptide discovery platforms such as phage and mRNA display [77,218,44]. Both natural and synthetic peptides can contain proteinogenic and non-proteinogenic amino acids as building blocks, which can improve the pharmacokinetic properties and enhance their efficacy and safety profiles [77,204] (Fig. 1 ).
Fig. 1.
Schematic of the natural and synthetic peptides. Natural and synthetic peptides can contain both proteinogenic and non-proteinogenic amino acids to achieve antiviral function.
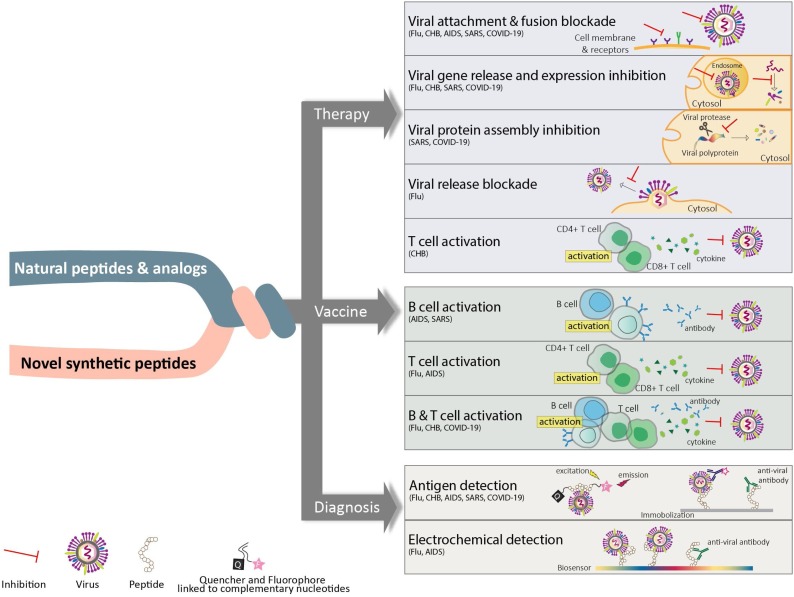
Natural or synthetic peptides have drawn great attention in a variety of applications as therapeutics, vaccines, and diagnostic agents. Due to their high specificity and ability to access hard to reach targets, peptides as therapeutics can offer the combined advantages of small and large molecules [139]. As vaccines, peptides are popular modalities for inducing anti-viral immune response driven by B and/or T cells. Peptide optimization such as multimerization and fusion to immunostimulatory adjuvants can be used to further enhance the immune response [140,107,135]. Moreover, peptides are employed in detection assays to diagnose various infectious diseases. Peptide-based diagnostic reagents are generally preferred due to their specificity, safety, suitability, and flexibility [107]. In this article, we provide an overview of the five infectious diseases; influenza (flu), chronic hepatitis B (CHB), AIDS, severe acute respiratory syndrome (SARS), and COVID-19. The current FDA approved therapeutics are reviewed for each disease and the applications of novel peptide-based agents are summarized. We highlight the discovery and development of peptide-based candidates under various development stages (pre-clinical and clinical) and their potentials as therapies, vaccines, and diagnostics. A summary of the strategies discussed in this article is depicted in Fig. 2 .
Fig. 2.
Schematic of peptide applications in targeting viral infectious diseases. Utilization of peptides as therapeutics, vaccines, or diagnostic reagents to combat viral diseases is illustrated here.
2. Influenza
Influenza (flu), a highly contagious respiratory disease caused by influenza virus, is classified into three types: influenza A, B, and C. Influenza types A and B are the major causes of the seasonal epidemics [92] and caused 36 million illnesses during the 2018–2019 flu season [30]. Two viral capsid proteins, Hemagglutinin (HA) and Neuraminidase (NA) play key roles during viral infection [82,187]. Based on the different combinations of HA and NA proteins, influenza A and B viruses are further classified into different groups and subtypes [31]. HA is presented on the viral surface as a homotrimer composed of HA1 and HA2 subunits linked by a disulfide bond. The infection event is triggered by HA1 binding to sialylated receptors on the host cell surface. Upon binding, the virus enters the host cell by endocytosis. The acidic environment of the endosome induces conformational changes in HA2, resulting in exposure of the fusion initiation region (FIR). This allows FIR to interact with the host endosomal membrane. This is followed by the interaction of matrix-2 (M2) ion channel with the viral envelope and release of viral ribonucleoproteins (RNPs) and genome into the host cell cytosol. The viral RNA is then replicated, translated, and assembled into viral particles. Virus particles then leave the host cells by budding out when NA cleaves sialic acid to enable viral release [8]. Due to their critical functions, viral HA, NA, and M2 proteins have been the focus of influenza therapies.
FDA approved influenza therapeutics targeting M2 and NA proteins shorten the disease time-course and reduce the severity of symptoms. Adamantane derivatives, amantadine (Symmetrel) and rimantadine (Flumadine), are small molecule inhibitors that target M2 proteins encoded by influenza A viruses. [111]. Orally delivered oseltamivir (Tamiflu), intranasally administered zanamivir (Relenza), and intravenously injected peramivir (Rapivab) are FDA approved NA inhibitors that prevent the virus release from infected host cells. In 2018, FDA approved Baloxavir marboxil (Xofluza) for influenza A and B. Xofluza inhibits polymerase acidic endonuclease, an enzyme essential for viral replication [112]. Favipiravir (Avigan), which also blocks viral replication is approved for influenza in Japan [67] and it is currently in clinical trials for the treatment of COVID-19. It is important to note that the use of adamantane is no longer recommended due to its poor tolerability and a high rate of drug resistance [151]. In addition, resistant viral mutants have shown decreased sensitivity to oseltamivir [28]. The emergence of resistant influenza viruses mandates the discovery of novel therapeutics with high specificity and efficacy. The developments of antiviral peptides for the treatment of influenza is discussed next.
Naturally occurring AMPs, such as defensins and cathelicidins, provide great starting scaffolds for drug development. Mouse β-defensin-4 (mBD4) has been shown to have antiviral activity against influenza A virus (H5N1 and H1N1). Truncations of mBD4 identified a 30-residue long peptide P9 that interacted with HA at high affinity. Pretreatment of P9 with the virus was shown to alter endosomal acidification and inhibit viral RNA release and replication. Potent antiviral activities of P9 against influenza viruses (H1N1, H3N2, H5N1, H7N7, H7N9) were reported in MDCK cells and mice [231]. θ-defensin, another member of defensins, is found in non-human primates. Synthetic θ-defensin, called retrocyclin neutralizes influenza virus by inducing its aggregation and enhancing its uptake by neutrophils [93]. Two families of cyclic peptides, Hapiviren and Diprovin, were designed based on retrocyclin and were shown to have neutralizing activity against H1N1 and H3N2 viruses [55]. Cathelicidin, another class of naturally occurring AMPs, can disrupt the viral envelope to achieve potent antiviral activity. Tripathi and colleagues have shown that the helical fragment of LL-37, a human cathelicidin, is essential for its antiviral activity. Helical fragment of LL-37 showed greater activity in viral reduction with H1N1 strain (IC50 3.2 μM) than the full-length LL-37 (IC50 11.3 μM) [197].
In addition to the naturally occurring peptides that are derived from the innate immune system, some proteins have shown potential as the starting point for the discovery and development of anti-viral peptides. Four examples will be discussed next. First, lactoferrin, found in bovine milk was shown to bind to HA and inhibit the infection of influenza subtypes H1N1 and H3N2. By molecular docking, three fragments on lactoferrin, SKHSSLDCVLRP (418–429), AGDDQGLDKCVPNSKEK (506–522), and NGESSADWAKN (552–563) were identified with the inhibitory activity against influenza virus at picomolar and femtomolar concentrations [7]. Scala and colleagues identified minimum pharmacophore of the most potent peptide (SKHSSLDCVLRP) and generated three tetrapeptides 14 (VLRP), 15 (SLDC) and 17 (SKHS) with broader anti-influenza activity. Tetrapeptide 17 was more active than the parental peptide in the neutralization assay in MDCK cells [184]. The second example, a 20-residue peptide EB derived from the signal sequence of fibroblast growth factor 4, was reported to exhibit antiviral activity against influenza viruses, including the H5N1 subtype. EB (RRKKAAVALLPAVLLALLAP) bound specifically to HA, prevented viral cell entry in the MDCK cell line, and had protective activity in mice at low micromolar concentration [106]. In the third example, a suppressor of cytokine signaling protein called tyrosine kinase inhibitor peptide was used to develop a family of 12–16 residue long anti-viral peptides termed FluPep (FP) for targeting influenza. FluPep inhibited HA binding to host cells and cell entry. One of the FP peptides (FP4: RRKKWLVFFVIFYFFR) showed nanomolar efficacy for Influenza A virus subtypes H1, H3, and H5 in a plaque-reduction assay. To compensate for the high hydrophobic content of FP peptides [149] and to enhance their solubility, Alghrair and team constructed gold and silver nanoparticle–FP conjugates. Interestingly, the peptide-nanoparticle conjugates showed enhanced solubility and increased activity in plaque assay using MDCK cells. The authors suggested that the use of FluPep-functionalized nanoparticles might be safe for delivery to target organs [5]. The last example includes the NA inhibitory peptide (PGEKGPSGEAGTAGPPGTPGPQGL) derived from cod skin hydrolysates. The peptide was reported to have considerable anti-viral activity and hence potential utility in preventing influenza virus infections. However, the peptide was shown to be highly susceptible to degradation in the presence of the simulated in vitro gastrointestinal fluid, suggesting that oral administration was not feasible [118].
The broadly neutralizing antibodies (bnAbs) targeting HA protein have also been utilized to generate peptides with improved potency. A linear peptide, P1 (SQLRSLEYFEWLSQ) was designed based on the complementarity-determining region loops of the heavy chain and framework region of two bnAbs called FI6v3 and CR9114 [108]. P1 sequence was optimized by incorporation of unnatural amino acids 5-phenyl-norvaline1 (Nva), ornithine2 (Orn), β-Alanine11, N-methylated Leu3, and dichloro-Phe6 (cyclization between position 2 and 11). This effort resulted in the discovery of an 11-residue cyclic peptide P7 (Nva-Orn-meLEYchlFEWLS-βAla) that targeted influenza A viruses (H1N1, H5N1) with a 100-fold increase in potency (IC50 30−70 nM and KD 17–37 nM). P7 inhibited HA conformational rearrangement in the endosome, preventing viral fusion with the host cell membrane [108].
In a parallel approach, viral proteins are used to develop anti-viral peptides. For example, a 16-residue long peptide, Flufirvitide-3, derived from the fusion initiation region of the HA protein, has shown high efficacy in plaque inhibition assay and in vivo studies against influenza virus and has completed phase I trials [11]. In silico and bioinformatic approaches were utilized to design peptides based on highly conserved regions of HA1 and HA2. As a result, nine peptides were identified that bound to HA stalk regions and had anti-viral activity at micromolar concentrations against H1N1 and H5N2 strains in MTT reduction assay using MDCK cells [130]. In a separate study, a HA-binding pentapeptide (ARLPR) was grafted into different carbohydrate scaffolds called carbosilane dendrimers. The scaffolds presented the HA-binding peptide with the needed flexibility, stability, and multivalency. In particular, the dumbbell-shaped carbosilane dendrimer had the strongest inhibitory activity of 0.72 μM and 0.6 μM against H1N1 and H3N2, respectively [83].
Novel peptide discoveries are facilitated by the use of in vitro peptide selection platforms. Peptide f1 (ARLSPTMVHPNGAQP) was identified from the phage library and inhibited the interaction of sialylglyco-conjugate receptors on the host cell surface with viral HA protein. Optimized f1 was an alkylated peptide (C-18-s2(1−5) C17H35CO-ARLPR-NH) with IC50 values of 1.9 μM and 1.6 μM for H1N1 and H3N2, respectively. The alkylation was implemented to enhance inhibitory activity through multivalency. The docking simulation suggested that the peptide makes the same hydrogen bond interactions with HA as the sialic acid [137]. Recently, a novel peptide inhibitor (errKPAQP) against influenza NA protein was identified from a peptide library, comprised of L and D amino acids. The peptide bound to NA with nanomolar affinity and inhibited H1N1 infection at μM concentration in MDCK cells. Administration of the peptide in infected mice reduced virus-induced inflammation, lung tissue damage, and mortality [34]. Potential peptide therapies are summarized in Table 1 .
Table 1.
Summary of potential peptide therapies for Influenza.
| Target protein | Peptide Name | Sequence | Derived from | IC50 | Assay Format Tested | Phase | Reference |
|---|---|---|---|---|---|---|---|
| HA | LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | Cathelicidin | 0.9−11.3 μM | Neutralization assay | Pre-clinical | [197] |
| GI-20 | GIKEFKRIVQRIKDFLRNLV | AMP (LL-37) | 1.6−3.2 μM | Neutralization assay | Pre-clinical | ||
| P1, P2, P3 | SKHSSLDCVLRP (1), AGDDQGLDKCVPNSKEK (2), and NGESSADWAKN (3) | Lactoferrin | PM-FM | Neutralization assay | Pre-clinical | [7] | |
| Tetrapeptides | Peptide 14 (VLRP) | Lactoferrin | fM range | Neutralization assay | Pre-clinical | [184] | |
| Peptide 15 (SLDC) and | SKHSSLDCVLRP (1), | ||||||
| Peptide 17 (SKHS) | |||||||
| P9 | NGAICWGPCPTAFRQIGNCGHFKVRCCKIR | mouse β-defensin-4 | 1.5−4.8 μg/mL | plaque reduction assay | Pre-clinical | [231] | |
| EB | RRKKAAVALLPAVLLALLAP | fibroblast growth factor 4 | 3−10 μM | hemagglutination inhibition assay | Pre-clinical | [106] | |
| FP-4 | RRKKWLVFFVIFYFFR | Tyrosine kinase inhibitor peptide | 0.05−0.13 μM | plaque-reduction assay | Pre-clinical | [5] | |
| Flufirvitide-3 | - * | fusion initiation region | nM range | plaque inhibition assay | Phase 1 | [67] | |
| P7 | (Nva-Orn-meLEYchlFEWLS-βAla | Neutralizing antibodies | 30−70 nM | AlphaLisa competition binding assay | Pre-clinical | [108] | |
| C-18-s2(1−5) | C17H35CO-ARLPR-NH | Phage library | 1.6−1.9 μM | plaque inhibition assay | Pre-clinical | [137] | |
| NA | peptide P | PGEKGPSGEAGTAGPPGTPGPQGL | Cod skin hydrolysates | 3.5 mg/mL | NA inhibitory assay | Pre-clinical | [118] |
| P2 | errKPAQP | Binding pockets of oseltamivir in NA | 4.25 μM | NA inhibitory assay Cytopathic effect (CPE) assay | Pre-clinical | [34] |
*not available in the literature.
