Abstract
Diatoms are the largest group of heterokont algae with more than 100,000 species. As one of the single-celled photosynthetic organisms that inhabit marine, aquatic and terrestrial ecosystems, diatoms contribute ~ 45% of global primary production. Despite their ubiquity and environmental significance, very few diatom plastid genomes (plastomes) have been sequenced and studied. This study explored patterns of nucleotide substitution rates of diatom plastids across the entire suite of plastome protein-coding genes for 40 taxa representing the major clades. The highest substitution rate was lineage-specific within the araphid 2 taxon Astrosyne radiata and radial 2 taxon Proboscia sp. Rate heterogeneity was also evident in different functional classes and individual genes. Similar to land plants, proteins genes involved in photosynthetic metabolism have lower synonymous and nonsynonymous substitutions rates than those involved in transcription and translation. Significant positive correlations were identified between substitution rates and measures of genomic rearrangements, including indels and inversions, which is a similar result to what was found in legume plants. This work advances the understanding of the molecular evolution of diatom plastomes and provides a foundation for future studies.
Subject terms: Genome-wide analysis of gene expression, Genomics
Introduction
Diatoms are photosynthetic, unicellular eukaryotes of the heterokont algal lineage. Two hundred fifty million years ago, diatom plastids were derived from a secondary endosymbiotic event, in which a non-photosynthetic eukaryote phagocytized a red alga1. Diatoms have since colonized freshwater, marine and terrestrial habitats contributing ~ 45% of global primary production2–4 and as much as 20% of global carbon fixation via photosynthesis5, 6.
Despite their ubiquity and the environmental significance of diatom photosynthesis, very few diatom plastid genomes (plastomes) have been sequenced and studied. More than 2,900 plant species with plastomes were represented in the public databases based on searches in the NCBI on the February 4, 2019, but just 40 diatom taxa have been sequenced thus far7. The study of the sequences of plastomes can potentially reveal novel insights on relationships between monophyletic diatom lineages7–11. Researchers have also found support for the theory of shared ancestry between diatoms and rhodophytes12. Furthermore, the availability of plastomes has enabled exploration of the variation in structure and gene content across orders, genera and species7, 9, 10, 13–16.
Within the diatom cytoplasm, there are numerous or singular plastids of variable shapes17, 18. Four previously examined diatom species showed each of their plastids contained a single nucleoid19, which comprises copies of the plastome monomer or unit-genome, RNA and proteins20. Diatom plastid genes are densely arrayed on both strands of the unit-genome, which represent one full complement of the gene space and intergenic regions. The plastome of an individual may include many copies of the unit-genome by repeating this complement pattern many times. Although this unit is often diagrammed as a circular molecule, the plastome more likely contains a collection of circular, linear and linear-branched molecules that each comprises two to many copies of the monomer21. All diatom plastomes sequenced to date include a large inverted repeat (IR) separated by large and small single-copy regions (LSC and SSC, respectively). Apart from the typical quadripartite structure, an extensive range of gene order arrangements, gains and losses of genes are exhibited in the diatom plastomes9, 10. Gene order changes not only arise through gene duplication by IR expansion, but also via inversions and insertions and deletions (indels) in both IR and SC regions.
Calculation of synonymous (dS) and nonsynonymous (dN) nucleotide substitution rates across individual genes and their functional groups between lineages provide insights into the plastome evolution22. In previous studies, genes encoding subunits that are integral to photosynthesis, such as cytochrome b6f. complex (PET) and photosystems I and II (PSA and PSB) have lower rates of nucleotide substitution than other functional groups in angiosperms and conifers23–26. Accelerated substitution rates have been detected in ribosomal protein (RPL and RPS) genes and RNA polymerase (RPO) genes24, 26–30. Besides differences in substitution rates, variation relative to genomic features such as rearrangements in gene order can also shape plastome evolution. Previous studies have identified a significant positive correlation between rates of nucleotide substitution and gene order changes in angiosperm plastid genomes30, 31, bacterial genomes32 and arthropod mitochondrial genomes33, 34.
In a previous study by Schwarz et al.26, both nonsynonymous and synonymous substitution rates were negatively correlated with plastome sizes and rearrangements such as the number of inversions and indels. The focus of these investigations was on three of the six subfamilies of flowering plant family Fabaceae. One of the subfamilies, papilionoids, has a wide diversity of plastome rearrangements including the loss of inverted repeats (IR) in one clade and relatively smaller plastomes than the other subfamilies. This study also found that genes in the IR show three to fourfold reduction in substitution rates compared to SC regions. Genes that used to be in IR showed accelerated rates compared to genes retained in the IR. A negative correlation between substitution rates and cupressophyte plastid DNA genome size has also been reported in conifers37.
Our hypothesis is that the relationship of nucleotide substitution rates between plastid genes and plastome size and architecture such as inversions, indels, and IR in diatoms are similar to what was observed with legumes and conifers. If correct, this reflects a fundamental aspect of how diatoms evolve. To date, no study has investigated the nucleotide substitution rates of all shared plastome protein-coding genes in diatoms. The present study explored the patterns of plastid nucleotide substitution rates across the entire suite of 103 shared genes for 40 species of diatoms. Correlations between plastome substitution rates and genome features, including plastome size, number of indels and genome rearrangement were examined. This work advances the current understanding of the molecular evolution of diatom plastomes.
Results
Phylogenetic relationships and branch lengths
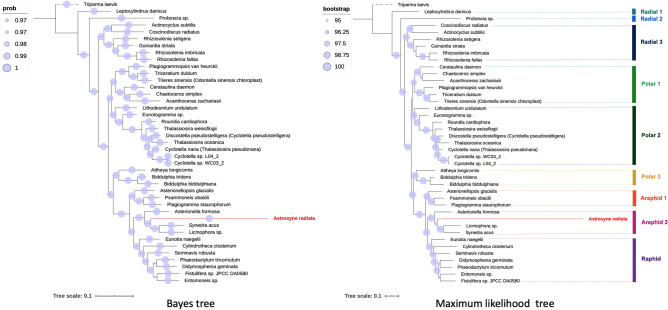
Phylogenetic analysis of 40 previously published diatom plastomes (Table S1) for the concatenated 103 gene data set (Table S2) generated Bayes and maximum likelihood (ML) trees with robust support of > 0.97 posterior probabilities and > 95% bootstraps for most of the branches, respectively (Fig. 1). The radial centrics of the Coscinodiscophyceae (radial 1, 2 and 3) formed a basal grade. The Mediophyceae including bi-polar and multi-polar diatoms plus the Thalassiosirales are paraphyletic and contained in three clades (polar 1, 2 and 3). Araphid 1 was sister to araphid 2, and phylogenetically close to another group, raphids. Raphid pennate diatoms were monophyletic. Within araphid 2, Astrosyne radiata showed an extremely long branch in both Bayes and ML trees (Fig. 1).
