Graphical abstract
Keywords: SARS-CoV-2, COVID-19, Oseltamivir
Highlight
-
•
The active center of 3CLpro of SARS-CoV-2 was similar to that of neuraminidase of influenza A.
-
•
Oseltamivir is ineffective against SARS-CoV-2 in vitro study.
-
•
The clinical use of oseltamivir did not improve the patients’ symptoms and signs.
Abstract
Background
Oseltamivir is a first-line antiviral drug, especially in primary hospitals. During the ongoing outbreak of coronavirus disease 2019 (COVID-19), most patients with COVID-19 who are symptomatic have used oseltamivir. Considering its popular and important role as an antiviral drug, it is necessary to evaluate oseltamivir in the treatment of COVID-19.
Objective
To evaluate the effect of oseltamivir against COVID-19.
Methods
Swiss-model was used to construct the structure of the N-terminal RNA-binding domain (NRBD) of the nucleoprotein (NC), papain-like protease (PLpro), and RNA-directed RNA polymerase (RdRp) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). TM-align program was performed to compare the structure of the viral proteins with the structure of the neuraminidase of influenza A. Molecular docking was used to analyze the theoretical possibility of effective binding of oseltamivir with the active centers of the viral proteins. In vitro study was used to evaluate the antiviral efficiency of oseltamivir against SARS-CoV-2. By clinical case analysis, we statistically evaluated whether the history of oseltamivir use influenced the progression of the disease.
Results
The structures of NRBD, PLpro, and RdRp were built successfully. The results from TM-align suggested that the S protein, NRBD, 3C-like protease (3CLpro), PLPrO, and RdRp were structurally similar to the influenza A neuraminidase, with TM-scores of 0.30077, 0.19254, 0.28766, 0.30666, and 0.34047, respectively. Interestingly, the active center of 3CL pro was found to be similar to the active center from the neuraminidase of influenza A. Through an analysis of molecular docking, we discovered that oseltamivir carboxylic acid was more favorable to bind to the active site of 3CLpro effectively, but its inhibitory effect was not strong compared with the positive group. Finally, we used in vitro study and retrospective case analysis to verify our speculations. We found that oseltamivir is ineffective against SARS-CoV-2 in vitro study and the clinical use of oseltamivir did not improve the patients’ symptoms and signs and did not slow the disease progression.
Conclusions
We consider that oseltamivir isn’t suitable for the treatment of COVID-19. During the outbreak of novel coronavirus, when oseltamivir is not effective for the patients after they take it, health workers should be highly vigilant about the possibility of COVID-19.
1. Introduction
As a neuraminidase inhibitor, oseltamivir was approved by the Food and Drug Admission (FDA) in 1999 [1]. Since then, it has played an essential role in treating against influenza A and influenza B and is becoming more widespread. The atypical pneumonia caused by severe acute respiratory syndrome coronavirus (SARS-CoV) that broke out in Guangzhou, China, in 2003, linked oseltamivir to coronavirus. Zhang et al. found that the active site of the Spike (S) 1 Protein of SARS is similar to that of neuraminidase, suggesting that neuraminidase inhibitors may be useful to treat SARS-CoV [2]. However, despite this similarity, no clinical data suggest that oseltamivir is effective in treating SARS-CoV. With the epidemic of coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2, oseltamivir has once again become a hot topic. A study from Thailand reported that a 71-year-old patient with severe COVID-19 on January 29, 2020 underwent a 48h treatment with lopinavir/ritonavir combined with oseltamivir, which improved the patient’s condition, and the throat swab test became negative [3]. Although a single case report does not prove the effectiveness of oseltamivir for COVID-19, it once again linked oseltamivir to the treatment of a coronavirus-induced disease. As frontline healthcare workers fighting against COVID-19, we were interested in the treatment regimen of this case and found most patients with COVID-19 who were symptomatic have used oseltamivir. We believed that it is of practical significance to further study the effect of oseltamivir on COVID-19. Because of its significant effectiveness on influenza and being sold over the counter, oseltamivir is not only stocked in common households but also a common antiviral drug in primary hospitals. Therefore, if oseltamivir is effective for COVID-19, the treatment could be family-oriented. If, instead, the treatment effectiveness is not significant, the use of oseltamivir should be stopped, which will avoid delaying other treatments and multiple oseltamivir adverse reactions, such as nausea, vomiting, epilepsy, elevated liver enzymes, and arrhythmias [4], [5]. Therefore, the study of the antiviral effect of oseltamivir against SARS-CoV-2 could have a positive effect on the treatment of COVID-19.
SARS-CoV-2 belongs to the β-genus of the coronavirus family and is the seventh coronavirus found to infect humans. Therefore, it has the structural features of coronaviruses, such as structural proteins, including S protein and Nucleoprotein (NC), and also non-structural proteins, including the main key enzymes 3C-like protease (3CLpro), papain-like protease (PLpro), and RNA-directed RNA polymerase (Pol/RdRp). These main structural proteins and key enzymes of SARS-CoV-2 are important targets for inhibiting SARS-CoV-2 infection [6], [7], [8], [9], [10]. Since the SARS outbreak in 2003, the S protein, NC, 3CLpro, PLpro, and Pol/RdRp of the coronavirus known to infect humans have been extensively studied due to their potential as therapeutic targets [11], [12], [13], [14], [15], [16].
Oseltamivir is a competitive inhibitor of neuraminidase designed by computer-assisted technology. In this context, the effective binding of oseltamivir to the active site of key proteins of SARS-CoV-2 is particularly important. Therefore, here we: 1. analyzed whether S protein, NC protein, 3CLpro, PLpro, and Pol/RdR have active centers similar to neuraminidase; 2. explored whether oseltamivir can form effective docking with key protein active centers of the virus using molecular simulation; 3. accessed the antiviral effect of oseltamivir against SARS-CoV-2 in vitro; 4. used case review to analyze whether oseltamivir is a factor to improve the conditions of patients with COVID-19.
2. Methods
2.1. Obtaining the viral protein structure
The structures of the S protein (PDB ID: 6VSB) and 3CL pro (PDB ID: 6LU7) were downloaded from the RCSB Protein Data Bank (http://www.rcsb.org). The structures of NC, PLpro, and Pol/RdRp were built by homology modeling. We first obtained the protein sequences of NC (YP_009724397.2), nsp3 (YP_009725299.1), and Pol/RdRp (YP_009725307.1) from the GenePept protein sequence database, and suitable templates were found for these protein sequences through the Swiss-model (www.swissmodel.org) for homology modeling. Pymol was used to generate model images [17].