The administration of influenza vaccines is recommended to prevent virus infection. During 2017–2018, the influenza vaccination prevented 91,000 influenza-related hospitalizations [27]. The current influenza vaccine induces a prophylactic antibody response against the HA and NA proteins. The preventive vaccines in the market include Trivalent inactivated influenza (TRI) and quadrivalent live attenuated influenza viruses (LAIV), such as FluMist and Fluzone. New variants of the virus emerge constantly, therefore, the type of vaccine preparation must be predicted and generated before strain identification for the upcoming year. It is often challenging to predict epidemics due to the high diversity of viral subtypes [29]. Next, we summarize a few peptide-based vaccines that have reached the advanced stages of clinical trials.
In one study, conserved sequences from influenza proteins that are targeted by T cells (human and mice) were identified using a proprietary epitope prediction algorithm. The mixture of chemically synthesized peptides (FLU-v) induced a specific HLA-A*0201-mediated CD8+ T cell response in immunized transgenic mice model and significantly increased their survival rate when the animals were challenged with a lethal dose of influenza [190]. Equimolar combination of four peptides (M1: DLEALMEWLKTRPILSPLTKGILGFVFTLTVP, NPA: DLIFLARSALILRGSVAHKSC, NPB: PGIADIEDLTLLARSMVVVRP, and M2: IIGILHLILWILDRLFFKCIYRLF) were tested in clinical trials and the safety and efficacy were reported. This suggested that FLU-v-like peptides activate the T cell arm of immune response and have potential as vaccines [163,164,190]. Multimeric-001 (M-001) is another peptide vaccine that has met its primary endpoint in phase II. It consists of three repetitions of nine conserved B and T cell epitopes from influenza HA, NP, and M1 proteins expressed as a single recombinant polypeptide. M-001 immunization was shown to induce B and T cell-specific immune responses and offers protection against infection with different influenza strains, including H5N1 strain. M-001 is the first universal vaccine, which is expected to protect against existing and future seasonal and pandemic virus strains [10]. Vacc-FLU by Bionor is another universal influenza peptide vaccine in pre-clinical development. A proprietary peptide design platform was used to identify the four peptide components of Vacc-Flu from the conserved regions of M2 and NP of influenza A. Immunization by Vacc-Flu results in the induction of IFNγ-producing T cells and antibody-producing B cells, both of which can protect from severe disease symptoms upon infection [90].
The currently used influenza diagnostic tests are based on molecular assays and antigen detection [202]. FDA-licensed tests are designed to detect and differentiate influenza A and B viruses but have a limited capability to identify subtypes of influenza A viruses. Therefore, newer approaches that are cost-effective, less labor-intensive, easy to perform, and able to detect and differentiate influenza subtypes are needed. A recently developed approach is a peptide-based molecular beacon (PEP-MB) for the detection of Influenza type A using fluorescence resonance energy transfer (FRET). A HA1-specific binding peptide is conjugated to two complementary nucleotides on each end. Sulfo-succinimidyl-4-(N-maleimidomethyl) cyclohexane-1-carboxylate (sulfo-SMCC) is used to cross-link the complementary nucleotides: a Cy3-modified oligonucleotide (Oligo-Cy3) and a black hole quencher 2 (BHQ2)-modified oligonucleotide (Oligo-BHQ2). In the absence of virus particles, PEP-MB constructs form hairpin structures that result in the quenching of donor Cy3 by BHQ2. In the presence of the H1N1 virus, PEP-MB undergoes a conformational change due to the binding to viral HA. As a result, strong fluorescence is emitted and detected by the beacon [122]. In another detection method, sialic acid-mimic pentapeptide (ARLPR) identified by phage display is immobilized on boron-doped diamond electrodes using click chemistry. Electrochemical detection of HA binding to the peptide indicates the presence of the Influenza virus [137,138]. Also, ARLPR peptide dimers can be conjugated to poly(glycidyl methacrylate) coated nanoparticles to detect the agglutination of the influenza virus. This method can be used as an alternative approach to agglutination assay using red blood cells since it is more stable with an extended storage period [136].
3. Chronic hepatitis B
Chronic Hepatitis B (CHB) is caused by the hepadnavirus hepatitis B virus (HBV), which targets the liver leading to a severe medical condition. Recent global statistics from the World Health Organization (WHO) estimated that 257 million people carried the virus in 2015 [208]. HBV is an enveloped virus that is coated with a lipid bilayer and packaged with viral polymerase, relaxed circular DNA (rcDNA), and three types of antigens: surface antigen (HBsAg), e antigen (HBeAg), and core antigen (HBcAg). Upon infection, rcDNA is converted into the host-specific covalently closed circular DNA (cccDNA) by the viral polymerase, which plays a critical role in HBV replication [165,146,75].
The current FDA-approved treatment for CHB is based on two main strategies: 1) block HBV replication using nucleoside or nucleotide analogs such as tenofovir and entecavir, which suppress polymerase activity, inhibit reverse transcription of viral RNAs, and stop the synthesis of cccDNA; 2) stimulate the immune response directly using Interferon α (IFN-α) or pegylated interferon. Nucleoside or nucleotide analogs are fast-acting and induce minimal drug resistance with a 90 % relapse rate within a year after treatment is stopped. IFN-α was expected to provide a gold standard therapy due to its immunomodulating function. However, only a moderate response rate (30–40 %) was observed in the clinic [121,45,192]. Although IFN-α therapy is still being used extensively, WHO has recommends the orally administrated tenofovir or entecavir for the treatment of CHB due to their minimal drug resistance and side effects [209]. Entecavir is an off-patent drug. The two tenofovir blockbusters for CHB approved by FDA are Viread (tenofovir disoproxil fumarate) and Vemlidy (tenofovir alafenamide) by Gilead.
In preclinical research, peptides derived from natural proteins have demonstrated promising potential for treating CHB. An example is a peptide called PTD-p37 that was designed to inhibit viral regeneration. The peptide contained an N-terminal cell-penetrating domain (YGRKKRRQRRR) fused to the residues 444–480 of the heat shock protein gp96 (p37). Typically, HBV activates the gp96 promoter through the NF-κB pathway. This results in an up-regulation of gp96 expression, causing positive feedback to stimulate HBV replication in cell culture [61]. Moreover, the molecular chaperone gp96 was shown to bind to both HBV repressor p53 and p53 E3 ligase Mdm2 to down-regulate p53 levels by enhancing its ubiquitination and degradation [217,154]. Peptide p37 binds to gp96 at the N-terminal helix-loop-helix region without interfering with the C-terminal dimerization domain. The peptide interrupts the helix-helix interaction and locks gp96 in an inactive "open" conformation. As a result, Mdm2 is disassociated and p53 levels are increased [113,120,170]. PTD-p37 treatment reduced HBsAg levels and viral proliferation in vitro and in mice [170,61,125].
Peptides have also demonstrated significant potential in interrupting virus infection in preclinical research. HBsAg is composed of three types of envelope glycoproteins: large (L) protein with pre-S1, pre-S2 and S domain; middle (M) protein with pre-S2 and S domain; and Small (S) protein with S domain only. The pre-S1 domain initiates virus infection by binding to the host receptor called sodium taurocholate co-transporting polypeptide (NTCP) and the low-affinity HBV co-receptor heparan sulfate proteoglycan (HSPG) [195,148,14]. Therefore viral cell entry can be blocked by targeting host receptors, NTCP and HSPG, or viral antigen HBsAg. These approaches will be discussed next.
Novel NTCP binding macrocycles of 6–17 residues were identified from a random peptide integrated discovery screening platform. Among the top ten candidates that were selected based on unique sequences, some showed sub-micromolar IC50 for inhibiting HBV cell entry in hepatocytes, five-fold more potent than CsA in the same assay [156]. Another example is a 47-mer lipopeptide derived from pre-S1, Myrcludex-B, that binds to NTCP and inhibit HBV infection in cell culture and animal models [203,76,159]. Liposomal formulation of Myrcludex B allows for oral administration and has been investigated as a hepatitis B peptide drug in the clinic [199]. A Phase IIa clinical evaluation of Myrcludex-B in patients with HBeAg Negative CHB was completed in 2018 (NCT02881008). The clinically used immunosuppressant, Cyclosporin A (CsA), has also been shown to bind to NTCP and efficiently block HBV in cell entry [207]. In the search of additional NTCP binders, Donkers and colleagues screened Prestwick Chemical Library, comprised of 1280 approved and off-patented drugs with known safety information. The five most potent drugs were tested in the HBV infection assay using human hepatic cell line HepaRG. They are the antidiabetic insulin sensitizer Rosiglitazone, the leukotriene inhibitor zafirlukast for asthma, antihistamines, and decongestants TRIAC targeting thyroid hormone receptor, glutathione transferase inhibitor sulfasalazine for inflammation, and pain and vital dye Chicago Sky Blue 6B. All five drugs were shown to block HBV infection at similar levels of potency. Although, Donker's unique approach suggested an efficient way for developing HBV prevention medicine [53], de novo peptides that specifically target NTCP may offer broader advantages than pre-existing drugs.
The host receptor, HSPG binds to pre-S1 to initiate the viral integration to host cells. This is followed by the interaction between HBV and the high-affinity receptor NTCP for endocytosis [127,84]. Liu and colleagues designed an array of pre-S1 truncations displayed on liposomes (LPs). The cellular attachment of the truncations was tested and the fragment pre-S1 (30–42) was identified with the highest affinity to mimic the binding of HBV to HSPG. It was shown that the complex targeted liver cells and induced endocytosis. The results also offered a promising approach for the specific delivery of HBV drugs to human liver cells [171,117].
The de novo peptide inhibitors of virus pre-S1 were also investigated for the treatment of CHB. A short peptide B10 (SGSGLRNIRST) was identified by the phage peptide library selection against pre-S1 (1–60). The peptide disrupted the interaction of HBV with human hepatocyte carcinoma cells [87]. B10 was optimized based on the key motif (LRNIR) to generate a concatemer 4B10 (LRNIRLRNIRLRNIRLRNIR) with three-fold stronger affinity to pre-S1 (28–42) of HBV. Peptide 4B10 was shown to inhibit HBV infection in both primary tupaia and human hepatocytes [225,71]. In a separate study, a 12-residue random phage library was selected against pre-S1 region (21–47) resulting in the discovery of peptides with a common linear motif (WTXWW) specific for pre-S1. The common motif was used to identify novel membrane and extracellular proteins that are involved in HBV infection such as lipoprotein lipase [50].
In addition to targeting virus antigens, peptides that block virus maturation have also been considered as antiviral therapeutics. A stretch of 13 residues corresponding to the C-terminus of the pre-S1 domain (PLSPPLRNTHPQA) and amino acids corresponding to the 56–80 region of S domain (PISNHSPTSCPPTCPGYRWMCLRRF) bind the core antigen, HBcAg. It was suggested that these fragments were responsible for virion maturation during which the pre-S domain of L protein is switched from inside to outside of the virion. This topological change allows HBV to become a "binder-type particle" and recognizes the receptors on the host cells [165,206]. Three β-turn structures in pre-S1 and one in S region were identified by NMR spectroscopy as critical regions for this process [146]. Although additional work is needed, this finding provided a new strategy for designing inhibitors that might prevent the conformational change in the pre-S domain and interrupt the HBV morphogenesis.
Another strategy for treating CHB patients is using vaccine-based therapies. Due to the immune tolerance of CHB patients, vaccine-based therapies can help restore the immune response for viral elimination [142]. A pilot study of a peptide-based T cell vaccine, CY-1899, was tested on 19 patients with CHB. This vaccine is composed of a palmitic acid linked peptide with the sequence of the T cell epitope. CY-1899 failed to induce vigorous cytotoxic T lymphocyte activity or viral clearance possibly due to T-cell exhaustion in CHB patients [89]. Other clinical studies using similar approaches have also failed to induce a sufficient antibody response in CHB patients [65,129,224]. In a separate study, Dou and colleagues designed a 37-residue peptide SLP (MDIDPYKEFGATVELLSFLPSDFFPSVRDLLDTASAL) based on a conserved region in HBcAg. The peptide is comprised of three CD4+ T-cell epitopes and three CD8+ T-cell epitopes. In a healthy state, CD4+ helper T-cells enhance the immune response regulated by CD8+ T-cells. Activated CD8+ T-cells release inflammatory cytokines resulting in killing the infected host cells. In CHB patients, CD8 + T-cells are not activated properly by mitogenic stimuli. However, the study showed that human monocyte-derived dendritic cells presented fragments of SLP as antigens to activate CD8+ and CD4+ T-cells ex vivo and to induce the immune response in CHB patients [56,19,115].
Peptide-based vaccines have been developed to prevent HBV infection in healthy individuals. It should be noted that in one study, HBsAg (113–135) derived peptides, a B-cell epitope, were displayed on a novel chimeric virus-like particle carrier that was derived from the truncated roundleaf bat HBcAg. The carrier was further optimized by the introduction of one CD8+ T cell epitope and two CD4+ T cell epitopes. The resulted construct was shown to induce a specific antibody response to the HBsAg (113–135) with enhanced T cell response. In addition, the particle induced a lasting response resulting in the virus DNA clearance in mouse model, and effective antibody production in rabbits and cynomolgus monkeys [229]. A grass pollen allergy vaccine, BM32, containing a pre-S domain (pre-S1 and pre-S2) fused with allergen-derived peptides, has previously been shown to induce preS-specific immune responses. It was tested for HBV-specific immune responses and was shown to be successful in inducing IgG antibodies against HBV. BM32 also protected rabbits from HIV infection as effectively as the approved vaccine Engerix-B [43]. Immunoinformatic approaches and bioinformatic tools have been employed to develop peptide-based vaccines for hepatitis B and might hold potentials in future pre-clinical studies [37,153]. While peptides-based vaccines have not reached the market yet, some FDA approved vaccines for HBV prevention are available. These include Pediarix (GlaxoSmithKline Biologicals), VAXELIS (MSP Vaccine Company), Twinrix (GlaxoSmithKline Biologicals), Recombivax HB (Merck & Co, Inc), ENGERIX-B (GlaxoSmithKline Biologicals), and HEPLISAV-B (Dynavax Technologies Corporation).
