Graphical abstract
Abbreviations: ACE2, angiotensin-converting enzyme 2; DFI, disease-free interval; DSS, disease-specific survival; EMT, epithelial-mesenchymal transition; FDR, false discovery rate; GO, gene ontology; GSEA, gene set enrichment analysis; OS, overall survival; PFI, progression-free interval; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TCGA, The Cancer Genome Atlas; TF, transcription factor; WGCNA, weighted gene co-expression network analysis; CESC, cervical squamous-cell carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; OV, ovarian carcinoma; PAAD, pancreatic adenocarcinoma; SKCM, skin cutaneous melanoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma
Keywords: ACE2 expression, Pan-cancer, Tumor immunity and immunotherapy, Tumor progression, Survival prognosis
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 29 million people and has caused more than 900,000 deaths worldwide as of September 14, 2020. The SARS-CoV-2 human cell receptor ACE2 has recently received extensive attention for its role in SARS-CoV-2 infection. Many studies have also explored the association between ACE2 and cancer. However, a systemic investigation into associations between ACE2 and oncogenic pathways, tumor progression, and clinical outcomes in pan-cancer remains lacking. Using cancer genomics datasets from the Cancer Genome Atlas (TCGA) program, we performed computational analyses of associations between ACE2 expression and antitumor immunity, immunotherapy response, oncogenic pathways, tumor progression phenotypes, and clinical outcomes in 13 cancer cohorts. We found that ACE2 upregulation was associated with increased antitumor immune signatures and PD-L1 expression, and favorable anti-PD-1/PD-L1/CTLA-4 immunotherapy response. ACE2 expression levels inversely correlated with the activity of cell cycle, mismatch repair, TGF-β, Wnt, VEGF, and Notch signaling pathways. Moreover, ACE2 expression levels had significant inverse correlations with tumor proliferation, stemness, and epithelial-mesenchymal transition. ACE2 upregulation was associated with favorable survival in pan-cancer and in multiple individual cancer types. These results suggest that ACE2 is a potential protective factor for cancer progression. Our data may provide potential clinical implications for treating cancer patients infected with SARS-CoV-2.
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 29 million people and has caused more than 900,000 deaths worldwide as of September 14, 2020 (https://coronavirus.jhu.edu/map.html). SARS-CoV-2 uses the angiotensin-converting enzyme 2 (ACE2) as a host cell receptor to infect humans [1], [2], [3], [4]. ACE2 plays an important role in regulating cardiovascular and renal function [5]. This protein has recently received extensive attention for its role in SARS-CoV-2 infection [1], [2], [4]. Our recent study revealed that ACE2 is expressed in various human tissues [6], suggesting that SARS-CoV-2 may invade various human organs besides the lungs. Moreover, SARS-CoV-2 infection may result in ACE2 upregulation [7]. However, ACE2 deficiency may exacerbate outcomes in patients with SARS-CoV-2 infection [8]. Indeed, a recent study showed that ACE2 was downregulated in virus-infected lung tissue [9], indicating a potential protective role of ACE2 in patients with SARS-CoV-2 infection. ACE2 also plays a protective role in hypertension and heart disease [8].
Many studies have investigated the association between ACE2 and cancer [10], [11], [12], [13], [14], [15], [16], [17], [18]. For example, Yu-Jun et al. analyzed ACE2 expression in various cancers and revealed a positive association between ACE2 expression and survival prognosis in liver cancer [10]. Cai et al. described the genetic alteration, mRNA expression, and DNA methylation of ACE2 in over 30 cancer types and revealed genetic and epigenetic variations of ACE2 in various cancers [11]. Several studies demonstrated that ACE2 had antitumor effects by inhibiting tumor angiogenesis [13], [14], [16]. Zhang et al. revealed that ACE2 expression was more highly expressed in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) than in normal tissues [17]. Huang et al. found that ACE2 expression had a significant association with immune cell infiltration in various normal and cancer tissues [18]. A recent study [9] showed that ACE2 expression was associated with increased tumor immune infiltration and was a positive prognostic factor in uterine corpus endometrial and renal papillary cell cancers. Despite these previous studies, a systemic investigation into the association between ACE2 expression and antitumor immunity, oncogenic pathways, tumor progression phenotypes, and clinical outcomes in pan-cancer remains lacking.
