Abstract
In the present study, specificity of laccase from Stropharia sp. ITCC-8422 against various substrates, i.e. 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), 2,6-dimethoxyphenol (DMP), guaiacol (GCL) and syringaldazine (SYZ) was determined. It exhibited maximum affinity against ABTS, followed by DMP and negligible activity for GCL and SYZ. As the concentration of substrate increased from 0.5 to 1.5 mM (ABTS) and 1 to 5 mM (DMP), the activity increased from 301.1 to 567.8 U/L and 254.4 to 436.2 U/L. Further, quadrupole time-of-flight liquid chromatography mass spectrometry (QTOF-LCMS) analysis of the extracellular proteome of Stropharia sp. ITCC-8422 identified eighty-four (84) extracellular proteins. The peptide sequence for the enzyme of interest exhibited sequence similarity with laccase-5 of Trametes pubescens. Using high molecular mass sequence of laccase-5, the protein structure of laccase was modelled and binding energy of laccase with four substrates, i.e. ABTS (− 5.65), DMP (− 4.65), GCL (− 4.66) and SYZ (− 5.5) was determined using autodock tool. The experimental and in silico analyses revealed maximum activity of laccase and lowest binding energy with ABTS. Besides, laccase was purified and it exhibited 2.1-fold purification with purification yield of 20.4% and had stability of 70% at pH 5–9 and 30–40 ℃. In addition, the bioremediation potential of laccase was explored by in silico analysis, where the binding energy of laccase with alizarin cyanine green was − 6.37 and both in silico work and experimental work were in agreement.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02399-8) contains supplementary material, which is available to authorized users.
Keywords: Laccase, Docking, In silico, Alizarin cyanine green, Bioremediation
Introduction
The multicopper oxidases (MCOs) consist of a family of enzymes which couples the reduction of molecular oxygen to water. The MCO superfamily consists of laccase (EC 1.10.3.2), ascorbate oxidase (EC 1.10.3.3), ferroxidase (EC 1.16.3.1), nitrite reductase (EC 1.7.2.1), bilirubin oxidase (EC 1.3.3.5) and ceruloplasmin (EC 1.16.3.1) where laccase has broad substrate specificity, e.g. polyamines and certain inorganic compounds (Baldrian 2005; Madhavi and Lele 2009; Janusz et al. 2020). Laccase is a glycosylated monomer or homodimer protein (Madhavi and Lele 2009) where 10–50% of the molecular mass of protein is due to glycosylation of which mannose is a major component attached to laccase (Madhavi and Lele 2009; Cannatelli and Ragauskas 2017).
Laccase consists of four copper atoms which are classified into three sites, i.e. Type 1 (T1 CuII), Type 2 (T2 CuII), Type 3 (T3 binuclear CuII–CuII cluster), where Type 2 and Type 3 form a trinuclear Cu cluster. The metallo-organic bond of T1 CuII consists of two histidines and one cysteine along with the side chain of methionine, leucine or isoleucine. On the other hand, T2 CuII and T3 CuII consist of two and six histidine ligands, respectively (Komori and Higuchi 2015). The mechanism of action involves the reduction of T1 Cu by substrate, followed by the transfer of electron over a distance of approximately 13 Å from T1 Cu to trinuclear cluster, and lastly oxygen reduction at the trinuclear cluster (Solomon 2001; Mukhopadhyay 2018). The oxidation of substrate occurs through T1 CuII due to electron transport and it has to be noted that the enzyme–substrate specificity is due to the structure–activity relationship at the site rather than the trinuclear cluster (Quintanar 2007).
The structure, function, and application of laccases in various sectors have been studied in detail; however, their potential is yet to be explored (Agrawal et al. 2018). The wide application of fungal laccase is due to its ability to oxidize a wide range of compounds as it has high redox potential. It is due to this fact that laccases have gained wide interest among the researchers and also have a wide range of application, e.g. dye decolorization, delignification and effluent treatment (Agrawal et al. 2019; Agrawal and Verma 2019a; 2019b; Huang et al. 2020). Laccase based on the redox potential (E0) has been classified laccase into two types, i.e. low (500 mV versus normal hydrogen electrode) and high (700–800 mV) E0 laccase (Xu et al. 1998; Garavaglia et al. 2004). The biological functions of laccase have been studied in detail, though the statistical (docking) studies of laccases with various substrates are still in the budding stage and the detailed study will enable the researchers for successful exploitation of laccases in various industrial and biotechnological sectors (Kameshwar et al. 2018; Hongyan et al. 2019).
As per our knowledge, the information on docking is scattered with respect to laccase. Thus, in the present study, a bioinformatic tool was used to analyse the substrate specificity via docking studies using laccase from Stropharia sp. ITCC-8422. In addition, the statistical (docking) studies were further extended to evaluate the bioremediation potential of laccase for the anthraquinone dye alizarin cyanine green. The sequence analysis of the extracellular peptide was carried out using quadrupole time-of-flight liquid chromatography mass spectrometry (QTOF-LCMS) and the substrate specificity was determined via docking of enzyme with four different substrates, i.e. 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), guaiacol (GCL), 2,6-dimethoxyphenol (DMP) and syringaldazine (SYZ). In addition, using in silico analysis, the chemical events underlying the higher specificity of ABTS with laccase were also inferred. Further, the potential application of laccase in dye decolorizing of anthraquinone dye alizarin cyanine green was also determined via experimental and in silico analysis.
Materials and methods
Chemicals and reagents
ABTS was purchased from Sigma; GCL and DMP were purchased from Hi-media; SYZ was purchased from Merck India. All other chemicals were of analytical grade and purchased from Hi-Media and Merck, India.
Sample preparation
The strain was previously isolated in our laboratory and identified at Indian Type Culture Collection (ITCC), Indian Agricultural Research Institute (IARI), New Delhi. Later, the identified strain Stropharia sp. was deposited under the accession number ITCC-8422 (Agrawal and Verma 2019a). The strain Stropharia sp. ITCC-8422 was maintained on malt extract agar (MEA) plates at 4 ℃ until further use. The media used for laccase production using Stropharia sp. ITCC-8422 consist of (g/L) KH2PO4 7, K2HPO4 2, MgSO4.7H2O 0.1, (NH4)2SO4 0.1, yeast extract 0.6, glucose 10 and pH 7. The fermentation was performed in 250-mL Erlenmeyer flask containing 50 mL media. The media was inoculated with two cubes (6 mm) of Stropharia sp. ITCC-8422 and incubated at 30 ℃ for 22 days. The crude supernatant was extracted on maximum day (16th day), centrifuged at 5850 g, and stored at 4 ℃ until further use.
Laccase activity and protein estimation
Laccase activity was determined quantitatively by measuring change in absorption of ABTS, GCL, DMP, and SYZ at 420 nm, ε420 = 36,000 M−1 cm− 1, 465 nm ε465 = 12,100 M− 1 cm− 1, 469 nm ε469 = 14,800 M− 1 cm− 1, 525 nm ε525 = 65,000 M− 1 cm− 1 for 5 min using a digital spectrophotometer (Labtronics; Model: LT-39). The concentration of the substrates was varied as follows: ABTS (0.5–1.5 mM), DMP (1–5 mM), GCL (20–30 mM) and SYZ (10–30 mM). ABTS, GCL and DMP were dissolved in sodium acetate buffer (0.1 M), pH 5, and SYZ was dissolved in 70% ethanol (Bourbonnais et al. 1995; Srinivasan et al. 1995; Jolivalt et al. 2005; Wu et al. 2010; Rajagopalu et al. 2016). One unit of enzyme activity is defined as the amount of enzyme oxidizing 1 μmol of substrate min− 1. The estimation of protein was done by Lowry’s method (Lowry et al. 1951) using bovine serum albumin (BSA) as the standard. All the experiments were performed in triplicates.
Molecular mass determination, sample preparation and analysis of peptides using QTOF-LC–MS
The protocol used for the determination of the approximate molecular mass of laccase from Stropharia sp. ITCC-8422 was as per Laemmli (1970). After electrophoresis, the gel was stained using silver staining technique. Silver staining was carried out for the determination of the crude protein from Stropharia sp. ITCC-8422. The gel was fixed using the fixative solution (100 mL-methanol 50%, acetic acid 10%, formaldehyde 50 μL) for 1 h. Then the gel was washed with wash solution (50% ethanol) three times for 20 min each, followed by treatment with hypo-solution (sodium thiosulfate 20 mg/100 mL). The gel was further washed with water three times for 20 s each and treated with silver nitrite solution (200 mg/100 mL) for 30 min followed by three times washing with water for 20 s each. Then the developing solution (100 mL-sodium carbonate 6 g, hypo-solution 2 mL, formaldehyde 50 μL) was added and incubated until the bands were observed. After the gel developed, stop solution (5% acetic acid) was added and the gel was stored in the above-mentioned fixative solution. The confirmation of the presence of laccase was done by zymography using ABTS as the substrate.