Figure 1.
Bayes and maximum likelihood (ML) trees are constructed using 103 concatenated protein-coding gene sequences from each diatom. There is strong bootstrap support for all branches (> 97%) in the ML tree. Astrosyne radiata has an unusually long branch length in both Bayes and ML trees. 40 species are grouped into nine morphological categories. Note that the outgroup, Triparma laevis, is not included into the nine categories.
Substitution rates in individual genes and functional groups
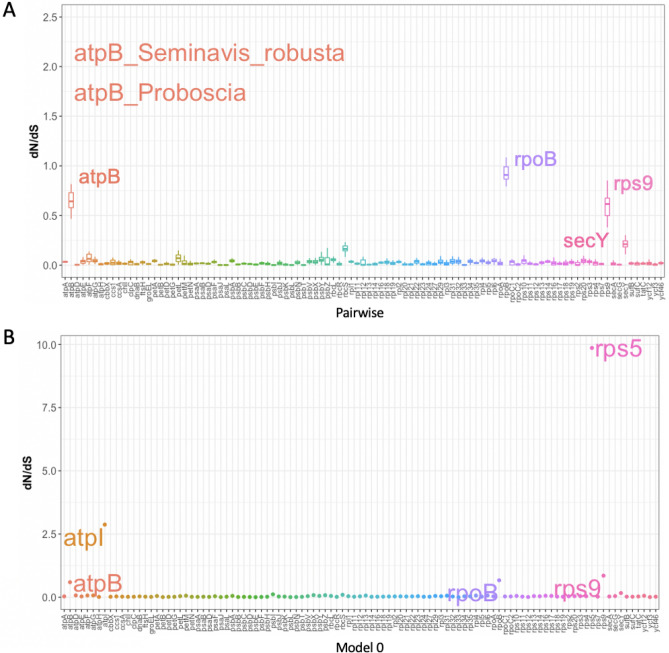
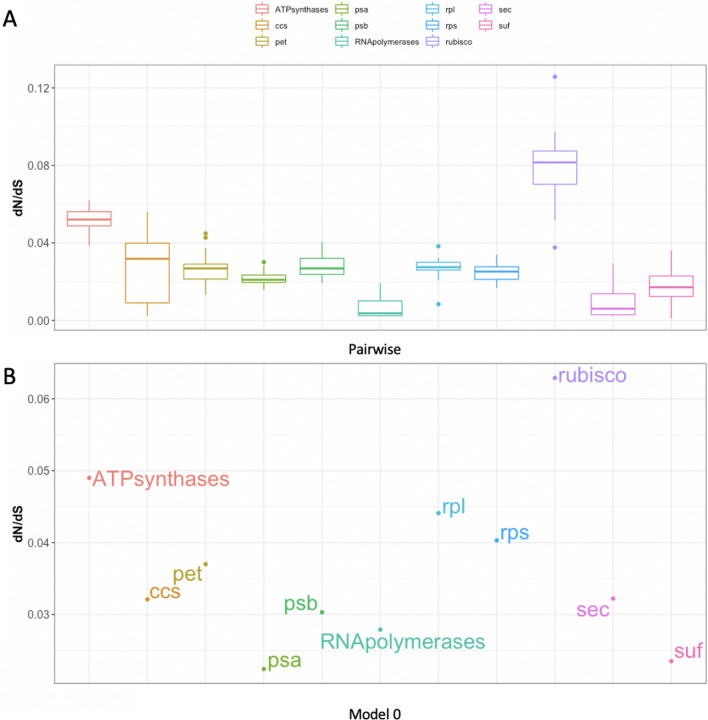
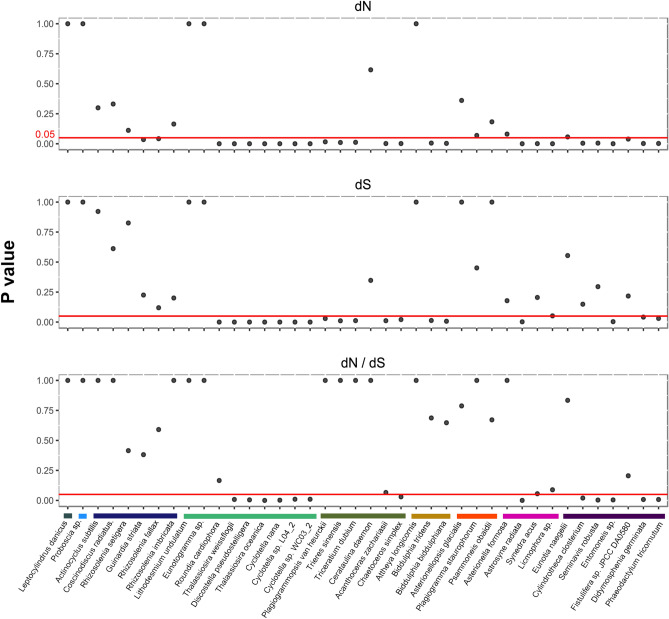
Estimations of dN, dS (Fig S1) and dN/dS (ω) (Figs. 2, 3) values were done for individual genes and different functional classes (Tables S1, S2). Two methods including pairwise and CODEML model 0 were used to estimate the nucleotide substitution rates. The dN/dS values were more widely distributed for individual genes (Fig. 2), whereas the ratio was limited to 0 to 0.13 for different functional classes (Fig. 3). Two genes, rps5 and atpI, have high dN/dS values > 1 and were subjected to positive selection analyses using CODEML model 8 versus 7. A total of 63 and 64 positively selected sites were detected in rps5 and atpI, respectively (Table 1). Four genes atpB, rpoB, rps9 and secY had higher dN/dS values, which suggested accelerated evolution of these genes. In particular, atpB in Proboscia sp. and Seminavis robusta showed dN/dS values of 1.75 and 2.15, respectively. When grouping genes into functional groups, all had dN/dS values close to 0 with the RubisCo subunit having median dN/dS values of only 0.08 (Fig. 3A), which suggested some genes with rapid evolution have their effects masked by other genes in the same functional group with purifying selection.
Figure 2.