In this study, we investigated associations between ACE2 expression and antitumor immune signatures in 13 human cancer cohorts from the Cancer Genome Atlas (TCGA) program (https://cancergenome.nih.gov/). We also explored associations between ACE2 expression and multiple tumor phenotypes, including cell proliferation, stemness, epithelial-mesenchymal transition (EMT), oncogenic signaling, and clinical outcomes in these cancer cohorts. We also investigated the association between ACE2 expression and immunotherapy response in four cancer cohorts receiving the immune checkpoint blockade therapy. This study aimed to provide new insights into the association between ACE2 and cancer and the potential association between cancer and SARS-CoV-2 infection.
2. Materials and methods
2.1. Datasets
From the genomic data commons data portal (https://portal.gdc.cancer.gov/), we obtained RNA-Seq gene expression profiling datasets (level 3 and RSEM normalized) for 13 TCGA cancer cohorts. The 13 cancer cohorts included cervical squamous-cell carcinoma (CESC), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), LUAD, LUSC, skin cutaneous melanoma (SKCM), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), ovarian carcinoma (OV), and pancreatic adenocarcinoma (PAAD). We log2-transformed all RSEM-normalized gene expression values before further analyses. Besides, we obtained gene expression profiling and clinical data in four cancer cohorts receiving anti-PD-1/PD-L1/CTLA-4 immunotherapy from their related publications, including Nathanson (melanoma) [19], Topalian (melanoma) [20], Ascierto (renal cell carcinoma) [21], and Snyder (bladder cancer) cohorts [22]. A summary of these datasets is presented in Supplementary Table S1.
2.2. Evaluating the enrichment levels of immune signatures, pathways, and tumor phenotypes
We evaluated the enrichment level of a pathway or tumor phenotype in a tumor sample by the single-sample gene-set enrichment analysis (ssGSEA) score [23]. The gene set included all marker genes of a pathway or tumor phenotype. A total of 14 cancer-associated pathways (cell cycle, mismatch repair, TGF-β, Wnt, VEGF, Notch signaling, axon guidance, renin angiotensin system, PPAR signaling, MAPK signaling, glycolysis, cell adhesion molecules, endocytosis, and calcium signaling) and three tumor phenotypes (cell proliferation, stemness, and EMT) were analyzed. We presented the marker genes of these pathways and tumor phenotypes in Supplementary Table S2.
2.3. Gene-set enrichment analysis
We defined high-ACE2-expression-level (upper third) and low-ACE2-expression-level (bottom third) tumors in each cancer type based on ACE2 expression profiles. We identified the KEGG [24] pathways highly enriched in both groups of tumors using GSEA [25] with a threshold of adjusted p-value < 0.05. Moreover, we used WGCNA [26] to detect the gene modules (gene ontology) differentially enriched between the high- and low-ACE2-expression-level tumors in pan-cancer. We identified the hub genes as the genes connected to at least 5 other genes with a connectedness weight greater than 0.25 in a gene module and built their co-expression network.
2.4. Statistical analysis
We used Spearman’s correlation test to evaluate the correlation (ρ) of ACE2 expression levels with the enrichment levels of pathways or tumor phenotypes, which were not normally distributed. We used Pearson's correlation test to evaluate the correlation (r) of ACE2 expression levels with the ratios of immune signatures, which was the log2-transformed values of the ratios between the mean expression levels of all marker genes in immune signatures and was normally distributed. We used the Benjamini and Hochberg method [27] to calculate the FDR for adjusting for multiple tests. We compared overall survival (OS), disease-specific survival (DSS), progression-free interval (PFI), and disease-free interval (DFI) between the high- and low-ACE2-expression-level tumors. We utilized Kaplan-Meier curves to display survival time differences and the log-rank test to evaluate the significance of survival time differences. The R package “survival” was used to perform the survival analyses.