Peptides are also used for the diagnosis of HBV. Traditionally, HBV DNA can be detected by polymerase chain reaction (PCR). HBsAg and HBcAg can be detected using immunostaining. Queiroz de Souza and colleagues designed a recombinant peptide complex (rMEHB) by assembling four conserved HBcAg epitopes (epitope 1: 61–83 aa; epitope 2: 107–118 aa; epitope 3: 128–133 aa; and epitope 4: 134–145 aa). rMEHB exhibited high sensitivity as a detection agent in ELISA by presenting multiple epitopes for binding to the anti-hepatitis B core antibody [48].
4. Acquired immunodeficiency syndrome
Acquired Immunodeficiency Syndrome (AIDS) is a disease caused by human immunodeficiency virus (HIV). HIV can directly weaken the immune system and make it vulnerable to other pathogens. The viral envelope (Env) protein is a key driver of the viral binding and fusion to the target cells and contains two major non-covalently associated glycoproteins, surface subunit gp120 and transmembrane gp41. These glycoproteins surround the nucleocapsid containing viral RNA and reverse transcriptase. The binding of viral gp120 to the CD4 receptor exposes its binding site to the chemokine coreceptor. Depending on the cell type, different chemokine receptors are recruited, for example, CXCR4 on T-cells and CCR5 on macrophages are identified as CD4 coreceptors. After binding of the virus to the host cells, gp41 undergoes a receptor-activated conformational change to form a six-helical bundle (6HB). The 6HB initiates viral entry by triggering the insertion of the fusion peptide (hydrophobic N-terminal fragment of gp41) into the host cell membrane [220,16,32]. Ultimately, the viral and host membrane fuse together followed by the insertion of viral genetic material in the cytosol. The viral RNA is transcribed to dsDNA. Integrase then carries dsDNA through the nucleopore to the nucleus and inserts it into the host chromosome causing lifelong infection. The viral DNA is replicated, translated, and assembled at the cell membrane as an immature polypeptide chain, which is cleaved by the viral proteases to form mature infectious virion [66].
Most of the FDA approved anti-HIV drugs target viral reverse transcription factor, proteases, integrases, and structural proteins to stop the fusion, entry, replication, or maturation of the virus. Since the first approved HIV drug azidothymidine in 1987, a reverse transcriptase inhibitor, more than 30 antivirals for AIDS have been approved [33]. Currently, the main antiviral therapies are small molecules and only a few are protein, peptide, or oligonucleotide-based therapies. In 2003, the FDA approved the first peptide-based fusion inhibitor, Enfuvirtide, a 36 residue long peptide designed to mimic the C-terminal heptad repeat (CHR) of the helical region of gp41 [102]. In 2018, Albuvirtide, a peptide fusion inhibitor, was approved in China [222]. Within the past decade, the development of peptide-based fusion/entry inhibitors is centered around targeting gp120 variable loops and gp41 functional regions [227,169]. Some of these efforts are summarized in Table 2 .
Table 2.
Peptide-based fusion inhibitors targeting gp41 and gp120.
| Peptide Name | Sequence | Derived from | Target | IC50 (nM) | Assay Format Tested | Phase | Reference |
|---|---|---|---|---|---|---|---|
| pV2α-Tys | KVQKEY(SO3H)ALFY(SO3H)-ELDIVPID | CCR5 N-terminus | Gp120 variable loops | 50,000 | CCR5-binding assay | Pre-clinical | [40] |
| pCCR5-Tys | MDYQVSSPIY(SO3H)DIANY(SO3H)YTSEPSQK | CCR5 N-terminus | Gp120 variable loops | 50,000 | CCR5-binding assay | Pre-clinical | [40] |
| E1P47 | WILEYLWKVPFDFWRGVI | GB virus C E1 protein | gp41 FP | 8200 | Competitive ELISA, fluorescence resonance energy transfer, haemolysis assays | Pre-clinical | [72] |
| SC29EK | WEEWDKKIEEYTKKIEELIKKSEEQQKKN | gp41 | Gp41 NHR | 9.6 | multinuclear activation of galactosidase indicator (MAGI) assays | Pre-clinical | [147] |
| FB006M | Ac-WEEWDREINNYTK (MPA)LIHELIEESQNQQEKNEQELL-CONH2 | Gp41 CHR | Gp41 NHR | 2.7 | peripheral blood mononuclear cell (PBMC) assay, ELISA | Approved | [222] |
| CP32M | VEWNEMTWMEWEREIENYTKLIYKILEESQEQ | gp41 CHR | Gp41 NHR | 65 | Native Polyacrylamide Gel Electrophoresis (N-PAGE) Assay, Cell–Cell Fusion Assay | Pre-clinical | [85] |
| HP23 | EMTWEEWEKK IEEYTKKIEEILK | Gp41 CHR | Gp41 NHR | 4.7 | Cell-cell fusion assay, Pierce firefly luciferase glow assay | Pre-clinical | [223] |
| AP3 | KKISEEQKKIQEEIKKILEESKKILEEIKKDWEEWTM | artificial peptide | Gp41 NHR | 13∼90 | ELISA | Pre-clinical | [234] |
| N36Fd | SGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILGYIPEAPRDGQAYVRKDGEWVLLSTFL | Gp41 NHR | Gp41 CHR | 99 | Cell-Cell Fusion, ELISA, luciferase assay | Pre-clinical | [100] |
| N28Fd | IEAQQHLLQLTVEGIKQLQARILAVERYGYIPEAPRDGQAYVRKDGEWVLISTFL | Gp41 NHR | Gp41 CHR | 39 | Cell-Cell Fusion, ELISA, luciferase assay | Pre-clinical | [100] |
A few natural peptides act as the first line of defense to protect against HIV infection. AMPs from bacteria, plants, animals, and humans have been extensively reviewed for their anti-HIV activities [205,41,144] and some have been considered as potential microbicide candidates [158]. Microbicides are a preventive option used to stop the transmission of HIV through sexual intercourse in women. As mentioned earlier, retrocyclins are natural θ-defensins produced by primates. Retrocyclins have been shown to be capable of binding to both HIV-1 and host cell glycoproteins to prevent HIV entry and have been considered as microbicides [158]. The only human cathelicidin-derived AMP, LL-37, has also shown anti-HIV activity. Three peptides, LL-37 and its two fragments LL13−37 and LL17−32, inhibit HIV-1 protease with weak potency with no reports of toxic effects [213].
Peptides that bind to gp41 have emerged as potential treatments for AIDS. An example is DP-178 (T-20), which binds to CHR and prevents the formation of 6HB. As a result, viral fusion to host cells is inhibited [104,211]. This peptide is currently marketed under the name Enfuvirtide [116]. Despite the safety profile and efficacy of Enfuvirtide, its poor stability in serum and large-dosage requirement have highlighted the need for other peptide-based drugs. Sifuvirtide (SFT), a second-generation HIV-1 gp41 fusion peptide developed by FusoGen Pharmaceutical, Inc. is currently in phase III clinical trials in China. SFT is an optimized version of Enfuvirtide with 10-fold enhanced efficacy and prolonged half-life [141]. Other advantages of SFT include stability and higher selectivity toward rigid membrane models, such as saturated dipalmitoylphosphatidylcholine (DPPC) and sphingomyelin (SM). The ability of SFT to strongly interact with rigid lipidic areas on the fusion site possibly contributes to its improved efficacy [86,26]. Another example of a peptide that interacts with gp41 is a PIE12 (Pocket-specific Inhibitor of Entry). PIE12 is a trimerized D-peptide identified using phage display and has a picomolar binding affinity to trimeric gp41 and a potency in the mid picomolar to low nanomolar range. Further, the half-life of PIE12-trimer was improved three-fold by cholesterol conjugation making the peptide less vulnerable to renal filtration in rat models. The optimized pegylated D-peptide (Cholesterol-PIE12-trimer-PEG31) with its sustained-release formulation has provided feasible dosing over an extended period in a non-human primate model [173]. Another example is the P3 peptide, which targets HIV-1 gp41 HR1 site. P3 is a 34 amino acid peptide derived from HIV-2 helical region 2 of gp41that blocks HIV infection at a sub-nanomolar concentration in both HIV-1 and HIV-2 primary isolates [21]. It was suggested that the alpha-helical structure of P3 provides resistance to proteases compared to the unstructured peptides like T-20. Due to the high stability in the human vaginal environment (pH 3.5–4.5) and the ability to retain its bioactivity at 25°, 37°, and 65 °C for a weeklong period, P3 can be a potential candidate for the development of microbicides [15]. A novel lipopeptide, LP-19 that is derived from the integration of multiple design approaches, targets the conserved pocket region of gp41 and prevents viral fusion to host cells. LP-19 with a membrane-anchoring lipid tail showed high affinity and potency in vitro for inhibiting HIV-1 and HIV-2 entry. LP-19 exhibited extended half-life in rhesus macaques and drastically reduced viral loads in infected monkeys [38]. Peptides derived from viral sequences, including gp41 can be potentially immunogenic. However, it is possible to lower the immunogenicity risk through peptide engineering. The novel HIV fusion inhibitor, V2o, is derived from the gp41 heptad repeat 2 region of HIV-2 and HIV-1. The removal of MHC-I epitopes from the naturally derived peptide reduced its immunogenicity and improved its antiviral activity profile. V2o efficiently inhibited the viral entry in PM-1 cells infected with lentiviral vector particles with the IC50 in the picomolar range [22]. It is worth mentioning that peptides derived from host cells that are involved in viral infection have also been considered. An example is peptide 2C which is derived from the C-terminal portion of the extracellular loop of CCR5. C2 competes with CCR5 for binding to gp120 [54] and inhibits HIV-1 entry at a double-digit micromolar concentration in vitro [52,18].
Despite over 30 years of effort, there is no available vaccine to prevent HIV infection and progression. Most vaccines that have been developed have failed to induce a protective immune response against HIV-1 infection in clinical trials. Eliciting neutralizing antibodies using gp120 as an immunogen is a less than ideal strategy for developing HIV vaccines. The VAX003 and VAX004 trials conducted by VaxGen Inc. determined the safety and efficacy of two bivalent subtypes B and E recombinant gp120 proteins, AIDSVAX(B/B) and AIDSVAX(B/E), respectively. The trials failed during phase III due to insufficient antibody induction and inability to maintain immune response after subsequent boosts [162,64,12]. The RV144 trial (Thia trial) that combined two failed vaccines, recombinant Canarypox vector ALVAC-HIV (vCP1521) and AIDSVAX (B/E), failed to show sufficient efficacy for approval by the Thailand Ministry of Health [174]. In 2019, the National Institutes of Health announced phase III clinical trials of Imbokodo (HVTN705/HPX2008) in women in Southern Africa and Mosaico (HVTN706/HPX3002) in men and transgender persons to evaluate vaccine efficacy [51]. The Ad26-based mosaic vaccine, containing Evn, reverse transcriptase Pol, and capsid protein Geg, showed 100 % immune response in humans when combined with high-dose of gp140 boost [13].
Despite the lack of success, tremendous effort has been made to develop vaccines. Some of this work that has employed peptides will be discussed next. The variable loops of gp120 (V1-V3) have shown promise as vaccines. Peptides corresponding to the variable loop 3 (V3) of gp120 are more accessible compared to other loops, hence have been marked as neutralizing epitopes. Immunization with the 31–39 amino acid long V3-peptides has induced generation of bnAbs in mice that could possibly block the interaction of gp120 with host co-receptors [96,60]. Cyclization of V3 has enhanced its immunogenicity, resulting in a 30-fold stronger HIV-1 neutralizing response in rabbits [145]. In addition to the loops of gp120, a minimal epitope derived from the external region of gp41 (EC26-2A4ΔM: ELLELDKM) has also shown potential in inducing HIV-neutralizing antibodies upon immunization of mice [177]. Another example is Vacc-4 × . Vacc-4x is comprised of four synthetic peptides derived from HIV capsid protein p24, which is present at high levels during the early and late stages of HIV infection. In REDUC clinical study, administration of Vacc-4x as part of combination therapy with an antiretroviral drug, Romidepsin, resulted in the activation of CD8+ T-cells and the reduction in HIV load in patients [191,212,178].
Peptides have also played a critical role in developing HIV diagnostic reagents. The US Centers for Disease Control and Prevention (CDC) recommends two types of diagnostic tests for AIDS; p24 antigen test for early-stage detection and HIV-1 and HIV-2 antibody test for early to chronic stage detection. Commonly used HIV tests range from immunofluorescence to nucleic acid-based assays [99]. Peptides can be useful tools to diagnose monoclonal antibodies and antigens in HIV patients. The FDA has approved OraQuickb HIV-1 rapid antibody test, which detects anti-gp41 antibodies in saliva, serum, plasma, or whole blood using synthetic gp41 peptide [152]. Another example is VIKIA®. This test uses HIV-1 gp41 and HIV-2 gp36 synthetic peptides immobilized on a strip. VIKIA® is sensitive and reliable, and it only requires a microliter volume sample size [73]. Another diagnostic assay called Alere Determine™ HIV 1/2 Ag/Ab Combo relies on a combination of synthetic peptides and recombinant antigens derived from gp41, gp120, and gp36 and can detect both HIV-1/2 antibodies and the HIV-1 p24 antigen [114]. Peptides are also used in applications with biosensors. Examples include an electrochemical peptide-based sensor developed for the detection of HIV anti-p24 antibodies [70] and a bifunctional colorimetric/fluorescence assay that uses short peptides derived from HIV-1 p17 protein tagged to a sensitive dye (P-17 antibodies are prevalent in the early stages of infection). Biosensors are advantageous due to their rapid turnover and minimal use of reagents and equipment [183]. Another sensor-based assay utilizes a 35-residue peptide fragment derived from gp41 (579–613) polymerized on a surface of quartz crystal microbalance to detect the majority of anti-HIV-antibodies in biological samples [132].