Distribution of dN/dS values for individual genes across all diatoms. Two types of dN/dS methods were used; (A) one based on pairwise dN/dS rates against the outgroup, Triparma laevis, and (B) the other method was based on PAML model 0 that produced a single dN/dS value for each orthologous set. The orthologous set has their gene names arranged in alphabetical order from left to right. Genes with higher dN/dS rates are indicated on the plot. In the pairwise model, the ratio was visualised by boxplot with each boxplot consisting of 40 species dN/dS values. rps5 and atpI were not shown on the pairwise dN/dS boxplots because these are special cases subjected to CODEML model 8 vs 7 comparison in Table 1.
Figure 3.
Distribution of dN/dS values for functional groups across all diatoms. The same pairwise and PAML model 0 dN/dS methods as described in the legend of Fig. 2 were used to calculate dN/dS values. The difference is functional groups consist of a set of genes. Table S2 shows how the genes were grouped into the 11 categories.
Table 1.
CODEML model 8 vs 7 comparison for rps5 and atpI and positively selected sites.
| Gene | Model | Log-likelihood | 2Δ (in L) | p-value | Positively selected sites | Tree length |
|---|---|---|---|---|---|---|
| rps5 | M7 | − 6,492.11 | ||||
| M8 | − 6,275.91 | 432.4 | 1.28e–94 | 3 T, 7 K , 8 T, 9 N, 10R, 11I, 14C, 15 N, 24S, 25 N, 28 N, 29 W, 31 W, 33 R, 34 N, 35 W, 36 N, 37C, 40C, 42F, 45 K, 48S, 50 N, 51S, 52S, 53Y, 54 N, 55I, 57S, 58 T, 59C, 60C, 61L, 63Y, 64Y, 65 T, 66C, 67Y, 70R, 71G, 74R, 75 W, 76C, 77S, 78 N, 79S, 80I, 82C, 83R, 84Y, 87 N, 92I, 97S, 98C, 99 N, 100 W, 101F, 102I, 104 N, 105I, 107 K, 108 P, 109S | 14.81 | |
| atpI | M7 | − 9,013.33 | ||||
| M8 | − 8,757.67 | 511.32 | 9.30e−112 | 1 K, 10L, 11 V, 13Y, 15 V, 18Y, 21Q, 22E, 23L, 24 K, 25I, 27L, 28 M, 30G, 36L, 37H, 38I, 39L, 41L, 43I, 48F, 53Y, 54H, 55I, 56 V, 57L, 63 V, 65I, 67 V, 68Q, 69F, 70L, 75Q, 77 V, 79Q, 80L, 81Q, 82L, 86L, 87L, 89R, 92P, 101D, 102 V, 104L, 107 M, 108L, 109 T, 110Q, 123L, 132, 133E, 136P, 139L, 140L, 142R, 144Y, 150Y, 151D, 154P, 163L, 164P, 166H, 170H | 10.92 |
Bayes Empirical Bayes (BEB) was used to calculate posterior probabilities and only those with Prob(ω > 1) > 0.99 were shown. The p-value was calculated with degree of freedom equals to 2.
Gene order in diatoms
Gene order analysis using MAUVE revealed substantial rearrangements of blocks of sequences in the 40 diatom species (Table 2). Only 14 species had identical gene order shared with at least one other species.
Table 2.
Local collinear blocks (LCBs) for each of the 40 diatom plastomes identified by Mauve.
| Species | Gene collinear block order |
|---|---|
| Leptocylindrus danicus | 27 26 − 28 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 1 2 3 4 − 16 − 15 − 23 21 20 19 13 25 24 42 39 33 34 35 36 37 32 − 31 − 30 − 29 − 38 − 40 − 41 |
| Probosica sp. | − 1 2 3 4 − 5 − 6 − 7 − 8 − 9 − 10 11 − 12 − 13 − 14 15 16 17 18 − 19 − 20 − 21 22 − 23 − 24 25 − 26 − 27 − 28 29 30 31 − 32 33 34 35 36 37 − 38 − 39 − 40 − 41 − 42 |
| aActinocyclus subtilis | 2 3 4 − 13 − 19 − 20 − 21 − 1 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 23 15 16 28 27 26 25 24 42 41 40 39 38 29 30 31 33 34 35 36 37 32 |
| aCoscinodiscus radiatus | 2 3 4 − 13 − 19 − 20 − 21 − 1 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 23 15 16 28 27 26 25 24 42 41 40 39 38 29 30 31 33 34 35 36 37 32 |
| Rhizosolenia setigera | 2 3 4 − 13 − 19 − 20 − 21 − 1 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 23 15 16 28 27 26 25 24 42 41 − 32 − 37 − 36 − 35 − 34 − 33 29 30 31 − 38 − 39 − 40 |
| Guinardia striata | 2 3 4 − 13 23 15 16 28 27 26 10 9 8 7 6 5 17 18 22 12 14 11 1 21 20 19 − 41 − 42 − 24 − 25 − 32 − 37 − 36 − 35 − 34 − 33 29 30 31 − 38 − 39 − 40 |
| Rhizosolenia fallax | 2 3 4 − 13 − 19 − 20 − 21 − 1 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 − 26 − 27 − 28 − 16 − 15 − 23 25 24 42 41 − 32 − 37 − 36 − 35 − 34 − 33 29 30 31 − 38 − 39 − 40 |
| Rhizosolenia imbricate | − 4 − 3 − 2 − 13 − 19 − 20 − 21 − 1 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 − 26 − 27 − 28 − 16 − 15 − 23 25 24 42 41 − 32 − 37 − 36 40 39 38 29 30 31 33 34 35 |
| Lithodesmium undulatum | 2 3 4 − 13 − 19 − 20 − 21 − 1 − 11 18 22 12 14 − 17 − 5 − 6 − 7 − 8 − 9 − 10 23 15 16 28 27 26 25 29 30 31 33 34 35 36 37 32 − 38 − 39 − 40 − 41 − 42 − 24 |
| Eunotogramma sp. | 22 12 14 11 16 28 27 26 2 3 4 − 13 − 19 − 20 − 21 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 15 − 23 1 25 24 42 41 40 39 38 30 31 33 34 35 36 37 32 − 29 |
| bRoundia cardiophora | 2 3 4 17 18 22 − 26 − 27 − 28 − 16 12 14 10 9 8 7 6 5 11 23 − 15 1 21 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| bThalassiosira weissflogii | 2 3 4 17 18 22 − 26 − 27 − 28 − 16 12 14 10 9 8 7 6 5 11 23 − 15 1 21 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| bDiscostella pseudostelligera | 2 3 4 17 18 22 − 26 − 27 − 28 − 16 12 14 10 9 8 7 6 5 11 23 − 15 1 21 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| Thalassiosira oceania | 2 3 4 17 18 28 27 26 16 22 23 − 15 − 1 21 20 19 − 5 − 6 − 7 − 8 − 9 − 10 − 14 − 12 11 13 25 − 38 − 39 24 42 41 40 32 − 31 − 30 − 29 33 34 35 36 37 |
