Abstract
Human rhinoviruses (HRVs) are the predominant infectious agents for the common cold worldwide. The HRV-C species cause severe illnesses in children and are closely related to acute exacerbations of asthma. 3C protease, a highly conserved enzyme, cleaves the viral polyprotein during replication and assists the virus in escaping the host immune system. These key roles make 3C protease an important drug target. A few structures of 3Cs complexed with an irreversible inhibitor rupintrivir have been determined. These structures shed light on the determinants of drug specificity. Here we describe the structures of HRV-C15 3C in free and inhibitor-bound forms. The volume-decreased S1’ subsite and half-closed S2 subsite, which were thought to be unique features of enterovirus A 3C proteases, appear in the HRV-C 3C protease. Rupintrivir assumes an “intermediate” conformation in the complex, which might open up additional avenues for the design of potent antiviral inhibitors. Analysis of the features of the three-dimensional structures and the amino acid sequences of 3C proteases suggest new applications for existing drugs.
Keywords: Human rhinoviruses (HRVs), 3C protease, Three-dimensional structures, Inhibitors, Rupintrivir (AG7088)
Introduction
Human rhinoviruses (HRVs), first discovered in the 1950s, are the predominant infectious agents of the common cold and the most common cause of upper respiratory tract infections worldwide (Arruda et al. 1997). In recent years, studies have shown that HRVs are responsible for the exacerbations of chronic pulmonary diseases and asthma (Johnston et al. 1995). Comprising about 160 serotypes, HRVs are divided into three groups HRV-A, HRV-B and HRV-C, utilizing intercellular adhesion molecule 1 (ICAM-1), low-density lipoprotein receptor (LDLR) and cadherin-related family member 3 (CDHR3) proteins, respectively, as receptors for host-cell entry (Greve et al. 1989; Hofer et al. 1994; Bochkov et al. 2015). HRV-C species were first discovered in 2006 and raised public concern quickly since they can cause more severe illnesses in children compared with HRV-A or HRV-B species, and are closely related to acute exacerbations of asthma (Arden et al. 2006; Lamson et al. 2006; Miller et al. 2009; Bizzintino et al. 2011). So far there are no effective protocols to culture HRV-C viruses in wild-type cells.
Rhinoviruses are members of the Picornaviridae family. Within this family, they belong to the Enterovirus genus, which also contains enterovirus A–D (EV A–D) (Palmenberg et al. 2010). Like all other picornaviruses, HRVs have a positive-sense, single-stranded RNA genome which is translated into a polyprotein first during replication. Two proteases 2A and 3C cleave the polyprotein to generate functional proteins and enzymes (Palmenberg 1990; Porter 1993). The 2A protease catalyzes the cleavage between the structural and nonstructural proteins only, while 3C protease catalyzes most of the internal cleavages (Palmenberg 1990; Porter 1993). In addition, 3C protease plays a key role in antagonizing antiviral immunity by cleaving the host complement C3 and retinoic acid-inducible gene I (RIG-I) to attenuate the innate immune response to viral infection (Barral et al. 2009; Tam et al. 2014). It is also reported that 3C protease cleaves the highly conserved receptor-interacting serine/threonine-protein kinase 1 (RIPK1) to suppress cell apoptosis and necroptosis (Croft et al. 2018; Lotzerich et al. 2018).
In the past decade, much effort has been put into finding ways to cure HRV infections. The highly conserved 3C protease is one of the most important targets for drug design and many inhibitors of this enzyme have been screened and tested (Binford et al. 2005; Wang and Chen 2007; Baxter et al. 2011; Mello et al. 2014). Rupintrivir (also known as AG7088) is an irreversible broad-spectrum inhibitor of 3C protease discovered via protein structure-based drug design methodologies. This inhibitor shows antiviral activity against a variety of HRV serotypes as well as some other picornaviruses, such as human enteroviruses (HEVs) (Dragovich et al. 1998, 1999a, 1999b; Binford et al. 2005). It has been reported that the 50% effective concentrations (EC50) of rupintrivir for EV71 and CVA16 is about two orders of magnitude higher than those of the compound for HRVs (Patick et al. 1999; Binford et al. 2005; Tsai et al. 2009). Complexes of rupintrivir and 3C protease of HRV-2, EV71 and CVA16 reveal the differences in the binding sites of 3C and the conformational changes of the inhibitor. The conformational differences of rupintrivir in these complexes explain the reduced binding affinities of the drug for EV71 and CVA16 (Matthews et al. 1999; Lu et al. 2011). However, a report by Hao et al. showed that the EC50 of rupintrivir for HRV-C15 is about 93 nmol/L, which is around 2 to sixfold higher than those observed for HRV-A and HRV-B serotypes (Patick et al. 1999; Hao et al. 2012). The reason behind the lower binding affinity of rupintrivir for HRV-C15 3C protease is still unclear.
