Abstract
Thickening and the subsequent invagination of the epithelium are an important initial step in ectodermal organ development. Ikkα has been shown to play a critical role in controlling epithelial growth, since Ikkα mutant mice show protrusions (evaginations) of incisor tooth, whisker and hair follicle epithelium rather than invagination. We show here that mutation of the Interferon regulatory factor (Irf) family, Irf6 also results in evagination of incisor epithelium. In common with Ikkα mutants, Irf6 mutant evagination occurs in a NF-κB-independent manner and shows the same molecular changes as those in Ikkα mutants. Irf6 thus also plays a critical role in regulating epithelial invagination. In addition, we also found that canonical Wnt signaling is upregulated in evaginated incisor epithelium of both Ikkα and Irf6 mutant embryos.
Keywords: Ikkα, Irf6, Wnt, Incisor, Invagination, Epithelium, Tooth development
Introduction
Many organs (e.g. lung, liver, kidney, glands, eyes, hair) develop through reciprocal epithelial–mesenchymal interactions and share similar signaling pathways such as Bmp, Shh, Fgf, Wnt and Tgf at early stages of their development. At the early stages, these organs also show similar morphological features, consisting of a thickening and subsequent invagination of the epithelium. Teeth arise from a series of reciprocal interactions between the oral epithelium and underlying neural crest-derived mesenchyme (for reviews, see Thesleff, 2006; Tucker and Sharpe, 2004). The first morphological sign of tooth development is a narrow band of thickened epithelium on the developing jaw primodia and subsequent localized invagination into underlying mesenchyme to form buds. The bud epithelium progressively takes the form of the cap and bell configurations as differentiation proceeds. Epithelial cells and mesenchymal cells (dental papilla) then differentiate into enamel-producing ameloblasts and dentin-producing odontoblasts, respectively.
The nuclear Factor kappa B (NF-κB) pathway plays a major role in many physiological and pathological process including immune response to infection and cancer (for review see Chariot, 2009; Häcker and Karin, 2006; Li and Verma, 2002; Sanz et al., 2010). In mammals, 12 NF-κB homo- or hetero-dimers are formed among the five proteins NF-κB1 (p50, generated from p105), NF-κB2 (p52, generated from p100), RelA (p65), RelB and c-Rel. NF-κB exists in the cytoplasm as an inactive form that is associated with inhibitory proteins termed Inhibitor of κB (IκB). Activation of the NF-κB pathway results in nuclear translocation of NF-κB proteins, and can proceed either through classical/canonical, alternative/noncanonical or hybrid pathways. Classical NF-κB activation is usually a rapid and transient response to a wide range of stimuli. In nonstimulated cells, IκB acts to retain NF-κB in the cytoplasm by masking the nuclear localization sequence. Exposure to stimuli results in rapid phosphorylation of IκB that leads to site-specific ubiquitination and degradation. The resulting free NF-κB dimers translocate to the nucleus and regulate target gene transcription. The protein kinase that phosphorylates IκB in response to stimuli is a multiprotein complex, IκB kinase (IKK), composed of two catalytic subunits, IKKα and IKKβ, and a regulatory subunit, IKKγ (also called NEMO). The alternative pathway involves slow activation of NF-κB leading to prolonged activation of NF-κB targets genes. NF-κB inducing kinase (NIK) recruits IKKα that phosphorylates p100, promoting p100 polyubiquitiation and subsequent proteasomal processing to p52, generating RelB/p52 dimers which facilitates full activation of the pathway. In addition, Ikkα is also known to function outside of the IKK complex in an NF-κB-independent manner. Ikkα is a critical regulator of keratinocyte differentiation by an NF-κB-independent manner, although the exact mechanism through which Ikkα acts is unclear. Taut undifferentiated epidermis, limb truncation, cleft palate, abnormal molar cusp shape and adhesion of oral epithelium are also observed in Ikkα mutant mice (Hu et al., 1999; Li et al., 1999; Takeda et al., 1999). In addition to these phenotypes, incisor tooth epithelium of Ikkα mutant mice fails to invaginate into the underlying mesenchyme and instead evaginates into the oral cavity, suggesting that Ikkα has a regulatory role in guiding directionality of developing tooth germs. This role of Ikkα is independent of the NF-κB (Ohazama et al., 2004).
Interferons (IFNS) play critical roles in many biological processes including the homeostasis and function of immune systems (Bonjardim et al., 2009; Hertzog et al., 2011). Ifn regulatory factor (Irf) genes regulate the transcription of interferons, proteins produced in response to the presence of pathogens, and function as an integral part of the immune system (Platanias, 2005). The Irf family is comprised of nine members (Irf1–Irf9) that share a highly conserved N-terminal, penta-tryptophan, helix-turn-helix DNA-binding domain and a less well-conserved protein-binding domain (Taniguchi et al., 2001). Many members of Irf family are known to activate the canonical NF-kB pathway (Hiscott, 2007; Taniguchi et al., 2001). Ikkα is also involved in Toll-like receptor (TLR)7- and TLR9-mediated IFNα induction in plasmacytoid dendritic cells via Irf7 phosphorylation as NF-kB independent manners (Hoshino et al., 2006). Mutation in IRF6 has previously been shown to cause Van der Woude syndrome (VWS) and poplyteal pterigium syndrome (PPS; Kondo et al., 2002). VWS is an autosomal dominant disorder of facial development that is characterized by cleft lip and palate and is the most common form of syndromic orofacial clefting (Van der Woude, 1954). PPS has a similar orofacial phenotype to VWS, but includes popliteal webbing, pterygia, oral synychiae, adhesions between the eyelids, syndactyly and genital anomalies (Bixler et al., 1973; Froster-Iskenius, 1990). Irf6 mutant mice show cleft palate, adhesion of oral epithelium, limb truncation and taut undifferentiated epidermis, all phenotypes also seen in Ikkα mutants (Hu et al., 1999; Ingraham et al., 2006; Li et al., 1999; Richardson et al., 2006; Takeda et al., 1999).
Thickening and the subsequent invagination of the epithelium is an important initial step in tooth development. Although several new insights have recently been revealed, the molecular mechanisms regulating epithelial invagination still remain unclear (Charles et al., 2011; Munne et al., 2009). We show here that null mutation of the Irf6 results in identical incisor phenotypes (evagination of incisor epithelium) and similar molecular changes to those in Ikkα mutants. Despite these shared phenotypes and associated molecular changes, crosses between Ikkα and Irf6 mutants failed to reveal any evidence of a genetic interaction in regulating tooth epithelial invagination.
Materials and methods
Production and analysis of transgenic mice
The production of mice with mutation of Ikkα, Irf6, Jagged2 (Jag2), Rip4 and Stratifin (Sfn; SfnEr/Er) has previously been described (Guenet et al., 1979; Holland et al., 2002; Hu et al., 1999; Jiang et al., 1998; Richardson et al., 2006). Mice overexpressing Ikkα under keratin (K) 5 promotor (K5-Ikkα) have been described previously (Lomada et al., 2007). Production of the NF-kB reporter [(Igκ)3conalacZ] and Axin2 reporter (Axin2lacZ) mice has also previously been described (Lustig et al., 2002; Schmidt-Ullrich et al., 1996). The Irf6 hypomorph allele (Irf6neo) was created as an intermediate step toward the construction of an Irf6 conditional knockout targeting vector. In addition to loxP sites, we inserted the neomycin resistance gene (neo), under the control of a constitutive promoter, as a selectable marker for the insertion event. To create this construct, we screened a BAC library derived from 129/SV strain and identified clone RPCI22-516G. From this BAC, we subcloned a 1.8 kb KpnI–BamHI fragment for the 5′-arm and a 3.9 kb BamHI–HindIII fragment for the 3′-arm into the KpnI and HindIII sites of pBluescript II SK(—) vector. The 3 kb BamHI fragment, located between the 5′- and 3′-arms was cloned into the SalI site in ploxP3NeopA vector (kind gift from Dr. Yagi and Dr. Hirabayashi, Osaka University). This vector was cut with XhoI, which liberated a 5.8 kb fragment that contained the floxed exons (3 and 4) and the Pgk-neo cassette, and was subcloned into the BamHI site located at the junction of the 5′ and 3′ arms. This gene targeting vector was digested with NotI and electroporated into ES cells derived from 129/SV strain. We screened 384 ES clones by PCR. After G418 selection, four clones were positive. The positive ES cells were microinjected into C57BL/6 blastocysts, and we obtained nine male chimeras that were mated with C57BL/6 mice. Germline transmission was verified as previously described (Ingraham et al., 2006).