| bCyclotella_nana | 2 3 4 17 18 22 − 26 − 27 − 28 − 16 12 14 10 9 8 7 6 5 11 23 − 15 1 21 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| cCyclotella sp. L04_2 | 2 3 4 16 28 27 26 − 22 − 18 − 17 12 14 10 9 8 7 6 5 11 23 − 15 1 21 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| cCyclotella sp. WC03_2 | 2 3 4 16 28 27 26 − 22 − 18 − 17 12 14 10 9 8 7 6 5 11 23 − 15 1 21 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| Plagiogrammopsis van heurckii | 2 3 4 10 9 8 7 6 5 17 18 22 12 14 1 21 20 19 13 15 16 23 − 26 − 27 − 28 11 25 24 42 41 40 39 38 − 29 30 31 33 34 35 36 37 32 |
| dTrieres sinensis | 2 3 4 10 9 8 7 6 5 17 18 22 12 14 1 21 20 19 13 15 16 28 27 26 23 11 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| dTriceratium dubium | 2 3 4 10 9 8 7 6 5 17 18 22 12 14 1 21 20 19 13 15 16 28 27 26 23 11 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| Cerataulina daemon | 2 3 4 − 23 10 9 8 7 6 5 17 18 22 12 14 1 21 20 19 13 15 16 28 27 26 11 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| Acanthoceras zachariasii | 10 9 8 2 3 4 7 6 5 17 18 22 12 14 1 21 20 19 13 15 16 28 27 26 23 11 25 29 30 31 33 34 35 36 37 32 24 42 41 40 39 38 |
| Chaetoceros simplex | 10 9 8 2 3 4 7 6 5 17 18 22 12 14 1 − 19 − 20 − 21 13 15 16 28 27 26 23 11 25 29 30 31 33 34 35 36 37 32 24 42 41 40 39 38 |
| Attheya logicornis | 2 3 4 13 − 19 − 20 − 21 23 15 16 1 28 27 26 22 12 14 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 11 25 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 38 |
| eBiddulphia tridens | 2 3 4 13 − 19 − 20 − 21 23 15 16 28 27 26 1 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 11 25 29 30 31 33 34 35 36 37 32 24 42 41 40 39 38 |
| eBiddulphia biddulphiana | 2 3 4 13 − 19 − 20 − 21 23 15 16 28 27 26 1 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 11 25 29 30 31 33 34 35 36 37 32 24 42 41 40 39 38 |
| Asterionellopsis glacialis | 2 3 4 13 − 23 − 1 21 20 19 15 16 − 11 − 14 − 12 − 22 − 18 − 17 10 9 8 7 6 5 − 26 − 27 − 28 25 − 38 33 34 35 36 37 − 39 − 40 − 41 − 42 − 24 − 31 − 30 − 29 32 |
| Plagiogramma staurophorum | − 21 1 23 − 4 − 3 − 2 10 9 8 7 6 5 17 18 22 12 14 11 − 16 − 15 28 27 26 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 − 29 30 31 33 34 35 36 37 32 |
| Psammoneis obaidii | 7 6 5 2 3 4 − 13 − 19 − 20 − 21 1 23 15 16 − 11 17 18 22 12 14 − 26 − 27 − 28 10 9 8 25 − 38 32 29 30 31 33 34 35 36 37 24 42 41 40 39 |
| Asterionella formosa | 2 3 4 − 26 − 27 − 28 10 9 8 7 6 5 17 18 22 12 14 23 15 16 − 11 1 21 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 − 32 |
| Astrosyne radiata | − 20 − 21 − 1 23 15 16 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 28 27 19 − 13 − 4 − 3 − 2 26 25 − 38 30 31 29 − 32 33 34 35 36 37 24 42 41 40 39 |
| Synedra acus | 2 3 4 − 13 − 19 − 20 − 21 1 23 15 16 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 28 27 26 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| Licmorphora sp. | − 4 − 3 − 2 − 26 − 27 − 28 10 9 8 7 6 5 17 18 22 12 14 11 − 16 − 15 − 23 − 1 21 20 19 13 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 − 32 − 37 − 36 − 35 − 34 − 33 |
| Eunotia naegelii | 2 3 4 − 13 − 19 − 20 − 21 − 1 23 15 16 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 − 26 − 27 − 28 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| Cylindrotheca closterium | − 2 − 23 9 8 7 6 5 17 18 22 12 14 11 − 16 − 15 − 13 − 19 − 20 − 21 1 − 26 − 27 − 28 − 10 25 − 32 33 34 35 36 37 29 30 31 24 42 41 40 39 38 − 4 − 3 |
| Seminavis robusta | − 4 − 3 − 2 − 13 − 19 − 20 − 21 − 1 23 15 16 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 26 − 27 − 28 25 − 38 − 39 − 40 − 41 − 42 − 24 − 37 − 36 − 35 − 34 − 33 − 31 − 30 − 29 32 |
| Entomoneis sp. | − 4 − 3 − 2 − 13 − 19 − 20 − 21 − 1 23 15 16 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 − 26 − 27 − 28 − 25 − 29 30 31 33 34 35 36 37 32 24 42 41 40 39 38 |
| Fistulifera sp JPCC DA0580 | − 4 − 3 − 2 − 13 − 19 − 20 − 21 − 1 23 15 16 − 26 − 27 − 28 10 9 8 7 6 5 17 18 22 12 14 11 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| fDidymosphenia geminata | − 4 − 3 − 2 − 13 − 19 − 20 − 21 − 1 23 15 16 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 − 26 − 27 − 28 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
| fPhaeodactylum tricornutum | − 4 − 3 − 2 − 13 − 19 − 20 − 21 − 1 23 15 16 − 11 − 14 − 12 − 22 − 18 − 17 − 5 − 6 − 7 − 8 − 9 − 10 − 26 − 27 − 28 25 − 38 − 39 − 40 − 41 − 42 − 24 29 30 31 33 34 35 36 37 32 |
Negative numbers indicate an inversion in a given LCB. Only one IR was included in this analysis. The same gene order is highlighted with the same superscript letter before the species name.