3. Results
3.1. Association of ACE2 expression with immune signatures and immunotherapy response in cancer
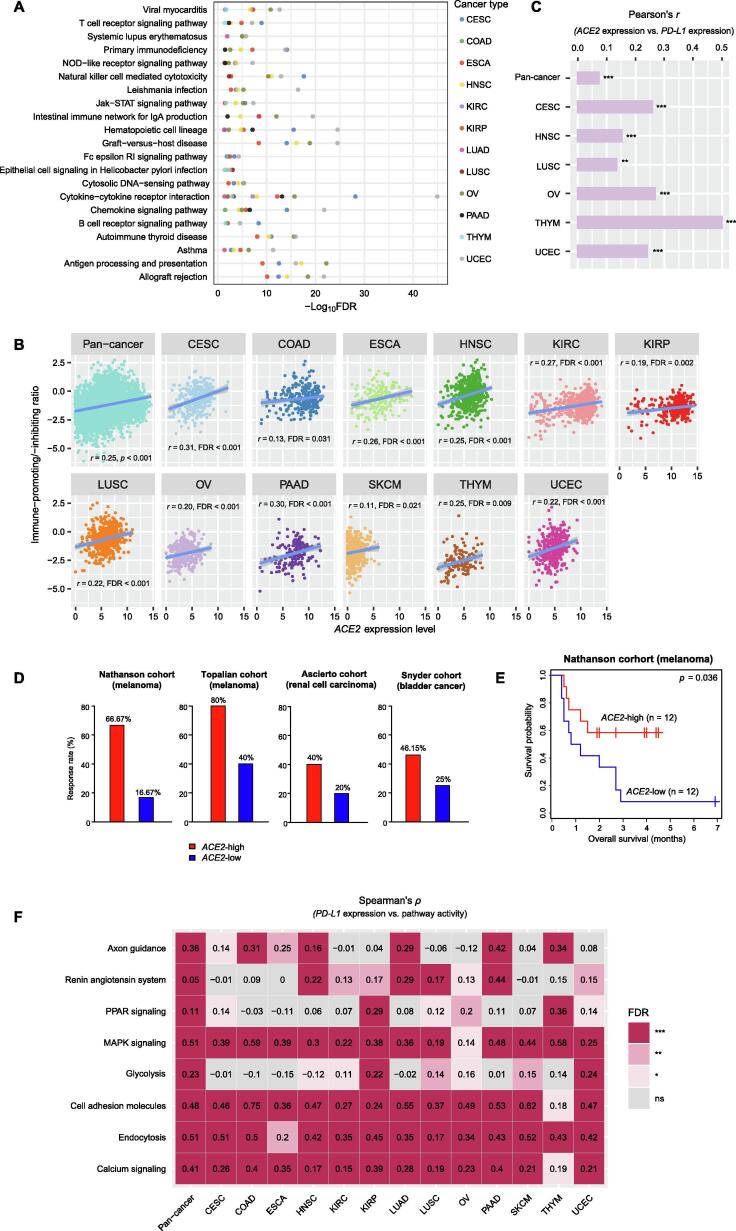
GSEA [25] identified many immune-related pathways highly enriched in the high-ACE2-expression-level tumors in at least 5 cancer types. These pathways included viral myocarditis, T cell receptor signaling, systemic lupus erythematosus, primary immunodeficiency, NOD-like receptor signaling, natural killer cell mediated cytotoxicity, Leishmania infection, Jak-STAT signaling, intestinal immune network for IgA production, hematopoietic cell lineage, graft-versus-host disease, Fc epsilon RI signaling, epithelial cell signaling in Helicobacter pylori infection, cytosolic DNA-sensing, cytokine-cytokine receptor interaction, chemokine signaling, B cell receptor signaling, autoimmune thyroid disease, asthma, antigen processing and presentation, and allograft rejection (Fig. 1A). Moreover, we found that ACE2 expression levels positively correlated with the immune-promoting/immune-inhibiting ratios in pan-cancer (Pearson’s correlation test, r = 0.25, p = 2.37 × 10−71) and in 12 individual cancer types (adjusted p-value (FDR) < 0.05) (Fig. 1B). This suggests that ACE2 expression has a stronger positive association with the immune-promoting signature than the immune-inhibiting signature in these cancer types. Altogether, these results suggest a prominent positive association between ACE2 expression and antitumor immune signatures in cancer. We found that ACE2 had a positive expression correlation with PD-L1 in pan-cancer and in 6 individual cancer types (FDR < 0.05) (Fig. 1C). We expected that the ACE2 expression would have a positive association with the response to anti-PD-1/PD-L1/CTLA-4 immunotherapy. We confirmed the anticipation in four cancer cohorts receiving immune checkpoint blockade therapy. In these cohorts, the high-ACE2-expression-level (> median) tumors displayed a higher rate of immunotherapy response than the low-ACE2-expression-level (< median) tumors (67% versus 17%, 80% versus 40%, 40% versus 20%, and 46% versus 25% for Nathanson (melanoma), Topalian (melanoma), Ascierto (renal cell carcinoma), and Snyder (bladder cancer) cohorts, respectively) (Fig. 1D). As a result, the former had better overall survival (OS) than the latter in the Nathanson cohort, which had related data available (log-rank test, p = 0.036) (Fig. 1E). These results suggest that the ACE2 expression is likely to be a positive predictor for anti-PD-1/PD-L1 immunotherapy.