Embryonic day 0 (E0) was taken to be midnight prior to finding a vaginal plug. Embryos were harvested at the appropriate time and genotyped using PCR analysis of genomic DNA extracted from unused embryonic tissue. Embryonic heads were fixed in 4% paraformaldehyde (PFA), wax embedded and serially sectioned at 7 μm. Sections were split over 5–10 slides and prepared for histology and radioactive in situ hybridisation.
In situ hybridisation
Radioactive in situ hybridisation with [35S]UTP-labeled riboprobes was carried out as described previously (Ohazama et al., 2008). Decalcification using 0.5 M EDTA (pH 7.6) was performed after fixation of newborn mice. The radioactive antisense probes were generated from mouse cDNA clones that were gifts from T.A. Mitsiadis (Notch1, Notch2), A. McMahon (Shh) or were obtained from RZPD (Irf1, Irf2, Irf3, Irf4, Irf5, Irf7, Irf8, Irf9) or Geneservice (Sfn).
Immunohistochemistry
After deparaffinization, sections were treated by proteinase K and then incubated with antibody to phosphorylated Smad1, Smad5 and Smad8 (p-Smad1/5/8; Cell signaling Technology) or CD44 (Chemicon). As a negative control, normal rabbit or rat serum was used instead of primary antibody. Tyramide signal amplification system was performed (Perkin Elmer Life Science or DAKO) for detecting primary antibody.
Apoptotic activity
For detecting apoptoptic cells, we used the Apoptag plus fluorescein in situ apoptosis detection kit (Chemicon), according to manufacturer's protocol.
β-galactosidase staining
Embryo heads of lacZ-reporter and reporter-crossed mice were fixed for 1 h at 4 °C in 1% PFA and 0.2% glutaraldehyde in nuclease-free phosphate buffered saline (PBS). X-gal staining was performed at 37 °C in 4 mM potassium ferrocyanide, 4 mM potassium ferricyanide, 2 mM MgCl2, and 400 μg ml−1 X-gal (4-chloro-5-bromo-3-indolyl-β-galactoside, Sigma) in 1x PBS for 4 h (Axin2lacZ mice) or 27 h [(Igκ)3conalacZ mice]. Afterwards, embryo heads were washed several times in 1x PBS and post-fixed in 2.5% glutaraldehyde. The embryo heads were vacuum wax embedded and 10 μm sections cut using a microtome. For analysis, sections were counterstained with eosin.
Scanning electron microscope (SEM) ultrastructure analysis
Heads were fixed with 2% glutaraldehyde in 0.1 M Na-cacodylate buffer with 0.1 M sucrose and postfixed with 1% osmium in 0.1 M coated Na-cacodylate buffer. After critical point drying, the samples were coated with gold and photographed using scanning electron microscopy.
Transmission electron microscope (TEM) ultrastructure analysis
Heads were fixed in 2.5% glutaraldehyde (phosphate buffer) overnight at 4 °C and postfixed in 2% osmium tetroxide (Millonigs buffer) for 90 mins at 4 °C after washing with phosphate buffer. Specimens were dehydrated through a graded series of ethanol and embedded in Epon 812-equivalent (TAAB Lab). Semi-thin sections (1 μm) were stained with toluidine blue for light microscopy analysis. Ultra-thin sections (40–90 nm) were cut, stained with uranyl acetate and lead citrate and examined with a Hitachi H7600 transmission electron microscope.
Explants culture
Mandibles from Axin2lacZ reporter or CD1 mouse embryos at E11.5 were dissected in D-MEM containing glutamax-1. The explants were cultured as previously described on membrane filters supported by metal grids (Ohazama et al., 2005). Explants were cultured in D-MEM including 10% FBS supplemented with 100 μM γ-secretase inhibitor (DAPT; LY-374973, N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester; Sigma), DMSO, 20 mM LiCl or H2O. After 1 day culture, the explants were fixed by 4% PFA and prepared for LacZ staining or protein was extracted from the explants.
Results
Irf6 expression in murine tooth development
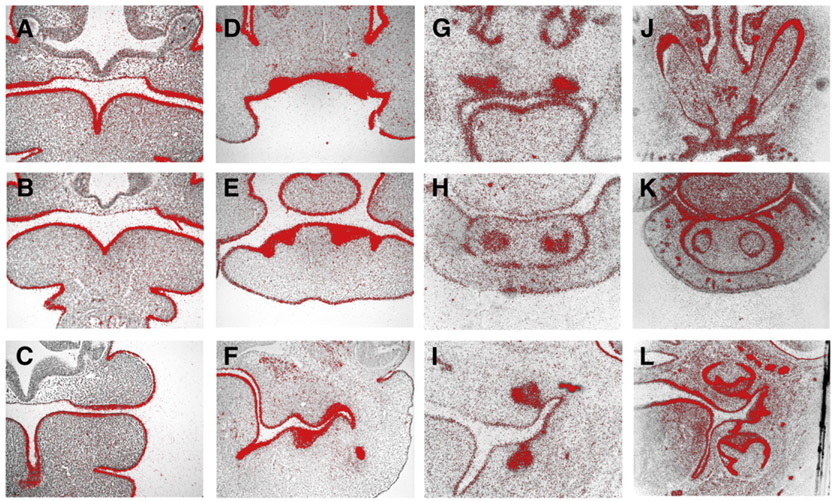
In order to elucidate whether Irf6 plays an integral role in tooth development, we first examined the Irf6 expression pattern during murine tooth development. Radioactive in situ hybridisation revealed that Irf6 was strongly expressed in dental and oral epithelium at all embryonic stages examined (Fig. 1). This included expression in both incisor and molar development.
Fig. 1.
Irf6 expression in murine tooth development. In situ hybridisation of Irf6 on frontal head sections at E10.5 (A–C), E12.5 (D–F), E14.5 (G–I) and E18.5 (J–L). Upper incisors (A, D, G, J); lower incisors (B, E, H, K); molars (C, F, I, L).
To ascertain if other Irf members were involved in incisor tooth development, we analyzed the expression of genes at the late bud stage of odontogenesis. Irf1, Irf2, Irf3, Irf4 and Irf8 were weakly expressed in upper incisor tooth germs whereas weak Irf1, Irf2, Irf3, Irf7 and Irf8 expression were observed in lower incisor tooth germs (Supplementary Fig. 1).