2.2. Evaluation of virus models
We used the SAVES v5.0 program (servicesn.mbi.ucla.edu/SAVES/) to carry out a model evaluation, and the evaluation criteria for a successful model were as follows: a. in ERRAT score result, the Overall Quality Factor was >85; b. in the Verify3D assessment result, >80% of the residues have average 3D-1D score ≥ 0.2; c. in the Procheck assessment result, ≥90% residues were in the most favored region [18], [19], [20], [21].
2.3. Alignment of the structure of a viral protein and the structure of influenza a neuraminidase
TM-align program (https://zhanglab.ccmb.med.umich.edu/TM-align/) was used to perform the structural alignment of PLpro, N-terminal RNA-binding Domain (NRBD) of NC, Pol/RdRp, 3CL pro, and S protein with influenza A neuraminidase (PDB ID: 3Ti6); 0.0 < TM-score < 0.30 was defined as random structural similarity, and 0.5 < TM-score < 1.00 was defined as in about the same fold [22]. The structural alignment between influenza B neuraminidase (PDB ID: 4CPY) and 3Ti6 was used as a positive control.
2.4. Molecular docking of oseltamivir with viral protein
We used ChemBioDraw Ultra 17.0 software to draw the structure of the active metabolite oseltamivir carboxylic acid and N3 inhibitor, then converted them into three-dimensional (3D) structure using ChemBio3D Ultra 17.0 software. Then the structures of agents were preprocessing by MMFF94 Force Field. AutoDock Tools 1.5.6 software was used to remove water molecules and ligands in the proteins, and then add hydrogen atoms, combine non-polar hydrogen atoms, add charges. And converted proteins and agents into the format of PDBQT. The GridBox module was used to generate a 3D ensemble around the active site, and the grid points in the ensemble were calculated for energy score evaluation. AutoDock Vina was used for molecular docking [23]. Exhaustiveness was set at 20 and other parameters were used default value. Finally, we selected the conformation with the highest docking score to analyze the result using Free Maestro 11.9.
2.5. In vitro study for accessing the antiviral effect of oseltamivir
The antiviral efficiency of oseltamivir against SARS-CoV-2 in vitro was evaluated using the method described in the previous work [24]. Briefly, Vero E6 cells (ATCC, no. 1586) were cultured in MEM medium with 10% fetal bovine serum (Gibco Invitrogen) at 37 °C in a 5% CO2 humidified incubator. And cells were precultured at 1 × 105/48-well plate overnight and pretreated with different doses of the oseltamivir carboxylic acid (CAS No. 187227-45-8/HY-13318) for 1h before infecting with the SARS-CoV-2 (nCoV-2019BetaCoV/Wuhan/WIV04/2019) at a multiplicity of infection (MOI) of 0.05 for 2 h. After that, cell culture supernatants were replaced with fresh complete medium containing different concentrations of oseltamivir. Virus-containing supernatants were collected at 24h after transfection for extracting RNA by the MiniBEST Viral RNA/DNA Extraction Kit (Takara, CAT No. 9766) according to the manufacturer’s instructions. Then cDNA synthesis was performed with a PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Cat no. RR047A). The qRT-PCR assay was conducted to evaluate the quantification of viral copy numbers in the cell supernatant with TB Green Premix Ex Taq II (Takara, CAT No. RR820A). The primers were targeted nCoV-2019 S gene, forward primer CAATGGTTTAACAGGCACAG; reverse primer CTCAAGTGTCTGTGGATCACG [25].
2.6. Retrospective analysis of clinical cases
2.6.1. Study design
We retrospectively analyzed 627 patients diagnosed with SARS-CoV-2 infection at the Second Hospital of Wuhan, Wuhan Union Hospital, and Wuhan Union Hospital West Campus, Hubei Province from January 19, 2020, to February 20, 2020, which was approved by the institutional ethics board of Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China. The enrollment criteria were: 1. patients were diagnosed with SARS-CoV-2 infection and negative for influenza A and B nucleic acid tests; 2. there were no other underlying diseases, such as hypertension, diabetes, cardiovascular and cerebrovascular diseases, malignant diseases, no respiratory tract diseases, such as COPD, asthma, and pulmonary heart disease, and no hypoproteinemia and hypokalemia. We defined the period from the start of antiviral therapy to clinical evaluation as a patients’ treatment period. We defined the patients with relieved symptoms and stable or absorbed lesions in CT during the treatment period as the remission group, and the patients without symptom improvement or no significant changes or progression of the lesions in CT as the non-remission group. They all were evaluated by front-line medical workers of union hospital based on Guidelines of the Diagnosis and Treatment of New Coronavirus Pneumonia (version 7) of National Health Commission of China. Whether the history of oseltamivir use influenced the progression of the disease was statistically evaluated.
2.6.2. Data collection
Patients’ information such as sex and age and their clinical data such as chest CT, the history of oseltamivir use, the usage and dosage of oseltamivir were collected from electronic medical records, including nursing records, clinical records and progress notes. Besides, pre- and post- antiviral treatment symptoms were collected and evaluated by front-line medical workers.
2.6.3. Statistical analysis
The difference of age between the remission group and the non-remission group was analyzed by independent samples t-test, while the difference of hospitalization days between the oseltamivir group and the Non-use oseltamivir group was analyzed by Mann Whitney tests. Pearson's chi-squared test was used to compare the difference of history of oseltamivir use and sex between the remission group and the non-remission group. Fisher's exact test was used to compare the difference of response rate at different course of using oseltamivir. P-value less than 0.05, 0.01 and 0.001 were considered significant.