The analysis of peptides from Stropharia sp. ITCC-8422 was done using Agilent 6546 QTOF-LCMS System (Agilent Technologies Singapore) at Agilent Technologies, Centre of Excellence Manesar, Gurugram, India. The strain Stropharia sp. ITCC-8422 was inoculated in the media and on the day of maximum laccase activity, the culture supernatant was centrifuged for 10 min at 5850 g at 4 ℃. To the cell-free supernatant, chilled acetone at − 20 ℃ was added in the ratio of 1:4 vortexed and incubated for 60 min for effective precipitation of the protein (Tiwari et al. 2016). After precipitation, the protein pellet was obtained after centrifugation for 15 min at 5850 g at 4 ℃. The protein sample (total protein 1 mg) was added to 6 M urea, 100 mM Tris buffer (pH 7.8), followed by the addition of a reducing agent (200 mM DTT dissolved in 100 mM Tris, pH 7.8). The protein was gently vortexed, and alkylated for 1 h at room temperature (28 ± 2 ℃). After the incubation, alkylating reagent (200 mM iodoacetamide in 100 mM Tris pH (7.8) was added, vortexed and incubated for 1 h at room temperature. The reducing agent was added again to remove the unreacted iodoacetamide, mixed and incubated for 1 h. The concentration of urea was reduced by diluting the reaction with water to approximately 0.6 M where chymotrypsin retains its activity. To the sample, chymotrypsin (20 μg) was added, gently vortexed and digested overnight at 37 ℃. Chymotrypsin provides protease-to-substrate ratio of 1:50. After the completion of incubation, pH was reduced to less than 6 using acetic acid (Kislinger and Emili 2003; Kinter and Sherman 2005; Ghosh et al. 2015). The peptides were analysed using Advance Biopeptide Column 2.1 × 100 mm, 2.7-micron (Agrawal and Verma 2020).
Three-dimensional (3D) structure of laccase
The homology modelling of laccase was performed to predict the 3D structure. The peptide sequence of laccase from QTOF-LCMS as mentioned previously (RLVSISCDPNFTF) was subjected to BlastP (Altschul et al. 1990). The amino acid sequence of laccase-5 from Trametes pubescens showed significant sequence alignment with queried peptide; thus, homology modelling was carried out from the amino acid sequence of laccase-5 using web-based server Phyre2 (https://www.sbg.bio.ic.ac.uk/phyre2), where hidden Markov method generated the alignments of laccase against the already existing template protein structure. The structure was obtained and used to produce a homology-based model of the desired protein sequence followed by the prediction of 3D structure. It also uses Poing (ab initio folding stimulation) model to detect minimum or no similarities with already known structures (Thakur and Shankar 2016). Initially, the FASTA sequence of laccase-5 was subjected to the Phyre2 server under an intensive mode, and template ‘c1gycA’ was (with confidence 100% and identities 73%) used. Further, the stereo-chemical quality of a protein structure was performed using the RAMPAGE server (https://mordred.bioc.cam.ac.uk/∼rapper/rampage.php). The phylogenetic analysis was performed from the peptide sequence of Stropharia sp. ITCC-8422 using BlastP. Multiple sequence alignment using Constraint-based multiple Alignment Tool (COBALT) at NCBI was done. Protein sequences retrieved in clustal format were subjected to MEGA (v10.1.8) software for the construction of evolutionary relation followed by phylogenetic analysis of the sequence (Kumar et al. 2018). Maximum-likelihood method in the MEGA (v10.1.8) software was used for the construction of the phylogenetic tree. The phylogenetic tree was constructed using bootstrap testing with 1000 replicates.
Molecular docking studies
The molecular docking was performed using Autodock tool-1.5.6 and the docking scores were calculated. The preparation of the ligand was done by adding Gasteiger charges, H-bonds (polar) along with well-defined rotatable bonds. The setting used was random for the initial position, orientation, and torsions of the ligand molecules. While the docking analysis was performed, the rotatable torsions were released and the parameters searched consisted of Lamarckian genetic algorithm followed by 50 runs. The Autogrid program was used to generate affinity (grid) maps of 90 × 860 × 104 xyz Å grid points and 0.636 Å spacing. The van der Waals and electrostatic terms were calculated using the Autodock parameter with distance-dependent dielectric functions. The docking experiment consisted of 50 different runs which were terminated after energy evaluation of 25 k and other docking parameters mentioned in the study by Zhou et al. (2014) were used where a translational step of 0.2 Å, and quaternion and torsion steps of 5 were applied during the search (Xiao et al. 2009; Lin and Lapointe 2013). From the docking analysis, binding energy and inhibition constant were inferred and the interactions of both the ligands were compared to determine the binding energy (Tiwari et al. 2019). The result was obtained from 50 runs, and the hydrophobic and H-bond interactions between the ligands and receptor of the docked complex were analysed using the Chimera 1.14 tool.
Determination of Michaelis–Menten constant (Km and Vmax)
The laccase was purified using the methodology used in our previous study (Agrawal et al. 2019). The kinetic constants Km and Vmax were calculated using standard reaction mixtures of the substrates and laccase. The assay reaction for laccase consisted of substrate ABTS, DMP and GCL (1–10 mM) dissolved in 0.1 M sodium acetate buffer pH 5 and 30 ℃. The Km and Vmax were calculated using a double-reciprocal plot method of Lineweaver and Burk plot of 1/[V] against 1/[S] giving intercepts where V is the enzyme activity and S substrate concentration.
pH and thermal stability of laccase
pH stability was determined by incubating laccase at a pH range of 2–11 and the thermal stability was performed at a temperature range of 30–60 ℃ under optimized conditions (Daroch et al. 2014; Fithri et al. 2020). The residual activity at pH and temperature were determined at a regular interval of 12 h using standard assay condition.
Effect of metal ion and inhibitor on laccase
Various metal ion and inhibitors were used, Zn2+, Cu2+, Ag2+, K+, Mg2+, Ni2+, Ca2+, Fe2+, SDS, sodium azide (NaN3) and EDTA, under the optimum condition to determine its effect on the residual activity of laccase at 1 and 5 mM concentrations (Mukhopadhyay and Banerjee 2015). The residual activity was determined after 12 h of incubation using standard assay conditions.
Experimental and in silico analysis of the dye decolorization potential of laccase
The dye alizarin cyanine green was selected and the decolorization potential at a regular interval of 6 h was determined in media as mentioned by Agrawal and Verma (2019a) for 48 h at 28 ± 2 ℃. The experimental analysis was performed in triplicate. The docking of alizarin cyanine green was done with laccase. For enzyme–dye interaction, the structure of the dye was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Docking analysis was carried to calculate binding energy and inhibition constant, and the enzyme–dye complex was analysed using Chimera 1.14 tool.
Results and discussion
Laccase activity of multicopper oxidases using various substrates
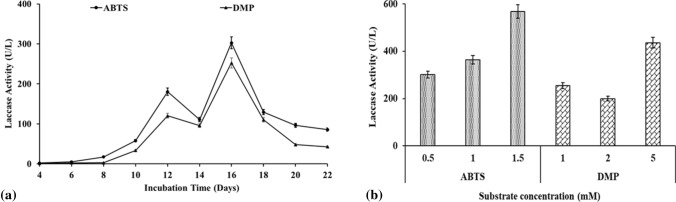
Laccase is able to oxidize phenolic substrates, mediated by a copper-containing active site; as a result, various substrates have been reported for the detection of laccase activity (Pundir et al. 2016; Kolomytseva et al. 2017; Mtibaà et al. 2018; Wang et al. 2018). Thus, the enzymatic profiling was done using various substrates for 22 days and the maximum affinity was exhibited by 0.5 mM ABTS (2.1, 180.4 U/L) followed by 1 mM DMP (1.1, 120.3 U/L) on the 4th and 12th days (Fig. 1a). The activity detected was negligible for GCL and SYZ. As the incubation time increased, the activity increased to 302.8 U/L (ABTS) and 252.3 (DMP) U/L on the 16th day. In Fig. 1a, first (trophophase) and second (idiophase) peaks were observed on the 14th and 16th day. It has been previously reported that the presence of two peaks indicates constitutive laccase production and it can be due to protease secretion which leads to proteolysis of laccase in culture media (Palmieri et al. 2001; Staszczak 2002; Staszczak and Jarosz-Wilkołazka 2005) or due to the instability of laccase in the growth conditions or due to the presence of independent genes of differential expression which allows the expression of various isoenzymes (Klonowska et al. 2001). However, as per our knowledge, and hypothesis regarding the physiological significance, repetitive laccase production has not been studied in detail.
Fig. 1.
The substrate specificity of laccase from Stropharia sp. ITCC-8422 with 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) and 2,6-dimethoxyphenol (DMP). a Enzymatic profiling of laccase using ABTS and DMP at regular intervals for 22 days; b effect of substrate concentration, i.e. ABTS (0.5–1.5 mM) and DMP (1–5 mM) on laccase activity (Note: as the activity was negligible for guaiacol (GCL) and syringaldazine (SYZ), it is not incorporated in the figure)
The preliminary screening of desired protein consists of laccase assay, an enzyme–substrate reaction that is a time-dependent process. The product formed as a result of oxidation during the assay is of prime importance for the detection of laccase (Agrawal and Verma 2019c). As the initial assay was performed using crude laccase from Stropharia sp. ITCC-8422, its affinity against various substrate was determined at a protein load of 0.16 mg/mL. The analysis of the specificity of enzymes against various substrates enables better detection of the desired protein in the extracellular crude extract with enhanced confirmation. The maximum specificity was with ABTS and as the concentration increased, the activity increased from 301.1 to 567.8 U/L at 0.5 and 1.5 mM, respectively. Similarly, in the case of DMP, the activity was 254.4, 199.9 and 436.2 U/L at 1, 2 and 5 mM, respectively (Fig. 1b). The results of the substrate affinity are in co-ordination with the previously reported data (Agrawal and Verma 2019c) where laccase from Myrothecium verrucaria ITCC-8447 and Pleurotus ostreatus showed maximum substrate affinity against ABTS, followed by DMP, GCL, and least affinity or negligible activity was detected with SYZ. The optimum pH of laccase is dependent on the substrate, where the optimum pH of fungal laccase for phenols and non-phenolic substrate ranges from 3 to 7 and < 4, respectively Thus, one main parameter which may have resulted in the detection of negligible activity in the case of GCL and SYZ might be attributed to the pH optimum of laccase (Madhavi and Lele 2009).