Evagination of incisor tooth in Irf6 mutant mice
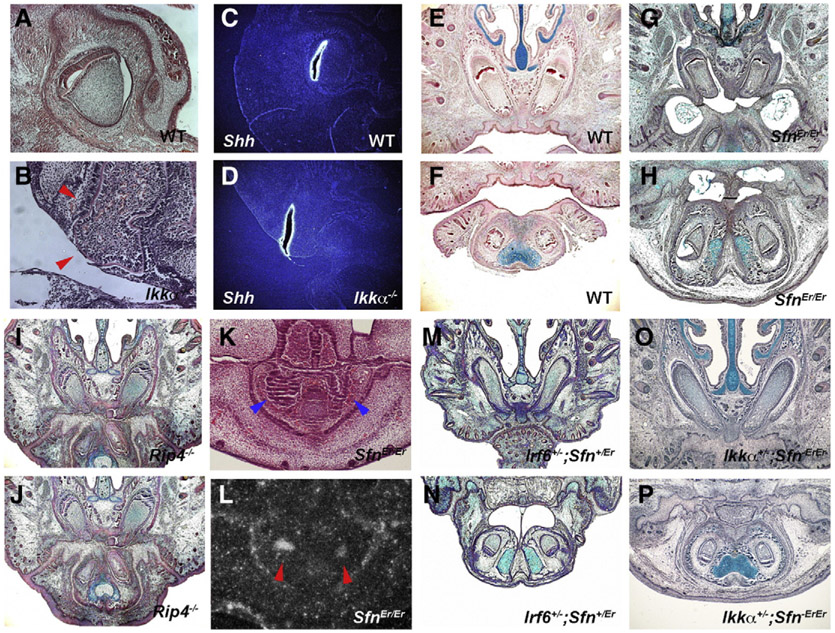
In order to investigate the role of Irf6 in tooth development, we examined sections of heads from newborn Irf6 null mutant mice. Newborn Irf6 mutant mice showed abnormal enlarged anterior extents of their incisors that were prematurely exposed to oral cavity compared to the incisors of wild-types that were covered with alveolar bone and connective tissue (Figs. 1A-D). Mutant incisor tooth germs thus lacked connective tissue, alveolar bone, oral epithelium, and dental lamina (epithelium which connect tooth germs to oral epithelium). To establish the timing of the first appearance of the incisor tooth anomalies, sections of mutant embryos from E10.5 to 14.5 were examined. The first distinguishable abnormalities were observed as abnormal protrusions of epithelium in presumptive incisor regions at E13.5 while wild-type incisors showed invaginating epithelium (Figs. 2E-H). Mutant molars showed inavaginated epithelium at the stage (data not shown). Expression of the primary enamel knot marker gene, Shh normally seen in the center of invaginated incisor epithelium (Fig. 2I), was localized at the outermost tip of the mutant protruded incisor epithelium (Fig. 2J). These results suggest that mutant incisor epithelium does not invaginate into the underlying mesenchyme but rather evaginates outward into the developing oral cavity as seen in Ikkα mutants (Ohazama et al., 2004).
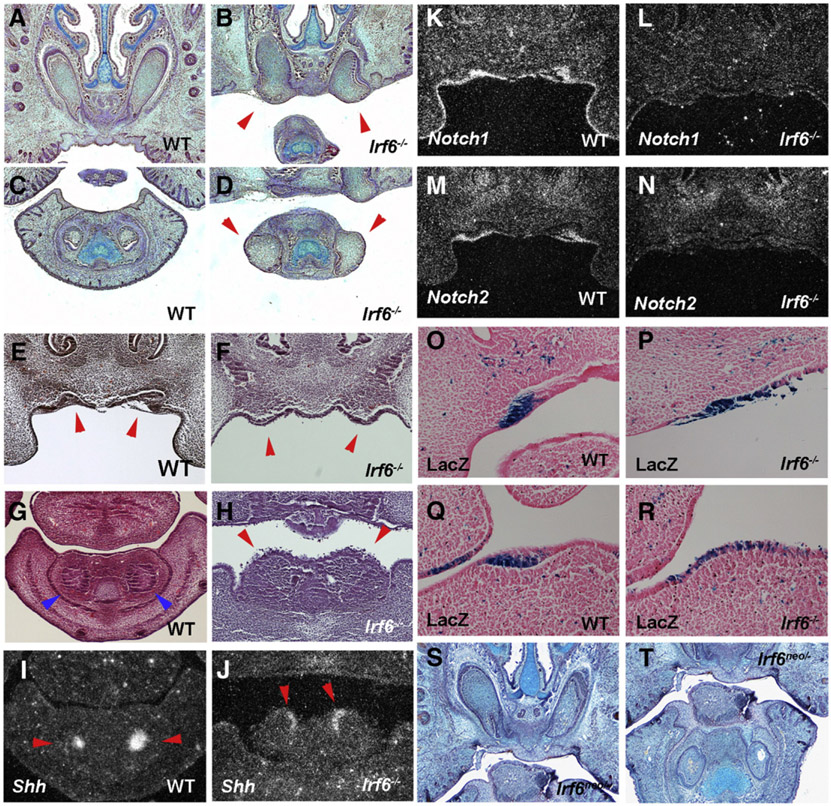
Fig. 2.
Incisor tooth development in Irf6 mutant mice. Anterior extents of incisors are enlarged and exposed to the oral cavity in newborn Irf6 mutants (arrowheads in B, D). Protrusion of incisor epithelium in Irf6 mutants at E13.5 (arrowheads in F, H). Arrowheads indicating wild-type incisors (E, G). Expression of Shh at the outermost tip of evaginating epithelium of incisors in mutant (arrowheads in J) whereas Shh expression at the bottom of invaginating epithelium in wild-type (arrowheads in I). Downregulation of Notch1 (L) and Notch2 (N) in evaginating epithelium of mutant incisors. In situ hybridisation on frontal head sections at E13.5 (I–N). LacZ expression in incisor epithelium in (Igκ)3conalacZ (O, Q) and Irf6−/−;(Igκ)3conalacZ (P, R) mice. Slight epithelial bulge and thickening were seen in mutant incisor (P, R). Incisors in Irf6neo/− (S, T) on frontal sections at E18.5. Upper (A, B, E, F, K-N, O, P, S) and lower (C, D, G–J, Q, R, T) incisors.
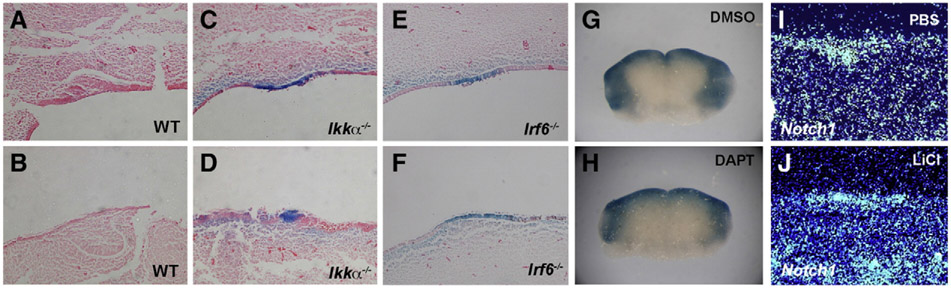
We previously identified changes in Notch1 and Notch2 expression in incisors of Ikkα mutant embryos (Ohazama et al., 2004). In Irf6 mutants, Notch1 and Notch2 expressions were also downregulated in incisor tooth germs (Figs. 2K-N). The Ikkα mutant incisor phenotype has previously been shown to occur independently of the NF-κB pathway (Ohazama et al., 2004). Members of Irf family are known to be involved in NF-κB pathway, although interaction between Irf6 and the NF-κB pathway remains unclear (Hiscott, 2007). In order to investigate whether Irf6 is involved in NF-κB pathway in incisor tooth development, we crossed the Irf6 mutants to an NF-κB reporter mouse [(Igκ)3conalacZ]. LacZ positive cells were slightly expanded in mutant incisor epithelium (Figs. 2O-R). The expression domain of Wnt7b in Ikkα mutant incisor epithelium was smaller than those in wild-types, consistent with the expansion of Shh expression, which were also observed in Irf6 mutants (Supplementary Fig. 2; Ohazama et al., 2004). Incisor tooth epithelial domains were thus expanded in Ikkα mutants, which was also observed in Irf6 mutant mice. It still however remains unclear how Ikkα/Irf6 regulates the size of tooth epithelium. Irf6 mutants also showed no significant changes of IκBα gene expression which is known to be led by NF-κB activation (Supplementary Fig. 3). No significant down- or up-regulation of NF-κB activity was thus observed in the Irf6 mutant embryo incisors, suggesting that, as with Ikkα mutant mice, the incisor evaginating epithelium seen in Irf6 mutant is likely to be independent of the NF-κB pathway (Figs. 2O-R). The Irf6 mutant incisors were thus identical to those of Ikkα mutant mice. No significant tooth anomalies could be detected in mice with null mutants of Irf1, Irf2, Irf3, Irf5, Irf7 and Irf9 (data not shown).