Here, we describe the structures of HRV-C15 3C protease both in free and inhibitor-bound forms. There is no obvious difference between the two forms of the enzyme. Surprisingly, the half-closed S2 subsite and the size-reduced S1’ subsite resemble those of EV71 more than those of HRV-2. The unique microenvironments of HRV-C15 3C protease led the rupintrivir to tilt the α, β-unsaturated ester away from the S1’ subsite while still inserting the fluoro-phenylalanine group into the S2 subsite of the protein. These conformational changes of the inhibitor explain why it exhibits lower binding affinity for HRV-C than HRV-A and HRV-B. Structure of the complex of HRV-C 3C protease with rupintrivir reveals the conformational flexibility of the inhibitor, which would be instructive in the structure-based broad-spectrum drug design. The features of the three-dimensional structure revealed in this study and the amino acid sequence of HRV-C 3C protease also suggest that anti HEV-A and HEV-B drugs which target the 3C proteases would probably work better in inhibiting HRV-C than anti HRV-A and HRV-B drugs.
Materials and Methods
Expression and Purification of 3C Protease
The DNA fragment encoding the full-length 3C protease was synthesized and the gene was then sub-cloned into pET-28a. Competent E. coli BL21 (DE3) cells were transformed with the recombinant plasmid. The bacterial cultures were grown at 37 °C to an OD600 of 0.6 and 0.5 mmol/L isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce the protein expression at 16 °C for 18 h. Cells were harvested and sonicated in the lysis buffer (50 mmol/L HEPES, 150 mmol/L NaCl and 10% (v/v) glycerol, pH 6.5). The lysate was centrifuged at 30,700 × g for 30 min to remove cell debris. The supernatant was added onto a Ni–NTA agarose column (Qiagen) equilibrated with the lysis buffer, and 3C protease was eluted using the lysis buffer supplemented with 250 mmol/L imidazole. The 3C protease samples were further purified by ion-exchange chromatography and gel filtration chromatography. The purified protein was concentrated to ~ 5 mg/mL for crystallization.
Crystallization, Data Collection, and Structure Determination
Crystals of 3C protease and the complex of 3C protease with rupintrivir were grown at 16 °C using the hanging drop vapor diffusion method over a reservoir solution consisting of 0.05 mol/L citric acid, 0.05 mol/L BIS–TRIS propane, pH 5.0, 16% (w/v) polyethylene glycol 3,350. Diffraction data sets were collected at beamline BL17U of the Shanghai synchrotron facility. Datasets were processed and scaled using the HKL2000 package (Otwinowski and Minor 1997). The initial structure solutions of 3C protease and the complex of 3C protease with rupintrivir were obtained by molecular replacement using the program Phaser v2.1 (Mccoy et al. 2007) with the crystal structure of 3C protease of human rhinovirus serotype 2 (Protein Data Bank [PDB] entry: 1CQQ (Matthews et al. 1999) and HRV-C15 3C protease as a search template, respectively. Manual model building and refinement were performed using COOT (Emsley and Cowtan 2004) and PHENIX (Adams et al. 2010). The structural figures were drawn with the program PyMOL (DeLano 2004).
Protein Structure Accession Numbers
Coordinates and structure factors have been deposited with RCSB accession codes: 6KU7, 6KU8.
Result
Overall Structure of the HRV-C15 3C Protease
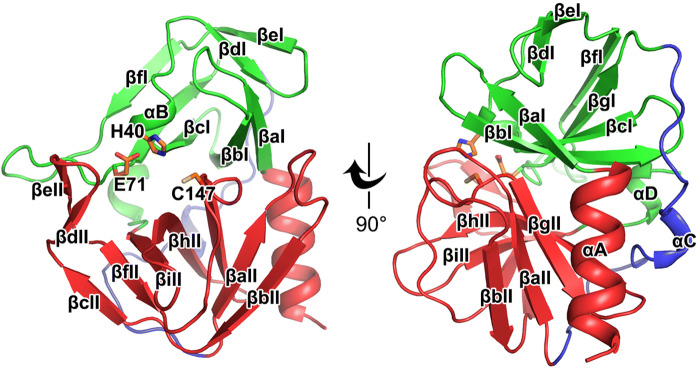
Similar to the previously reported structures of the 3C proteases, the HRV-C15 3C protease contains a chymotrypsin-like fold that can be divided into two major domains and one soft linker (Table 1) (Mosimann et al. 1997; Matthews et al. 1999; Anand et al. 2002; Yang et al. 2003; Lee et al. 2009; Lu et al. 2011). In domain I, seven β-strands (βaI–βgI) surround the αB short helix and make up the core of the domain (Fig. 1). The domain II is mainly composed of nine β-strands (βaII–βiII), which is further stabilized by the neighboring N-terminal helix αA (Fig. 1). The two main domains are linked by αC and a long loop which are positioned on the opposite side of the catalytic center (Fig. 1). Three highly conserved residues, H40, E71 and C147, located in the center of the substrate-binding groove formed by the two major domains, play key roles in the catalysis (Fig. 1). The long soft linker situated on the opposite side enables the substrate-binding groove to be more flexible in terms of size and orientation (Fig. 1).
Table 1.