3. Result
3.1. The obtainment of the viral protein structure
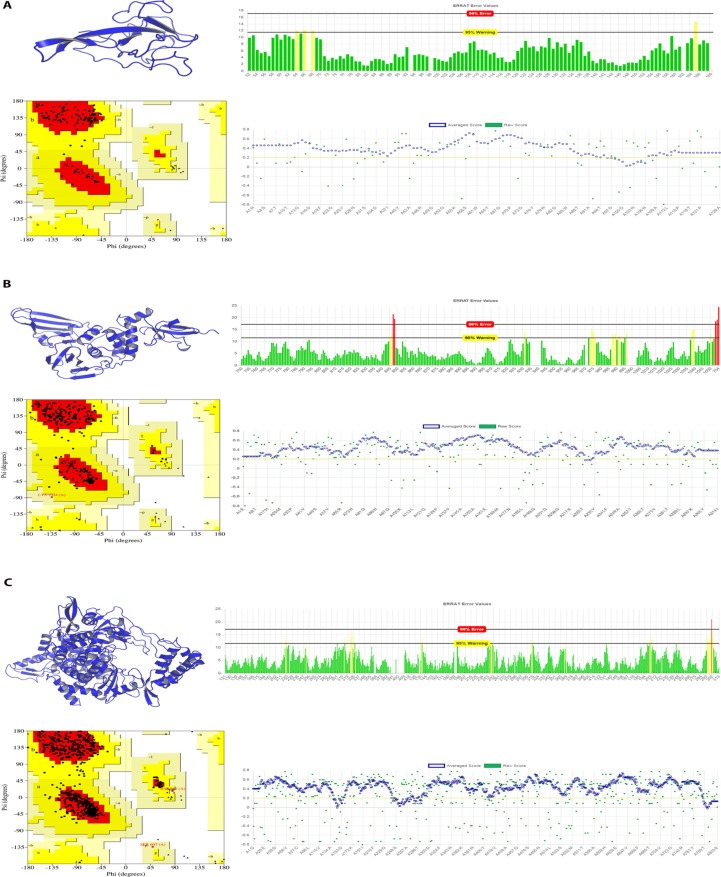
The structures of the S protein (PDB ID: 6VSB) and 3CL pro (PDB ID: 6LU7) of SARS-CoV-2 were obtained from the RCSB Protein Data Bank (http://www.rcsb.org). Then the structures of NC, PLpro, and Pol/RdRp were built through the method of homology modeling. The models were constructed successfully and shown in Fig. 1 A–C. Among them, the NC of SARS-CoV-2 contains 419 amino acid residues. The search of a homology template using the Swissmodel found that the 35–174 residues of NC have an identity of 92.37% with the NRBD of NC from SARS-CoV(PDB ID: 1ssk), which is the active region for the binding of NC of SARS-CoV with RNA. Inhibiting NRBD activity can affect coronavirus RNA transcription and replication, and virion assembly [26]. Thus, it is one of the targets for inhibiting coronaviruses. Therefore, NRBD is the main activity center of NC, and this model can be used for the active site of NC from SARS-CoV-2 for further research.
Fig. 1.
The evaluation of the model of NRBD, PLpro and Pol/RdRp from SARS-CoV-2 constructed through homology modeling. (A) The model of NRBD from SARS-CoV-2 with the evaluation of EBRAT, Verify3D, and Ramachandran plot; (B) The model of PLpro from SARS-CoV-2 with the evaluation of EBRAT, Verify3D, and Ramachandran plot; (C) The model of Pol/RdRp of SARS-CoV-2 with the evaluation of EBRAT, Verify3D, and Ramachandran plot. Abbreviations: NRBD: N-terminal RNA-binding domain; Pol/RdRp: RNA-directed RNA polymerase; PLpro: papain-like protease;
3.2. Evaluation of virus models
Subsequently, we used the SAVES v5.0 program (servicesn.mbi.ucla.edu/SAVES/) to carry out a model evaluation. The results are shown in Table 1 , suggesting all three models passed the evaluation. Among them, the ERRAT score was based on whether the error value of the alignment of the non-bonding interaction among the atoms in the structure and the high-resolution crystal structure was within a reasonable confidence interval. If the percentage of the reasonable confidence interval is higher, the quality is higher. Almost all residues of NRBD, Pol/RdRp, and PLpro were within reasonable confidence intervals, and the final scores were 96.4602, 95.9157, and 92.7586, respectively (Fig. 1A–C). Verify3D was used to analyze the compatibility of the 3D structure and the primary sequence in the model. A 3D-1D score ≥ 0.2 of a residue is reasonable. The higher the ratio of reasonable residues, the higher the quality of the model. The majority of residues of NRBD, Pol/RdRp, and PLpro belonged to reasonable residues (green), and there were 91.27%, 89.91%, and 98.73% reasonable residues in NRBD, Pol/RdRp, and PLpro respectively (Fig. 1A–C). The results of Procheck were mainly shown by Ramachandran plots. As can be seen in the Ramachandran plot, there were A, B, L, a, b, l, p, ~a, ~b, ~p, and ~l labeled areas. In Porcheck, A, B, and L are classified as reasonable areas; a, b, l are relatively reasonable regions, whereas ~a, ~b, ~p, and ~l are acceptable regions. The distributions of amino acid residues in the structure were observed. The large proportion of amino acids in reasonable regions proves that the model is of high quality. It can be seen that most of the residues of NRBD, Pol/RdRp, and PLpro were in A, B, and L, and the reasonable region residues accounted for 92.9%, 92.5%, and 90.0%, respectively (Fig. 1A–C). The above results suggested that the three modeled structures passed the evaluation, were of good quality, and could be used for further research.
Table 1.
The model of NRBD, PLpro and Pol/RdRp evaluated through SAVES program.
| Protein Residues have average 3D-1D score>=0.2 | Overall Quality Factor | Residues in most favored region | Final | |
|---|---|---|---|---|
| NRBD | 91.27% | 96.4602 | 92.90% | PASS |
| Pol/RdRp | 89.91% | 95.9157 | 92.50% | PASS |
| PLpro | 98.73% | 92.7586 | 90.00% | PASS |
Abbreviations: NRBD: N-terminal RNA-binding domain; Pol/RdRp: RNA-directed RNA polymerase; PLpro: papain-like protease.