Fig. 1.
Association of ACE2 expression with immune signatures and immunotherapy response in cancer. (A) Immune-related pathways upregulated in high- (upper third) versus low-ACE2-expression-level (bottom third) tumors in at least 5 cancer types identified by GSEA [25] (adjusted p-value (FDR) < 0.05). (B) Significant positive correlations of ACE2 expression levels with the ratios of immune-promoting/immune-inhibiting cytokines in pan-cancer and in 12 individual cancer types. The Pearson correlation coefficient (r) and p- or FDR-value are shown. (C) The positive expression correlation between ACE2 and PD-L1 in pan-cancer and in 6 individual cancer types. (D) Higher rate of immunotherapy response in the high-ACE2-expression-level (> median) than in the low-ACE2-expression-level (< median) tumors in four cancer cohorts receiving immune check point blockade th.erapy. (E) Kaplan-Meier survival curves showing better survival in high-ACE2-expression-level (> median) than in low-ACE2-expression-level (< median) cancer patients with immune checkpoint blockade therapy. The log-rank test p-value is shown. (F) Correlations between pathway activity and PD-L1 expression levels in pan-cancer and in 13 individual cancer types. The Spearman correlation coefficient (ρ) and FDR-value are shown. FDR: false discovery rate. * FDR < 0.05; ** FDR < 0.01; *** FDR < 0.001; ns: not significant. They also apply to the following figures.
GSEA [25] also identified several cancer-associated pathways highly enriched in the high-ACE2-expression-level tumors in at least 5 cancer types. These pathways included axon guidance, renin angiotensin system, PPAR signaling, MAPK signaling, glycolysis, cell adhesion molecules, endocytosis, and calcium signaling. We found that the activities of these pathways were significantly and positively associated with PD-L1 expression levels in pan-cancer (FDR < 0.001) (Fig. 1F). In individual cancer types, the elevated activities of these pathways were also likely to correlate with increased PD-L1 expression levels (FDR < 0.05) (Fig. 1F). The elevated activities of these pathways could be responsible for the more active immunotherapy response in cancer.
3.2. Association of ACE2 expression with oncogenic pathways and tumor phenotypes in cancer
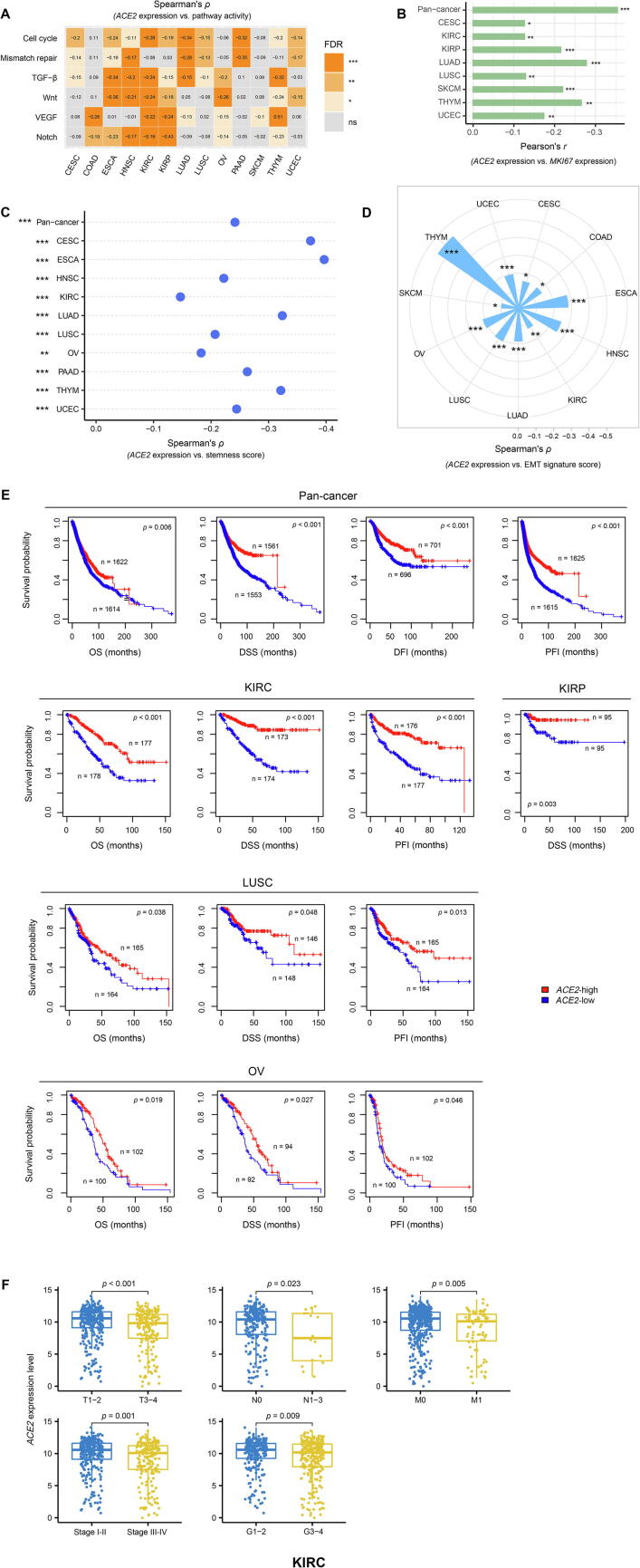