Correlation of substitution rates and plastome characteristics
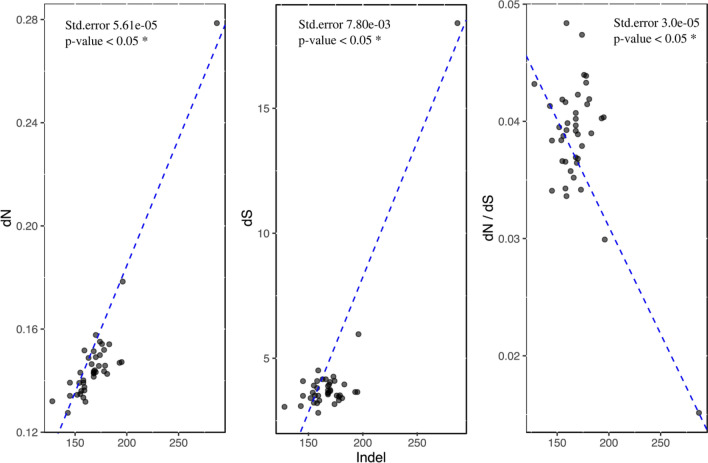
All correlations of the parameters of interest were visualised in Fig. S2. Significant correlation was observed between the number of indels and dN, dS and dN/dS (p < 0.05; Fig. 4). No significant correlation was found between the substitution rates and plastome size (Fig. S3). Astrosyne radiata, which has a relatively small plastome among diatoms, had the highest overall dN and dS (Fig. S3, Table S5). Significant correlations were found between plastome size and the length of the inverted repeat (IR), and the length of the small/large single copy region (SSC, LSC respectively) (Fig. S4).
Figure 4.
Relationship between the number of indels and substitution rates. Scatterplots were constructed and the regression line (dashed blue) and statistical values are shown. X-axis gives the number of indels in each species.
Correlation of pairwise substitution rates and inversion distance (Table S6) was tested in the 40 diatom plastomes. Significant correlation (p < 0.05) was found between dN and inversion distance in 25 out of 40 pairwise comparisons (Fig. 5; Table S6). Among the 40 plastomes, dS was significantly correlated with inversion distance in 18 pairwise comparisons, whereas the number of significant pairwise comparisons reduced to 13 for dN/dS values. The polar 1 group had the largest proportion of significant correlations between substitution rates and inversion distances. Seven of nine sampled taxa were significantly correlated in both dN and dS, and six of nine taxa were significantly correlated in dN/dS. Astrosyne radiata, which produced the longest branch in the diatom phylogeny (Fig. 1), also showed significant correlation of dN (p-value = 2.41e−06), dS (p-value = 2.23e−03), and dN/dS (p-value = 2.54e−03) with inversion distance (Fig. 5; Table S6).
Figure 5.
Pairwise correlation of substitution rate and plastome inversion distance in diatoms. 0.05 (red horizontal line) was used to assess the level of significance; P-values were plotted on the X-axis. Coloured bars indicate different clades of diatoms and correspond to Fig. 1. From left to right: radial 1, radial 2, radial 3, polar 1, polar 2, polar 3, araphid 1, araphid 2, raphid.
Discussion
Only limited studies have been performed using plastome protein-coding sequences from diatoms and not much is known about their molecular evolution. In this study, 103 plastid genes were examined across 40 diatom species, most of which were recently published by our group7, 9, 10. The ribosomal subunit and RNA polymerase genes have higher nucleotide substitution rates than other functional groups. Positive correlations are evident between dN and dS values and number of indels and inversion distances, which are proxies of genome rearrangements. Unlike the studies on legumes and conifers, we found no strong correlation between nucleotide substitution rates and diatom plastome size. The reason for the differences between diatoms and plants with respect to substitution rates may be attributed to fundamental differences in their genome content. Diatom plastomes are gene dense, with very little space dedicated to non-coding sequences and most are devoid of large repeat sequences61. However, the average diatom plastomes size is close to those of seed plants because they encode for more genes7, 9, 10, 62. Previous studies in diatoms also showed that variation in the unit-genome size is mainly due to expansion and contraction of the IR, gene loss and the introduction of foreign DNA of unknown origin7, 9, 10.
Astrosyne radiata, which is known to have undergone many gene loss events7, had the highest dN and dS but a relatively small plastome. This finding is in agreement with the legume plants26, 37. Perhaps species closely related to A. radiata will show the same negative correlation between substitution rates and plastome size but more sampling of related species is needed to confirm this observation. Astrosyne radiata is highly unusual from a morphological perspective, which perhaps explains its unusually long branch length in the phylogenetic tree. Although it is placed among araphid pennates, this diatom has elongated sternum and bilateral symmetry, and they have fully reverted to the ancestral radial symmetry (where all structures are rotationally arranged and symmetric around a single point in the centre) of diatoms in radial 3, such as Coscinodiscus and Actinocyclus.
Significant positive correlations were identified between substitution rates and two measures of genomic rearrangements, indels and inversions. This result is similar to legumes26 but not to conifers37. A recent study has also found significant correlations between branch length and gene order changes17 for two of the taxa in this study, Astrosyne radiata and Proboscia sp. This suggests that the evolutionary forces shaping the structural rearrangements between plants and diatoms are similar. Disruption of DNA repair, recombination and replication (DNA-RRR) systems has been suggested to cause highly elevated nucleotide substitution rates and genome rearrangements24, 35. A recent study revealed the potential correlation between dN rates of nuclear encoded DNA-RRR genes of plastomes and measures of plastome complexity in one angiosperm family36.
Like land plants, diatom plastid genes mainly fall into two general classes, those encoding proteins involved in photosynthetic metabolism (PSA, PSB, PET, ATP) and those with roles in transcription and translation (RPS, RPL, RPO). The finding that genes involved in photosynthesis had relatively lower overall substitution rates than genes in transcription-translation apparatus confirms that rate heterogeneity by functional class is a shared feature of diatoms and land plant plastomes.
Upon inspection of the protein alignments of the two positively selected genes, rps5 and atpI, we found that Thalassiosira oceania and A. radiata have very divergent sequences but were annotated with the correct gene symbols. The possibility of annotation errors leading to statistically significant positive selection cannot be discounted.