5. Severe acute respiratory syndrome
Severe acute respiratory syndrome (SARS) is caused by SARS coronavirus 1 (SARS-CoV-1), a subgroup B betacoronavirus. The SARS-CoV-1 genome consists of 29.7 Kb positive-stranded RNA composed of replicase and structural regions [47]. The replicase region encodes two polyproteins that contain 16 non-structural proteins (nsp1 through nsp16) required for viral replication and transcription. The polyproteins are cleaved into individual functional proteins by two proteases, papain-like protease (PLpro) and 3C-like protease (3CLpro or Mpro) [189]. The structural region encodes four primary structural proteins; spike protein (S), envelope protein (E), membrane glycoprotein (M), and nucleocapsid protein (N) along with several accessory proteins that interfere with the host innate immune response with unknown or poorly understood function. The viral infection begins when the S protein binds to the host receptor, angiotensin-converting enzyme 2 (ACE2). This allows cellular entry where the viral RNA genome is released, transcribed, and translated. The newly formed viral proteins and genomic RNA assemble in the endoplasmic reticulum-golgi intermediate compartment and fuse to the plasma membrane releasing the virus [49].
The first known case of SARS occurred in southern China in November 2002 [49]. The disease spread to more than 30 countries, infected over 8000 individuals, and resulted in about 800 deaths [80]. SARS-CoV-1 was identified as the causative agent [58,157] and the pandemic was controlled in less than four months through the measures of infection control instead of drug prevention or therapy [214]. During the SARS outbreak, ribavirin was most frequently used to inhibit viral RNA synthesis [74]. Ribavirin is a guanosine analog that inhibits guanosine-triphosphate (GTP) synthesis, viral mRNA capping, and viral RNA-dependent RNA polymerase [49,20]. At the time, ribavirin was used for Hepatitis C virus (HCV) therapy [233] and pediatric respiratory syncytial viral (RSV) infection [42]. The reported uses of ribavirin in SARS cases showed conflicting results with numerous methodological issues and its effect could not be distinguished from other treatments such as corticosteroids or other antivirals. Ribavirin treatment for SARS was not tested in clinical trials, so its efficacy remains to be assessed [20,167]. Since the SARS outbreak 17 years ago, all antiviral drugs or vaccines tested in controlled trials have shown to be ineffective [235] and lack translation in antiviral efficacy from pre-clinical studies to human patients [36]. Below we discuss the current developments in peptide-based therapies, vaccines, and detection methods specific to SARS-CoV-1 and how they can help fill the 17-year void.
Discovery efforts have been focused on targeting SARS-CoV-1 structural proteins. In one study, bioinformatic analysis was used to identify 10 peptides from the S protein sequence. Two peptides (P8: PSSKRFQPFQQFGRDVSDFT and P9: CANLLLQYGSFCTQLNRALSGIA) blocked the interaction of viral S protein with the host ACE2 receptor and showed promising viral neutralization in syncytia formation models at nearly 50 % [133]. In a separate study, a set of 20-residue peptides derived from S protein fragments were investigated. Four peptides (P2: PTTFMLKYDENGTITDAVDC, P6: YQDVNCTDVSTAIHADQLTP, P8: QYGSFCTQLNRALSGIAAEQ and P10: IQKEIDRLNEVAKNLNESLI) showed significant antiviral effects in cytopathic effect (CPE)-based assays, a reduction in viral titer in TCID50 assays, as well as a reduction in intracellular viral RNA levels as determined by quantitative real-time PCR (qRT-PCR). The antiviral effects of P8 were further confirmed by electron microscopy, which showed the absence of virus in the P8-treated cells and the presence of SARS-CoV-1 particles in the untreated cells [232]. It should be noted that the sequence corresponding to 737–753 (QYGSFCTQLNRALSGIA) of the S protein was shared in both studies mentioned above, warranting further work to optimize this peptide. In another study, S protein derived peptide S471−503 (ALNCYWPLNDYGFYTTTGIGYQPYRWVLSFEL) was identified and shown to block the interaction between receptor binding domain (RBD) and ACE2 in a competition ELISA and inhibit plaque formation of SARS-CoV in Vero E6 cells [95]. Additional viral structural proteins have been studied as potential targets. N protein from the nucleocapsid forms a complex with the viral RNA [128]. A 15-mer peptide phage display library was panned against purified N protein. As a result, six phage clones that bound to the N protein in ELISA were identified. The selected peptides had high homology with the peptide SNA5 (GGGWFCPIVRGRVSC) that appeared at the highest frequency among the selected peptides.
Targeting viral proteases has been exploited for peptide antiviral drug therapies. Molecular modeling and docking studies were used to identify an octapeptide AVLQSGFR that docked to the viral Mpro protease [39,59]. The peptide acted as an enzyme inhibitor with confirmed antiviral properties against SARS-CoV-1 [68]. The octapeptide showed a dose-dependent inhibition of SARS-CoV-1 in Vero E6 cell-based assays with low toxicity, suggesting that it can serve as a starting scaffold for an effective SARS-CoV-1 protease inhibitor. In addition, the peptide P9 (NGAICWGPCPTAFRQIGNCGHFKVRCCKIR), mentioned earlier, exhibits antiviral effects against multiple respiratory viruses including influenza viruses and SARS-CoV-1. By binding to the viral glycoprotein, P9 was able to alter endosomal acidification, block viral membrane fusion, and RNA viral release [231]. P9 showed efficacy in cell-based assays and viral load reduction in small animal models when used as prophylaxis against SARS-CoV-1. The potential peptide-based therapies for SARS are summarized in Table 3 .
Table 3.
Summary of potential peptide therapies for SARS.
| Peptide Name | Sequence | Derived from | Target | IC50 | Assay Format Tested | Phase | Reference |
|---|---|---|---|---|---|---|---|
| P8 | PSSKRFQPFQQFGRDVSDFT | S protein | ACE2 receptor | * | Syncytia inhibition assay | Pre-clinical | [133] |
| P9 | CANLLLQYGSFCTQLNRALSGIA | S protein | ACE2 receptor | * | Syncytia inhibition assay | Pre-clinical | [133] |
| P2 | PTTFMLKYDENGTITDAVDC | S protein | ACE2 receptor | 112.5 μg /mL, (IC90) | Cytopathic effect (CPE)-based assay | Pre-clinical | [232] |
| P6 | YQDVNCTDVSTAIHADQLTP | S protein | ACE2 receptor | 113.0 μg /mL, (IC90) | Cytopathic effect (CPE)-based assay | Pre-clinical | [232] |
| P8 | QYGSFCTQLNRALSGIAAEQ | S protein | ACE2 receptor | 24.9 μg /mL, (IC90) | Cytopathic effect (CPE)-based assay | Pre-clinical | [232] |
| P10 | IQKEIDRLNEVAKNLNESLI | S protein | ACE2 receptor | 73.5 μg /mL, (IC90) | Cytopathic effect (CPE)-based assay | Pre-clinical | [232] |
| S471−503 | ALNCYWPLNDYGFYTTTGIGYQPYRWVLSFEL | S protein | ACE2 receptor | * | Competition ELISA, plaque assay | Pre-clinical | [95] |
| SNA5 | GGGWFCPIVRGRVSC | Phage display library | N protein | n/a | Monoclonal phage ELISA | Pre-clinical | [128] |
| octapeptide | AVLQSGFR | Structure-based drug design and modeling | Mpro | 2.7 × 10−2 mg/L | Cytopathic effect (CPE)-based assay | Pre-clinical | [39] |
| P9 | NGAICWGPCPTAFRQIGNCGHFKVRCCKIR | β-defensin-4 | Endosome acidification | 5 μg/mL | plaque reduction assay | Pre-clinical | [231] |
*not available in the literature.
SARS-CoV-1 vaccines can be used to prevent viral infection and transmission thereby aid in the control of outbreaks. Inactive virus particles generated by formaldehyde, UV light, and β-propiolactone were common strategies to serve as vaccines [103] and tested for safety in Phase I clinical trials in China [123]. Despite the promising safety results, these candidate vaccines induced an immunopathologic respiratory disease in small animal studies when the immunized mice were challenged with SARS-CoV-1 [198]. Consequently, attention was diverted to other approaches. Synthetic peptides that overlapped the entire S protein were tested against antisera from small animals immunized with inactive SARS-CoV-1 sera and from SARS patients [88]. The results revealed a major immunodominant epitope of S protein at residues 528−635. Therefore, the synthetic peptide S603−634 was generated and showed to react with sera from all SARS patients, but not with control serum samples from healthy donors or hepatitis patients. This approach was shown to be effective to design and optimize peptide-based vaccines against SARS and to provide a potential antigen for diagnostic application. In another study, bioinformatic analysis was employed for vaccine development using peptides from the major structural N protein [124]. Two peptides, N1 and N2, were predicted to have similar Antigenic Index (AI) to induce antibody generation. Rabbits immunized with synthetic N1 and N2 peptides produced specific antibodies against both peptides. Only N1 peptide-induced antiserum showed strong binding to recombinant SARS-CoV-1 N protein by western blot. The immunogen N1 peptide reacted to sera of SARS patients in ELISA, suggesting that N1 peptide could serve as the epitope of the N protein to induce an immune response.
Currently, SARS-CoV-1 infection is diagnosed by detection of viral RNA in clinical samples by qRT-PCR, serological detection of specific antibody to SARS-CoV-1, and virus growth in cell culture, which requires a biosafety level III facility [176]. SARS-CoV-1 specific peptides can be a much safer and cost-effective alternate. In the studies above, the S603−634 and N1 peptide immunogens could also be used as a tool for detecting SARS-CoV-1 antibodies in patient serological samples. PL8 (PSSKRFQPFQQFGRDVSDFTDSVRDPKTSE) showed a 70 % positive response rate to sera from diagnosed SARS patients and 5% false-positive response to uninfected individuals, suggesting that further development is needed to optimize the peptide sequence for accurate diagnosis [133].
The surveillance of bat coronaviruses in the years since the 2003 SARS outbreak revealed a diverse family of SARS-related coronaviruses (SARSr-CoVs), with warnings that spillover into humans was already happening and that additional surveillance and precautions were necessary to prevent a likely future pandemic [94,150,105].
6. Coronavirus disease 2019
Coronavirus disease 2019 (COVID-19) is a novel infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The virus shares 79 % genetic similarity with SARS-CoV-1 and its 29.9 Kb genome consists of the same replicase and structural regions [216,62]. SARS-CoV-2 utilizes the host ACE2 receptor for cellular entry with higher affinity compared to its other family members [35,215]. The S protein contains a furin cleavage site which could contribute to the increased infectivity of SARS-CoV-2 relative to SARS-CoV-1 [91,155].
A series of COVID-19 cases were first detected in Wuhan China in December 2019, WHO declared a global health emergency by January 30, 2020, and a global pandemic two months later [9,193]. As of August 7th, 2020, over 19 million confirmed cases and 715,163 deaths worldwide have been reported [2]. In a situation similar to the SARS outbreak of 2003, no FDA approved vaccines or antivirals were available to combat the disease [188]. With an immediate need for COVID-19 therapies, attention quickly turned to existing drugs. Liu et al. created an extensive overview of notable published scientific articles, key viral target proteins, existing drugs with potential therapeutic use, select patents for drug repurposing, small molecules in development, and patents on therapeutic antibodies against SARS-CoV-2 [126]. In addition, Gordon et al. used affinity-purification mass spectrometry to identify 66 druggable human proteins that interact with 26 SARS-CoV-2 proteins, and reported 69 compounds at various stages (approved, clinical or preclinical) that target these interactions [46]. Of the potential therapeutics, almost none are peptide-based drugs. Here we discuss some of the current peptides under development as possible future therapeutics for SARS-CoV-2, summarized in Table 4 .
Table 4.
Summary of potential peptide therapies for COVID-19.
| Peptide Name | Sequence | Derived from | Target | IC50 | Assay Format Tested | Phase | Reference |
|---|---|---|---|---|---|---|---|
| SBP1 | IEEQAKTFLDKFNHEAEDLFYQS | ACE2 receptor | S protein | * | Kinetic binding assay using bio-layer interferometry (BLI) | Pre-clinical | [228] |
| EK1C4 | SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL-GSGSG-PEG4-Chol | S protein, HR2 domain | S protein, HR1 domain | 15.8 nM | cell-cell fusion assay, plaque reduction assays | Pre-clinical | [221] |
| P9R | NGAICWGPCPTAFRQIGNCGRFRVRCCRI | β-defensin-4 | Endosome acidification | 0.9 μg/mL | plaque assay | Pre-clinical | [230] |
| QS1 | Ac-Abu-Tle-Leu-Gln-VS | Hybrid combinatorial substrate library (HyCoSuL) | Mpro | n/a | Kinetic analysis | Pre-clinical | [179] |
| VIR251 | Ac-hTyr-Dap-Gly-Gly-VME | Hybrid combinatorial substrate library (HyCoSuL) | PLpro | n/a | Kinetic analysis | Pre-clinical | [180] |
| VIR250 | Ac-Abu(Bth)-Dap-Gly-Gly-VME | Hybrid combinatorial substrate library (HyCoSuL) | PLpro | n/a | Kinetic analysis | Pre-clinical | [180] |
*not available in the literature.
The co-crystal structure of S protein RBD with ACE2 led to the identification of a 23-residue peptide (SBP1) that disrupts the protein-protein interaction between the virus and host receptor and prevents viral cell entry. SBP1 (IEEQAKTFLDKFNHEAEDLFYQS) was derived from α1 helix region of the human ACE2 and bound to SARS-CoV-2 RBD with a KD of 47 nM determined by biolayer interferometry [228]. SBP1 has yet to be tested in cell-based assays and animal models of SARS-CoV-2 infection. Another peptide in development is EK1 (SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL), which is derived from the heptad repeat 2 (HR2) domain of the S protein. After binding to the RBD, heptad repeat 1 (HR1) and HR2 interact with each other to form a 6HB fusion core bringing the viral and host membrane in close proximity for fusion. EK1 peptide binds to the HR1 domain inhibiting the viral infection as shown in S-mediated cell-cell fusion assays [221]. A similar lipidation strategy that was used to develop anti-HIV-1 peptide LP-19, was implemented to improve the antiviral activity and half-life of the EK1 peptide. EK1 was linked to cholesterol or palmitic acid through either a glycine-serine linker, polyethylene glycol (PEG) spacer, or both combination [38,221]. EK1C4 (EK1-GSGSG-PEG4-Chol) was found to be the most potent inhibitory peptide in cell-cell fusion assays and inhibition plaque reduction assays. In another study, the aforementioned P9 peptide [231] was used as the parent peptide to develop P9R with antiviral activity against SARS-CoV-2 [230]. Three weakly positive-charged amino acids in the P9 (H21, K23, and K28) were substituted with arginine to increase the overall net positive charge from +4.7 to +5.6 (P9R: NGAICWGPCPTAFRQIGNCGRFRVRCCRI). It was hypothesized that a peptide with a stronger net positive charge could further neutralize protons in the endosome to inhibit endosomal acidification, thereby preventing viral RNA release. P9R showed 1000-fold enhanced potency compared to its parent for inhibition of SARS-CoV-2 replication in a multicycle growth assay.