We quantified the activity of a pathway using the single-sample gene-set enrichment analysis (ssGSEA) [23] score of the set of genes included in the pathway. We found that ACE2 expression levels inversely correlated with the activity of cell cycle, mismatch repair, TGF-β, Wnt, VEGF, and Notch signaling pathways in 10, 7, 9, 7, 6, and 7 individual cancer types, respectively (Spearman’s correlation test, FDR < 0.05) (Fig. 2A). Moreover, we found that ACE2 expression levels had a significant inverse correlation with the expression levels of MKI67, which is a tumor proliferation index marker, in pan-cancer and 8 individual cancer types (Pearson’s correlation test, FDR < 0.05) (Fig. 2B). Tumor stemness represents a stem cell-like tumor phenotype associated with tumor progression, metastasis, immune evasion, and drug resistance. We found that ACE2 expression levels showed a marked negative correlation with tumor stemness scores (ssGSEA scores) in pan-cancer and in 10 individual cancer types (FDR < 0.05) (Fig. 2C). EMT plays an outstanding role in facilitating malignant transformation, tumor progression, and metastasis. We observed a marked negative correlation between ACE2 expression levels and EMT signature scores (ssGSEA scores) in 11 individual cancer types (FDR < 0.05) (Fig. 2D). Overall, these data indicate that ACE2 could be a protective factor for cancer progression. Indeed, survival analyses showed that ACE2 upregulation was associated with favorable survival in pan-cancer (log-rank test, p < 0.01 for OS, DSS, PFI, and DFI) and in KIRC, KIRP, LUSC, and OV (log-rank test, p < 0.05 for OS, DSS, PFI, and/or DFI) (Fig. 2E). Furthermore, we found that ACE2 expression levels significantly decreased with the tumor advancement in KIRC (two-sided Student’s t test, p < 0.05, fold change > 1.5 for high-grade (G3-4) versus low-grade (G1-2), late-stage (stage III-IV) versus early-stage (stage I-II), large tumor size (T3-4) versus small tumor size (T1-2), with lymph nodes (N1-3) versus without regional lymph nodes (N0), and metastasis (M1) versus no metastasis (M0)) (Fig. 2F).
Fig. 2.
Association of ACE2 expression with oncogenic pathways and tumor phenotypes in cancer. ACE2 expression levels are likely to inversely correlate with the activity of oncogenic pathways (A), MKI67 expression levels (B), stemness scores (C), and EMT signature scores (D) in cancer. EMT: epithelial-mesenchymal transition; (E) Kaplan-Meier survival curves showing that ACE2 upregulation is associated with favorable survival in pan-cancer and multiple individual cancer types. Log-rank test p-values are shown. OS: overall survival; DSS: disease-specific survival; PFI: progression-free interval; DFI: disease-free interval. (F) ACE2 expression levels significantly decrease with tumor advancement in KIRC. KIRC: kidney renal clear cell carcinoma.