Also, high specificity of laccase against ABTS has been reported from Maugniella sp. (Palonen et al. 2003), Trichophyton rubrum LKY-7 (Jung et al. 2002) and Fomitella fraxinea (Park and Park 2008). As per previously reported data and kinetics study, ABTS has been reported to be a suitable substrate for laccase from Pleurotus ostreatus as compared to DMP (Tlecuitl-Beristain et al. 2008). Xiao et al. (2003) suggest that ABTS was the best substrate followed by DMP for laccase from Trametes sp. strain AH28-2. Ramírez-Cavazos et al (2014) used purified laccase from Pycnoporus sanguineus, a basidiomycete, and it exhibited a high binding affinity for ABTS followed by DMP and GCL. Thus, it can also be inferred that the crude and purified laccase exhibits similar substrate affinity, i.e. ABTS followed by DMP (Tlecuitl-Beristain et al. 2008; Ramírez-Cavazos et al 2014; Agrawal and Verma 2019c). In the study by Park and Park (2008), laccase showed high activity with DMP which has a double methoxy group as compared to GCL which has one methoxy group, whereas laccase activity was very low with SYZ. Similarly, in the present study, laccase from Stropharia sp. ITCC-8422 exhibited highest activity with ABTS followed by DMP and negligible for SYZ.
Harkin et al. (1974) has stated SYZ, was a laccase-specific substrate; however, as per the study by Solano et al. (2001) among various substrates, DMP has been regarded as a best substrate as it has wide oxidation capacity, rather high molar coefficient and stability of its oxidized dimeric coloured form, i.e. 3.3′,5,5′-tetramethoxydiphenylquinone. The coefficient of quinone at 468 nm is 14,800 M− 1 cm− 1 which is nearly three times higher than the one obtained by the oxidation of o-methoxyphenol (GCL) to tetraguaiacol (5570 M− 1 cm− 1) (Solano et al. 1997, 2001). In the study by Nyanhongo et al. (2006), the substrate oxidation by laccase LTM1 did not follow a specific pattern for substrate oxidation where it had high affinity for SYZ and low for DMP, and ABTS was observed to be over GCL. Thus, it was stated that the range of substrates oxidized by laccases from various sources varies significantly due to differences in redox potentials of the laccases from different sources as well as the redox potential of the substrate (Shin and Lee 2000; Nyanhongo et al. 2006).
Further, multicopper oxidase was classified into three groups, first consisted of real laccase, which oxidized DMP in the absence of copper supplementation that was characterized by the linear accumulation of the oxidized product of DMP. The second group, pseudo-laccase oxidized DMP after the addition of copper supplementation and consists of copper-tolerance proteins. The last group showed very slow DMP oxidation and the reaction lasted for hours in the presence of copper whereas the oxidation was not detected if copper were absent. Also, DMP is more preferred as it is more easily oxidized than GCL and the coloured product formed as a result of the oxidation of DMP is more stable than SYZ, and most importantly as stated above, the other two groups of multicopper oxidase required copper for the reaction to occur and peroxidases require hydrogen peroxide (Solano et al. 1997, 2001; Nakatani et al. 2010). Thus, on the basis of the above result and reported data, it was confirmed that the enzyme produced by Stropharia sp. ITCC-8422 was laccase (Agrawal and Verma 2019a) and similar result has also been reported by previously by Daroch et al. (2014) in Stropharia aeruginosa.
Analysis of peptides using QTOF-LCMS
The molecular mass of crude glycosylated yellow laccase from Stropharia sp. ITCC-8422 was determined using silver staining and the molecular mass ranged from approximately 100 to 135 kDa (Fig. 2a-b). The strain has been reported for the production of yellow laccase as it lacks absorption at 610 nm (Agrawal and Verma 2019a). Baldrian (2005) had reported variation in molecular mass of laccases and the typical fungal laccase has a molecular mass of 60–70 kD. However, few strains i.e. Phellinus ribis (Min et al. 2001) and Cantharellus cibarius (Ng and Wang 2004) with a higher molecular mass of 152 and 92 kDa have also been reported. It was also stated that these were homodimeric structures implying that the enzyme has two identical subunits with molecular mass typical of monomeric laccases (Baldrian 2005). It has also been observed that the molecular mass of glycosylated laccase is high as compared to de-glycosylated laccase. The pure laccase from strain Pycnoporus sanguineus CeIBMD001 has a molecular mass of 68 kDa whereas after N-deglycosylation decrease of molecular mass by approximately 8–10 kDa was observed (Vite-Vallejo et al. 2009). Thus, the high molecular mass of laccase is due to glycosylation and is in accordance with the previous literature. Further, in the case of zymography (Fig. 2c), oxidation of ABTS to form green band confirmed the presence of laccase in the crude protein. As per Murugesan et al. (2007), native-PAGE (zymography) of crude enzyme helped in the identification of the ligninolytic enzyme produced by Ganoderma lucidum. The crude enzyme showed single laccase activity against ABTS and GCL, thus confirming the presence of laccase from Ganoderma lucidum. Also, in the study by Li et al. (2009), the crude extract from Rigidoporus lignosus W1 was run on SDS-PAGE where multiple bands were observed. Further, after incubating the renaturation gel with guaiacol, a single band was obtained due to the oxidation GCL, thereby confirming the presence of laccase and its role in dye removal. Thus, considering the previous concept, the present study was designed where the silvers staining of crude laccase showed the presence of multiple bands, whereas in zymography, staining of only single band was observed due to the oxidation of ABTS, thus, confirming the production of laccase by Stropharia sp. ITCC-8422. Also, QTOF-LCMS was performed to further confirm the presence of laccase in the extracellular proteome of Stropharia sp. ITCC-8422. Thus, the data, i.e. laccase assay, zymography and QTOF-LCMS, were in correlation with each other confirming the presence of laccase in crude extract of Stropharia sp. ITCC-8422.
Fig. 2.
Silver staining and zymography analysis of crude protein from Stropharia sp. ITCC-8422. a Protein ladder, b silver staining of crude glycosylated laccase, c zymography of the crude glycosylated laccase with 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS)
The QTOF-LCMS was carried out to determine the amino acid sequence of laccase from Stropharia sp. ITCC-8422. The extracellular proteome of Stropharia sp. ITCC-8422 identified 84 different proteins among which laccase was one of the identified proteins. Among all the proteins transpeptidase, protein kinases, sulfatase, thymidylate kinase, and many more proteins belonging to various protein families were identified. The glycosylated yellow laccase exhibited maximum sequence similarity with laccase-5 from Trametes pubescens/Trametes versicolor (white-rot fungus; Coriolus versicolor) with the sequence RLVSISCDPNFTF (Agrawal and Verma 2020). Similarly, in the work carried out by Daroch et al. (2014), Stropharia aeruginosa was identified to produce glycosylated yellow laccases Yel1p and Yel3p. The Yel1p protein contained a unique peptide sequence whereas may be due to the glycosylation, no such peptide was found in Yel3p protein sequence data. Thus, a more detailed study is required for a better understanding as well as more molecular data need to be updated in the database.
Sequence alignment of a peptide sequence
The BlastP of the 13 aa sequence RLVSISCDPNFTF obtained from QTOF-LCMS was used for the analysis. The laccase from Stropharia sp. ITCC-8422 showed sequence similarity (E value 1e-04) with laccase-5 from Trametes pubescens with sequence ID OJT07652.1 followed by laccase-4 from Trametes versicolor sequence ID BAD98308.1 and so on. The strain Trametes pubescens was considered for further study as it has high molecular mass as compared to Trametes versicolor. Further, “Stropharia sp. ITCC-8422” experimentally showed laccase activity, and considerably significant E value (1e-04) of the peptide exhibited identity with laccase-5 from Trametes pubescens. In addition, 13 out 13 amino acids showed 100% identities between query and subject (a high Scoring Segment). Thus, considering the experimental data along with a significant E value, the amino acid sequence of laccase-5 from Trametes pubescens was used as a template sequence for structure analysis. The phylogenetic relationship was determined for the peptide sequence laccase from Stropharia sp. ITCC-8422. It was observed from the evolutionary tree results that exhibited sequence similarity with other fungal species from Class Agaricomycetes (Basidiomycetes) (Supplementary Fig. 1). It is worth mentioning that strain Stropharia sp. also belongs to the Class Agaricomycetes.
Three-dimensional (3D) structure of laccase
As the activities of extracellular laccase with four selected substrates were determined, the molecular docking studies were further carried out to understand the stability of the enzyme–substrate complex. The hydrophilic and hydrophobic interaction between the enzyme–substrate complexes may shed light on the chemical event taking place during the interaction. The 3D structure of laccase from Stropharia sp. ITCC-8422 was a model for the docking with four different substrates, i.e. ABTS, DMP, GCL and SYZ (Table 1). The ribbon representation of laccase has been represented in Fig. 3. Laccase is a multicopper oxidase that consists of multi-domain cupredoxins, e.g. nitrite reductase and ceruloplasmin consisting of two and six domains, respectively (Zaitseva et al. 1996; Murphy et al. 1997). The common laccases have the presence of three similar cupredoxin domains where at least seven β-strands form a β-barrel structure (Hakulinen and Rouvinen 2015). The three-domain laccase has been reported in various strains Lentinus tigrinus (Ferraroni et al. 2007), Rigidoporus lignosus (Garavaglia et al. 2004), Trametes hirsuta (Polyakov et al. 2009), etc. among which Trametes versicolor (Bertrand et al. 2002; Piontek et al. 2002) also has the presence of three domains which is in correlation with the present study.