Data collection and refinement statistics.
| Name | 3C | 3C in complex with rupintrivir |
|---|---|---|
| Data collection | ||
| Resolution (Å) | 50–2.15 (2.23–2.15) | 50.00–2.05 (2.12–2.05) |
| Unique reflections | 13,578 (1332) | 16,114 (1496) |
| Space group | C2221 | C2221 |
| Cell dimensions | ||
| a (Å) | 52.1 | 52.9 |
| b (Å) | 94.7 | 95.6 |
| c (Å) | 98.1 | 98.9 |
| α (°) | 90 | 90 |
| β (°) | 90 | 90 |
| γ (°) | 90 | 90 |
| Redundancy | 7.5 (6.4) | 3.1 (3.0) |
| Completeness (%) | 99.8 (99.5) | 92.1 (90.2) |
| aRmerge | 0.069 (0.328) | 0.060 (0.263) |
| I/σ(I) | 35.6 (8.1) | 22.8 (7.0) |
| Refinement | ||
| Resolution(Å) | 2.15 | 2.05 |
| No. reflections | 13,546 | 14,812 |
| bRwork/cRfree | 0.210/0.253 | 0.184/0.223 |
| No. of non-H atoms | ||
| Protein | 1498 | 1,419 |
| Mean B-factor (Å2) | 31.5 | 28.0 |
| Ramachandran statistics (%) | ||
| Most favored | 97.2 | 98.3 |
| Allowed | 2.8 | 1.7 |
| Outliers | 0.0 | 0.0 |
| R.m.s.deviations | ||
| Bond lengths (Å) | 0.010 | 0.008 |
| Bond angles (°) | 1.089 | 1.086 |
Values in parentheses are for highest-resolution shell.
a
b
cRfree was calculated for a test set of reflections (5%) omitted from the refinement.
Fig. 1.
Overall structure of the HRV-C15 3C protease. Cartoon representation of the structure of HRV-C15 3C protease is shown with domain I, domain II and the linker colored green, red and blue, respectively. The α-helices are marked αA to αD and the β-strands are labeled βaI to βgI in domain I and βaII to βiII in domain II according to their occurrence along with the primary structure. The catalytic triad (H40, E71, and C147) is shown as orange sticks and labeled.
Structure of the Complex of HRV-C15 3C Protease and Rupintrivir
Rupintrivir is a specific 3C inhibitor which was designed based on the protein structure (Matthews et al. 1999). As reported previously, this irreversible inhibitor could be divided into five groups—a lactam (P1), a fluorophenylalanine (P2), a Val (P3), a 5-methyl-3-isoxazole (P4), and an α, β-unsaturated ester (P1′) (Matthews et al. 1999; Lu et al. 2011). Rupintrivir fills itself into the catalytic pocket of 3C protease and further stabilizes the binding with a covalent linkage to C147, one of the three key catalytic residues (Fig. 2A and Table 1). Four of the five groups of the inhibitor: P1, P2, P3 and P1′, and the covalent bond can be traced based on clear and unambiguous density, while the P4 group of rupintrivir is totally invisible in the complex, suggesting that a simplified inhibitor lacking the P4 group could still possibly block the cleavage efficiently (Fig. 2A).
Fig. 2.
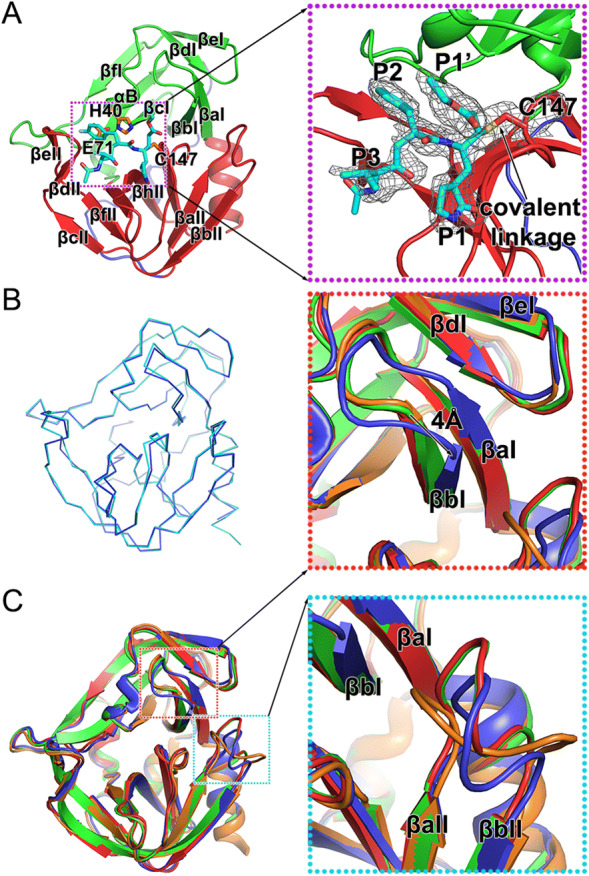
Structure of the HRV-C15 3C-rupintrivir complex. A Overall structure of the 3C-rupintrivir complex. 3C protease is colored and labeled as Fig. 1. The rupintrivir is shown as sticks. The covalent bond formed between the inhibitor and the protease is highlighted with an arrow. The inset corresponds to zoomed-in view of boxed region and illustrates the features of the inhibitor. B Line representation of the superimposition of the 3C protease in free and inhibitor-bound forms colored in cyan and blue, respectively. C Cartoon representation of the superimposition of the 3C proteases of HRV-2, HRV-C15, EV71 and CVA16, colored orange, blue, red and green, respectively. Insets correspond to zoomed-in views of boxed regions and illustrate the structural differences with secondary elements labeled.