Gene essentiality is a widely studied factor in substitution rate variation, with the idea that essential genes are subject to stronger selective constraints than non-essential genes45–47. Several studies utilizing nuclear sequences have demonstrated that rates of nucleotide substitution are associated with gene expression levels where highly or more widely expressed genes evolve at slower rates in plants48–50 and animals51–53 supporting the notion that these genes may evolve under greater selective constraints. The slow rates of evolution in most of the genes examined in our study suggests they are essential genes.
In summary, positive correlations between nucleotide substitution rates and plastome rearrangements in both diatoms and legume plants motivate further studies to explore causal relationships between rates and plastome features. This will require expanded plastome sampling, both within and between diatom lineages. Future diatom studies should also consider the aspect of coevolution between nuclear and plastome genes, which has been done in several plant lineages43, 44, 54, 55.
Methods
Sequence alignment and phylogenetic analysis
Plastid protein-coding genes were extracted from all available complete diatom plastomes (40 taxa) together with the outgroup species Triparma laevis (Bolidophyceae) (Table S1). If similar sequences were annotated with the same gene names (i.e. isoforms) orthologs were selected using a phylogenetic tree-based approach57. Protein-coding genes were partitioned by functional category following Yu et al.7 The gene sequences were translated with the transeq function in EMBOSS v6.5.758. Both gene and protein sequences were aligned with Multiple Alignment using Fast Fourier Transform (MAFFT v7.305)38. The aligned FASTA files of gene sequences were altered to PHYLIP format with ALTER v1.3.459, and matched with the aligned protein sequences with PAL2NAL v14.160. The Bayesian phylogenetic analysis was conducted under the GTR + G model using MrBayes v3.2.656 with aligned protein sequences. The Markov chain Monte Carlo (mcmc) with default chain temperatures were run for 50,000 generations in two runs. The maximum likelihood trees were constructed with RAxML 7.2.939, with the substitution model GTR + G and -f option. One thousand bootstrap replicates were performed to assess strength of support for clades. The maximum likelihood trees of individual genes and functional groups were then used as the constraint trees to estimate the substitution rates from individual-gene and functional-group levels, respectively.
Nucleotide substitution rates
Nucleotide substitution rates (dN and dS) were estimated using the CODEML function implemented in PAML v4.840. Gapped regions were excluded with the parameter “-nogap” flag in PAL2NAL to avoid spurious rate inference. Pairwise rates were calculated relative to the outgroup species Triparma laevis and estimated with the parameter runmode = − 2. All shared plastome genes (103) were concatenated for nucleotide substitution rate estimation and separate estimations were calculated on individual genes or concatenated sequences of genes in different functional groups as listed in Table S2. CODEML model 0 was also used to estimate dN/dS values at the level of individual genes and functional groups. For genes with dN/dS > 1 in model 0, these genes were tested further with CODEML model 7 (neutral) and model 8 (positive selection) to uncover potential positively selected sites using similar methodology as described previously61.
Plastome features for correlation analyses
The number of indels for the concatenated 103 protein-coding genes was calculated using a custom Python script. Triparma laevis (Bolidophyceae) was used as a reference. Indels within aligned protein-coding regions were summed using a custom Python script resulting in a single value for each taxon; only intact genes were included (in-frame indels). Whole genome alignment among the 40 diatom species was performed using the ProgressiveMauve algorithm in Mauve v2.3.141. The same IR copy (IRb) was removed from all plastomes. The locally collinear blocks (LCBs) identified by Mauve were numbered with positive or negative sign based on strand orientation to estimate genome rearrangement distance (Table S3). Pairwise inversion (IV) distances were estimated using Genome Rearrangements In Man and Mouse (GRIMM; Table S4)42. The feature ‘plastome size’ excludes one copy of the IR for each taxon.
Correlation between substitution rates and genome characteristics
Pairwise dN and dS values were calculated for the 103 shared genes from each taxon relative to the outgroup Triparma laevis. Correlation of dN and dS with plastome size and indel number for each plastome was tested. Phylogenetic Generalized Least Squares was performed using the ape v5.462 and nlme v3.163 packages in R. The ML constraint tree was utilized with outgroup taxa pruned. The correlation between dN and dS with IV distance was tested using the Pearson test64. The resulting p-values were Bonferroni65 corrected using the built-in p.adjust function to account for the effect of multi-hypothesis testing.
Supplementary information
Acknowledgements
We are grateful to the President of King Abdulaziz University, Prof. Abdulrahman O. Alyoubi, for funding support, the Genome Sequencing and Analysis Facility (GSAF) at the University of Texas at Austin (UT Austin) for Illumina sequencing, the Texas Advanced Computing Center (TACC) at UT Austin for access to supercomputers and Erika Schwarz, Mao-Lun Weng and Jin Zhang for advice on rate analyses.
Author contributions
Conceived and designed the experiments: M.Y., R.K.J., E.C.T., J.S.M.S., N.H.H.; Performed analyses and interpreted results: Y.R., W.Y.L., M.Y., T.A.R., M.J.S., A.M.A., M.K.A., E.C.T., A.E.O., M.R.K., R.K.J.; Secured funding for project: J.S.M.S., I.A.R., N.H.H., A.E.O., M.K.A.; Wrote the paper: Y.R., W.Y.L., T.A.R., M.Y., R.K.J., J.S.M.S., I.A.R., E.C.T.
Data availability
The NCBI accession numbers of the diatoms used in this study: NC_024084.1, MG755791.1, MG755799.1, NC_024081.1, MG755793.1, MG755796.1, MG755802.1, NC_025311.1, NC_024085.1, MG755797.1, NC_025312.1, NC_025314.1, MG755804.1, NC_014808.1, NC_008589.1, KJ958480.1, KJ958481.1, MG755794.1, NC_001713.1, MG755801.1, NC_025313.1, MG755808.1, NC_025310.1, MG755798.1, MG755806.1, MG755805.1, NC_024080.1, MG755792.1, MG755803.1, NC_024079.1, MG755807.1, NC_016731.1, MG755795.1, NC_024928.1, NC_024082.1, MH356727.1, MG755800.1, NC_015403.1, NC_024083.1, NC_008588.1, NC_027746.1.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Robert K. Jansen, Email: jansen@austin.utexas.edu
Irfan A. Rather, Email: erfaan21@gmail.com
Supplementary information
is available for this paper at 10.1038/s41598-020-71473-1.