3.3. Identifying interaction networks of ACE2 in cancer
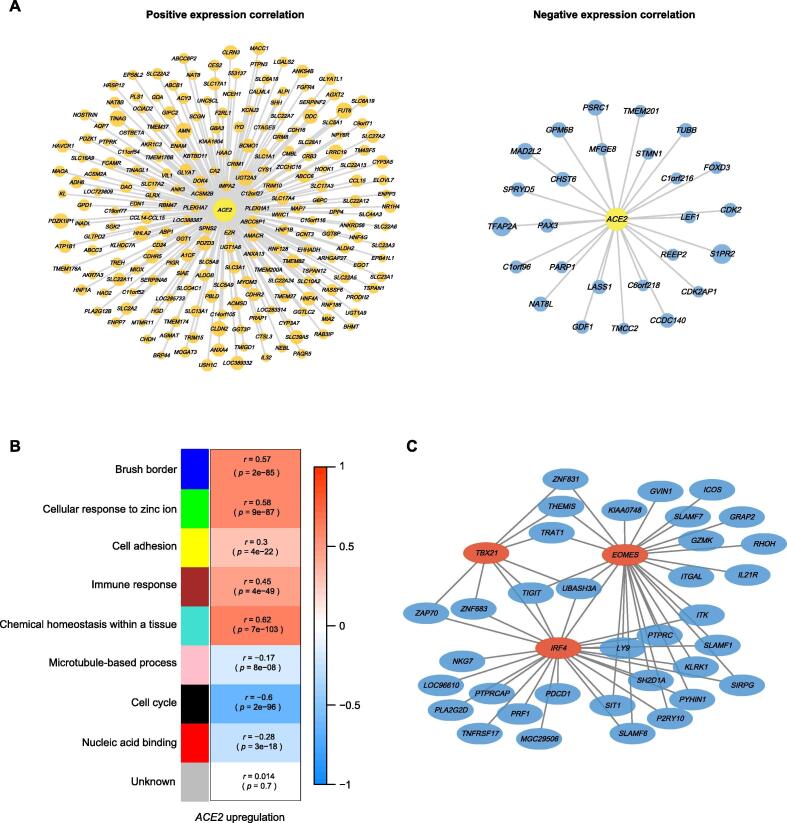
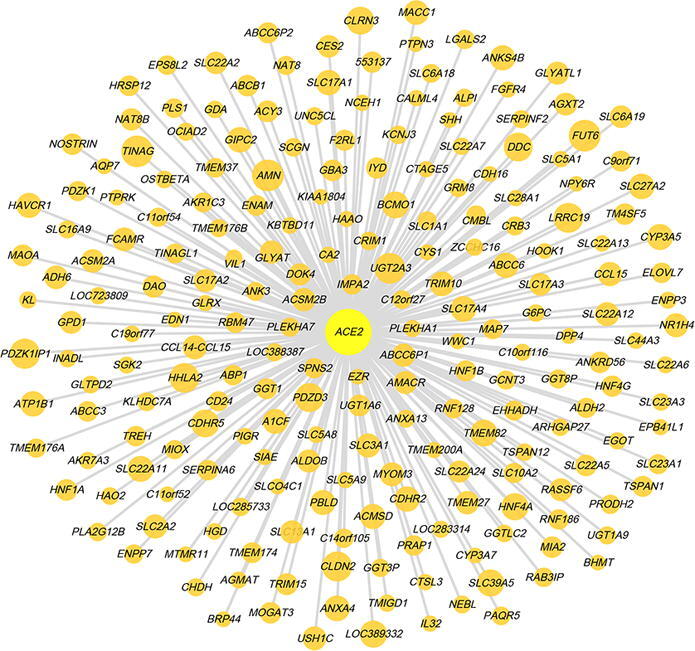
We identified 217 and 26 genes having marked positive and negative expression correlations with ACE2 in pan-cancer, respectively (|r| > 0.5) (Fig. 3A and Supplementary Table S3). WGCNA [26] identified five gene modules (indicated in blue, green, yellow, brown, and turquoise color, respectively) highly enriched in the high-ACE2-expression-level tumors and three gene modules (indicated in pink, black, and red color, respectively) highly enriched in the low-ACE2-expression-level tumors (Fig. 3B). The GO terms highly enriched in the high-ACE2-expression-level tumors mainly included brush border, cellular response to zinc ion, cell adhesion, immune response, and chemical homeostasis within a tissue. In contrast, the GO terms highly enriched in the low-ACE2-expression-level tumors mainly included microtubule-based process, cell cycle, and nucleic acid binding (Fig. 3B). Again, these results indicate that ACE2 expression has a significant positive association with antitumor immune response and a significant negative association with the cell cycle in cancer, suggesting the potential protective role of ACE2 from cancer progression.
Fig. 3.