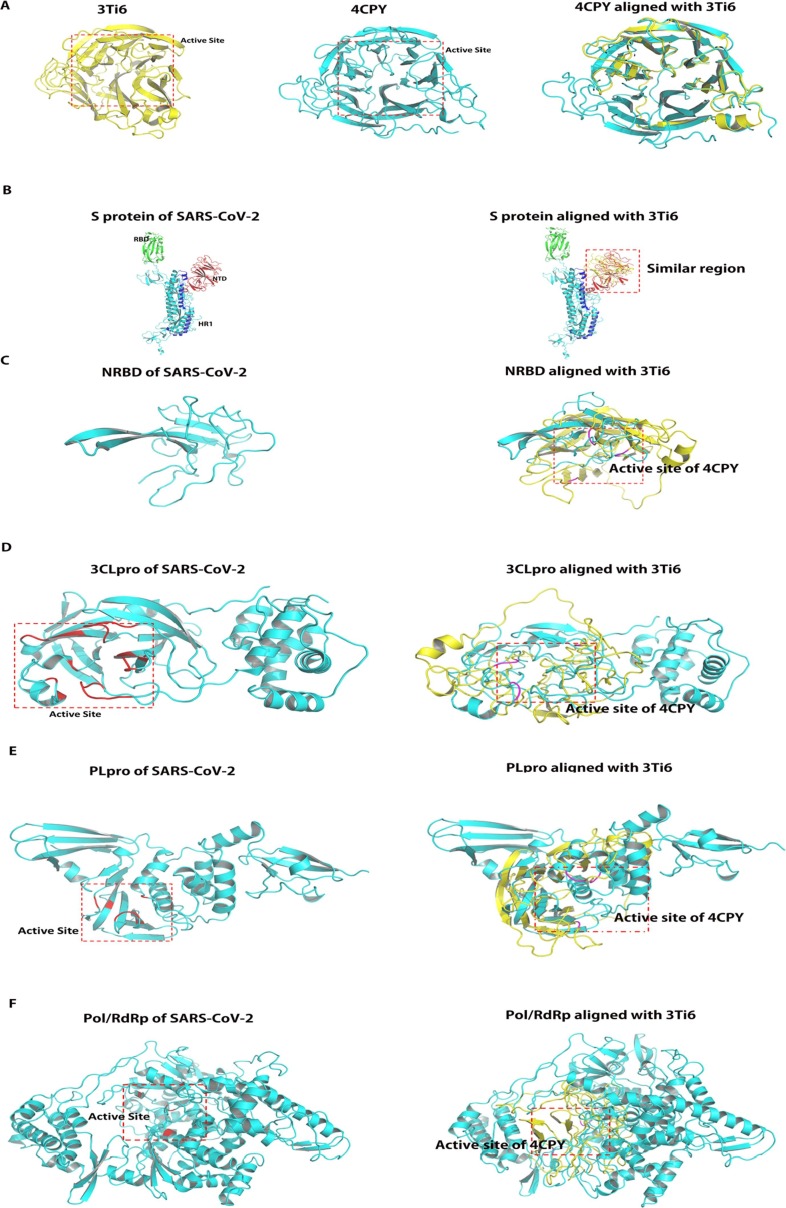
3.3. Alignment of the structure of viral proteins and influenza A neuraminidase
To explore whether oseltamivir is effective for SARS-CoV-2, we first analyzed whether there was structural similarity between the structure of viral protein and influenza A neuraminidase (PDB ID: 3Ti6), especially whether there was a similarity in the active center. TM-align program (https://zhanglab.ccmb.med.umich.edu/TM-align/) was used for structural alignment [27]. The alignment of the structure of influenza B neuraminidase (PDB ID: 4CPY) with 3Ti6 was selected as a positive control. TM-score was used to analyze the structural similarity. The active site of influenza A and B neuraminidase were determined based on the location of ligand in crystal structure (Fig. 2 A) [28], [29]. By comparing 4CPY with 3Ti6, the TM-score of the two was found to be 0.94460, which proved that the two were structurally homologous. Subsequently, we aligned the structures of S protein, NRBD, 3CL pro, PLpro, and Pol/RdRp with 3Ti6; the TM-scores were 0.30077, 0.19254, 0.28766, 0.30666, and 0.34047, respectively (Table 2 ). The results suggested that there was only a structural similarity between the protein structure of the above SARS-CoV-2 and 3Ti6. The active center of the S protein was mainly the RBD domain (Fig. 2B green region) and S2 subunit heptad-repeat 1 domain (HR1) (Fig. 2B blue region), which were also two important targets for studying the inhibition of S protein [30], [31]. We found that the similar structure of 3Ti6 and S protein is located in the N-terminal domain (NTD) region of S protein. As the active center of NC, we found that NRBD did not match the active center of 3Ti6 (Fig. 2C). The active center of 3CLpro was determined based on that it is homologous to the 3CLpro from SARS-CoV. Previous studies have confirmed the active center of 3CLpro of SARS-CoV. Thus, this region was used as the active center of 3CLpro of SARS-CoV-2 [13]. By alignment, we found that the active centers of the two proteins were similar (Fig. 2D). Subsequently, we also determined the active center of the PLpro and Pol/RdRp based on the homology of the proteins of SARS-CoV-2 and those of SARS-CoV. And we found that the active center of PLpro did not match the active center from 3Ti6 through the alignment of structure (Fig. 2E) [32]. In studies focused on the Pol/RdRp of SARS-CoV, it has found that Thr680, Asn691, and Asp623 can bind nucleotides and promote viral gene transcription, and Val557 also plays an important role in the replication of the virus [29]. Thus, the region where the above residues are located was determined as the active center of SARS-CoV-2 (Fig. 2F). It was found that the similar structure between Pol/RdRp and neuraminidase was not located in the active center. The above results suggested that there was a theoretical possibility of effective binding of oseltamivir with the active center of 3CLpro because it was found to be similar to the active center from the neuraminidase.
Fig. 2.