The overall structures of 3C protease of HRV-C15 in free and inhibitor-bound forms are very similar (Fig. 2B). Superimposition of the two structures of 3C protease yields an RMSD of 0.239 Å for the overlapping Cα atoms and reveals that rupintrivir blocks the catalytic activity of 3C without changing the overall conformation of the protease. Interestingly, although the overall structures of complexes of 3C protease and rupintrivir of HRV-2, HRV-C15, EV71 and CVA16 are very similar, subtle deviations in the structures have been observed during the superposition (Fig. 2C). Two antiparallel β-strands (βaI and βbI) of domain I move “down” towards domain II by about 4 Å when compared with the same two β-strands of the other three complexes (Fig. 2C). The βaII-βbII loop of domain II (the loop connecting the βaII and βbII strands) varies in conformation and the βaII-βbII loops of HRV-2 and HRV-C15 tend to bend towards the catalytic center, which is different from those observed for 3C proteases from EV71 and CVA16 (Fig. 2C). These conformational changes of HRV-C15 3C protease lead to a relatively more closed S1’ subsite (the binding site of rupintrivir P1′ group) and reduce the volume of this substrate-binding pocket.
Rupintrivir Adopts an “Intermediate” Conformation
3C proteases of HRV-2, HRV-C15, EV71 and CVA16 share a chymotrypsin-like fold in the overall structure and a highly conserved catalytic center. Rupintrivir binds to the different proteases in a similar manner. Besides the obvious covalent linkage, some distinct characteristics have been observed from the conformation of the inhibitor while comparing these complexes. In different complexes, P1 and P3 groups of the inhibitors are in the same position while the P1′ and P2 groups rotate to adapt to the different microenvironments of the binding subsites (Fig. 3). Rupintrivir shows a new conformation upon binding to HRV-C15 3C protease when compared to its binding to 3C proteases of HRV-2, EV71 and CVA16. The P1′ group rotates ~ 54° relative to its position in the complex with HRV-2 and stretches out into the solvent, which is similar to that observed in the complexes of the inhibitor with EV71 and CVA16 (Fig. 3). The P2 group sticks into the S2 subsite of HRV-C15 in a manner similar to that observed for HRV-2 3C-rupintrivir complex and is tilted 21° from the P2 groups of rupintrivir binding with 3C proteases of EV71 and CVA16 (Fig. 3). An “intermediate” conformation of rupintrivir appears in the complex of HRV-C15 and displays some characteristics of both conformers. The new conformation of rupintrivir and the differences in various binding modes might help explain the differences in the inhibitor’s binding affinities and EC50 values for different viruses.
Fig. 3.
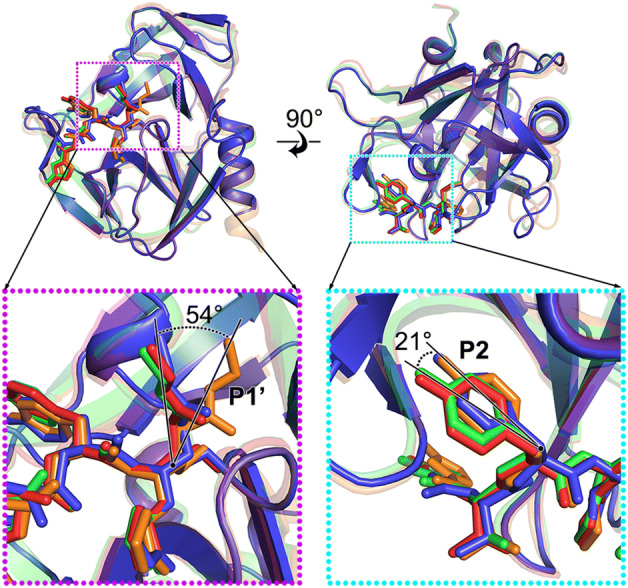
Rupintrivir adopts a new conformation. The inhibitor (shown in sticks) and the 3C proteases (shown in cartoon) of HRV-2, HRV-C15, EV71 and CVA16 are colored orange, blue, red and green, respectively. The transparency of the cartoon representation of 3C proteases of HRV-2, EV71 and CVA16 is 0.7. Insets correspond to zoomed-in views of boxed regions and illustrate the conformational changes of the inhibitor with the rotation angles labeled.