In order to determine whether epithelial invagination is controlled by dosage of Irf6, we generated a series of hypomorphic alleles. The evaginating incisor phenotype was absent in Irf6neo/neo or Irf6neo/− mice (Figs. 2S, T; data not shown). The adhesion of oral epithelium which is one of phenotypes in Irf6 mutants also disappeared in Irf6neo/neo or Irf6neo/− mice. Analysis of this allelic series of Irf6 mutants revealed that the evaginating incisor phenotype only occurs in the complete absence of Irf6 activity.
It is known that Bmp signaling plays critical roles in regulating early stages of incisor tooth development (Tucker and Sharpe, 2004). In order to investigate whether impaired Bmp signaling results in epithelial evagination, phosphorylated Smad1, Smad5 and Smad8 (p-Smad1/5/8) were examined in Irf6 mutants. Significant changes of p-Smad1/5/8 could not be detected in incisor tooth germs of Irf6 mutant mice (Supplementary Fig. 4). In addition to growth factors, cell adhesion is also known to be important in regulating epithelial invagination (Schock and Perrimon, 2002). To understand whether the disruption of cell-adhesion results in evagination, CD44 (one of the epithelial cell-adhesion molecules) was examined in mutants (Goodison et al., 1999). CD44-positive cells were found in wild-type tooth epithelium, which were also observed in mutant evaginated epithelium (Supplementary Fig. 4). Highly coordinated cellular proliferation and apoptosis has been shown to be key factor for controlling early stages of incisor tooth development (Charles et al., 2011; Munne et al., 2009). Abnormalities of cellular proliferation or apoptosis could not be detected in mutants (Supplementary Fig. 4).
Tissue specificity in regulating epithelial invagination
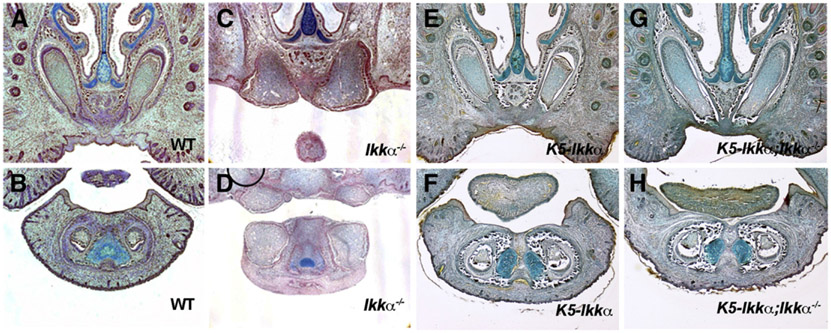
Tooth formation requires epithelial–mesenchymal interaction throughout development. It has been shown that signaling from either epithelium or mesenchyme plays a critical role in controlling early stages of incisor tooth development (Charles et al., 2011; Munne et al., 2009). In order to determine whether incisor tooth invagination is regulated by epithelium and/or mesenchyme, we crossed Ikkα mutants with mice over-expressing Ikkα under the Keratin 5 (K5) promotor (K5-Ikkα). K5 is expressed in oral and dental epithelium from early stage of development (data not shown). K5-Ikkα mice yielded no tooth phenotype or oral epithelial adhesion (one of the phenotypes of Ikkα mutants) but it did completely rescue the incisor phenotype when crossed to Ikkα mutant mice (Figs. 3E-H). Epithelial Ikkα is thus required to control the direction of incisor tooth epithelial growth. In addition to evagination of incisor epithelium, the adhesion of oral epithelium were also rescued in K5-Ikkα; Ikkα−/− mice, suggesting that the adhesion is also caused by the disruption of epithelial Ikkα function (data not shown; Ohazama et al., 2004).
Fig. 3.
Rescue of incisor epithelial evagination. Incisors in wild-type (A, B), Ikkα−/− (C, D), K5-Ikkα (E, F) and K5-Ikkα;Ikkα−/− (G, H) on frontal sections at E18.5. Upper (A, C, E, G) and lower (B, D, F, H) incisors.
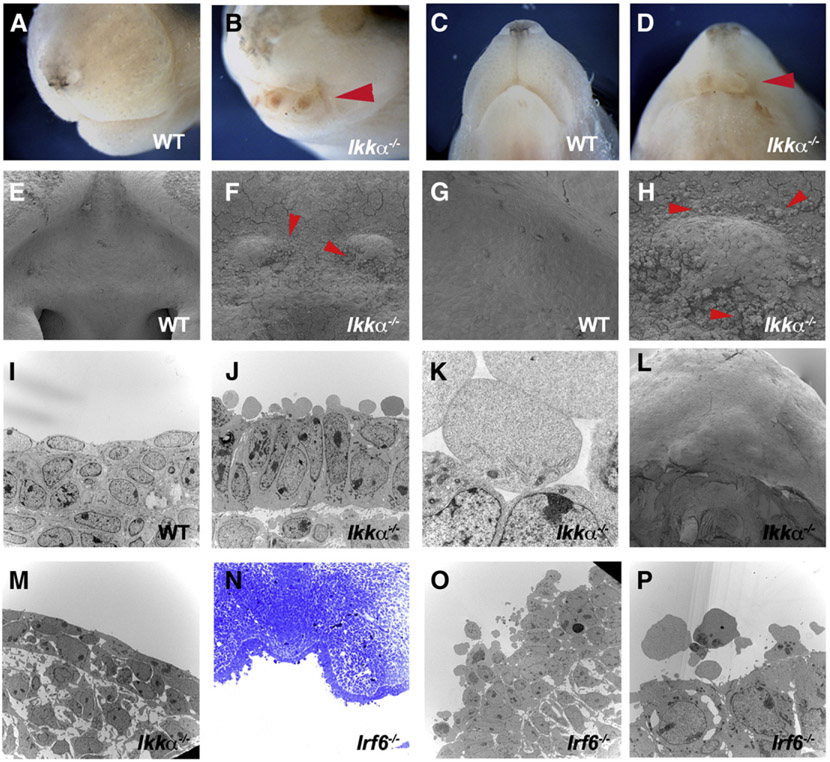
Since both Ikkα and Irf6 expressions in epithelium thus play critical roles in the development of incisors and oral mucosa development, we carried out further analysis on mutant epithelium. Irf6 has previously been shown to be involved in the formation of periderm which is a superficial monolayer of embryonic epithelium (Thomason et al., 2010). Both Ikkα and Irf6 mutant mice showed no epithelial adhesion in anterior parts of the mouth, including incisor regions, which allowed us to perform SEM analysis on the surface of mutant incisor epithelium (Figs. 4A-D). SEM showed the presence of cobblestone-like structures on the surface of anterior regions of Ikkα mutant mouth epithelium at E13.5, whereas these could not be detected at E10.5 or E11.5 or at birth (Figs. 4E-H, L; data not shown). To examine whether the cobblestone-like structures were cells, TEM assay was undertaken on mutant embryos. TEM showed that the cobblestone-like structures contained fragments of organelles, suggesting that these were disintegrated cells (Figs. 4J, K). The cobblestone-like cells were present only on the surface of the epithelium, not deeper inside. Similar cells were also observed in anterior regions of Irf6 mutant mouth epithelium, although these cells retained more organelles than those in Ikkα mutants (Figs. 4N-P). Cobblestone-like cells are known to be present along the medial epithelial edge of palatal shelves just before palatal fusion (Charoenchaikorn et al., 2009). It is possible that the anterior part of the mouth where incisors develop in Ikkα and Irf6 mutants shows initial signs of adhesion or fusion, but do not adhere.