The PLpro and Mpro enzymes of SARS-CoV-2 are also targets for peptide-based inhibitors. Through the use of a combinatorial library, one group has developed peptide inhibitors against Mpro and PLpro [180,179]. Substrates for both proteases were identified using a library that contained proteogenic and non-proteogenic amino acids. The best substrate candidates were then converted into peptide inhibitors through the addition of irreversible reactive group at the C-terminus, resulting in Mpro inhibitor QS1 (Ac-Abu-Tle-Leu-Gln-VS), and PLpro inhibitors VIR251 and VIR250 (Ac-hTyr-Dap-Gly-Gly-VME and Ac-Abu(Bth)-Dap-Gly-Gly-VME, respectively). Kinetic analysis of these peptides against the SARS- CoV-2 Mpro and PLpro indicated inhibition.
With the increasing number of infected individuals and lack of effective antiviral therapies, the need for SARS-CoV-2 vaccine development has been an international priority. The current vaccine developments include RNA vaccines, DNA vaccines, recombinant protein vaccines, viral vector-based vaccines, live attenuated vaccines, and inactivated vaccines [6]. An mRNA-based vaccine co-developed by Moderna and the Vaccine Research Center at the National Institutes of Health has begun dosing patients with mRNA-1273 in their Phase III study, COVE (Coronavirus Efficacy) [143]. The results from their Phase I study showed that mRNA-1273 was safe and well-tolerated. It elicited Th1-biased CD4 T cell responses, produced neutralizing antibody titers in 100 % of evaluated participants at the 100 μg dose [101]. Another mRNA based vaccine, co-developed by Pfizer and BioNTech, has advanced the candidate BNT162b2 into Phase II/III study [161]. Phase I/II results showed that BNT162b2 was safe and well-tolerated and stimulated the production of neutralizing antibodies and elicited CD4+ and CD8+ T cell responses against the RBD [160]. While both aforementioned mRNA vaccines encode for the SARS-CoV-2 S proteins, other viral recombinant proteins are being investigated [126]. One group has employed immune-informatics to identify and characterize SARS-CoV-2 S protein-based epitopes in silico for vaccine development [119]. Li et al. used a myriad of online tools to analyze the structure of SARS-CoV-2 S protein to predict and identify highly antigenic B-cell and T-cell epitopes [219,69,201,23,63,57]. The peptide epitopes were further characterized for allergenicity, physiochemical features, toxicity, and stability [3,196,185]. The immuno-informatic approach has identified four peptide epitopes (VRQIAPGQTGKIAD, VLGQSKRVDFCGKG, GLTGTGVLTESNKK, and KIADYNYKLPDDFT). Unlike mRNA vaccines, peptide-based vaccines require careful design to identify epitopes that are capable to induce a strong immune response against the desired pathogen and the strategies reported by Li and colleagues can be used for other targets of interest.
Kalita and team utilized a similar in silico approach to design a multi-peptide subunit-based epitope vaccine against SARS-CoV-2. The vaccine contained human β-defensin as an adjuvant and 33 highly antigenic epitopes of cytotoxic T-lymphocyte, helper T-lymphocyte, and B-cell linked by specific linkers [110]. The highly antigenic epitopes were derived from N, M, and S protein as determined by prediction tools [79,175,97,17,182,81]. Further, computational analysis suggested that the multi-epitope subunit vaccine was non-toxic, thermostable, and immunogenic [57,168,4,181,166,24,172,210]. Molecular docking and dynamics studies validated the stability of the vaccine construct [110,109] and in silico cloning proposed potential expression in microbial expression system [78]. The extensive bioinformatics data suggest that the multi-epitope based subunit vaccine has a high probability of protective efficacy and safety against SARS-CoV-2. However, its synthesis and experimental evaluation have to be conducted to validate the predictions. Additionally, it is important to note that there are on-going efforts to generate peptide-based vaccines specific to SARS-CoV-2 with the internal discovery platforms from Generex, Vaxil, IMV Inc., and Axon Neuroscience.
Current laboratory tests for SARS-CoV-2 diagnostics involve the detection of viral RNA by qRT-PCR and the detection of anti-SARS-CoV-2 antibodies [186]. Tests based on qRT-PCR are commonly used with samples collected from the nasopharyngeal and throat swabs or saliva. The qRT-PCR targets one or more SARS-CoV-2 genes from the E, N, S, polymerase, or open reading frame 1 (ORF1). Serological testing is utilized for patients with mild symptoms or who are asymptomatic. In a peptide-based diagnostic, 20 biotinylated synthetic SARS-CoV-2 antigens from the ORF1a/b, S, and N proteins were studied [25]. Among the peptides tested, a peptide from the S protein showed high specificity to SARS-CoV-2 with no cross-reactivity. Sera samples were added to the biotinylated S peptide in complex with streptavidin-coated magnetic beads and the presence of SARS-CoV-2 antibodies was detected by luminescence. In this assay IgG and IgM from COVID-19 patients were detected with an accuracy of 71.4 % and 57.2 %, respectively.
7. Conclusion
Infectious diseases are an ongoing global health concern with a significant consequence of morbidity and mortality. The COVID-19 pandemic is the most recent example that has caused unprecedented global initiatives to expedite therapy, vaccine, and diagnostic development. Challenges such as rising microbial resistance and poor specificity and efficacy of the anti-viral compounds demand the discovery of novel molecules. In this review, we have highlighted the potential peptide-based anti-viral therapeutics and vaccines in early-development and their application in the diagnosis of viral infections. Peptide-based therapeutics hold great promises as a novel approach due to their high specificity, efficacy, and safety. However, the use of naturally and synthetically derived peptides is limited by their short in vivo half-lives caused by proteolysis and renal clearance. Incorporation of D-amino acid in peptide sequences has been shown to increase peptide potency and proteolytic stability. The small size of peptides results in renal filtration; therefore, lipid moieties are introduced to add to their size and improve their half-life of peptides. Peptide-based therapies with extended-release dosing hold potential in the future for HIV drugs and can alleviate daily drug administration. Nanoparticle conjugation of peptides has been shown to enhance their solubility and efficacy and provide delivery to the targeted cell. In summary, peptide-based therapies such as Enfuvirtide is FDA approved for AIDS, while others such as Flufirvitide-3 (phase I) and Myrcludex-B (phase IIa) for influenza, and Sifuvirtide (phase III in China) for AIDS are ongoing clinical trials.
Peptide-based vaccines are considered as a novel means for the induction of a robust immune response. A few peptide-based vaccines have shown great potential in the clinical studies, including FLU-v-like peptides (phase I), Multimeric-001, and Vacc-4 × . The use of minimal microbial components in peptide-based vaccines can eliminate some of the problems associated with the use of whole organisms or proteins, such as allergic and autoimmune responses. However, low immunogenicity can be a limiting factor for peptide-based vaccines and may require the use of adjuvants [140]. Lastly, peptides have been used in innovative technologies, such as biosensors, to diagnose viral infections. Overall, as discussed herein, peptides can be used to help combat different viral diseases.
Funding
Eurofins Lancaster Laboratories provided support in the form of salary for author SAA. Eli Lilly and Company provided support in the form of salaries for authors YD, JL, PP, JPT, and SA. The funders had no additional role in the manuscript preparation
CRediT authorship contribution statement
Shams Al-Azzam: Writing - original draft, Writing - review & editing. Yun Ding: Writing - original draft, Writing - review & editing. Jinsha Liu: Writing - original draft, Writing - review & editing. Priyanka Pandya: Writing - original draft, Writing - review & editing. Joey Paolo Ting: Writing - original draft, Writing - review & editing. Sepideh Afshar: Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors report no declarations of interest.
References
- 1.Preventing emerging infectious diseases: a strategy for the 21st century. Overview of the updated CDC plan. MMWR Recomm. Rep. 1998;47(Rr-15):1–14. [PubMed] [Google Scholar]
- 2.JHUoMCr center, Journal. 2020 https://coronavirus.jhu.edu/map.html [Google Scholar]
- 3.1.0 AF. http://ddg-pharmfac.net/AllergenFP/.
- 4.2020. 2.0 Av. Bioinformatics Tool For Allergenicity Prediction.http://www.ddg-pharmfac.net/AllerTOP/index.html [Google Scholar]
- 5.Alghrair Z.K., Fernig D.G., Ebrahimi B. Enhanced inhibition of influenza virus infection by peptide-noble-metal nanoparticle conjugates. Beilstein J. Nanotechnol. 2019;10:1038–1047. doi: 10.3762/bjnano.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52(4):583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ammendolia M.G., Agamennone M., Pietrantoni A., Lannutti F., Siciliano R.A., De Giulio B., et al. Bovine lactoferrin-derived peptides as novel broad-spectrum inhibitors of influenza virus. Pathog. Glob. Health. 2012;106(1):12–19. doi: 10.1179/2047773212Y.0000000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arbeitskreis Blut U. Influenza virus. Transfus Med. Hemother. 2009;36(1):32–39. doi: 10.1159/000197314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ysrafil Astuti I. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Atsmon J., Caraco Y., Ziv-Sefer S., Shaikevich D., Abramov E., Volokhov I., et al. Priming by a novel universal influenza vaccine (multimeric-001)-a gateway for improving immune response in the elderly population. Vaccine. 2014;32(44):5816–5823. doi: 10.1016/j.vaccine.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 11.Badani H. 2014. Understanding The Mechanism Of Action Of Flufirvitide-3 A. Peptide Based Inhibitor Of Influenza Virus. [Google Scholar]
- 12.Balasubramanian P., Williams C., Shapiro M.B., Sinangil F., Higgins K., Nadas A., et al. Functional antibody response against V1V2 and V3 of HIV gp120 in the VAX003 and VAX004 vaccine trials. Sci. Rep. 2018;8(1):542. doi: 10.1038/s41598-017-18863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barouch D.H., Tomaka F.L., Wegmann F., Stieh D.J., Alter G., Robb M.L., et al. Evaluation of a mosaic HIV-1 vaccine in a multicentre, randomised, double-blind, placebo-controlled, phase 1/2a clinical trial (APPROACH) and in rhesus monkeys (NHP 13-19) Lancet. 2018;392(10143):232–243. doi: 10.1016/S0140-6736(18)31364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrera A., Guerra B., Notvall L., Lanford R.E. Mapping of the hepatitis B virus pre-S1 domain involved in receptor recognition. J. Virol. 2005;79(15):9786–9798. doi: 10.1128/JVI.79.15.9786-9798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bártolo I., Diniz A.R., Borrego P., Ferreira J.P., Bronze M.R., Barroso H., et al. Evaluation of the fusion inhibitor P3 peptide as a potential microbicide to prevent HIV transmission in women. PLoS One. 2018;13(4) doi: 10.1371/journal.pone.0195744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger E.A., Murphy P.M., Farber J.M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 17.Bioinformatics D. 2017. NetCTL 1.2 Server. [Google Scholar]
- 18.Bobyk K.D., Mandadapu S.R., Lohith K., Guzzo C., Bhargava A., Lusso P., et al. Design of HIV coreceptor derived peptides that inhibit viral entry at submicromolar concentrations. Mol. Pharm. 2017;14(8):2681–2689. doi: 10.1021/acs.molpharmaceut.7b00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boni C., Fisicaro P., Valdatta C., Amadei B., Di Vincenzo P., Giuberti T., et al. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J. Virol. 2007;81(8):4215–4225. doi: 10.1128/JVI.02844-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Booth C.M., Matukas L.M., Tomlinson G.A., Rachlis A.R., Rose D.B., Dwosh H.A., et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA. 2003;289(21):2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 21.Borrego P., Calado R., Marcelino J.M., Pereira P., Quintas A., Barroso H., et al. An ancestral HIV-2/simian immunodeficiency virus peptide with potent HIV-1 and HIV-2 fusion inhibitor activity. Aids. 2013;27(7):1081–1090. doi: 10.1097/QAD.0b013e32835edc1d. [DOI] [PubMed] [Google Scholar]
- 22.Brauer F., Schmidt K., Zahn R.C., Richter C., Radeke H.H., Schmitz J.E., et al. A rationally engineered anti-HIV peptide fusion inhibitor with greatly reduced immunogenicity. Antimicrob. Agents Chemother. 2013;57(2):679–688. doi: 10.1128/AAC.01152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buchan D.W., Minneci F., Nugent T.C., Bryson K., Jones D.T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41(Web Server issue):W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchan D.W.A., Jones D.T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47(W1):W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai X. 2020. A Peptide-Based Magnetic Chemiluminescence Enzyme Immunoassay for Serological Diagnosis of Corona Virus Disease 2019 (COVID-19) medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao P., Dou G., Cheng Y., Che J. The improved efficacy of sifuvirtide compared with enfuvirtide might be related to its selectivity for the rigid biomembrane, as determined through surface plasmon resonance. PLoS One. 2017;12(2) doi: 10.1371/journal.pone.0171567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.CDC; 2018. What Are the Benefits of Flu Vaccination? [Google Scholar]
- 28.CDC; 2019. Influenza Antiviral Drug Resistance.https://www.cdc.gov/flu/treatment/antiviralresistance.htm [Google Scholar]
- 29.CDC; 2020. CDC Seasonal Flu Vaccine Effectiveness Studies, 2004-2019. [Google Scholar]
- 30.CDC; 2020. Estimated Influenza Illnesses, Medical Visits, Hospitalizations, and Deaths in the United States — 2018–2019 Influenza Season. [Google Scholar]
- 31.CDC; 2020. Influenza A Subtypes and the Species Affected. [Google Scholar]