Structural Basis for the Transformation of Rupintrivir
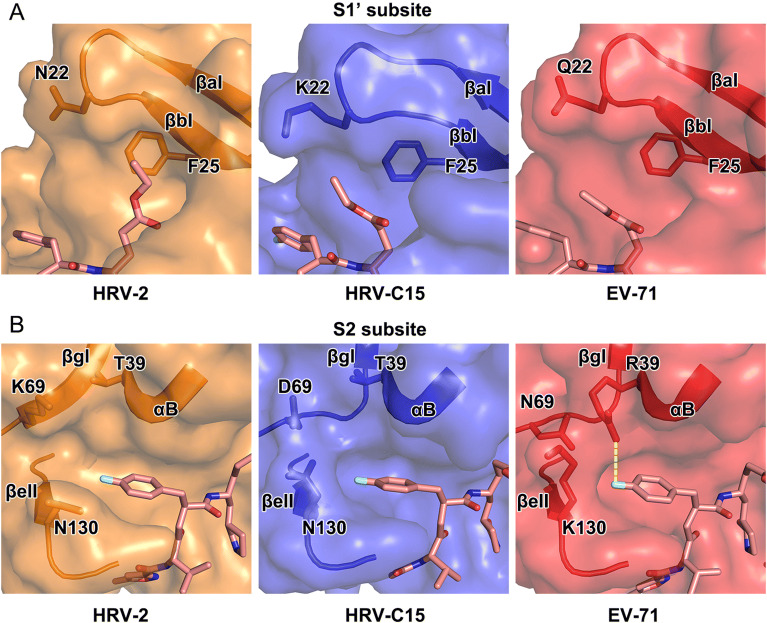
In the complex of HRV-C15 3C protease with rupintrivir, microenvironment in the pocket somewhat impacts the inhibitor’s conformation, especially while binding to S1’ and S2 subsites. In the S1’ subsite of HRV-C15 3C-rupintrivir complex, the 4 Å-movement of βaI and βbI reduces the volume of the subsite remarkably when compared with those of HRV-2, EV71 and CVA16 (the structure of CVA16 3C protease is almost identical to that of EV71 and therefore omitted in the following structural analysis) (Figs. 2 and 4A). Residues with long side chains in the β-strands, such as K22 and F25, compress the S1’ subsite further and leave insufficient space for holding the P1′ group of rupintrivir. Steric clashes would have occurred if the P1′ group inserted itself into the subsite in HRV-C15 3C-rupintrivir complex just like that observed in the complex of HRV-2. Residue N22 of HRV-2 3C protease and Q22 of EV71 3C protease stretch their side chains out to the solvent and bring much less stress than K22 of HRV-C15 3C protease does (Fig. 4A). The highly conserved residue F25 follows the movement of the two β-strands to encroach on the subsite with its long side chain (Fig. 4A). Interestingly, although the S1 pocket of HRV-C15 3C protease is smaller than that of EV71, the rotation angle of the inhibitor’s P1′ group in its complex with HRV-C15 3C is smaller than that observed for EV71 3C-rupintrivir complex (Figs. 3 and 4A).
Fig. 4.
Structural comparison of the S1’ and S2 subsites of 3C proteases of HRV-2, HRV-C15 and EV71. In A and B, rupintrivir is shown as sticks and the 3C proteases are shown in surface representations with the key residues and secondary structural elements shown as sticks and cartoon, respectively. The polar interaction between R39 and P2 group of the inhibitor in the complex of EV71 is colored yellow in B.
Two states of the S2 subsite, fully open and half-closed, have been reported for the 3C protease of HRV-2 and EV71, respectively. Accordingly, the P2 group of rupintrivir has been observed to insert itself into S2 subsite when open, and reach out of the subsite when half-closed (Matthews et al. 1999; Lu et al. 2011). Interestingly, the P2 group of rupintrivir still inserts itself into the pocket while the S2 subsite of HRV-C15 3C protease stays half-closed, which indicates that the binding mode of P2 group of rupintrivir is not determined by the state of the subsite (Figs. 3 and 4B). N130 is conserved in HRV-2 and HRV-C15, and the orientations of its side chain control the state of the S2 subsite together with K69/D69 located on the other side. The state of the subsite switches from fully open to half-closed as the side chain of N130 swaps from “downside” (like that observed for N130 of HRV-2 3C protease) to “upside” (as observed for K130 of EV71 3C protease) (Fig. 4B). The polar interaction between R39 and rupintrivir P2 group makes the P2 group turn ~ 21° and extend into the solvent since the long side chain of R39 stretches out of the subsite (Figs. 3 and 4B). The conserved T39 residues in rhinoviruses do not interact with rupintrivir so that the P2 group could remain inserted inside the subsite (Figs. 3 and 4B).
The size-reduced S1’ and the half-closed S2 subsites are not the unique features of the enterovirus A 3C proteases (such as EV71 and CVA16). The S1’ subsite of HRV-C protease is even smaller than that of EV71 as well as CVA16 and the half-closed S2 subsite is also observed in the protease of HRV-C. Unexpectedly, the states of the S2 subsite have no impact on the conformation of the P2 group of rupintrivir. Although most 3C proteases share a highly conserved overall structure, the differences in the residues could result in variations in the local microenvironments of the subsites. Consequently, the conformation of the inhibitor changes to adapt to the variations in the subsites.
Discussion
3C protease, a highly conserved enzyme of picornaviruses, not only cleaves the viral polyprotein during replication, but also helps the virus escape the host immune system (Palmenberg 1990; Porter 1993; Barral et al. 2009; Tam et al. 2014). The three amino acid residues that form the catalytic triad (H40, E71 and C147) are highly conserved in rhinoviruses A–C as well as enteroviruses A–D (Fig. 5A). Here, we present the structures of HRV-C15 3C protease both in free and inhibitor-bound forms. Rupintrivir adopts a new “intermediate” conformation in the HRV-C15 3C protease-rupintrivir complex when compared with the conformation of the inhibitor observed in complex with 3C proteases of HRV-2, EV71 and CVA16 (Matthews et al. 1999; Lu et al. 2011). Notably, the “intermediate” conformation for rupintrivir observed in HRV-C15 are not possibly caused by low pH since that all three complexes (HRV-C15 3C-rupintrivir, EV71 3C-rupintrivir and CVA16 3C-rupintrivir) were crystallized at pH < 5, suggesting the specific interactions between the HRV-C15 3C and rupintrivir. The P1′ group of the inhibitor is oriented towards the solvent after binding with the HRV-C15 3C protease. Such mode of binding of rupintrivir is similar to that observed in its complex with EV71. The P2 group stays embedded in the S2 subsite, which is akin to that observed in the complex of HRV-2 with rupintrivir. These conformational changes could provide some explanation to the differences in EC50 values of rupintrivir for HRV-2, HRV-C15 and EV71.