Fig. 4.
Ultrastructure of mutant epithelium. Anterior heads in wild-type (A, C) and Ikkα−/− (B, D) at E18.5 (A–D). Cobblestone-like cells in Ikkα−/− (arrowheads in F, H, J, K) and Irf6−/− (N–P) at E13.5. No cobblestone-like cells in Ikkα−/− (L, M) at E18.5. SEM (E–H, L) and TEM (I–K, M, O, P).
In order to investigate the relationship between incisor epithelial evagination and epithelial adhesion, we examined mice that have previously been reported to have oral epithelial adhesion. Stratifin (Sfn; also known as 14-3-3 isoform σ) is known to have a pivotal role in cell cycle regulation and Receptor interacting protein 4 (Rip4; DIK, PKK) plays an important role during cellular stress (Hermeking and Benzinger, 2006; Janssens and Beyaert, 2003; Wang et al., 2000). Mice with mutation of either these molecules exhibit thickened epithelium, palatal cleft and adhesion of oral epithelium; identical phenotypes to those seen in Ikkα and Irf6 mutant mice (Bhr et al., 2000; Chen et al., 2001; Guenet et al., 1979; Holland et al., 2002). Evagination of incisor epithelium is characterized by abnormal enlargement of the anterior extent of incisors, a lack of associated dental lamina, alveolar bone and connective tissue, and displacement of enamel knot marker gene expression (Figs. 2, 5A-D; Ohazama et al., 2004). Rip4 and Sfn mutant (SfnEr/Er) mice however did not show any of these incisor phenotypes (Figs. 5G-L). Both mutants revealed no significant abnormalities in molar tooth germs (data not shown). Although both Rip4 and Sfn were expressed in tooth and oral epithelium, it is possible that null mutation of Rip4 or Sfn is not enough to induce evagination of incisor epithelium. To address this question, we generated Ikkα+/−;SfnEr/Er and Irf6+/−;SfnEr/Er compound mice but none presented with any obvious tooth anomalies (Figs. 5M-P; data not shown). Sfn expression showed no significant changes in incisors of Irf6 or Ikkα mutant mice while Irf6 and Ikkα expression were also normal in SfnEr/Er and Rip4 mutant mice, suggesting that there is unlikely to be interaction between these molecules in incisor tooth development (Supplementary Fig. 5E–5R; data not shown). Ikkα+/−;SfnEr/Er and Irf6+/−;SfnEr/Er compound mice showed no significant anomalies in molar tooth germs (data not shown).
Fig. 5.
Incisors in mutant mice with oral epithelial adhesion. Sagittal sections of incisors in wildtype (A) and Ikkα−/− (B) mice. Arrowheas indicating evaginated incisor (B). In situ hybridisation of Shh on sagittal head sections at E18.5 in wildtype (C) and Ikkα−/− (D) mice. Incisors of wild-type (E, F), SfnEr/Er (G, H), Rip4−/− (I, J), Irf6+/−;Sfn+/Er (M, N) and Ikkα+/−;SfnEr/Er (O, P) mice. Arrowheads indicating incisor tooth germs in SfnEr/Er (K). In situ hybridisation of Shh on frontal head sections at E14.5 in SfnEr/Er mice (arrowheads in L). Upper (A–E, G, I, M, O) and lower (F, H, J–L, N, P) incisors.
Canonical Wnt signaling in incisor evagination
Wnt signaling is known to play a critical role in determining the direction of cell growth (Barth et al., 2008; Killeen and Sybingco, 2008). Conditional increased activation of the canonical Wnt pathway in epithelial tissues results in the abnormal enlarged anterior extents of incisors exposing to oral cavity; a similar phenotype to those observed in the Ikkα and Irf6 mutant embryos (Liu et al., 2008b). To investigate whether the incisor epithelial evagination is the result of aberrant canonical Wnt signaling, we crossed both the Ikkα and Irf6 mutant mice to reporter mice for canonical Wnt signaling, Axin2lacZ. Strong activity was seen at E12.5 specifically in the incisor dental epithelium of both Ikkα and Irf6 mutant embryos (Figs. 6C-F; Supplementary Fig. 6). This suggests that canonical Wnt signaling may play a role in orienting the direction of tooth germ formation.
Fig. 6.
Wnt and Notch signalling in evaginated incisor tooth germs. LacZ expression in thickened incisor epithelium in Axin2lacZ (A, B), Ikkα−/−;Axin2lacZ (C, D) and Irf6−/−;Axin2lacZ (E, F) mice, and DMSO (G) and DAPT (H) treated mandible explants from Axin2lacZ mice. Notch1 expression in LiCl treated- (I) and nontreated- (J) mandible explants. Upper (A, C, E) and lower (B, D, F) incisors at E12.5.1 day cultured mandible from E11.5 (G–J).
In addition to alteration of Wnt signaling, downregulation of Notch1 and Notch2 expressions was also observed in incisors of both Ikkα mutant and Irf6 mutant embryos (Fig. 3; Ohazama et al., 2004). In order to investigate the interaction between Notch and canonical Wnt signaling in incisor development at an early stage of development, we cultured E11.5 mandibles in the presence of γ-secretase inhibitor (DAPT), an inhibitor of cleavage of Notch1/2 to inhibit Notch signaling or GSK3β inhibitor, LiCl to increase canonical Wnt signaling (Grosveld, 2009; Klein and Melton, 1996; Shih and Wang, 2007). Reduced Notch signaling led to no significant changes in Axin2 reporter expression in DAPT-treated Axin2lacZ mouse mandibles compared to those in control explants (Figs. 6G, H). Upregulation of Wnt signaling resulted in no significant changes of Notch1 expression in LiCl treated explants compared to those seen in control explants (Figs. 6I, J). These results suggested that an obvious relationship between Notch and canonical Wnt signaling is unlikely to be present in incisor development.