Alignment of the structure of viral proteins and influenza A neuraminidase analyzed by TM-align program. (A) Alignment of the structure of influenza A neuraminidase and influenza B neuraminidase. Left panel shows structure of influenza A neuraminidase (PDB ID: 3Ti6) and its active center is framed by red dotted line, Middle panel shows structure of influenza B neuraminidase (PDB ID: 4CPY) and its active center is framed by red dotted line; Right panel: Alignment of 3Ti6 and 4CPY revealed that the active sites of the both structures are the same. (B): Alignment of the structure of S protein and influenza A neuraminidase; Left panel shows structure of S protein and its active sites were RBD domain (Green region) and HR1 (Blue region). Right panel: Alignment of S protein and 4CPY revealed that the active sites of 4CPY is similar with NTD (Red region), suggesting the active sites of the two proteins are not identical. (C) Alignment of the structure of NRBD and influenza A neuraminidase; Left panel shows structure of NRBD, which is the active site of Nucleoprotein. Right panel: Alignment of NRBD and 4CPY revealed that the active sites of the two proteins do not coincide. The key residues of active site of 4CPY are marked in magentas. (D) Alignment of the structure of 3CLpro and influenza A neuraminidase; Left panel shows structure of 3CLpro and its active site is framed by red dotted line, which was based on the key residues marked in red. Right panel: Alignment of 3CLpro and 4CPY revealed that the active sites of the two proteins are similar. The key residues of active site of 4CPY are marked in magentas. (E) Alignment of the structure of PLpro and influenza A neuraminidase; Left panel shows structure of PLpro and its active site is framed by red dotted line. The key residues of active site are marked in red. Right panel: Alignment of PLpro and 4CPY revealed that the active sites of the two proteins do not coincide. The key residues of active site of 4CPY are marked in magentas. F: Alignment of the structure of Pol/RdRp and influenza A neuraminidase; Left panel shows structure of Pol/RdRp and its active site is framed by red dotted line. The key residues of active site are marked in red. Right panel: Alignment of Pol/RdRp and 4CPY revealed that the active sites of the two proteins are not similar. The key residues of active site of 4CPY are marked in magentas. NRBD: N-terminal RNA-binding domain; Pol/RdRp: RNA-directed RNA polymerase; PLpro: papain-like protease; 3CLpro: 3C-like protease; S protein: Spike Protein; RBD: RNA-binding domain; HR1: heptad-repeat 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Structural alignment analyzed by TM-align program.
| Protein | TM-score | Final |
|---|---|---|
| Influenza B neuraminidase | 0.94460 | In about the same fold |
| S protein | 0.30077 | Random structural similarity |
| NRBD | 0.19254 | Random structural similarity |
| 3CL pro | 0.28766 | Random structural similarity |
| PLpro | 0.30666 | Random structural similarity |
| Pol/RdRp | 0.34047 | Random structural similarity |
Abbreviations: S protein: Spike Protein.; NRBD: N-terminal RNA-binding domain; 3CLpro:3C-like protease; PLpro: papain-like protease; Pol/RdRp: RNA-directed RNA polymerase.
3.4. Molecular docking studies for predicting the antiviral effect of oseltamivir
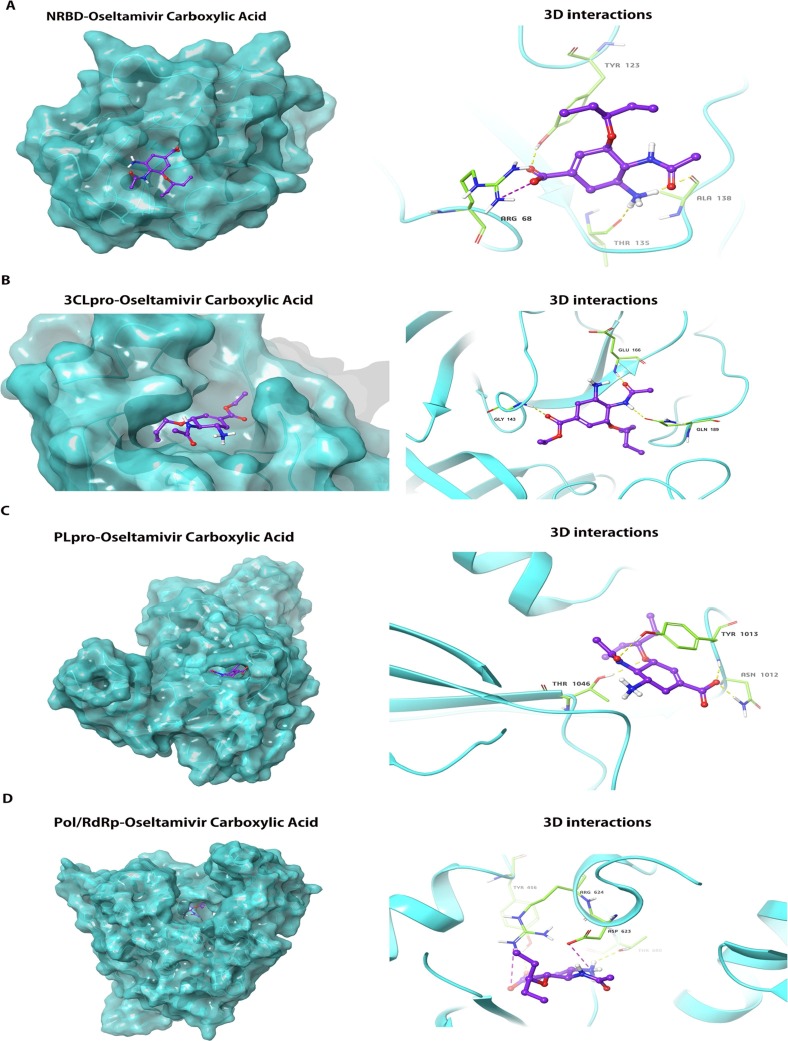
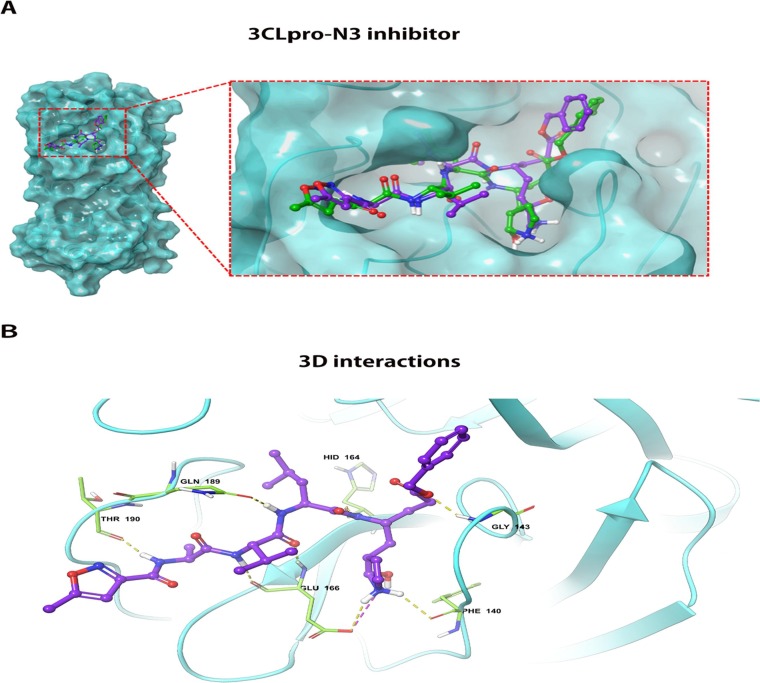
To pursue our hypothesis, we used molecular docking to analyze whether oseltamivir carboxylic acid could stably bind to the active center of the viral protein structure. The docking conformations were described in Fig. 3 A–D. The results showed that the docking energies of oseltamivir carboxylic acid with NRBD, 3CLpro, PLpro, and Pol/RdRp were −4.7 kcal/mol, −7.0 kcal/mol, −5.8 kcal/mol, and −5.4 kcal/mol, respectively (Table 3 ). The lower the binding energy, the more suitable the binding of the inhibitor molecule to the receptor. In terms of binding energy, oseltamivir carboxylic acid was more favorable to bind to the active site of 3CLpro effectively. To further analyze the ability of oseltamivir to inhibit 3CLpro, we selected N3 inhibitor as the positive group, which exhibited strong inhibition for 3CLpro of SARS-CoV-2 in vitro [33] . The result showed that its docking energy was −8.5 kcal/mol, which was lower than that of oseltamivir carboxylic acid. The docking conformation was shown in Fig. 4 A and B. It was found that the conformation of ligand (Purple) docked into the active site was almost identical to the original ligand (Green), suggesting the reliability of this docking program. Based on the results described above, we considered that the ability of oseltamivir to inhibit 3CLpro is not strong compared with N3 inhibitor.