Fig. 5.
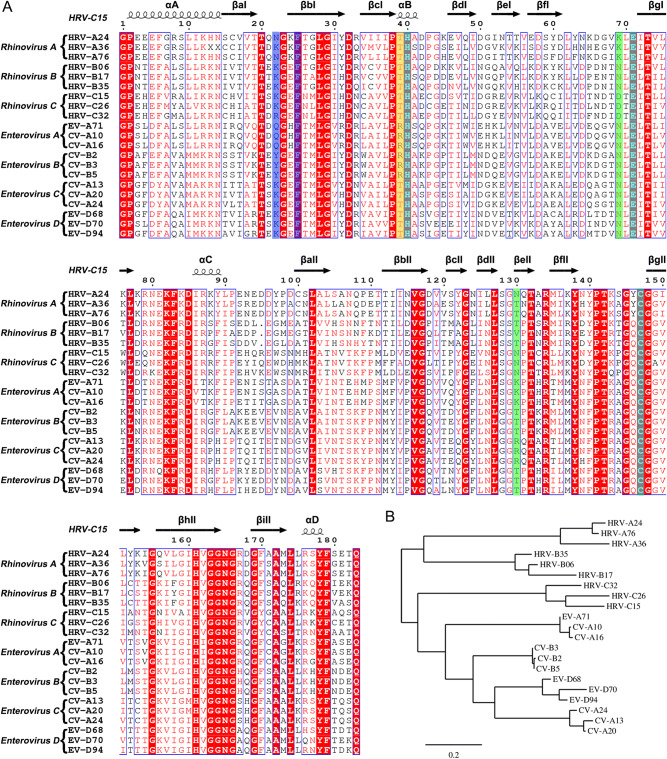
Sequence analysis of the 3C proteases from rhinovirus A–C and enterovirus A–D. A Multiple-sequence alignment of the 3C proteases using ESPript (Robert and Gouet 2014). The spiral lines indicate α-helices and the horizontal arrows represent β-strands. The catalytic triad is colored cyan. The residues which play key roles in decreasing the S1’ subsite volume are highlighted with a blue background. The residues that control the state of the S2 subsite are colored green. The residue which interacts with the P2 group of rupintrivir is highlighted with a yellow background. B Phylogenetic tree of the 3C proteases generated using the online tool—Phylogeny (Dereeper et al. 2008, 2010).
Based on the structures of the complexes of 3C protease with rupintrivir, an in-depth characterization of the S1’ and S2 subsite could provide potential guidelines for rational design of broad-spectrum inhibitors. In the S1’ subsite of HRV-C15 3C, the two antiparallel β-strands (βaI and βbI) with long side-chain residues (K22 and F25) move about 4 Å and reduce the volume of this pocket. Interestingly, F25 is highly conserved in rhinovirus A–C and enterovirus A–D while N22/K22/Q22/Y22 are conserved in each subgroup only (Fig. 5A). Based on the structures and analysis of the amino acid sequences, it is quite likely that the S1’ subsite is not large enough to hold the inhibitor’s P1′ group and therefore a smaller group at this position is suggested for designing new broad-spectrum inhibitors. In the S2 subsite, positions of two residues, N130 and D69/K69, determine whether the pocket is fully open or half-closed. Interestingly, the states of the subsite do not influence the insertion of the P2 group of rupintrivir. The residue R39 which forms polar interaction with the P2 group stretches out to the solvent and makes the P2 group turn outside. Surprisingly, R39 is highly conserved just in enterovirus A and B, whereas in rhinovirus A–C and enterovirus C–D, residue 39 is occupied by threonine (Fig. 5A). Without the polar interaction, the P2 group would probably insert into the S2 subsite in rhinovirus A–C and enterovirus C–D, regardless of whether the pocket is fully open or half-closed. Therefore, a nonpolar P2 group would be an appropriate choice for designing broad-spectrum inhibitors. Given that K130 and R130 are conserved in enterovirus A and C, respectively, the long side chains block the S2 subsite more easily than the short side chains of T130/V130/N130 of rhinoviruses A–C, as well as enteroviruses B and D (Fig. 5A). Therefore, the P2 group is suggested not to be too long in the broad-spectrum inhibitor design. Otherwise, the binding of the inhibitor would probably be affected by the half-closed S2 pocket.