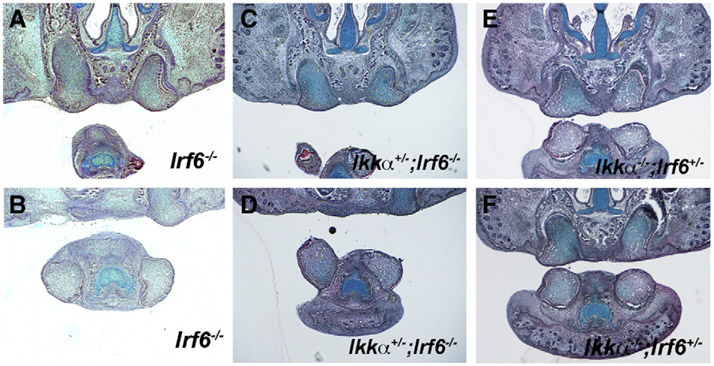
Irf6 mutant incisors thus phenocopy those of Ikkα mutants. Both Ikkα and Irf6 are expressed in the developing incisor epithelium (Fig. 1; Ohazama et al., 2004). In order to investigate potential genetic interactions between Ikkα and Irf6 in incisor tooth development, we intercrossed Irf6 mutants with Ikkα mutant mice. Ikkα+/−;Irf6+/− compound heterozygotes were viable (data not shown) but neither Ikkα+/−;Irf6−/− nor Ikkα−/−;Irf6+/− compound mice showed exacerbation or attenuation of the evaginating incisor phenotype (Figs. 7A-F). Irf6 expression showed no significant changes in incisors of Ikkα mice and Ikkα expression was also normal in Irf6 mice (Supplementary Fig. 5A-5D; data not shown). The results thus fail to show any evidence of direct genetic interaction between Ikkα and Irf6 in incisor development.
Fig. 7.
Interaction between Ikkα and Irf6 in incisor tooth development. Incisors in Irf6−/− (A, B), Ikkα+/−;Irf6−/− (C, D), Ikkα−/−;Irf6+/− (E, F) on frontal sections at E18.5. Upper (A, C, E) and lower (B, D, F) incisors.
Discussion
Irf6 mutant mice show identical phenotypes including tooth, oral mucosa and skin to those described in Ikkα mutants (Hu et al., 1999; Ingraham et al., 2006; Li et al., 1999; Ohazama et al., 2004; Richardson et al., 2006; Takeda et al., 1999). Irf6 is a transcription factor that belongs to the Irf family which is known to be involved in the NF-κB pathway (Taniguchi et al., 2001). Although other Irf family members are known to be related to the NF-kB pathway, any interaction between the NF-κB pathway and Irf6 remains unclear. Since no change in NF-κB pathway activity was observed in Irf6 mutant embryos, in common with Ikkα mutants, the incisor evagination seen is likely to occur independently of the NF-κB pathway (Fig. 2; Ohazama et al., 2004). Based on the phenotypes and common downstream molecular changes, one might predict that these proteins interact, either by Ikkα phosphorylating Irf6, or Irf6 transcriptionally regulating Ikkα. Ikkα has previously been shown to activate Irf7 by phosphorylation (Hoshino et al., 2006). However, although it is feasible that Ikkα could phosphorylate other members of the Irf protein family (including Irf6), the C-terminal protein-binding domains are less well conserved between members and only Irf1, Irf3, Irf5 and Irf7 are currently recognized as possessing phosphorylation sites (Lohoff and Mak, 2005). A lack of physical interaction between Ikkα and Irf6 functional proteins is supported by previous co-immunoprecipitation experiments (Ingraham et al., 2006). In addition, the loss of one gene does not appear to affect the expression of the other (Supplementary Fig. 5) and compound homozygote–heterozygotes show no exacerbation or attenuation of phenotype (Fig. 7). This is further supported by micro-array data that shows no change in Ikkα gene expression in Irf6 mutant skin (Ingraham et al., 2006). Identical phenotypes with the same downstream molecular changes but lack of direct evidence of genetic interaction in incisor tooth development between Irf6 and Ikkα mutants suggested that Irf6 and Ikkα are likely to function in separate (parallel) pathways involved in the same processes. Our data also show no link between the oral fusion and epithelial envagination phenotypes, suggesting that the invagination of incisor epithelium is unlikely to be regulated by the same molecular pathways as those in palate and oral mucosa development. Irf6 and Ikkα thus appear to be potentially involved in multiple pathways in epithelial cell biology.
All Irf proteins contain a carboxy (C)-terminal Irf-association domain, which facilitates homodimerization and heterodimerization between family members (Lohoff and Mak, 2005). Null mutations of Irf genes 1 to 9 are viable and fertile with the exception of Irf6, although cause of the differences still remained unclear (Holtschke et al., 1996; Honda et al., 2005; Kimura et al., 1996; Matsuyama et al., 1993; Mittrücker et al., 1997; Richardson et al., 2006; Sato et al., 2000; Takaoka et al., 2005). It is feasible that there is a level of redundancy within the Irfs and that in the null condition, other family members can compensate for the loss of one gene through heterodimerization. This possibility is obviously excluded in the case of Irf6 mutant. The Irf6 gene is thus unique within the Irf family and this is reflected not only in its expression pattern, but also in the severity of the phenotype.
Irf6 is also known to be responsible for Van der Woude syndrome that often features congenital missing teeth such as mandibular second premolars (Arangannal et al., 2002; Oberoi and Vargervik, 2005; Schneider, 1973). In addition to the hypodontia, taurodontism and dental pulp stones have also been shown in Van der Woude syndrome (Kantaputra et al., 2002; Nawa et al., 2008). Although crown shape is probably most affected by evagination of tooth epithelium, no significant anomalies of crown shape have been reported in Van der Woude syndrome. In mice, only incisor tooth epithelium was altered by Irf6 mutation. Molecular mechanisms of epithelial invagination are thus likely to be different between humans and mice, possibly related to the continuous growth of mouse incisors.
Wnt signaling plays critical roles in many biological processes including development (Pellón-Cárdenas et al., 2011; Sylvie et al., 2011). Many Wnt signaling related molecules are known to be expressed during tooth development (Sarkar et al., 2000). We show strong canonical Wnt activation in mutant evaginating incisor epithelium (Fig. 6). The conditionally increased activation of the canonical Wnt signaling pathway in epithelial tissues shows enlarged misshapen incisors which exposed to oral cavity, suggesting that Wnt signaling may regulate the direction of epithelial growth at early stages of tooth development (Liu et al., 2008b). Ikkα has been shown to interact with canonical Wnt signaling to positively regulate β-catenin-dependent transcriptional activity by increasing the cytosolic levels of β-catenin (Albanese et al., 2003; Lamberti et al., 2001). Ikkα has also been shown to physically interact with β-catenin, inhibiting Axin/APC/GSK-3β and Siah-1 pathway-mediated β-catenin ubiquitination and subsequent degradation, a process that occurs independently of the NF-κB pathway (Carayol and Wang, 2006). P63 function is known to be required at early stages of tooth development, since p63 mutant mice show no epithelial thickening of tooth epithelium which is accompanied by downregulation of Notch1 expression (Laurikkala et al., 2006). P63 has been shown to be associated with Ikkα and Irf6 in skin and palate, respectively (Candi et al., 2006; Ferretti et al., 2011; Thomason et al., 2010). Since the interaction of p63 with Irf6 is known to be regulated by Wnt signaling, it is possible that Ikkα and Irf6 are involved in p63 through Wnt signaling in tooth epithelial invagination (Ferretti et al., 2011; Thomason et al., 2010).
Wnt signaling is known to play a critical role in tooth initiation and Notch signaling is believed to regulate the direction of murine incisor rotation at early stages of tooth development (Järvinen et al., 2006; Liu et al., 2008a, 2008b; Mucchielli and Mitsiadis, 2000). Both Ikkα and Irf6 null mutant incisors showed alteration of Notch and Wnt signaling (Figs. 2, 6; Ohazama et al., 2004). Interactions between Wnt and Notch signaling have been reported in intestine, somatogenesis and keratinocytes (Dunty et al., 2008; Nicolas et al., 2003; van Es et al., 2005). Our results however showed no significant relationship between Notch and Wnt in incisor development. Regulating the direction of incisor epithelial invagination thus appears to involve multiple signaling networks.