- 32.Chan D.C., Kim P.S. HIV entry and its inhibition. Cell. 1998;93(5):681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 33.Chaudhuri S., Symons J.A., Deval J. Innovation and trends in the development and approval of antiviral medicines: 1987-2017 and beyond. Antiviral Res. 2018;155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J., Feng S., Xu Y., Huang X., Zhang J., Chen J., et al. Discovery and characterization of a novel peptide inhibitor against influenza neuraminidase. RSC Med. Chem. 2020;11(1):148–154. doi: 10.1039/c9md00473d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. 2020 doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng V.C., Lau S.K., Woo P.C., Yuen K.Y. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin. Microbiol. Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choga W.T., Anderson M., Zumbika E., Phinius B.B., Mbangiwa T., Bhebhe L.N., et al. In silico prediction of human leukocytes antigen (HLA) class II binding hepatitis B virus (HBV) peptides in Botswana. Viruses. 2020;12(7) doi: 10.3390/v12070731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chong H., Xue J., Xiong S., Cong Z., Ding X., Zhu Y., et al. A lipopeptide HIV-1/2 fusion inhibitor with highly potent in vitro, ex vivo, and in vivo antiviral activity. J. Virol. 2017;91(11) doi: 10.1128/JVI.00288-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou K.C., Wei D.Q., Zhong W.Z. Binding mechanism of coronavirus main proteinase with ligands and its implication to drug design against SARS. Biochem. Biophys. Res. Commun. 2003;308(1):148–151. doi: 10.1016/S0006-291X(03)01342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cimbro R., Peterson F.C., Liu Q., Guzzo C., Zhang P., Miao H., et al. Tyrosine-sulfated V2 peptides inhibit HIV-1 infection via coreceptor mimicry. EBioMedicine. 2016;10:45–54. doi: 10.1016/j.ebiom.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conlon J.M., Mechkarska M., Lukic M.L., Flatt P.R. Potential therapeutic applications of multifunctional host-defense peptides from frog skin as anti-cancer, anti-viral, immunomodulatory, and anti-diabetic agents. Peptides. 2014;57:67–77. doi: 10.1016/j.peptides.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Cooper A.C., Banasiak N.C., Allen P.J. Management and prevention strategies for respiratory syncytial virus (RSV) bronchiolitis in infants and young children: a review of evidence-based practice interventions. Pediatr. Nurs. 2003;29(6):452–456. [PubMed] [Google Scholar]
- 43.Cornelius C., Schöneweis K., Georgi F., Weber M., Niederberger V., Zieglmayer P., et al. Immunotherapy with the PreS-based grass pollen allergy vaccine BM32 induces antibody responses protecting against hepatitis B infection. EBioMedicine. 2016;11:58–67. doi: 10.1016/j.ebiom.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cotten S.W., Zou J., Wang R., Huang B.C., Liu R. mRNA display-based selections using synthetic peptide and natural protein libraries. Methods Mol. Biol. 2012;805:287–297. doi: 10.1007/978-1-61779-379-0_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Craxi A., Di Bona D., Camma C. Interferon-alpha for HBeAg-positive chronic hepatitis B. J. Hepatol. 2003;39(Suppl 1):S99–105. doi: 10.1016/s0168-8278(03)00154-5. [DOI] [PubMed] [Google Scholar]
- 46.Gordon David E., GMJ, Krogan Nevan J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Clercq E. Potential antivirals and antiviral strategies against SARS coronavirus infections. Expert Rev. Anti Infect Ther. 2006;4(2):291–302. doi: 10.1586/14787210.4.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Souza M.Q., Galdino A.S., dos Santos J.C., Soares M.V., de Nobrega Y.C., Alvares Ada C., et al. A recombinant multiepitope protein for hepatitis B diagnosis. Biomed Res. Int. 2013;2013 doi: 10.1155/2013/148317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng Q., Zhai J.W., Michel M.L., Zhang J., Qin J., Kong Y.Y., et al. Identification and characterization of peptides that interact with hepatitis B virus via the putative receptor binding site. J. Virol. 2007;81(8):4244–4254. doi: 10.1128/JVI.01270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dieffenbach C.W., Fauci A.S. The search for an HIV vaccine, the journey continues. J. Int. AIDS Soc. 2020;23(5) doi: 10.1002/jia2.25506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dogo-Isonagie C., Lam S., Gustchina E., Acharya P., Yang Y., Shahzad-ul-Hussan S., et al. Peptides from second extracellular loop of C-C chemokine receptor type 5 (CCR5) inhibit diverse strains of HIV-1. J. Biol. Chem. 2012;287(18):15076–15086. doi: 10.1074/jbc.M111.332361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donkers J.M., Zehnder B., van Westen G.J.P., Kwakkenbos M.J., AP I.J., Oude Elferink R.P.J., et al. Reduced hepatitis B and D viral entry using clinically applied drugs as novel inhibitors of the bile acid transporter NTCP. Sci. Rep. 2017;7(1):15307. doi: 10.1038/s41598-017-15338-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dorr P., Westby M., Dobbs S., Griffin P., Irvine B., Macartney M., et al. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 2005;49(11):4721–4732. doi: 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doss M., Ruchala P., Tecle T., Gantz D., Verma A., Hartshorn A., et al. Hapivirins and diprovirins: novel θ-defensin analogs with potent activity against influenza a virus. J. Immunol. 2012;188(6):2759–2768. doi: 10.4049/jimmunol.1101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dou Y., van Montfoort N., van den Bosch A., de Man R.A., Zom G.G., Krebber W.J., et al. HBV-derived synthetic long peptide can boost CD4+ and CD8+ T-cell responses in chronic HBV patients Ex vivo. J. Infect. Dis. 2018;217(5):827–839. doi: 10.1093/infdis/jix614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 59.Du Q.S., Wang S.Q., Zhu Y., Wei D.Q., Guo H., Sirois S., et al. Polyprotein cleavage mechanism of SARS CoV mpro and chemical modification of the octapeptide. Peptides. 2004;25(11):1857–1864. doi: 10.1016/j.peptides.2004.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eda Y., Takizawa M., Murakami T., Maeda H., Kimachi K., Yonemura H., et al. Sequential immunization with V3 peptides from primary human immunodeficiency virus type 1 produces cross-neutralizing antibodies against primary isolates with a matching narrow-neutralization sequence motif. J. Virol. 2006;80(11):5552–5562. doi: 10.1128/JVI.02094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fan H., Yan X., Zhang Y., Zhang X., Gao Y., Xu Y., et al. Increased expression of Gp96 by HBx-induced NF-kappaB activation feedback enhances hepatitis B virus production. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0065588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferre F., Clote P. DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Nucleic Acids Res. 2006;34(Web Server issue):W182–W185. doi: 10.1093/nar/gkl189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Flynn N.M., Forthal D.N., Harro C.D., Judson F.N., Mayer K.H., Para M.F. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 2005;191(5):654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 65.Fontaine H., Kahi S., Chazallon C., Bourgine M., Varaut A., Buffet C., et al. Anti-HBV DNA vaccination does not prevent relapse after discontinuation of analogues in the treatment of chronic hepatitis B: a randomised trial--ANRS HB02 VAC-ADN. Gut. 2015;64(1):139–147. doi: 10.1136/gutjnl-2013-305707. [DOI] [PubMed] [Google Scholar]
- 66.Frankel A.D., Young J.A. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 67.Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93(7):449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gan Y.R., Huang H., Huang Y.D., Rao C.M., Zhao Y., Liu J.S., et al. Synthesis and activity of an octapeptide inhibitor designed for SARS coronavirus main proteinase. Peptides. 2006;27(4):622–625. doi: 10.1016/j.peptides.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gasteiger E., Gattiker A., Hoogland C., Ivanyi I., Appel R.D., Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerasimov J.Y., Lai R.Y. An electrochemical peptide-based biosensing platform for HIV detection. Chem. Commun. (Camb.) 2010;46(3):395–397. doi: 10.1039/b919070h. [DOI] [PubMed] [Google Scholar]
- 71.Glebe D., Urban S., Knoop E.V., Cag N., Krass P., Grun S., et al. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129(1):234–245. doi: 10.1053/j.gastro.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 72.Gómara M.J., Sánchez-Merino V., Paús A., Merino-Mansilla A., Gatell J.M., Yuste E., et al. Definition of an 18-mer synthetic peptide derived from the GB virus C E1 protein as a New HIV-1 entry inhibitor. Biochim. Biophys. Acta. 2016;1860(6):1139–1148. doi: 10.1016/j.bbagen.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 73.Gomes C., Azevedo-Pereira J.M. The performance of the VIKIA(®) HIV1/2 rapid test-evaluation of the reliability and sensitivity. J. Virol. Methods. 2011;173(2):353–356. doi: 10.1016/j.jviromet.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 74.Graci J.D., Cameron C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006;16(1):37–48. doi: 10.1002/rmv.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grimm D., Thimme R., Blum H.E. HBV life cycle and novel drug targets. Hepatol. Int. 2011;5(2):644–653. doi: 10.1007/s12072-011-9261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gripon P., Cannie I., Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J. Virol. 2005;79(3):1613–1622. doi: 10.1128/JVI.79.3.1613-1622.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Groß A., Hashimoto C., Sticht H., Eichler J. Synthetic peptides as protein mimics. Front. Bioeng. Biotechnol. 2015;3:211. doi: 10.3389/fbioe.2015.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Grote A., Hiller K., Scheer M., Munch R., Nortemann B., Hempel D.C., et al. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33(Web Server issue):W526–W531. doi: 10.1093/nar/gki376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.2020. Group I. Tools: Predicting Antigenic Peptides.http://imed.med.ucm.es/Tools/antigenic.pl [Google Scholar]
- 80.Guan Y., Peiris J.S., Zheng B., Poon L.L., Chan K.H., Zeng F.Y., et al. Molecular epidemiology of the novel coronavirus that causes severe acute respiratory syndrome. Lancet. 2004;363(9403):99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R. Open source drug Discovery C, et al. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hamilton B.S., Whittaker G.R., Daniel S. Influenza virus-mediated membrane fusion: determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses. 2012;4(7):1144–1168. doi: 10.3390/v4071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hatano K., Matsubara T., Muramatsu Y., Ezure M., Koyama T., Matsuoka K., et al. Synthesis and influenza virus inhibitory activities of carbosilane dendrimers peripherally functionalized with hemagglutinin-binding peptide. J. Med. Chem. 2014;57(20):8332–8339. doi: 10.1021/jm5007676. [DOI] [PubMed] [Google Scholar]
- 84.Hayes C.N., Zhang Y., Makokha G.N., Hasan M.Z., Omokoko M.D., Chayama K. Early events in hepatitis B virus infection: from the cell surface to the nucleus. J. Gastroenterol. Hepatol. 2016;31(2):302–309. doi: 10.1111/jgh.13175. [DOI] [PubMed] [Google Scholar]
- 85.He Y., Cheng J., Lu H., Li J., Hu J., Qi Z., et al. Potent HIV fusion inhibitors against enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. U. S. A. 2008;105(42):16332–16337. doi: 10.1073/pnas.0807335105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.He Y., Xiao Y., Song H., Liang Q., Ju D., Chen X., et al. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 2008;283(17):11126–11134. doi: 10.1074/jbc.M800200200. [DOI] [PubMed] [Google Scholar]
- 87.He Y., Ye X., Tiollais P., Zhang J., Zhang J., Liu J., et al. Selection of HBV preS1-binding penta-peptides by phage display. Acta Biochim. Biophys. Sin. (Shanghai) 2014;46(8):691–698. doi: 10.1093/abbs/gmu049. [DOI] [PubMed] [Google Scholar]
- 88.He Y., Zhou Y., Wu H., Luo B., Chen J., Li W., et al. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J. Immunol. 2004;173(6):4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 89.Heathcote J., McHutchison J., Lee S., Tong M., Benner K., Minuk G., et al. A pilot study of the CY-1899 T-cell vaccine in subjects chronically infected with hepatitis B virus. The CY1899 T cell vaccine study group. Hepatology. 1999;30(2):531–536. doi: 10.1002/hep.510300208. [DOI] [PubMed] [Google Scholar]
- 90.Herrera-Rodriguez J., Meijerhof T., Niesters H.G., Stjernholm G., Hovden A.O., Sorensen B., et al. a novel peptide-based vaccine candidate with protective efficacy against influenza a in a mouse model. Virology. 2018;515:21–28. doi: 10.1016/j.virol.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 91.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. 271-80 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Horimoto T., Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 2005;3(8):591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 93.Hsieh I.N., Hartshorn K.L. The role of antimicrobial peptides in influenza virus infection and their potential as antiviral and immunomodulatory therapy. Pharmaceuticals (Basel) 2016;9(3) doi: 10.3390/ph9030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu B.Z.L., Yang X.L., Ge X.Y., Zhang W. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 2017;13(11) doi: 10.1371/journal.ppat.1006698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu H., Li L., Kao R.Y., Kou B., Wang Z., Zhang L., et al. Screening and identification of linear B-cell epitopes and entry-blocking peptide of severe acute respiratory syndrome (SARS)-associated coronavirus using synthetic overlapping peptide library. J. Comb. Chem. 2005;7(5):648–656. doi: 10.1021/cc0500607. [DOI] [PubMed] [Google Scholar]
- 96.Huang C.C., Tang M., Zhang M.Y., Majeed S., Montabana E., Stanfield R.L., et al. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310(5750):1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.IFNepitope; 2020. Epitope Prediction.http://crdd.osdd.net/raghava/ifnepitope/ [Google Scholar]
- 98.Irwin K.K., Renzette N., Kowalik T.F., Jensen J.D. Antiviral drug resistance as an adaptive process. Virus Evol. 2016;2(1):vew014. doi: 10.1093/ve/vew014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iweala O.I. HIV diagnostic tests: an overview. Contraception. 2004;70(2):141–147. doi: 10.1016/j.contraception.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 100.Izumi K., Nakamura S., Nakano H., Shimura K., Sakagami Y., Oishi S., et al. Characterization of HIV-1 resistance to a fusion inhibitor, N36, derived from the gp41 amino-terminal heptad repeat. Antiviral Res. 2010;87(2):179–186. doi: 10.1016/j.antiviral.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 101.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jamjian M.C., McNicholl I.R. Enfuvirtide: first fusion inhibitor for treatment of HIV infection. Am. J. Health Syst. Pharm. 2004;61(12):1242–1247. doi: 10.1093/ajhp/61.12.1242. [DOI] [PubMed] [Google Scholar]