References
- 1.Sorhannus U. A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Mar. Micropaleontol. 2007;65:1–12. [Google Scholar]
- 2.Nelson DM, Treguer P, Brzezinski MA, Leynaert A, Queguiner B. Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cy. 1995;9:359–372. [Google Scholar]
- 3.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 4.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 5.Mann DG. The species concept in diatoms. Phycologia. 1999;38:437–495. [Google Scholar]
- 6.Bowler C, Allen AE, Badger JH, Grimwood J, Jabbari K, Kuo A, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 7.Yu M, Ashworth M, Hajrah NH, Khiyami MA, Sabir MJ, Alhebshi AM, et al. Evolution of the plastid genomes in diatoms. Adv. Bot. Res. 2018;85:129–155. [Google Scholar]
- 8.Theriot EC, Ashworth M, Nakov T, Ruck EC, Jansen RK. A preliminary multigene phylogeny of the diatoms (Bacillariophyta): challenges for future research. Plant Ecol. Evol. 2010;143:278–296. [Google Scholar]
- 9.Ruck EC, Nakov T, Jansen RK, Theriot EC, Alverson AJ. Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol. Evol. 2014;6:644–654. doi: 10.1093/gbe/evu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabir JS, Yu M, Ashworth MP, Baeshen NA, Baeshen MN, Bahieldin A, et al. Conserved gene order and expanded inverted repeats characterize plastid genomes of Thalassiosirales. PLoS ONE. 2014;9:e107854. doi: 10.1371/journal.pone.0107854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theriot EC, Ashworth M, Nakov T, Ruck EC, Jansen RK. Dissecting signal and noise in diatom chloroplast protein encoding genes with phylogenetic information profiling. Mol. Phylogenet. Evol. 2015;89:28–36. doi: 10.1016/j.ympev.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Martin W, Stoebe B, Goremykin V, Hapsmann S, Hasegawa M, Kowallik KV. Gene transfer to the nucleus and the evolution of chloroplasts. Nat. 1998;393:162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 13.Kowallik KV, Stoebe B, SchaVran I, Kroth-Pancic P, Freier U. The chloroplast genome of a chlorophyll a + c- containing alga Odontella sinensis. Plant Mol. Biol. Rep. 1995;13:336–342. [Google Scholar]
- 14.Oudot-Le Secq MP, Grimwood J, Shapiro H, Armbrust EV, Bowler C, Green BR. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana and comparison with other plastid genomes of the red lineage. Mol. Genet. Genom. 2007;277:427–439. doi: 10.1007/s00438-006-0199-4. [DOI] [PubMed] [Google Scholar]
- 15.Lommer M, Roy AS, Schilhabel M, Schreiber S, Rosenstiel P, LaRoche J. Recent transfer of an iron-regulated gene from the plastid to the nuclear genome in an oceanic diatom adapted to chronic iron limitation. BMC Genom. 2010;11:718. doi: 10.1186/1471-2164-11-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka T, Fukuda Y, Yoshino T, Maeda Y, Muto M, Matsumoto M, et al. High-throughput pyrosequencing of the chloroplast genome of a highly neutral-lipid-producing marine pennate diatom, Fistulifera sp. strain JPCC DA0580. Photosynth. Res. 2011;109:223–229. doi: 10.1007/s11120-011-9622-8. [DOI] [PubMed] [Google Scholar]
- 17.Bedoshvili YD, Popkova TP, Likhoshway YV. Chloroplast structure of diatoms of different classes. Cell Tissue Biol. 2009;3:297–310. [Google Scholar]
- 18.Cooper JT, Malsy JP. Speciation in diatoms: patterns, mechaninsms and environmental change. In: Pawel M, editor. Speciation: Natural Processes, Genetics and Biodiversity. New York: Nova Science Publishers; 2013. pp. 1–6. [Google Scholar]
- 19.Kuroiwa T, Suzuki T, Ogawa K, Kawano S. The chloroplast nucleus: Distribution, number, size, and shape, and a model for the multiplication of the chloroplast genome during chloroplast development. Plant Cell Physiol. 1981;22:381–396. [Google Scholar]
- 20.Sato N. Origin and evolution of plastids: genomic view on the unification and diversity of plastids. In: Wise RR, Hoober JK, editors. The Structure and Function of Plastids, Advances in Photosynthesis and Respiration. Springer: Dordrecht; 2007. pp. 75–102. [Google Scholar]
- 21.Oldenburg DJ, Bendich AJ. DNA maintenance in plastids and mitochondria of plants. Front. Plant Sci. 2015;6:883. doi: 10.3389/fpls.2015.00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bromham L, Hua X, Lanfear R, Cowman PF. Exploring the relationships between mutation rates, life history, genome size, environment, and species richness in flowering plants. Am. Nat. 2015;185:507–524. doi: 10.1086/680052. [DOI] [PubMed] [Google Scholar]
- 23.Chang CC, Lin HC, Lin IP, Chow TY, Chen HH, Chen WH, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol. Biol. Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- 24.Guisinger MM, Kuehl JV, Boore JL, Jansen RK. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Natl. Acad. Sci. USA. 2008;105:18424–18429. doi: 10.1073/pnas.0806759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J. Mol. Evol. 2010;70:149–166. doi: 10.1007/s00239-009-9317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz EN, Ruhlman TA, Weng ML, Khiyami MA, Sabir JSM, Hajarah NH, et al. Plastome-wide nucleotide substitution rates reveal accelerated rates in Papilionoideae and correlations with genome features across legume subfamilies. J. Mol. Evol. 2017;84:187–203. doi: 10.1007/s00239-017-9792-x. [DOI] [PubMed] [Google Scholar]
- 27.Sloan DB, Alverson AJ, Wu M, Palmer JD, Taylor DR. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol. Evol. 2012;4:294–306. doi: 10.1093/gbe/evs006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong W, Xu C, Cheng T, Zhou S. Complete chloroplast genome of Sedum sarmentosum and chloroplast genome evolution in Saxifragales. PLoS ONE. 2013;8:e77965. doi: 10.1371/journal.pone.0077965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S, Ruhlman TA, Weng ML, Hajrah NH, Sabir JSM, Jansen RK. Contrasting patterns of nucleotide substitution rates provide insight into dynamic evolution of plastid and mitochondrial genomes of Geranium. Genome Biol. Evol. 2017;9:1766–1780. doi: 10.1093/gbe/evx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng ML, Blazier JC, Govindu M, Jansen RK. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats and nucleotide substitution rates. Mol. Biol. Evol. 2013;31:645–659. doi: 10.1093/molbev/mst257. [DOI] [PubMed] [Google Scholar]