Fig. 3.
The docking conformation and 3D interactions of oseltamivir carboxylic acid inside the active centre of NRBD, 3CLpro, PLpro and Pol/RdRp. (A) Left panel: Docking conformation of NRBD and oseltamivir carboxylic acid; Right panel:3D interactions of oseltamivir carboxylic acid inside the binding pocket of NRBD revealed binding energy of −4.7 Kcal/mol, 3H-bonds(yellow dotted line) and 1 salt bridge(purple dotted line); (B) Left panel: Docking conformation of 3CLpro and oseltamivir carboxylic acid; Right panel:3D interactions of oseltamivir carboxylic acid inside the binding pocket of 3CLpro revealed binding energy of −7.5 Kcal/mol and 3H-bonds(yellow dotted line); (C) Left panel: Docking conformation of PLpro and oseltamivir carboxylic acid; Right panel:3D interactions of oseltamivir carboxylic acid inside the binding pocket of PLpro revealed binding energy of −5.8 Kcal/mol and 4H-bonds (yellow dotted line); (D) Docking conformation of Pol/RdRp and oseltamivir carboxylic acid; Right panel:3D interactions of oseltamivir carboxylic acid inside the binding pocket of Pol/RdRp revealed binding energy of −5.4 Kcal/mol, 2H-bonds (yellow dotted line) and 2 salt bridges(purple dotted line); NRBD: N-terminal RNA-binding domain; Pol/RdRp: RNA-directed RNA polymerase; PLpro: papain-like protease; 3CLpro: 3C-like protease. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 3.
The calculated binding energy of binding for oseltamivir carboxylic acid.
| Protein | Binding energy (Kcal/mol) |
|---|---|
| NRBD | −4.7 |
| 3CLpro | −7.0 |
| PLpro | −5.8 |
| Pol/RdRp | −5.4 |
Abbreviations: NRBD: N-terminal RNA-binding domain; 3CLpro:3C-like protease; PLpro: papain-like protease; Pol/RdRp: RNA-directed RNA polymerase.
Fig. 4.
The docking conformation and 3D interactions of N3 inhibitor inside the active centre of 3CLpro. (A) Docking conformation of 3CLpro and N3 inhibitor revealed the conformation of ligand (Purple) docked into the active site was almost identical to the original ligand (Green), suggesting the reliability of this docking program. (B) 3D interactions of N3 inhibitor inside the binding pocket of 3CLpro revealed binding energy of −8.5 Kcal/mol, 5 H-bonds (yellow dotted line) and 1 salt bridge(purple dotted line). 3CLpro: 3C-like protease. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. In vitro study for accessing the antiviral effect of oseltamivir
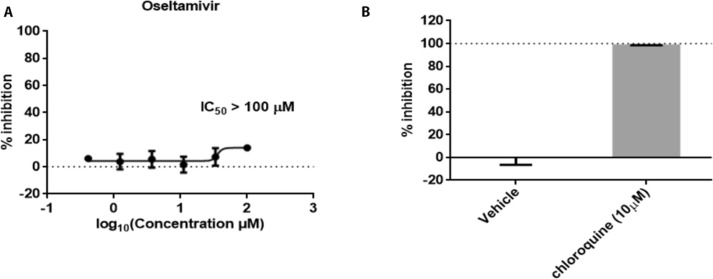
To further verify our hypothesis, we evaluated the antiviral efficiency of oseltamivir against SARS-CoV-2 in vitro as described in the previous work [24], [25]. Fig. 5 A showed that treatment of oseltamivir did not inhibit SARS-CoV-2 replication, with half-inhibitory concentration (IC50) value above 100 μM. Chloroquine was selected as a positive control, which can inhibit SARS-CoV-2 replication (Fig. 5B). Therefore, we concluded that oseltamivir is ineffective against SARS-CoV-2.
Fig. 5.
The antiviral activities of oseltamivir against SARS-CoV-2 in vitro. A-B. Antiviral activities of oseltamivir. Vero E6 cells infected with SARS-CoV-2 were cultured with the treatment of tested drugs for 24 h. Viral copy numbers in the cell supernatant were quantified by qRT-PCR. Chloroquine was selected as the positive group. Data from the experiments performed in triplicate with two independent repeats represented the mean ± SD.