- 103.Jiang S., He Y., Liu S. SARS vaccine development. Emerg. Infect. Dis. 2005;11(7):1016–1020. doi: 10.3201/eid1107.050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang S., Lin K., Strick N., Neurath A.R. HIV-1 inhibition by a peptide. Nature. 1993;365(6442):113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 105.Jie Cui F.L.Z.-L.S. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jones J.C., Turpin E.A., Bultmann H., Brandt C.R., Schultz-Cherry S. Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J. Virol. 2006;80(24):11960–11967. doi: 10.1128/JVI.01678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Joshi V.G., Dighe V.D., Thakuria D., Malik Y.S., Kumar S. Multiple antigenic peptide (MAP): a synthetic peptide dendrimer for diagnostic, antiviral and vaccine strategies for emerging and re-emerging viral diseases. Indian J. Virol. 2013;24(3):312–320. doi: 10.1007/s13337-013-0162-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kadam R.U., Juraszek J., Brandenburg B., Buyck C., Schepens W.B.G., Kesteleyn B., et al. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358(6362):496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kalita P., Lyngdoh D.L., Padhi A.K., Shukla H., Tripathi T. Development of multi-epitope driven subunit vaccine against fasciola gigantica using immunoinformatics approach. Int. J. Biol. Macromol. 2019;138:224–233. doi: 10.1016/j.ijbiomac.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 110.Kalita P., Padhi A.K., Zhang K.Y.J., Tripathi T. Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb. Pathog. 2020;145 doi: 10.1016/j.micpath.2020.104236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kamali A., Holodniy M. Influenza treatment and prophylaxis with neuraminidase inhibitors: a review. Infect. Drug Resist. 2013;6:187–198. doi: 10.2147/IDR.S36601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kikuchi T., Watanabe A. Baloxavir heralds a new era in influenza virus biology. Respir. Investig. 2019;57(1):1–2. doi: 10.1016/j.resinv.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 113.Kliger Y., Levy O., Oren A., Ashkenazy H., Tiran Z., Novik A., et al. Peptides modulating conformational changes in secreted chaperones: from in silico design to preclinical proof of concept. Proc. Natl. Acad. Sci. U. S. A. 2009;106(33):13797–13801. doi: 10.1073/pnas.0906514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kshatriya R., Cachafeiro A.A., Kerr R.J., Nelson J.A., Fiscus S.A. Comparison of two rapid human immunodeficiency virus (HIV) assays, determine HIV-1/2 and OraQuick advance rapid HIV-1/2, for detection of recent HIV seroconversion. J. Clin. Microbiol. 2008;46(10):3482–3483. doi: 10.1128/JCM.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kulinski J.M., Tarakanova V.L., Verbsky J. Regulation of antiviral CD8 T-cell responses. Crit. Rev. Immunol. 2013;33(6):477–488. doi: 10.1615/critrevimmunol.2013007909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lambert D.M., Barney S., Lambert A.L., Guthrie K., Medinas R., Davis D.E., et al. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. U. S. A. 1996;93(5):2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Leistner C.M., Gruen-Bernhard S., Glebe D. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell. Microbiol. 2008;10(1):122–133. doi: 10.1111/j.1462-5822.2007.01023.x. [DOI] [PubMed] [Google Scholar]
- 118.Li J., Chen Y., Yuan N., Zeng M., Zhao Y., Yu R., et al. a novel natural influenza a H1N1 virus neuraminidase inhibitory peptide derived from cod skin hydrolysates and its antiviral mechanism. Mar. Drugs. 2018;16(10) doi: 10.3390/md16100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li L. Epitope-based peptide vaccines predicted against novel coronavirus disease caused by SARS-CoV-2. bioRxiv. 2020 doi: 10.1016/j.virusres.2020.198082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li X., Wang B., Liu W., Gui M., Peng Z., Meng S. Blockage of conformational changes of heat shock protein gp96 on cell membrane by a alpha-helix peptide inhibits HER2 dimerization and signaling in breast cancer. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0124647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Liaw Y.F., Chu C.M. Hepatitis B virus infection. Lancet. 2009;373(9663):582–592. doi: 10.1016/S0140-6736(09)60207-5. [DOI] [PubMed] [Google Scholar]
- 122.Lim E.K., Guk K., Kim H., Chung B.H., Jung J. Simple, rapid detection of influenza a (H1N1) viruses using a highly sensitive peptide-based molecular beacon. Chem. Commun. (Camb.) 2016;52(1):175–178. doi: 10.1039/c5cc05684e. [DOI] [PubMed] [Google Scholar]
- 123.Lin J.T., Zhang J.S., Su N., Xu J.G., Wang N., Chen J.T., et al. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir. Ther. 2007;12(7):1107–1113. [PubMed] [Google Scholar]
- 124.Lin Y., Shen X., Yang R.F., Li Y.X., Ji Y.Y., He Y.Y., et al. Identification of an epitope of SARS-coronavirus nucleocapsid protein. Cell Res. 2003;13(3):141–145. doi: 10.1038/sj.cr.7290158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Linderoth N.A., Popowicz A., Sastry S. Identification of the peptide-binding site in the heat shock chaperone/tumor rejection antigen gp96 (Grp94) J. Biol. Chem. 2000;275(8):5472–5477. doi: 10.1074/jbc.275.8.5472. [DOI] [PubMed] [Google Scholar]
- 126.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu Q., Somiya M., Kuroda S. Elucidation of the early infection machinery of hepatitis B virus by using bio-nanocapsule. World J. Gastroenterol. 2016;22(38):8489–8496. doi: 10.3748/wjg.v22.i38.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Liu Z., Wang Z., Liu Y., Dong W., Qi Y. Analysis of proteins that interact with nucleocapsid protein of SARS-CoV using 15-mer phage-displayed library. Chin. Sci. Bull. 2007;52(15):2072–2080. doi: 10.1007/s11434-007-0303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lok A.S., Pan C.Q., Han S.H., Trinh H.N., Fessel W.J., Rodell T., et al. Randomized phase II study of GS-4774 as a therapeutic vaccine in virally suppressed patients with chronic hepatitis B. J. Hepatol. 2016;65(3):509–516. doi: 10.1016/j.jhep.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 130.Lopez-Martinez R., Ramirez-Salinas G.L., Correa-Basurto J., Barron B.L. Inhibition of influenza a virus infection in vitro by peptides designed in silico. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0076876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lou Z., Sun Y., Rao Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014;35(2):86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lu C.H., Zhang Y., Tang S.F., Fang Z.B., Yang H.H., Chen X., et al. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 2012;31(1):439–444. doi: 10.1016/j.bios.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 133.Lu W., Wu X.D., Shi M.D., Yang R.F., He Y.Y., Bian C., et al. Synthetic peptides derived from SARS coronavirus S protein with diagnostic and therapeutic potential. FEBS Lett. 2005;579(10):2130–2136. doi: 10.1016/j.febslet.2005.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mahlapuu M., Håkansson J., Ringstad L., Björn C. antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 2016;6(194) doi: 10.3389/fcimb.2016.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malonis R.J., Lai J.R., Vergnolle O. Peptide-based vaccines: current progress and future challenges. Chem. Rev. 2020;120(6):3210–3229. doi: 10.1021/acs.chemrev.9b00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Matsubara T., Kubo A., Sato T. Detection of influenza virus by agglutination using nanoparticles conjugated with a sialic acid-mimic peptide. Polym. J. 2020;52(2):261–266. [Google Scholar]
- 137.Matsubara T., Onishi A., Saito T., Shimada A., Inoue H., Taki T., et al. Sialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J. Med. Chem. 2010;53(11):4441–4449. doi: 10.1021/jm1002183. [DOI] [PubMed] [Google Scholar]
- 138.Matsubara T., Ujie M., Yamamoto T., Akahori M., Einaga Y., Sato T. Highly sensitive detection of influenza virus by boron-doped diamond electrode terminated with sialic acid-mimic peptide. Proc. Natl. Acad. Sci. U. S. A. 2016;113(32):8981–8984. doi: 10.1073/pnas.1603609113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.McGregor D.P. Discovering and improving novel peptide therapeutics. Curr. Opin. Pharmacol. 2008;8(5):616–619. doi: 10.1016/j.coph.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 140.Meloen R.H., Langeveld J.P., Schaaper W.M., Slootstra J.W. Synthetic peptide vaccines: unexpected fulfillment of discarded hope? Biologicals. 2001;29(3-4):233–236. doi: 10.1006/biol.2001.0298. [DOI] [PubMed] [Google Scholar]
- 141.Meng Q., Dong T., Chen X., Tong B., Qian X., Che J., et al. Pharmacokinetics of sifuvirtide in treatment-naive and treatment-experienced HIV-infected patients. J. Pharm. Sci. 2014;103(12):4038–4047. doi: 10.1002/jps.24174. [DOI] [PubMed] [Google Scholar]
- 142.Michel M.L., Bourgine M., Fontaine H., Pol S. Therapeutic vaccines in treating chronic hepatitis B: the end of the beginning or the beginning of the end? Med. Microbiol. Immunol. 2015;204(1):121–129. doi: 10.1007/s00430-014-0381-y. [DOI] [PubMed] [Google Scholar]
- 143.Moderna I. 2020. Moderna Announces Phase 3 COVE Study of mRNA Vaccine Against COVID-19 (mRNA-1273) Begins. [Google Scholar]
- 144.Mookherjee N., Anderson M.A., Haagsman H.P., Davidson D.J. Antimicrobial host defence peptides: functions and clinical potential. Nat. Rev. Drug Discov. 2020;19(5):311–332. doi: 10.1038/s41573-019-0058-8. [DOI] [PubMed] [Google Scholar]
- 145.Moseri A., Tantry S., Sagi Y., Arshava B., Naider F., Anglister J. An optimally constrained V3 peptide is a better immunogen than its linear homolog or HIV-1 gp120. Virology. 2010;401(2):293–304. doi: 10.1016/j.virol.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Muhamad A., Ho K.L., Rahman M.B., Tejo B.A., Uhrin D., Tan W.S. Hepatitis B virus peptide inhibitors: solution structures and interactions with the viral capsid. Org. Biomol. Chem. 2015;13(28):7780–7789. doi: 10.1039/c5ob00449g. [DOI] [PubMed] [Google Scholar]
- 147.Naito T., Izumi K., Kodama E., Sakagami Y., Kajiwara K., Nishikawa H., et al. SC29EK, a peptide fusion inhibitor with enhanced alpha-helicity, inhibits replication of human immunodeficiency virus type 1 mutants resistant to enfuvirtide. Antimicrob. Agents Chemother. 2009;53(3):1013–1018. doi: 10.1128/AAC.01211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ni Y., Sonnabend J., Seitz S., Urban S. The pre-s2 domain of the hepatitis B virus is dispensable for infectivity but serves a spacer function for L-protein-connected virus assembly. J. Virol. 2010;84(8):3879–3888. doi: 10.1128/JVI.02528-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nicol M.Q., Ligertwood Y., Bacon M.N., Dutia B.M., Nash A.A. a novel family of peptides with potent activity against influenza a viruses. J. Gen. Virol. 2012;93(Pt 5):980–986. doi: 10.1099/vir.0.038679-0. [DOI] [PubMed] [Google Scholar]
- 150.Ning Wang S.-Y.L., Yang Xing-Lou, Huang Hui-Min, Zhang Yu-Ji, Guo Hua, Luo Chu-Ming, Miller Maureen, Zhu Guangjian, Chmura Aleksei A., Hagan Emily, Zhou Ji-Hua, Zhang Yun-Zhi, Wang Lin-Fa, Daszak Peter, Shi Zheng-Li. Serological evidence of Bat SARS-related coronavirus infection in humans, China. Virol. Sin. 2018;33(1) doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nyanguile O. Peptide antiviral strategies as an alternative to treat lower respiratory viral infections. Front. Immunol. 2019;10:1366. doi: 10.3389/fimmu.2019.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.O’Connell R.J., Merritt T.M., Malia J.A., VanCott T.C., Dolan M.J., Zahwa H., et al. Performance of the OraQuick rapid antibody test for diagnosis of human immunodeficiency virus type 1 infection in patients with various levels of exposure to highly active antiretroviral therapy. J. Clin. Microbiol. 2003;41(5):2153–2155. doi: 10.1128/JCM.41.5.2153-2155.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ondieki M.C., Nyaribari M.C., Ejekwumadu N.J., Nyakang’o M.J. Design of a recombinant hepatitis B vaccine based on stably binding HLA-I peptides. J. Biomol. Res. Ther. 2015;4(120):2. [Google Scholar]
- 154.Ori A., Zauberman A., Doitsh G., Paran N., Oren M., Shaul Y. p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998;17(2):544–553. doi: 10.1093/emboj/17.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1):1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Passioura T., Watashi K., Fukano K., Shimura S., Saso W., Morishita R., et al. De Novo macrocyclic peptide inhibitors of hepatitis B virus cellular entry. Cell Chem. Biol. 2018;25(7) doi: 10.1016/j.chembiol.2018.04.011. 906-15 e5. [DOI] [PubMed] [Google Scholar]
- 157.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Penberthy W.T., Chari S., Cole A.L., Cole A.M. Retrocyclins and their activity against HIV-1. Cell. Mol. Life Sci. 2011;68(13):2231–2242. doi: 10.1007/s00018-011-0715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Petersen J., Dandri M., Mier W., Lutgehetmann M., Volz T., von Weizsacker F., et al. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat. Biotechnol. 2008;26(3):335–341. doi: 10.1038/nbt1389. [DOI] [PubMed] [Google Scholar]
- 160.2020. Pfizer. Pfizer and BioNTech Announce Early Positive Update from German Phase 1/2 Covid-19 Vaccine Study, Including First T Cell Response Data. [Google Scholar]
- 161.2020. Pfizer. Pfizer And BioNTech Choose Lead mRNA Vaccine Candidate Against Covid-19 and Commence Pivotal Phase 2/3 Global Study. [Google Scholar]
- 162.Pitisuttithum P., Gilbert P., Gurwith M., Heyward W., Martin M., van Griensven F., et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J. Infect. Dis. 2006;194(12):1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 163.Pleguezuelos O., James E., Fernandez A., Lopes V., Rosas L.A., Cervantes-Medina A., et al. Efficacy of FLU-v, a broad-spectrum influenza vaccine, in a randomized phase IIb human influenza challenge study. NPJ Vaccines. 2020;5:22. doi: 10.1038/s41541-020-0174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pleguezuelos O., Robinson S., Stoloff G.A., Caparros-Wanderley W. Synthetic influenza vaccine (FLU-v) stimulates cell mediated immunity in a double-blind, randomised, placebo-controlled phase I trial. Vaccine. 2012;30(31):4655–4660. doi: 10.1016/j.vaccine.2012.04.089. [DOI] [PubMed] [Google Scholar]