Interaction networks of ACE2 in cancer. (A) 217 and 26 genes having marked positive and negative expression correlations with ACE2 in pan-cancer, respectively (|r| > 0.5). The size of nodes is proportional to the absolute values of the expression correlation coefficients. (B) Gene modules (gene ontology) enriched in high-ACE2-expression-level and low-ACE2-expression-level pan-cancer. (C) Co-expression subnetwork of the immune response module (in brown) enriched in high-ACE2-expression-level pan-cancer centered on three transcription factor genes (EOMES, IRF4, and TBX21).
From the brown gene module, we identified 82 hub genes mainly associated with immune-related pathways. Among the 82 hub genes, three transcription factor (TF) genes, including EOMES, IRF4, and TBX21, were co-expressed with many other immune-related genes, such as PDCD1, TIGIT, GZMK, IL21R, and PRF1 (Fig. 3C). The association between these TFs and immune regulation has been well recognized, such as EOMES (Eomesodermin) mediating the CD8+ T cell differentiation [28], IRF4 (interferon regulatory factor 4) regulating immune cell development [29], and TBX21 (T-bet) playing a pivotal role in regulating Th1 cell development [30].
4. Discussion
We investigated the association of ACE2 expression with immune signatures, oncogenic pathways, and tumor phenotypes in diverse cancer cohorts. Our results indicate that ACE2 is a potential protective factor for cancer progression. In particular, the ACE2 downregulation correlates with worse survival and tumor advancement in KIRC, also known as clear cell renal cell carcinoma (ccRCC). Previous studies demonstrated that ACE2 exerts antitumor effects by inhibiting tumor angiogenesis [13] and promoting tumor immune infiltration [9]. Our results are consistent with these previous findings. Besides, we found that ACE2 upregulation was associated with reduced cell proliferation, stemness, and EMT, as well as the downregulation of oncogenic pathways, such as cell cycle, mismatch repair, TGF-β, Wnt, and Notch signaling. Moreover, we found that ACE2 had a positive expression correlation with PD-L1, a predictive marker for an active response to immune checkpoint inhibitors. As a result, ACE2 upregulation correlates with a favorable response to anti-PD-1/PD-L1/CTLA-4 immunotherapy.
Our and others’ studies indicate that ACE2 plays a protective role in cancer, hypertension, heart disease, and COVID-19 patients. An intriguing phenomenon is that ACE2 as a SARS-CoV-2 human host cell receptor is crucial for SARS-CoV-2 to invade human cells, while its deficiency may exacerbate outcomes in COVID-19 patients [8]. A potential explanation is that the ACE2 deficiency could worsen outcomes in the people with underlying conditions, such as hypertension, heart disease, and cancer, who are infected with COVID-19. Thus, using ACE2 inhibitors for preventing and treating COVID-19 may not be an advisable strategy for individuals with certain underlying conditions, such as hypertension, heart disease, and cancers.
It should be noted that the negative correlation between ACE2 and cancer progression presented in this study is an association but not a causation. To prove their causal relationship, further experiments are necessary. This would be an important direction for further studies.
5. Conclusions
ACE2 upregulation was associated with increased antitumor immunity and immunotherapy response, reduced tumor malignancy, and favorable survival in cancer, suggesting that ACE2 is a potential protective factor for cancer progression. Our data may provide potential clinical implications for treating cancer patients infected with SARS-CoV-2.
Funding
This work was supported by the China Pharmaceutical University (grant numbers 3150120001 to XW).