Table 1.
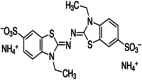
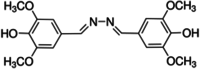
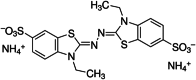
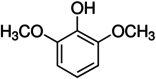
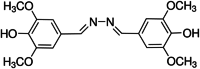
The structure and CID number of ABTS, DMP, GCL and SYZ
| Sl. no. | Substrate | Structure | CID number |
|---|---|---|---|
| 1 | ABTS |
|
16,240,279 |
| 2 | DMP |
|
7041 |
| 3 | GCL |

|
460 |
| 4 | SYZ |
|
135,401,234 |
Fig. 3.

The ribbon representation of laccase predicted using the sequence obtained after quadrupole time-of-flight liquid chromatography mass spectrometry (QTOF-LC–MS)
Homology modelling and its validation using various substrates
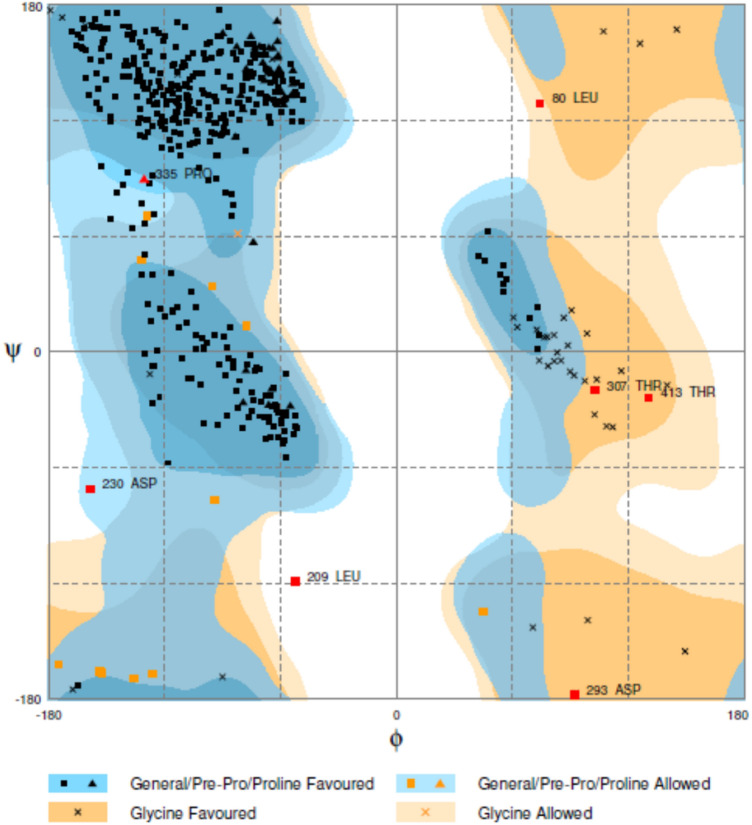
The docking of laccase from Stropharia sp. ITCC-8422 was carried out with four different substrates, i.e. ABTS, DMP, GCL, and SYZ. The docking analysis revealed the binding energy and it was observed that laccase exhibited the lowest binding energy of − 5.65 with ABTS (Fig. 4a). On the other hand, the binding energy of laccase with the other three substrates was also determined, where with DMP, GCL and SYZ, the binding energies were − 4.65, − 4.66, and − 5.5 (Fig. 4b-d) (Table 2). Thus, the present study is in correlation with the experimental analysis from the previously reported data where higher substrate specificity with other laccases from Myrothecium verrucaria ITCC-8447, Pleurotus ostreatus (Agrawal et al. 2019; Agarwal and Verma 2019c), Pycnoporus cinnabarinus (Eggert et al. 1996) and Mauginiella sp. (Palonen et al. 2003) was with ABTS. The interactions of laccase with these substrates were varied due to the formation of H-bonds at different residues of amino acids (Table 3). The lowest binding energy (− 5.65) for ABTS substrate forms H-bonds at residue ALA23 and THR38 with the enzyme. However, a substrate having binding − 5.5 forms H-bonds at LEU57 and THR38. The formation of H-bonds at common residue THR38 indicates the active site for these two substrates (ABS/SYZ). For the other two substrates (DMP/GCL), we observed a common residue HIS133 of laccase interacting with these two substrates (Table 4). Our in silico analysis correlates with experimental data having better enzyme activity with ABTS than these substrates, e.g. SYZ/DMP/GCL. It also indicates that laccase enzymes have more than one active site depending on the substrate (Quintanar et al. 2007). The Ramachandran plot analysis inferred that 96.1%, i.e. 465 residues were in the favoured regions, 2.5%, i.e. 12 in the allowed region and 1.4%, i.e. 7 residues (Fig. 5) in outlier region (Lovell et al. 2002).
Fig. 4.
The 3D representation of the interaction of the amino acid residues of laccase with various substrates via in silico analysis. a 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS); b 2,6-dimethoxyphenol (DMP); c guaiacol (GCL); d syringaldazine (SYZ)
Table 2.
The binding energy and inhibition constant of laccase with ABTS, DMP, GCL and SYZ
| Sl. no. | Substrate | Binding energy | Inhibition constant |
|---|---|---|---|
| 1 | ABTS | − 5.65 | 72.23 |
| 2 | DMP | − 4.65 | 392.43 |
| 3 | GCL | − 4.66 | 383.64 |
| 4 | SYZ | − 5.5 | 92.52 |
Table 3.
The inter- and intra-model H-bonds of laccase with 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), 2,6-dimethoxyphenol (DMP), guaiacol (GCL), and syringaldazine (SYZ)
| Substrate | Donor | Acceptor | Hydrogen | D..A dist | D-H..A dist |
|---|---|---|---|---|---|
|
ABTS
|
ALA 23 N | UNK 0.A O2 | ALA 23 HN1 | 3.17 | 2.292 |
| THR 38 N | UNK 0.A O1 | THR 38 HN | 3.21 | 2.23 | |
DMP
|
HIS 133 ND1 | UNK 0.A O1 | HIS 133 HD1 | 3.07 | 2.12 |
| UNK 0.A O3 | HIS 133 O | UNK 0.A H10 | 3.389 | 2.451 | |
|
GCL
|
HIS 133 ND1 | UNK 0.A O2 | HIS 133 HD1 | 3.121 | 2.161 |
| SER 135 N | UNK 0.A O1 | SER 135 HN | 2.935 | 1.919 | |
|
SYZ
|
THR 38 N | UNK 0.A O5 | THR 38 HN | 3.111 | 2.104 |
| LEU 57 N | UNK 0.A O4 | LEU 57 HN | 2.972 | 2.003 |
Table 4.
The interaction of amino acid residues of laccase with four substrates, i.e. 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), 2,6-dimethoxyphenol (DMP), guaiacol (GCL), and syringaldazine (SYZ)
| Substrate | ABTS | DMP | GCL | SYZ | ||||
|---|---|---|---|---|---|---|---|---|
| Amino acid | Amino acid | Amino acid | Amino acid | |||||
| Laccase | ||||||||
| Laccase from Stropharia sp. ITCC-8422 |
THR38 VAL 37 PRO 39 PHL 53 PRO 54 ASN 194 THR 169 LEU 57 ALA 23 ASP 164 |
UNK 0.A |
ALA 101 ALA 102 HIS 133 LEU 134 SER 135 LEU 486 GLU 487 PHE 477 |
UNK 0.A |
LEU 486 HIS 133 SER 135 LEU 134 PHE 477 ALA 102 PHE 103 |
UNK 0.A |
THR 38 VAL 37 PHE 53 VAL 52 PRO 54 THR 169 PRO 56 PRO 145 LEU 57 LEU 196 LEU 48 |
UNK 0.A |
Fig. 5.
The Ramachandran plot analysis of the modelled protein showed 465 residues in the favoured region, 12 residues in the allowed region and 7 residues in the outlier region
Determination of Michaelis–Menten constant (Km and Vmax)
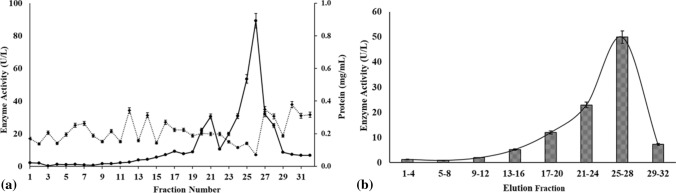
Laccase activity in the extracellular proteome of Stropharia sp. ITCC-8422 was confirmed by laccase assay, zymography and QTOF-LCMS analysis. Thus, to further broaden the study, the kinetic parameters were determined using purified laccase from Stropharia sp. ITCC-8422 for ABTS, DMP, and GCL using Lineweaver–Burk plot. The crude laccase was purified using ammonium sulphate precipitation and ion exchange as summarized in Table 5. The protein during ion exchange was eluted with NaCl gradient (0.1-1 M) and one prominent fraction was obtained in the 25–28th fraction (Fig. 6a, b). The maximum activity in 25–28th fraction gave 2.1-fold purification with 20.4% purification yield. As per our knowledge, the purification fold after ion exchange in the case of laccase varies (Ko et al. 2001; Junghanns et al. 2009) and the variation is observed as the purification depends upon the purification conditions (Zhao et al. 2012; Zhou et al. 2016).