The antiviral effects of rupintrivir against HRV-C15 and other viruses could validate the potential relationship between the “intermediate” conformation and the lower binding affinity. However, lack of HRV-C15 efficiently infected cell model limit this exploration in this study. To utilize existing drugs for curing the viral infection is as important as designing new broad-spectrum inhibitors. Intriguingly, phylogenetic analysis of 3C proteases based on the amino acid sequences reveals that 3C proteases of HRV-C resemble the enzymes of HEV-A and HEV-B more than those of HRV-A and HRV-B (Fig. 5B). Taking into consideration the phylogenetic analysis and the reduced volume of S1’ subsites of 3C proteases of HRV-C and HEV-A, it seems that anti HEV-A and HEV-B drugs which target the 3C proteases would probably work better in inhibiting HRV-C than anti HRV-A and HRV-B drugs targeting the 3C protease.
Acknowledgements
We gratefully acknowledge the assistance of the staff of the beamline BL17U and BL19U at Shanghai Synchrotron Radiation Facility (SSRF) with the X-ray diffraction data collection. Work was supported by the National Key Research and Development Program (2018YFA0900801), National Science Foundation of China (31800145), the State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences and the seed Foundation of Innovation and Creation for Graduate Students in Northwestern Polytechnical University ZZ2019279. LZ was sponsored by the Youth Innovation Promotion Association at the Chinese Academy of Sciences.
Author Contributions
LZ and HH conceived and designed the experiment. SY performed the experiments. SY, KF, ZC, YS analyzed the data. SY, LZ and HH wrote the manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Kaiyue Fan and Zhonghao Chen have contributed equally to this work.
Contributor Information
Hai Hou, Email: houhai@nwpu.edu.cn.
Ling Zhu, Email: lingzhu@ibp.ac.cn.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D-Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand K, Palm GJ, Mesters JR, Siddell SG, Ziebuhr J, Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda E, Pitkaranta A, Witek TJ, Jr, Doyle CA, Hayden FG. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol. 1997;35:2864–2868. doi: 10.1128/JCM.35.11.2864-2868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral PM, Sarkar D, Fisher PB, Racaniello VR. RIG-I is cleaved during picornavirus infection. Virology. 2009;391:171–176. doi: 10.1016/j.virol.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A, Chambers M, Edfeldt F, Edman K, Freeman A, Johansson C, King S, Morley A, Petersen J, Rawlins P, Spadola L, Thong B, Van de Poel H, Williams N. Non-covalent inhibitors of rhinovirus 3C protease. Bioorg Med Chem Lett. 2011;21:777–780. doi: 10.1016/j.bmcl.2010.11.110. [DOI] [PubMed] [Google Scholar]
- Binford SL, Maldonado F, Brothers MA, Weady PT, Zalman LS, Meador JW, Matthews DA, Patick AK. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob Agents Chemother. 2005;49:619–626. doi: 10.1128/AAC.49.2.619-626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzintino J, Lee WM, Laing IA, Vang F, Pappas T, Zhang G, Martin AC, Khoo SK, Cox DW, Geelhoed GC, McMinne PC, Goldblatt J, Gern JE, Le Soue PN. Association between human rhinovirus C and severity of acute asthma in children. Eur Respir J. 2011;37:1037–1042. doi: 10.1183/09031936.00092410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov YA, Watters K, Ashraf S, Griggs TF, Devries MK, Jackson DJ, Palmenberg AC, Gern JE. Cadherin-related family member 3, a childhood asthma susceptibility gene product, mediates rhinovirus C binding and replication. Proc Natl Acad Sci USA. 2015;112:5485–5490. doi: 10.1073/pnas.1421178112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft SN, Walker EJ, Ghildyal R. Human Rhinovirus 3C protease cleaves RIPK1, concurrent with caspase 8 activation. Sci Rep. 2018;8:1569. doi: 10.1038/s41598-018-19839-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. Use of PYMOL as a communications tool for molecular science. Abstracts P Am Chem Soc. 2004;228:U313–U314. [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovich PS, Webber SE, Babine RE, Fuhrman SA, Patick AK, Matthews DA, Reich SH, Marakovits JT, Prins TJ, Zhou R, Tikhe J, Littlefield ES, Bleckman TM, Wallace MB, Little TL, Ford CE, Meador JW, Ferre RA, Brown EL, Binford SL, DeLisle DM, Worland ST. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 2. Peptide structure-activity studies. J Med Chem. 1998;41:2819–2834. doi: 10.1021/jm9800696. [DOI] [PubMed] [Google Scholar]
- Dragovich PS, Prins TJ, Zhou R, Fuhrman SA, Patick AK, Matthews DA, Ford CE, Meador JW, Ferre RA, Worland ST. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 3. Structure - Activity studies of ketomethylene-containing peptidomimetics. J Med Chem. 1999;42:1203–1212. doi: 10.1021/jm980537b. [DOI] [PubMed] [Google Scholar]