3.6. Evaluating the antiviral effect of oseltamivir against SARS-CoV-2 in case review
To further verify the efficacy of oseltamivir on SARS-CoV-2, we retrospectively analyzed a total of 627 patients from the Second Hospital of Wuhan, Wuhan Union Hospital, and Wuhan Union Hospital West Campus, Hubei Province. Finally, 79 patients were included for analysis. Among them, there were 31 patients in remission group, aged 50.68 ± 14.912. There were 48 non-remission patients, and their age was 50.33 ± 15.099. There was no significant difference in age between the two groups (P = 0.921). In the remission group, there were 12 males (with a proportion of 38.71%), and in the non-remission group there were 25 males (52.10%); there was no statistical difference in sex between the two groups (P = 0.261). We analyzed the use of oseltamivir in the two groups. In the remission group, a total of seven patients used oseltamivir with a proportion of 22.58%. In the non-remission group, a total of 35 patients used oseltamivir, with a proportion of 72.92%. The chi-square value was 19.166, and there was a significant statistical difference between the two groups. The number of patients receiving oseltamivir in the non-remission group was significantly higher than the number in the remission group (P < 0.001) (Table 4 ). To compare the difference of response rate at different course of using oseltamivir, we divided the patients who had history of time of taking oseltamivir being recorded into groups according to the course of using oseltamivir: group A: 75 mg Bis In Die (bid, twice daily) 1–3 days; group B: 75 mg bid 3–5 days; group C:75 mg bid 5-7days. We found that the response rate of three group were 0%, 31%, 29%, respectively. (P = 0.405) (Table 5 ), suggesting that in term of standard quantity of influenza (75 mg bid), the course of using oseltamivir didn’t seem to improve the response rate. We next compared the difference of hospitalization days between the oseltamivir group and the Non-use oseltamivir group. The mean hospital stay for oseltamivir group was 9 days, and the days of Non-use oseltamivir group were 12. There was no statistical difference in hospitalization days between the two groups (P = 0.506) (Table 6 ). In combination with clinical practice, we believe that oseltamivir had no inhibitory effect on COVID-19 and could not effectively slow the progression of the disease with early use.
Table 4.
Comparison of the difference of history of oseltamivir use between the remission group and the progressive group.
| Variables | Remission (n = 31) | Non-remission (n = 48) | p value |
|---|---|---|---|
| Age, years Mean ± SD |
50.68 ± 14.912 | 50.33 ± 15.099 | 0.921 |
| Male sex | 12 (38.71) | 25 (52.10) | 0.261 |
| Oseltamivir | 7 (22.58) | 35 (72.92) | <0.001*** |
| Non-use Oseltamivir | 24 (77.42) | 13 (27.08) |
*** means p value < 0.001 analyzed using Pearson's chi-squared test.
Table 5.
Compare the difference of response rate at different course of using oseltamivir.
| Variables | Remission | Non-remission | p value |
|---|---|---|---|
| 75 mg bid 1–3 days (n = 6) | 0 (0%) | 6 (100%) | 0.405 |
| 75 mg bid 3–5 days (n = 13) | 4 (30.77%) | 9 (69.23%) | |
| 75 mg bid 5–7 days (n = 7) | 2 (28.57%) | 5 (71.43%) |
Table 6.
Compare the difference of hospitalization days between the oseltamivir group and the Non-use oseltamivir group.
| Variables | Oseltamivir (n = 42) | Non-use Oseltamivir (n = 37) | p value |
|---|---|---|---|
| Hospitalization days |
0.506 |
||
| Median (IQR) | 9(12) | 12(21) |
4. Discussion
In this article, we constructed protein models of NRBD, PLpro, and Pol/RdRp of SARS-CoV-2. We used structural alignment to find that the active center of 3CLpro was similar to that of influenza A neuraminidase. Subsequently, we used a molecular docking method to explore whether oseltamivir could effectively bind to the NRBD, 3CLPro, PLpro, and Pol/RdRp of SARS-CoV-2. It was found that oseltamivir carboxylic acid was more favorable to bind to the active site of 3CLpro effectively, but its inhibitory effects was not strong compared with the positive group. Finally, we used in vitro study and retrospective case analysis to verify our speculations. We found that oseltamivir is ineffective against SARS-CoV-2 in vitro study and the clinical use of oseltamivir did not improve the patients’ symptoms and signs and did not slow the disease progression.
As a neuraminidase inhibitor, oseltamivir has effectively combated the pandemic influenza A and B, so it is a first-line commonly used antiviral drug, especially in primary hospitals. At the same time, oseltamivir, as an over-the-counter drug, is also a popular antiviral drug. As healthcare workers fighting against COVID-19, we have found that many patients experiencing discomfort or considered to be infected with a virus take oseltamivir. From SARS-CoV in 2003 to MERS- CoV in 2012, and now the current SARS-CoV-2 epidemic, there is not plenty of evidence showing that oseltamivir is effective against coronavirus. Still, there is also no sufficient evidence to refute its ineffectiveness. We cannot predict whether there will be a pandemic of respiratory coronavirus in the future, so we hope to initiate such research and preliminarily explore whether oseltamivir is effective for COVID-19, which can better guide healthcare workers in the selection of appropriate antiviral drugs in the face of coronavirus epidemics. Although there was no expression of neuraminidase in SARS-CoV-2, Zhang et al, who discovered through homology modeling that the S1 protein of SARS-CoV had a similar active center as neuraminidase and considered that neuraminidase inhibitors were likely to inhibit SARS-CoV through targeting S1 protein activity, which provided us inspiration for research the structural similarity between SARS-CoV-2 and neuraminidase [2]. Considering the homology of the S protein of SARS-CoV-2 and SARS-CoV, we conducted a structural alignment of the crystal structure of S protein with influenza A neuraminidase. We found that the active center of neuraminidase was not similar to the RBD domain on the S1 subunit and the HR1 domain on the S2 subunit, which were the active sites of S protein. We were not surprised by these findings because the sequence conservation of S1 in both SARS-CoV-2 and SARS-CoV was only 71%, and there were structural variations [34]. We, therefore, focused our attention on the NC protein, 3CLPro, PLpro, and Pol/RdRp, which have been widely studied in SARS-CoV, are highly conserved with SARS-CoV-2, and are the main proteins targeted by drugs. We used structural alignment to discover that, except for 3CLpro, the active centers of the other three proteins did not match that of neuraminidase. We observed that in the reported regimen in Thailand, the dosage of oseltamivir was 300 mg/day, and our patients’ treatment dose was still based on the conventional dose of 150 mg/day for flu (news link: https://xw.qq.com/cmsid/20200221A0HN1400?f=newdc), so we next studied the relationship between dose of oseltamivir and therapeutic effect. However, the data suggested that oseltamivir failed to fight against SARS-CoV-2 in vitro, which was consistent with the result of our retrospective case analysis. Xi Wang et al and Choy et al also found that oseltamivir was ineffective in inhibiting SARS-CoV-2 in vitro [25], [35].