Table 5.
Purification profile of laccase produced from Stropharia sp. ITCC-8422
| Purification steps | Enzyme activity (U/L) | Enzyme activity (U/mL) | Protein (mg/mL) | Specific activity (U/mg) | Purification fold | Yield (%) |
|---|---|---|---|---|---|---|
| Crude extract | 302.8 | 0.3 | 0.8 | 0.4 | 1.0 | 100.0 |
| Ammonium sulphate precipitation | 437.8 | 0.4 | 0.7 | 0.6 | 1.5 | 144.6 |
| DEAE sephadex | 89.2 | 0.09 | 0.07 | 1.3 | 2.1 | 20.4 |
Fig. 6.
Purification of laccase from Stropharia sp. ITCC-8422 by ion exchange chromatography. a Chromatogram representing laccase activity and protein concentration and b laccase activity obtained at various fractions after ion exchange chromatography
The Km of the purified laccase was 0.74 (ABTS), 9.5 (DMP), and 1.9 (GCL) mM for ABTS, DMP, and GCL whereas the Vmax was 42.0, 185.2 and 36.4 μmol/min/L, respectively. The lower the Km, higher is the affinity of laccase against the substrate. Thus, maximum affinity was observed for ABTS, followed by GCL and least by DMP (Fig. 7a-c). The data are in correlation with the previously reported data where laccase exhibits maximum specificity against ABTS (Palonen et al. 2003; Zhao et al 2012; Ramírez-Cavazos et al 2014). In the study by More et al. (2011), the Km and Vmax of laccase from Basidiomycetes Pleurotus sp. was 250 (mM) and 0.33 (μmol/min) using ABTS as the substrate whereas for DMP it was 38.46 (mM) and 20 (μmol/min), respectively. Similarly, in the other study, the Km of G. lucidum using ABTS was 0.114 mM. Thus, variation exists from strain to strain with respect to Km and Vmax values of laccase; however, the substrate affinity is more against ABTS which has been supported by Palonen et al. (2003), Xiao et al (2003), Tlecuitl-Beristain et al. (2008) and Zhao et al (2012). Further, the enzyme is laccase as the enzyme acts on DMP in the absence of copper supplementation (Solano et al. 2001).
Fig. 7.

The determination of Michaelis–Menten and Lineweaver–Burk plot of purified laccase from Stropharia sp. ITCC-8422 using a 2,2-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), b 2,6-dimethoxyphenol (DMP) and c guaiacol (GCL) as the substrates
pH and thermal stability of laccase
The effect of pH on laccase was determined at a pH (2–11). The enzyme retained more than 70% stability at 5–9 and retained 67.8 and 56.8% residual activity at pH 10 and 11 after 48 h of incubation, respectively (Fig. 8a). Laccase from Stropharia sp. ITCC-8422 unlike others reported exhibited stability towards a wide range of pH (5–11). However, at acidic pH of 2, the activity diminished completely after 12 h of incubation and retained less than 10% activity at pH 3–4 after 48 h of incubation. The results are in accordance with Daroch et al (2014) where yellow laccase from Stropharia aeruginosa was stable at pH 4–12 (Yel1p) and (5–9) Yel3p. Similarly, in the study by Nakatani et al. (2010), Lac I from Trametes sp. Ha1 retained stability at a pH range of 7–10, laccase (Lcc3) from Cerrena sp. WR1 retained maximum activity at pH 5 whereas the activity decreased at pH 3 and 4 (Chen et al. 2012). Thus, as per the reported literature with respect to basidiomycetes, laccase stability has been reported at a wide range of pH. The thermostability of laccase from Stropharia sp. ITCC-8422 was determined where it exhibited maximum thermostability (80.5%) at 30 ℃ followed (74.4%) at 40 ℃ (Fig. 8b). The activity diminished significantly at 50 and 60 ℃ retaining 19.8 and 11.4% residual activity after 48 h of incubation. In the study by Yuan et al (2016), laccase Lac1 and Lac3 showed stability at 50–60 ℃ whereas Lac2 exhibited stability at 60 ℃; however, at comparatively lower temperatures, i.e. 30 ℃ or 40 ℃, Lac1 and Lac3 had more activity compared to Lac2. As per Nakatani et al. (2010), Lac I from Trametes sp. Ha1 was stable up to 65 ℃ with more than 50% of the initial activity. As laccases have been reported to be stable at higher temperatures laccase from Stropharia aeruginosa exhibited maximum stability at 40 ℃ (Daroch et al. 2014) which is in accordance with the present study. Thus, as in the case of pH, the thermal stability also differs from strain to strain.
Fig. 8.
Determination of residual activity of laccase from Stropharia sp. ITCC-8422 at regular intervals for 12 h at various pH and temperature values. a Effect of pH on laccase in the range of 2–11. b Effect of temperature on laccase in the range of 30–60 ℃
Effect of metal ion and inhibitor on laccase
The residual activity of laccase at various concentrations (1 and 5 mM) of metal ions and inhibitors was determined. Laccase retained more than 90% residual activity with Zn2+, Ag2+, Ni2+ and more than 60% with Cu2+, K+, Mg2+ and Fe2+ at 1 mM. As the concentration of metal increased, the activity decreased from 1 to 5 mM (Table 6) and the residual activity decreased after 12 h of incubation. In the other study by Lorenzo et al. (2005), it was observed that laccase from Trametes versicolor exhibited 40% inhibition with Cu and Cd whereas complete inhibition was observed when the concentration increased to 80 mM. On the other hand, yellow laccase from Lentinus squarrosulus MR13 retained more than 95% relative activity with NiSO4 and more than 80% with MgSO4 (Mukhopadhyay and Banerjee 2015) which is in accordance with the present study. Further, the residual activity of laccase was significantly inhibited by NaN3 (2.8%) which suggests the oxidase nature of laccase (Zhao et al. 2012). The laccase from Trichoderma harzianum WL1 too was completely inhibited by NaN3 (20 µM) and it was stated that NaN3 binds to T2 and T3 Cu sites affecting the electron transfer thereby leading to the inhibition of laccase activity (Ryan et al. 2003; Sadhasivam et al. 2008). Laccase from Stropharia sp. ITCC-8422 in the present study and laccase from Trichoderma harzianum WL1 both were mildly inhibited by EDTA, thereby being in accordance with the previously reported literature (Sadhasivam et al. 2008).
Table 6.
The effect of metal ion and inhibitor on laccase activity at 1 and 5 mM after 12 h of incubation under standard assay condition
| Effect of metal ion/inhibitors | Residual activity (%) | |
|---|---|---|
| 1 mM | 5 mM | |
| Metal ion | ||
| Control | 100 | 100 |
| Zn2 + | 99.6 | 95.2 |
| Cu2 + | 69.5 | 65.4 |
| Ag2 + | 97.9 | 67.9 |
| K + | 75.6 | 71.6 |
| Mg2 + | 81.2 | 78.9 |
| Ni2 + | 93.9 | 85.3 |
| Ca2 + | 91.1 | 84.7 |
| Fe2 + | 72.7 | 86.5 |
| Inhibitor | ||
| SDS | 73.7 | 59.4 |
| Azide | 2.7 | 2.8 |
| EDTA | 63.2 | 74.3 |
Experimental and in silico analyses of dye decolorization potential of laccase
The strain effectively decolorized alizarin cyanine green and it was observed that with the increase in incubation, decolorization increased from 9.8% (6 h) to 88.5% (18 h) and compete removal was obtained after 48 h of incubation (Supplementary Table 1). The removal of alizarin cyanine green by yellow laccase from Stropharia sp. ITCC-8422 has been previously reported in our study (Agrawal et al 2019) where more than 90% decolorization was obtained at 600 ppm within 48 h of incubation. In the study by Tišma et al. (2020), crude and partially purified laccase from Trametes versicolor was used for dye removal and it was observed that partially purified laccase had enhanced dye removal potential and stability. However, at a higher temperature of 55 ℃, overall best result was attained with crude enzyme, though the stability was higher for partially purified laccase that was due to stable pH. Thus, it can be inferred that though partially purified laccase may have certain advantages as stated above, crude laccase seems to have an upper hand as it reduces the capital cost associated with purification; further, the removal efficiency was also high (Tišma et al. 2020).The strain Trametes sp. SQ01 effectively decolorized Congo red, acid red, amido black, orange G, Fast Blue RR salt and Remazol Brilliant Blue R and triphenylmethane dyes. It, however, has to be noted that 97–99% of azo dyes and RBBR, an anthraquinonic dye, were degraded in 7 days, whereas only 30–70% triphenylmethane dyes were removed after 7 days of incubation except for bromphenol blue (Yang et al. 2009). Similarly, other fungal strains too have been reported for the decolorization of the dye. Armillaria sp. strain F022 has been reported for effective decolourization of various synthetic dyes, e.g. azo, triphenylmethane as well as anthraquinone dyes (Hadibarata et al. 2012). The docking study revealed that the binding energy of laccase was − 6.37 with alizarin cyanine green (Fig. 9). The negative binding energy of laccase with dyes suggests that the degradation is spontaneous (Hongyan et al. 2019) and the dye formed H-bonds at residues ALA23 (Table 7) with laccase. This was also observed in “homology modeling and its validation using various substrates”. Laccase formed H-bond with two residues, of which ALA23 is common for both the substrate and alizarin cyanine green. The in silico analysis is in correlation with our previously published experimental work where alizarin cyanine green was effectively decolorized by laccase from Stropharia sp. ITCC-8422.