Supplementary Material
Acknowledgments
We would like to thank Amgen for Rip4 mutant mice, Ken Brady for TEM analysis, Dr. Koyama for HistoneH4C plasmids (Ochiai et al., 2010; Yasuda et al., 2010) and Tony Brain for SEM analysis. This work was supported by the MRC and was partly funded by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (No. 16592080). K.K. is supported by JSPS International Program for Young Researcher Overseas Visits. Y.O.-K. is supported by Nihon University. S.C.A. and A.K. were supported by NIH DE13513.
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.ydbio.2012.02.009
References
- Albanese C, Wu K, D'Amico M, Jarrett C, Joyce D, Hughes J, Hulit J, Sakamaki T, Fu M, Ben-Ze'ev A, Bromberg JF, Lamberti C, Verma U, Gaynor RB, Byers SW, Pestell RG, 2003. IKKalpha regulates mitogenic signaling through transcriptional induction of cyclin D1 via Tcf. Mol. Biol. Cell 14, 585–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arangannal P, Muthu MS, Nirmal L, 2002. Van der Woude syndrome: a case report. J. Indian Soc. Pedod. Prev. Dent 20, 102–103. [PubMed] [Google Scholar]
- Barth AI, Caro-Gonzalez HY, Nelson WJ, 2008. Role of adenomatous polyposis coli (APC) and microtubules in directional cell migration and neuronal polarization. Semin. Cell Dev. Biol 19, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhr C, Rohwer A, Stempka L, Rincke G, Marks F, Gschwendt M, 2000. DIK, a novel protein kinase that interacts with protein kinase Cdelta. Cloning, characterization, and gene analysis. J. Biol. Chem 275, 36350–36357. [DOI] [PubMed] [Google Scholar]
- Bixler D, Poland C, Nance WE, 1973. Phenotypic variation in the popliteal pterygium syndrome. Clin. Genet 4, 220–228. [DOI] [PubMed] [Google Scholar]
- Bonjardim CA, Ferreira PC, Kroon EG, 2009. Interferons: signaling, antiviral and viral evasion. Immunol. Lett 122,1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Terrinoni A, Rufini A, et al. , 2006. p63 is upstream of IKK alpha in epidermal development. J. Cell Sci 119 (Pt 22), 4617–4622. [DOI] [PubMed] [Google Scholar]
- Carayol N, Wang CY, 2006. IKKalpha stabilizes cytosolic beta-catenin by inhibiting both canonical and non-canonical degradation pathways. Cell. Signal 18, 1941–1946. [DOI] [PubMed] [Google Scholar]
- Chariot A, 2009. The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol. 19, 404–413. [DOI] [PubMed] [Google Scholar]
- Charles C, Hovorakova M, Ahn Y, et al. , 2011. Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling. Development 138 (18), 4063–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoenchaikorn K, Yokomizo T, Rice DP, Honjo T, Matsuzaki K, Shintaku Y, Imai Y, Wakamatsu A, Takahashi S, Ito Y, Takano-Yamamoto T, Thesleff I, Yamamoto M, Yamashiro T, 2009. Runx1 is involved in the fusion of the primary and the secondary palatal shelves. Dev. Biol 326, 392–402. [DOI] [PubMed] [Google Scholar]
- Chen L, Haider K, Ponda M, Cariappa A, Rowitch D, Pillai S, 2001. Protein kinase C-associated kinase (PKK), a novel membrane-associated, ankyrin repeat-containing protein kinase. J. Biol. Chem 276, 21737–21744. [DOI] [PubMed] [Google Scholar]
- Dunty WC, Bins KK, Chalamalasetty RB, Taketo MM, Lewandoski M, Yamaguchi TP, 2008. Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development 135, 85–94. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Li B, Zewdu R, et al. , 2011. A conserved Pbx-Wnt-p63-Irf6 regulatory module controls face morphogenesis by promoting epithelial apoptosis. Dev. Cell 21 (4), 627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froster-Iskenius UG, 1990. Popliteal pterygium syndrome. J. Med. Genet 27, 320–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodison S, Urquidi U, Tarin D, 1999. CD44 cell adhesion molecules. J. Clin. Pathol. Mol. Pathol 52, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld GC, 2009. Gamma-secretase inhibitors: notch so bad. Nat. Med 15, 20–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenet JL, Salzgeber B, Tassin MT, 1979. Repeated epilation: a genetic epidermal syndrome in mice. J. Hered 70, 90–94. [DOI] [PubMed] [Google Scholar]
- Häcker H, Karin M, 2006. Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Benzinger A, 2006. 14-3-3 proteins in cell cycle regulation. Semin. Cancer Biol 16, 183–192. [DOI] [PubMed] [Google Scholar]
- Hertzog P, Forster S, Samarajiwa S, 2011. Systems biology of interferon responses. J. Interferon Cytokine Res 31, 5–11. [DOI] [PubMed] [Google Scholar]
- Hiscott J, 2007. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18, 483–490. [DOI] [PubMed] [Google Scholar]
- Holland P, Willis C, Kanaly S, Glaccum M, Warren A, Charrier K, Murison J, Derry J, Virca G, Bird T, Peschon J, 2002. RIP4 is an ankyrin repeat-containing kinase essential for keratinocyte differentiation. Curr. Biol 12, 1424–1428. [DOI] [PubMed] [Google Scholar]
- Holtschke T, Löhler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch KP, Gabriele L, Waring JF, Bachmann MF, Zinkernagel RM, Morse HC, Ozato K, Horak I, 1996. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell 87, 307–317. [DOI] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T, 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777. [DOI] [PubMed] [Google Scholar]
- Hoshino K, Sugiyama T, Matsumoto M, Tanaka T, Saito M, Hemmi H, Ohara O, Akira S, Kaisho T, 2006. IkappaB kinase-alpha is critical for interferon-alpha production induced by Toll-like receptors 7 and 9. Nature 440, 949–953. [DOI] [PubMed] [Google Scholar]
- Hu Y, Baud V, Delhase M, Zhang P, Deerinck T, Ellisman M, Johnson R, Karin M, 1999. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKalpha subunit of IkappaB kinase. Science 284, 316–320. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC, 2006. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat. Genet 38, 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Beyaert R, 2003. Functional diversity and regulation of different interleukin-1 receptor-associated kinase (IRAK) family members. Mol. Cell 11, 293–302. [DOI] [PubMed] [Google Scholar]
- Järvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I, 2006. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. U. S. A 103,18627–18632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Lan Y, Chapman HD, Shawber C, Norton CR, Serreze DV, Weinmaster G, Gridley T, 1998. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 12, 1046–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantaputra PN, Sumitsawan Y, Schutte BC, Tochareontanaphol C, 2002. Van der Woude syndrome with sensorineural hearing loss, large craniofacial sinuses, dental pulp stones, and minor limb anomalies: report of a four-generation Thai family. Am. J. Med. Genet 108, 275–280. [DOI] [PubMed] [Google Scholar]
- Killeen MT, Sybingco SS, 2008. Netrin, Slit and Wnt receptors allow axons to choose the axis of migration. Dev. Biol 323, 143–151. [DOI] [PubMed] [Google Scholar]
- Kimura T, Kadokawa Y, Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Tarutani M, Tan RS, Takasugi T, Matsuyama T, Mak TW, Noguchi S, Taniguchi T, 1996. Essential and non-redundant roles of p48 (ISGF3 gamma) and IRF-1 in both type I and type II interferon responses, as revealed by gene targeting studies. Genes Cells 1, 115–124. [DOI] [PubMed] [Google Scholar]