However, the limitation of our study shouldn't be neglected. First, the sample size from our study was relatively small because of our strict inclusion criteria. Second, introduced bias of retrospective design was inevitable due to that all factors correlated with efficacy of oseltamivir were unable to be controlled. Because we faced to the newly virus and attempted several courses of antiviral treatments. Currently, a phase III trial and a phase IV trial are underway by Tongji hospital, another affiliated hospital of Huazhong University of Science and Technology to evaluate the safety and efficacy of oseltamivir in patients with COVID-19 (ClinicalTrials.gov Identifier: NCT04255017 and NCT04261270).
5. Conclusions
In summary, we consider that oseltamivir isn’t suitable for the treatment of COVID-19. During the outbreak of novel coronavirus, when oseltamivir is not effective for the patients after they take it, health workers should be highly vigilant about the possibility of COVID-19. At the same time, it should be noted that we are still in the influenza season, and we have also found five cases of influenza A in combined with COVID-19. Therefore, oseltamivir should be combined in the treatment regimen in a timely manner when influenza occurs.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Guorong Hu for participating in the design of our paper. She is from Department of Respiratory Medicine, Second Hospital of Wuhan. We thank Leike Zhang and his colleagues for assistance with the in vitro experiments. They are from State Key Laboratory of Virology, Wuhan Institute of Virology, Center for Biosafety Mega-Science, Chinese Academy of Sciences. We show our acknowledgments to the medical workers, people ’s liberation army, community workers, police officers, volunteers on the front line. This work was supported by Novel coronavirus pneumonia emergency project, China (no. 2020YFC084430), National Major Science and Technology Projects of China (CN):2019ZX09301001, Ministry of Science and Technology of the People's Republic of China (CN):2020YFC0844300, and the Fundamental Research Funds for the Central Universities, HUST: 2020kfyXGYJ011, State Key Laboratory of Drug Research, China (no. 2019ZX09301001) and the National Natural Science Foundation of China (no. 82041018, no. 81770096 and no. 81700091).
References
- 1.Hayden F.G., Treanor J.J., Fritz R.S. Use of the oral neuraminidase inhibitor oseltamivir in experimental human influenza: randomized controlled trials for prevention and treatment. Jama. 1999;282(13):1240–1246. doi: 10.1001/jama.282.13.1240. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X.W., Yap Y.L. The 3D structure analysis of SARS-CoV S1 protein reveals a link to influenza virus neuraminidase and implications for drug and antibody discovery. Theochem. 2004;681(1):137–141. doi: 10.1016/j.theochem.2004.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. Offord, Combining the medications improved conditions in patients with severe 2019-nCoV infections, say doctors in Thailand, 2020. https://www.the-scientist.com/news-opinion/flu-and-anti-hiv-drugs-show-efficacy-against-coronavirus-67052 (accessed February 3 2020).
- 4.Rossi S. Australian medicines handbook: Sydney, 2006.
- 5.Jefferson T., Jones M.A., Doshi P. Neuraminidase inhibitors for preventing and treating influenza in healthy adults and children. Cochrane Database Syst. Rev. 2014;4:Cd008965. doi: 10.1002/14651858.CD008965.pub3. [DOI] [PubMed] [Google Scholar]
- 6.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu X., Chen P., Wang J. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehr A.R., Perlman S. Springer; 2015. Coronaviruses: An Overview of their Replication and Pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canrong W., Yang L., Yueying Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuate S., Cinatl J., Doerr H.W., Überla K. Exosomal vaccines containing the S protein of the SARS coronavirus induce high levels of neutralizing antibodies. Virology. 2007;362(1):26–37. doi: 10.1016/j.virol.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu M.-S., Pan Y., Chen H.-Q. Induction of SARS-nucleoprotein-specific immune response by use of DNA vaccine. Immunol. Lett. 2004;92(3):237–243. doi: 10.1016/j.imlet.2004.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H., Yang M., Ding Y. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 2003;100(23):13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Báez-Santos Y.M., John S.E.S., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng B., Lui Y.-w., Meng S., Cao C., Hu Y. Identification of effective siRNA blocking the expression of SARS viral envelope E and RDRP genes. Mol. Biotechnol. 2006;33(2):141–148. doi: 10.1385/MB:33:2:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Y.-S. Wu, W.-H. Lin, T.-A. Hsu, H.-P. Hsieh, Antiviral drug discovery against SARS-CoV, Curr. Med. Chem. 13(17) (2006) 2003–2020. [DOI] [PubMed]
- 17.Waterhouse A., Bertoni M., Bienert S. SWISS-MODEL: homology modelling of protein structures and complexes. Nucl. Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laskowski R.A., Macarthur M.W., Moss D.S., Thornton J.M.J. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26(2):283–291. [Google Scholar]
- 19.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowie J.U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 21.Lüthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356(6364):83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucl. Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31(2) doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Cao R., Zhang H. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6(1):28. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Q., Yu L., Petros A.M. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43(20):6059–6063. doi: 10.1021/bi036155b. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y., Jeffrey S. TM-align: a protein structure alignment algorithm based on the TM-score. Nucl. Acids Res. 2005;33(7):2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vavricka C.J., Li Q., Wu Y. Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition. PLoS Pathogens. 2011;7(10) doi: 10.1371/journal.ppat.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Escuret V., Collins P.J., Casalegno J.-S. A novel I221L substitution in neuraminidase confers high-level resistance to oseltamivir in influenza B viruses. J. Infect. Dis. 2014;210(8):1260–1269. doi: 10.1093/infdis/jiu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia S., Yan L., Xu W. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5(4):eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wrapp D., Wang N., Corbett K.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratia K., Pegan S., Takayama J. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. 2008;105(42):16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin Z., Du X., Xu Y., Deng Y., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020:1–9. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 34.Wu C., Yang Y., Liu Y., Zhang P., Wang Y., Wang Q., Xu Y., Li M., Zheng M., Chen L., Li H. Furin, a potential therapeutic target for COVID-19. ChinaXiv. 2020 doi: 10.1016/j.isci.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choy K.T., Wong A.Y., Kaewpreedee P. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]