- 165.Poisson F., Severac A., Hourioux C., Goudeau A., Roingeard P. Both pre-S1 and S domains of hepatitis B virus envelope proteins interact with the core particle. Virology. 1997;228(1):115–120. doi: 10.1006/viro.1996.8367. [DOI] [PubMed] [Google Scholar]
- 166.Portal E.B.R. 2020. ProtParam Tool.https://web.expasy.org/protparam/ [Google Scholar]
- 167.Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., et al. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 2003;348(20):1995–2005. doi: 10.1056/NEJMoa030634. [DOI] [PubMed] [Google Scholar]
- 168.Predictor S.P. 2020. ANTIGENpro. [Google Scholar]
- 169.Pu J., Wang Q., Xu W., Lu L., Jiang S. Development of protein- and peptide-based HIV entry inhibitors targeting gp120 or gp41. Viruses. 2019;11(8) doi: 10.3390/v11080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Qian L., Fan H., Ju Y., Chen L., Li X., Ye X., et al. A peptide-based inhibitor of gp96 suppresses HBsAg expression and HBV replication by upregulation of p53. J. Gen. Virol. 2019;100(8):1241–1252. doi: 10.1099/jgv.0.001289. [DOI] [PubMed] [Google Scholar]
- 171.Qiushi Liu M.S., Iijima Masumi, Tatematsua Kenji, Kuroda Shun’ichi. A hepatitis B virus-derived human hepatic cellspecific heparin-binding peptide: identification and application to a drug delivery system. Biomater. Sci. 2019;7:322. doi: 10.1039/c8bm01134f. [DOI] [PubMed] [Google Scholar]
- 172.Laskowski MWM R.A., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;(26):283–291. [Google Scholar]
- 173.Redman J.S., Francis J.N., Marquardt R., Papac D., Mueller A.L., Eckert D.M., et al. Pharmacokinetic and chemical synthesis optimization of a potent d-peptide HIV entry inhibitor suitable for extended-release delivery. Mol. Pharm. 2018;15(3):1169–1179. doi: 10.1021/acs.molpharmaceut.7b01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Rerks-Ngarm S., Pitisuttithum P., Nitayaphan S., Kaewkungwal J., Chiu J., Paris R., et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 175.Resource I.A. 2020. MHC-II Binding Predictions. [Google Scholar]
- 176.Richardson S.E., Tellier R., Mahony J. The laboratory diagnosis of severe acute respiratory syndrome: emerging laboratory tests for an emerging pathogen. Clin. Biochem. Rev. 2004;25(2):133–141. [PMC free article] [PubMed] [Google Scholar]
- 177.Ringel O., Muller K., Koch J., Brill B., Wolf T., Stephan C., et al. Optimization of the EC26-2A4 epitope in the gp41 membrane proximal external region targeted by neutralizing antibodies from an elite controller. AIDS Res. Hum. Retroviruses. 2018;34(4):365–374. doi: 10.1089/aid.2017.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Rockstroh J.K., Asmuth D., Pantaleo G., Clotet B., Podzamczer D., van Lunzen J., et al. Re-boost immunizations with the peptide-based therapeutic HIV vaccine, vacc-4x, restores geometric mean viral load set-point during treatment interruption. PLoS One. 2019;14(1) doi: 10.1371/journal.pone.0210965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rut W. Substrate specificity profiling of SARS-CoV-2 main protease enables design of activity-based probes for patient-sample imaging. bioRxiv. 2020 [Google Scholar]
- 180.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., et al. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. bioRxiv. 2020 doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Saha S., Raghava G.P. AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res. 2006;34(Web Server issue):W202–W209. doi: 10.1093/nar/gkl343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Saha S., Raghava G.P. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 183.Sapsford K.E., Blanco-Canosa J.B., Dawson P.E., Medintz I.L. Detection of HIV-1 specific monoclonal antibodies using enhancement of dye-labeled antigenic peptides. Bioconjug. Chem. 2010;21(2):393–398. doi: 10.1021/bc9003712. [DOI] [PubMed] [Google Scholar]
- 184.Scala M.C., Sala M., Pietrantoni A., Spensiero A., Di Micco S., Agamennone M., et al. Lactoferrin-derived peptides active towards influenza: identification of three potent tetrapeptide inhibitors. Sci. Rep. 2017;7(1):10593. doi: 10.1038/s41598-017-10492-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.server Pd. http://db.systemsbiology.net:8080/proteomicsToolkit/proteinDigest.html.
- 186.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 187.Shtyrya Y.A., Mochalova L.V., Bovin N.V. Influenza virus neuraminidase: structure and function. Acta Nat. 2009;1(2):26–32. [PMC free article] [PubMed] [Google Scholar]
- 188.Sisk J.M., Frieman M.B. Screening of FDA-approved drugs for treatment of emerging pathogens. ACS Infect. Dis. 2015;1(9):401–402. doi: 10.1021/acsinfecdis.5b00089. [DOI] [PubMed] [Google Scholar]
- 189.Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H.D., et al. SARS--beginning to understand a new virus. Nat. Rev. Microbiol. 2003;1(3):209–218. doi: 10.1038/nrmicro775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Stoloff G.A., Caparros-Wanderley W. Synthetic multi-epitope peptides identified in silico induce protective immunity against multiple influenza serotypes. Eur. J. Immunol. 2007;37(9):2441–2449. doi: 10.1002/eji.200737254. [DOI] [PubMed] [Google Scholar]
- 191.Tapia G., Højen J.F., Ökvist M., Olesen R., Leth S., Nissen S.K., et al. Sequential vacc-4x and romidepsin during combination antiretroviral therapy (cART): immune responses to vacc-4x regions on p24 and changes in HIV reservoirs. J. Infect. 2017;75(6):555–571. doi: 10.1016/j.jinf.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 192.Tassopoulos N.C., Volpes R., Pastore G., Heathcote J., Buti M., Goldin R.D., et al. Efficacy of lamivudine in patients with hepatitis B e antigen-negative/hepatitis B virus DNA-positive (precore mutant) chronic hepatitis B. lamivudine precore mutant study group. Hepatology. 1999;29(3):889–896. doi: 10.1002/hep.510290321. [DOI] [PubMed] [Google Scholar]
- 193.Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020 doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tinoco A.D., Saghatelian A. Investigating endogenous peptides and peptidases using peptidomics. Biochemistry. 2011;50(35):7447–7461. doi: 10.1021/bi200417k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Tiollais P., Pourcel C., Dejean A. The hepatitis B virus. Nature. 1985;317(6037):489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 196.ToxinPred. https://webs.iiitd.edu.in/raghava/toxinpred/index.html.
- 197.Tripathi S., Wang G., White M., Qi L., Taubenberger J., Hartshorn K.L. Antiviral activity of the human cathelicidin, LL-37, and derived peptides on seasonal and pandemic influenza a viruses. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0124706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tseng C.T., Sbrana E., Iwata-Yoshikawa N., Newman P.C., Garron T., Atmar R.L., et al. Immunization with SARS coronavirus vaccines leads to pulmonary immunopathology on challenge with the SARS virus. PLoS One. 2012;7(4) doi: 10.1371/journal.pone.0035421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Uhl P., Helm F., Hofhaus G., Brings S., Kaufman C., Leotta K., et al. A liposomal formulation for the oral application of the investigational hepatitis B drug myrcludex B. Eur. J. Pharm. Biopharm. 2016;103:159–166. doi: 10.1016/j.ejpb.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 200.UNAIDS; 2020. AIDS Data. [Google Scholar]
- 201.v2.0 T. http://www.cbs.dtu.dk/services/TMHMM/.
- 202.Vemula S.V., Zhao J., Liu J., Wang X., Biswas S., Hewlett I. Current approaches for diagnosis of influenza virus infections in humans. Viruses. 2016;8(4):96. doi: 10.3390/v8040096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Volz T., Allweiss L., Ben M.M., Warlich M., Lohse A.W., Pollok J.M., et al. The entry inhibitor myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J. Hepatol. 2013;58(5):861–867. doi: 10.1016/j.jhep.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 204.Walsh C.T., O’Brien R.V., Khosla C. Nonproteinogenic amino acid building blocks for nonribosomal peptide and hybrid polyketide scaffolds. Angew. Chem. Int. Ed. Engl. 2013;52(28):7098–7124. doi: 10.1002/anie.201208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Wang G. Natural antimicrobial peptides as promising anti-HIV candidates. Curr. Top. Pept. Protein Res. 2012;13:93–110. [PMC free article] [PubMed] [Google Scholar]
- 206.Watashi K. HBV slow maturation process leads to infection. Trends Microbiol. 2016;24(8):597–599. doi: 10.1016/j.tim.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 207.Watashi K., Sluder A., Daito T., Matsunaga S., Ryo A., Nagamori S., et al. Cyclosporin a and its analogs inhibit hepatitis B virus entry into cultured hepatocytes through targeting a membrane transporter, sodium taurocholate cotransporting polypeptide (NTCP) Hepatology. 2014;59(5):1726–1737. doi: 10.1002/hep.26982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.WHO. (https://www.who.int/en/news-room/fact-sheets/detail/hepatitis-b) 2015.
- 209.WHO. Hepatitis B, https://www.who.int/en/news-room/fact-sheets/detail/hepatitis-b#:-:text=Hepatitis%20B%20is%20a%20potentially%20life-threatening%20liver%20infection,risk%20of%20death%20from%20cirrhosis%20and%20liver%20cancer; 2020.
- 210.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407–W4010. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Wild C., Oas T., McDanal C., Bolognesi D., Matthews T. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. U. S. A. 1992;89(21):10537–10541. doi: 10.1073/pnas.89.21.10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Winckelmann A., Morcilla V., Shao W., Schleimann M.H., Hojen J.F., Schlub T.E., et al. Genetic characterization of the HIV-1 reservoir after Vacc-4x and romidepsin therapy in HIV-1-infected individuals. Aids. 2018;32(13):1793–1802. doi: 10.1097/QAD.0000000000001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wong J.H., Legowska A., Rolka K., Ng T.B., Hui M., Cho C.H., et al. Effects of cathelicidin and its fragments on three key enzymes of HIV-1. Peptides. 2011;32(6):1117–1122. doi: 10.1016/j.peptides.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 214.World Health Organization Multicentre Collaborative Network for Severe Acute Respiratory Syndrome D A multicentre collaboration to investigate the cause of severe acute respiratory syndrome. Lancet. 2003;361(9370):1730–1733. doi: 10.1016/S0140-6736(03)13376-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microb. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Wu B., Chu X., Feng C., Hou J., Fan H., Liu N., et al. Heat shock protein gp96 decreases p53 stability by regulating Mdm2 E3 ligase activity in liver cancer. Cancer Lett. 2015;359(2):325–334. doi: 10.1016/j.canlet.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 218.Wu C.H., Liu I.J., Lu R.M., Wu C.H. Advancement and applications of peptide phage display technology in biomedical science. J. Biomed. Sci. 2016;23:8. doi: 10.1186/s12929-016-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wyatt R., Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280(5371):1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 221.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Xie D., Yao C., Wang L., Min W., Xu J., Xiao J., et al. An albumin-conjugated peptide exhibits potent anti-HIV activity and long in vivo half-life. Antimicrob. Agents Chemother. 2010;54(1):191–196. doi: 10.1128/AAC.00976-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Xiong S., Borrego P., Ding X., Zhu Y., Martins A., Chong H., et al. A helical short-peptide fusion inhibitor with highly potent activity against human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus. J. Virol. 2017;91(1) doi: 10.1128/JVI.01839-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Xu D.Z., Wang X.Y., Shen X.L., Gong G.Z., Ren H., Guo L.M., et al. Results of a phase III clinical trial with an HBsAg-HBIG immunogenic complex therapeutic vaccine for chronic hepatitis B patients: experiences and findings. J. Hepatol. 2013;59(3):450–456. doi: 10.1016/j.jhep.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 225.Ye X., Zhou M., He Y., Wan Y., Bai W., Tao S., et al. Efficient inhibition of hepatitis B virus infection by a preS1-binding peptide. Sci. Rep. 2016;6:29391. doi: 10.1038/srep29391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yoshikawa M. Bioactive peptides derived from natural proteins with respect to diversity of their receptors and physiological effects. Peptides. 2015;72:208–225. doi: 10.1016/j.peptides.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 227.Yu F., Lu L., Du L., Zhu X., Debnath A.K., Jiang S. Approaches for identification of HIV-1 entry inhibitors targeting gp41 pocket. Viruses. 2013;5(1):127–149. doi: 10.3390/v5010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhang G. The first-in-class peptide binder to the SARS-CoV-2 spike protein. bioRxiv. 2020 [Google Scholar]
- 229.Zhang T.Y., Guo X.R., Wu Y.T., Kang X.Z., Zheng Q.B., Qi R.Y., et al. A unique B cell epitope-based particulate vaccine shows effective suppression of hepatitis B surface antigen in mice. Gut. 2020;69(2):343–354. doi: 10.1136/gutjnl-2018-317725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Zhao H. A broad-spectrum virus- and host-targeting antiviral peptide against SARS-CoV-2 and other respiratory viruses. Nat. Res. 2020 doi: 10.1038/s41467-020-17986-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zhao H., Zhou J., Zhang K., Chu H., Liu D., Poon V.K., et al. A novel peptide with potent and broad-spectrum antiviral activities against multiple respiratory viruses. Sci. Rep. 2016;6:22008. doi: 10.1038/srep22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zheng B.J., Guan Y., Hez M.L., Sun H., Du L., Zheng Y., et al. Synthetic peptides outside the spike protein heptad repeat regions as potent inhibitors of SARS-associated coronavirus. Antivir. Ther. 2005;10(3):393–403. [PubMed] [Google Scholar]
- 233.Zhou S., Liu R., Baroudy B.M., Malcolm B.A., Reyes G.R. The effect of ribavirin and IMPDH inhibitors on hepatitis C virus subgenomic replicon RNA. Virology. 2003;310(2):333–342. doi: 10.1016/s0042-6822(03)00152-1. [DOI] [PubMed] [Google Scholar]
- 234.Zhu X., Zhu Y., Ye S., Wang Q., Xu W., Su S., et al. Improved pharmacological and structural properties of HIV fusion inhibitor AP3 over enfuvirtide: highlighting advantages of artificial peptide strategy. Sci. Rep. 2015;5:13028. doi: 10.1038/srep13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]