- Klein PS, Melton DA, 1996. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. U. S. A 93, 8455–8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C, Houdayer C, Bahuau M, Moretti-Ferreira D, Richieri-Costa A, Dixon MJ, Murray JC, 2002. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet 32, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti C, Lin KM, Yamamoto Y, Verma U, Verma IM, Byers S, Gaynor RB, 2001. Regulation of beta-catenin function by the IkappaB kinases. J. Biol. Chem 276, 42276–42286. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola ML, James M, Tummers M, Mills AA, Thesleff I, 2006. p63 regulates multiple signalling pathways required for ectodermal organogenesis and differentiation. Development 133 (8), 1553–1563. [DOI] [PubMed] [Google Scholar]
- Li Q, Verma IM, 2002. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734. [DOI] [PubMed] [Google Scholar]
- Li Q, Lu Q, Hwang JY, Bóscher D, Lee KF, Izpisua-Belmonte JC, Verma IM, 1999. IKK1-deficient mice exhibit abnormal development of skin and skeleton. Genes Dev. 13, 1322–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Xia X, Zhu F, Park E, Carbajal S, Kiguchi K, DiGiovanni J, Fischer SM, Hu Y, 2008a. IKKalpha is required to maintain skin homeostasis and prevent skin cancer. Cancer Cell 14, 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Chu EY, Watt B, Zhang Y, Gallant NM, Andl T, Yang SH, Lu MM, Piccolo S, Schmidt-Ullrich R, Taketo MM, Morrisey EE, Atit R, Dlugosz AA, Millar SE, 2008b. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol 313, 210–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff M, Mak TW, 2005. Roles of interferon-regulatory factors in T-helper-cell differentiation. Nat. Rev. Immunol 5, 125–135. [DOI] [PubMed] [Google Scholar]
- Lomada D, Liu B, Coghlan L, Hu Y, Richie ER, 2007. Thymus medulla formation and central tolerance are restored in IKKalpha−/− mice that express an IKKalpha transgene in keratin 5+ thymic epithelial cells. J. Immunol 178, 829–837. [DOI] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, Behrens J, 2002. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol 22, 1184–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kóndig TM, Amakawa R, Kishihara K, Wakeham A, 1993. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75, 83–97. [PubMed] [Google Scholar]
- Mittrócker HW, Matsuyama T, Grossman A, Kóndig TM, Potter J, Shahinian A, Wakeham A, Patterson B, Ohashi PS, Mak TW, 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science 275, 540–543. [DOI] [PubMed] [Google Scholar]
- Mucchielli ML, Mitsiadis TA, 2000. Correlation of asymmetric Notch2 expression and mouse incisor rotation. Mech. Dev 91, 379–382. [DOI] [PubMed] [Google Scholar]
- Munne PM, Tummers M, Jarvinen E, Thesleff I, Jernvall J, 2009. Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development 136 (3), 393–402. [DOI] [PubMed] [Google Scholar]
- Nawa H, Oberoi S, Vargervik K, 2008. Taurodontism and Van der Woude syndrome. Is there an association? Angle Orthod. 78, 832–837. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F, 2003. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet 33, 416–421. [DOI] [PubMed] [Google Scholar]
- Oberoi S, Vargervik K, 2005. Hypoplasia and hypodontia in Van der Woude syndrome. Cleft Palate Craniofac. J 42, 459–466. [DOI] [PubMed] [Google Scholar]
- Ochiai T, Shibukawa Y, Nagayama M, Mundy C, Yasuda T, Okabe T, Shimono K, Kanyama M, Hasegawa H, Maeda Y, Lanske B, Pacifici M, Koyama E, 2010. Indian hedgehog roles in post-natal TMJ development and organization. J. Dent. Res 89, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohazama A, Hu Y, Schmidt-Ullrich R, Cao Y, Scheidereit C, Karin M, Sharpe PT, 2004. A dual role for Ikk alpha in tooth development. Dev. Cell 6, 219–227. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Tucker A, Sharpe PT, 2005. Organized tooth-specific cellular differentiation stimulated by BMP4. J. Dent. Res 84, 603–606. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Johnson EB, Ota MS, Choi HY, Choi HJ, Porntaveetus T, Oommen S, Itoh N, Eto K, Gritli-Linde A, Herz J, Sharpe PT, 2008. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One 3, e4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellón-Cárdenas O, Schweitzer J, D'Souza-Schorey C, 2011. Endocytic trafficking and Wnt/β-catenin signaling. Curr. Drug Targets 12, 1216–1222. [DOI] [PubMed] [Google Scholar]
- Platanias LC, 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol 5, 375–386. [DOI] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ, 2006. Irf6 is a key determinant of the keratinocyte proliferation–differentiation switch. Nat. Genet 38, 1329–1334. [DOI] [PubMed] [Google Scholar]
- Sanz AB, Sanchez-Niño MD, Ramos AM, Moreno JA, Santamaria B, Ruiz-Ortega M, Egido J, Ortiz A, 2010. NF-kappaB in renal inflammation. J. Am. Soc. Nephrol 21, 1254–1262. [DOI] [PubMed] [Google Scholar]
- Sarkar L, Cobourne M, Naylor S, Smalley M, Dale T, Sharpe PT, 2000. Wnt/Shh interactions regulate ectodermal boundary formation during mammalian tooth development. Proc. Natl. Acad. Sci. U. S. A 97, 4520–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T, 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13, 539–548. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R, Mémet S, Lilienbaum A, Feuillard J, Raphaël M, Israel A, 1996. NF-kappaB activity in transgenic mice: developmental regulation and tissue specificity. Development 122, 2117–2128. [DOI] [PubMed] [Google Scholar]
- Schneider EL, 1973. Lip pits and congenital absence of second premolars: varied expression of the Lip Pits syndrome. J. Med. Genet 10, 346–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schock F, Perrimon N, 2002. Molecular mechanisms of epithelial morphogenesis. Annu. Rev. Cell Dev. Biol 18, 463–493. [DOI] [PubMed] [Google Scholar]
- Shih IM, Wang TL, 2007. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 67, 1879–1882. [DOI] [PubMed] [Google Scholar]
- Sylvie J, Ellen C, Kris V, 2011The role of Wnt in cell signaling and cell adhesion during early vertebrate development. Front. Biosci 17, 2352–2366. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T, 2005. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434, 243–249. [DOI] [PubMed] [Google Scholar]
- Takeda K, Takeuchi O, Tsujimura T, Itami S, Adachi O, Kawai T, Sanjo H, Yoshikawa K, Terada N, Akira S, 1999. Limb and skin abnormalities in mice lacking IKKalpha. Science 284, 313–316. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N, 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol 19, 623–655. [DOI] [PubMed] [Google Scholar]
- Thesleff I, 2006. The genetic basis of tooth development and dental defects. Am. J. Med. Genet. A 140, 2530–2535. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H, Dixon J, 2010. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J. Clin. Invest 120, 1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker A, Sharpe P, 2004. The cutting-edge of mammalian development; how the embryo makes teeth. Nat. Rev. Genet 5, 499–508. [DOI] [PubMed] [Google Scholar]
- Van der Woude A, 1954. Fistula labii inferioris congenita and its association with cleft lip and palate. Am. J. Hum. Genet 6, 244–256. [PMC free article] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H, 2005. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963. [DOI] [PubMed] [Google Scholar]
- Wang AH, Kruhlak MJ, Wu J, Bertos NR, Vezmar M, Posner BI, Bazett-Jones DP, Yang XJ, 2000. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol. Cell. Biol 20, 6904–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Mundy C, Kinumatsu T, Shibukawa Y, Shibutani T, Grobe K, Minugh-Purvis N, Pacifici M, Koyama E, 2010. Sulfotransferase Ndst1 is needed for mandibular and TMJ development. J. Dent. Res 89, 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.