Fig. 9.

The in silico analysis showing the 3D representation of the interaction of laccase Stropharia sp. ITCC-8422 with the dye alizarin cyanine green
Table 7.
The inter- and intra-model H-bonds of laccase with alizarin cyanine green
| Donor | Acceptor | Hydrogen | D..A dist | D-H..A dist |
|---|---|---|---|---|
| ALA 23 N | UNK 0.A O8 | ALA 23 HN3 | 2.631 | 1.737 |
| UNK 0.A N1 | ASN 194 O | UNK 0.A H3 | 3.103 | 2.521 |
| UNK 0.A N2 | UNK 0.A O7 | UNK 0.A H4 | 2.503 | 1.796 |
| UNK 0.A O4 | LEU 57 O | UNK 0.A H22 | 2.862 | 2.172 |
Conclusion
The production of laccase by Stropharia sp. ITCC-8422 was analysed using ABTS and DMP as the substrate. The maximum affinity against ABTS and oxidation of DMP in the absence of copper confirmed the presence of laccase. The zymography using ABTS also confirmed the presence of laccase in the extracellular proteome of Stropharia sp. ITCC-8422. In addition, the presence of laccase in the extracellular proteome of Stropharia sp. ITCC-8422 was also confirmed by QTOF-LCMS analysis. The QTOF-LCMS of the extracellular proteome of Stropharia sp. ITCC-8422 identified 84 different proteins of which laccase exhibited maximum peptide sequence similarity with laccase from Trametes spp. The sequence enabled the prediction of the 3D structure and three domains of laccase were obtained. The experimental data suggested that laccase exhibited maximum affinity against ABTS, which is in correlation with the docking studies where the binding energy was lowest with ABTS. Thus, the present study enabled a better understanding of the mechanism in which laccase binds with various substrates thereby explaining its better affinity against ABTS in comparison to other substrates. Laccase finds its application in the removal of various pollutants, e.g. dyes. The in silico analysis helps to give an insight into the impact of laccase on various pollutants and the binding energy via docking will help in establishing whether the system of ligand and enzyme is effective or not. Thus, in the present work, in silico analysis confirmed spontaneous removal of alizarin cyanine green which further helped design the experimental work, and both in silico and experimental work were in correlation. Thus, the use of a similar concept can open a wide dimension of possibilities along with easier and faster analysis of various environmental and biotechnological applications exhibited by industrially important enzymes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors acknowledge the support of Dr. Jata Shankar, Genomics Laboratory, Department of Biotechnology and Bioinformatics, Jaypee University of Information Technology, Solan, India, for Bioinformatics studies. The authors are also thankful to Department of Biotechnology, Ministry of Science and Technology, Government of India (Grant No. BT/304/ NE/TBP/2012 and BT/PR7333/PBD/26/373/2012). The authors are also thankful to DST-FIST (Grant No. SR/FST/LSI-676/2016) for the infrastructure facility at the Department of Microbiology, CURAJ. The authors would also thank Agilent Technologies, Centre of Excellence Manesar, Gurugram, India, for QTOF-LC–MS. The authors would also like to acknowledge the financial support provided by CURAJ, Ajmer, India.
Author contributions
KA: methodology, formal analysis and investigation, writing—original draft preparation; JS: methodology, formal analysis and investigation, writing—review and editing and PV: conceptualization; methodology; writing—review and editing; funding acquisition; resources; supervision.
Funding
The authors are also thankful to Department of Biotechnology, Ministry of Science and Technology, Government of India (Grant No. BT/304/ NE/TBP/2012 and BT/PR7333/PBD/26/373/2012).
Compliance with ethical standards
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Agrawal K, Verma P. Biodegradation of synthetic dye Alizarin Cyanine Green by yellow laccase producing strain Stropharia sp. ITCC-8422. Biocatal Agric Biotechnol. 2019;21:101291. doi: 10.1016/j.bcab.2019.101291. [DOI] [Google Scholar]
- Agrawal K, Verma P. Column bioreactor of immobilized Stropharia sp. ITCC 8422 on natural biomass support of L. cylindrica for biodegradation of anthraquinone violet R. Biores Technol Rep. 2019;8:100345. doi: 10.1016/j.biteb.2019.100345. [DOI] [Google Scholar]
- Agrawal K, Verma P. Laccase: addressing the ambivalence associated with the calculation of enzyme activity. 3 Biotech. 2019;9:365. doi: 10.1007/s13205-019-1895-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal K, Verma P. Multicopper oxidase laccases with distinguished spectral properties: a new outlook. Heliyon. 2020;6:e03972. doi: 10.1016/j.heliyon.2020.e03972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal K, Chaturvedi V, Verma P. Fungal laccase discovered but yet undiscovered. Bioresour Bioprocess. 2018 doi: 10.1186/s40643-018-0190-z. [DOI] [Google Scholar]
- Agrawal K, Bhardwaj N, Kumar B, Chaturvedi V, Verma P. Process optimization, purification and characterization of alkaline stable white laccase from Myrothecium verrucaria ITCC-8447 and its application in delignification of agroresidues. Int J Biol Macromol. 2019;125:1042–1055. doi: 10.1016/j.ijbiomac.2018.12.108. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baldrian P. Fungal laccases–occurrence and properties. FEMS Microbiol Rev. 2005;30:215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- Bertrand T, Jolivalt C, Briozzo P, Caminade E, Joly N, Madzak C, Mougin C. Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics. Biochemistry. 2002;41:7325–7333. doi: 10.1021/bi0201318. [DOI] [PubMed] [Google Scholar]
- Bourbonnais R, Paice MG, Reid ID, Lanthier P, Yaguchi M. Lignin oxidation by laccase isozymes from Trametes versicolor and role of the mediator 2, 2'-azinobis (3-ethylbenzthiazoline-6-sulfonate) in kraft lignin depolymerization. Appl Environ Microbiol. 1995;61:1876–1880. doi: 10.1128/AEM.61.5.1876-1880.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannatelli MD, Ragauskas AJ. Two decades of laccases: advancing sustainability in the chemical industry. Chem Rec. 2017;17:122–140. doi: 10.1002/tcr.201600033. [DOI] [PubMed] [Google Scholar]
- Chen SC, Wu PH, Su YC, Wen TN, Wei YS, Wang NC, Hsu CA, Wang AHJ, Shyur LF. Biochemical characterization of a novel laccase from the basidiomycete fungus Cerrena sp. WR1. Protein Eng Des Sel. 2012;25:761–769. doi: 10.1093/protein/gzs082. [DOI] [PubMed] [Google Scholar]
- Daroch M, Houghton CA, Moore JK, Wilkinson MC, Carnell AJ, Bates AD, Iwanejko LA. Glycosylated yellow laccases of the basidiomycete Stropharia aeruginosa. Enzyme Microb Technol. 2014;58:1–7. doi: 10.1016/j.enzmictec.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Eggert C, Temp U, Eriksson KE. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl Environ Microbiol. 1996;62:1151–1158. doi: 10.1128/AEM.62.4.1151-1158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraroni M, Myasoedova NM, Schmatchenko V, Leontievsky AA, Golovleva LA, Scozzafava A, Briganti F. Crystal structure of a blue laccase from Lentinus tigrinus: evidences for intermediates in the molecular oxygen reductive splitting by multicopper oxidases. BMC Struct Biol. 2007;7:60. doi: 10.1186/1472-6807-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fithri L, Puspaningsih NNT, Asmarani O, Dewi GDF, Arizandy RY. Characterization of fungal laccase isolated from oil palm empty fruit bunches (OPEFB) and its degradation from the agriculture waste. Biocatal Agric Biotechnol. 2020 doi: 10.1016/j.bcab.2020.101676. [DOI] [Google Scholar]
- Garavaglia S, Cambria MT, Miglio M, Ragusa S, Iacobazzi V, Palmieri F, D'Ambrosio C, Scaloni A, Rizzi M. The structure of Rigidoporus lignosus laccase containing a full complement of copper ions, reveals an asymmetrical arrangement for the T3 copper pair. J Mol Biol. 2004;342:1519–1531. doi: 10.1016/j.jmb.2004.07.100. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Nagpal S, Bhat MA, Anupa G, Srivastava A, Sharma JB, Sengupta J. Gel-free proteomics reveals neoplastic potential in endometrium of infertile patients with stage IV ovarian endometriosis. J Reprod Health Med. 2015;1:83–95. doi: 10.1016/j.jrhm.2015.06.003. [DOI] [Google Scholar]
- Hadibarata T, Yusoff ARM, Aris A, Hidayat T, Kristanti RA. Decolorization of azo, triphenylmethane and anthraquinone dyes by laccase of a newly isolated Armillaria sp. F022. Water Air Soil Pollut. 2012;223:1045–1054. doi: 10.1007/s11270-011-0922-6. [DOI] [Google Scholar]