- 31.Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leeben MJ, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belda E, Moya A, Siva FJ. Genome rearrangement distances and gene order phylogeny in gamma-Proteobacteria. Mol. Biol. Evol. 2005;22:1456–1467. doi: 10.1093/molbev/msi134. [DOI] [PubMed] [Google Scholar]
- 33.Shao R, Dowton M, Murrell A, Barker SC. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 2003;20:1612–1619. doi: 10.1093/molbev/msg176. [DOI] [PubMed] [Google Scholar]
- 34.Xu W, Jameson D, Tang B, Higgs PG. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J. Mol. Evol. 2006;63:375–392. doi: 10.1007/s00239-005-0246-5. [DOI] [PubMed] [Google Scholar]
- 35.Jansen RK, Ruhlman TA. Plastid genomes of seed plants. In: Bock R, Knoop V, editors. Advances in Photosynthesis and Respiration, Volume 35: Genomics of Chloroplasts and Mitochondria. Dordrecht: Springer; 2012. pp. 103–126. [Google Scholar]
- 36.Zhang J, Ruhlman TA, Sabir JSM, Blazier JC, Weng ML, Park S, et al. Coevolution between nuclear-encoded DNA replication, recombination, and repair genes and plastid genome complexity. Genome Biol. Evol. 2016;8:622–634. doi: 10.1093/gbe/evw033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu CS, Chaw SM. Highly rearranged and size-variable chloroplast genomes in conifers II clade (cupressophytes): evolution towards shorter intergenic spacers. Plant Biotechnol. J. 2014;12:344–353. doi: 10.1111/pbi.12141. [DOI] [PubMed] [Google Scholar]
- 38.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 41.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tesler G. GRIMM: genome rearrangements web server. Bioinformatics. 2002;18:492–493. doi: 10.1093/bioinformatics/18.3.492. [DOI] [PubMed] [Google Scholar]
- 43.Sloan DB, Triant DA, Wu M, Taylor DR. Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol. Biol. Evol. 2014;3:673–682. doi: 10.1093/molbev/mst259. [DOI] [PubMed] [Google Scholar]
- 44.Weng ML, Ruhlman TA, Jansen RK. Plastid–nuclear interaction and accelerated coevolution in plastid ribosomal genes in Geraniaceae. Genome Biol. Evol. 2016;8:1824–1838. doi: 10.1093/gbe/evw115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson AC, Carlson SS, White TJ. Biochemical evolution. Annu. Rev. Biochem. 1997;46:573–639. doi: 10.1146/annurev.bi.46.070177.003041. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Yang JR. Determinants of the rate of protein sequence evolution. Nat. Rev. Genet. 2015;16:409–420. doi: 10.1038/nrg3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Havird JC, Sloan DB. The roles of mutation, selection, and expression in determining relative rates of evolution in mitochondrial versus nuclear genomes. Mol. Biol. Evol. 2016;11:3042–3053. doi: 10.1093/molbev/msw185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright SI, Yau CB, Looseley M, Meyers BC. Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol. Biol. Evol. 2004;21:1717–1726. doi: 10.1093/molbev/msh191. [DOI] [PubMed] [Google Scholar]
- 49.Ingvarsson PK. Gene expression and protein length influence codon usage and rates of sequence evolution in Populus tremula. Mol. Biol. Evol. 2007;24:836–844. doi: 10.1093/molbev/msl212. [DOI] [PubMed] [Google Scholar]
- 50.De La Torre AR, Lin YC, Van de Peer Y, Ingvarsson PK. Genome-wide analysis reveals diverged patterns of codon bias, gene expression, and rates of sequence evolution in Picea gene families. Genome Biol. Evol. 2015;7:1002–1015. doi: 10.1093/gbe/evv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shields DC, Sharp PM, Higgins DG, Wright F. Silent sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol. Biol. Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 52.Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol. Biol. Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- 53.Shen Y, Lv Y, Huang L, Liu W, Wen M, Tang T, Zhang R, et al. Testing hypotheses on the rate of molecular evolution in relation to gene expression using microRNAs. Proc. Natl. Acad. Sci. USA. 2011;108:15942–15947. doi: 10.1073/pnas.1110098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang J, Ruhlman TA, Sabir JSM, Blazier JC, Jansen RK. Coordinated rates of evolution between interacting plastid and nuclear genes in Geraniaceae. Plant Cell. 2015;27:563–573. doi: 10.1105/tpc.114.134353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rockenbach K, Havird JC, Monroe JG, Triant DA, Taylor DR, Sloan DB. Positive selection in rapidly evolving plastid-nuclear enzyme complexes. Genetics. 2016;204:1507–1522. doi: 10.1534/genetics.116.188268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 57.David MK, Yuri IW, Arcady RM, Eugene VK. Computational methods for Gene Orthology inference. Brief. Bioinform. 2011;12:379–391. doi: 10.1093/bib/bbr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 59.Glez-Peña D, Gómez-Blanco D, Reboiro-Jato M, Fdez-Riverola F, Posada D. ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010;38:W14–W18. doi: 10.1093/nar/gkq321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan HM, Low WY. Rapid birth-death evolution and positive selection in detoxification-type glutathione S-transferases in mammals. PLoS ONE. 2018;13:12. doi: 10.1371/journal.pone.0209336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 63.Venables WN, Ripley BD. Modern Applied Statistics with S. 4. New York.: Springer; 2002. [Google Scholar]
- 64.Benesty J, Chen J, Huang Y, Cohen I. Pearson Correlation Coefficient. Noise Reduction in Speech Processing. New York: Springer; 2009. pp. 37–40. [Google Scholar]
- 65.Hochberg Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika. 1988;75:800–803. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NCBI accession numbers of the diatoms used in this study: NC_024084.1, MG755791.1, MG755799.1, NC_024081.1, MG755793.1, MG755796.1, MG755802.1, NC_025311.1, NC_024085.1, MG755797.1, NC_025312.1, NC_025314.1, MG755804.1, NC_014808.1, NC_008589.1, KJ958480.1, KJ958481.1, MG755794.1, NC_001713.1, MG755801.1, NC_025313.1, MG755808.1, NC_025310.1, MG755798.1, MG755806.1, MG755805.1, NC_024080.1, MG755792.1, MG755803.1, NC_024079.1, MG755807.1, NC_016731.1, MG755795.1, NC_024928.1, NC_024082.1, MH356727.1, MG755800.1, NC_015403.1, NC_024083.1, NC_008588.1, NC_027746.1.