- Dragovich PS, Prins TJ, Zhou R, Webber SE, Marakovits JT, Fuhrman SA, Patick AK, Matthews DA, Lee CA, Ford CE, Burke BJ, Rejto PA, Hendrickson TF, Tuntland T, Brown EL, Meador JW, Ferre RA, Harr JEV, Kosa MB, Worland ST. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P-1 lactam moieties as L-glutamine replacements. J Med Chem. 1999;42:1213–1224. doi: 10.1021/jm9805384. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D-Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Greve JM, Davis G, Meyer AM, Forte CP, Yost SC, Marior CW, Kamarck ME, Mcclelland A. The major human rhinovirus receptor Is Icam-1. Cell. 1989;56:839–847. doi: 10.1016/0092-8674(89)90688-0. [DOI] [PubMed] [Google Scholar]
- Hao W, Bernard K, Patel N, Ulbrandt N, Feng H, Svabek C, Wilson S, Stracener C, Wang K, Suzich J, Blair W, Zhu Q. Infection and propagation of human rhinovirus C in human airway epithelial cells. J Virol. 2012;86:13524–13532. doi: 10.1128/JVI.02094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer F, Gruenberger M, Kowalski H, Machat H, Huettinger M, Kuechler E, Blaas D. Members of the low-density-lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc Natl Acad Sci USA. 1994;91:1839–1842. doi: 10.1073/pnas.91.5.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SL, Pattemore PK, Sanderson G, Smith S, Lampe F, Josephs L, Symington P, Otoole S, Myint SH, Tyrrell DAJ, Holgate ST. Community study of role of viral-infections in exacerbations of asthma in 9–11 year-old children. BMJ. 1995;310:1225–1229. doi: 10.1136/bmj.310.6989.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, Dean A, George KS, Briese T, Lipkin WI. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CC, Kuo CJ, Ko TP, Hsu MF, Tsui YC, Chang SC, Yang S, Chen SJ, Chen HC, Hsu MC, Shih SR, Liang PH, Wang AH. Structural basis of inhibition specificities of 3C and 3C-like proteases by zinc-coordinating and peptidomimetic compounds. J Biol Chem. 2009;284:7646–7655. doi: 10.1074/jbc.M807947200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotzerich M, Roulin PS, Boucke K, Witte R, Georgiev O, Greber UF. Rhinovirus 3C protease suppresses apoptosis and triggers caspase-independent cell death. Cell Death Dis. 2018;9:272. doi: 10.1038/s41419-018-0306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu GW, Qi JX, Chen ZJ, Xu X, Gao F, Lin DZ, Qian WK, Liu H, Jiang HL, Yan JH, Gao GF. Enterovirus 71 and coxsackievirus A16 3C proteases: binding to rupintrivir and their substrates and anti-hand, foot, and mouth disease virus drug design. J Virol. 2011;85:10319–10331. doi: 10.1128/JVI.00787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DA, Dragovich PS, Webber SE, Fuhrman SA, Patick AK, Zalman LS, Hendrickson TF, Love RA, Prins TJ, Marakovits JT, Zhou R, Tikhe J, Ford CE, Meador JW, Ferre RA, Brown EL, Binford SL, Brothers MA, DeLisle DM, Worland ST. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc Natl Acad Sci USA. 1999;96:11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C, Aguayo E, Rodriguez M, Lee G, Jordan R, Cihlar T, Birkus G. Multiple classes of antiviral agents exhibit In Vitro activity against human rhinovirus type C. Antimicrob Agents Chemother. 2014;58:1546–1555. doi: 10.1128/AAC.01746-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EK, Khuri-Bulos N, Williams JV, Shehabi AA, Faouri S, Al Jundi I, Chen QX, Heil L, Mohamed Y, Morin LL, Ali A, Halasa NB. Human rhinovirus C associated with wheezing in hospitalised children in the Middle East. J Clin Virol. 2009;46:85–89. doi: 10.1016/j.jcv.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann SC, Cherney MM, Sia S, Plotch S, James MN. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Palmenberg AC. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- Palmenberg AC, Rathe JA, Liggett SB. Analysis of the complete genome sequences of human rhinovirus. J Allergy Clin Immunol. 2010;125:1190–1199. doi: 10.1016/j.jaci.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patick AK, Binford SL, Brothers MA, Jackson RL, Ford CE, Diem MD, Maldonado F, Dragovich PS, Zhou R, Prins TJ, Fuhrman SA, Meador JW, Zalman LS, Matthews DA, Worland ST. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob Agents Chemother. 1999;43:2444–2450. doi: 10.1128/AAC.43.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter AG. Picornavirus nonstructural proteins—emerging roles in virus-replication and inhibition of host-cell functions. J Virol. 1993;67:6917–6921. doi: 10.1128/JVI.67.12.6917-6921.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam JCH, Bidgood SR, McEwan WA, James LC. Intracellular sensing of complement C3 activates cell autonomous immunity. Science. 2014;345:1134. doi: 10.1126/science.1256070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MT, Cheng YH, Liu YN, Liao NC, Lu WW, Kung SH. Real-time monitoring of human enterovirus (HEV)-infected cells and anti-HEV 3C protease potency by fluorescence resonance energy transfer. Antimicrob Agents Chemother. 2009;53:748–755. doi: 10.1128/AAC.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QM, Chen SH. Human rhinovirus 3C protease as a potential target for the development of antiviral agents. Curr Protein Pept Sci. 2007;8:19–27. doi: 10.2174/138920307779941523. [DOI] [PubMed] [Google Scholar]
- Yang HT, Yang MJ, Ding Y, Liu YW, Lou ZY, Zhou Z, Sun L, Mo LJ, Ye S, Pang H, Gao GF, Anand K, Bartlam M, Hilgenfeld R, Rao ZH. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]