CRediT authorship contribution statement
Zhilan Zhang: Software, Validation, Formal analysis, Investigation, Data curation, Writing - review & editing, Visualization. Lin Li: Software, Formal analysis, Investigation, Visualization. Mengyuan Li: Software, Formal analysis, Investigation, Visualization. Xiaosheng Wang: Conceptualization, Methodology, Investigation, Resources, Writing - original draft, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.08.024.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2) doi: 10.1016/j.cell.2020.02.052. 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894–904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan R., Zhang Y. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan J., Ge J., Yu J. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 5.Danilczyk U., Penninger J.M. Angiotensin-converting enzyme II in the heart and the kidney. Circ Res. 2006;98(4):463–471. doi: 10.1161/01.RES.0000205761.22353.5f. [DOI] [PubMed] [Google Scholar]
- 6.Li M.-Y., Li L., Zhang Y., Wang X.-S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9(1):45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith J.C., Sausville E.L., Girish V., Yuan M.L., Vasudevan A., John K.M., Sheltzer J.M. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE2 in the respiratory tract. Dev Cell. 2020;53(5) doi: 10.1016/j.devcel.2020.05.012. 514–529.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J., Li H., Hu S., Zhou Y. ACE2 correlated with immune infiltration serves as a prognostic biomarker in endometrial carcinoma and renal papillary cell carcinoma: implication for COVID-19. Aging (Albany NY) 2020;12(8):6518–6535. doi: 10.18632/aging.103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai Y.J., Hu F., Li H., Huang H.Y., Wang D.W., Liang Y. A profiling analysis on the receptor ACE2 expression reveals the potential risk of different type of cancers vulnerable to SARS-CoV-2 infection. Ann Transl Med. 2020;8(7):481. doi: 10.21037/atm.2020.03.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai P., Yu J., Ge S., Jia R., Fan X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis. J Hematol Oncol. 2020;13(1):43. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winkler T., Ben-David U. Elevated expression of ACE2 in tumor-adjacent normal tissues of cancer patients. Int J Cancer. 2020 doi: 10.1002/ijc.33145. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q.i., Lu S., Li T., Yu L., Zhang Y., Zeng H. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J Exp Clin Cancer Res. 2019;38(1):173. doi: 10.1186/s13046-019-1156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y., Wan H., Liu J., Zhang R., Ma Q., Han B. The angiotensin-converting enzyme 2 in tumor growth and tumor-associated angiogenesis in non-small cell lung cancer. Oncol Rep. 2010;23(4):941–948. doi: 10.3892/or_00000718. [DOI] [PubMed] [Google Scholar]
- 15.Xu J., Fan J., Wu F., Huang Q., Guo M., Lv Z. The ACE2/angiotensin-(1–7)/Mas receptor axis: pleiotropic roles in cancer. Front Physiol. 2017;8:276. doi: 10.3389/fphys.2017.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Y., Ni L., Wan H., Fan L., Fei X., Ma Q. Overexpression of ACE2 produces antitumor effects via inhibition of angiogenesis and tumor cell invasion in vivo and in vitro. Oncol Rep. 2011;26(5):1157–1164. doi: 10.3892/or.2011.1394. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H., Quek K., Chen R., Chen J., Chen B. Expression of the SAR2-Cov-2 receptor ACE2 reveals the susceptibility of COVID-19 in non-small cell lung cancer. J Cancer. 2020;11(18):5289–5292. doi: 10.7150/jca.49462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X., He C., Hua X., Kan A., Sun S., Wang J. Bioinformatic analysis of correlation between immune infiltration and COVID-19 in cancer patients. Int J Biol Sci. 2020;16(13):2464–2476. doi: 10.7150/ijbs.48639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nathanson T., Ahuja A., Rubinsteyn A., Aksoy B.A., Hellmann M.D., Miao D. Somatic mutations and neoepitope homology in melanomas treated with CTLA-4 blockade. Cancer Immunol Res. 2017;5(1):84–91. doi: 10.1158/2326-6066.CIR-16-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ascierto M.L., Makohon-Moore A., Lipson E.J., Taube J.M., McMiller T.L., Berger A.E. Transcriptional mechanisms of resistance to anti-PD-1 therapy. Clin Cancer Res. 2017;23(12):3168–3180. doi: 10.1158/1078-0432.CCR-17-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ascierto M.L., McMiller T.L., Berger A.E., Danilova L., Anders R.A., Netto G.J. The intratumoral balance between metabolic and immunologic gene expression is associated with anti-PD-1 response in patients with renal cell carcinoma. Cancer Immunol Res. 2016;4(9):726–733. doi: 10.1158/2326-6066.CIR-16-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder A., Nathanson T., Funt S.A., Ahuja A., Buros Novik J., Hellmann M.D. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med. 2017;14(5) doi: 10.1371/journal.pmed.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B. 1995;57:289–300. [Google Scholar]
- 28.Knox J.J., Cosma G.L., Betts M.R., McLane L.M. Characterization of T-bet and eomes in peripheral human immune cells. Front Immunol. 2014;5:217. doi: 10.3389/fimmu.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam S., Lim J.-S. Essential role of interferon regulatory factor 4 (IRF4) in immune cell development. Arch Pharm Res. 2016;39(11):1548–1555. doi: 10.1007/s12272-016-0854-1. [DOI] [PubMed] [Google Scholar]
- 30.Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., Glimcher L.H. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.