- Hakulinen N, Rouvinen J. Three-dimensional structures of laccases. Cell Mol Life Sci. 2015;72:857–868. doi: 10.1007/s00018-014-1827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkin JM, Larsen MJ, Obst JR. Use of syringaldazine for detection of laccase in sporophores of wood rotting fungi. Mycologia. 1974;66:469–476. doi: 10.1080/00275514.1974.12019628. [DOI] [PubMed] [Google Scholar]
- Hongyan L, Zexiong Z, Shiwei X, He X, Yinian Z, Haiyun L, Zhongsheng Y. Study on transformation and degradation of bisphenol A by Trametes versicolor laccase and simulation of molecular docking. Chemosphere. 2019;224:743–750. doi: 10.1016/j.chemosphere.2019.02.143. [DOI] [PubMed] [Google Scholar]
- Huang Q, Wang C, Zhu L, Zhang D, Pan C. Purification, characterization, and gene cloning of two laccase isoenzymes (Lac1 and Lac2) from Trametes hirsuta MX2 and their potential in dye decolorization. Mol Biol Rep. 2020;47:477–488. doi: 10.1007/s11033-019-05154-2. [DOI] [PubMed] [Google Scholar]
- Janusz G, Pawlik A, Świderska-Burek U, Polak J, Sulej J, Jarosz-Wilkołazka A, Paszczyński A. Laccase properties, physiological functions, and evolution. Int J Mol Sci. 2020;21:966. doi: 10.3390/ijms21030966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivalt C, Madzak C, Brault A, Caminade E, Malosse C, Mougin C. Expression of laccase IIIb from the white-rot fungus Trametes versicolor in the yeast Yarrowia lipolytica for environmental applications. Appl Microbiol Biotechnol. 2005;66:450–456. doi: 10.1007/s00253-004-1717-0. [DOI] [PubMed] [Google Scholar]
- Jung H, Xu F, Li K. Purification and characterization of laccase from wood-degrading fungus Trichophyton rubrum LKY-7. Enzyme Microb Technol. 2002;30:161–168. doi: 10.1016/S0141-0229(01)00485-9. [DOI] [Google Scholar]
- Junghanns C, Pecyna MJ, Böhm D, Jehmlich N, Martin C, Von Bergen M, Schauer F, Hofrichter M, Schlosser D. Biochemical and molecular genetic characterisation of a novel laccase produced by the aquatic ascomycete Phoma sp. UHH 5–1-03. Appl Microbiol Biotechnol. 2009;84:1095–1105. doi: 10.1007/s00253-009-2028-2. [DOI] [PubMed] [Google Scholar]
- Kameshwar AKS, Barber R, Qin W. Comparative modeling and molecular docking analysis of white, brown and soft rot fungal laccases using lignin model compounds for understanding the structural and functional properties of laccases. J Mol Graph Model. 2018;79:15–26. doi: 10.1016/j.jmgm.2017.10.019. [DOI] [PubMed] [Google Scholar]
- Kinter M, Sherman NE. Protein sequencing and identification using tandem mass spectrometry. 9. New Jersey: John Wiley & Sons; 2005. [Google Scholar]
- Kislinger T, Emili A. Going global: protein expression profiling using shotgun mass spectrometry. Curr Opin Mol Ther. 2003;5:285–293. [PubMed] [Google Scholar]
- Klonowska A, Le Petit J, Tron T. Enhancement of minor laccases production in the basidiomycete Marasmius quercophilus C30. FEMS Microbiol Lett. 2001;200:25–30. doi: 10.1111/j.1574-6968.2001.tb10687.x. [DOI] [PubMed] [Google Scholar]
- Ko EM, Leem YE, Choi H. Purification and characterization of laccase isozymes from the white-rot basidiomycete Ganoderma lucidum. Appl Microbiol Biotechnol. 2001;57:98–102. doi: 10.1007/s002530100727. [DOI] [PubMed] [Google Scholar]
- Kolomytseva M, Myasoedova N, Samoilova A, Podieiablonskaia E, Chernykh A, Classen T, Pietruszka J, Golovleva L. Rapid identification of fungal laccases/oxidases with different pH-optimum. Process Biochem. 2017;62:174–183. doi: 10.1016/j.procbio.2017.07.027. [DOI] [Google Scholar]
- Komori H, Higuchi Y. Structural insights into the O2 reduction mechanism of multicopper oxidase. The J Biochem. 2015;158:293–298. doi: 10.1093/jb/mvv079. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4, Nature 227: 680–685. https://www.nature.com/articles/227680a0 [DOI] [PubMed]
- Li L, Dai W, Yu P, Zhao J, Qu Y. Decolorisation of synthetic dyes by crude laccase from Rigidoporus lignosus W1. J Chem Technol Biot. 2009;84:399–404. doi: 10.1002/jctb.2053. [DOI] [Google Scholar]
- Lin SX, Lapointe J (2013) Theoretical and experimental biology in one—A symposium in honour of Professor Kuo-Chen Chou’s 50th anniversary and Professor Richard Giegé’s 40th anniversary of their scientific careers. doi:10.4236/jbise.2013.64054
- Lorenzo M, Moldes D, Couto SR, Sanromán MAA. Inhibition of laccase activity from Trametes versicolor by heavy metals and organic compounds. Chemosphere. 2005;60:1124–1128. doi: 10.1016/j.chemosphere.2004.12.051. [DOI] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB, III, et al. Structure validation by Calpha geometry: phi, psi and C beta deviation. Proteins Struct Funct Genet. 2002;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Madhavi V, Lele SS. Laccase: properties and applications. BioResources. 2009;4:1694–1717. [Google Scholar]
- Min KL, Kim YH, Kim YW, Jung HS, Hah YC. Characterization of a novel laccase produced by the wood-rotting fungus Phellinus ribis. Arch Biochem Biophys. 2001;392:279–286. doi: 10.1006/abbi.2001.2459. [DOI] [PubMed] [Google Scholar]
- More SS, Renuka PS, Malini S. Isolation, purification, and characterization of fungal laccase from Pleurotus sp. Enzyme Res. 2011 doi: 10.4061/2011/248735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mtibaà R, Barriuso J, de Eugenio L, Aranda E, Belbahri L, Nasri M, Martínez MJ, Mechichi T. Purification and characterization of a fungal laccase from the ascomycete Thielavia sp. and its role in the decolorization of a recalcitrant dye. Int J Biol Macromol. 2018;120:1744–1751. doi: 10.1016/j.ijbiomac.2018.09.175. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay BP. Recognition dynamics of trinuclear copper cluster and associated histidine residues through conserved or semi-conserved water molecules in human ceruloplasmin: the involvement of aspartic and glutamic acid gates. J Biomol Struct Dyn. 2018;36:3829–3842. doi: 10.1080/07391102.2017.1401003. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M, Banerjee R. Purification and biochemical characterization of a newly produced yellow laccase from Lentinus squarrosulus MR13. 3 Biotech. 2015;5:227–236. doi: 10.1007/s13205-014-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ME, Turley S, Adman ET. Structure of nitrite bound to copper-containing nitrite reductase from Alcaligenes faecalis mechanistic implications. J Biol Chem. 1997;272:28455–28460. doi: 10.1074/jbc.272.45.28455. [DOI] [PubMed] [Google Scholar]
- Murugesan K, Nam IH, Kim YM, Chang YS. Decolorization of reactive dyes by a thermostable laccase produced by Ganoderma lucidum in solid state culture. Enzyme Microb Technol. 2007;40:1662–1672. doi: 10.1016/j.enzmictec.2006.08.028. [DOI] [Google Scholar]
- Nakatani M, Hibi M, Minoda M, Ogawa J, Yokozeki K, Shimizu S. Two laccase isoenzymes and a peroxidase of a commercial laccase-producing basidiomycete, Trametes sp. Ha1. New Biotechnol. 2010;27:317–323. doi: 10.1016/j.nbt.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Ng TB, Wang HX. A homodimeric laccase with unique characteristics from the yellow mushroom Cantharellus cibarius. Biochem Bioph Res Co. 2004;313:37–41. doi: 10.1016/j.bbrc.2003.11.087. [DOI] [PubMed] [Google Scholar]
- Nyanhongo GS, Couto SR, Guebitz GM. Coupling of 2, 4, 6-trinitrotoluene (TNT) metabolites onto humic monomers by a new laccase from Trametes modesta. Chemosphere. 2006;64:359–370. doi: 10.1016/j.chemosphere.2005.12.034. [DOI] [PubMed] [Google Scholar]
- Palmieri G, Bianco C, Cennamo G, Giardina P, Marino G, Monti M, Sannia G. Purification, characterization, and functional role of a novel extracellular protease from Pleurotus ostreatus. Appl Environ Microbiol. 2001;67:2754–2759. doi: 10.1128/AEM.67.6.2754-2759.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palonen H, Saloheimo M, Viikari L, Kruus K. Purification, characterization and sequence analysis of a laccase from the ascomycete Mauginiella sp. Enzyme Microb Technol. 2003;33:854–862. doi: 10.1016/S0141-0229(03)00247-3. [DOI] [Google Scholar]
- Park KM, Park SS. Purification and characterization of laccase from basidiomycete Fomitella fraxinea. J Microbiol Biotechnol. 2008;18:670–675. [PubMed] [Google Scholar]
- Piontek K, Antorini M, Choinowski T. Crystal structure of a laccase from the fungus Trametes versicolor at 1.90-Å resolution containing a full complement of coppers. J Biol Chem. 2002;277:37663–37669. doi: 10.1074/jbc.M204571200. [DOI] [PubMed] [Google Scholar]
- Polyakov KM, Fedorova TV, Stepanova EV, Cherkashin EA, Kurzeev SA, Strokopytov BV, Lamzin VS, Koroleva OV. Structure of native laccase from Trametes hirsuta at 1.8 Å resolution. Acta Crystallogr D Biol Crystallogr. 2009;65:611–617. doi: 10.1107/S0907444909011950. [DOI] [PubMed] [Google Scholar]
- Pundir RK, Mishra VK, Rana S, Lakhani M. Screening of laccase producing fungi from soil samples—an in vitro study. Electron J Biol. 2016;12:254–257. [Google Scholar]
- Quintanar L, Stoj C, Taylor AB, Hart PJ, Kosman DJ, Solomon EI. Shall we dance? How a multicopper oxidase chooses its electron transfer partner. Acc Chem Res. 2007;40:445–452. doi: 10.1021/ar600051a. [DOI] [PubMed] [Google Scholar]
- Rajagopalu D, Show PL, Tan YS, Muniandy S, Sabaratnam V, Ling TC. Recovery of laccase from processed Hericium erinaceus (Bull.: Fr) Pers. fruiting bodies in aqueous two-phase system. J Biosci Bioeng. 2016;122:301–306. doi: 10.1016/j.jbiosc.2016.01.016. [DOI] [PubMed] [Google Scholar]
- Ramírez-Cavazos LI, Junghanns C, Ornelas-Soto N, Cárdenas-Chávez DL, Hernández-Luna C, Demarche P, Enaud E, García-Morales R, Agathos SN, Parra R. Purification and characterization of two thermostable laccases from Pycnoporus sanguineus and potential role in degradation of endocrine disrupting chemicals. J Mol Catal B-Enzym. 2014;108:32–42. doi: 10.1016/j.molcatb.2014.06.006. [DOI] [Google Scholar]
- Ryan S, Schnitzhofer W, Tzanov T, Cavaco-Paulo A, Gübitz GM. An acid-stable laccase from Sclerotium rolfsii with potential for wool dye decolourization. Enzyme Microb Technol. 2003;33:766–774. doi: 10.1016/S0141-0229(03)00162-5. [DOI] [Google Scholar]
- Sadhasivam S, Savitha S, Swaminathan K, Lin FH. Production, purification and characterization of mid-redox potential laccase from a newly isolated Trichoderma harzianum WL1. Process Biochem. 2008;43:736–742. doi: 10.1016/j.procbio.2008.02.017. [DOI] [Google Scholar]
- Shin KS, Lee YJ. Purification and characterization of a new member of the laccase family from the white-rot basidiomycete Coriolus hirsutus. Arch Biochem Biophys. 2000;384:109–115. doi: 10.1006/abbi.2000.2083. [DOI] [PubMed] [Google Scholar]
- Solano F, Garcia E, Perez D, Sanchez-Amat A. Isolation and characterization of strain MMB-1 (CECT 4803), a novel melanogenic marine bacterium. Appl Environ Microbiol. 1997;63:3499–3506. doi: 10.1128/AEM.63.9.3499-3506.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano F, Lucas-Elío P, López-Serrano D, Fernández E, Sanchez-Amat A. Dimethoxyphenol oxidase activity of different microbial blue multicopper proteins. FEMS Microbiol Lett. 2001;204:175–181. doi: 10.1111/j.1574-6968.2001.tb10882.x. [DOI] [PubMed] [Google Scholar]
- Solomon EI, Chen P, Metz M, Lee SK, Palmer AE. Oxygen binding, activation, and reduction to water by copper proteins. Angew Chem Int Ed. 2001;40:4570–4590. doi: 10.1002/1521-3773(20011217)40:24<4570:AID-ANIE4570>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Srinivasan C, Dsouza TM, Boominathan K, Reddy CA. Demonstration of Laccase in the White rot Basidiomycete Phanerochaete chrysosporium BKM-F1767. Appl Environ Microbiol. 1995;61:4274–4277. doi: 10.1128/AEM.61.12.4274-4277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staszczak M. Proteasomal degradation pathways in Trametes versicolor and Phlebia radiata. Enzym Microb Technol. 2002;30:537–541. doi: 10.1016/S0141-0229(02)00010-8. [DOI] [Google Scholar]
- Staszczak M, Jarosz-Wilkołazka A. Inhibition of the proteasome strongly affects cadmium stimulated laccase activity in Trametes versicolor. Biochimie. 2005;87:755–762. doi: 10.1016/j.biochi.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Thakur R, Shankar J. In silico analysis revealed high-risk single nucleotide polymorphisms in human pentraxin-3 gene and their impact on innate immune response against microbial pathogens. Front Microbiol. 2016;7:192. doi: 10.3389/fmicb.2016.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tišma M, Šalić A, Planinić M, Zelić B, Potočnik M, Šelo G, Bucić-Kojić A. Production, characterisation and immobilization of laccase for an efficient aniline-based dye decolourization. J Water Process Eng. 2020;36:101327. doi: 10.1016/j.jwpe.2020.101327. [DOI] [Google Scholar]
- Tiwari S, Thakur R, Goel G, Shankar J. Nano-LC-Q-TOF analysis of proteome revealed germination of Aspergillus flavus conidia is accompanied by MAPK signalling and cell wall modulation. Mycopathologia. 2016;181:769–786. doi: 10.1007/s11046-016-0056-x. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Shishodia SK, Shankar J. Docking analysis of hexanoic acid and quercetin with seven domains of polyketide synthase A provided insight into quercetin-mediated aflatoxin biosynthesis inhibition in Aspergillus flavus. 3 Biotech. 2019;9:149. doi: 10.1007/s13205-019-1675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlecuitl-Beristain S, Sánchez C, Loera O, Robson GD, Díaz-Godínez G. Laccases of Pleurotus ostreatus observed at different phases of its growth in submerged fermentation: production of a novel laccase isoform. Mycol Res. 2008;112:1080–1084. doi: 10.1016/j.mycres.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Vite-Vallejo O, Palomares LA, Dantán-González E, Ayala-Castro HG, Martínez-Anaya C, Valderrama B, Folch-Mallol J. The role of N-glycosylation on the enzymatic activity of a Pycnoporus sanguineus laccase. Enzyme Microb Technol. 2009;45:233–239. doi: 10.1016/j.enzmictec.2009.05.007. [DOI] [Google Scholar]
- Wang SN, Chen QJ, Zhu MJ, Xue FY, Li WC, Zhao TJ, Li GD, Zhang GQ. An extracellular yellow laccase from white rot fungus Trametes sp. F1635 and its mediator systems for dye decolorization. Biochimie. 2018;148:46–54. doi: 10.1016/j.biochi.2018.02.015. [DOI] [PubMed] [Google Scholar]
- Wu J, Kim KS, Lee JH, Lee YC. Cloning, expression in Escherichia coli, and enzymatic properties of laccase from Aeromonas hydrophila WL-11. J Environ Sci. 2010;22:635–640. doi: 10.1007/s00253-004-1717-0. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Tu X, Wang J, Zhang M, Cheng Q, Zeng W, Shi Y. Purification, molecular characterization and reactivity with aromatic compounds of a laccase from basidiomycete Trametes sp. strain AH28-2. Appl Microbiol Biot. 2003;60:700–707. doi: 10.1007/s00253-002-1169-3. [DOI] [PubMed] [Google Scholar]
- Xiao X, Wang P, Chou KC. GPCR-CA: A cellular automaton image approach for predicting G-protein–coupled receptor functional classes. J Comput Chem. 2009;30:1414–1423. doi: 10.1002/jcc.21163. [DOI] [PubMed] [Google Scholar]
- Xu F, Berka RM, Wahleithner JA, Nelson BA, Shuster JR, Brown SH, Palmer AE, Solomon EI. Site-directed mutations in fungal laccase: effect on redox potential, activity and pH profile. Biochem J. 1998;334:63–70. doi: 10.1042/bj3340063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XQ, Zhao XX, Liu CY, Zheng Y, Qian SJ. Decolorization of azo, triphenylmethane and anthraquinone dyes by a newly isolated Trametes sp. SQ01 and its laccase. Process Biochem. 2009;44:1185–1189. doi: 10.1016/j.procbio.2009.06.015. [DOI] [Google Scholar]
- Yuan X, Tian G, Zhao Y, Zhao L, Wang H, Ng TB. Biochemical characteristics of three laccase isoforms from the Basidiomycete Pleurotus nebrodensis. Molecules. 2016;21:203. doi: 10.3390/molecules21020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva I, Zaitsev V, Card G, Moshkov K, Bax B, Ralph A, Lindley P. The X-ray structure of human serum ceruloplasmin at 3.1 Å: nature of the copper centres. J Biol Inorg Chem. 1996;1:15–23. doi: 10.1007/s007750050018. [DOI] [Google Scholar]
- Zhao D, Zhang X, Cui D, Zhao M. Characterisation of a novel white laccase from the deuteromycete fungus Myrothecium verrucaria NF-05 and its decolourisation of dyes. PLoS ONE. 2012 doi: 10.1371/journal.pone.0038817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Fu C, Fu S, Zhan H (2014) Purification and characterization of white laccase from the white-rot fungus Panus conchatus. BioResources 9: 1964–1976. https://ojs.cnr.ncsu.edu/index.php/BioRes/article/view/3874.
- Zhou X, Yu S, Su J, Sun L. Computational study on new natural compound inhibitors of pyruvate dehydrogenase kinases. Int J Mol Sci. 2016;17:340. doi: 10.3390/ijms17030340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.